Background: Regulation of PHD expression and their function in hypoxia is unknown in nucleus pulposus.
Results: Expression of PHD1–3 is regulated differentially by HIF-1/2. Under hypoxia, PHD2 degrades HIF-1α, whereas PHD3 promotes its activity.
Conclusion: In hypoxic nucleus pulposus, HIFs and PHDs constitute a regulatory network.
Significance: PHD2 and 3 may serve as oxygen sensors in the intervertebral disc.
Keywords: Cartilage Biology, Chondrocytes, Gene Regulation, Hypoxia, Hypoxia-inducible Factor (HIF), Intervertebral Disc, Nucleus Pulposus, Prolyl Hydroxylase
Abstract
Adaptive response to hypoxia in nucleus pulposus cells of the intervertebral disc is regulated by the hypoxia-inducible factors, HIF-1α and HIF-2α. Moreover, oxygen-dependent turnover of HIF-1α in these cells is controlled by the prolyl-4-hydroxylase domain (PHD) family of proteins. Whether HIF homologues control expression of PHDs and whether PHDs control hypoxia-inducible factor (HIF) turnover and/or activity under hypoxia is not known. Here, we show that in nucleus pulposus cells, hypoxia robustly induces PHD3 expression and, to a lesser extent, of PHD2 and PHD1. Reporter analysis shows that the hypoxic induction of the PHD2 promoter is HIF-1α dependent, whereas PHD3 promoter/enhancer activity is dependent on both HIF-1α and HIF-2α. Lentiviral delivery of HIF-1α, ShHIF-1α, and ShHIF-1β confirmed these observations. Noteworthy, HIF-1α maintains basal expression of PHD1 in hypoxia at the posttranscriptional level. Finally, loss of function studies using lentiviral transduction of ShPHDs clearly shows that even at 1% O2, PHD2 selectively degrades HIF-1α. In contrast, in hypoxia, PHD3 enhances HIF-1α transcriptional activity without affecting protein levels. To correlate these observations with disc disease, a condition characterized by tissue vascularization, we analyzed human tissues. Increased PHD1 mRNA expression but decreased PHD2 and PHD3 expression is observed in degenerate tissues. Interestingly, the hypoxic responsiveness of all the PHDs is maintained in isolated nucleus pulposus cells regardless of the disease state. We propose that PHD2 and PHD3 can be used as a biomarker of tissue oxygenation in the disc and that, as such, it may have important clinical implications.
Introduction
Intervertebral disc disease is a major cause of disability and has been associated with low back and neck pain (1, 2). The disc is a complex tissue that permits a range of motions between adjacent vertebrae and accommodates high biomechanical forces. The annulus fibrosus and the cartilagenous end plates enclose a central aggrecan-rich gel-like tissue, the nucleus pulposus, that is sparsely populated with cells. These cells are removed from the systemic circulation. Thus, blood vessels originating in the vertebral body penetrate the superficial region of the endplates. None of these vessels infiltrate the nucleus pulposus. With respect to the annulus, Gruber et al. (3–5) pointed out that that this tissue is avascular except for small discrete capillary beds in the dorsal and ventral surfaces. In no case does the annulus vasculature enter the nucleus pulposus. Because the vasculature is limited, it is concluded that nucleus pulposus cells reside in a hypoxic environment (6). However, during degeneration, there is vascular ingrowth into the tissue, altering its oxemic status (7).
To understand how pulposus cells respond to and survive in the low-oxygen tension of the disc, we have previously examined the expression of the transcription factors HIF-1 and HIF-2. Τhese molecules are a member of the bHLH-PER-ARNT-SIM (PAS)2 family of proteins and are composed of a constitutively expressed β subunit and regulatory α subunit (8). We have reported previously that HIF-1α and HIF-2α are expressed in nucleus pulposus cells and play an important role in regulating energy metabolism and matrix synthesis (9–12). Recent evidence suggests that HIF-1α and HIF-2α are not redundant and that the relative importance of each of the homologues, in response to hypoxia, varies among different cell types (13). Moreover, the target genes are different. For example, HIF-2 regulates expression of a number of unique genes, including SOD2, catalase, frataxin and cited2, whereas HIF-1 regulates a plethora of genes, including those concerned with energy generation, vascularization, and survival (14–16). It is known that HIF-1α and HIF-2α are regulated by prolyl-4-hydroxylase domain (PHD) proteins, members of the 2-oxoglutarate/Fe2+-dependent dioxygenase superfamily. These proteins hydroxylate specific prolyl residues in the oxygen-dependent degradation domain of HIF-α subunits. The hydroxylated proteins are bound by the ubiquitin ligase von Hippel-Lindau tumor suppressor protein (pVHL), which targets them for rapid ubiquitination and 26 S proteasomal degradation (17).
Because the activity of PHDs depends on the tissue oxygen tension, these molecules serve as oxygen sensors that control the cellular abundance of HIF-α proteins. Importantly, the expression of PHD2 and PHD3 is induced by hypoxia in a few cell types (18, 19), whereas PHD1 expression is shown to be hypoxia-independent (20–22). We reported recently that expression of PHD1–3 is higher in cells of the nucleus pulposus than in cells of the annulus fibrosus and that PHD2 controls degradation of HIF-1α in an oxygen-dependent manner (23). However, the mechanism of their expression and their function in the hypoxic niches of the nucleus pulposus is not understood.
One of the major objectives of this study is to determine whether PHD expression is dependent on oxemic tension and whether regulation is HIF-1α- and/or HIF-2α-dependent in cells of the nucleus pulposus. We show that in nucleus pulposus cells, PHD2 is selectively regulated by HIF-1α, whereas PHD3 is regulated by both the HIF-1α and HIF-2α at the transcript level. Noteworthy, unlike other tissues, hypoxic expression of PHD1 is also dependent on HIF-1α activity. Finally, for the first time, we demonstrate that PHD2 selectively promotes HIF-1α degradation, even under hypoxia. On the other hand, in hypoxia, PHD3 enhances HIF-1 transcriptional activity. Thus, both PHD2 and PHD3 form a regulatory feedback loop with HIF-1 in nucleus pulposus cells, suggesting that they may serve as the oxygen sensors in the nucleus pulposus.
EXPERIMENTAL PROCEDURES
Plasmids and Reagents
The plasmid HA-HIF-1α with a double mutation (P402A/P564A) (catalog no. 18955) and the plasmid siHIF-2α (catalog no. 22131), developed by Dr. William G. Kaelin; human PHD2 reporter (1454/3172) (catalog no. 21404) and HRE-mutated promoter constructs (catalog no. 21405), developed by Dr. Eric Metzen; the plasmid siHIF-1α (catalog no. 21103), developed by Dr. Connie Cepko; psPAX2 (catalog no. 12260) and pMD2G (catalog no. 12259), developed by Didier Trono; and the plasmid 3xHRE-Luc (catalog no. 26731), developed by Dr. Navdeep Chandel, were obtained from the Addgene plasmid repository. The plasmid CA-HIF-2α with a triple mutation (P405A/P530A/N851A) (11) was provided by Dr. Celeste Simon, University of Pennsylvania, Philadelphia, PA. Human PHD3 reporter plasmids were kind gifts from Dr. Luis Del Peso, Universidad Autónoma de Madrid, Spain (19). Lentiviral ShPHD2 and ShPHD3 constructs were from Dr. Kenneth Thirstrup, H. Lundbeck A/S, Denmark (24). The lentiviral CA-HIF-1α vector, GFP control vector, and TAT expression plasmid were provided by Dr. Karen Westerman, Harvard Medical School. The lentiviral ShHIF-1β construct and control vector were from Dr. John Basile, University of Maryland, MD. Lentiviral ShHIF-1α was from Dr. Andree Yeramian, University of Lleida, Spain. As an internal transfection control, vector pRL-TK (Promega) containing the Renilla reniformis luciferase gene was used. HEK293T cells were provided by Dr. Aviva Symes, and TC28 chondrocytes were from Dr. Mary Goldring. The amount of transfected plasmid, the pretransfection period after seeding, and the posttransfection period before harvesting have been optimized for rat nucleus pulposus cells using the pSV β-galactosidase plasmid (Promega) (11).
Isolation of Nucleus Pulposus Cells and Hypoxic Culture
Rat and human nucleus pulposus cells were isolated using a method reported earlier by Risbud et al. (9). Nucleus pulposus, HEK293T, and TC28 cells were maintained in DMEM and 10% FBS supplemented with antibiotics. Cells were cultured in a hypoxia workstation (In vivo2 300, Ruskinn, UK) with a mixture of 1% O2, 5% CO2, and 94% N2 for 8–72 h.
Transfections and Dual Luciferase Assay
Cells were transferred to 48-well plates at a density of 2 × 104 cells/well one day before transfection. Lipofectamine 2000 (Invitrogen) was used as a transfection reagent. For each transfection, plasmids were premixed with the transfection reagent. For measuring the effect of hypoxia on PHD2 or PHD3 reporter activity 24 h after transfection, the cells in some wells were moved to the hypoxia workstation. The next day, the cells were harvested, and a dual luciferase reporter assay system (Promega) was used for sequential measurements of firefly and Renilla luciferase activities. Quantification of luciferase activities and calculation of relative ratios were carried out using a luminometer (TD-20/20, Turner Designs, CA).
Real Time RT-PCR Analysis
Total RNA was extracted from rat nucleus pulposus cells using RNAeasy mini columns (Qiagen). Before elution from the column, RNA was treated with RNase free DNase I (Qiagen). The purified, DNA-free RNA was converted to cDNA using Superscript III reverse transcriptase (Invitrogen). Reactions were set up in triplicate in a 96-well plate using 1 μl of cDNA with SYBR Green PCR Master Mix (Applied Biosystems) to which gene-specific forward and reverse PCR primers were added (see supplemental Table 1, synthesized by Integrated DNA Technologies, Inc.). PCR reactions were performed in a StepOnePlus real-time PCR system (Applied Biosystems) according to the instructions of the manufacturer. β-actin was used to normalize. Melting curves were analyzed to verify the specificity of the RT-PCR reaction and the absence of primer dimer formation. Amplification efficiencies of the PHD primer sets were determined by generating individual standard curves and were found to be similar (94–96% efficient).
Immunofluorescence Microscopy
Cells were plated in flat-bottom 96-well plates (5 × 103/well) for 24 h, fixed with 4% paraformaldehyde, permeabilized with 0.2% Triton X-100 in PBS for 10 min, blocked with PBS containing 5% FBS, and incubated with antibodies against PHD1, PHD2, and PHD3 (1:200) (Novus) at 4 °C overnight. As a negative control, cells were reacted with isotype IgG under similar conditions. After washing, the cells were incubated with Alexa Fluor 488-conjugated appropriate secondary antibody (Invitrogen) at a dilution of 1:50 for 1 h at room temperature. Cells were imaged using a laser scanning confocal microscope (Olympus Fluoview, Japan).
Protein Extraction and Western Blotting
Cells were placed on ice immediately and washed with ice-cold Hanks' balanced salt solution. Nuclear and cytosolic proteins were prepared using the CellLytic NuCLEAR extraction kit (Sigma-Aldrich). All wash buffers and the final resuspension buffer included 1× protease inhibitor mixture (Pierce), NaF (5 mm), and Na3VO4 (200 μm). Proteins were resolved on 8–12% SDS-polyacrylamide gels and transferred by electroblotting to nitrocellulose membranes (Bio-Rad). The membranes were blocked with 5% nonfat dry milk in TBST (50 mm Tris (pH 7.6), 150 mm NaCl, 0.1% Tween 20) and incubated overnight at 4 °C in 3% nonfat dry milk in TBST with the anti-HIF-1α or anti-HIF-2α (1:1000, R&D Systems), HIF-1β (1:1000, BD Biosciences), anti-PHD1, or anti-PHD3 antibody (1:1000, Novus); anti-PHD2 (1:1000, Cell Signaling Technology), anti-β-tubulin (1:3000, Developmental Studies Hybridoma Bank), and anti-Lamin A/C (1:1000, Cell Signaling Technology). Immunolabeling was detected using the ECL reagent (Amersham Biosciences). Relative expression levels were determined by quantitative densitometric analysis using one-dimensional image analysis software (Quantity One, Bio-Rad).
Lentiviral Production and Transduction
HEK 293T cells were seeded in 10-cm plates (60 × 106 cells/plate) in DMEM with 10% heat-inactivated FBS one day before transfection. Cells were transfected with 9 μg of ShHIF-1α, ShPHD2, and ShPHD3 plasmids, along with 6 μg of psPAX2 and 3 μg of pMD2G using Lipofectamine 2000. CA-HIF-1α and shHIF-1β were used following a method reported earlier (25, 26). After 16 h, transfection media were removed and replaced with DMEM with 5% heat-inactivated FBS and penicillin-streptomycin. Lentiviral particles were harvested at 48 and 60 h post-transfection. Nucleus pulposus cells were plated in DMEM with 5% heat-inactivated FBS one day before transduction. Cells in 10-cm plates were transduced with 5 ml of conditioned media containing viral particles along with 6 μg/ml polybrene. After 24 h, conditioned media was removed and replaced with DMEM with 5% heat-inactivated FBS. Cells were harvested for protein extraction 5 days after viral transduction. A transduction efficiency of 80–90% was achieved as determined from the number of GFP/YFP-positive cells.
Human Tissue Collection and Grading
Lumbar disc tissues were collected as surgical waste from individuals undergoing elective spinal surgical procedures. In line with the Institutional Review Board guidelines of Thomas Jefferson University, informed consent for sample collection was obtained from each patient. Assessment of the disease state was performed using Pfirrmann grading (27).
Statistical Analysis
All measurements were performed in triplicate. Data is presented as mean ± S.E. Differences between groups were analyzed by Student's t test and one way analysis of variance (*, p < 0.05).
RESULTS
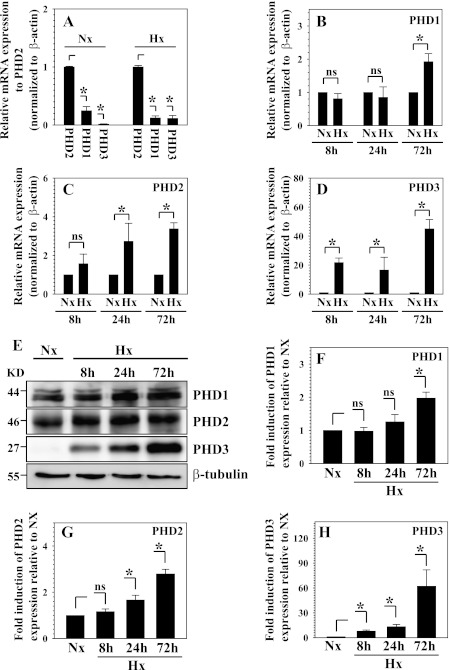
Real-time RT-PCR analysis shows that PHD2 is the most abundant isoform in nucleus pulposus cells (Fig. 1A). To evaluate whether the expression of PHD1–3 is regulated by hypoxia, we measured their expression in the nucleus pulposus cells under normoxic and hypoxic condition with real-time RT-PCR and Western blot analysis. Although mRNA expression of PHD1 showed an increase only at 72 h of hypoxia, PHD3 and, to a lesser extent, PHD2 are significantly up-regulated by 24 h (Fig. 1, B–D). Remarkably, the induction of PHD3 mRNA expression is about 50-fold at 72 h in hypoxia. Western blot analysis and subsequent densitometric analysis confirmed that protein expression is influenced by hypoxia. Thus, the protein level of PHD3 exhibits the most striking hypoxic induction as early as 8 h (Fig. 1, E and H). Although the PHD2 protein level is induced significantly at 24 h (Fig. 1, E–H), an increase in PHD1 is not evident until 72 h (Fig. 1, E and F).
FIGURE 1.
Hypoxic regulation of PHD expression in nucleus pulposus (NP) cells. A, real-time RT-PCR analysis of PHD mRNA expression in NP cells after 72 h of normoxic and hypoxic conditions. Expression of PHD1 and PHD3 is shown relative to PHD2 expression in normoxia or hypoxia, respectively. Note that PHD2 showed the highest relative expression under both normoxia and hypoxia. B, C, and D, real-time RT-PCR analysis of hypoxic change of PHD mRNA expression in NP cells. B, PHD1 mRNA expression was significantly up-regulated at 72 h under hypoxia. ns, not significant. C, the PHD2 expression level was induced significantly by 24 h in hypoxia. D, PHD3 was induced robustly by hypoxia at all time points. E, Western blot analysis of PHDs expression in NP cells cultured under hypoxia. The protein level of all PHDs is induced by hypoxia. PHD3 shows a robust response to hypoxia, whereas the PHD1 response was not as pronounced. KD, kilodalton. F, G, and H, multiple blots were quantified by densitometric analysis. β-tubulin expression was used as a loading control and to calculate relative expression level in hypoxia compared with normoxic level. PHD3 showed the most striking hypoxic induction as early as 8 h. The PHD2 protein level was also significantly increased at 24 h. PHD1 showed induction only at 72 h. Data are represented as mean ± S.E. of three independent experiments performed in triplicate (n = 3). *, p < 0.05.
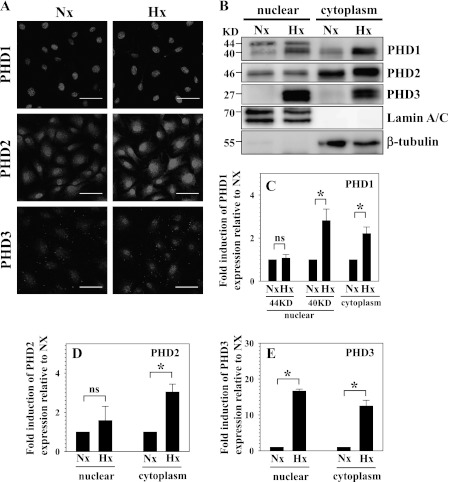
We next studied the cellular localization of PHDs in nucleus pulposus cells under normoxia and hypoxia. Immunofluorescence studies indicate that PHD1 is mainly localized to the nucleus, whereas, depending on the pO2, PHD2 and PHD3 are present in both the nucleus and cytoplasm (Fig. 2A). Next, using Western blot analysis, we evaluated the expression of PHDs in cytoplasmic and nuclear fractions of nucleus pulposus cells following 72 h of culture under normoxia or hypoxia. Fig. 2B shows that, in agreement with a previous report, two splice variants of PHD1 are present in the nuclear fraction, whereas only the low molecular weight (40-kDa) variant is detected in the cytoplasm (27). Noteworthy, only the 40-kDa band evidenced hypoxic induction (Fig. 2, B and C). Fig. 2, B and D, shows that PHD2 is present in the nucleus and cytoplasm. In hypoxia, there is an increased level of PHD2 in the cytoplasmic fraction. In contrast, PHD3 levels are higher in the nucleus than the cytoplasm. The level in both compartments is robustly induced by hypoxia (Fig. 2, B and E).
FIGURE 2.
Cellular localization of PHDs in NP cells under normoxia and hypoxia. A, immunofluorescence analysis of cellular localization of PHDs in NP cells. PHD1 is localized in the nucleus, whereas PHD2 and PHD3 are present in both the nucleus and cytoplasm. Magnification ×20. B, Western blot analysis of cellular localization of PHDs in NP cells. Two splice variants of PHD1 (44 and 40 kDa) are present in the nuclear fraction, whereas only the 40-kDa variant is detected in the cytoplasm (27). The 40-kDa band shows hypoxic induction. PHD2 is localized in the nucleus and cytoplasm, and its expression is increased by hypoxia in cytoplasmic fraction. The PHD3 level is higher in the nucleus than cytoplasm. Expression in both compartments is robustly induced by hypoxia. KD, kilodalton. C, D, and E, multiple blots were quantified by densitometric analysis. Expression of β-tubulin for cytoplasmic and Lamin A/C for nuclear protein was used as a loading control and to calculate relative expression levels. C and D, the expression of PHD1 (40 kDa) was induced in both the nuclear and cytoplasmic fraction by hypoxia. PHD2 only in the cytoplasmic fraction was significantly induced by hypoxia. E, PHD3 expression was robustly increased in both nucleus and cytoplasm under hypoxia. Data is represented as mean ± S.E. of three independent experiments performed in triplicate (n = 3). *, p < 0.05. ns, not significant.
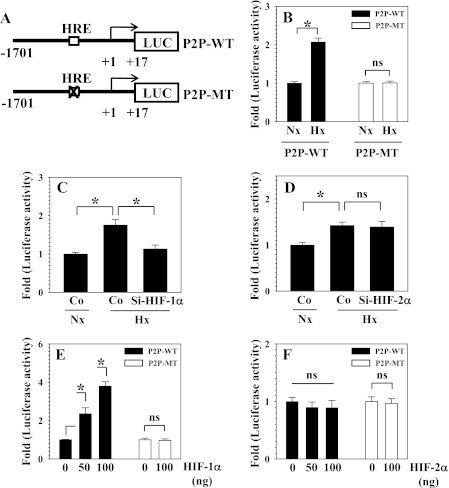
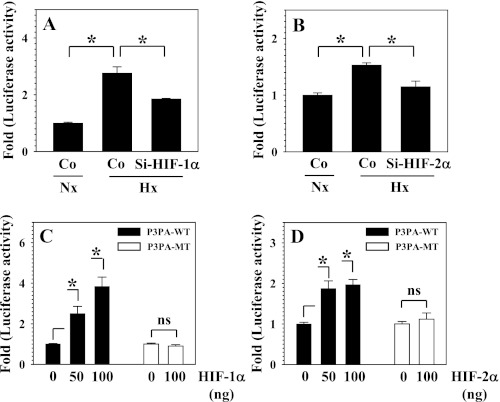
We further evaluated the relationship between PHD2 transcription and oxygen tension by measuring the activity of the PHD2 promoter under normoxic and hypoxic conditions. Also, to evaluate the role of HIF in oxemic regulation of promoter activity, a mutation in the hypoxia responsive element (HRE) is introduced (Fig. 3A). Fig. 3B shows that in nucleus pulposus cells, wild-type promoter activity is significantly induced under hypoxic conditions, whereas the HRE mutation completely abolishes this induction. To evaluate the role of HIF-α homologues in transcriptional regulation, loss and gain of function studies were performed. When HIF-1α expression was suppressed using siRNA, the hypoxic induction of PHD2 promoter activity was abolished completely (Fig. 3C). On the other hand, HIF-2α siRNA has no effect on hypoxic induction of PHD2 promoter activity (Fig. 3D). Similarly, when HIF-1α was cotransfected, wild-type promoter activity is significantly induced, whereas the mutant promoter is unaffected (Fig. 3E). In contrast, overexpression of HIF-2α does not affect either wild-type or mutant promoter activities (Fig. 3F).
FIGURE 3.
The role of hypoxia and HIF-1α and HIF-2α in controlling PHD2 promoter activity in NP cells. A, schematic of PHD2 reporters used for transfections (P2P-WT and P2P-MT). HRE site “CGTGCA” is muted to “ATAATA” in P2P-MT. B, although P2P-WT shows hypoxic induction in activity, the HRE mutation abolishes hypoxic induction in activity (P2P-MT). C and D, hypoxic induction of PHD2 promoter activity was suppressed by siRNA silencing of HIF-1α (C) but not HIF-2α (D). Co, Si-control. E, overexpression of HIF-1α induced PHD2 promoter activity (P2P-WT) in a dose-dependent fashion, P2P-MT activity was not affected. F, overexpression of HIF-2α did not increase the activity both P2P-WT and P2P-MT. Data represent mean ± S.E. of three independent experiments performed in triplicate (n = 3). *, p < 0.05. ns, not significant.
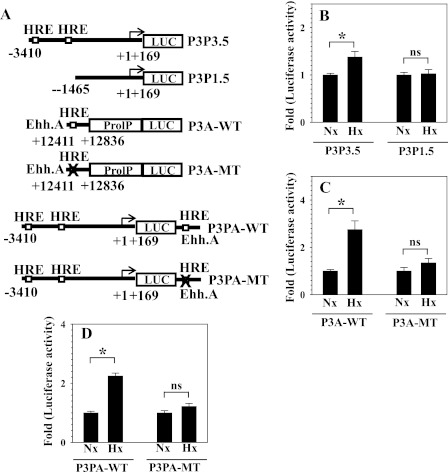
To study regulation of PHD3 transcription by hypoxia in nucleus pulposus cells, we measured the activity of the PHD3 promoter and/or enhancer. We used 3.5-kb (P3P3.5) and a 1.5-kb (P3P1.5) promoter fragments of PHD3 that contained either two or no HRE sites and an enhancer region of PHD3 (P3A-WT) 12 kb upstream of the transcription site containing a single HRE (19). A reporter containing both the promoter and enhancer as well as a reporter with mutant HRE enhancer (P3A-MT, P3PA-MT) were also used in the analysis (Fig. 4A). Fig. 4B shows that the promoter activity of P3P3.5 with two putative HREs is induced by hypoxia. There is no change in activation of the P3P1.5 with no putative HRE. Fig. 4C shows that the activity of PHD3 enhancer (P3A-WT) is significantly up-regulated and that this induction is abolished by mutation of single HRE. Furthermore, when we measured the activity of PHD3 promoter in combination with a wild-type or a mutant enhancer (P3PA-WT or P3PA-MT), we found that P3PA-WT activity is significantly induced by hypoxia. In contrast, P3PA-MT is not affected by the low pO2 (Fig. 4D).
FIGURE 4.
The effect of hypoxia on the promoter and/or enhancer activity of PHD3 in NP cells. A, schematic of PHD3 reporters used for transfections. B, hypoxia significantly induced the activity of the 3.5-kb promoter with two putative HRE sites (P3P3.5), whereas the activity of the 1.5-kb promoter lacking HRE sites was not affected by hypoxia (P3P1.5). C, the enhancer activity was significantly increased by hypoxia (P3A-WT), whereas the mutation of HRE abolished this induction (P3A-MT). D, the activity of the 3.5-kb promoter with wild-type enhancer A was significantly increased by hypoxia (P3PA-WT), whereas the induction was suppressed by mutagenesis of HRE in enhancer A (P3PA-MT). Data are represented as mean ± S.E. of three independent experiments performed in triplicate (n = 3). *, p < 0.05. ns, not significant.
Next, we evaluated the role of HIF-1α and HIF-2α in the transcriptional regulation of PHD3 expression by performing loss and gain-of-function analysis with the wild-type and mutant PHD3 promoter/enhancer. Fig. 5, A and B, shows that the hypoxic induction of P3PA-WT reporter activity is significantly suppressed by HIF-1α and HIF-2α siRNA in nucleus pulposus cells. On the other hand, the activity of P3PA-WT was significantly induced by cotransfection of both HIF-1α (Fig. 5C) and HIF-2α (D) in a dose-dependent manner. Again, P3PA-MT is unaffected by both the HIF-α homologues (Fig. 5, C and D). To determine whether PHD2 and PHD3 regulation by HIF is cell-type specific, we measured the reporter activities in 293T cells and T/C28 chondrocytes. Supplemental Fig. S1, A and B shows that, similar to nucleus pulposus cells, in both cell types, PHD2 reporter activity is induced by cotransfection of HIF-1α but not HIF-2α, whereas PHD3 reporter activity is up-regulated by both the HIF-α homologues. Moreover, silencing of HIF-2α resulted in suppression of PHD3 but not PHD2 promoter activity in 293T cells under hypoxia (Supplemental Fig. S1, C and D).
FIGURE 5.
The role of HIF-1α and HIF-2α in regulating PHD3 promoter activity in NP cells. A and B, hypoxic induction of PHD3 promoter/enhancer activity (P3PA-WT) was suppressed by SiRNA-mediated silencing both HIF-1α (A) and HIF-2α (B). Co, Si-control. C, cotransfection of HIF-1α significantly induced the activity of PHD3 reporter (P3PA-WT) in a dose-dependent fashion. The activity of the HRE mutant PHD3 reporter (P3PA-MT) was unaffected. D, overexpression of HIF-2α significantly increased the activity of PHD3 reporter (P3PA-WT) in a dose-dependent manner, the activity of the HRE mutant PHD3 reporter (P3PA-MT) did not change. Data represent mean ± S.E. of three independent experiments performed in triplicate (n = 3). *, p < 0.05. ns, not significant.
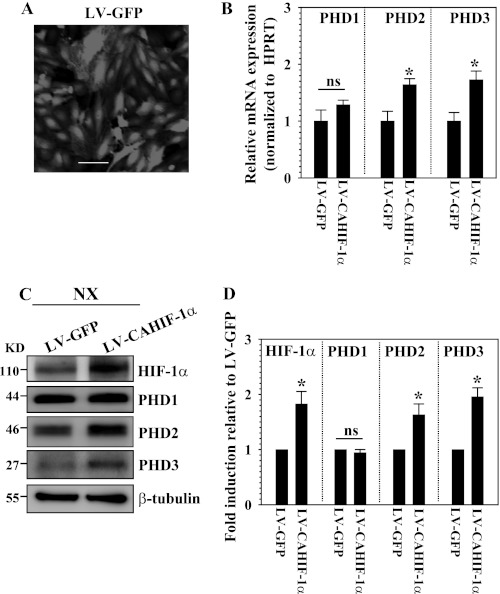
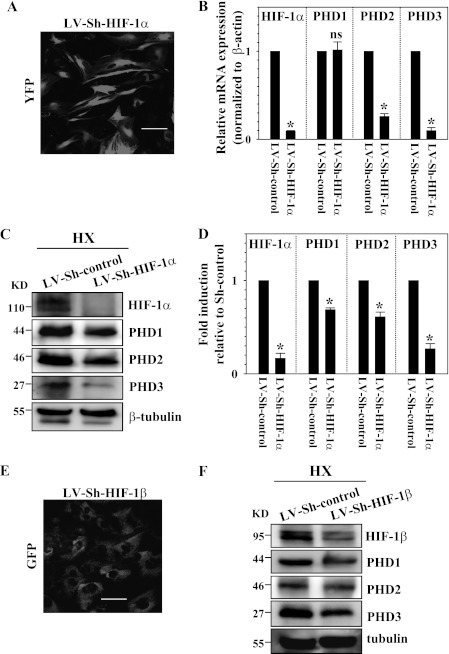
To measure the effect of stable overexpression of HIF-1α on PHD protein levels, we transduced nucleus pulposus cells with lentivirus expressing CA-HIF-1α. By assessment of GFP-positive cells, we confirmed that the transduction efficiency is more than 80% (Fig. 6A). Real-time RT-PCR analysis shows that mRNA expression of PHD2 and PHD3 but not PHD1 is significantly induced in the cells transduced by LV-CAHIF-1α compared with LV-GFP (Fig. 6B). As expected, Western blot analysis confirms that there is an increased level of HIF-1α protein expression in the transduced cells (Fig. 6C). Moreover, we found that although the HIF-1α-transduced cells show increased PHD2 and PHD3 expression, no change in PHD1 expression in normoxia is seen. Densitometry analysis confirmed that the increase in PHD2 and PHD3 level but not PHD1 is correlated with the accumulation of HIF-1α (Fig. 6D). We next stably suppressed the activity of HIF-1α or HIF-1β by transducing nucleus pulposus cells with a lentivirus expressing HIF-1α or HIF-1β shRNA. Because functional HIF-1 and HIF-2 proteins require heterodimerization of the HIF-1α/2α and HIF-1β subunits, lack of a β inhibits both HIF-1 and HIF-2 transcriptional activity. Fig. 7A shows that we achieved high transduction efficiency of lentiviral sh-HIF-1α in nucleus pulposus cells. Real-time RT-PCR analysis confirmed that mRNA expression of HIF-1α is robustly suppressed in the cells transduced by Sh-HIF-1α compared with Sh-control (Fig. 7B). HIF-1α-suppressed cells evidence a significant decrease in PHD2 and PHD3 mRNA expression but no change in PHD1 expression (Fig. 7B). Interestingly, Western blot analysis shows that hypoxic suppression of HIF-1α results in decreased PHD isoform protein levels, including PHD1, in nucleus pulposus cells (Fig. 7, C and D). In the same study, we also silenced HIF-1β with high transduction efficiency (Fig. 7E). Western blot analysis confirms that hypoxic suppression of HIF-1β results in decreased levels of PHD1, PHD2, and PHD3 (Fig. 7F).
FIGURE 6.
Role of HIF-1α in controlling PHD expression. A, immunofluorescence analysis of GFP in NP cells transduced with control lentivirus expressing GFP (LV-GFP) shows high transduction efficiency. Magnification ×20. B, real-time RT-PCR analysis of PHD expression in NP cells transduced with LV-GFP or LV-CA-HIF-1α. Note that the significant induction of PHD2 and PHD3 was seen in the cells transduced by LV-CAHIF-1α, whereas PHD1 expression was not affected. HPRT, hypoxanthine phosphoribosyl transferase. C, Western blot analysis of cells transduced with LV-GFP or LV-CAHIF-1α. HIF-1α was accumulated in LV-CAHIF-1α-transduced cells compared with controls (LV-GFP). Note that the induction of PHD2 and PHD3 was seen in the cells transduced by LV-CAHIF-1α, whereas PHD1 expression was not affected. D, densitometric analysis of multiple blots from the experiment described in C above. As expected, relative HIF-1α level in LV-CAHIF-1α group was higher compared with LV-GFP. Expression of PHD2 and PHD3 was significantly increased by LV-CAHIF-1α. PHD1 expression remained unchanged. Data are represented as mean ± S.E. of three independent experiments performed in triplicate (n = 3). *, p < 0.05. ns, not significant.
FIGURE 7.
Role of HIF-1α in controlling PHD expression. A, immunofluorescence analysis of NP cells transduced with lentivirus coexpressing Sh-HIF-1α (LV-Sh-HIF-1α) with YFP shows high transduction efficiency. Magnification ×20. B, real-time RT-PCR analysis of HIF-1α and PHD expression in nucleus pulposus cells transduced with LV-Sh-control or LV-Sh-HIF-1α under hypoxia. Note that the significant reduction of HIF-1α was seen in the cells transduced by LV-Sh-HIF-1α. HIF-1α-suppressed cells evidenced significantly decreased mRNA expression of PHD2 and PHD3 but not of PHD1. C, Western blot analysis of cells transduced with LV-Sh-control or LV-Sh-HIF-1α under hypoxia. The HIF-1α level was suppressed by LV-Sh-HIF-1α compared with cells transduced with control lentivirus (LV-Sh-control). Note that the reduction of all PHDs was seen in the cells transduced by LV-Sh-HIF-1α. KD, kilodalton. D, densitometric analysis of multiple blots from the experiment described in C above. Protein expression of all PHDs is decreased significantly, correlating with suppression of HIF-1α. E, immunofluorescence analysis of NP cells transduced with lentivirus coexpressing GFP with Sh-HIF-1β shows high transduction efficiency. Magnification ×20. F, Western blot analysis of cells transduced with LV-Sh-control or LV-Sh-HIF-1β. HIF-1β was suppressed by LV-Sh-HIF-1β compared with cells transduced with control lentivirus (LV-Sh-control). Note that the reduction of all PHDs was seen in the cells transduced by LV-Sh-HIF-1β. Data are represented as mean ± S.E. of three independent experiments performed in triplicate (n = 3). *, p < 0.05. ns, not significant.
To confirm the role of PHDs in HIF-1α and HIF-2α turnover and activity in nucleus pulposus cells, we transduced cells with lentivirus expressing ShPHD2 and ShPHD3 under both normoxia and hypoxia. Assessment of GFP-positive cells confirmed that there is high transduction efficiency (Fig. 8A). Real-time RT-PCR analysis indicates that shRNA selectively suppresses the expression of either PHD2 or PHD3 without affecting the expression of the other PHD isoforms (Fig. 8B). Western blot analysis confirmed that shRNAs suppress PHD2 and PHD3 protein expression (Fig. 8, C and D). Noteworthy, silencing of PHD2 but not PHD3 results in increased accumulation of HIF-1α even under hypoxic conditions, whereas HIF-2α is unaffected by either PHD2 or PHD3 suppression, regardless of the oxemic state (Fig. 8, C and D). Densitometric analysis of multiple blots confirms that accumulation of HIF-1α is significantly correlated with silencing of only PHD2 in hypoxia as well as normoxia, whereas the accumulation HIF-2α is unaffected by silencing of both PHD2 and PHD3 (Fig. 8E). To assess the role of PHD3 in controlling HIF-1α transcriptional activity, we measured the activity of a well characterized HIF-1 responsive HRE-luc reporter in PHD silenced cells. Fig. 8F shows that in hypoxia, silencing of PHD3 results in a significant decrease in HRE-luc reporter activity, whereas in normoxia, silencing has little effect on reporter activity. As expected, PHD2 silencing increased HRE-luc activity (not shown).
FIGURE 8.
Effect of PHD2 and PHD3 silencing on the stability of HIF-1α and HIF-2α in NP cells under normoxia and hypoxia. A, immunofluorescence analysis of NP cells transduced with lentivirus coexpressing GFP and ShRNA of either PHD2 (LV-Sh-PHD2) or PHD3 (LV-Sh-PHD3) shows good transduction efficiency. Magnification ×20. B, real-time RT-PCR analysis of cells transduced with LV-Sh-PHD2/3. The mRNA expression of PHD2 or PHD3 was suppressed robustly by respective ShRNAs compared with cells transduced with control lentivirus (LV-Sh-control) under both normoxic and hypoxic conditions. C and D, Western blot analysis of cells transduced with LV-Sh-PHD2/3. The expression of PHD2 or PHD3 was suppressed by respective ShRNAs compared with cells transduced with control lentivirus (LV-Sh-control). Note that accumulation of HIF-1α was seen in PHD2- but not PHD3-silenced cells under both normoxia (C) and hypoxia (D). KD, kilodalton. Accumulation of HIF-2α was not affected by silencing of either PHD2 or PHD3. E, densitometric analysis of multiple blots from experiment described in C and D above. Note that Relative HIF-1α level compared with LV-Sh-control was significantly increased with LV-Sh-PHD2 but not LV-Sh-PHD3 irrespective oxemic tension. Relative HIF-2α level was not affected by either LV-Sh-PHD2 and LV-Sh-PHD3. F, role of PHD3 on maintenance of HIF-1 transcriptional activity. NP cells were transfected with the HRE-luc reporter along with the Sh-PHD3 construct, and activity was measured under normoxic or hypoxic conditions. Note that hypoxic induction in HRE activity is significantly blocked by silencing of PHD3 expression. Data are represented as mean ± S.E. of three independent experiments performed in triplicate (n = 3). *, p < 0.05. ns, not significant.
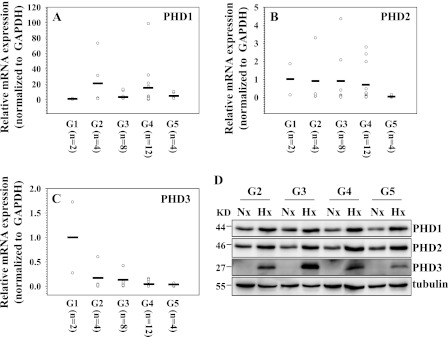
We next evaluated the expression of PHDs in human disc tissues with different stages of degeneration. Assessment of the disease state was performed using Pfirrmann grading (28). Grade 5 (G5) is the most severe stage. Real-time RT-PCR analysis shows that there is a trend toward increased mRNA expression of PHD1 in degenerate tissues when compared with normal tissues (G1), whereas mRNA expression of PHD2 and PHD3 exhibits the opposite tendency (Fig. 9, B and C). As expected with human discal tissues, there was sample-to-sample variability within the same degenerative grade (29). To determine whether the hypoxic responsiveness of PHDs is altered in human cells during degeneration, we cultured nucleus pulposus cells from tissues of different grades of degeneration and evaluated the PHD expression following 72 h in hypoxia. Western blot analysis shows that hypoxic responsiveness of PHD1, PHD2, and PHD3 is maintained regardless of the severity of disc degeneration (Fig. 9D).
FIGURE 9.
Expression of PHDs in degenerate human NP tissues and hypoxic response of PHDs in NP cells isolated from these tissues. G1-G5 represent grades of increasing severity of disc degeneration. (A, B, and C). Real-time RT-PCR analysis of PHD1 (A), PHD2 (B), and PHD3 (C) expression. Note that PHD1 mRNA levels were higher in degenerate tissues, whereas PHD2 and PHD3 levels decreased. Expression is shown relative to the mean expression of G1 samples. The horizontal line represents the mean expression of all the samples in the same grade. KD, kilodalton. D, Western blot analysis of PHD expression in NP cells isolated from human tissues with different grades of degeneration. Note that the expression of PHDs increased under hypoxia in all the samples.
DISCUSSION
The mechanism by which cells adapt to the local oxemic environment is incompletely understood. In the case of the intervertebral disc, the nucleus pulposus cells maintain their biosynthetic activities by constitutive expression of HIF-1α and HIF-2α (10–12). It is understood that HIF activity is dependent on the activity of the PHD1–3 which, in most other tissues, promotes HIF-α turnover and degradation. Interestingly, we noted that degradation of HIF-2α was not mediated by the classical O2-dependent PHD pathway. It was also evident that, although PHD2 can mediate limited HIF-1α turnover in nucleus pulposus cells, it is primarily controlled by O2 independent proteasomal and lysosomal pathways (23). Thus, the absence of clear-cut regulatory function calls into question the role of the PHDs as HIF regulatory proteins. Here we show for the first time that PHD2 is selectively regulated by HIF-1α, whereas PHD3 expression is regulated by both HIF-1 and HIF-2 at the transcript level. Interestingly, unlike other cells, basal PHD1 expression was HIF-1 sensitive at the posttranscriptional level. On the basis of these findings, we put forward the hypothesis that PHDs and HIF form a self-regulating loop so that a fall in pO2 would be expected to increase HIF-1α activity which, in turn, would increase PHD2 expression. When there is a shift in O2 levels, PHD2 can hydroxylate HIF-1α and promote partial degradation of this transcription factor, thereby down-regulating its own expression. These results also suggest that activities of PHD2 may serve functions other than that of regulating HIF transcription.
We noted that hypoxia robustly promotes induction of PHD3 expression and, to a lesser extent, the expression of PHD2 and PHD1. Moreover, the effect of hypoxia influences cellular localization of members of the PHD family. For example, PHD1 appears to be evenly divided between the nucleus and cytoplasm in normoxia and hypoxia. If the sole function of this protein was to hydroxylate HIF, a change in distribution and expression would have been expected. In contrast, hypoxia causes a redistribution of PHD2 from the nucleus and the cytoplasmic compartments of the nucleus pulposus cells. In this case, hypoxia promoted a significant increase in this PHD isoform in the cytoplasmic compartment of the cell. The most dramatic change was evidenced by PHD3, which was exquisitely sensitive to hypoxia. However, in hypoxia, both nuclear and cytoplasmic expression was robustly elevated. The variable nature of PHDs was further highlighted by studies that examined the role of these isoforms on the regulation of HIF-α. We found that PHD3 does not regulate HIF-α degradation, whereas PHD2 can degrade only HIF-1α. This raises the question concerning their physiological function in the nucleus pulposus. The literature suggests that PHD1 may serve to regulate cyclinD1 levels and play a role in proliferation (30, 31). Other studies show that it stabilizes ATF4 and suppresses its transcriptional activity independent of hydroxylation (32). Interestingly, PHD3 promotes ATF4 degradation and also regulates PAX2 and IKK-β stability, an important mediator of NF-κB signaling (33–35). Two recent observations are relevant to the intervertebral niche. Anderson et al. (36) showed that PHDs mediate hydroxylation and degradation of Spry2, a modulator of receptor tyrosine kinases that, in this case, may control growth factor signaling. In another study, Walmsley et al. (37) noted that PHD3 regulated neutrophil survival by modulating antiapoptotic molecules in hypoxia. It is, therefore, not unreasonable to assume that in the niche of the nucleus pulposus in addition to modulating HIF-1α activity, PHDs may contribute to several other physiological functions both dependent and independent of their hydroxylation activity. Experiments are in progress to test this premise.
Both PHD2 and PHD3 were sensitive to hypoxia, with PHD3 exhibiting the highest level of induction. To elucidate the mechanism of expression, we analyzed the regulation of PHD2 and PHD3 promoter/enhancer activities in hypoxia. Our findings clearly show that in nucleus pulposus cells, PHD2 and PHD3 are regulated positively by hypoxia at the transcriptional level. Unlike HeLa cells, where the activity of the 3.5-kb PHD3 promoter is shown to be hypoxia-insensitive, in nucleus pulposus cells there is a small but significant increase in promoter activity, with enhancer displaying the higher sensitivity, suggesting that hypoxic regulation of the PHD3 promoter/enhancer is cell type-specific (19). Noteworthy, in nucleus pulposus cells, PHD2 was responsive only to HIF-1α, whereas PHD3 was responsive to both HIF-1α and HIF-2α. These results may explain the differences in hypoxic responsiveness between PHD2 and PHD3.
Differences in hypoxic sensitivity between PHD1 and other PHD isoforms in nucleus pulposus were evident from the expression data. PHD1 expression evidenced hypoxic induction only after a prolonged culture period. This finding is unique as PHD1 is not reported to be a hypoxia sensitive gene in other cell types and regulation of its expression remains unclear (20–22). Our gain of function studies clearly indicated that although HIF-1α controlled the hypoxic induction of both PHD2 and PHD3 at the transcript level, it failed to increase PHD1 expression at both the mRNA and protein level. This result suggests that hypoxic induction of PHD1 may involve factors other than HIF-1α. Moreover, a lack of change in mRNA expression following HIF-1α silencing, a decrease in PHD1 protein level in both HIF-1α- and HIF-1β-silenced cells indicates that the regulation is at the posttranscriptional level. Importantly, these functional studies suggest that HIF-1α is concerned with maintenance of basal PHD1 protein levels.
The observation that in nucleus pulposus cells, even under hypoxic conditions (1% O2), PHD2 selectively mediates HIF-1α turnover clearly indicates preservation of PHD2 enzymatic activity. This finding is in marked contrast with the previous reports demonstrating HIF-1α stabilization at low oxygen tensions because of the inhibition of PHD enzymatic activity (38). Moreover, this observation highlights the unique physiology of the nucleus pulposus cells and suggests very low cellular utilization of O2, an adaptive response to their hypoxic niche (39). Noteworthy, the finding that in hypoxia, PHD3 promotes HIF-1α transcriptional activity is in line with a recent report that describes a crucial role of the PKM2·PHD3 complex in enhancing HIF-1α interaction with p300, an important transcriptional coactivator (40). Taken together, these observations clearly indicate the existence of a regulatory feedback loop between PHD2, PHD3, and HIF-1α in the hypoxic nucleus pulposus cells. This scenario is distinct from the articular cartilage, a tissue functionally similar to the nucleus pulposus, where PHD2 also regulates HIF-2α degradation (41). These results highlight the unique nature and control of the HIF-PHD system in nucleus pulposus cells. From this perspective, in nucleus pulposus cells, the regulatory system is tissue-specific and more complex than has hitherto been considered. Moreover, analysis of human tissues and cultured cells showed that hypoxic response of all the PHD isoforms is maintained regardless of the disease state. On the basis of all these findings and cognizant of the fact that there is an increase in tissue vascularization and oxygenation during disc degeneration, we forward the notion that PHD2 and PHD3 can be used as biomarkers for tissue oxygenation in the degenerate disc. We are currently examining the clinical implications of these findings.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants R01-AR050087 and R01-AR055655.

This article contains supplemental Fig. S1.
- PAS
- PER-ARNT-SIM
- PHD
- prolyl-4-hydroxylase domain
- HRE
- hypoxia responsive element
- NP
- nucleus pulposus
- Luc
- luciferase
- Nx
- normoxia
- Hx
- hypoxia.
REFERENCES
- 1. Livshits G., Popham M., Malkin I., Sambrook P. N., Macgregor A. J., Spctor T., Williams F. M. (2011) Lumbar disc degeneration and genetic factors are the main risk factors for low back pain in women. The UK Twin Spine Study. Ann. Rheum. Dis. 70, 1740–1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Arana E., Marti-Bonmati L., Molla E., Costa S. (2004) Upper thoracic-spine disc degeneration in patients with cervical pain. Skeletal. Radiol. 33, 29–33 [DOI] [PubMed] [Google Scholar]
- 3. Gruber H. E., Ashraf N., Kilburn J., Williams C., Norton H. J., Gordon B. E., Hanley E. N. (2005) Vertebral endplate architecture and vascularization. Application of micro-computerized tomography, a vascular tracer, and immunocytochemistry in analyses of disc degeneration in the aging sand rat. Spine 30, 2593–2600 [DOI] [PubMed] [Google Scholar]
- 4. Hassler O. (1969) The human intervertebral disc. A micro-angiographical study on its vascular supply at various ages. Acta Orthop. Scand. 40, 765–772 [DOI] [PubMed] [Google Scholar]
- 5. Rudert M., Tillmann B. (1993) Lymph and blood supply of the human intervertebral disc. Cadaver study of correlations to discitis. Acta Orthop. Scand. 64, 37–40 [DOI] [PubMed] [Google Scholar]
- 6. Bartels E. M., Fairbank J. C., Winlove C. P., Urban J. P. (1998) Oxygen and lactate concentrations measured in vivo in the intervertebral discs of patients with scoliosis and back pain. Spine 23, 1–7 [DOI] [PubMed] [Google Scholar]
- 7. Kauppila L. I. (1995) Ingrowth of blood vessels in disc degeneration. Angiographic and histological studies of cadaveric spines. J. Bone Jt. Surg. Am. 77, 26–31 [DOI] [PubMed] [Google Scholar]
- 8. Semenza G. L., Roth P. H., Fang H. M., Wang G. L. (1994) Transcriptional regulation of genes encoding glycolytic enzymes by hypoxia-inducible factor 1. J. Biol. Chem. 269, 23757–23763 [PubMed] [Google Scholar]
- 9. Rajpurohit R., Risbud M. V., Ducheyne P., Vresilovic E. J., Shapiro I. M. (2002) Phenotypic characteristics of the nucleus pulposus. Expression of hypoxia inducing factor-1, glucose transporter-1 and MMP-2. Cell Tissue Res. 308, 401–407 [DOI] [PubMed] [Google Scholar]
- 10. Risbud M. V., Guttapalli A., Stokes D. G., Hawkins D., Danielson K. G., Schaer T. P., Albert T. J., Shapiro I. M. (2006) Nucleus pulposus cells express HIF-1 α under normoxic culture conditions. A metabolic adaptation to the intervertebral disc microenvironment. J. Cell Biochem. 98, 152–159 [DOI] [PubMed] [Google Scholar]
- 11. Agrawal A., Guttapalli A., Narayan S., Albert T. J., Shapiro I. M., Risbud M. V. (2007) Normoxic stabilization of HIF-1α drives glycolytic metabolism and regulates aggrecan gene expression in nucleus pulposus cells of the rat intervertebral disk. Am. J. Physiol. Cell Physiol. 293, C621–631 [DOI] [PubMed] [Google Scholar]
- 12. Agrawal A., Gajghate S., Smith H., Anderson D. G., Albert T. J., Shapiro I. M., Risbud M. V. (2008) Cited2 modulates hypoxia-inducible factor-dependent expression of vascular endothelial growth factor in nucleus pulposus cells of the rat intervertebral disc. Arthritis Rheum. 58, 3798–3808 [DOI] [PubMed] [Google Scholar]
- 13. Sowter H. M., Raval R. R., Moore J. W., Ratcliffe P. J., Harris A. L. (2003) Predominant role of hypoxia-inducible transcription factor (HIF)-1α versus Hif-2α in regulation of the transcriptional response to hypoxia. Cancer Res. 63, 6130–6134 [PubMed] [Google Scholar]
- 14. Scortegagna M., Ding K., Oktay Y., Gaur A., Thurmond F., Yan L. J., Marck B. T., Matsumoto A. M., Shelton J. M., Richardson J. A., Bennett M. J., Garcia J. A. (2003) Multiple organ pathology, metabolic abnormalities and impaired homeostasis of reactive oxygen species in Epas1−/− mice. Nat. Genet. 35, 331–340 [DOI] [PubMed] [Google Scholar]
- 15. Oktay Y., Dioum E., Matsuzaki S., Ding K., Yan L. J., Haller R. G., Szweda L. I., Garcia J. A. (2007) Hypoxia-inducible factor 2α regulates expression of the mitochondrial aconitase chaperone protein frataxin. J. Biol. Chem. 282, 11750–11756 [DOI] [PubMed] [Google Scholar]
- 16. Aprelikova O., Wood M., Tackett S., Chandramouli G. V., Barrett J. C. (2006) Role of ETS transcription factors in the hypoxia-inducible factor-2 target gene selection. Cancer Res. 66, 5641–5647 [DOI] [PubMed] [Google Scholar]
- 17. Maxwell P. H., Wiesener M. S., Chang G. W., Clifford S. C., Vaux E. C., Cockman M. E., Wykoff C. C., Pugh C. W., Maher E. R., Ratcliffe P. J. (1999) The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature 399, 271–275 [DOI] [PubMed] [Google Scholar]
- 18. Metzen E., Stiehl D. P., Doege K., Marxsen J. H., Hellwig-Bürgel T., Jelkmann W. (2005) Regulation of the prolyl hydroxylase domain protein 2 (phd2/egln-1) gene. Identification of a functional hypoxia-responsive element. Biochem. J. 387, 711–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pescador N., Cuevas Y., Naranjo S., Alcaide M., Villar D., Landázuri M. O., Del Peso L. (2005) Identification of a functional hypoxia-responsive element that regulates the expression of the EGL nine homologue 3 (egln3/phd3) gene. Biochem. J. 390, 189–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. D'Angelo G., Duplan E., Boyer N., Vigne P., Frelin C. (2003) Hypoxia up-regulates prolyl hydroxylase activity. A feedback mechanism that limits HIF-1 responses during reoxygenation. J. Biol. Chem. 278, 38183–38187 [DOI] [PubMed] [Google Scholar]
- 21. Marxsen J. H., Stengel P., Doege K., Heikkinen P., Jokilehto T., Wagner T., Jelkmann W., Jaakkola P., Metzen E. (2004) Hypoxia-inducible factor-1 (HIF-1) promotes its degradation by induction of HIF-α-prolyl-4-hydroxylases. Biochem. J. 381, 761–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Henze A. T., Riedel J., Diem T., Wenner J., Flamme I., Pouyseggur J., Plate K. H., Acker T. (2010) Prolyl hydroxylases 2 and 3 act in gliomas as protective negative feedback regulators of hypoxia-inducible factors. Cancer Res. 70, 357–366 [DOI] [PubMed] [Google Scholar]
- 23. Fujita N., Chiba K., Shapiro I. M., Risbud M. V. (2012) HIF-1α and HIF-2α degradation is differentially regulated in nucleus pulposus cells of the intervertebral disc. J. Bone Miner. Res. 27, 401–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Johansen J. L., Sager T. N., Lotharius J., Witten L., Mørk A., Egebjerg J., Thirstrup K. (2010) HIF prolyl hydroxylase inhibition increases cell viability and potentiates dopamine release in dopaminergic cells. J. Neurochem. 115, 209–219 [DOI] [PubMed] [Google Scholar]
- 25. Sun Q., Zhou H., Binmadi N. O., Basile J. R. (2009) Hypoxia-inducible factor-1-mediated regulation of semaphorin 4D affects tumor growth and vascularity. J. Biol. Chem. 284, 32066–32074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kong T., Westerman K. A., Faigle M., Eltzschig H. K., Colgan S. P. (2006) HIF-dependent induction of adenosine A2B receptor in hypoxia. FASEB J. 20, 2242–2250 [DOI] [PubMed] [Google Scholar]
- 27. Tian Y. M., Mole D. R., Ratcliffe P. J., Gleadle J. M. (2006) Characterization of different isoforms of the HIF prolyl hydroxylase PHD1 generated by alternative initiation. Biochem. J. 397, 179–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pfirrmann C. W., Metzdorf A., Zanetti M., Hodler J., Boos N. (2001) Magnetic resonance classification of lumbar intervertebral disc degeneration. Spine 26, 1873–1878 [DOI] [PubMed] [Google Scholar]
- 29. Wang J., Markova D., Anderson D. G., Zheng Z., Shapiro I. M., Risbud M. V. (2011) TNF-α and IL-1β promote a disintegrin-like and metalloprotease with thrombospondin type I motif-5-mediated aggrecan degradation through syndecan-4 in intervertebral disc. J. Biol. Chem. 286, 39738–39749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Seth P., Krop I., Porter D., Polyak K. (2002) Novel estrogen and tamoxifen induced genes identified by SAGE (Serial Analysis of Gene Expression). Oncogene 21, 836–843 [DOI] [PubMed] [Google Scholar]
- 31. Zhang Q., Gu J., Li L., Liu J., Luo B., Cheung H. W., Boehm J. S., Ni M., Geisen C., Root D. E., Polyak K., Brown M., Richardson A. L., Hahn W. C., Kaelin W. G., Jr., Bommi-Reddy A. (2009) Control of cyclin D1 and breast tumorigenesis by the EglN2 prolyl hydroxylase. Cancer Cell 16, 413–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hiwatashi Y., Kanno K., Takasaki C., Goryo K., Sato T., Torii S., Sogawa K., Yasumoto K. (2011) PHD1 interacts with ATF4 and negatively regulates its transcriptional activity without prolyl hydroxylation. Exp. Cell Res. 317, 2789–2799 [DOI] [PubMed] [Google Scholar]
- 33. Köditz J., Nesper J., Wottawa M., Stiehl D. P., Camenisch G., Franke C., Myllyharju J., Wenger R. H., Katschinski D. M. (2007) Oxygen-dependent ATF-4 stability is mediated by the PHD3 oxygen sensor. Blood 110, 3610–3617 [DOI] [PubMed] [Google Scholar]
- 34. Yan B., Jiao S., Zhang H. S., Lv D. D., Xue J., Fan L., Wu G. H., Fang J. (2011) Prolyl hydroxylase domain protein 3 targets Pax2 for destruction. Biochem. Biophys. Res. Commun. 409, 315–320 [DOI] [PubMed] [Google Scholar]
- 35. Fu J., Taubman M. B. (2010) Prolyl hydroxylase EGLN3 regulates skeletal myoblast differentiation through an NF-κB-dependent pathway. J. Biol. Chem. 285, 8927–8935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Anderson K., Nordquist K. A., Gao X., Hicks K. C., Zhai B., Gygi S. P., Patel T. B. (2011) Regulation of cellular levels of Sprouty2 protein by prolyl hydroxylase domain and von Hippel-Lindau proteins. J. Biol. Chem. 286, 42027–42036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Walmsley S. R., Chilvers E. R., Thompson A. A., Vaughan K., Marriott H. M., Parker L. C., Shaw G., Parmar S., Schneider M., Sabroe I., Dockrell D. H., Milo M., Taylor C. T., Johnson R. S., Pugh C. W., Ratcliffe P. J., Maxwell P. H., Carmeliet P., Whyte M. K. (2011) Prolyl hydroxylase 3 (PHD3) is essential for hypoxic regulation of neutrophilic inflammation in humans and mice. J. Clin. Invest. 121, 1053–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Epstein A. C., Gleadle J. M., McNeill L. A., Hewitson K. S., O'Rourke J., Mole D. R., Mukherji M., Metzen E., Wilson M. I., Dhanda A., Tian Y. M., Masson N., Hamilton D. L., Jaakkola P., Barstead R., Hodgkin J., Maxwell P. H., Pugh C. W., Schofield C. J., Ratcliffe P. J. (2001) C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell 107, 43–54 [DOI] [PubMed] [Google Scholar]
- 39. Bibby S. R., Jones D. A., Ripley R. M., Urban J. P. (2005) Metabolism of the intervertebral disc: effects of low levels of oxygen, glucose, and pH on rates of energy metabolism of bovine nucleus pulposus cells. Spine 30, 487–496 [DOI] [PubMed] [Google Scholar]
- 40. Luo W., Hu H., Chang R., Zhong J., Knabel M., O'Meally R., Cole R. N., Pandey A., Semenza G. L. (2011) Pyruvate kinase M2 is a PHD3-stimulated coactivator for hypoxia-inducible factor 1. Cell 145, 732–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Thoms B. L., Murphy C. L. (2010) Inhibition of hypoxia-inducible factor-targeting prolyl hydroxylase domain-containing protein 2 (PHD2) enhances matrix synthesis by human chondrocytes. J. Biol. Chem. 285, 20472–20480 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.