Background: Nuclear and cytosolic poly(ADP-ribose) metabolism is established but debated in mitochondria.
Results: Novel mitochondrial and cytosolic poly(ADP-ribose) glycohydrolase splice variants are inactive for poly(ADP-ribose) degradation.
Conclusion: Degradation of mitochondrial matrix-accumulated poly(ADP-ribose) can be catalyzed only by ADP-ribosylhydrolase 3, whereas small poly(ADP-ribose) glycohydrolase isoforms may have functions different from poly(ADP-ribose) degradation.
Significance: Important insights into the regulation of subcellular poly(ADP-ribose) metabolism are provided.
Keywords: ADP-ribosylation, Alternative Splicing, Mitochondria, Mitochondrial Metabolism, NAD, ADP-ribosylhydrolase 3, Poly(ADP-ribose) Glycohydrolase, Poly(ADP-ribose) metabolism
Abstract
Important cellular processes are regulated by poly(ADP-ribosyl)ation. This protein modification is catalyzed mainly by nuclear poly(ADP-ribose) polymerase (PARP) 1 in response to DNA damage. Cytosolic PARP isoforms have been described, whereas the presence of poly(ADP-ribose) (PAR) metabolism in mitochondria is controversial. PAR is degraded by poly(ADP-ribose) glycohydrolase (PARG). Recently, ADP-ribosylhydrolase 3 (ARH3) was also shown to catalyze PAR-degradation in vitro. PARG is encoded by a single, essential gene. One nuclear and three cytosolic isoforms result from alternative splicing. The presence and origin of a mitochondrial PARG is still unresolved. We establish here the genetic background of a human mitochondrial PARG isoform and investigate the molecular basis for mitochondrial poly(ADP-ribose) degradation. In common with a cytosolic 60-kDa human PARG isoform, the mitochondrial protein did not catalyze PAR degradation because of the absence of exon 5-encoded residues. In mice, we identified a transcript encoding an inactive cytosolic 52-kDa PARG lacking the mitochondrial targeting sequence and a substantial portion of exon 5. Thus, mammalian PARG genes encode isoforms that do not catalyze PAR degradation. On the other hand, embryonic fibroblasts from ARH3−/− mice lack most of the mitochondrial PAR degrading activity detected in wild-type cells, demonstrating a potential involvement of ARH3 in PAR metabolism.
Introduction
ADP-ribosylation is an NAD+-dependent protein modification in which the ADP-ribose moiety of NAD+ is transferred to a protein with release of nicotinamide. Mono- and poly(ADP-ribosyl)ation reactions have been described (reviewed in Refs. 1–5). Mono-ADP-ribosyltransferases catalyze the transfer of a single ADP-ribose (ADPR)3 moiety from NAD+ onto acceptor proteins. In poly(ADP-ribosyl)ation reactions, proteins are modified by long, branched polymers composed of up to 200 ADP-ribose units (poly(ADP-ribose)) (PAR) (6). Poly(ADP-ribosyl)ation is catalyzed by poly(ADP-ribose) polymerases (PARPs), of which nuclear PARP1 is the most abundant and best characterized protein. PARP1 is activated in response to DNA single-strand breaks and regulates important cellular processes such as DNA repair (7–9), transcription (4, 10, 11), and maintenance of genomic stability. Strong activation of PARP1 in response to genotoxic stress is accompanied by depletion of cellular NAD (12–15) and translocation of proapoptotic factors from the mitochondria to the nucleus, eventually leading to cell death (16–18). In addition to PARP1, other nuclear and cytosolic PARP isoforms have been identified (19–23), and still other genes have sequence similarity (5, 24) and await better characterization.
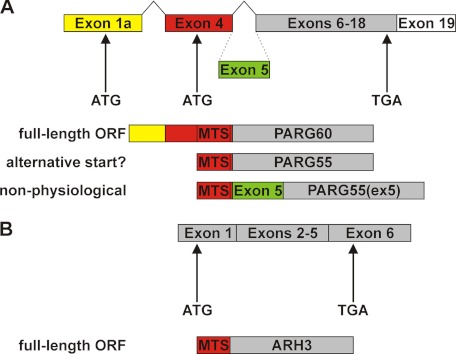
Poly(ADP-ribosyl)ation is reversed by PAR-degrading enzymes, which hydrolyze the polymers to ADPR. Poly(ADP-ribose) glycohydrolase (PARG) was initially identified as PAR-degrading enzyme (reviewed in Ref. 25). Subsequently, ADP-ribosylhydrolase 3 (ARH3) was shown to degrade PAR in vitro (26) and in cells (27), but its specific activity was significantly less than that of PARG (26). PARG is encoded by a single gene, which is essential, because PARG−/− mice are embryonic lethal (28). The human protein expressed from the full-length ORF, PARG111, is found in the nucleus because of a nuclear localization signal encoded by exon 1 (29). However, this isoform accounts for only a minor fraction of the total cellular PARG activity. In contrast to the nuclear localization of PARP1, PARG can be detected primarily in the cytosol (30). Cytosolic PARG activities were shown to be catalyzed by the PARG102 and PARG99 isoforms in humans. These isoforms result from alternative splicing of the primary PARG transcript and are translated from start codons in exons 2 and 3, respectively (29). The finding that transgenic mice expressing a PARG gene lacking exons 2 and 3 (PARGΔexon2–3/Δexon2–3) were viable (31) led to the identification of a fourth splice variant of the primary murine PARG transcript, where exon 1a is linked to exon 4, thereby skipping exons 2 and 3 and parts of exon 1 (32). In the human counterpart of this isoform, an additional splicing event occurs that links exon 4 to exon 6 (32). The ORF of this human transcript encodes a 60-kDa protein, PARG60 (Fig. 1A). It was postulated that this transcript may encode another PARG isoform, PARG55, which would result from alternative translation initiation from the second in-frame ATG triplet. In the putative PARG55, a canonical mitochondrial targeting sequence (MTS) would constitute the N terminus and therefore mediate import into the organelle (Ref. 32) and Fig. 1A). Both PARG60 and the hypothetical PARG55 were reported to be associated with mitochondria (32). The absence of the three large PARG isoforms in hypomorphic PARGΔexon2–3/Δexon2–3 mice suggests a vital function of the small PARG isoforms that may have to be carried out in mitochondria. Although several reports suggest that PAR is present in mitochondria, neither a mitochondrial PAR acceptor nor a mitochondrial PARP has been identified.
FIGURE 1.
Proposed origin and molecular architecture of small human PARG isoforms. A, the PARG60 mRNA results from alternative splicing events that link exon 1a with exon 4 and exon 4 with exons 6–19 of the human PARG gene. PARG55 was suggested to result from utilization of an alternative start codon in exon 4. In humans, there is no genetic evidence for the existence of PARG55(ex5) (also known as PARG59) containing exon 5-encoded amino acids. B, the ARH3 mRNA encodes a protein with a predicted N-terminal mitochondrial targeting sequence.
ARH3 is a member of the family of dinitrogenase reductase-activating glycohydrolase-related proteins and shares only little similarity with PARG in the catalytic domain sequence (26, 33). The ORFs of the human and murine ARH3 transcripts are encoded by six exons (Fig. 1B). The ARH3 primary structure predicts a mitochondrial targeting sequence at the N terminus of the protein (Fig. 1B). The protein was localized to this organelle (27) but is also present in the nucleus (26, 27) and the cytosol (26). ARH3 catalyzes hydrolysis of both PAR (26) and O-acetyl-ADP-ribose (OAADPR) (34), a metabolite that results from the activity of NAD+-dependent protein deacetylases of the sirtuin family (35).
We have demonstrated recently that PAR is accumulated within mitochondria upon targeted expression of the catalytic domain of PARP1 (PARP1cd). These artificial polymers are subject to turnover, indicating the presence of PAR-degrading activity within mitochondria. Although the degradation of matrix-accumulated PAR proceeded slowly, two different enzymes were identified to potentially account for this activity (27). One enzyme was ARH3, the second represented a hypothetical human PARG isoform, PARG59, which is an N-terminally truncated version of full-length PARG111. Translation of PARG59 was initiated from an ATG triplet in exon 4 of the human PARG full-length cDNA, thereby constituting an N-terminal MTS. However, the absence of exon 5 in the human PARG60 mRNA indicated that PARG59, which contains exon 5-encoded amino acids, is not physiological. In the context of this study, PARG59 is therefore termed PARG55(ex5) (Fig. 1A). Moreover, there is no evidence that PARG55, which would carry the same MTS as PARG55(ex5), is indeed generated from an alternative translational start codon. Consequently, the existence of a mitochondrial matrix PARG isoform remains an open question.
In this study, we identified the genetic background for human PARG55 and established the protein as the only intramitochondrial PARG isoform. We found that the lack of amino acids encoded by exon 5 in both small human PARG isoforms precluded their PAR-degrading activities. In mice, we detected a novel, catalytically inactive cytosolic PARG isoform. This study establishes ARH3 as the only enzyme that catalyzes the degradation of matrix-accumulated PAR.
EXPERIMENTAL PROCEDURES
Antibodies
Rabbit polyclonal ARH3 antibody (HPA027141), mouse (M2) and rabbit (SIG1–25) FLAG antibodies, mouse β-tubulin antibody (2-28-33) and mouse α-tubulin antibody (DM1A) were from Sigma-Aldrich. Rabbit (96-10−04) and mouse (10H) PAR antibodies were from Enzo Life Sciences or Abcam. Mouse myc antibody (9E10) was from Abcam. Mouse GFP antibody (JL-8) was from Clontech. Rabbit polyclonal GFP antibody was from Santa Cruz Biotechnology. Mouse GAPDH antibody (6C5) was from HyTest. Fluorescence-conjugated secondary antibodies (Alexa Fluor 594-conjugated goat anti-rabbit and Alexa Fluor 647-conjugated goat anti-mouse) were from Invitrogen. HRP goat anti-mouse antibody was from Pierce.
Cell Culture
HeLaS3 cells were cultivated in Ham's F-12 medium supplemented with 10% FCS, penicillin (10,000 units/ml), and streptomycin (10 mg/ml). 293 (HEK-293) cells were cultivated in high-glucose DMEM supplemented with 10% FCS, 2 mm l-glutamine, penicillin, and streptomycin. Cells were incubated at 37 °C in a humidified atmosphere with 5% CO2. Mouse embryonic fibroblasts (MEFs) were prepared from embryonic day 14.5 embryos and incubated in high-glucose DMEM supplemented with 10% FCS at 37 °C in a humidified atmosphere with 5% CO2. Cells from passages 2 to 20 were frozen in Cell Banker 2 (Wako Chemical) and stored at −80 °C. For experiments, thawed cells were grown in high-glucose DMEM supplemented with 10% FCS, penicillin, and streptomycin at 37 °C in a humidified atmosphere with 5% CO2. To establish stably transfected MEFs, 1.0 × 105 cells were cultured on a 96-well plate for 2 days prior to transfection. Two days post-transfection, G418 (1 mg/ml) was added to the medium to select and maintain stably transfected cells.
RT-PCR Analyses
Reverse transcription of total RNA, isolated from HeLaS3, 293, HepG2, and SH-SY5Y cells as well as brain tissue from BALB/c mice and 3T3 cells was performed according to standard procedures. Isoform-specific cDNA fragments of the individual 5′ UTRs of the PARG55 and the PARG60 transcripts were initially amplified from 293 cDNA using primer pair 5′GCGGGAATTCCTTTCCGGTGGTGGGAAAGTG (exon-1/1a) and 5′CGCAGTTCGCTCACCATTCC (exon 4/6) (Fig. 2A). Subsequently, detection of PARG55 and PARG60 transcripts in cDNA preparations from various human cell lines (supplemental Fig. 1B) and human peripheral blood mononuclear cells4 (supplemental Fig. 1C) was done using two exon 1/1a-specific forward primers (5′AATTGCAGAAGCAGGCAGCGG and 5′ATGGTGCAGGCAGGCGCTGAG) in combination with an exon 6-specific reverse primer (5′GTGCAGTCTGAATGAGCTCCC). Detection of ARH3 mRNA was done using primer pair 5′CATGGAGGAGCGTCCATACT (exon 5) and 5′GGGCCAGGATGTCTGTCTC (exon 6).
FIGURE 2.
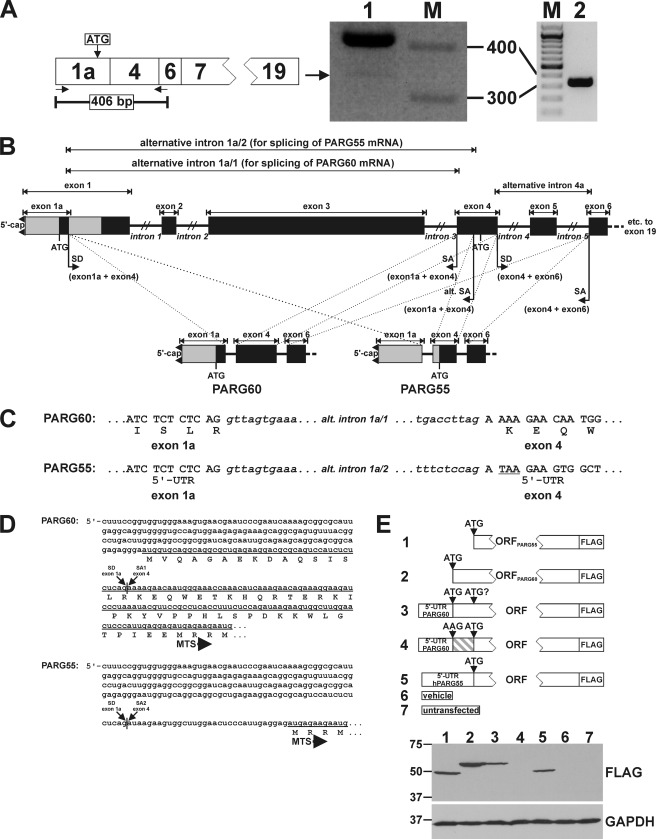
Human PARG55 originates from an alternatively spliced PARG mRNA. A, PCR analysis with primers that bind to the 5′ end and to the transition of exons 4 and 6 of the PARG cDNA (left panel). Fragments amplified from 293 cDNA were separated in a 2.5% ethidium bromide-stained agarose gel (center panel). Two specific DNA fragments were amplified (lane 1). The major band (406 bp) corresponded to PARG60 mRNA. The lower band (arrow) was reamplified (right panel, lane 2). Sequence analysis revealed a novel splicing event between exons 1a and 4. The size of selected marker bands (M) is indicated. B, overview of alternative splicing events leading to the generation of PARG55 and PARG60 mRNAs. Exon 1a is linked via a common splice donor (SD) site with two alternative splice acceptor (SA) sites in exon 4. Common SD and SA sites in exons 4 and 6 lead to skipping of exon 5. C, the PARG60 ORF starts in exon 1a. For PARG55, the use of an alternative SA site in exon 4 results in the generation of a stop codon (TAA, underlined), thereby converting the upstream region (exon 1a and part of exon 4) into the 5′ UTR. D, 5′ mRNA and N-terminal amino acid sequences of PARG60 and PARG55. Translational start of PARG60 in exon 1a masks the MTS that follows 52 amino acid residues downstream of the start codon. In contrast, translation of PARG55 starts in exon 4 at the ATG of the MTS. E, the ORFs of PARG55 and PARG60 were expressed as C-terminally FLAG-tagged proteins in 293 cells in the absence (lanes 1 and 2) or presence (lanes 3 and 5) of their endogenous 5′ UTRs. Expression of PARG60 in the context of its endogenous 5′ UTR (lane 3) did not lead to detection of an additional band resulting from an alternative translation site. No protein was detected upon expression of PARG60 in the context of its endogenous 5′ UTR harboring a mutated start codon in exon 1a (lane 4). Lysates from untransfected cells (lane 7) and cells transfected with the vector backbone (lane 6) were used as control. GAPDH was used as a loading control.
Isoform-specific cDNA fragments of the individual 5′ UTRs of the murine PARG transcripts were amplified using primer pair 5′GGGAAAGTGAGCCTGGAGCC (exon 1/1a) and 5′GTGGCATATTCTAAGAAATGGG (exon 7) in primary PCR reactions and primer pair 5′ATCTCGAGCCGAGTGGAAGC (exon 1/1a) and 5′GGGAGCTCATTCAGACTGCAC (exon 6) in nested PCR reactions.
qRT-PCR Analyses
qRT-PCR analyses of cDNA from HeLaS3, 293, HepG2, and SH-SY5Y cells were done using Universal ProbeLibrary (UPL, Roche) hydrolysis probes on a LightCycler 480 instrument (Roche). Actin β mRNA was detected using UPL probe 64 with primers 5′CCAACCGCGAGAAGATGA and 5′CCAGAGGCGTACAGGGATAG. Total PARG mRNA was detected using UPL probe 45 with primers 5′TCAGAGAATCTTTCTGCAGTGG and 5′CAGCAGCTGCCAATATCTGTAT. Human PARG55 and PARG60 mRNAs were detected using UPL probe 34 with primers 5′GGAGATGAGAAGAATGCCTC and 5′CAGTTCGCTCACCATTCCA. Human ARH3 mRNA was detected using UPL probe 50 with primers 5′GACGTCAGTCCTGCGTCAT and 5′TCATCTGTGTAGTACAAGGCTTCTG. Relative expression was calculated using the ΔΔCt method.
qRT-PCR analyses of cDNA preparations from stably transfected MEFs expressing shRNAs were done using a Taqman expression assay with a 7900HT fast real-time PCR system. PARG mRNA levels were normalized to GAPDH mRNA. Predesigned 6-carboxyfluorescein-labeled probes and primers to detect PARG and GAPDH mRNAs were purchased from ABI. PARG-specific primers recognized exons 6–7.
Vector Construction
A eukaryotic expression vector encoding C-terminally FLAG-tagged PARG55(ex5) was described in (27). The ORFs encoding PARG55 and PARG60 were amplified from HeLaS3 cDNA using forward primers 5′GCGGGAATTCCACCATGAGAAGAATGCCTCGGTG (for PARG55) and 5′GCGGGAATTCCACCATGGTGCAGGCAGGCGCTGAG (for PARG60) along with reverse primer 5′GCGGGTCGACGGTCCCTGTCCTTTGCCCTG and ligated into pCMV-FLAG5a (Sigma) via the EcoRI and SalI sites. The 5′ UTRs specific for PARG55 and PARG60 were introduced into these vectors by a PCR-based approach. Mutation of the start codon of the ORF encoding PARG60 in the context of its endogenous ORF was done by PCR-based site-directed mutagenesis.
For expression of PARG-PARP1cd fusion proteins, ORFs encoding PARG55, PARG55(ex5), and PARG60 were amplified using forward primers 5′GCGGGCTAGCCCACCATGAGAAGAATGCCTC (for PARG55 and PARG56(ex5)) or 5′GCGGGCTAGCCCACCATGGTGCAGGCAGGCGC (for PARG60) along with reverse primer 5′GCGGGAATTCGGTCCCTGTCCTTTGCCCTG. The PCR products were ligated into the NheI and EcoRI sites of a vector harboring the sequence encoding the catalytic domain of PARP1 (36). Preparation of vectors coding for PARP1cd fusion constructs with glutamate dehydrogenase (GDH) and GDHΔ1–53 was described previously (36). To generate vectors encoding MTS-PARG55, MTS-PARG55(ex5), and MTS-PARG60, we modified the vector pCMV/myc/mito (Invitrogen) by inserting a DNA sequence coding for the FLAG epitope to yield pCMV/FLAG/mito. Next, the ORFs encoding PARG55, PARG55(ex5), and PARG60 were ligated into pCMV/FLAG/mito via the SalI and XhoI sites. Exon 5 in MTS-PARG60(ex5) was introduced into the vector encoding MTS-PARG60 by a PCR-based approach.
Vectors encoding shRNA along with nuclear EGFP were generated from pCMV/myc/nuc/GFP (Invitrogen). The GFP-ORF was replaced by that of EGFP. Subsequently, a gene cassette allowing for expression of shRNA under control of the H1 RNA promoter (37) was introduced into the DraIII site of the vector. The PARG-specific shRNA sequence was derived from a reported short interfering RNA (38). Vectors encoding an irrelevant shRNA and harboring only the H1 RNA promoter were used as controls.
Transfection and Immunoblot Analysis
293 and HeLaS3 cells were transfected using Effectene reagent (Qiagen). After 24 h, cells were washed and lysed by adding SDS sample buffer. Cell lysates were subjected to SDS-PAGE in 6 and 10% gels prior to transfer to nitrocellulose membranes. MEFs were transfected with Lipofectamine 2000 reagent (Invitrogen). After 2 days, cell lysates were prepared with radioimmune precipitation assay buffer (Sigma-Aldrich) supplemented with complete protease inhibitor mixture (Roche) for detection of ARH3 and with 2% SDS in 20 mm Tris-HCl (pH 7.4) for detection of PAR, respectively. For ARH3 detection, cell lysates were subjected to SDS-PAGE in 10% gels prior to transfer to nitrocellulose membranes. For PAR detection, cell lysates were subjected to SDS-PAGE in 3–8% gradient gels prior to transfer to nitrocellulose membranes. Protein detection was done by enhanced chemiluminescence.
Immunocytochemistry and Confocal Laser Scanning Microscopy
Transfected HeLaS3 cells were fixed after 24 h and MEFs after 2 days with ice-cold 4% (v/v) formaldehyde in PBS for 20–30 min and subsequently permeabilized with 0.5% (v/v) Triton X-100 in PBS for 15 min. After blocking for 1 h at room temperature and incubation with primary antibodies for 3 h at room temperature or overnight at 4 °C, cells were washed twice with PBS and once with 0.1% (v/v) Triton X-100 in PBS prior to adding secondary antibodies. Following 1-h incubation at room temperature, nuclei were stained with DAPI, and the cells were subjected to confocal laser scanning microscopy.
Images from HeLaS3 cells were taken using the 405-, 488-, 594-, and 633-nm laser lines of a Leica TCS SP2 confocal laser scanning microscope (Leica Microsystems) equipped with a ×63 oil immersion objective (numerical aperture, 1.40). Images from MEFs were taken using the 405-, 488-, and 594-nm laser lines of a Zeiss LSM 510 Meta confocal laser scanning microscope (Carl Zeiss, Inc.) equipped with a ×40 oil immersion objective (numerical aperture, 1.3). Fluorescence data were processed with ImageJ 1.37a (National Institutes of Health) and calculated as a ratio of PAR to EGFP.
Statistical Analysis
Statistical analysis was performed using GraphPad Prism 4 (GraphPad Software, Inc., La Jolla, CA). Significance was determined using Student's t test and one-way or two-way analysis of variance with Bonferroni post-hoc test. Values are means ± S.E. p values < 0.05 were considered significant.
RESULTS
Human PARG55 Originates from Alternative Splicing of the Primary PARG Transcript
We first addressed the question of the existence and origin of human PARG55. Amplification of cDNA from human 293 cells using primers that bind to the 5′ end and the transition of exon 4 and exon 6 of the PARG60 mRNA (indicated by arrows in Fig. 2A) resulted in a ∼400 bp DNA fragment and an additional shorter DNA fragment (A). The longer fragment resulted from the known splicing events leading to PARG60 mRNA. The shorter DNA fragment derived from linkage of exon 1a via its known splice donor site to an alternative splice acceptor site in exon 4, downstream of that used for splicing of the PARG60 mRNA (Fig. 2, B and C). As a result, the reading frame that starts from the ATG triplet in exon 1a that is used to initiate translation of PARG60 is interrupted. The ORF starts at the ATG in exon 4 that is followed by the mitochondrial targeting sequence and encodes PARG55 (Fig. 2, C and D). Both exon 1a and the sequence of exon 4 upstream of the ATG constitute the 5′ UTR.
To investigate whether PARG55 may also result from the PARG60 mRNA, we generated vectors encoding C-terminally FLAG-tagged PARG55 and PARG60, including their endogenous 5′ UTRs (Fig. 2E). For PARG60, an additional vector was generated harboring the endogenous 5′ UTR along with a mutated (ATG = > AAG) start codon (Fig. 2E). Expression of the PARG60 ORF in the presence of its 5′ UTR led to expression of a single 60-kDa protein (Fig. 2E, lane 3). No additional band was detected that might have been derived from bypassing the first ATG of the PARG60 ORF. Moreover, no protein was detected when the start codon was mutated (Fig. 2E, lane 4). Importantly, expression of the PARG55 ORF in the context of the newly identified 5′ UTR led to detection of the same protein that was expressed from the PARG55 ORF (Fig. 2E, lanes 1 and 5). Taken together, the data shown in Fig. 2 demonstrate the existence of a human PARG55 isoform that exclusively results from a hitherto unrecognized splicing event.
We verified the presence of the PARG55, PARG60, and ARH3 transcripts in other cells. For the small PARG isoforms, we additionally included cDNA from human peripheral blood mononuclear cells in our studies. Primers that allow for coamplification of the PARG55 and PARG60 mRNAs as well as primers specific for the ARH3 transcript were used. DNA fragments that derived from these three transcripts could be amplified from cDNA from 293, HepG2, SH-SY5Y, and HeLaS3 cells (supplemental Fig. 1, A and B). Importantly, our RT-PCR analyses confirmed the splicing event leading to the PARG55 transcript in human peripheral blood mononuclear cells (indicated by black arrows in supplemental Fig. 1C). The nucleotide sequences of selected PARG55-specific PCR products were confirmed by DNA sequence analysis (asterisks in supplemental Fig. 1, B and C).
Next, we determined the relative transcript levels of the mRNAs for PARG55 and PARG60 by qRT-PCR and compared them with the expression levels of all known PARG isoforms in four human cell lines. The PARG55- and PARG60-specific mRNAs accounted in total for 15–18% of all known PARG mRNAs (supplemental Fig. 1D). Interestingly, in all cell lines, the expression levels of the ARH3 mRNA were up to 4-fold higher than the total PARG mRNA levels. (supplemental Fig. 1D).
Human PARG55 but Not PARG60 Localizes to the Mitochondrial Matrix
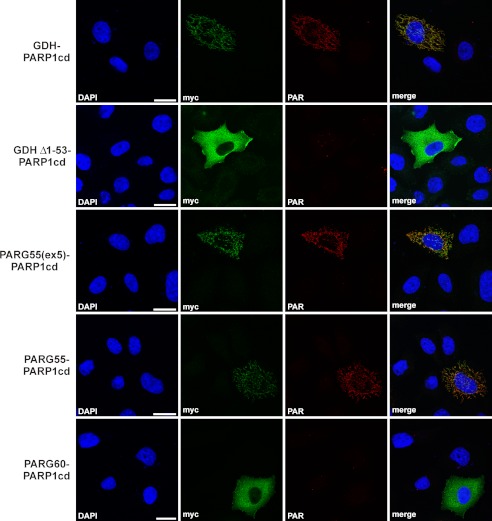
We then subjected the human isoforms PARG55 and PARG60 to in-depth subcellular localization analyses. To establish whether PARG55 and, possibly, a fraction of PARG60 are located in the mitochondrial matrix, we subjected them to poly(ADP-ribose)-assisted localization assay analysis. This assay conclusively establishes matrix localization (36). The protein of interest is fused to the PARP1cd. Although the targeting of the fusion protein is fully dependent on the protein, whose localization is the subject of investigation, the activity of the PARP1cd is used to monitor the localization of the protein of interest by using the product of its activity, PAR, as readout. In the context of this study, immunodetection of PAR is possible if the PARP1cd is directed to the mitochondrial matrix, whereas PAR formation cannot be observed when the PARP1cd is present within the cytosol (36). Human PARG55 and PARG60 fused to the N terminus of PARP1cd (PARG55-PARP1cd and PARG60-PARP1cd) were expressed in HeLaS3 cells. As a positive control for matrix localization, we used GDH as well as PARG55(ex5) in fusion with PARP1cd. GDH-PARP1cd lacking the endogenous MTS encoded by amino acids 1–53 (GDHΔ1–53-PARP1cd) served as a negative control. Fusion of the PARG isoforms to PARP1cd did not change the cellular distribution (as detected by the myc-epitope in Fig. 3) that was observed for the C-terminally FLAG-tagged PARG isoforms (supplemental Fig. 2). Both FLAG-tagged PARG55 and PARG55(ex5) colocalized with mitochondrially targeted EGFP, whereas PARG60 showed the same distribution as the cytosolic portion of untargeted EGFP (supplemental Fig. 2). For the PARP1cd fusion constructs, PAR was detected in cells expressing PARG55-PARP1cd, GDH-PARP1cd, and PARG55(ex5)-PARP1cd, demonstrating their matrix localization (Fig. 3). No polymer formation could be detected upon expression of PARG60-PARP1cd and GDHΔ1–53-PARP1cd. PARG60 is absent from the mitochondrial matrix (Fig. 3). It is important to note that accumulation of immunodetectable PAR in mitochondria is prevented when PARG55(ex5) is coexpressed in cells along with mitoPARP1cd (Ref. 27) and this study). That is, in accordance with the reported catalytic inactivity of PARG in PARG-GFP fusion proteins (39), C-terminal fusion of PARP1cd to hPARG55(ex5) also disrupted the catalytic activity of the PARG portion within the hPARG55(ex5)-PARP1cd construct.
FIGURE 3.
Human PARG55 localizes to the mitochondrial matrix as revealed by poly(ADP-ribose)-assisted protein localization assay. The PARP1cd along with a myc epitope were fused to the C terminus of PARG55, PARG55(ex5), and PARG60. The fusion proteins were expressed in HeLaS3 cells and detected by myc immunocytochemistry. PAR immunodetection was used as readout for matrix localization of the fusion proteins. Fusion proteins composed of GDH-PARP1cd-myc and GDHΔ1–53-PARP1cd-myc (lacking the N-terminal MTS) were used as controls. Scale bars = 20 μm.
Human PARG55 Does Not Degrade Matrix-accumulated PAR
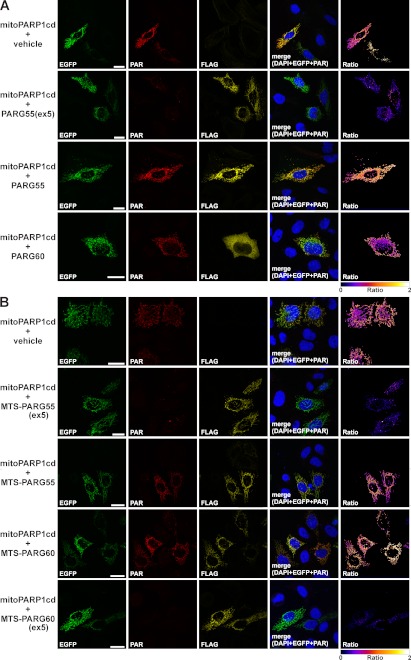
Next, we investigated the PAR-degrading activities of the small human PARG isoforms on PAR accumulated in the mitochondrial matrix. We expressed a fusion construct composed of enhanced green fluorescent protein and the catalytic domain of PARP1 targeted to the mitochondrial matrix (referred to as mitoPARP1cd) along with C-terminally FLAG-tagged PARG55, PARG60, and PARG55(ex5). Transfected cells, identified by FLAG immunocytochemistry and the green fluorescence of mitoPARP1cd, were analyzed for mitochondrial PAR. As shown in Fig. 4, A and B, cells expressing mitoPARP1cd in the absence of an overexpressed PARG isoform displayed robust PAR formation within mitochondria. The PAR immunoreactivity increased with the EGFP fluorescence intensity, reflecting the expression level of mitoPARP1cd (shown for MEFs in supplemental Fig. 3). In presence of PARG55(ex5), mitochondrial PAR was strongly reduced (Fig. 4A), as expected (27). The expression of human PARG60 did not change the amount of polymers generated by mitoPARP1cd (Fig. 4A), which is in accordance with the cytosolic localization of this isoform (Fig. 3 and supplemental Fig. 2). Surprisingly, we still observed accumulation of PAR within mitochondria in the presence of the matrix-localized isoform PARG55 (Fig. 4A). That is, in contrast to PARG55(ex5), PARG55 appears to be inactive for PAR degradation.
FIGURE 4.
Lack of PAR-degrading activity of human PARG55 and PARG60 is due to the absence of exon 5-encoded amino acids. The catalytic domain of PARP1 expressed in fusion with EGFP (mitoPARP1cd) was targeted to the mitochondrial matrix and coexpressed with C-terminally FLAG-tagged PARG constructs. PARG expression was detected by FLAG immunocytochemistry. Cotransfection of vectors encoding mitoPARP1cd and the empty vector served as controls. MitoPARP1cd expression was visualized by its green fluorescence. Ratio images were created by dividing PAR images (red) by EGFP images (green) and color-coded using the indicated scale. A, matrix-accumulated PAR was almost absent only in cells coexpressing mitoPARP1cd and PARG55(ex5). B, mitoPARP1cd was coexpressed with C-terminally FLAG-tagged MTS-PARG55, MTS-PARG55(ex5), MTS-PARG60, and MTS-PARG60(ex5) (endowed with an authentic N-terminal MTS). Matrix-accumulated PAR was almost absent in the presence of PARG isoforms harboring the amino acids encoded by exon 5 (MTS-PARG55(ex5) and MTS-PARG60(ex5)). Scale bars = 20 μm. Degradation of matrix-accumulated PAR by ARH3 was shown in Ref. 27.
Both Small Human PARG Isoforms Are Catalytically Inactive because of the Absence of Exon 5-encoded Amino Acids
Besides PARG55, PARG60 also lacks exon 5-encoded amino acids. The absence of these amino acids may impair the PAR-degrading capacity of this isoform, as observed for PARG55. On the other hand, the additional N-terminal residues in PARG60 may restore its catalytic function. To address these questions, we forced PARG60 into the mitochondrial matrix by fusion of a bona fide MTS to the N terminus (MTS-PARG60). To control for the possibility that this modification affects the ability of PARG55 and PARG55(ex5) to degrade matrix-accumulated PAR, the same MTS was N-terminally fused to these isoforms. Mitochondrial localization of the proteins was confirmed in cells that coexpressed mitochondrial EGFP (supplemental Fig. 4). The MTS-PARG isoforms were coexpressed with mitoPARP1cd and cells analyzed for PAR. Fusion of the MTS to PARG55(ex5) resulted in a strong decrease in matrix-accumulated PAR (Fig. 4B). In contrast, mitochondrial PAR was detectable after expression of both MTS-PARG55 and MTS-PARG60, along with mitoPARP1cd (Fig. 4B). Thus, the lack of the amino acids encoded by exon 5 also abolishes the PAR-degrading activity of PARG60. We reintroduced exon 5 in the ORF of MTS-PARG60 to determine whether this could restore the PAR-degrading activity of the resulting isoform, MTS-PARG60(ex5). Indeed, coexpression of MTS-PARG60(ex5) and mitoPARP1cd caused a strong decrease in PAR immunoreactivity of transfected cells (Fig. 4B). Thus, absence of exon 5-encoded amino acids dramatically impairs the catalytic activity of both physiological small PARG isoforms. These findings were further substantiated by calculating the ratios of PAR and EGFP signal intensities of fluorescence micrographs (supplemental Fig. 5A) and PAR immunoblot analysis of lysates from cells coexpressing mitoPARP1cd along with PARG55, PARG55(ex5), MTS-PARG60, and MTS-PARG60(ex5) (supplemental Fig. 5B). Although expressed at far lower levels, PARG55(ex5) and MTS-PARG60(ex5) caused a noticeable reduction of matrix-accumulated PAR compared with their corresponding counterparts lacking exon 5-encoded residues (supplemental Fig. 5B). Our data suggest that the human PARG gene encodes cytosolic and mitochondrial isoforms that do not have PAR-degrading activity.
The Murine PARG Gene Encodes a Catalytically Inactive Cytosolic PARG Isoform
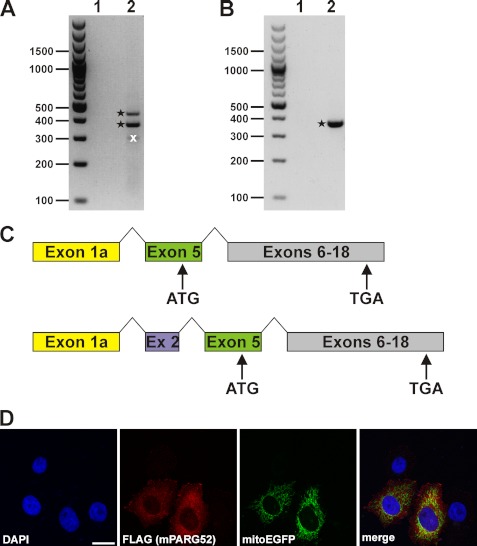
In mice, the reported counterpart of PARG60 contains exon 5. Nevertheless, a similar splicing event that leads to the PARG55 mRNA in humans could also occur in mice. We conducted RT-PCR analyses using cDNA from mouse 3T3 fibroblasts (Fig. 5A) and brain (B) as templates. Primary PCR reactions performed in presence of primers that bind to the 5′-UTR and exon 7 (present in all known murine PARG mRNAs) did not lead to the amplification of specific cDNA fragments of any known PARG isoform (lane 1 in Fig. 5A and 5B). Secondary, nested PCR reactions using primers specific for exon 1/1a and exon 6 resulted in specific PARG cDNA fragments (asterisks in Fig. 5, A and B). These fragments originated from fusion of exon 1a and exon 5 (for cDNA from 3T3 cells and brain) or of exon 1a, exon 2, and exon 5 (for 3T3 cells only). Both splicing events predict the existence of a murine 52-kDa PARG isoform (mPARG52) whose translation is initiated from a start codon in exon 5 (Fig. 5C). This isoform lacks a substantial portion of exon 5-encoded residues, and the MTS is completely absent. Indeed, recombinant mPARG52 localized in the cytosol of HeLaS3 cells (Fig. 5D). Because the primer combinations used in our analyses should also enable the amplification of cDNA sequences of the longer murine PARG mRNAs, we scrutinized the specificity of the reaction conditions. Indeed, by extending the elongation time, the same RT-PCRs led to the amplification of the cDNA sequences of mPARG101 and mPARG110 (supplemental Fig. 6). We could not detect any cDNA sequence compatible with the reported isoform mPARG63 as well as mPARG58, the putative murine counterpart of human PARG55.
FIGURE 5.
The murine PARG gene encodes a cytosolic 52-kDa PARG isoform. RT-PCR analyses of cDNA preparations from 3T3 cells (A) and brain tissue (B). Primary PCRs were performed using primers binding to exon 1/1a and exon 7 (lanes 1 in A and B), and secondary PCRs were done with nested primers annealing to exon 1/1a and exon 6 (lanes 2 in A and B). PCR products are indicated by asterisks derived from mPARG transcripts. An additional prominent PCR product (crossed out (x) in lane 2 of A) did not contain any mPARG-specific DNA sequence. C, DNA sequence analyses of the PCR products shown in A and B revealed alternative splicing of the primary murine PARG transcript via established splice acceptor and donor sites in exon 1a and exon 5 (for brain tissue and 3T3 cells) as well as in exon 1a, exon 2, and exon 5 (for 3T3 cells). Both splicing events predicted a 52-kDa mPARG isoform. Translation is initiated from a start codon in exon 5. D, overexpressed C-terminally FLAG-tagged mPARG52 was detected in the cytosol and did not colocalize with mitochondrial structures (as detected by coexpressed mitochondrial EGFP).
ARH3 Knockout Results in Significantly Reduced Turnover of Matrix-accumulated PAR
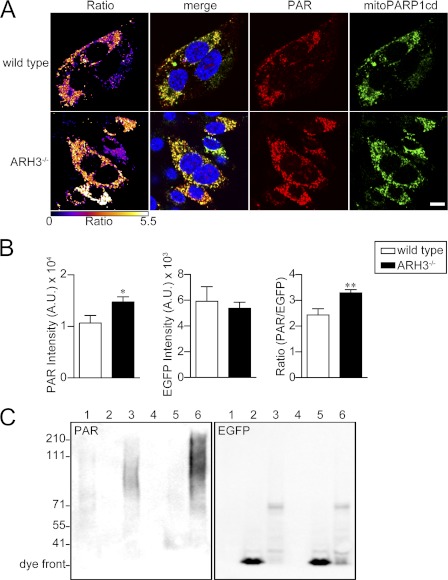
These genetic and functional data suggested that degradation of matrix-accumulated PAR does not involve a physiological PARG isoform but may be primarily borne by ARH3. To address this hypothesis, we made use of MEFs from ARH3−/− mice. Immunoblot analyses of MEF lysates confirmed the absence of the ARH3 protein in ARH3−/− mice (supplemental Fig. 7). We compared mitoPARP1cd-mediated PAR accumulation in wild-type and ARH3−/− MEFs. Expression of mitoPARP1cd in ARH3−/− cells resulted in a significantly increased mitochondrial PAR accumulation, as revealed by PAR immunocytochemistry (Fig. 6, A and B) and PAR immunoblot analysis of cell lysates (C). Further, the polymers were longer in the ARH3-deficient cells.
FIGURE 6.
Expression of mitoPARP1cd in ARH3−/− cells leads to increased mitochondrial PAR accumulation. A, fluorescence micrographs of wild-type and ARH3−/− MEFs expressing mitoPARPcd1. Cells were subjected to PAR immunocytochemistry 2 days post-transfection. Ratio images were created by dividing PAR images (red) by EGFP images (green) and color-coded using the indicated scale. Scale bar = 20 μm. B, quantification of fluorescent micrographs. The bar charts show the average intensities of PAR and EGFP and the PAR/EGFP ratio in wild-type (white column) and ARH3−/−cells (black column). Data are mean ± S.E. *, p < 0.05; **, p < 0.01 (Student's t test). Data are representative of three experiments. C, MEFs from wild-type (lanes 1–3) and ARH3−/− (lanes 4–6) mice were transfected with mitoPARP1cd (lanes 3 and 6), mitochondrial EGFP (lanes 2 and 5) or left untransfected (lanes 1 and 4). Cell lysates were subjected to PAR and EGFP immunoblot analyses. Data are representative of three experiments.
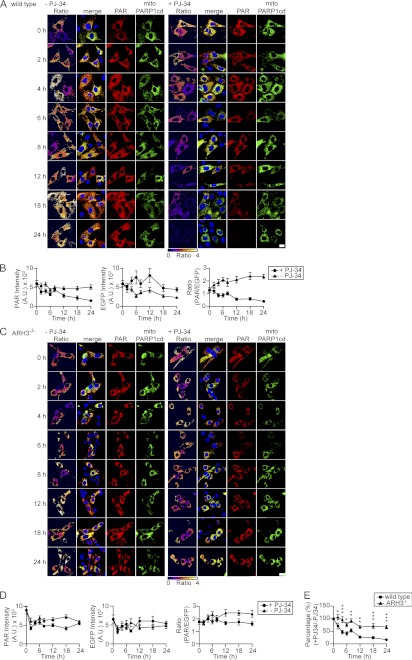
We then investigated whether the difference between wild-type and ARH3−/− cells in the capacity to accumulate mitochondrial PAR could be attributed to differences in PAR-degrading activities. The PARP inhibitor PJ-34 was added to mitoPARP1cd-expressing cells 24 h after transfection. Then, the decay of mitochondrial PAR content was monitored over time. The PAR/EGFP ratio of mitoPARP1cd-expressing cells from wild-type mice was reduced by half after ∼12 h of incubation with PJ-34 (Fig. 7, A and B). In contrast, PARP inhibition did not alter the ratio of PAR and EGFP signal intensities in mitoPARP1cd-expressing ARH3−/− cells (Fig. 7, C and D). In the absence of PJ-34, both cell types showed an increase in PAR intensity and PAR/EGFP ratio over time. Comparing the ratios of PAR/EGFP obtained in the presence or absence of PJ-34, the ratio of ARH3−/− cells was greater than that of wild-type cells (Fig. 7E). A similar result was obtained by immunoblot analyses (supplemental Fig. 8). These data indicate that degradation of mitochondrial PAR proceeds much more slowly in ARH3−/− cells than in wild-type cells.
FIGURE 7.
Absence of ARH3 abolishes degradation of matrix-accumulated PAR. A and C, wild-type and ARH3−/− MEFs transiently expressing mitoPARP1cd were incubated in the absence (left panels) or presence (right panels) of 5 μm PJ-34 for the indicated times and then subjected to PAR immunocytochemistry. Upon PARP inhibition, the mitochondrial PAR content decreased over time in mitoPARP1cd-expressing wild-type MEFs but persisted in ARH3−/− MEFs. Scale bar = 20 μm. B and D, graphs summarize the time course of PAR signal and EGFP fluorescence intensities and PAR/EGFP signal ratio in the presence (●) and absence (▴) of 5 μm PJ-34 (n = 17–50 cells for each time point). Data are representative of three experiments. E, summarized data of relative PAR/EGFP signal ratios in wild-type (●) and ARH3−/− (▴) cells. Values were calculated by dividing the PAR/EGFP signal ratio in the presence of PJ-34 by that in its absence. Mean ± S.E. *, p < 0.05; **, p ≤ 0.01; ***, p ≥ 0.001 (two-way repeated measures analysis of variance with post hoc Bonferroni test).
Degradation of Matrix-accumulated PAR Does Not Involve a PARG Isoform
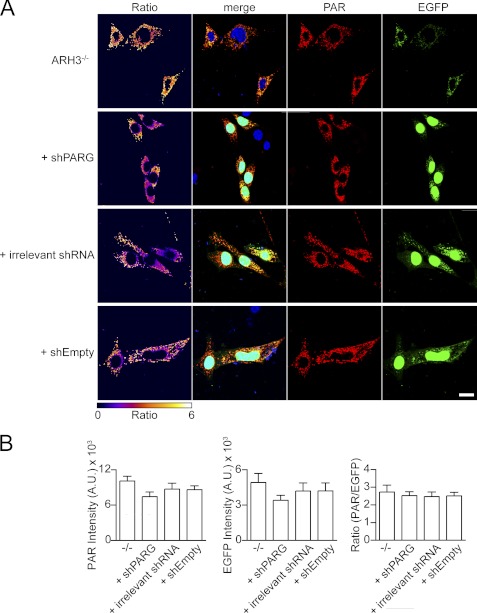
Finally, we tested whether, in addition to ARH3, a putative murine mitochondrial PARG isoform contributes to mitochondrial PAR degradation. To this end, we down-regulated PARG expression levels in ARH3−/− cells by stable transfection of a plasmid encoding shRNA specific for all known mPARG transcripts. Expression of nuclear EGFP from the same plasmid allowed for the selection and identification of shRNA-positive cells. qRT-PCR analyses revealed down-regulation of PARG gene expression by 35% in stably transfected ARH3−/− cells (supplemental Fig. 9). We determined the EGFP and PAR fluorescence intensities in the mitochondria of transfected cells. As shown in Fig. 8, A and B, expression of mitoPARP1cd in these cells did not affect the PAR/EGFP ratio, indicating that the degradation of matrix-accumulated PAR is not catalyzed by a PARG isoform.
FIGURE 8.
Silencing of PARG gene expression does not affect PAR accumulation in mitochondria. A, mitoPARP1cd expression in the context of PARG knockdown did not lead to increased PAR accumulation within mitochondria. Control ARH3−/− MEFs, ARH3−/− MEFs stably expressing PARG-specific shRNA, irrelevant shRNA, or an empty vector were transfected with the mitoPARP1cd-encoding plasmid. Cells were subjected to PAR immunocytochemistry 2 days post-transfection. Nuclear EGFP encoded by the shRNA vectors was used to identify shRNA-expressing cells. Ratio images were created by dividing PAR images (red) by EGFP images (green) and color-coded using the indicated scale. Scale bar = 20 μm. B, quantification of fluorescent micrographs. The average intensities of PAR signal and EGFP fluorescence and the ratio of PAR/EGFP signal intensities in the ARH3−/− cells from A are shown. Data are mean ± S.E. Data are representative of three experiments.
DISCUSSION
This study establishes the genetic background of a human mitochondrial PARG isoform, PARG55 (Fig. 2), and a cytosolic murine PARG isoform, mPARG52 (Fig. 5), which result from hitherto unrecognized alternative splicing events of the corresponding primary PARG transcripts. The primary structures of PARG55, PARG60, and mPARG52 contain the three acidic residues that were reported previously to be essential for catalysis (40), but they do not cleave PAR. The lack of their enzymatic activity is due to the absence of amino acids encoded by exon 5, which results from these splicing events. Consequently, even though PARG55 localizes to the mitochondrial matrix, it cannot contribute to a putative PAR metabolism within this organelle. Rather, our experiments demonstrate that ARH3 accounts for the residual PAR-degrading activity that is detectable in mitochondria of PARGΔexon2–3/Δexon2–3 mice (31) and is responsible for the degradation of matrix-accumulated PAR (27).
Our observations thus significantly extend the current knowledge on PAR metabolism and resolve issues that have been controversial. First, human PARG55(ex5) does not represent a physiological isoform because it contains exon 5-encoded amino acids. In both human PARG60 (32) and human PARG55 (this study, Fig. 2), exon 5 is removed from the primary PARG transcript upon maturation of the corresponding mRNAs. Second, for PARG60, association with mitochondria was suggested on the basis of indirect immunocytochemistry (32). However, the poly(ADP-ribose)-assisted protein localization assay performed here excludes PARG60 as a matrix protein and confirms matrix localization of PARG55 (Fig. 3). Third, it was postulated that PARG55 might result from alternative utilization of the second in-frame ATG within the ORF of the transcript encoding PARG60 (32). We excluded this possibility because the second in-frame ATG within the PARG60 ORF was not recognized as initiation site, even in the context of the endogenous 5′ UTR and an ATG = > AAG mutation of the nucleotide triplet constituting the start codon in the PARG60 mRNA (Fig. 2E).
For a long time, small PARG proteins were thought to result from proteolytic cleavage of the full-length protein. For instance, PARG is subject to caspase 3-mediated cleavage during apoptosis (41). Moreover, purification of PARG from tissue or cells as well as Western blot analyses indicated the presence of small PARG proteins (reviewed in Ref. 25). In fact, the first full-length PARG cDNA encoding bovine PARG111 was identified using peptide sequences from a purified ∼60-kDa protein (42). However, our data suggest that small PARG isoforms originate from posttranscriptional regulation. It cannot be excluded that the primary PARG transcript may encode even more, yet unidentified, PARG transcripts that have not been detected by classical Northern blot analyses because of low expression levels. In this regard, the data shown in Fig. 2A and supplemental Fig. 1, B–D, indicate that PARG transcript levels, in particular of PARG55, are very low. In addition to the identification of a splicing event leading to expression of PARG55, we provide evidence for another alternatively spliced murine PARG isoform, mPARG52, whose ORF starts in exon 5 (Fig. 5). Interestingly, a splicing event between exon 1a and exon 5 has already been reported in PARGΔexon2–3/Δexon2–3 mice (32) but was not demonstrated in wild-type mice.
Both PARG55(ex5) and PARG55 were localized to the mitochondrial matrix as revealed by poly(ADP-ribose)-assisted localization assay (Fig. 3). However, lack of exon 5 affected the catalytic activity, not only of PARG55 but also of PARG60. In our experimental system, PAR artificially accumulated within mitochondria was only degraded by PARG55(ex5) or after reintroduction of exon 5-encoded amino acids into PARG60 that was forced into the mitochondrial matrix. These findings extend the critical role of amino acids within the MTS for catalysis (43) by the adjacent amino acids encoded by exon 5. Amino acid exchanges within the MTS affected the catalytic activity of PARG55(ex5) as well as of N-terminally extended PARG variants (43). However, the MTS is part of the primary structures of both PARG55 and PARG60, and deletion of the MTS caused PARG55(ex5) to become inactive. Therefore, cleavage of the MTS upon import of PARG55(ex5) into mitochondria is unlikely because its catalytic activity is retained. In addition, it is unlikely that PARG60 is subject to N-terminal processing. It does not localize to mitochondria, and the amino acids constituting an MTS are masked by 52 N-terminal residues. Consequently, loss of PAR-degrading activity of PARG55 and PARG60 is due to the lack of amino acids encoded by exon 5. This conclusion is further substantiated by the identification of a splicing event leading to the cytosolic murine PARG isoform mPARG52 (Fig. 5). This isoform completely lacks exon 4 (and thus, the MTS) as well as a substantial amount of exon 5-encoded amino acids, whose importance for efficient PAR-degradation was demonstrated in this study. Indeed, lack of PAR-degrading activity was shown for an N-terminally truncated human PARG isoform that would reflect mPARG52 with regard to translational initiation and exon composition (43). Taken together, these findings suggest that the mammalian PARG gene encodes cytosolic and mitochondrial isoforms that do not display PAR-degrading activity. Interestingly, the catalytic deficiency is on the basis of alterations of the primary structure that are remote from the catalytic site of PARG enzymes. Therefore, the loss of PAR-degrading activity may be accompanied by changes of specificity that are not revealed by PARG-specific enzyme assays.
The realization that PARG55 was catalytically inactive necessitated the investigation of alternative activities that would account for mitochondrial PAR degradation. Using cells from ARH3−/− mice, we provide evidence that the degradation of PAR artificially accumulated within mitochondria is primarily mediated by ARH3. First, overexpression of mitoPARP1cd in ARH3−/− cells resulted in a greater accumulation of mitochondrial PAR than in wild-type cells. Second, there was no significant decrease in mitochondrial PAR content upon PARP inhibition in ARH3−/− cells expressing mitoPARP1cd in contrast to wild-type MEFs. Third, no further increase in mitochondrial PAR accumulation could be detected after additional down-regulation of PARG expression in mitoPARP1cd-expressing ARH3−/− cells (Fig. 8). Our data support a possible role of cytosolic and nuclear ARH3 in PAR metabolism. That is, ARH3 may compensate to some extent in PARGΔexon2–3/Δexon2–3 mice for the lack of nuclear mPARG110 and possibly also for the lack of the cytosolic isoforms mPARG101 and mPARG98. In addition, because complete PARG knockout is lethal (28), our findings raise the question whether the viability of PARGΔexon2–3/Δexon2–3 mice is due to a residual PAR-degrading activity of the small PARG isoforms.
In summary, our results support the conclusion that both ARH3 and PARG may carry out dual functions within the cells of higher eukaryotes. For ARH3, these functions include hydrolysis of O-acetyl-ADP-ribose, which is generated by NAD+-dependent protein deacetylases in the nucleus, the cytosol, and mitochondria. Because of its capacity to hydrolyze PAR, ARH3 may assist PARG in degrading polymers generated by nuclear and cytosolic PARP isoforms. In mitochondria, ARH3 appears to be the only candidate enzyme responsible for putative PAR degradation. The primary function of PARG is to catalyze PAR degradation in the nucleus and cytosol, but another hypothetical activity may be borne by the cytosolic and mitochondrial isoforms that do not display PAR-degrading activity.
Supplementary Material
Acknowledgments
The confocal imaging was performed in part at the Molecular Imaging Center (Fuge, Norwegian Research Council), University of Bergen.
This study was supported, in whole or in part, by the Intramural Research Program, National Heart, Lung, and Blood Institute, National Institutes of Health. This work was also supported by the Norwegian Cancer Society.

This article contains supplemental Figs. 1–9.
Human peripheral blood mononuclear cells were kindly provided by Dr. Silke Appel, University of Bergen. The use of human samples was approved by the Ethical Committee of the University in Bergen, Norway (242.06).
- ADPR
- ADP-ribose
- PAR
- poly(ADP-ribose)
- PARP
- poly(ADP-ribose) polymerase
- PARG
- poly(ADP-ribose) glycohydrolase
- ARH
- ADP-ribosylhydrolase
- PARP1cd
- catalytic domain of poly(ADP-ribose) polymerase 1
- MEF
- mouse embryonic fibroblast
- qRT-PCR
- quantitative RT-PCR
- UPL
- Universal ProbeLibrary
- GDH
- glutamate dehydrogenase
- EGFP
- enhanced green fluorescent protein
- MTS
- mitochondrial targeting sequence.
REFERENCES
- 1. Corda D., Di Girolamo M. (2003) Functional aspects of protein mono-ADP-ribosylation. EMBO J. 22, 1953–1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Seman M., Adriouch S., Haag F., Koch-Nolte F. (2004) Ecto-ADP-ribosyltransferases (ARTs). Emerging actors in cell communication and signaling. Curr. Med. Chem. 11, 857–872 [DOI] [PubMed] [Google Scholar]
- 3. Di Girolamo M., Dani N., Stilla A., Corda D. (2005) Physiological relevance of the endogenous mono(ADP-ribosyl)ation of cellular proteins. FEBS J. 272, 4565–4575 [DOI] [PubMed] [Google Scholar]
- 4. Hassa P. O., Haenni S. S., Elser M., Hottiger M. O. (2006) Nuclear ADP-ribosylation reactions in mammalian cells. Where are we today and where are we going? Microbiol. Mol. Biol. Rev. 70, 789–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schreiber V., Dantzer F., Ame J. C., de Murcia G. (2006) Poly(ADP-ribose): novel functions for an old molecule. Nat. Rev. Mol. Cell Biol. 7, 517–528 [DOI] [PubMed] [Google Scholar]
- 6. Alvarez-Gonzalez R., Jacobson M. K. (1987) Characterization of polymers of adenosine diphosphate ribose generated in vitro and in vivo. Biochemistry 26, 3218–3224 [DOI] [PubMed] [Google Scholar]
- 7. Masson M., Niedergang C., Schreiber V., Muller S., Menissier-de Murcia J., de Murcia G. (1998) XRCC1 is specifically associated with poly(ADP-ribose) polymerase and negatively regulates its activity following DNA damage. Mol. Cell. Biol. 18, 3563–3571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Malanga M., Althaus F. R. (2005) The role of poly(ADP-ribose) in the DNA damage signaling network. Biochem. Cell Biol. 83, 354–364 [DOI] [PubMed] [Google Scholar]
- 9. Dantzer F., Amé J. C., Schreiber V., Nakamura J., Ménissier-de Murcia J., de Murcia G. (2006) Poly(ADP-ribose) polymerase-1 activation during DNA damage and repair. Methods Enzymol. 409, 493–510 [DOI] [PubMed] [Google Scholar]
- 10. Hassa P. O., Hottiger M. O. (2002) The functional role of poly(ADP-ribose)polymerase 1 as novel coactivator of NF-κB in inflammatory disorders. Cell. Mol. Life Sci. 59, 1534–1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kraus W. L. (2008) Transcriptional control by PARP-1. Chromatin modulation, enhancer-binding, coregulation, and insulation. Curr. Opin. Cell Biol. 20, 294–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chiarugi A. (2002) Poly(ADP-ribose) polymerase. Killer or conspirator? The “suicide hypothesis” revisited. Trends Pharmacol. Sci. 23, 122–129 [DOI] [PubMed] [Google Scholar]
- 13. Alano C. C., Ying W., Swanson R. A. (2004) Poly(ADP-ribose) polymerase-1-mediated cell death in astrocytes requires NAD+ depletion and mitochondrial permeability transition. J. Biol. Chem. 279, 18895–18902 [DOI] [PubMed] [Google Scholar]
- 14. Cipriani G., Rapizzi E., Vannacci A., Rizzuto R., Moroni F., Chiarugi A. (2005) Nuclear poly(ADP-ribose) polymerase-1 rapidly triggers mitochondrial dysfunction. J. Biol. Chem. 280, 17227–17234 [DOI] [PubMed] [Google Scholar]
- 15. Ying W., Alano C. C., Garnier P., Swanson R. A. (2005) NAD+ as a metabolic link between DNA damage and cell death. J. Neurosci. Res. 79, 216–223 [DOI] [PubMed] [Google Scholar]
- 16. Yu S. W., Wang H., Poitras M. F., Coombs C., Bowers W. J., Federoff H. J., Poirier G. G., Dawson T. M., Dawson V. L. (2002) Mediation of poly(ADP-ribose) polymerase-1-dependent cell death by apoptosis-inducing factor. Science 297, 259–263 [DOI] [PubMed] [Google Scholar]
- 17. Yu S. W., Andrabi S. A., Wang H., Kim N. S., Poirier G. G., Dawson T. M., Dawson V. L. (2006) Apoptosis-inducing factor mediates poly(ADP-ribose) (PAR) polymer-induced cell death. Proc. Natl. Acad. Sci. U.S.A. 103, 18314–18319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. David K. K., Andrabi S. A., Dawson T. M., Dawson V. L. (2009) Parthanatos, a messenger of death. Front. Biosci. 14, 1116–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Amé J. C., Rolli V., Schreiber V., Niedergang C., Apiou F., Decker P., Muller S., Höger T., Ménissier-de Murcia J., de Murcia G. (1999) PARP-2, a novel mammalian DNA damage-dependent poly(ADP-ribose) polymerase. J. Biol. Chem. 274, 17860–17868 [DOI] [PubMed] [Google Scholar]
- 20. Kickhoefer V. A., Siva A. C., Kedersha N. L., Inman E. M., Ruland C., Streuli M., Rome L. H. (1999) The 193-kD vault protein, VPARP, is a novel poly(ADP-ribose) polymerase. J. Cell Biol. 146, 917–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rouleau M., McDonald D., Gagné P., Ouellet M. E., Droit A., Hunter J. M., Dutertre S., Prigent C., Hendzel M. J., Poirier G. G. (2007) PARP-3 associates with polycomb group bodies and with components of the DNA damage repair machinery. J. Cell. Biochem. 100, 385–401 [DOI] [PubMed] [Google Scholar]
- 22. Smith S., de Lange T. (1999) Cell cycle dependent localization of the telomeric PARP, tankyrase, to nuclear pore complexes and centrosomes. J. Cell Sci. 112, 3649–3656 [DOI] [PubMed] [Google Scholar]
- 23. Lyons R. J., Deane R., Lynch D. K., Ye Z. S., Sanderson G. M., Eyre H. J., Sutherland G. R., Daly R. J. (2001) Identification of a novel human tankyrase through its interaction with the adaptor protein Grb14. J. Biol. Chem. 276, 17172–17180 [DOI] [PubMed] [Google Scholar]
- 24. Amé J. C., Spenlehauer C., de Murcia G. (2004) The PARP superfamily. BioEssays 26, 882–893 [DOI] [PubMed] [Google Scholar]
- 25. Bonicalzi M. E., Haince J. F., Droit A., Poirier G. G. (2005) Regulation of poly(ADP-ribose) metabolism by poly(ADP-ribose) glycohydrolase: where and when? Cell. Mol. Life Sci. 62, 739–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Oka S., Kato J., Moss J. (2006) Identification and characterization of a mammalian 39-kDa poly(ADP-ribose) glycohydrolase. J. Biol. Chem. 281, 705–713 [DOI] [PubMed] [Google Scholar]
- 27. Niere M., Kernstock S., Koch-Nolte F., Ziegler M. (2008) Functional localization of two poly(ADP-ribose)-degrading enzymes to the mitochondrial matrix. Mol. Cell. Biol. 28, 814–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Koh D. W., Lawler A. M., Poitras M. F., Sasaki M., Wattler S., Nehls M. C., Stöger T., Poirier G. G., Dawson V. L., Dawson T. M. (2004) Failure to degrade poly(ADP-ribose) causes increased sensitivity to cytotoxicity and early embryonic lethality. Proc. Natl. Acad. Sci. U.S.A. 101, 17699–17704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Meyer-Ficca M. L., Meyer R. G., Coyle D. L., Jacobson E. L., Jacobson M. K. (2004) Human poly(ADP-ribose) glycohydrolase is expressed in alternative splice variants yielding isoforms that localize to different cell compartments. Exp. Cell Res. 297, 521–532 [DOI] [PubMed] [Google Scholar]
- 30. Winstall E., Affar E. B., Shah R., Bourassa S., Scovassi I. A., Poirier G. G. (1999) Preferential perinuclear localization of poly(ADP-ribose) glycohydrolase. Exp. Cell Res. 251, 372–378 [DOI] [PubMed] [Google Scholar]
- 31. Cortes U., Tong W. M., Coyle D. L., Meyer-Ficca M. L., Meyer R. G., Petrilli V., Herceg Z., Jacobson E. L., Jacobson M. K., Wang Z. Q. (2004) Depletion of the 110-kilodalton isoform of poly(ADP-ribose) glycohydrolase increases sensitivity to genotoxic and endotoxic stress in mice. Mol. Cell. Biol. 24, 7163–7178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Meyer R. G., Meyer-Ficca M. L., Whatcott C. J., Jacobson E. L., Jacobson M. K. (2007) Two small enzyme isoforms mediate mammalian mitochondrial poly(ADP-ribose) glycohydrolase (PARG) activity. Exp. Cell Res. 313, 2920–2936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mueller-Dieckmann C., Kernstock S., Lisurek M., von Kries J. P., Haag F., Weiss M. S., Koch-Nolte F. (2006) The structure of human ADP-ribosylhydrolase 3 (ARH3) provides insights into the reversibility of protein ADP-ribosylation. Proc. Natl. Acad. Sci. U.S.A. 103, 15026–15031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ono T., Kasamatsu A., Oka S., Moss J. (2006) The 39-kDa poly(ADP-ribose) glycohydrolase ARH3 hydrolyzes O-acetyl-ADP-ribose, a product of the Sir2 family of acetyl-histone deacetylases. Proc. Natl. Acad. Sci. U.S.A. 103, 16687–16691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Imai S., Guarente L. (2010) Ten years of NAD-dependent SIR2 family deacetylases. Implications for metabolic diseases. Trends Pharmacol. Sci. 31, 212–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dölle C., Niere M., Lohndal E., Ziegler M. (2010) Visualization of subcellular NAD pools and intra-organellar protein localization by poly(ADP-ribose) formation. Cell. Mol. Life Sci. 67, 433–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Brummelkamp T. R., Bernards R., Agami R. (2002) A system for stable expression of short interfering RNAs in mammalian cells. Science 296, 550–553 [DOI] [PubMed] [Google Scholar]
- 38. Blenn C., Althaus F. R., Malanga M. (2006) Poly(ADP-ribose) glycohydrolase silencing protects against H2O2-induced cell death. Biochem. J. 396, 419–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Haince J. F., Ouellet M. E., McDonald D., Hendzel M. J., Poirier G. G. (2006) Dynamic relocation of poly(ADP-ribose) glycohydrolase isoforms during radiation-induced DNA damage. Biochim. Biophys. Acta 1763, 226–237 [DOI] [PubMed] [Google Scholar]
- 40. Patel C. N., Koh D. W., Jacobson M. K., Oliveira M. A. (2005) Identification of three critical acidic residues of poly(ADP-ribose) glycohydrolase involved in catalysis. Determining the PARG catalytic domain. Biochem. J. 388, 493–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Affar E. B., Germain M., Winstall E., Vodenicharov M., Shah R. G., Salvesen G. S., Poirier G. G. (2001) Caspase-3-mediated processing of poly(ADP-ribose) glycohydrolase during apoptosis. J. Biol. Chem. 276, 2935–2942 [DOI] [PubMed] [Google Scholar]
- 42. Lin W., Amé J. C., Aboul-Ela N., Jacobson E. L., Jacobson M. K. (1997) Isolation and characterization of the cDNA encoding bovine poly(ADP-ribose) glycohydrolase. J. Biol. Chem. 272, 11895–11901 [DOI] [PubMed] [Google Scholar]
- 43. Botta D., Jacobson M. K. (2010) Identification of a regulatory segment of poly(ADP-ribose) glycohydrolase. Biochemistry 49, 7674–7682 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.