Summary
Background
Neurons require highly specialized intracellular membrane trafficking, especially at synapses. Rab GTPases are considered master regulators of membrane trafficking in all cells and only very few Rabs have known neuron-specific functions. Here, we present the first systematic characterization of neuronal expression, subcellular localization and function of Rab GTPases in an organism with a brain.
Results
We report the surprising discovery that half of all Drosophila Rabs function specifically or predominantly in distinct subsets of neurons in the brain. Furthermore, functional profiling of the GTP/GDP-bound states reveals that these neuronal Rabs are almost exclusively active at synapses and the majority of these synaptic Rabs specifically mark synaptic recycling endosomal compartments. Our profiling strategy is based on Gal4 knock-ins in large genomic fragments that are additionally designed to generated mutants by ends-out homologous recombination. We generated 36 large genomic targeting vectors and transgenic rab-Gal4 fly strains for 25 rab genes. Proof-of-principle knock-out of the synaptic rab27 reveals a sleep phenotype that matches its cell-specific expression.
Conclusions
Our findings suggest that up to half of all Drosophila Rabs exert specialized synaptic functions. The tools presented here allow systematic functional studies of these Rabs and provide a method that is applicable to any large gene family in Drosophila.
Introduction
Rab GTPases were first described in 1987 when Touchot and colleagues isolated four family members from a rat brain cDNA library and consequently named them ‘rab’ for ‘ras gene from rat brain’ [1]. Subsequent work over the last 24 years has firmly established rab GTPases as key regulators of membrane organization and intracellular membrane trafficking in all eukaryotic cells [2-5]. The majority of Rab GTPases are thought to serve ubiquitous cell biological functions.
Rab proteins cycle between active and inactive states. In response to signal stimuli, guanine nucleotide exchange factors (GEFs) interact with Rab GTPases and trigger their binding to GTP. In the GTP-bound active form, each Rab interacts with a different complex of proteins (effectors) to facilitate the delivery of transport vesicles to different acceptor membranes [6, 7]. GTPase-activating proteins (GAPs) work in the opposite direction and accelerate Rab GTP hydrolysis, typically rendering the subsequently GDP-bound Rab proteins inactive.
Neurons have specialized demands on membrane trafficking both during development (wiring-specific extensive arborizations) and function (neurotransmitter release). The importance of Rab function in the nervous system is highlighted by the observation that mutations in rab genes and their regulators cause several hereditary and neurological diseases including Griscelli syndrome (Rab27), Charcot-Marie-Tooth type 2B disease (Rab7), Warburg Micro Syndrome (a GTPase activating protein for Rab3), X-linked mental retardation (RabGDI - a Rab GTP dissociation inhibitor) and Hermansky-Pudlak syndrome (a Rab geranylgeranyl transferase) [8-11]. Recently, Rab8-dependent trafficking was identified as a key mechanism underlying Bardet-Biedl syndrome, which causes retinopathy and blindness [12]. In Drosophila, Rab11 is required for post-Golgi trafficking of rhodopsin [13] and guidance receptors during brain wiring [14]. Lastly, active zone assembly at synapses requires Rab3 [15]. While these examples underscore the importance of Rab-dependent trafficking in neurons, comprehensive profiling of rab GTPase function in a brain at cellular and subcellular resolutions in vivo has not been attempted.
Drosophila is ideally suited for the systematic study of rab GTPases in the nervous system. There are 31 potential rab or rab-related genes in the fly genome of which at least 6 have no clear vertebrate ortholog [16]. 23 rab have direct orthologs in human that are at least 50% identical at the protein level (see also Suppl. Fig. 4). Hence, Rab proteins in Drosophila exhibit high evolutionary conservation and low redundancy compared to over 70 rabs in vertebrates [16]. For example, the best-characterized neuronal rab GTPase, rab3, exists as four partially redundant genes in vertebrates, but as a single gene in Drosophila. Yet, clear orthologs exist and serve as gold standard markers for many intracellular compartments across species. These include Rab1 for the endoplasmic reticulum, Rab5 for early endosomes, Rab6 for the Golgi, Rab7 for multivesicular bodies and Rab11 for recycling endosomal compartments [7, 17, 18].
Recently developed tools combined with established Drosophila genetics allow for the systematic characterization of this complete gene family in vivo [16, 19]. We generated ends-out homologous recombination competent targeting vectors of typically >50kb for 25 conserved rab loci using a BAC-mediated recombineering. Within these vectors we replaced the rab genes with the yeast transcription factor Gal4, yielding ‘Gal4 drivers’ and inserted them at a predefined landing site within the genome prior to labor-intensive homologous recombination experiments. This enabled us to perform a systematic profiling of the cellular and subcellular expression of Rab GTPases in Drosophila in combination with an existing collection of UAS-YFP-Rabs transgenic fly lines. Our profiling reveals that half of all rab GTPases are neuron-specific or strongly enriched in varying and specific subsets of neurons in the brain and that all neuronal rabs encode synaptic proteins. Finally, we perform a proof-of-principle knock-out screen for the neuron-specific rab27 gene and describe a specific behavioral sleep defect for the rab27 null mutant that matches its cell-specific expression.
Results
Generation of 36 rab-Gal4 knock-in vectors and transgenic fly strains for ends-out homologous recombination using BAC recombineering and PhiC31 transgenesis
In order to genetically characterize and manipulate all members of the rab gene family in parallel, we devised a modification to P[acman] technology by incorporating an optimized ends-out homologous recombination cassette [20], thereby generating the P[acman]-KO vector (Figure 1A). Using recombineering [21], we generated 41 P[acman]-KO targeting vectors of typically more than 50kb, centered on individual rab loci. A key advantage of using large genomic fragments is that they are predicted to contain all regulatory elements; fully functional genomic rescue fragments in Drosophila are traditionally between 5-10kb. In these large genomic regions, we used a positively marked knock-in cassette (Figure 1B) to replace rab genes with the yeast transcription factor Gal4, yielding ‘Gal4 driver lines’. These Gal4 lines are functional after integration in a ‘landing site’ and can be utilized for expression and functional profiling prior to labor-intensive homologous recombination experiments. All technical details and protocols are available in the Supplemental Materials.
Figure 1. Design of P[acman]-KO: Combining BAC Recombineering, PhiC31 transgenesis, and ends-out homologous recombination.
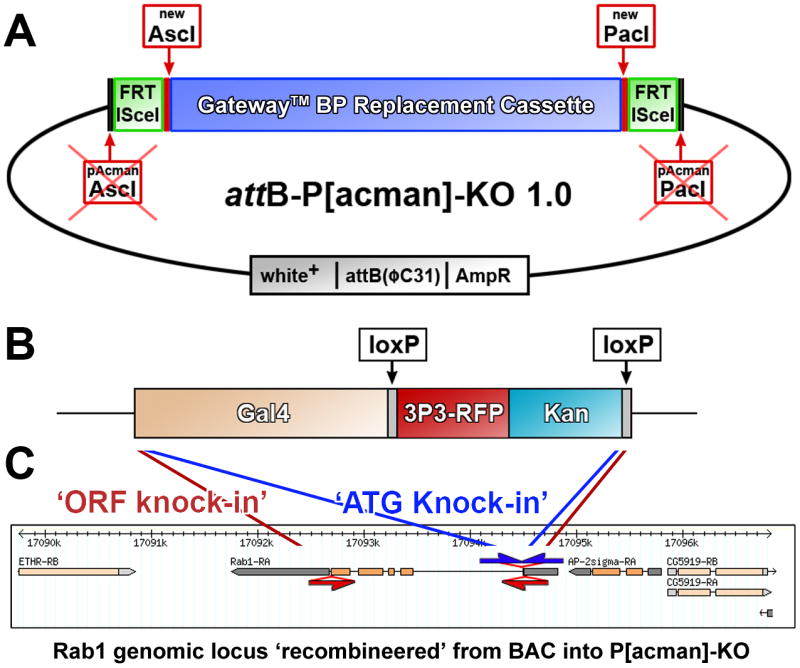
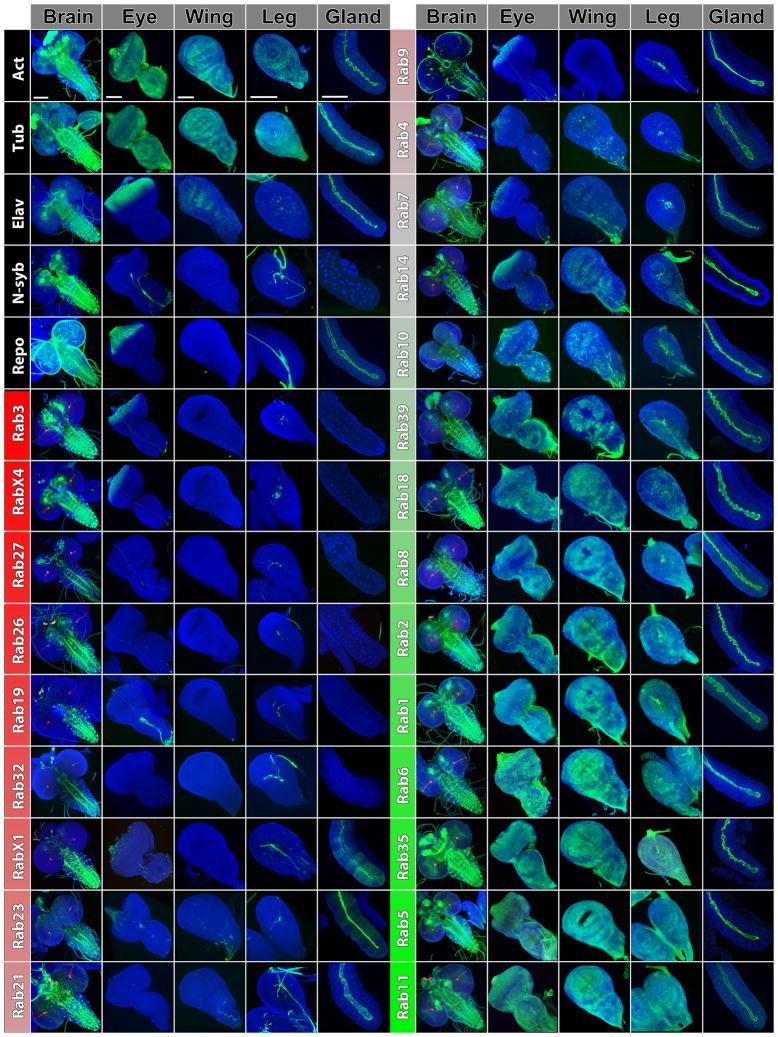
(A) Vector design. First, a [Frt, ISce1] cassette was inserted into the AscI and PacI sites of attB-P[acman]-ApR to add mobilization capability for endogenous targeting. Second, a GatewayTM cassette for bacteriophage λ–mediated BP recombination was introduced with new AscI and PacI sites. Hence, homology arm cassettes can either be integrated using AscI, PacI conventional cloning or by including GatewayTM attB sites into the primers used to create such cassettes. See Supplemental Materials for more details. All other features of the vector are described in Venken et al., (2006). (B) Design of the Gal4 knock-in cassette. This 6.7kb cassette is adapted for desired loci by including gene-specific 100bp homology arms in the primers used to amplify the cassette. Note that optimized PCR conditions are identical for any primer pair (see Suppl. Materials). The floxed 3×P3-RFP, Kan cassette serves for selection during bacterial recombineering cloning (Kan) and positive selection of the targeting cassette in a homologous recombination experiment in vivo in the fly (3×P3-RFP) and can be removed in transgenic or gene-targeted flies easily by crossing to available Cre strains. (C) Strategies for ORF and ATG knock-ins. The Gal4 knock-in cassette replaces the complete open reading frame in ORF knock-ins, whereas in ATG knock-ins only the start codon and the remaining part of the start-codon containing exon are replaced. Note that in both cases the ATG of Gal4 replaces the ATG of the rab gene. Brain: L3 larval brain; Eye: L3 eye disc; wing: L3 wing disc; leg: L3 leg disc; Gland: L3 salivary gland. Scale bar for each tissue: 100μm.
We designed targeting vectors for two types of knock-ins: First, we replaced complete open reading frames (ORFs) with Gal4 for 27 rab loci (red arrows/primer pairs in Figure 1C and Figure S1). These targeting vectors are designed for the generation of unequivocal null mutants and are referred to as ‘ORF knock-ins’. For two additional loci (rab26 and rab32) we chose to only replace short coding regions of the first exons because of the large size of the genomic loci (Figure S1). Second, we replaced only the short coding regions starting with the ATG to the end of the ATG-containing exon for 12 rab loci in cases where the ORF knock-ins delete potential regulatory sequences (blue arrows/primer pairs in Figure 1C and Figure S1). These Gal4 knock-ins are designed to ensure that the resulting lines express Gal4 in the endogenous gene expression pattern. We refer to these as ‘ATG knock-ins’. The generation of these 12 alternative targeting vectors was straightforward, since only one round of recombineering is required to replace a different sequence in the targeting vectors that already contain the genomic DNA, highlighting the efficiency of the recombineering approach. Figure 1C shows the ‘ORF knock-in’ and ‘ATG knock-in’ for the rab1 locus; all loci are shown in Figure S1. In most cases the Gal4 cassette was knocked into a genomic fragment of 40kb, with few exceptions where cloning of the large region proved difficult (rab5: 30kb; rab18, rab19: 20kb). In total, we generated 41 targeting vectors for the generation of transgenic flies to create Gal4 driver lines under the endogenous regulatory elements of 29 rab loci. (Suppl. Fig. 1).
Recovery of transgenic flies for the large vectors (>55kb) was challenging with typically less than one transformant in 10,000 progeny; however, recent efforts show that this can be substantially improved [22, 23]. For five genomic constructs, we could not obtain transgenics after injection of more than 1,500 embryos (‘ORF knock-ins’ of rab4, rab30, rab40, rabX5, and rabX6). We obtained transgenic flies for 24 ‘ORF knock-ins’ and all 12 ‘ATG knock-ins’ (including rab4). In total, we obtained 36 transgenic fly strains for 25 rab loci.
To determine the expression patterns of these rab genes, we crossed the rab-Gal4 lines to UAS-CD8-GFP and analyzed the brains, eye discs, wing discs, leg discs and salivary glands of rab-Gal4>UAS-CD8-GFP third instar larvae and obtained confocal high-resolution 3D datasets in duplicate or triplicate for each line and each tissue. Antibodies or GFP tags in the genomic loci exist for some rabs providing independent verification of the fidelity of our Gal4 lines (Figure S2A-H). In addition, we performed rescue experiments for existing mutants or knock-outs generated with the technique presented here to verify the accuracy of the Gal4 lines including rab3 and rab6 (Figure S2I-N) and the rab27 knock-out presented below. By these various methods we verified rab2, rab3, rab5, rab6, rab7, rab11 and rab27, and in no case did we find evidence for significant expression differences between the rab-Gal4 knock-ins and endogenous rabs. For the detailed characterizations in the following sections we selected the 12 ATG knock-ins, additional ATG/exon 1 knock-ins for rab26 and rab32, and ORF knock-ins for the remaining nine rab loci.
Cellular expression profiling reveals that half of all rabs are neuron-specific or strongly neuron-enriched with highly variable expression patterns in the brain
The selected rab lines exhibit a surprising variety of expression patterns with a strong bias for the nervous system (Figure 2). Specifically, we identified six lines with expression exclusively in neurons and possibly glia in the larval brain (rab3, rab19, rab26, rab27, rab32 and rabX4). Furthermore, rab9-Gal4, rab21-Gal4, rab23-Gal4, and rabX1-Gal4 exhibit expression exclusive to neurons, glia and salivary glands (a non-neuronal secretory tissue). rab4-Gal4, rab7-Gal4, rab10-Gal4, rab14-Gal4 and rab39-Gal4 exhibit their strongest expression in neurons, but also diverse patterns of expression in other tissues. Finally, we found ubiquitous expression patterns, albeit with different expression levels in different tissues and neurons, for only eight lines (rab1, rab2, rab5, rab6, rab8, rab11, rab18, and rab35). Hence, half of the rab-Gal4 lines analyzed here exhibit neuron-specific or strongly neuron-enriched expression. Neuronal Rabs are present in all major branches of the phylogenetic tree and protein similarity does not correlate with neuronal expression (Fig. S3A).
Figure 2. Systematic Expression Profiling in larval tissues reveals that half of all rab GTPases in Drosophila are neuron-specific or neuron-enriched.
Shown are the five larval tissues brain, eye disc, wing disc, leg disc and salivary gland (from left to right) for a total of 23 rab-Gal4 lines crossed to UAS-CD8-GFP (green). Toto-3 labels cell bodies/nuclei (blue), and the 3×P3-RFP cassette from our knock-in cassette marks the termini of the larval photoreceptor organs in the brain (red). On the top left five control Gal4 lines are shown: act-Gal4 and tub-Gal4 (both showing ubiquitous expression), elav-Gal4 (showing expression in developing and functional neurons as well as low levels in some other cells), n-syb-Gal4 (showing panneurononal expression), and repo-Gal4 (showing expression in all glial cells). All rabs are sorted from the most neuron-specific in red (starting with rab3 and rabX4), via lines with somewhat specialized patterns in grey to the most ubiquitous Gal4 driver lines in green (rab5-Gal4 and rab11-Gal4).
Of the ten rab-Gal4 lines that exclusively express in neurons, glia and salivary glands only rab3-Gal4 and rabX4-Gal4 exhibit broad neural expression (Figure 2). In contrast, all other lines express in surprisingly specific and varying subsets of neurons. The highest expression overlap is in the ventral ganglion, where the motor neurons that innervate the body wall musculature reside; the highest diversity of expression patterns is apparent in the functioning and developing central brain. Developing photoreceptor neurons only show strong and neuron-specific expression in the eye discs of rab3-Gal4, rabX4-Gal4, rab9-Gal4, rab14-Gal4 (with some non-neuronal cells) and, surprisingly, rab7-Gal4, which is not neuron-specific in other tissues and later during development expresses more ubiquitously. Taken together, our findings suggest that rab GTPases have highly diverse expression patterns in active neurons in particular.
All rab GTPases are strongly expressed in functional neurons, and none exclusively in developing neurons
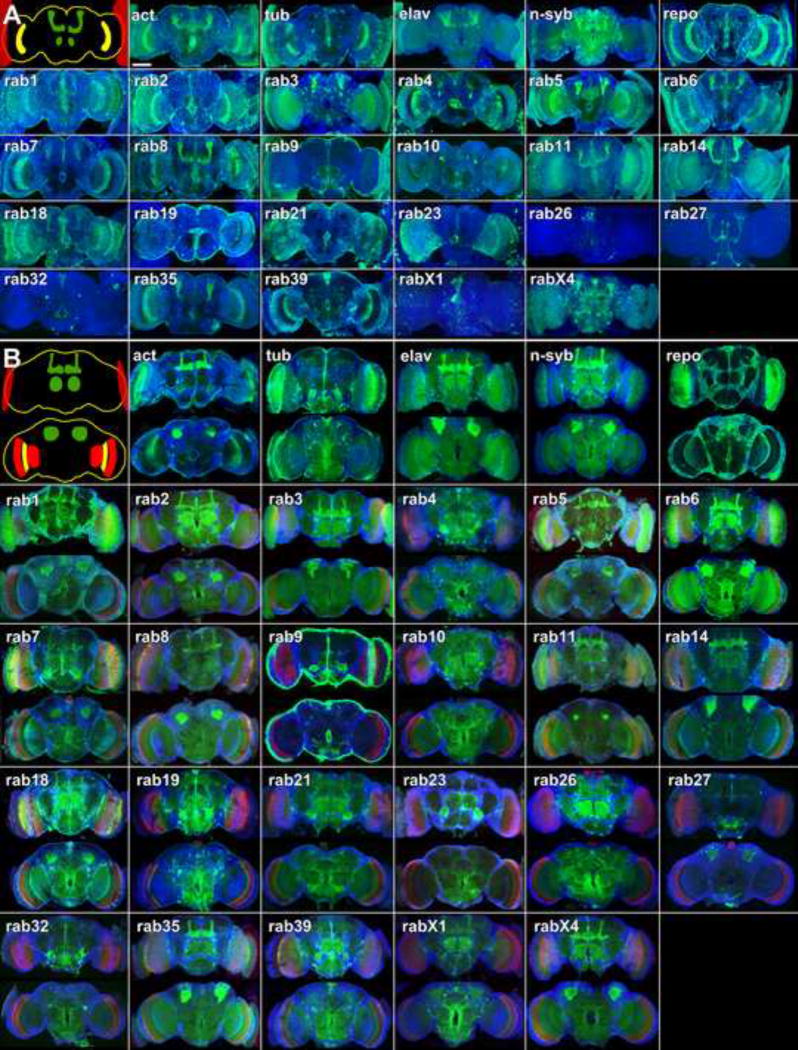
While all rab-Gal4 lines exhibit expression in at least some active neurons of the larval ventral ganglion, expression levels in the developing pupal brain (30-40% of pupal development) vary greatly (Figure 3A). Expression in different subsets of developing neurons is apparent for all rab-Gal4 lines. The strongest glial driver is rab9-Gal4. None of the rab-Gal4 lines drive expression that increases during development and decreases in the adult. The most prominent hallmark of the expression differences is the cell type specificity, suggesting neuronal sub-type-specific employment of Rab-mediated membrane trafficking during brain development.
Figure 3. rab-Gal4 expression patterns in the pupal and adult brain.
(A) Pupal brains (30-40% pupal development). Shown are maximum projections with CD8-GFP driven by the denoted rab-Gal4 lines (green) and nuclear labeling with Toto-3 (blue). The top left corner shows a schematic with a few prominently labeled landmark structures: the developing eyes (red), glia (yellow), and developing mushroom bodies and antennal lobes (green). (B) Adult brains. Shown are partial maximum projections of 20μm depth of the anterior adult brain on top and 20μm depth of the posterior brain on the bottom. CD8-GFP is driven by the denoted rab-Gal4 line (green), the 3×P3-RFP marker from the knock-in cassette labels the photoreceptor projections(red) and Toto-3 labels nuclei (blue). The top left corner shows an anterior brain schematic with the lamina (red) and the mushroom bodies and antennal lobes (green), and a posterior brain schematic showing the cell bodies of the Kenyon cells that form the mushroom body (green) and the optic neuropils medulla and lobula complex (red). Glia is marked in yellow. The different rab-Gal4 lines drive expression in these structures with highly varying intensity. Scale bar for all panels: 100μm.
Adult brains (1 day old) exhibit a similar variety of expression patterns (Figure 3B). Four rabs exhibit sparse expression patterns: rab9-Gal4, which is almost exclusively expressed in glia and neurons of the olfactory system, rab19-Gal4, which is strong in a subset of central brain neurons, rab23-Gal4, which is strong in the antennal lobes, and rab27-Gal4, which is highly restricted to few central brain neurons including Kenyon cells of the mushroom bodies. For most of the other lines strong differences can be observed in expression levels between different neuronal cell types, suggesting differential employment in the various neuronal sub-types. Similar to pupal brain, we find no obvious adult glial expression for the Gal4 lines of the neuronal rab3, rab23, rab26, rab27, rab32, rabX1 and rabX4. Strong expression in mushroom bodies is apparent for an increased number of lines compared to pupal brain, including rab1, rab2, rab3, rab5, rab6, rab7, rab8, rab11, rab14, rab18, rab27, rab35, rabX1 and rabX4. Closer investigation of the high-resolution data further yields a wealth of data, e.g. lines with high level expression in mushroom bodies also exhibit high levels of expression in photoreceptors (note that photoreceptors are co-labeled the 3×P3-RFP of the knock-in cassettes in Figure 3B). We conclude that rab GTPases exhibit highly dynamic and variable expression in the different neuronal subtypes of the developing and functional brain. However, the adult expression patterns do not change notably with age, as shown for one week and three week old brains of eight neuronal rabs in Figure S4A.
All neuronal rabs encode synaptic proteins
Next, we investigated the subcellular localization of the different Rabs in neurons in which they are endogenously expressed by expressing YFP-tagged versions of the wild type proteins [16]. We expect that the rab-Gal4>UAS-YFP-Rab expression system recapitulates spatiotemporal expression dynamics; however, the amplification effect of the Gal4/UAS system may cause overexpression of YFP-Rab proteins compared to endogenous expression levels. Importantly, wild type Rab proteins typically do not have overexpression phenotypes and expression of fluorescently tagged Rab proteins is an accepted standard for the study of wild type function in cell culture. We tested the effect of YFP-Rab overexpression in vivo in a characterized neuronal cell type by expressing all YFP-Rab proteins in photoreceptor neurons using the strong GMR-Gal4 driver line [24]. Indeed, none of the wild type YFP-Rab proteins cause developmental defects based on eye morphology or photoreceptor functional defects based on electroretinogram recordings (data not shown). Similarly, no deleterious effects were observed for any UAS-YFP-Rab expression when expressed with the corresponding rab-Gal4 line (see below). We conclude that Gal4/UAS expression of YFP-tagged Rabs does not cause obvious developmental or functional defects. However, it has long been known that overexpression of several wild type Rab proteins causes enlargements of the intracellular organelles they mark [e.g. [25]]. Indeed, we have observed this phenomenon for several of the Rab GTPases, as presented below. The enlarged wild type compartments provide an advantage for the profiling presented here, as size- and fluorescence-increased compartments are easier identified and co-labeled with other compartment markers.
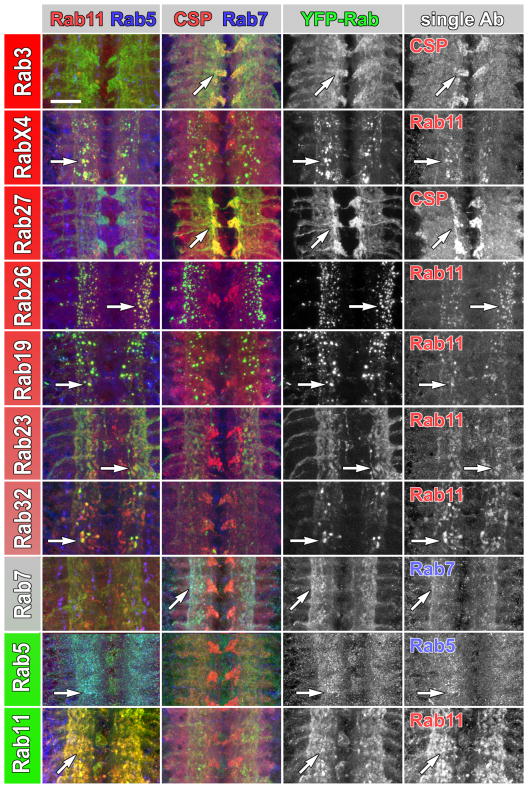
We chose the posterior segments of the ventral ganglion with high commonality between the rab lines to study the subcellular distribution of all Rabs expressed under control of their own regulatory elements (Figure 4; complete dataset in Figure S5A). Strikingly, all neuronal Rabs exhibit either synapse-specific localization (Rab3, RabX4, Rab27, Rab26, RabX1) or strongly synaptic enriched localization if some compartments are discernible also in cell bodies (Rab9, Rab19, Rab21, Rab23, Rab32) (Figure S5A). The synaptic localization is specific to the YFP-Rab proteins, since both CD8-GFP (Figure 2) and cytosolic GFP (Figure S4B) expressed using the same rab-Gal4 lines exhibit evenly cell body and synaptic distribution. In contrast to the neuronal rab-Gal4 lines, Rabs that exhibit both synaptic and cell body localization are not neuron specific (Rab5, Rab7, Rab11, Rab35). Finally, only five Rab proteins exhibit mostly cell body localization, all of which are ubiquitously expressed (Rab1, Rab2, Rab14, Rab18, Rab39). Hence, all neuronal Rabs display synapse-specific or synapse-enriched localization (Figure 4; Figure S5A; Table 1). These observations suggest that approximately half of all Rab proteins not only serve neuron-specific tasks, but function to meet the demands of synapse-specific membrane trafficking.
Figure 4. Subcellular Localization Profiling of YFP-tagged Rab proteins expressed under control of their endogenous regulatory elements.
All neuronal rabs encode synaptic proteins that colocalize with recycling endosome or synaptic vesicle markers. Double immunolabelings of the posterior larval brain ventral ganglion at high resolution are shown for selected YFP-Rabs driven by their respective rab-Gal4 lines (green), anti-Rab11 (red, recycling endosomes) and anti-Rab5 (blue, early endosomes) labeling in the first column and anti-CSP (red, synaptic vesicles) and anti-Rab7 (blue, late endosomes) in the second column. Cell bodies are peripherally and synaptic neuropils centrally located. Single channels of the colocalizing labels are depicted in the two columns on the right. Shown are only the seven most neuron-specific lines and rab5, rab7 and rab11 as controls; see Figure S5A for the complete dataset. Arrows point to colocalizing compartments. Scale bar for all panels: 20μm.
Table 1. Cellular and Subcellular Localization Profiling Summary.
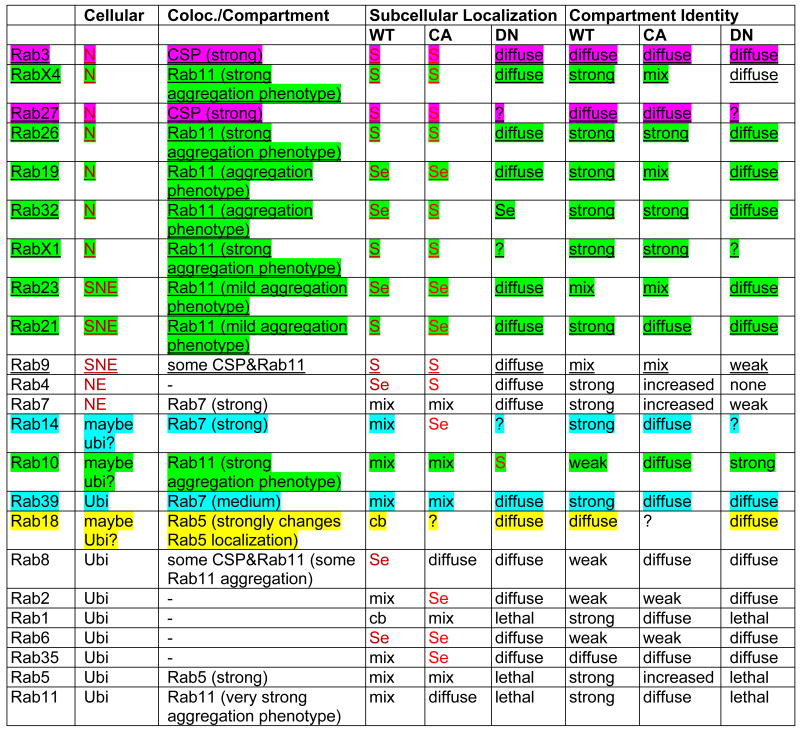 |
Legend:
underlined: neuronal Rab;
 ,
,
 ,
,
 ,
,
 colocalization
colocalization
 neuron-specific;
neuron-specific;
 strongly neuron-enriched;
strongly neuron-enriched;
 : neuron-enriched
: neuron-enriched
 synaptic;
synaptic;
 synapse-enriched; cb: cell body; mix: cell body and synapses
synapse-enriched; cb: cell body; mix: cell body and synapses
“?” indicates the YFP signal was too weak.
“Compartment Identity” refers to how punctate the YFP signal appears.
The high-resolution confocal analysis of the synaptic region of the ventral ganglion does not reveal pre- versus postsynaptic localization. We therefore analyzed the larval neuromuscular junction (NMJ) for neuronal Rabs. As shown in Figure S6A, Rab3, RabX4, Rab26 and Rab19 exhibit clear localization inside presynaptic boutons. In contrast, Rab23 is strongly enriched on the outside of boutons, suggesting postsynaptic localization, and Rab21 is mostly localized to compartments in the muscle. Rab27, Rab32 and RabX1 are not detected at the NMJ, either because the protein is not synaptic in motorneurons, or because these more restrictively expressed rabs are not expressed in motorneurons (Figure S6A).
Several Rabs mark large, distinct subcellular compartments, while others exhibit more diffuse localization. To assess the molecular nature of the compartments marked by Rab proteins in the neurons where they are endogenously expressed, we co-labeled YFP-Rabs driven by their corresponding rab-Gal4 lines with antibodies against early (Rab5), late (Rab7) and recycling endosome (Rab11) markers and the synaptic vesicle marker Cysteine-String Protein (CSP). Seven Rabs exhibit strong colocalization with large, distinct Rab11-positive recycling endosomes specifically at synapses. Interestingly, six of these seven are neuronal Rabs (Rab19, Rab21, Rab26, Rab32, RabX1 and RabX4) (Table 1). We further observed that expression of all seven Rab11-positive YFP-Rabs caused larger Rab11-positive compartments than observed in the Rab11 immunolabeling expressing other YFP-Rabs (Figure S5B). This suggests that the overexpression of these Rabs causes an increase in types of recycling endosomal compartments at synapses. Only Rab3 and Rab27 exhibit strong colocalization with the synaptic vesicle marker CSP. YFP-Rab8 and YFP-Rab9 exhibit partial colocalization with both CSP and Rab11. YFP-Rab14 and YFP-Rab39 exhibit significant colocalization with the late endosomal marker Rab7. YFP-Rab18 is the only Rab protein (other than YFP-Rab5) that exhibits strong colocalization with anti-Rab5 labeling and also clearly changes Rab5 localization. Finally, five YFP-Rabs (Rab1, Rab2, Rab4, Rab6, Rab35) exhibit no significant colocalization with any of the four immunolabels (Figure S5A; Table 1). The subcellular localization profiling of wild type Rabs thus reveals that the majority of novel neuronal rabs encode synaptic proteins that mark Rab11-positive synaptic recycling endosomal compartments.
Expression of constitutively active and dominant negative Rab GTPases in their endogenous expression patterns
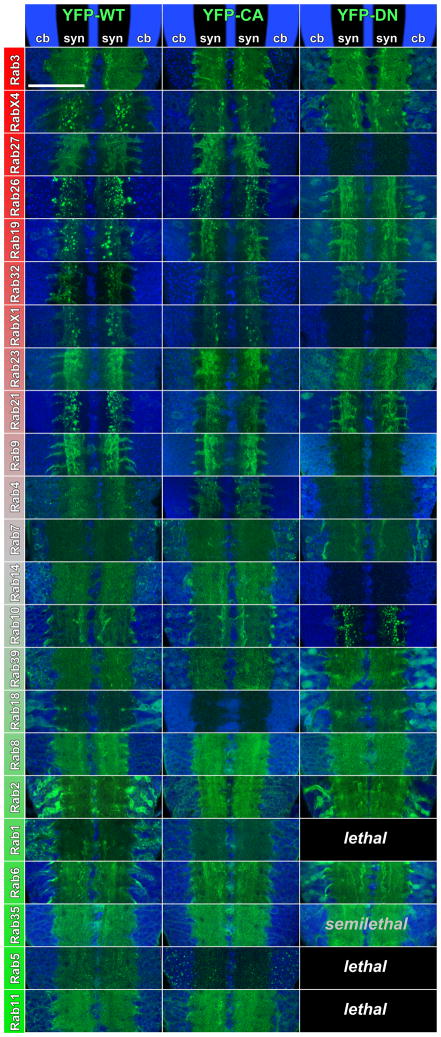
Functional analyses of rab GTPases can be performed using the GTP bound (constitutively active; CA) or GDP bound (dominant negative; DN) mutants [16]. We performed a functional profiling of all Rab proteins analyzed here by investigating the CA and DN variants expressed in their endogenous expression patterns (Figure 5). Remarkably, only the DN variants of the ubiquitous Rab1, Rab5, Rab11 and Rab35 cause lethality. None of the CA or DN variants of the neuronal Rabs caused obvious developmental or functional defects. We also tested dominant negative expression for the nine most neuronal Rabs at the neuromuscular junction and did not observe any obvious morphological defects (Figure S6A,B). These data suggest that neuronal rabs may serve modulating or partially redundant functions. This idea is consistent with the findings that a mutant of the best characterized pan-neuronal rab3 is viable in Drosophila [15] and a quadruple knock-out for all four vertebrate rab3 isoforms in mice develops normally, is born alive, and has a surprisingly mild defect on neuronal function [26]. In addition, the functional profiling of CA and DN Rabs revealed that the neuronal Rabs that mark synaptic Rab11-positive compartments (RabX4, Rab26, Rab19, Rab32, RabX1, Rab21) again show a common behavior: Both the WT and CA variants mark distinct compartments at synapses, while the DN variants exhibit no preferential synaptic localization and are diffusely distributed throughout the neurons (Figure 5). This is in accordance with the observation that GTP-bound Rabs exhibit increased labeling of specific compartments. Similarly, the well characterized endosomal marker proteins Rab5 and Rab7 label increased endosomal compartments as CA variants (Figure 5). A notable exception is Rab10, which exhibits increased synaptic aggregations when the dominant negative version is expressed and a more diffuse neuronal labeling when the constitutively active form is expressed. Amongst the ubiquitous Rabs, Rab2 shows the most ‘neuronal’ subcellular relocalization behavior, i.e. synaptic enrichment of the Rab2CA and loss of localization for Rab2DN. Taken together, our findings suggest that synapses are the principle site of action for neuronal Rabs based on RabCA protein localization.
Figure 5. Subcellular Localization Profiling as a function of GTP- and GDP-bound states.
Proximal ventral ganglion sections are shown sorted for all Rabs from most neuronal (top, red) to most ubiquitously expressed (bottom, green) similar to Figure 4. Corresponding Gal4-lines drive the expression of wild type YFP-tagged Rabs in the left column, constitutively active (GTP-bound) YFP-tagged Rabs in the middle column and dominant negative (GDP-bound) YFP-tagged Rabs in the right column. Toto-3 labels nuclei (blue). Note that most neuronal Rabs show synaptic localization and little or no cell body localization that is maintained and further enriched as constitutively active, but lost as dominant negative versions. Further note that only expression of the dominant negatives of Rab1, Rab5 and Rab11 cause embryonic or early larval lethality, while Rab35 dominant negative is semilethal with few adult escapers. A high-resolution version of this Figure is available online. Scale bar for all panels: 50 μm.
Generation of a rab27 knock-out by ends-out homologous recombination of the Gal4 knock-in cassette reveals a specific sleep phenotype
The objective of our profiling effort was to identify synaptic Rabs that potentially play roles in brain development and function and at the same time provide the tools to generate knock-outs in these genes. Our Gal4 knock-in cassettes can be mobilized in vivo and targeted to endogenous loci using ends-out homologous recombination [20] (Figure S6). This technique is, to our knowledge, the first application of BAC Recombineering to utilize genomic fragments as large (10-20kb) homology arms in Drosophila gene targeting. Furthermore, our strategy differs from published techniques in that the targeting cassette is positively marked with 3×P3-RFP and can be separated from the differently marked landing site (yellow +) and P[acman] backbone (white +) (Figure S6 and Supplemental Materials)
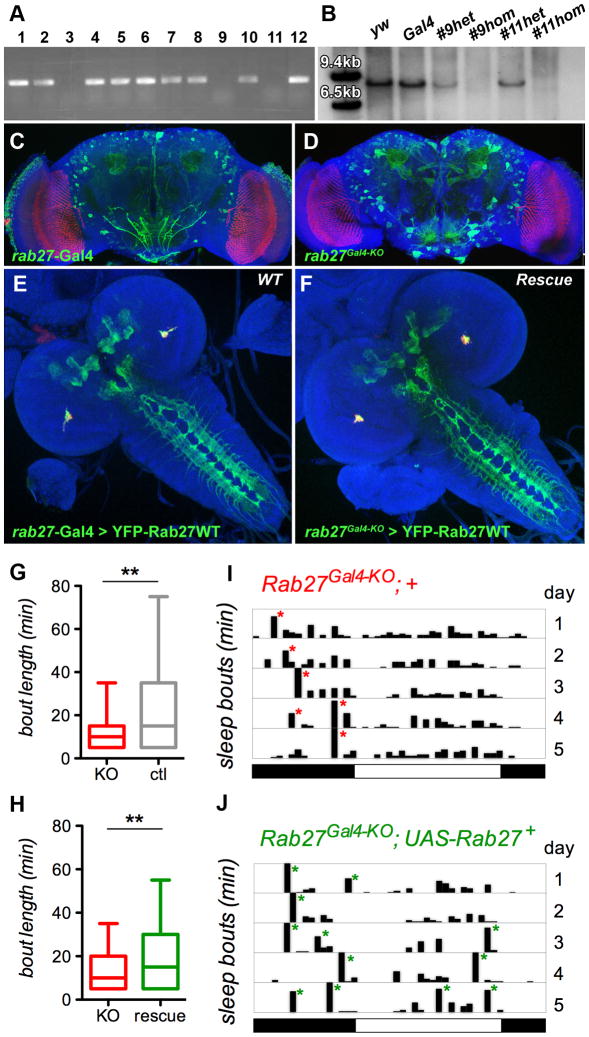
We chose rab27 for a ‘proof-of-principle’ knock-out screen for two reasons: First, it is the only strong synaptic vesicle marker other than the well-characterized rab3, consistent with a recent characterization of its role in synaptic vesicle exocytosis [27]; second, rab27 exhibits a highly specific expression pattern in brain structures with known functions. We screened approximately 30,000 F2 progeny for separation of the targeting cassette from the mobilization site and identified 37 re-integrations of the targeting cassette away from the mobilization site. 32 of the 37 genomic integration events occurred on the X chromosome, where the endogenous rab27 gene maps. 24 of the 32 lines were subsequently tested by PCR (12 shown in Figure 6A), indicating that the rab27 open reading frame was correctly replaced with the Gal4 cassette in 6 lines. The resulting recombination frequency is 1 in 4000 or 2.5 * 10−4, which is higher than the homologous recombination efficiency reported in a comparable recent report [28]. Finally, we tested and confirmed two lines by Southern blotting with a DNA probe against the rab27 open reading frame (Figure 6B). UAS-YFP-Rab27 driven by rab27-Gal4 in the landing site (Figure 6C, E) or rab27-Gal4 knocked into the endogenous locus (Figure 6D, F) exhibit identical Rab27 expression, corroborating that the 40kb genomic targeting cassette likely contained all the functionally significant regulatory elements of the endogenous locus.
Figure 6. Generation of a rab27 knock-out by ends-out homologous recombination of the Gal4 knock-in cassette reveals a specific sleep phenotype.
(A) PCR verification of 3×P3-RFP positive targeting cassette mobilizations and re-integrations in the genome indicate loss of the endogenous rab27 locus for three out of 12 potential knock-out lines. (B) Verification of two of the knock-outs by Southern with an ORF probe (comp. Suppl. Fig. 6). (C, D) rab27-Gal4 in the landing site and rab27Gal4-KO in the endogenous site exhibit very similar expression patterns (CD8-GFP in green) specific to the mushroom bodies in the adult brain. (E, F) YFP-tagged Rab27 expression is identical between YFP-Rab27 in wild type using rab27-Gal4 and YFP-Rab27 rescuing expression in a homozygous null mutant with rab27Gal4-KO knock-in. (G, H) Sleep phenotype in rab27Gal4-KO flies. rab27Gal4-KO flies show decreased daytime sleep bout length (median, quartiles, 90th percentile) compared to controls (G), a phenotype rescued by introducing the UAS-Rab27-YFP transgene (H). (I, J) Representative single fly sleepograms showing decreased bout length. Each bar represents a sleep bout, with the height indicating the duration (Y-axis =120 min, X-axis = 24 hours, light/dark cycle indicated by white/black boxes). rab27Gal4-KO flies have a decreased number of long, >60 min sleep bouts (indicated by asterisk), especially during the daytime, compared to rab27Gal4-KO;UAS-rab27+-YFP rescue flies.
rab27 homozygous mutant adults are viable and fertile. Previous results demonstrated that mushroom bodies, which exhibit specific expression of rab27, regulate sleep in flies [29, 30]. To determine whether loss of rab27 causes a behavioral phenotype, we assayed activity, circadian rhythm and sleep behavior of rab27Gal4-KO. rab27Gal4-KO flies displayed normal activity levels and rhythm strength in a 12 hrs/12 hrs light/dark (LD) cycle (Fig. S7A-C). Measurements of the duration of the longest sleep-bout for each day and night, showed a significant reduction during the daytime for rab27Gal4-KO. Analysis of all bouts revealed a more than 30% reduction in median and top quartile bout length for rab27Gal4-KO homozygotes compared to controls in the same y−w− genetic background (Figure 6G-J). This phenotype can be rescued by expressing UAS-YFP-rab27 driven by rab27Gal4-KO (Figure 6H). Hence, loss of rab27 leads to less consolidated daytime sleep, consistent with its cell-specific expression in mushroom bodies. This cell-specificity is preserved for two isoforms of rab27, both of which are knocked out in rab27Gal4-KO (Fig. S7D-F). Our data suggest that the cell-specific expression of neuronal rabs in different parts of the brain may relate to surprisingly specialized functions.
Discussion
In this paper, we present a novel approach to functional profiling of gene families in Drosophila by combining BAC recombineering with ends-out homologous recombination. We used this approach to generate 36 transgenic rab-Gal4 lines and performed systematic cellular and subcellular expression profiling of 23 Rab proteins. We report the surprising findings that (1) half of all Rabs are neuronal-specific or neuron-enriched, (2) different neuronal Rabs function in distinct subsets of neurons in the brain, (3) all neuronal Rabs localize to synapses and (4) synaptic Rabs predominantly recruit and mark Rab11-positive synaptic recycling endosomal compartments. Finally, we demonstrate the mobilization and homologous recombination of the Gal4 knock-in cassette by generating a knock-out for the neuron-specific rab27 gene. This rab27Gal4-KO null mutant exhibits a specific behavioral sleep phenotype that matches the cell-specific expression pattern.
An Improved Transgenesis Platform for Drosophila
Homologous recombination techniques in Drosophila have hitherto been limited by difficult vector construction and inefficient in vivo recombination. BAC recombineering allows streamlined cloning independent of restriction enzymes or other sequence-specific restrictions. We modified the existing BAC recombineering-based P[acman] vector [31] by incorporating a cassette for ends-out homologous recombination [20]. The underlying idea is to utilize large genomic fragments as homology arms for gene targeting instead of conventional PCR-based and size-restricted homology arms. We find that the key advantage of this technique lies in the ease of base-pair precise manipulation of genomic fragments in a single round of recombineering once the parent vector with genomic region has been generated. This is demonstrated in this paper for numerous alternative Gal4 knock-ins for the various rab loci. In contrast, PCR-based cloning often requires complete redesign for each targeting vector. The ability to systematically characterize cellular and subcellular expression patterns prior to performing an ends-out homologous recombination screen represents a second key strategic advantage for the present study. The large genomic fragments are several times larger than traditional genomic rescue constructs in Drosophila, thereby ensuring the expression accuracy of the Gal4 knock-ins. We chose Gal4 knock-ins as highly versatile tools, especially in light of the earlier generation of a complete collection of UAS-YFP-tagged rabs strains [16]. Gal4 knock-ins thus provide means for subcellular profiling and rescue using YFP-tagged proteins expressed under their own regulatory elements. In summary, the vectors and protocols generated here may provide a widely applicable method to modify large genomic constructs that can be mobilized for homologous recombination in a separate experimental step.
Our mobilization of the 40kb rab27 Gal4 targeting cassette by standard heat shock activation of the FLP and ISce1 enzymes was efficient. In contrast to previous implementations of ends-out homologous recombination in Drosophila, our knock-in cassette is positively marked with eye-specific RFP expression, thereby providing a simple means to follow the separation of the targeting cassette from the landing site and re-integration somewhere else in the genome with a rate of 1 in ∼800 progeny. More than 85% of the re-integration events were on the correct target chromosome, and 25% of these correct target chromosome insertions were correct gene replacements. This brings the final recombination frequency to 1 in 4,000 or 2.5 * 10−4, which compares favorably to rates between 7 * 10−6 and 1.9 * 10−4 in a recent report on improved ends-out homologous recombination for six different loci [28]. Since homologous recombination is highly locus-specific it is too early to quantitatively assess our method. In particular, a systematic test of the effect of homology arm length on recombination frequency has not been performed in Drosophila. P[acman]-KO may provide an effective means to test and implement further improvements of almost restriction free genomic gene manipulation in Drosophila on a medium to large scale.
Novel Insights into Rab Function in the Nervous System
The development of a BAC recombineering-based gene targeting technique was motivated by the desire to systematically study a large gene family in vivo. rab GTPases have been at the focal point of several systematic profiling efforts and dubbed the ‘membrome’ due to their common and yet diversified functional significance for all intracellular membrane trafficking [32]. Many aspects of the earlier microarray-based expression profiling are consistent with our finding, e.g. the neuronal expression of rab3 and rab26. However, a previous microarray-based profiling study did not observe the overall bias toward nervous system expression [32]. For example, both vertebrate rab27 isoforms were found to be expressed at low levels in the brain, yet we identify rab27 as a neuron-specific rab with highly restricted expression in the brain. Such cell-type specific expression is likely obscured in any microarray study, which by necessity assays a heterogeneous population of cells. Importantly, a partially overlapping role of the molecular functions of rab27 and rab3 was recently described for synaptic neurotransmitter release [27]. While this finding is consistent with our identification of rab27 and rab3 as the strongest synaptic vesicle co-localizing rabs, the restricted rab27 expression pattern makes it an unlikely general regulator of exocytosis in Drosophila. We surmise that the cellular and subcellular resolution profiling presented here captures important information about Rab protein localization that was not attainable in earlier studies on homogenized tissues.
Our functional profiling with wild type, constitutively active and dominant negative Rabs expressed in their endogenous expression patterns suggest that few, if any, of the neuronal rab GTPases are required for neuronal viability. One possible explanation is that dominant negative Rab expression may be poor substitutes for genetic loss of function alleles. An alternative or additional explanation may be partially redundant functions, which can be investigated with the tools presented here. Indeed, the best characterized neuronal rab3 is viable in Drosophila [15]. Similarly, a quadruple knock-out for all four vertebrate rab3 isoforms in mice develops normally, is born alive, and has a surprisingly mild defect on neuronal function [26]. In addition, we show here that a null mutant for the second synaptic vesicle-associated Rab, rab27, is viable. Interestingly, both loss of rab3 and rab27 cause mild and specific neuronal phenotypes [15, 26]. Taken together, these data lead us to speculate that neuronal rabs may serve specialized, modulatory functions in neurons.
Our subcellular localization profiling revealed that all neuronal Rab proteins (Rab3, RabX4, Rab27, Rab26, Rab19, Rab23, Rab32, RabX1, Rab21, Rab9) localize highly preferentially or exclusively to synapses. This observation suggests that the specialized or modulatory functions of neuronal rabs serve specific demands on membrane trafficking at synapses. This observation may not be too surprising, given that the axon termini are arguably the most specialized and distinct cellular compartments of neurons in comparison with other cell types. However, the identification of six of these 10 neuronal and synaptic Rab proteins as markers of Rab11-positive compartments is remarkable. Rab11 is a “gold-standard” marker for recycling endosomal compartments [33]. The highly regulated recycling of presynaptic release machinery, AMPA receptors and guidance receptors, to name but a few, may provide an explanation for the existence of such extensive, specialized synaptic membrane trafficking machinery. In this paper, we provide the tools and techniques to dissect this machinery in vivo.
Materials and Methods
Drosophila Strains and Genetics
For all rab-Gal4 transgenic strains we used the landing site attP-3B (Bloomington Stock #24871). The following mutant chromosomes for rescue experiments were obtained: rab3rup (gift from Aaron DiAntonio); rab6ASI, Frt40A (gift from Spyros Artivanis-Tsakonis); rab6[D23D], Frt40A (gift from Hugo Bellen); stocks #8907 and #25729 from the Bloomington Stock Center. Rescue experiments were set up as follows: Rab3: Df(2R)BSC639/CyO; UAS-YFP-Rab3-WT/TM3 × rab3rup/CyO; rab3-ATG-Gal4/TM3. Rab6: rab6[D23D], UAS-YFP-Rab6-WT/CyO × Df(2L)ED775/CyO; rab6-Gal4/TM3. Rab6: rab6ASI, FRT40A/CyO; UAS-YFP-Rab6-WT/TM3 × rab6ASI, FRT40A/CyO; rab6-Gal4/TM3. See Supplemental Experimental Procedures for full description of genetics and genotypes used.
Molecular Biology and Recombineering
attB-P[acman]-KO was generated by inserting Frt/ISce1 site into the existing Pac1 and Asc1 sites of attB-P[acman] [31] such that the original Pac1 and Asc1 sites were destroyed and new Asc1 and Pac1 sites generated proximally (Fig. 1). Second, the GatewayTM BP recombination cassette from pDONR221 was inserted to facilitate cloning proximally of the Frt/ISce1 sites independent of the Asc1 and Pac1 sites. The Gal4 knock-in cassette is described in detail in the Supplemental Experimental Procedures. The recombineering protocol was adapted from Venken et al. (2006) with the following modifications: For first round recombineering, two 500bp homology arms (LA and RA) flanking the 40kb fragment were PCR soe'd, with a BamH1 site added in the middle and attB1/attB2 site at the ends. The 1kb PCR products were cloned into P[acman]-KO using the Gateway BP reaction following manufacturer's instruction (Invitrogen BP clonase II 11789-020). 10 ug of the resulting P[acman]-KO-1kb was digested with BamH1-HF (NEB R3136S) at 37°C for 6 hours. After gel electrophoresis, DNA was extracted with Zymoclean Gel DNA recovery kit (Zymoresearch D4008). 5ul of total 12ul DNA eluate was electroporated into recombineering-competent DY380 cells. To verify the colonies, both LA and RA junctions were verified by PCR using primers outside of the 500bp regions, and then sequenced. Second round recombineering: Two 100bp sequences flanking the target region were added to the Gal4-RFP-Kan cassette as homology arms by PCR using Phusion® High-Fidelity DNA Polymerase (Finnzymes, Cat # F-530S) kit with the following conditions and cycling: Sterile Water 35.5μL, 5X Phusion HF Buffer 10 μL, MgCl2 (50 mM) 1 μL, dNTPs (10 mM TOTAL) 1.5 μL, Forward Primer (10 uM) 0.5 μL, Reverse Primer (10 uM) 0.5 μL, p-ENTR-Gal4 (45 ng / μL) 0.5 μL, Phusion DNA polymerase 0.5 μL. After gel electrophoresis, a 6.7kb band was excised and extracted with the Zymoclean kit. 100ng of DNA was transformed into recombineering-competent DY380 cells containing P[acman]-KO-40kb. The transformants were selected from Tet-Amp-Kan triple selective LB plates, and colonies were verified by PCR and confirmed by sequencing (P[acman]-Gal4). DNA from confirmed colonies was extracted and transformed into EPI300 cells for copy number induction and subsequent injection (Rainbow Transgenics). We note that the preparation of high quality DNA proved a key requirement for all steps from recombineering to transgenesis. The detailed recombineering protocol is available in Supplemental Experimental Procedures.
Immunohistochemistry, Microscopy, and Image Processing
Adult brains, eyes, and eye-lamina complexes as well a pupal brains and eye-brain complexes were dissected as reported [34]. The tissues were fixed in phosphate buffered saline (PBS) with 3.5% formaldehyde for 15 min and washed in PBS with 0.4% Triton X-100. High-resolution light microscopy was performed using the Resonance Scanning Confocal Microscope Leica SP5. Imaging data was processed and quantified using Amira 5.2 (Indeed, Berlin, Germany) and Adobe Photoshop CS4 as described in [35]. The following antibodies were used at 1:500: rabbit anti-rab5, rabbit anti-rab7, mouse anti-rab11. A mouse monoclonal antibody against CSP was used at 1:50.
Knock-out Screen
A full crossing scheme is depicted in Suppl. Fig. 6. In brief, rab27-Gal4 male transformants containing the Gal4 cassette in the landing site were crossed to females with hs-FLP and hs-ISceI (Bloomington stock number 6935). 48 hours after egg laying, embryos were heat-shocked at 37°C (twice per day for 3 consecutive days). 24 of 32 lines were subsequently verified by PCR for both 3′ and 5′ junctions of the open reading frame. 6 of 24 are negative in PCR, indicating the replacement of rab27 open reading frame with the cassette. The primers for PCR are listed below: 5′ junction fwd: 5-TCGCAGATTCCTTCCAGATC-3. 5′ junction rev: 5-CAATTAGGAGCAAACCACAA-3. 3′ junction fwd: 5-ATGGGTTTCCTGCTCATCTT-3. 3′ junction rev: 5-GCAGGCATCGCGACTGGGTC-3
Southern blot
Genomic DNA was prepared following Quick Fly Genomic DNA Prep from BDGP (http://www.fruitfly.org/about/methods/inverse.pcr.html). 20 ug of DNA from each strain were digested with Nhe1 -HF (NEB R3131S, 25U) at 37C for overnight, and then separated using 4% DNA gel at 35V, 4°C for overnight. The gel was subsequently incubated in Denaturing solution (1.5 M NaCl, 0.5M NaOH) for 45 minutes; Depurination solution (0.2N HCl) for 15 minutes; Neutralization Solution (1M Tris pH7.4, 1.5M NaCl) twice for 30 minutes; and then transferred onto a membrane (Amersham Hybond-N RPN303N) using 10% SSC buffer at room temperature for overnight. The membrane was crosslinked by UV, and incubated in pre-heated hybridization buffer (Roche 11796895001) for 30 minutes at 42°C. Dig-labeled Rab27 ORF probe was generated by PCR using the following primers: 5′-TTGACGTTGGCGCCGGTGCA-3′, 5′-TGAGCCTCTGCAATTAGCCGGAT-3′, labeled with Dig using Klenow (NEB) with labeling mix (NEB), boiled for 5 minutes to denature, and added to the membrane for hybridization overnight at 45°C. At room temperature, the membrane was washed twice in 2× SSC, 0.1% SDS for 20 minutes each, twice in 0.5× SSC, 0.1% SDS at 68C for 30 minutes each, rinsed in maleic acid buffer (100mM Maleic acid, 150 mM NaCl, pH7.5) with shaking for 5 minutes, blocked in block reagent (Roche 11096176001) for 3 hours, incubated in block solution with anti-Dig antibody (1:1000, 11093274910) for 30 minutes, washed twice in wash buffer (30ml maleic acid buffer with 90 ul Tween 20) for 15 minutes each, rinsed in detection buffer (0.1M Tris, 0.1M NaCl pH9.5) for 5 minutes, immersed in CDP-star solution, and then exposed to film for visualization.
Behavioral Analysis
Flies were assayed in a y w genetic background, and experimental and controls always were in identical, though sometimes hybrid backgrounds. Flies were entrained for three days, and assayed in a 12hr/12hr light/dark incubator and monitored with the Drosophila Activity Monitor system (TriKinetics, Waltham, Ma). Sleep times were determined as described [30].
Supplementary Material
Highlights.
Up to half of all Drosophila Rabs exhibit neuron-specific or highly enriched expression
All neuronal rabs encode synaptic proteins, most often labeling recycling endosomes
P[acman]-KO provides a new method combining recombineering and gene targeting
Proof-of-principle knock-out of rab27 reveals a cell-specific sleep phenotype
Acknowledgments
We would like to thank Matthew Scott, Hugo Bellen, Aaron DiAntonio, Spyros-Artavanis-Tsakonas, Patrick Dolph and the Bloomington Stock Center, and the University of Iowa Developmental Studies Hybridoma Bank for reagents. We further thank Koen Venken, Hugo Bellen, Nevine Shalaby, Tong-Wey Koh and all members of the Buszczak and Hiesinger labs for discussion and comments on the manuscript. We are especially grateful to Susanne Eaton for discussion and communication of results prior to publication. Technical assistance by Elzi Volk in the early stages of this project and by Ossama Saladin during revision is gratefully acknowledged. This work was supported by grants from the National Institute of Health to PRH (RO1EY018884), to MB (RO1GM086647) and AR (R01AA019526), a grant by the Cancer Prevention Research Institute of Texas to MB and PRH (RP100516), the Whitehall Foundation to PRH and the Welch Foundation (I-1657) to PRH. Adrian Rothenfluh is an Effie Marie Cain Scholar in Biomedical Research, Mike Buszczak is an E.E. and Greer Garson Fogelson Scholar in Biomedical Research and P.R. Hiesinger is a Eugene McDermott Scholar in Biomedical Research at UT Southwestern Medical Center.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Touchot N, Chardin P, Tavitian A. Four additional members of the ras gene superfamily isolated by an oligonucleotide strategy: molecular cloning of YPT-related cDNAs from a rat brain library. Proc Natl Acad Sci U S A. 1987;84:8210–8214. doi: 10.1073/pnas.84.23.8210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zerial M, McBride H. Rab proteins as membrane organizers. Nat Rev Mol Cell Biol. 2001;2:107–117. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
- 3.Pfeffer SR. Unsolved mysteries in membrane traffic. Annu Rev Biochem. 2007;76:629–645. doi: 10.1146/annurev.biochem.76.061705.130002. [DOI] [PubMed] [Google Scholar]
- 4.Pfeffer SR. Rab GTPases: master regulators of membrane trafficking. Curr Opin Cell Biol. 1994;6:522–526. doi: 10.1016/0955-0674(94)90071-x. [DOI] [PubMed] [Google Scholar]
- 5.Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol. 2009;10:513–525. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- 6.Molendijk AJ, Ruperti B, Palme K. Small GTPases in vesicle trafficking. Curr Opin Plant Biol. 2004;7:694–700. doi: 10.1016/j.pbi.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 7.Pfeffer S, Aivazian D. Targeting Rab GTPases to distinct membrane compartments. Nat Rev Mol Cell Biol. 2004;5:886–896. doi: 10.1038/nrm1500. [DOI] [PubMed] [Google Scholar]
- 8.Aligianis IA, Johnson CA, Gissen P, Chen D, Hampshire D, Hoffmann K, Maina EN, Morgan NV, Tee L, Morton J, et al. Mutations of the catalytic subunit of RAB3GAP cause Warburg Micro syndrome. Nat Genet. 2005;37:221–223. doi: 10.1038/ng1517. [DOI] [PubMed] [Google Scholar]
- 9.Menasche G, Pastural E, Feldmann J, Certain S, Ersoy F, Dupuis S, Wulffraat N, Bianchi D, Fischer A, Le Deist F, et al. Mutations in RAB27A cause Griscelli syndrome associated with haemophagocytic syndrome. Nat Genet. 2000;25:173–176. doi: 10.1038/76024. [DOI] [PubMed] [Google Scholar]
- 10.Verhoeven K, De Jonghe P, Coen K, Verpoorten N, Auer-Grumbach M, Kwon JM, FitzPatrick D, Schmedding E, De Vriendt E, Jacobs A, et al. Mutations in the small GTP-ase late endosomal protein RAB7 cause Charcot-Marie-Tooth type 2B neuropathy. Am J Hum Genet. 2003;72:722–727. doi: 10.1086/367847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mitra S, Cheng KW, Mills GB. Rab GTPases implicated in inherited and acquired disorders. Semin Cell Dev Biol. 2010;22:57–68. doi: 10.1016/j.semcdb.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nachury MV, Loktev AV, Zhang Q, Westlake CJ, Peranen J, Merdes A, Slusarski DC, Scheller RH, Bazan JF, Sheffield VC, et al. A Core Complex of BBS Proteins Cooperates with the GTPase Rab8 to Promote Ciliary Membrane Biogenesis. Cell. 2007;129:1201–1213. doi: 10.1016/j.cell.2007.03.053. [DOI] [PubMed] [Google Scholar]
- 13.Satoh AK, O'Tousa JE, Ozaki K, Ready DF. Rab11 mediates post-Golgi trafficking of rhodopsin to the photosensitive apical membrane of Drosophila photoreceptors. Development. 2005;132:1487–1497. doi: 10.1242/dev.01704. [DOI] [PubMed] [Google Scholar]
- 14.Mehta SQ, Hiesinger PR, Beronja S, Zhai RG, Schulze KL, Verstreken P, Cao Y, Zhou Y, Tepass U, Crair MC, et al. Mutations in Drosophila sec15 reveal a function in neuronal targeting for a subset of exocyst components. Neuron. 2005;46:219–232. doi: 10.1016/j.neuron.2005.02.029. [DOI] [PubMed] [Google Scholar]
- 15.Graf ER, Daniels RW, Burgess RW, Schwarz TL, DiAntonio A. Rab3 dynamically controls protein composition at active zones. Neuron. 2009;64:663–677. doi: 10.1016/j.neuron.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang J, Schulze KL, Hiesinger PR, Suyama K, Wang S, Fish M, Acar M, Hoskins RA, Bellen HJ, Scott MP. Thirty-one flavors of Drosophila rab proteins. Genetics. 2007;176:1307–1322. doi: 10.1534/genetics.106.066761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ali BR, Seabra MC. Targeting of Rab GTPases to cellular membranes. Biochem Soc Trans. 2005;33:652–656. doi: 10.1042/BST0330652. [DOI] [PubMed] [Google Scholar]
- 18.Jordens I, Marsman M, Kuijl C, Neefjes J. Rab proteins, connecting transport and vesicle fusion. Traffic. 2005;6:1070–1077. doi: 10.1111/j.1600-0854.2005.00336.x. [DOI] [PubMed] [Google Scholar]
- 19.Venken KJ, Bellen HJ. Transgenesis upgrades for Drosophila melanogaster. Development. 2007;134:3571–3584. doi: 10.1242/dev.005686. [DOI] [PubMed] [Google Scholar]
- 20.Gong WJ, Golic KG. Ends-out, or replacement, gene targeting in Drosophila. Proc Natl Acad Sci U S A. 2003;100:2556–2561. doi: 10.1073/pnas.0535280100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharan SK, Thomason LC, Kuznetsov SG, Court DL. Recombineering: a homologous recombination-based method of genetic engineering. Nat Protoc. 2009;4:206–223. doi: 10.1038/nprot.2008.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Venken KJ, Popodi E, Holtzman SL, Schulze KL, Park S, Carlson JW, Hoskins RA, Bellen HJ, Kaufman TC. A molecularly defined duplication set for the X chromosome of Drosophila melanogaster. Genetics. 2010;186:1111–1125. doi: 10.1534/genetics.110.121285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Venken KJ, Carlson JW, Schulze KL, Pan H, He Y, Spokony R, Wan KH, Koriabine M, de Jong PJ, White KP, et al. Versatile P[acman] BAC libraries for transgenesis studies in Drosophila melanogaster. Nat Methods. 2009;6:431–434. doi: 10.1038/nmeth.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Freeman M. Reiterative use of the EGF receptor triggers differentiation of all cell types in the Drosophila eye. Cell. 1996;87:651–660. doi: 10.1016/s0092-8674(00)81385-9. [DOI] [PubMed] [Google Scholar]
- 25.Zerial M. Regulation of endocytosis by the small GTP-ase rab5. Cytotechnology. 1993;11(1):S47–49. doi: 10.1007/BF00746052. [DOI] [PubMed] [Google Scholar]
- 26.Schluter OM, Schmitz F, Jahn R, Rosenmund C, Sudhof TC. A complete genetic analysis of neuronal Rab3 function. J Neurosci. 2004;24:6629–6637. doi: 10.1523/JNEUROSCI.1610-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pavlos NJ, Gronborg M, Riedel D, Chua JJ, Boyken J, Kloepper TH, Urlaub H, Rizzoli SO, Jahn R. Quantitative analysis of synaptic vesicle Rabs uncovers distinct yet overlapping roles for Rab3a and Rab27b in Ca2+-triggered exocytosis. J Neurosci. 2010;30:13441–13453. doi: 10.1523/JNEUROSCI.0907-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang J, Zhou W, Dong W, Watson AM, Hong Y. Directed, efficient, and versatile modifications of the Drosophila genome by genomic engineering. Proc Natl Acad Sci U S A. 2009;106:8284–8289. doi: 10.1073/pnas.0900641106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Joiner WJ, Crocker A, White BH, Sehgal A. Sleep in Drosophila is regulated by adult mushroom bodies. Nature. 2006;441:757–760. doi: 10.1038/nature04811. [DOI] [PubMed] [Google Scholar]
- 30.Pitman JL, McGill JJ, Keegan KP, Allada R. A dynamic role for the mushroom bodies in promoting sleep in Drosophila. Nature. 2006;441:753–756. doi: 10.1038/nature04739. [DOI] [PubMed] [Google Scholar]
- 31.Venken KJ, He Y, Hoskins RA, Bellen HJ. P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science. 2006;314:1747–1751. doi: 10.1126/science.1134426. [DOI] [PubMed] [Google Scholar]
- 32.Gurkan C, Lapp H, Alory C, Su AI, Hogenesch JB, Balch WE. Large-scale profiling of Rab GTPase trafficking networks: the membrome. Mol Biol Cell. 2005;16:3847–3864. doi: 10.1091/mbc.E05-01-0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ullrich O, Reinsch S, Urbe S, Zerial M, Parton RG. Rab11 regulates recycling through the pericentriolar recycling endosome. J Cell Biol. 1996;135:913–924. doi: 10.1083/jcb.135.4.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Williamson WR, Hiesinger PR. Preparation of developing and adult Drosophila brains and retinae for live imaging. J Vis Exp. 2010 doi: 10.3791/1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Williamson WR, Yang T, Terman JR, Hiesinger PR. Guidance receptor degradation is required for neuronal connectivity in the Drosophila nervous system. PLoS Biol. 2010;8:e1000553. doi: 10.1371/journal.pbio.1000553. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.