Abstract
The localization of the planar cell polarity proteins Vang12, frizzled-3, Vang11, and Celsr1 in the rat incisors was examined using immunocytochemistry. The results showed that Vang12 was localized at two regions of the Tomes’ processes of inner enamel–secretory ameloblasts in rat incisors: a proximal and a distal region. In contrast, frizzled-3 was localized at adherens junctions of the proximal and distal areas of inner enamel– and outer enamel–secretory ameloblasts, where N-cadherin and β-catenin were localized. frizzled-3 was also localized in differentiating inner enamel epithelial cells. Vang11 was localized sparsely in differentiating preameloblasts and extensively at the cell boundary of stratum intermedium. Celsr1 was not localized in ameloblasts but localized in odontoblasts extensively. These results suggest the involvement of planar cell polarity proteins in odontogenesis.
Keywords: ameloblasts, Vang12, frizzled-3, Vang11, Celsr1, planar cell polarity, immunofluorescence, enamel, rat incisor
Epithelial cells are known to be polarized along the apical-basal axis. In addition to this apical-basal polarity, most epithelial cells have another polarity, which is referred to as planar cell polarity (PCP). PCP genes and their expression have been extensively studied using Drosophila wings and eyes (Klein and Mlodzik 2005; Wu and Mlodzik 2009). Drosophila wing cells are polarized along the distal-proximal axis, and Drosophila eye cells are polarized along the dorsal-ventral axis. PCP proteins are located in the non-canonical Wnt signaling pathway (Klein and Mlodzik 2005; van Amerongen and Berns 2006). The core PCP genes are frizzled, disheveled, Van Gogh (Vang) (also known as strabismus), prickle, diego, and flamingo. Disheveled is also shown to have a role in the canonical Wnt/β-catenin pathway. These core PCP genes are present in all organisms from Drosophila to mammals and are therefore evolutionarily conserved (Fanto and McNeill 2004; Klein and Mlodzik 2005; van Amerongen and Berns 2006; Lawrence et al. 2007; Seifert and Mlodzik 2007; Wang and Nathans 2007; Vlader et al. 2009; Wu and Mlodzik 2009; McNeill 2010; Skoglund and Keller 2010; Wallingford 2010; Goodrich and Strutt 2011; Wallingford and Mitchell 2011; Wansleeben and Meijlink 2011). The presence of Vang12 and frizzled-3, which are vertebrate genes of Vang and frizzled, has been previously detected in dental tissue by using in situ hybridization and microarray gene expression analysis (Tissir and Goffinet 2006). However, incisor, which has been long studied because of examination of whole amelogenesis in a tooth, has not been examined. Also, no information of protein distribution of PCP in dental tissues is available.
PCP proteins are also involved in convergent extension of spinal cord epithelia and notochordal and somatic tissues in Xenopus and zebrafish (Keller et al. 2000; Keller 2002; Wallingford et al. 2002; Torban et al. 2004; Seifert and Mlodzik 2007). In these notochord or somite elongation processes, cells with lamellipodia at their medial or lateral end, where the actin cytoskeleton is abundant, are intercalated by pulling each other medially or laterally.
An enamel rod is a basic unit of enamel structure. The course along which an enamel rod runs varies between different regions of enamel and also between different animal teeth, including human teeth (Kawai 1955; Osborn 1968a, 1968b; Boyde 1971). As expected, the movement of secretory ameloblasts during enamel secretion dictates enamel rod arrangement and orientation (Nishikawa 1992; Hanaizumi et al. 2010). Actin and actin-related cytoskeletal components such as myosin and tropomyosin are abundant at ameloblast junctions (Nishikawa and Kitamura 1985a, 1986; Nishikawa et al. 1988; Nishikawa 1992). The localization of adherens junction proteins such as N-cadherin (A-CAM), E-cadherin, β-catenin, and p120-catenin at these ameloblast junctions was previously examined (Nishikawa et al. 1990; Obara et al. 1998; Sorkin et al. 2000; Bartlett et al. 2010). However, directional movement of ameloblasts may require ameloblast planar cell polarization as well as a distinct localization of actin-based filament bundles. In fact, polarized distribution of actin filament bundles has been reported (Nishikawa and Kitamura 1985b, 1986; Nishikawa et al. 1988). Because the downstream signaling pathway of planar cell polarity involves actin cytoskeleton organization, it was therefore of interest to examine the localization of PCP proteins in ameloblasts to determine if they play a role in odontogenesis.
Materials and Methods
Seventeen male Jcl Wistar rats (4–5 weeks old, 73–145 g, obtained from CLEA Japan, Tokyo, Japan) were used for the present study. Institutional guidelines for animal care were followed for all experimental procedures. The animals were sacrificed by decapitation under deep anesthesia. The maxillae and mandibles were dissected and were then fixed with 4% paraformaldehyde at 4C overnight. Subsequently, the maxillae and mandibles were demineralized for 3 weeks at 4C with 5% EDTA, pH 7.2 (adjusted with concentrated NaOH solution). After washing with PBS and infusing with a 25% sucrose solution in PBS, the tissues were rapidly frozen, and cryosections were made using a cryotome (HM505E, Microm, Walldorf, Germany).
Cryosections were labeled with rabbit polyclonal antibodies against a synthetic peptide corresponding to the N-terminal extracellular domain of human frizzled-3 (FZD3, LS-A4434; MBL International, Woburn, MA) and goat polyclonal antibodies against a synthetic peptide corresponding to a region within the N-terminal domain of Vang12 (N-13) (sc-46561; Santa Cruz Biotechnology, Santa Cruz, CA). Cryosections were incubated with anti–frizzled-3 Ab diluted 1:10 with 1% bovine serum albumin in PBS (BSA-PBS) at room temperature for 30 min, followed by labeling with Alexa 488–conjugated donkey anti-rabbit IgG or Alexa 555–conjugated donkey anti-rabbit IgG (Invitrogen, Camarillo, CA) diluted 1:100 at room temperature for 30 min. Sections as negative control for stainings were incubated with normal rabbit immunoglobulin (Dako, Glostrup, Denmark) instead of the primary antibody, using an equivalent dilution, and were processed in the same way as described above. Some cryosections were labeled with anti-Vang12 Ab at a concentration of 20 mg/ml diluted with 1% BSA-PBS, at 4C overnight, followed by labeling with FITC-conjugated anti-goat IgG (Santa Cruz Biotechnology) diluted 1:20 with 1% BSA-PBS at room temperature for 30 min. For staining control on sections, the anti-Vang12 antibody solution was first incubated with excess blocking peptide (sc-46561P; Santa Cruz Biotechnology) for 2 hr, according to the manufacturer’s instructions. Sections as negative control for stainings were then incubated with the resulting antibody-absorbed solution and processed in the same way as described above. Some other sections were labeled with anti-Vang12 antibody, followed by ImmunoCruz Staining System (goat LSAB, sc-2053; Santa Cruz Biotechnology) and developed by diaminobenzidine (DAB) as chromogen.
Other cryosections were labeled with antibodies against adherens junction proteins: rabbit monoclonal anti–N- cadherin antibody (C-terminus clone EPR1792Y; Millipore, Temecula, CA) and rabbit polyclonal anti-β-catenin antibody (Millipore). Sections were labeled with anti-N- cadherin and anti-β-catenin Abs and then diluted 1:50 or 1:100 with 1% BSA-PBS, at 4C for 30 min, followed by labeling with Alexa 488–conjugated anti-rabbit IgG (Invitrogen).
For F-actin detection, sections were labeled with rhodamine-phalloidin (Invitrogen), diluted 1:50, at room temperature for 30 min. Cell nuclei were labeled by incubation of sections with 1 mg/ml Hoechst 33342 (Invitrogen) at room temperature for 30 min.
Some of the animals (nine-day-old Jcl Wistar rats; CLEA Japan) were sacrificed under deep anesthesia and rapidly frozen, and freeze-dried sections were obtained without fixation and demineralization according to the method of Kawamoto (2003). Freeze-dried sections (5 µm thick) of mandibles, including incisors, were labeled with anti– frizzled-3 antibodies followed by Alexa 488–conjugated anti-rabbit IgG in the manner described above. Freeze-dried sections were also labeled with anti-Vang11 (G-17, sc-46557; Santa Cruz Biotechnology) or anti-Celsr1 (M-125, sc-99198; Santa Cruz) diluted 1:20, followed by labeling with FITC-conjugated anti-goat IgG (Santa Cruz Biotechnology) or Alexa 488–conjugated anti-rabbit IgG (Invitrogen), respectively. Some sections were also labeled with Alexa Fluor 647 phalloidin (Invitrogen) diluted 1:50 for F-actin detection. Sections as negative control for stainings were incubated with 1% BSA-PBS alone for anti-Vang11 antibody and normal rabbit IgG in 1% BSA-PBS for anti-Celsr1 antibody, resulting in no specific labelings in the dental tissues of incisors.
Fluorescent images were acquired using an Olympus AX80 fluorescence microscope equipped with a CCD camera (Quantix KAF1401E; Photometrics, Tucson, AZ) and using MetaMorph software (Universal Imaging; Downington, PA).
Results
Adherens Junction Protein Localization in Ameloblasts
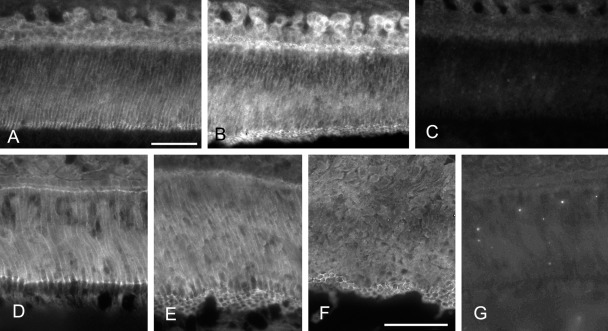
Because the downstream effector of the non-canonical pathway of Wnt signaling is actin cytoskeleton, actin-associated adherens junction proteins were examined. Adherens junctions of secretory ameloblasts were localized by staining for the junction markers β-catenin and N-cadherin. An N-cadherin signal was detected at the proximal and distal ends of inner enamel–secretory and outer enamel–secretory ameloblasts (Fig. 1A–C). Inner enamel–secretory and outer enamel–secretory ameloblasts were classified by the shape of the distal junction of each cell and arrangement of Tomes’ processes of cells: The former had a rectangular junction and herringbone pattern of Tomes’ processes, and the latter had a flattened hexagonal junction and uniformly running pattern of Tomes’ processes (Nishikawa et al. 1988). Based on this labeling pattern, the labeled structures were considered to be adherens junctions located at proximal and distal junctional complexes (Nishikawa et al. 1990; Bartlett et al. 2010). Another adherens junction protein, β-catenin, was localized at the proximal and distal junctions of inner enamel–secretory, outer enamel–secretory, and early maturation ameloblasts (Figs. 1D–G and 2B,C).
Figure 1.
Immunofluorescent localization of anti–N-cadherin and anti–β-catenin antibody signals in the ameloblasts of rat mandibular incisors. Sections of rat mandibular incisors were immunohistochemically stained with anti–N-cadherin (A, B) or anti–β-catenin (D–F) antibodies. Sections as negative control for anti–N-cadherin (C) and anti–β-catenin (G) are also shown. Inner enamel–secretory (A, C, D, G), outer enamel–secretory (E), outer enamel–secretory to transitional (B), and transitional (F) ameloblasts are shown. An anti–N-cadherin antibody signal was detected at the proximal and distal junctions of inner enamel– and outer enamel–secretory ameloblasts (A, B). The sections as negative control show no specific labeling (C). An anti–β-catenin antibody signal was also detected at the proximal and distal junctional complexes in inner enamel– and outer enamel–secretory ameloblasts as well as at the distal cell end of transitional amelobasts (D, E, G). No specific reactivity was observed in the section as the negative control (G). Bars = 50 µm.
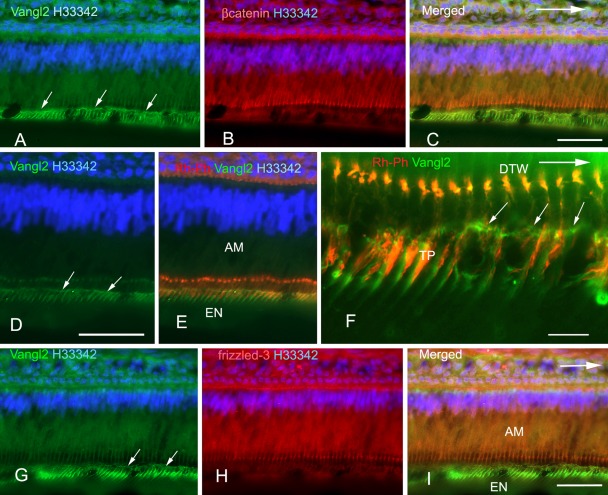
Figure 2.
Triple labeling of inner enamel–secretory ameloblasts with anti-Vang12 and Hoechst 33342 (H33342) and anti–β-catenin, rhodamine-phalloidin, or anti–frizzled-3. Sections of rat incisors were stained with anti-Vang12 Ab (A, C–G, I) (green), anti–β-catenin (B, C) (red), rhodamine-phalloidin (Rh-Ph) (E, F) (red), or anti–frizzled-3 (H, I) (red), and nuclei were stained with Hoechst 33342 (H33342) (A–E, G–I) (blue). A merged image of (A) and (B) is shown in (C). An anti-Vang12 Ab-labeled horizontal fluorescent line (arrows in A, D, F, G) was localized at the proximal part of Tomes’ processes. This region was located 5 to 10 µm below the distal junctional complexes that were labeled with anti–β-catenin (B, C) or F-actin (E, F). Anti–frizzled-3 antibodies labeled distal junctional complexes (H, I). Anti-Vang12-labeled vertical fluorescent lines were also seen below the horizontal fluorescent line in Tomes’ processes (A, C–G, I). Double-labeling of Tomes’ processes (TP in F) with anti-Vang12 Ab and rhodamine-phalloidin showed that these vertical fluorescent lines were localized at the lateral surface of Tomes’ process on the apical side of the incisor (F). The arrows indicate the incisal direction. AM, ameloblast; EN, enamel; DTW, distal terminal web; TP, Tomes’ process. (C, D, I) Bars = 50 µm. (F) Bar = 10 µm.
Localization of frizzled-3, Vang12, and Vang11 in Incisor Enamel Organ
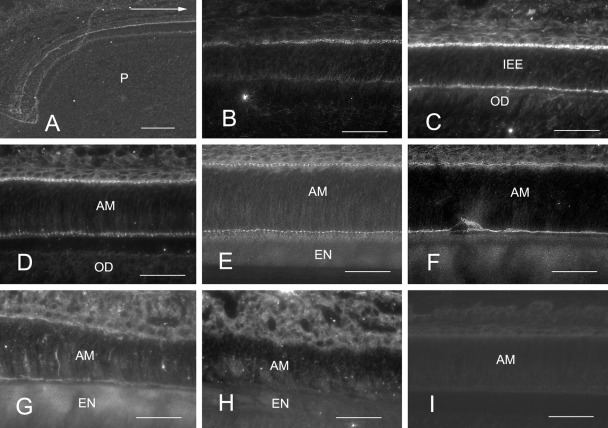
The localization of two PCP proteins, Vang12 and frizzled-3, in rat incisor sections was then analyzed by immunofluorescence (Figs. 2–4). The anti–frizzled-3 Ab labeled proximal and distal junctional complexes in inner enamel–secretory ameloblasts (Figs. 2H,I, 3A,C, and 4), whereas the anti-Vang12 Ab labeled only secretory ameloblasts. The localization of frizzled-3 in freeze-dried specimens that were not fixed or demineralized was then analyzed, and frizzled-3 localization at different stages of ameloblast differentiation and enamel secretion was determined. An anti–frizzled-3 Ab signal was localized at the proximal junctional area of differentiating inner enamel epithelial cells (Fig. 4A,B). At a later stage of differentiation, it became localized at the distal junctional area in addition to the proximal junctional area (Fig. 4C). In secretory ameloblasts, the anti–frizzled-3 Ab clearly labeled both the proximal and distal junctional areas (Fig. 4D–F), whereas it did not label ameloblasts at the transition zone or at maturation (Fig. 4G,H). Sections as negative control for stainings showed no specific labeling (Fig. 4I). The anti-Vang12 antibodies strongly labeled Tomes’ processes of inner enamel–secretory ameloblasts, displaying a linear staining pattern, but only weakly labeled outer enamel–secretory ameloblasts (Figs. 2A,D,G and 3B,D,F,G).
Figure 4.
Frizzled-3 labeling of freeze-dried sections of rat incisors without fixation and demineralization. Anti–frizzled-3 labeling (A–H) of inner enamel epithelia and ameloblasts at the following stages are shown: proliferation (A), early differentiation (B), late differentiation (C), early inner enamel secretion (D), middle inner enamel secretion (E), outer enamel secretion (F), transition (G), and early maturation (H). A section for negative control is also shown (I). Initially, the proximal part of the inner enamel epithelia was anti–frizzled-3 Ab-positive (A, B). The distal part then became positive in late differentiation (C). Both proximal and distal junctional complexes in the region of inner and outer enamel secretion were positive (D–F), but fluorescence gradually diminished at the transition zone (G) and disappeared in the zone of maturation (H). The section for negative control showed no specific labeling (I). Odontoblasts showed no labeling with the anti–frizzled-3 Ab (C, D). The arrow in (A) shows incisal direction. IEE, inner enamel epithelial cell; AM, ameloblast; EN, enamel; OD, odontoblast; P, dental pulp. Bars = 50 µm.
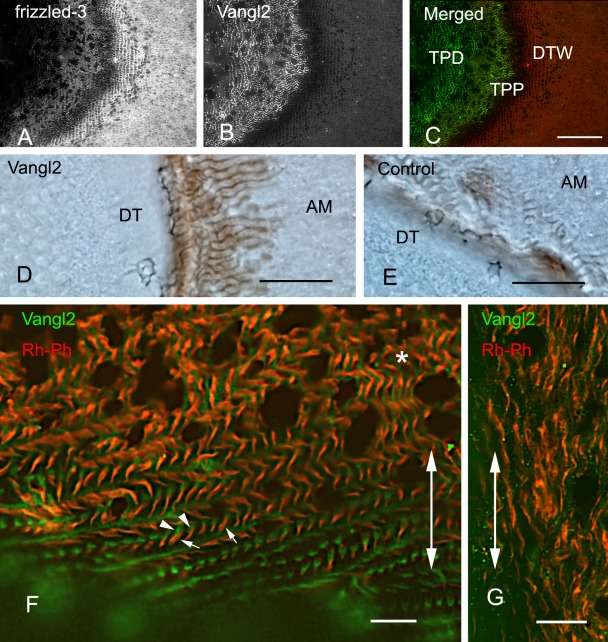
Figure 3.
Labeling of Vang12, frizzed-3, and actin in tangential sections of ameloblasts. Tangential sections of rat incisors at the level of Tomes’ processes and the distal terminal web at the zone of inner enamel secretion (A–F) and outer enamel secretion (G) are shown. (A–C) Double labeling with anti–frizzled-3 and anti-Vang12. The anti–frizzled-3 Ab labeled distal junctional complexes (DTW) but not Tomes’ processes (A, C), whereas the anti-Vang12 Ab labeled proximal and distal parts of Tomes’ processes (TPP and TPD, respectively) but not distal junctional complexes (B, C). (D, E) Anti-Vang12 localization in early enamel formation visualized by immunoperoxidase followed by DAB staining showed brown positive reactivity around the newly formed Tomes’ processes (D). The staining control on the section that was incubated with anti-Vang12 antibodies preabsorbed with excess antigen showed no specific reactivity (E). DT, dentin; AM, ameloblast. (F, G) Double labeling with the anti-Vang12 antibody and with rhodamine-phalloidin (Rh-Ph) for F-actin detection. Whole Tomes’ processes were labeled with rhodamine-phalloidin (red). Distal parts of Tomes’ processes are shown by small arrows (F). In the area of inner enamel secretion (F), the anti-Vang12 Ab (green) labeled the periphery of the proximal part of Tomes’ processes (F, asterisk) and also one side of the distal part of Tomes’ processes (F, arrowheads), whereas in the area of outer enamel secretion (G), only long, slender fluorescent Tomes’ processes (red) and weak anti-Vang12 Ab fluorescence (green) were observed. Large double-pointed arrows (F, G) show the long axis of the incisor. (C) Bar = 50 µm. (D, E) Bars = 20 µm. (F, G) Bars = 10 µm.
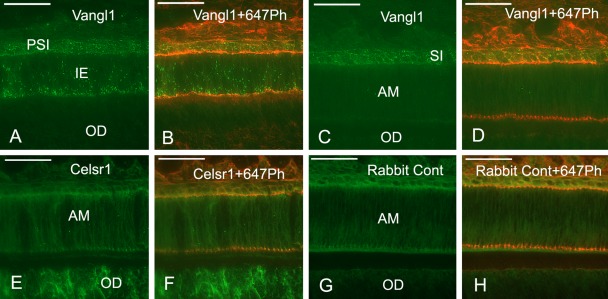
Double labeling of ameloblasts with anti-Vang12 and anti–β-catenin Abs indicated that these proteins were localized in different regions of ameloblasts. When incisors were longitudinally sectioned, the anti-Vang12 Ab signal was localized 6 to 8 µm below the anti–β-catenin–positive signal. This anti–β-catenin–positive signal was detected as a distal dotted line and indicated adherens junctions at the distal end of the cell body (Fig. 2B,C). The anti-Vang12 reactive signal appeared as a horizontally extended fluorescent line (Fig. 2A,C). Vertical fluorescent lines extended downwards from the horizontal linear fluorescence, toward the dentino-enamel junction, and reached up to 15 µm in height (Fig. 2A,C). Longitudinal sections were also double labeled with anti-Vang12 and rhodamine- phalloidin (Rh-Ph), which stains F-actin at the adherens junction and at the Tomes’ processes of the ameloblasts. This staining indicated a horizontal fluorescent anti-Vang12 signal at the proximal region of Tomes’ processes that was similar in appearance to the staining at the distal region. This anti-Vang12 signal was located 6 µm below the Rh-Ph–labeled distal junctional area, which appeared as a dotted line (Fig. 2D–F). Furthermore, vertical fluorescent lines were again detected with the anti-Vang12 Ab. These lines were inclined in the apical direction of the incisor and were always attached to Rh-Ph–positive Tomes’ processes (Fig. 2E,F). Double labeling of ameloblasts with anti-Vang12 and anti–frizzled-3 antibodies confirmed that these antibodies labeled different parts of the ameloblasts; the anti-Vang12 Ab labeled Tomes’ processes as vertical and horizontal fluorescent lines, and the anti–frizzled-3 Ab labeled the distal junctional complexes as dotted lines (Fig. 2G–I). Tangential sections of the incisor were also examined by double labeling with anti–frizzled-3 and anti-Vang12 Abs. These sections provided profiles of transverse sections of ameloblasts, which allowed analysis of inner enamel–secretory ameloblasts. The antibody signals were clearly detected at different locations in these sections: The anti–frizzled-3 Ab signal was localized at the distal junctional complexes in the form of rectangular outlines, and the anti-Vang12 Ab signal was localized at Tomes’ processes (Fig. 3A–C). In regions of the section in which enamel secretion was being initiated, an anti-Vang12 antibody signal that had the appearance of a brown deposit was localized at the periphery of newly formed Tomes’ processes (Fig. 3D). Staining control on sections that were incubated with antibody that had been preabsorbed with excess antigen showed no specific reactivity (Fig. 3E). Double labeling with anti-Vang12 and Rh-Ph showed that, in the middle region of the area of inner enamel secretion, anti-Vang12 was localized both around the middle part and at the end of the distal part of Tomes’ processes that were labeled with Rh-Ph (Fig. 3F). On the other hand, little anti-Vang12 signal could be detected at Tomes’ processes in the outer enamel–secretory ameloblasts (Fig. 3G). As another Vang gene of vertebrates, Vang11 is known (Wu and Mlodzik 2009). Anti-Vang11 localization was, therefore, examined to know whether Vang11 and Vang12 play a redundant role in odontogenesis, using the freeze-dried specimens that were not fixed or demineralized. Anti-Vang11 Ab was localized in the stratum intermedium from differentiating enamel epithelia to the papillary layer of the enamel maturation zone in the incisors (Fig. 5A–D). In close examination, the boundary of stratum intermedium cells was labeled as bright fluorescent dots (Fig. 5B,D). Inner enamel epithelial cells in the differentiating zone were also labeled as sparsely fluorescent dots (Fig. 5A,B).
Figure 5.
Vang11 and Celsr1 labeling of freeze-dried sections of rat incisors without fixation and demineralization. Double labeling of differentiating inner epithelia (IE) and provisional stratum intermedium (PSI) (A, B) and inner enamel–secretory ameloblasts (AM) and stratum intermedium (SI) (C, D) with anti-Vang11 (A–D, green) and Alexa 647 phalloidin (B, D, red). Double labeling of inner enamel–secretory ameloblasts (AM) and odontoblasts (OD) with anti-Celsr1 (E, F, green) and Alexa 647 phalloidin (F, red). The section as the negative control for staining was incubated with normal rabbit IgG followed with Alexa 488–conjugated anti-rabbit IgG (G, H, green) and Alexa 647 phalloidin (H, red). Anti-Vang11 labels differentiating preameloblasts and provisional stratum intermedium as bright fluorescent dots (A, B), and stratum intermedium but not secretory ameloblasts (C, D). Anti-Celsr1 labels odontoblasts extensively but does not label ameloblasts (E, F), whereas the section as the negative control for staining incubated with normal rabbit antibody shows no specific labeling in odontoblasts or ameloblasts (G, H). Bars = 50 µm.
Immunofluorescent Localization of Celsr1 in Dental Tissues of the Incisors
As Celsr1 and Vang12 are known to be necessary for orienting hair follicles and to be interdependent for their polarized hair follicle distribution (Devenport and Fuchs 2008), Celsr1 localization was examined using the freeze-dried, unfixed, and non-demineralized specimens. Anti–Celsr1-derived fluorescence was brightly localized in the cell bodies of odontoblasts but not in the ameloblasts (Fig. 5E–H).
Discussion
The presence of Vang12 and frizzled-3 in dental tissue has been previously detected by using in situ hybridization and microarray gene expression analysis. Tissir and Goffinet (2006) reported strong expression of frizzled-3 and Vang12 by using oligonucleotide probes in an in situ hybridization study of mouse molar tooth germ. Pemberton et al. (2007) reported that the developing mouse molar tooth showed increased expression of Vang12 along with increased frizzled-6 expression, compared with control tissues. Thus, the mRNA expression of PCP genes, including Vang12, frizzled-3, and frizzled-6, in tooth germ has been shown. In the present study, the localization of Vang12 and frizzled-3 protein was examined in rat incisor ameloblasts by immunohistochemistry. The results showed that an anti-Vang12 Ab signal was localized at Tomes’ processes of secretory ameloblasts but not in presecretory or maturation ameloblasts. In contrast, an anti–frizzled-3 Ab signal was localized at both presecretory and secretory ameloblasts but not at maturation ameloblasts. The localization of the anti–frizzled-3 Ab signal at presecretory and secretory ameloblasts was similar to the localization previously reported for F-actin (Nishikawa and Kitamura 1986). F-actin first appears at the proximal junctional area of differentiating ameloblasts and later appears at the distal junctional area of differentiating ameloblasts (Nishikawa and Kitamura 1986). In secretory ameloblasts, F-actin is continuously localized both at the proximal and distal junctional areas (Nishikawa and Kitamura 1986). The anti–frizzled-3 Ab showed a similar localization pattern in the present study. Furthermore, F-actin filament bundles are components of adherens junctions. N-cadherin and β-catenin, which are adherens junction proteins, have been previously shown to be localized in ameloblasts (Nishikawa et al. 1990; Bartlett et al. 2010). In the present study, frizzled-3 is localized both at proximal and distal junctions, which are stained positive by antibodies of adherens junction proteins, anti–N- cadherin, and anti–β-catenin.
Vang12 and actin filament bundles are abundant in Tomes’ processes of secretory ameloblasts, but the frizzled-3 protein has not been localized in these processes. In the present study, Vang12 was localized in two different regions of Tomes’ processes: a proximal area and a distal area. Both of these regions appear to be associated with the secretory surface of the membrane of Tomes’ processes, although ultrastructural localization of Vang12 is necessary to clarify the stained structure. Thus, PCP proteins appear to be involved in enamel rod formation. Vangl- and frizzled-type proteins are known to antagonize each other and to function at different cell membrane areas of the cell (Seifert and Mlodzik 2007). PCP proteins have been shown to play roles in tissue formation such as in alignment of mouse inner ear hair cells and Drosophila eyes (Seifert and Mlodzik 2007). All PCP proteins examined in this study were localized in the incisors. Because Wnt5a is known to be a representative of the non-canonical Wnt pathway, which is the upstream signaling pathway of PCP proteins (Moon et al. 1993; Kilian et al. 2003), and Wnt5a-deficient mice exhibit small tooth germ (Lin et al. 2011), it is likely that PCP proteins are deeply related to tooth formation.
Mouse body hairs incline in a head-to-tail direction. The underlying molecular basis of this inclination was shown to depend on PCP proteins such as Vang12 and the atypical cadherin Celsr1 (Devenport and Fuchs 2008). Another member of frizzled, frizzled-6, is localized in a pattern indistinguishable from those of Vang12 and Celsr1 in the hair follicles (Devenport and Fuchs 2008). In incisors of rats, anti-Celsr1 was localized in different tooth-forming cells, odontoblasts. Thus, it is possible that some family proteins associated with enamel and dentin formation remain to be clarified. To examine the possibility of another Vang protein in the ameloblast junctions, we examined Vang11. Anti-Vang11 Ab was localized in the stratum intermedium. This suggests that Vang11 may function in the formation and maintenance of the cell-to-cell junction in the stratum intermedium. In amelogenesis, anti-Vang11 Ab labeled differentiating inner enamel epithelial cells as sparse bright dots but rarely was labeled in secretory ameloblasts. These distribution patterns are different from that of the frizzled-3 in this study. Thus, interaction of Vang11 and frizzled-3 proteins is unlikely in ameloblasts. It would be interesting to know the odontogenesis of knockout mice of PCP genes. Although many kinds of knockout mice have been shown (van Amerongen and Berns 2006), looptail and crash mutant mice for Vang12 and Celsr1, respectively, are not viable after birth (Devenport and Fuchs 2008). Knockout mice of the frizzled-3 gene are also not viable after birth (van Amerongen and Berns 2006). Experiments of odontogenesis using these mutant mice thus may be limited.
In Drosophila wing cells, small vesicles containing frizzled are transported to adherens junctions at the cell boundary via microtubules that run in a horizontal direction (Shimada et al. 2006), suggesting that frizzled proteins are transported by microtubules that are oriented horizontally to the cell membrane. In secretory ameloblasts, most microtubules run along the long axis of the cell, which is the apical-basal axis, but some microtubules do run horizontally—that is, along the orthogonal plane of the apical-basal axis at the level of the distal junctional complex (Nishikawa and Kitamura 1985a). It is therefore possible that material important for junctional membranes, including frizzled-3, may be transported by microtubules in secretory ameloblasts. Because frizzled is a Wnt receptor, and a non-canonical downstream signaling pathway of frizzled is actin cytoskeleton reorganization (van Amerongen and Berns 2006; Klein and Mlodzik 2005), the presence of frizzled in ameloblast junctional complexes may induce the formation of, and maintain, actin cytoskeleton structure via the regulation of a small G-protein such as RhoA. The small G-proteins RhoA and Rac, as well as the Rho kinase Rock, are known downstream components of frizzled receptor signaling (Klein and Mlodzik 2005; van Amerongen and Berns 2006). Inhibition of Rho kinase was reported to mimic PCP-linked defects (Yates et al. 2010). Furthermore, Rho GTPases such as RhoA and Rac, as well as their regulator molecules, have been shown to function in amelogenesis (Li et al. 2005; Hatakeyama et al. 2009; Biz et al. 2010; Otsu et al. 2010; Li et al. 2011).
Although the characteristic localization of frizzled-3 and Vang12 in ameloblasts strongly suggests the involvement of PCP proteins in tooth enamel formation, further studies are obviously needed to confirm their involvement and to understand their mechanism of action.
Footnotes
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
The authors received no financial support for the research, authorship, and/or publication of this article.
References
- Bartlett JD, Dobeck JM, Tye CE, Perez-Moreno M, Stokes N, Reynolds AB, Fuchs E, Skobe Z. 2010. Targeted p120-catenin ablation disrupts dental enamel development. PLoS One. 5(9):e12703 doi:10.1371/journal.pone.0012703 10.1371/journal.pone.0012703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biz MT, Marques MR, Crema VA, Moriscot AS, dos Santos MF. 2010. GTPases RhoA and Rac1 are important for amelogenin and DSPP expression during differentiation of ameloblasts and odontoblasts. Cell Tissue Res. 340:459–470 [DOI] [PubMed] [Google Scholar]
- Boyde A. 1971. Comparative histology of mammalian teeth. In: Dahlberg AA, editor. Dental morphology and evolution. Chicago: University of Chicago Press; pp. 81–94 [Google Scholar]
- Devenport D, Fuchs E. 2008. Planar polarization in embryonic epidermis orchestrates global asymmetric morphogenesis of hair follicles. Nat Cell Biol. 11:1257–1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanto M, McNeill H. 2004. Planar polarity from flies to vertebrates. J Cell Sci. 117:527–533 [DOI] [PubMed] [Google Scholar]
- Goodrich LV, Strutt D. 2011. Principles of planar polarity in animal development. Development. 138:1877–1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanaizumi Y, Yokota R, Domon T, Wakita M, Kozawa Y. 2010. The initial process of enamel prism arrangement and its relation to the Hunter-Schreger bands in dog teeth. Arch Histol Cytol. 73:23–36 [DOI] [PubMed] [Google Scholar]
- Hatakeyama J, Fukumoto S, Nakamura T, Haruyama N, Suzuki S, Hatakeyama Y, Shum L, Gibson CW, Yamada Y, Kulkarni AB. 2009. Synergistic roles of amelogenin and ameloblastin. J Dent Res. 88:318–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamoto T. 2003. Use of a new adhesive film for the preparation of multi-purpose fresh-frozen sections from hard tissues, whole-animals, insects and plants. Arch Histol Cytol. 66:123–143 [DOI] [PubMed] [Google Scholar]
- Kawai N. 1955. Comparative anatomy of the bands of Schreger. Okajimas Folia Anat Jpn. 27:115–131 [DOI] [PubMed] [Google Scholar]
- Keller R. 2002. Shaping the vertebrate body plan by polarized embryonic cell movements. Science. 298:1950–1954 [DOI] [PubMed] [Google Scholar]
- Keller R, Davidson L, Edlund A, Elul T, Ezin M, Shook D, Skoglund P. 2000. Mechanisms of convergence and extension by cell intercalation. Phil Trans R Soc Lond B. 355: 897–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilian B, Mansukoski H, Barbosa FC, Ulrich F, Tada M, Heisenberg CP. 2003. The role of Ppt/Wnt5a in regulating cell shape and movement during zebrafish gastrulation. Mech Dev. 120:467–476 [DOI] [PubMed] [Google Scholar]
- Klein TJ, Mlodzik M. 2005. Planar cell polarization: an emerging model points in the right direction. Annu Rev Cell Dev Biol. 21:155–176 [DOI] [PubMed] [Google Scholar]
- Lawrence PA, Struhl G, Casal J. 2007. Planar cell polarity: one or two pathways? Nat Rev Genet. 8:555–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Decker S, Yuan Z, DenBesten PK, Aragon MA, Jordan-Sciutto K, Abrams WR, Huh J, McDonald C, Chen E, et al. 2005. Effects of sodium fluoride on the actin cytoskeleton of murine ameloblasts. Archs Oral Biol. 50:681–688 [DOI] [PubMed] [Google Scholar]
- Li Y, Pugach MK, Kuehl MA, Peng L, Bouchard J, Hwang SY, Gibson CW. 2011. Dental enamel structure is altered by expression of dominant negative RhoA in ameloblasts. Cells Tissues Organs. 194:2–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M, Li L, Liu C, Liu H, He F, Yan F, Zhang Y, Chen Y. 2011. Wnt5a regulates growth, patterning, and odontoblast differentiation of developing mouse tooth. Dev Dyn. 240:432–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeill H. 2010. Planar cell polarity: keeping hairs straight is not so simple. Cold Spring Harb Perspect Biol. 2:a003376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon RT, Campbell RM, Christian JL, McGrew LL, Shih J, Fraser S. 1993. Xwnt-5a: a maternal Wnt that affects morphogenetic movements after overexpression in embryos of Xenopus laevis. Development. 119:97–111 [DOI] [PubMed] [Google Scholar]
- Nishikawa S. 1992. Correlation of the arrangement pattern of enamel rods and secretory ameloblasts in pig and monkey teeth: a possible role of the terminal webs in ameloblast movement during secretion. Anat Rec. 232:466–478 [DOI] [PubMed] [Google Scholar]
- Nishikawa S, Fujiwara K, Kitamura H. 1988. Formation of the tooth enamel rod pattern and the cytoskeletal organization in secretory ameloblasts of the rat incisor. Eur J Cell Biol. 47:222–232 [PubMed] [Google Scholar]
- Nishikawa S, Kitamura H. 1985a. Three-dimensional network of microtubules in secretory ameloblasts of rat incisors. Archs Oral Biol. 30:1–11 [DOI] [PubMed] [Google Scholar]
- Nishikawa S, Kitamura H. 1985b. Actin filaments in the terminal webs of secretory ameloblasts of rats. Archs Oral Biol. 30:13–21 [DOI] [PubMed] [Google Scholar]
- Nishikawa S, Kitamura H. 1986. Localization of actin during differentiation of the ameloblast, its related epithelial cells and odontoblasts in the rat incisor using NBD-phallacidin. Differentiation. 30:237–243 [DOI] [PubMed] [Google Scholar]
- Nishikawa S, Tsukita S, Tsukita S, Sasa S. 1990. Localization of adherens junction proteins along the possible sliding interface between secretory ameloblasts of the rat incisor. Cell Struct Funct. 15:245–249 [DOI] [PubMed] [Google Scholar]
- Obara N, Suzuki Y, Nagai Y, Takeda M. 1998. Expression of E- and P-cadherin during tooth morphogenesis and cytodifferentiation of ameloblasts. Anat Embryol. 197:469–475 [DOI] [PubMed] [Google Scholar]
- Osborn JW. 1968a. Evaluation of previous assessments of prism directions in human enamel. J Dent Res. 47:217–222 [DOI] [PubMed] [Google Scholar]
- Osborn JW. 1968b. Directions and interrelationships of enamel prisms from the sides of human teeth. J Dent Res. 47:223–232 [DOI] [PubMed] [Google Scholar]
- Otsu K, Kishigami R, Fujiwara N, Ishizeki K, Harada H. 2010. Functional role of Rho-kinase in ameloblast differentiation. J Cell Physiol. 226:2527–2534 [DOI] [PubMed] [Google Scholar]
- Pemberton TJ, Li F-Y, Oka S, Mendoza-Fandino GA, Hsu Y-H, Bringas P, Jr, Chai Y, Snead ML, Mehrian-Shai R, Patell PI. 2007. Identification of novel genes expressed during mouse tooth development by microarray gene expression analysis. Dev Dyn. 236:2245–2257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert JRK, Mlodzik M. 2007. Frizzled/PCP signaling: a conserved mechanism regulating cell polarity and directed motility. Nat Rev Genet. 8:126–138 [DOI] [PubMed] [Google Scholar]
- Shimada Y, Yonemura S, Ohkura H, Strutt D, Uemura T. 2006. Polarized transport of Frizzled along the planar microtubule arrays in Drosophila wing epithelium. Dev Cell. 10:209–222 [DOI] [PubMed] [Google Scholar]
- Skoglund P, Keller R. 2010. Integration of planar cell polarity and ECM signaling in elongation of the vertebrate body plan. Curr Opin Cell Biol. 22:589–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorkin BC, Wang MY, Dobeck JM, Albergo KL, Skobe Z. 2000. The cadherin-catenin complex is expressed alternately with the adenomatous polyposis coli protein during rat incisor amelogenesis. J Histochem Cytochem. 48:397–406 [DOI] [PubMed] [Google Scholar]
- Tissir F, Goffinet AM. 2006. Expression of planar cell polarity genes during development of the mouse CNS. Eur J Neurosci. 25:597–607 [DOI] [PubMed] [Google Scholar]
- Torban E, Kor C, Gros P. 2004. Van Gogh–like2 (Strabismus) and its role in planar cell polarity and convergent extension in vertebrates. Trends Genet. 20:570–577 [DOI] [PubMed] [Google Scholar]
- van Amerongen R, Berns A. 2006. Knockout mouse models to study Wnt signal transduction. Trends Genet. 22:678–689 [DOI] [PubMed] [Google Scholar]
- Vladar EK, Antic D, Axelrod JD. 2009. Planar cell polarity signaling: the developing cell’s compass. Cold Spring Harb Perspect Biol. 1:a002964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallingford JB. 2010. Planar cell polarity signaling, cilia and polarized ciliary beating. Curr Opin Cell Biol. 22:597–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallingford JB, Fraser SE, Harland RM. 2002. Convergent extension: the molecular control of polarized cell movement during embryonic development. Dev Cell. 2:695–706 [DOI] [PubMed] [Google Scholar]
- Wallingford JB, Mitchell B. 2011. Strange as it may seem: the many links between Wnt signaling, planar cell polarity, and cilia. Genes Dev. 25:201–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Nathans J. 2007. Tissue/planar cell polarity in vertebrates: new insights and new questions. Development. 134:647–658 [DOI] [PubMed] [Google Scholar]
- Wansleeben C, Meijlink F. 2011. The planar cell polarity pathway in vertebrate development. Dev Dyn. 240:616–626 [DOI] [PubMed] [Google Scholar]
- Wu J, Mlodzik M. 2009. A quest for the mechanism regulating global planar cell polarity of tissues. Trends Cell Biol. 19:295–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates LL, Schnatwinkel C, Murdoch JN, Bogani D, Formstone CJ, Townsend S, Greenfield A, Niswander LA, Dean CH. 2010. The PCP genes Celsr1 and Vang12 are required for normal lung branching morphogenesis. Hum Mol Genet. 19:2251–2267 [DOI] [PMC free article] [PubMed] [Google Scholar]