Background: Lysine acetylation is a major post-translational modification of proteins.
Results: Human glycine N-acyltransferase-like 2 is regulated by acetylation/deacetylation of lysine 19.
Conclusion: Reversible lysine acetylation is important in regulating the activity and function of human glycine N-acyltransferases.
Significance: Our study links post-translational modification of proteins with the production of lipid signaling molecules, the N-acyl glycines.
Keywords: Coenzyme A, Endocannabinoids, Enzyme Kinetics, Enzyme Mechanisms, Enzyme Mutation, Acyl-CoA, Glycine N-Acyltransferase, Lipid Signaling
Abstract
Lysine acetylation is a major post-translational modification of proteins and regulates many physiological processes such as metabolism, cell migration, aging, and inflammation. Proteomic studies have identified numerous lysine-acetylated proteins in human and mouse models (Kim, S. C., Sprung, R., Chen, Y., Xu, Y., Ball, H., Pei, J., Cheng, T., Kho, Y., Xiao, H., Xiao, L., Grishin, N. V., White, M., Yang, X. J., and Zhao, Y. (2006) Mol. Cell 23, 607–618). One family of proteins identified in this study was the murine glycine N-acyltransferase (GLYAT) enzymes, which are acetylated on lysine 19. Lysine 19 is a conserved residue in human glycine N-acyltransferase-like 2 (hGLYATL2) and in several other species, showing that this residue may be important for enzyme function. Mutation of lysine 19 in recombinant hGLYATL2 to glutamine (K19Q) and arginine (K19R) resulted in a 50–80% lower production of N-oleoyl glycine and N-arachidonoylglycine, indicating that lysine 19 is important for enzyme function. LC/MS/MS confirmed that Lys-19 is not acetylated in wild-type hGLYATL2, indicating that Lys-19 requires to be deacetylated for full activity. The hGLYATL2 enzyme conjugates medium- and long-chain saturated and unsaturated acyl-CoA esters to glycine, resulting in the production of N-oleoyl glycine and also N-arachidonoyl glycine. N-Oleoyl glycine and N-arachidonoyl glycine are structurally and functionally related to endocannabinoids and have been identified as signaling molecules that regulate functions like the perception of pain and body temperature and also have anti-inflammatory properties. In conclusion, acetylation of lysine(s) in hGLYATL2 regulates the enzyme activity, thus linking post-translational modification of proteins with the production of biological signaling molecules, the N-acyl glycines.
Introduction
Lysine acetylation is a major post-translational modification of histones and other proteins. The occurrence of acetyl groups located on the N terminus of histones was first reported in 1963 (1). More recent reports have also demonstrated that numerous proteins located outside the nucleus are also acetylated, strongly suggesting that this protein modification plays major regulatory roles in eukaryotes (for review, see Ref. 2). Acetylation of proteins may modulate protein-protein, protein-ligand, protein-DNA interactions, protein stability, or intracellular localization (3) and plays a crucial role in many physiological processes such as migration, metabolism, and aging as well as in pathological diseases such as cancer and neurodegenerative disorders (for review, see Ref. 4). Lysine acetylation is a post-translational modification that is highly regulated by enzymes called acetyltransferases and deacetylases. Acetylation is catalyzed by the (histone) acetyltransferase enzyme family (HATs), and deacetylation is carried out by the (histone) deacetylase class of enzymes (HADC) (5–8) or by one of the seven sirtuins or sirt (silent information regulators) deacetylase enzymes identified to date (7–9). Eleven HDAC enzymes have been identified, with HDAC1, -2, -8, and -11 localized in the nucleus and HDAC3, -4, -5, -7, -9, and -10 distributed in nucleus and cytoplasm (10). Deacetylase enzymes belonging to the mammalian SIRT family are localized in mitochondria (SIRT3, -4, -5) and cytoplasm (SIRT1 and SIRT2), although SIRT1 together with SIRT6 and -7 have been localized in the nucleus (7, 9).
A proteomics survey by Kim et al. (11) focused on non-nuclear acetylated proteins, identifying new potential substrates and a diversity of lysine-acetylated proteins in mouse and human models. Their study revealed 195 acetylated proteins from HeLa cells and mouse liver mitochondria, with more than 20% of total mitochondrial proteins being lysine-acetylated, including metabolic enzymes. Two of these enzymes identified in mouse mitochondria were members of the gene family of glycine N-acyltransferases (GLYATs).2 GLYATs are a family of enzymes that conjugate various xenobiotics and acyl-CoA esters to glycine (12–14). Lysine 19 of the murine GLYAT was shown to be acetylated (11), and this lysine is conserved in GLYAT enzymes in several different species (including human GLYATL2). We have recently characterized the hGLYATL2 enzyme and showed that it conjugates medium- and long-chain saturated and unsaturated acyl-CoA esters to glycine, resulting in the production of mainly N-oleoyl glycine but also other N-acyl glycines including N-arachidonoyl glycine (14). N-Arachidonoyl glycine and N-oleoyl glycine are structurally and functionally related to endocannabinoids and have been identified as signaling molecules that regulate functions like the perception of pain and body temperature and also have anti-inflammatory properties (15–17).
In this study we examined if lysine acetylation of hGLYATL2 could regulate enzyme activity. We replaced lysine at position 19 with arginine K19R (which retains a positive charge and is thus considered a conserved substitution) or glutamine K19Q (which abolishes the positive charge and, therefore, may “mimic” the effect of acetylation) in the hGLYATL2 protein. We found that the K19Q had a markedly reduced enzymatic activity, indicating that a positive charge is important at position 19 in hGLYATL2 and that the active wild-type enzyme requires deacetylation for full activity. The mutation K19R also resulted in lowered enzyme activity showing the importance of a lysine at position 19. Moreover, our tandem mass spectrometry (MS/MS) results shows that hGLYATL2 is not acetylated on lysine 19 in mammalian or bacterial systems, but treatment with nicotinamide (NAM, an inhibitor of the SIRT family of deacetylases and, therefore, resulting in an acetylated enzyme) resulted in acetylation of lysine 19, supporting an inhibitory effect of acetylation on enzyme activity in hGLYATL2.
EXPERIMENTAL PROCEDURES
Cloning and Expression of Recombinant Human GLYATL2 Proteins
The hGLYATL2 open reading frame was previously cloned into the pMal-c2x vector (New England Biolabs, Beverly, MA) (14). Two mutations were introduced into the hGLYATL2 protein; K19R, which is a conserved substitution in the sense that the Arg has the same charge and is about the same size as Lys but cannot be acetylated. This results in a mutant protein with approximately the same properties as the wild type, except that it cannot be acetylated on Lys-19. The second mutation that was introduced was K19Q, the uncharged residue glutamine, which may mimic acetylation of lysine.
The QuikChange II site-directed mutagenesis kit (Stratagene) was used for mutagenic PCR. Mutagenic primers were designed as follows: K19R, 5′-CTGTATAAATCCTTAGAAAGGAGCATCCCTGAATCC-3′ and 5′-GGATTCAGGGATGCTCCTTTCTAAGGATTTATACAG-3′; K19Q, 5′-CTGTATAAATCCTTAGAACAGAGCATCCCTGAATCC-3′ and 5′-GGATTCAGGGATGCTCTGTTCTAAGGATTTATACAG-3′ (Cybergene AB, Stockholm, Sweden), with the mutated codon in bold and the base pair change underlined. The PCR was prepared according to the manufacturer's instructions and carried out as follows: 95 °C for 30 s, 16 cycles of 95 °C for 30 s and 55 °C for 60 s, and finally 68 °C for 7 min. The PCR products were incubated for 1 h at 37 °C with Dpn1, and the digested samples were transformed into XL-1 Blue bacteria (Stratagene) and fully sequenced. Purified, sequenced plasmids were introduced into BL21(DE3) pLysS cells (Novagen Inc., Madison, WI), and overnight cultures were transferred to 250-ml Rich Medium containing 10 mg/ml Tryptone, 5 mg/ml yeast, 5 mg/ml NaCl, and 2 mg/ml glucose. Induction of recombinantly expressed proteins was carried out as described by the addition of isopropyl-1-thio-β-d-galactopyranoside (0.3 mm) (14). Treatment with NAM, a deacetylase inhibitor, was carried out by adding 5 mm NAM into the culture media together with the isopropyl-1-thio-β-d-galactopyranoside. The purified recombinant proteins were analyzed on SDS/PAGE gel and stained with Coomassie Brilliant Blue (data not shown).
Identification of N-Acyl Glycines by Electrospray Mass Spectrometry (ESI-MS) Analysis
Incubation mixtures were set up containing acyl-CoAs (50 μm), 1 μg of recombinant hGLYATL2 proteins, and glycine (50 mm) in 50 mm potassium phosphate buffer, pH 7.4. Bovine serum albumin (BSA) was added in a molar ratio of 1:5.5 BSA:acyl-CoA to all samples. The reactions were incubated for 5 min at 37 °C in a water bath. Samples were purified using EVOLUTE™ ABN 25 mg, 1-ml SPE columns (Biotage AB, Uppsala, Sweden) and analyzed by Quattro Micro triple quadrupole mass spectrometer (Micromass, Manchester, UK) essentially as described in Waluk et al. (14).
Sample Preparation for LC/MS/MS Analysis
A HEK293 cell lysate overexpressing recombinant hGLYATL2 containing a C-terminal myc/DDK tag was purchased (Origene Technologies, Rockville, MD). A plasmid expressing hGLYATL2 as a green fluorescent fusion protein was expressed in HepG2 cells as described previously (14). HepG2 cells were treated with 5 mm nicotinamide (a deacetylase inhibitor) for 24 h. Cells were collected by scraping, resuspended in 500 ml of PBS, sonicated in pulses 3 × 10 s at 5-s intervals, and incubated 5 min at 95 °C. Cell lysates were centrifuged at 13,000 rpm for 5 min, and the supernatant collected and frozen at −20 °C for LC/MS/MS analysis as described below.
100 μg of cell lysate or recombinant wild-type hGLYATL2, wild-type treated with NAM, or mutant (K19Q, K19R) proteins were prepared in 20 mm Tris-HCl, pH 8.3, 2.5 mm EDTA and heated at 95 °C for 5 min. TCEP (tris(2-carboxyethyl)phosphine) (Thermo Scientific) was added to a final concentration of 10 mm for 1 h at 37 °C to reduce disulfide bonds. Samples were treated with iodoacetamide (final concentration 30 mm) for 1 h at ambient temperature. 5 μg of chymotrypsin (Pierce) was added to each sample (1:20 v/v), and digestion was performed overnight at 37 °C. Samples were desalted on Solid Phase Extraction Sep-Pack Vac C18 100-mg columns (Waters) and dried in a SpeedVac.
Nano-High Performance Liquid Chromatography Combined with Tandem Mass Spectrometry (Nano-HPLC/MS/MS) and Data Base Analysis
All data were acquired on an Amazon ETD mass spectrometer (Bruker Daltonik, Bremen, Germany). Chromatographic separation of peptides was achieved on a Proxeon EASY-nLC system equipped with a 10-cm-long, 3-μm ReproSil-Pur C18 resin, 100-μm fused silica column, and 2-cm-long, 5-μm ReproSil-Pur C18 resin, 100-μm ID precolumn. The LC system operated with mobile phases, solvent A (98:2 H2O:acetonitrile (ACN)) (v:v) and solvent B (20:80 H2O:ACN) (v:v), both supplemented with 0.1% formic acid. Samples were loaded from a cooled (7 °C) auto sampler and separated with a linear gradient that was formed at a flow rate of 300 nl/min. It consisted of a 100-min linear ramp up to 30% solvent B, subsequently 70% B for 10 min, 10 min of isocratic run at 70% B, and eventually a 10-min isocratic run at 100% solvent A. The scan range was set to 150–2400 m/z in MS mode and increased in MS2 mode up to 3000 m/z. The instrument was operated in auto-MSn mode. The three highest peaks within the range of 450–2400 were automatically fragmented when their intensity exceeded threshold value. Each precursor ion was fragmented with collision-induced dissociation (CID) and (electron transfer dissociation (ETD) method. LC-MS/MS spectra were analyzed using Proteinscape Software (Bruker Daltonik). Peak lists were submitted to the Mascot Server 2.3 (Matrix Science, London, UK). Searches were performed using a data base containing three sequences of hGLYATL2 proteins: wild type, K19R mutant, and K19Q mutant. Chymotrypsin was selected as a site-specific enzyme for digestion. The precursor ion tolerance was set to 0.5 Da, and fragment mass tolerance was set to 0.6 Da. In addition, cysteine was defined with fixed carboxyamidomethylation modification (+57.0214 Da), lysine was defined with variable acetylation modification (+42.0106 Da), and serine/threonine were set with phosphorylation as variable modification (+79.9663 Da). Finally, y and b fragment ions were defined for all CID data, whereas c and z fragment ions were defined for ETD. Annotation of MS2 (MS/MS) spectra was done using Proteinscape, and mass chromatograms were manually inspected with Data Analysis 4.0 (Bruker Daltonik).
RESULTS
Lysine 19 of GLYATL2 Is a Conserved Residue in Several Species
hGLYATL2 is a member of a gene family of acyltransferases with four genes identified in human, hGLYAT, hGLYATL1, hGLYATL2, and hGLYATL3 (14), with a similar gene family existing in mouse. The human gene products show ∼40% sequence identity to each other, suggesting that they have different substrate specificities and functions. GLYAT is involved in the detoxification of endogenous and exogenous xenobiotic acyl-CoAs and in the metabolism of short chain fatty acids in mammals (for review, see Ref. 18). hGLYATL2 conjugates long chain acyl-CoAs such as oleoyl-CoA, stearoyl-CoA, and arachidonoyl-CoA to glycine (14), whereas the substrate(s) for hGLYATL1 and hGLYATL3 remains unidentified. A recent proteomics study identified lysine 19 in murine GLYAT proteins that was acetylated (11). The lysine 19 residue identified in the murine GLYAT enzymes is also conserved in the hGLYATL2. Alignment of the amino acid sequences of GLYATs from various species (Homo sapiens, Equus caballus, Canis lupus familiaris, Macaca mulatta, Bos taurus, and Mus musculus) shows that lysine 19 is conserved in these species (Fig. 1). However, in hGLYATL1 this lysine is replaced by an arginine. Interestingly, in most species a serine is the residue adjacent to the Lys-19 (Ser-20), and cross-talk between lysine acetylation and phosphorylation on adjacent serines has been described (2).
FIGURE 1.
Lysine 19 is a conserved residue in GLYATs in several species. Alignment of the first ∼24 amino acids of GLYATs in the indicated species was carried out using the ClustalW method. The conserved lysine 19 residue is shown in bold (note this residue is an arginine in hGLYATL1). There is a serine (Ser-20) adjacent to lysine 19 in most species. L1, like-1; L2, like-2.
Wild-type hGLYATL2 Is Not Acetylated on Lysine 19
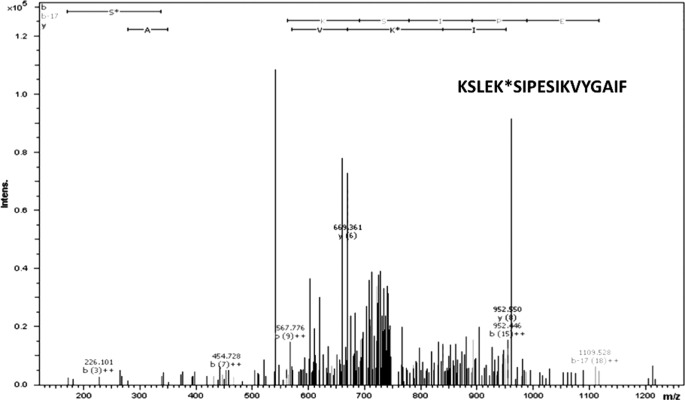
To confirm the acetylation status of Lys-19 in hGLYATL2 expressed in a mammalian expression system, hGLYATL2 was overexpressed in HEK293 cells. The cell lysate was digested with chymotrypsin after LC-MS/MS including ETD and CID fragmentation. The MS/MS analysis confirmed that the generated triply ionized KSLEKSIPESIKVYGAIF peptide containing Lys-19 (underlined) with a corresponding m/z 751.9 was not acetylated at Lys-19 in hGLYATL2 (Fig. 2). Similarly, HepG2 cells overexpressing a hGLYATL2/GFP fusion protein (14) were treated with NAM (a deacetylase inhibitor), and this treatment resulted in the acetylation of Lys-19, which was again confirmed using LC-MS/MS including ETD and CID fragmentation (data not shown). These results confirm that the acetylation of Lys-19 is a reversible process, and the presence of the deacetylase inhibitor NAM resulted in acetylation of Lys-19.
FIGURE 2.
Lysine 19 in human GLYATL2 expressed in HEK293 cells is not acetylated. Fragmentation CID mass spectrum of the triply ionized KSLEK*SIPESIKVYGAIF (751.9 m/z) peptide (K* indicates lysine at position 19 in the protein sequence) of the hGLYATL2 wild-type protein expressed in HEK293 cells.
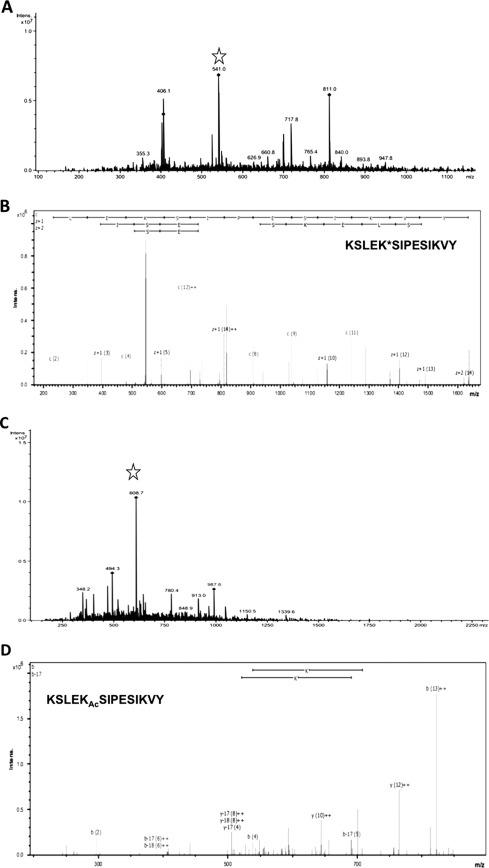
To examine the effect of acetylation/deacetylation on the enzyme activity of hGLYATL2, we expressed the protein in Escherichia coli and purified by affinity chromatography. To identify if lysine 19 is an acetylated residue in hGLYATL2 expressed in E. coli (a prokaryotic system), LC/MS/MS analysis was carried out after digestion of recombinantly expressed hGLYATL2 (purified from BL21(DE3) pLysS cells) with MS grade chymotrypsin. The sequences obtained were searched for a well identified peptide that included lysine at position 19 (Lys-19). The triple-ionized KSLEKSIPESIKVY peptide with Lys-19 (underlined) was selected, and the corresponding m/z 541 was directed for ETD and CID MS/MS analysis (Fig. 3A). When this peptide was fragmented using ETD and CID, peptide backbone fragmentations formed a complete or almost complete series of c and z ions and thus extensive peptide sequence information, as presented in the ETD MS/MS spectrum of wild-type hGLYATL2. The fragmentation spectrum of the peptide KSLEKSIPESIKVY identified Lys-19 in hGLYATL2 as a non-acetylated residue (Fig. 3B). The hGLYATL2 produced in the presence of the deacetylase inhibitor NAM was analyzed using nano-liquid chromatography tandem mass spectrometry (LC/MS/MS). From the MS spectrum a triple-ionized KSLEKSIPESIKVY peptide corresponding to m/z 608.7 was selected (Fig. 3C) and analyzed by ETD and CID MS/MS. Lysine 19 was acetylated in the NAM-treated hGLYATL2 protein as shown in the CID MS/MS spectrum (Fig. 3D). Additionally, two other lysine residues in the KSLEKSIPESIKVY (K indicates lysine at position 19 in the protein sequence) peptide were acetylated (Lys-15, Lys-26), with three adjacent serines phosphorylated (Ser-16, Ser-20, Ser-24) (Fig. 3D). Wild-type hGLYALT2 contained only one phosphorylated serine (Ser-16) and no modified lysine residues in this peptide (Fig. 3B). These results explain the mass differences between corresponding peptides from wild-type hGLYATL2 and NAM-treated hGLYATL2.
FIGURE 3.
Lysine 19 in human GLYATL2 is acetylated/deacetylated in response to treatment with the deacetylase inhibitor NAM. A, recombinant hGLYATL2 was produced in E. coli and affinity-purified as outlined under “Experimental Procedures.” Mass spectrum of the KSLEKSIPESIKVY peptide (K indicates lysine at position 19 in the protein sequence) from the wild-type hGLYATL2 protein, with different levels of ionization. The peptide targeted for MS/MS analysis is marked with a star. B, fragmentation ETD tandem mass spectrum of the KSLEK*SIPESIKVY peptide from the hGLYATL2 wild-type protein; lysine 19 is not acetylated. C, recombinant hGLYATL2 was produced in the presence of 5 mm deacetylase inhibitor NAM, and ETD MS/MS analysis was carried out on the KSLEKSIPESIKVY peptide from the NAM-treated hGLYATL2. The peptide chosen for further MS/MS analysis is marked with a star. D, CID tandem mass spectrum of the peptide shows an acetylated peptide with the sequence KSLEKAcSIPESIKVY, where KAc indicates an acetylated lysine at position 19 (Lys-19).
Interestingly, NAM treatment significantly enhanced the overall acetylation of hGLYATL2. Nine acetylated lysine residues in hGLYATL2 were identified when protein expression was induced in the presence of NAM, compared with four acetylated lysines in hGLYATL2 produced in the absence of NAM (data not shown).
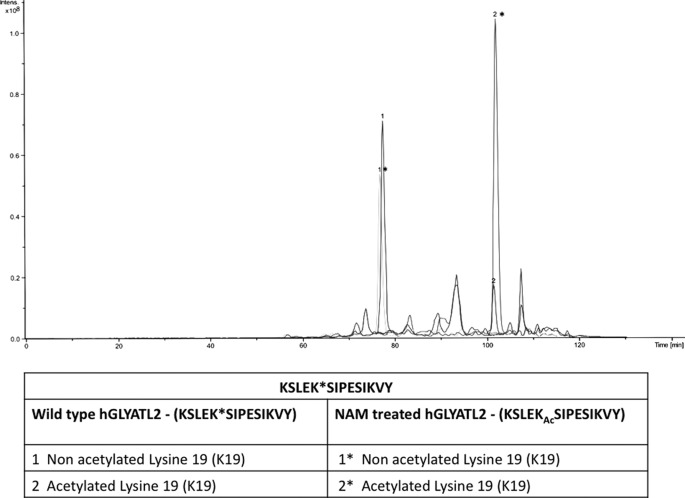
Quantitative Analysis of Acetylation Levels in hGLYATL2 KSLEKSIPESIKVY Peptide Analyzed by LC/MS/MS
The acetylation of Lys-19 in the KSLEKSIPESIKVY peptide generated by proteolytic digestion of wild-type hGLYATL2 with chymotrypsin was compared with non-acetylated lysine 19 and resulted in a quantitative ratio of 1:4 (acetylated:non-acetylated). In contrast, Lys-19 in KSLEKSIPESIKVY peptide from NAM-treated hGLYATL2 resulted in a quantitative ratio of 2:1 (acetylated:non-acetylated) (Fig. 4). As these data are obtained for peptides arising from the same protein, they may be considered as quantitative. We have observed different retention times for selected peptides containing the Lys-19 residue. Distinctive retention times are due to the level of acetylation occurring on lysine residues in the KSLEKSIPESIKVY peptide. Introduction of an acetyl group into the ϵ-amino site of lysine residues abolishes the positive charge and, therefore, may result in an interaction between the chromatographic column and the peptide and in turn influence the retention times of KSLEKSIPESIKVY peptides with either acetylated lysines or non-acetylated lysines (Fig. 4).
FIGURE 4.
Quantitative analysis of acetylation levels in the KSLEKSIPESIKVY peptide of hGLYATL2 by LC/MS/MS. The peptides KSLEKSIPESIKVY where lysine 19 is not acetylated elute earlier than the same peptides with modified lysine 19. The ratio of acetylated/non-acetylated lysines in the KSLEKSIPESIKVY peptide in wild-type hGLYATL2 and NAM-treated hGLYATL2 were calculated by measuring the area under the peaks.
Acetylation of Lysine 19 in hGLYATL2 Regulates Enzyme Activity
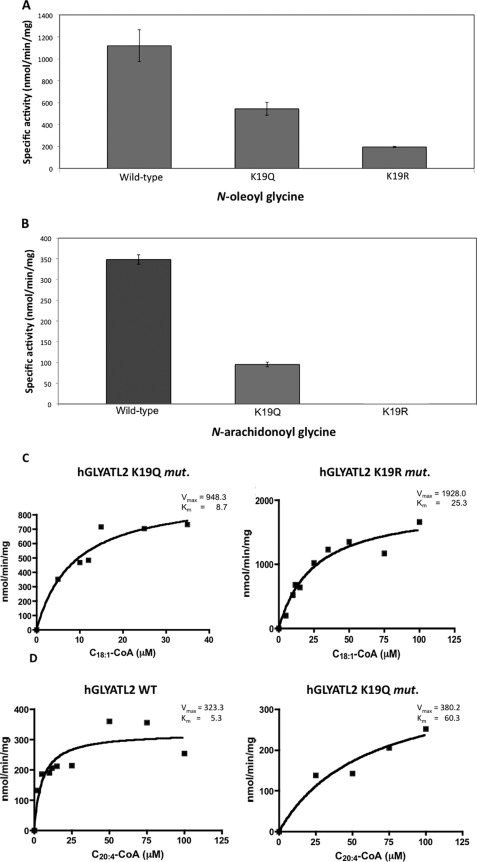
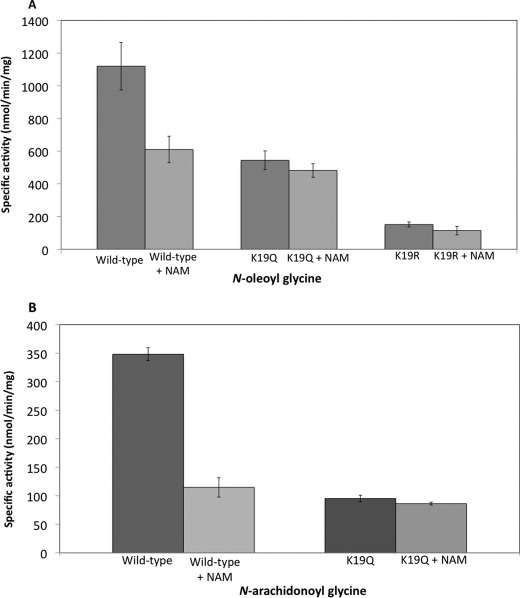
Reversible lysine acetylation has been reported to regulate the enzyme activity of the ornithine carbamoyltransferase (19) and the mitochondrial acetyl-CoA synthetase 2 (20). To investigate if lysine acetylation is involved in regulation of hGLYATL2, two mutations were introduced into the recombinant protein, K19R and K19Q. The K19R is a conserved mutation in that arginine has the same charge and is approximately the same size as lysine but cannot be acetylated. This results in a mutant protein with approximately the same properties as the wild type except for lack of acetylation on lysine 19. The K19Q mutation introduces an uncharged glutamine that abolishes the positive charge and, therefore, may mimic acetylation. The wild-type protein and the K19Q and K19R mutants were expressed in pLys bacteria, and the mutations did not affect the levels of recombinant protein expression (data not shown). To determine whether Lys-19 is important for hGLYATL2 enzyme activity, hGLYATL2 activity was measured with wild type, K19Q, and K19R using oleoyl-CoA (C18:1-CoA) and arachidonoyl-CoA (C20:4-CoA), which were previously shown to be substrates for hGLYATL2 (14). The wild-type enzyme activity with oleoyl-CoA was calculated to be ∼1100 nmol/min/mg of protein (Fig. 5A). Mutation of the lysine 19 to glutamine (K19Q) resulted in a 50% decrease of acyltransferase activity, whereas K19R resulted in an 80% reduction in enzyme activity (Fig. 5A). Similarly, the wild-type enzyme activity with C20:4-CoA was calculated to be ∼350 nmol/min/mg of protein (Fig. 5B). Mutation of the lysine 19 to glutamine (K19Q) resulted in a 72% reduction in enzyme activity, with almost no detectable activity remaining when Lys-19 was mutated to arginine (K19R). These results show the importance of the lysine residue in position 19 of hGLYATL2 for the enzyme function.
FIGURE 5.
Lysine 19 is an important residue in regulation of hGLYATL2 enzyme activity. A, ∼1 μg of hGLYATL2 (wild type), K19Q, and K19R mutant proteins was incubated with 50 μm C18:1-CoA, 50 μm C20:4-CoA, 50 mm glycine with the addition of BSA in a molar ratio of 1:5.5 BSA:acyl-CoA as outlined under “Experimental Procedures.” A, N-oleoyl glycine conjugates formed. B, N-arachidonoyl glycine conjugates formed and were quantified using ESI-MS. The experiment was repeated four times (three times for the K19Q mutant), and the mean ± S.D. is shown. C, recombinant K19Q and K19R proteins (∼1 μg) were incubated for 2 min at various concentrations of oleoyl-CoA (5–100 μm) (C) or N-arachidonoyl-CoA (5–100 μm) (D) with the addition of BSA in a molar ratio of 1:5.5 BSA:acyl-CoA in the presence of glycine (50 mm). N-Arachidonoyl glycine was added as an internal standard (5 μm) to reactions where N-oleoyl glycine is the product formed, and N-oleoyl glycine was added as an internal standard (5 μm) to reactions where N-arachidonoyl glycine is the product formed. Samples were purified on Evolute columns, analyzed by ESI-MS, and quantified according to the internal standard. Km (μm) and Vmax (nmol/min/mg) were calculated using Sigma Plot Enzyme Kinetics program. The experiments were repeated twice, and one representative experiment is shown.
The wild-type enzyme has a calculated Km of 4.4 μm and a Vmax of 933 nmol/min/mg (14). We first measured the Km and Vmax with the K19Q and K19R mutants using oleoyl-CoA as substrate. These mutations resulted in an increase of the Km value (8.7 μm) and a similar Vmax 948.3 nmol/min/mg for the K19Q mutant with oleoyl-CoA, whereas the K19R mutant showed a high increase in Km to 25.3 μm as well as Vmax 1928 nmol/min/mg compared with wild type (Fig. 5C and mean calculated values shown in Table 1). The Km and Vmax was also measured using arachidonoyl-CoA for wild-type hGLYATL2 and the K19Q mutant; however, it was not possible to determine Km and Vmax values for the K19R mutant due to undetectable activity with arachidonoyl-CoA. The wild-type enzyme had a calculated Km value of 5.3 μm with arachidonoyl-CoA and a Vmax of 323.3 nmol/min/mg, and the K19Q mutation resulted in an increase of Km to 60.3 μm with the Vmax increasing slightly to 380.2 nmol/min/mg (Fig. 5D and mean calculated values shown in Table 1).
TABLE 1.
The calculated Km and Vmax of human GLYATL2 wild type and the K19Q and K19R mutants with N-oleoyl-CoA, N-arachidonoyl-CoA, and glycine
Km and Vmax values for N-oleoyl-CoA, N-arachidonoyl-CoA, and glycine were calculated based on two or three independent protein purifications using the Sigma Plot Enzyme Kinetics program. Data shown are the mean ± S.E. The ratio of Vmax/Km is shown. ND, not determined.
| hGLYATL2 |
N-Oleoyl-CoA (C18:1-CoA) |
N-Arachidonoyl-CoA (C20:4-CoA) |
Glycine |
|||||
|---|---|---|---|---|---|---|---|---|
| Km | Vmax | Vmax/Km | Km | Vmax | Vmax/Km | Km | Vmax/Km | |
| μm | nmol/min/mg | μm | nmol/min/mg | mm | mmol/min/mg | |||
| Wild type | 4.3 ± 0.3 | 929.1 ± 18.6 | 215.8 | 6.8 ± 1.0 | 333.1 ± 15.8 | 49.0 | 11.2 ± 2.4 | 1230.0 ± 88.0 |
| K19Q mutant | 9.6 ± 0.9 | 1102.2 ± 153.8 | 114.3 | 62.0 ± 16.7 | 390.0 ± 61.4 | 6.3 | ND | ND |
| K19R mutant | 28.7 ± 2.0 | 2037.7 ± 91.3 | 71.3 | ND | ND | ND | ND | |
Overall, these data indicate the importance of the conservation of a lysine at position 19 in hGLYATL2, as mutation of this residue results in reduced enzyme activity. The mass spectrometry data (Figs. 2 and 3B) support that the deacetylation of lysine 19 is necessary for full activity of hGLYATL2, and it may be that the enzyme is regulated by acetylation/deacetylation.
Treatment with Deacetylase Inhibitor NAM Reduces hGLYATL2 Enzymatic Activity
Our results presented above show that lysine 19 in wild-type hGLYATL2 is not acetylated (Figs. 2 and Fig. 3B), and therefore, deacetylation is likely important for hGLYATL2 enzyme activity. Recombinant wild-type hGLYATL2 was, therefore, produced in the presence of the deacetylase inhibitor NAM. Mass spectrometry analysis of wild-type hGLYATL2 revealed four acetylated lysine residues in the protein, whereas analysis of NAM-treated wild-type hGLYATL2 showed the acetylation of nine residues (data not shown); thus, as expected, NAM treatment increased the overall acetylation status of hGLYATL2, including acetylation of lysine 19 (Fig. 3D). Treatment of wild-type hGLYATL2 with NAM decreased the enzyme activity with oleoyl-CoA by ∼45% (Fig. 6A) and with N-arachidonoyl glycine by ∼65% (Fig. 6B). These data together with the mass spectrometry data support that deacetylation of lysine 19 is important for enzyme function, as treatment with NAM results in acetylation of lysine 19 and a reduced enzyme activity. However, it may also be that the overall acetylation status of hGLYATL2 could affect the enzyme activity.
FIGURE 6.
Acetylation/deacetylation of hGLYATL2 regulates enzyme activity. Wild-type hGLYATL2, K19Q, and K19R mutant proteins were produced in BL21(DE3) pLysS cells in the absence/presence of 5 mm NAM, a deacetylase inhibitor. ∼1 μg of hGLYATL2 (wild type), K19Q, and K19R mutant proteins was incubated with 50 μm oleoyl-CoA, arachidonoyl-CoA, 50 mm glycine with the addition of BSA in a molar ratio of 1:5.5 BSA:acyl-CoA as outlined under “Experimental Procedures,” and the N-oleoyl glycine (A) or N-arachidonoyl glycine (B) conjugates formed were quantified using ESI-MS. The experiment was repeated four times (three times for NAM-treated K19Q and K19R mutants), and the mean ± S.D. is shown.
Therefore, to examine if lysine 19 is the important lysine residue in regulating hGLYATL2 or if the overall acetylation status of hGLYATL2 affects enzyme activity, the K19Q and K19R proteins were produced in the presence of the deacetylase inhibitor NAM. K19Q cannot be acetylated at lysine 19, but substitution of lysine with glutamine (K19Q) mimics acetylation. As shown in Fig. 5, A and B, mutation of K19Q reduced enzyme activity. Interestingly, the enzyme activities for the K19Q mutant (with 9 acetylated lysines; data not shown) and NAM-treated K19Q were similar, and both activities were about 45–50% lower than wild-type hGLYATL2 activity (Fig. 6A), showing that the overall acetylation status of hGLYATL2 by NAM treatment does not affect enzyme activity but that the reduction in enzyme activity is due to mutation of lysine 19. The K19R mutant retains a positive charge, but again, the residue cannot be acetylated. Treatment of the K19R mutant protein with NAM resulted in a large increase in overall protein acetylation (18 acetylated lysine residues; data not shown), and determination of enzyme activity of K19R in the absence and presence of NAM showed a similar activity with oleoyl-CoA, again showing that the overall acetylation status does not affect enzyme activity but that lysine is an important residue at position 19 in hGLYATL2. Due to the low activity of K19R with arachidonoyl-CoA, we could not determine the activity of K19R in the presence of NAM. Similar experiments were carried out with arachidonoyl-CoA on wild-type and the K19Q mutant. The NAM-treated wild-type and K19Q mutant protein activity were both reduced by ∼70/75%. This data strongly support that lysine 19 is the most important lysine residue for regulating hGLYATL2 activity and that deacetylation of this residue also is necessary for full enzyme activity.
DISCUSSION
Lysine residues in proteins may be acetylated, methylated, ubiquitinated, biotinylated, lipoylated, and hydroxylated (21). Lysine acetylation of non-histone proteins has recently emerged as an important post-translational modification in the regulation of metabolic enzymes (2). However, post-translational modifications are mutually exclusive on the same lysine and generate a great potential for cross-regulation. Interestingly cross-talk between lysine acetylation and other modifications such as phosphorylation occur, as has been shown on histone H3 at Lys-9 and Lys-27, with phosphorylation of Ser-10 and Ser-28, respectively (22). Yang et al. (for review, see Ref. 2) also pointed out in a recent review that histone H3, p53, and several transcription factors such as FoxO1 and C/EBPb all contain serine residues adjacent to acetylated lysines. In a similar way, the hGLYATL2 amino acid sequence contains a serine at position 20 (Ser-20), which is adjacent to Lys-19 (Fig. 1), which could indicate a potential cross-talk between acetylation and phosphorylation of this protein. Our results show that serine 20 is phosphorylated in hGLYATL2 when lysine 19 is acetylated (following NAM treatment), but in wild-type hGLYATL2 not treated with NAM, both residues are unmodified. The regulation of hGLYATL2 by acetylation/deacetylation and/or phosphorylation suggests a link between post-translational modifications and the production of bioactive signaling molecules, especially N-oleoyl glycine (C18:1-glycine) and N-arachidonoyl glycine (C20:4-glycine). Signaling pathways activated by N-oleoyl glycine are not fully understood, but administration of N-oleoyl glycine to rats resulted in reduced motor activity and lowered body temperature (23). N-Oleoyl glycine was first identified in N18TG2 cells by Merkler et al. (24) and was proposed to be involved in the biosynthesis of oleamide, a primary fatty acid amide. Acetylation/deacetylation of hGLYATL2 would thus result in a rapid regulation of N-oleoyl glycine levels by modulating the activity of hGLYATL2 and may also further regulate the production of oleamide from N-oleoyl glycine. Oleamide has cannabinoid-like actions and is involved in sleep induction (25) and acts as a vasodilator in the cardiovascular system (26). N-Arachidonoyl glycine has been identified in rat brain and other tissues (17) and is synthesized by several pathways including an enzymatic conjugation of arachidonoyl-CoA with glycine (14), synthesis by cytochrome c in the presence of arachidonoyl-CoA and hydrogen peroxide (27), and finally by oxidation of anandamide (an endocannabinoid) by alcohol and aldehyde dehydrogenases (28). N-Arachidonoyl glycine is a ligand for G protein-coupled receptors GPR18 (29), GPR92 (30), and GPR72 (31) and is antiproliferative in mouse macrophage RAW cells (32).
Recent studies show that most intermediate metabolic enzymes are acetylated and that acetylation can directly affect the enzyme activity, translocation, or stability (33, 34). Lysine acetylation effects can be classified into three groups: (a) single residue acetylation, where acetylation acts as a simple on/off switch such as in the inactivation of the acetyl-CoA synthetase-2 (20); (b) where acetylation may cover charged patches, where it is the number of acetylated residues that count and not which particular residue in the charged patch is modified, e.g. cortactin (a protein that regulates cell motility) is acetylated on about 10 lysine residues in a repeat domain, with each repeat having at least one lysine (35); (c) where acetylation may result in a site-specific effect that influences the affinity for interacting partners, e.g. the activity of the ornithine carbamoyltransferase toward one of its substrates, carbamoyl phosphate, is regulated by acetylation of Lys-88 in the enzyme (19). Lysine 19 in GLYAT enzymes is highly conserved through many species, and mutation of lysine 19 (K19Q and K19R) in hGLYATL2 significantly affected the enzyme activity and production of N-oleoyl glycine and N-arachidonoyl glycine. Our results based on tandem mass spectrometry analysis indicate that Lys-19 is not the only acetylated lysine residue in hGLYATL2. This protein contains almost 10% lysine residues: 28 of 294 amino acids, with a number of these residues being acetylated. The highest enzymatic activity for N-oleoyl glycine and N-arachidonoyl glycine production was observed with wild-type hGLYATL2, where 4 lysines are acetylated, with Lys-19 not being acetylated. However, mutation of Lys-19 confirmed the importance of this residue for the function of hGLYATL2. Interestingly, treatment of the K19Q and K19R mutants of hGLYATL2 with NAM did not further affect the enzyme activity, showing that Lys-19 is the dominant lysine involved in regulation and that acetylation/deacetylation of Lys-19 is important but that the overall acetylation status does not affect enzyme function. The crystal structure of GLYAT proteins in any species has yet to be solved, making it difficult to predict where Lys-19 would be located/oriented in the three-dimensional structure of the protein, e.g. if it is located close to the active site or is involved in substrate (acyl-CoA) or acceptor (glycine) binding. A recent study by Badenhorst et al. (36) has revealed glutamic acid 226 as being catalytically important in bovine GLYAT, and the suggested catalytic mechanism is that Glu-226 functions to deprotonate glycine, facilitating a nucleophilic attack on the acyl-CoA.
In conclusion, we identified that acetylation/deacetylation of lysine 19 of hGLYATL2, which is conserved in most species, is important for the enzyme function. This study suggests a potential mechanism by which cells could regulate production of N-acyl glycines through acetylation/deacetylation as a posttranslational process that is a highly conserved modification on proteins throughout many organisms (37). Our study also links the post-translational modification of proteins with the production of N-acyl glycines, thus allowing a rapid regulation of hGLYATL2 in response to lipid signaling requirements.
Acknowledgment
We gratefully acknowledge Professor Stefan Alexson, Karolinska Institutet, for use of the electrospray mass spectrometer.
This work was supported by the Swedish Research Council, Carl Tryggers Foundation, Professor Nanna Svartz Fond, and Åke Wibergs Stiftelse and by the fellowship Doctus-Lesser Poland donation program for PhD students.
- GLYAT
- glycine N-acyltransferase
- hGLYATL2
- human glycine N-acyltransferase-like 2
- NAM
- nicotinamide
- ESI
- electrospray ionization
- CID
- collision-induced dissociation
- ETD
- electron transfer dissociation.
REFERENCES
- 1. Phillips D. M. (1963) The presence of acetyl groups of histones. Biochem. J. 87, 258–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yang X. J., Seto E. (2008) Lysine acetylation. Codified cross-talk with other posttranslational modifications. Mol. Cell 31, 449–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Polevoda B., Sherman F. (2002) The diversity of acetylated proteins. Genome Biol. 3, 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Close P., Creppe C., Gillard M., Ladang A., Chapelle J. P., Nguyen L., Chariot A. (2010) The emerging role of lysine acetylation of non-nuclear proteins. Cell. Mol. Life Sci. 67, 1255–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yang X. J., Seto E. (2007) HATs and HDACs. From structure function and regulation to novel strategies for therapy and prevention. Oncogene 26, 5310–5318 [DOI] [PubMed] [Google Scholar]
- 6. Brownell J. E., Allis C. D. (1995) An activity gel assay detects a single, catalytically active histone acetyltransferase subunit in Tetrahymena macronuclei. Proc. Natl. Acad. Sci. U.S.A. 92, 6364–6368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Haigis M.C., Guarente L. P. (2006) Mammalian sirtuins-emerging roles in physiology, aging, and calorie restriction. Genes Dev. 20, 2913–2921 [DOI] [PubMed] [Google Scholar]
- 8. Michan S., Sinclair D. (2007) Sirtuins in mammals. Insights into their biological function. Biochem. J. 404, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Michishita E., Park J. Y., Burneskis J. M., Barrett J. C., Horikawa I. (2005) Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol. Biol. Cell 16, 4623–4635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Riccio A. (2010) New endogenous regulators of Class I histone deacetylases. Sci. Signal. 3, pe1-pe1 [DOI] [PubMed] [Google Scholar]
- 11. Kim S. C., Sprung R., Chen Y., Xu Y., Ball H., Pei J., Cheng T., Kho Y., Xiao H., Xiao L., Grishin N. V., White M., Yang X. J., Zhao Y. (2006) Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol. Cell 23, 607–618 [DOI] [PubMed] [Google Scholar]
- 12. Schachter D., Taggart J. V. (1954) Glycine N-acylase. Purification and properties. J. Biol. Chem. 208, 263–275 [PubMed] [Google Scholar]
- 13. Gregersen N., Kølvraa S., Mortensen P. B. (1986) Acyl-CoA:glycine N-acyltransferase. In vitro studies on the glycine conjugation of straight- and branched-chain acyl-CoA esters in human liver. Biochem. Med. Metab. Biol. 35, 210–218 [DOI] [PubMed] [Google Scholar]
- 14. Waluk D. P., Schultz N., Hunt M. C. (2010) Identification of glycine N-acyltransferase-like 2 (GLYATL2) as a transferase that produces N-acyl glycines in humans. FASEB J. 24, 2795–2803 [DOI] [PubMed] [Google Scholar]
- 15. Bradshaw H. B., Rimmerman N., Hu S. S., Burstein S., Walker J. M. (2009) Novel endogenous N-acyl glycines. Identification and characterization. Vitam. Horm. 81, 191–205 [DOI] [PubMed] [Google Scholar]
- 16. McHugh D., Hu S. S., Rimmerman N., Juknat A., Vogel Z., Walker J. M., Bradshaw H. B. (2010) N-Arachidonoyl glycine, an abundant endogenous lipid, potently drives directed cellular migration through GPR18, the putative abnormal cannabidiol receptor. BMC Neurosci. 11, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Huang S. M., Bisogno T., Petros T. J., Chang S. Y., Zavitsanos P. A., Zipkin R. E., Sivakumar R., Coop A., Maeda D. Y., De Petrocellis L., Burstein S., Di Marzo V., Walker J. M. (2001) Identification of a new class of molecules, the arachidonoyl amino acids, and characterization of one member that inhibits pain. J. Biol. Chem. 276, 42639–42644 [DOI] [PubMed] [Google Scholar]
- 18. Knights K. M., Sykes M. J., Miners J. O. (2007) Amino acid conjugation. Contribution to the metabolism and toxicity of xenobiotic carboxylic acids. Expert Opin. Drug Metab. Toxicol. 3, 159–168 [DOI] [PubMed] [Google Scholar]
- 19. Yu W., Lin Y., Yao J., Huang W., Lei Q., Xiong Y., Zhao S., Guan K. L. (2009) Lysine 88 acetylation negatively regulates ornithine carbamoyltransferase activity in response to nutrient signals. J. Biol. Chem. 284, 13669–13675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schwer B., Bunkenborg J., Verdin R. O., Andersen J. S., Verdin E. (2006) Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. Proc. Natl. Acad. Sci. U.S.A. 103, 10224–10229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Walsh C. T., Garneau-Tsodikova S., Gatto G. J., Jr. (2005) Protein posttranslational modifications. The chemistry of proteome diversifications. Angew. Chem. Int. Ed. Engl. 44, 7342–7372 [DOI] [PubMed] [Google Scholar]
- 22. Latham J. A., Dent S. Y. (2007) Cross-regulation of histone modifications. Nat. Struct. Mol. Biol. 14, 1017–1024 [DOI] [PubMed] [Google Scholar]
- 23. Chaturvedi S., Driscoll W. J., Elliot B. M., Faraday M. M., Grunberg N. E., Mueller G. P. (2006) In vivo evidence that N-oleoylglycine acts independently of its conversion to oleamide. Prostaglandins Other Lipid Mediat. 81, 136–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Merkler D. J., Chew G. H., Gee A. J., Merkler K. A., Sorondo J. P., Johnson M. E. (2004) Oleic acid-derived metabolites in mouse neuroblastoma N18TG2 cells. Biochemistry 43, 12667–12674 [DOI] [PubMed] [Google Scholar]
- 25. Cravatt B. F., Prospero-Garcia O., Siuzdak G., Gilula N. B., Henriksen S. J., Boger D. L., Lerner R. A. (1995) Chemical characterization of a family of brain lipids that induce sleep. Science 268, 1506–1509 [DOI] [PubMed] [Google Scholar]
- 26. Hiley C. R., Hoi P. M. (2007) Oleamide. A fatty acid amide signaling molecule in the cardiovascular system? Cardiovasc. Drug Rev. 25, 46–60 [DOI] [PubMed] [Google Scholar]
- 27. McCue J. M., Driscoll W. J., Mueller G. P. (2008) Cytochrome c catalyzes the in vitro synthesis of arachidonoyl glycine. Biochem. Biophys. Res. Commun. 365, 322–327 [DOI] [PubMed] [Google Scholar]
- 28. Burstein S. H., Rossetti R. G., Yagen B., Zurier R. B. (2000) Oxidative metabolism of anandamide. Prostaglandins Other Lipid Mediat. 61, 29–41 [DOI] [PubMed] [Google Scholar]
- 29. Kohno M., Hasegawa H., Inoue A., Muraoka M., Miyazaki T., Oka K., Yasukawa M. (2006) Identification of N-arachidonoylglycine as the endogenous ligand for orphan G-protein-coupled receptor GPR18. Biochem. Biophys. Res. Commun. 347, 827–832 [DOI] [PubMed] [Google Scholar]
- 30. Oh D. Y., Yoon J. M., Moon M. J., Hwang J. I., Choe H., Lee J. Y., Kim J. I., Kim S., Rhim H., O'Dell D. K., Walker J. M., Na H. S., Lee M. G., Kwon H. B., Kim K., Seong J. Y. (2008) Identification of farnesyl pyrophosphate and N-arachidonylglycine as endogenous ligands for GRP92. J. Biol. Chem. 283, 21054–21064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Roy M. O., Hannedouche S. (November 2, 2010) U.S. Patent 7824866
- 32. Burstein S. (2008) The elmiric acids. Biologically active anandamide analogs. Neuropharmacology 55, 1259–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhao S., Xu W., Jiang W., Yu W., Lin Y., Zhang T., Yao J., Zhou L., Zeng Y., Li H., Li Y., Shi J., An W., Hancock S. M., He F., Qin L., Chin J., Yang P., Chen X., Lei Q., Xiong Y., Guan K. L. (2010) Regulation of cellular metabolism by protein lysine acetylation. Science 327, 1000–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ventura M., Mateo F., Serratosa J., Salaet I., Carujo S., Bachs O., Pujol M. J. (2010) Nuclear translocation of glyceraldehyde-3-phosphate dehydrogenase is regulated by acetylation. Int. J. Biochem. Cell Biol. 42, 1672–1680 [DOI] [PubMed] [Google Scholar]
- 35. Zhang X., Yuan Z., Zhang Y., Yong S., Salas-Burgos A., Koomen J., Olashaw N., Parsons J. T., Yang X. J., Dent S. R., Yao T. P., Lane W. S., Seto E. (2007) HDAC6 modulates cell motility by altering the acetylation level of cortactin. Mol. Cell 27, 197–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Badenhorst C. P., Jooste M., van Dijk A. A. (2012) Enzymatic characterization and elucidation of the catalytic mechanism of a recombinant bovine glycine N-acyltransferase. Drug Metab. Dispos. 40, 346–352 [DOI] [PubMed] [Google Scholar]
- 37. Bienvenut W. V., Espagne C., Martinez A., Majeran W., Valot B., Zivy M., Vallon O., Adam Z., Meinnel T., Giglione C. (2011) Dynamics of post-translational modifications and protein stability in the stroma of Chlamydomonas reinhardtii chloroplasts. Proteomics 11, 1734–1750 [DOI] [PubMed] [Google Scholar]