Background: Glycosylphosphatidylinositol (GPI) anchor addition occurs in the endoplasmic reticulum (ER).
Results: Efficient GPI signals have marginal hydrophobicity, intermediate between transmembrane helices and secreted proteins.
Conclusion: Proteins released into the ER lumen and those retained in the ER membrane are both bona fide substrates of the GPI anchoring reaction.
Significance: The results resolve a long-standing issue regarding the processing of GPI signals.
Keywords: Endoplasmic Reticulum (ER), Glycosylphosphatidylinositol Anchors, Membrane Proteins, Post-translational Modification, Protein Translocation, Transmembrane Helix
Abstract
Many plasma membrane proteins are anchored to the membrane via a C-terminal glycosylphosphatidylinositol (GPI) moiety. The GPI anchor is attached to the protein in the endoplasmic reticulum by transamidation, a reaction in which a C-terminal GPI-attachment signal is cleaved off concomitantly with addition of the GPI moiety. GPI-attachment signals are poorly conserved on the sequence level but are all composed of a polar segment that includes the GPI-attachment site followed by a hydrophobic segment located at the very C terminus of the protein. Here, we show that efficient GPI modification requires that the hydrophobicity of the C-terminal segment is “marginal”: less hydrophobic than type II transmembrane anchors and more hydrophobic than the most hydrophobic segments found in secreted proteins. We further show that the GPI-attachment signal can be modified by the transamidase irrespective of whether it is first released into the lumen of the endoplasmic reticulum or is retained in the endoplasmic reticulum membrane.
Introduction
10–20% of eukaryotic membrane proteins are anchored in the outer lipid leaflet of the plasma membrane by a C-terminal glycosylphosphatidylinositol (GPI)2 anchor (1). GPI addition takes place in the lumen of the endoplasmic reticulum (ER) and is catalyzed by a multisubunit GPI transamidase. The transamidation reaction occurs soon after translation of the pro-protein and its translocation across the ER membrane are completed (2). The signal for GPI-anchor addition (the “GPI signal”) consists of a carboxyl-terminal hydrophobic domain separated from the upstream GPI-attachment site (the “ω site”) by a short stretch of hydrophilic amino acids (Fig. 1A). Because the hydrophobic domain displays many properties of a typical transmembrane α-helix, it has been postulated that this segment partitions into the lipid bilayer before being cleaved during the GPI-addition reaction (3, 4). Other reports challenge the classical membrane-integration model, however, and claim that translocation of the C-terminal hydrophobic domain into the ER lumen is a prerequisite for GPI addition (5, 6).
FIGURE 1.
A, schematic representation of the GPI signal architecture and the GPI-attachment reaction. B, a dataset of putative GPI-anchored proteins was downloaded from navet.ics.hawaii.edu/∼fraganchor/data.zip and analyzed using the ΔG prediction server (dgpred.cbr.su.se). Distributions for mammalian secreted proteins (ΔGapp value of the most hydrophobic segment in each protein, excluding the N-terminal signal peptide) as well as single-spanning membrane proteins (7) are shown for comparison. Data points represent the relative frequency of proteins with ΔGapp values within ±0.5 kcal mol−1 of the corresponding value on the x axis.
The recent development of a “biological” hydrophobicity scale based on experimental measurements of the Sec61-mediated integration of transmembrane α-helices into the ER membrane makes it possible to predict the membrane insertion efficiency of hydrophobic polypeptide segments with good precision (7). This inspired us to revisit the issue of whether the hydrophobic part of the GPI signal is embedded in or translocated across the ER membrane prior to the transamidation reaction. To this end, we have analyzed a collection of GPI signals, both with the ΔG predictor software (7) that uses the biological hydrophobicity scale to predict the apparent free energy of membrane insertion (ΔGapp), and by in vitro translation of model protein constructs in the presence of ER-derived rough microsomes (RMs). Our results show that the hydrophobicity of natural GPI signals falls in a rather narrow range around ΔGapp = 0 kcal mol−1, and that this is a requirement for efficient GPI anchoring. Moreover, we have identified natural GPI signals that are not released from the ER membrane prior to transamidation, and others that are. These observations suggest that the transamidase has a binding site for the hydrophobic tail of the GPI signal and can only recognize substrates that are not too firmly embedded in the ER membrane.
EXPERIMENTAL PROCEDURES
Enzymes and Chemicals
Unless stated otherwise, all chemicals were from Sigma. Oligonucleotides were purchased from MWG Biotech AG (Ebersberg, Germany). Pfu Turbo DNA polymerase was purchased from Agilent Technologies, endoglycosidase H (EndoH), and USERTM enzyme from New England Biolabs, phosphatidylinositol-specific phospholipase C (PI-PLC) from Invitrogen, and PfuX7 polymerase was a kind gift from Dr. M. H. Norholm (8). All other enzymes were from Fermentas. The plasmid pGEM-1 and the TnT® SP6 Transcription/Translation System were from Promega, [35S]Met was from PerkinElmer Life Sciences.
DNA Manipulations
The Lep constructs used carried one acceptor site for N-linked glycosylation in positions 97–99 (Asn-Ser-Thr) and the wild-type acceptor site in positions 215–217 (Asn-Glu-Thr). For the glycosylation mapping assay, the latter sequon was mutated (N215Q) and a second glycosylation acceptor site (Asn-Ser-Thr) was introduced closer to the C terminus (supplemental Fig. S2) either within the annealed oligonucleotide sequence or by site-directed mutagenesis.
GPI signals were constructed by using two double-stranded oligonucleotides (30–74 nucleotides long) with overlapping overhangs at the ends. The four oligonucleotides (20 μm) were mixed and incubated at 90 °C for 10 min in buffer O (20 mm Tris-HCl, pH 7.4, 2 mm MgCl2, 50 mm NaCl2). After slow cooling to 30 °C, the annealed oligonucleotides were ligated as a SpeI-KpnI fragment into the vector pGEM1 containing the Lep construct. Alternatively, uracil excision-based cloning was used (9). All inserts and mutagenesis were confirmed by sequencing of plasmid DNA at Eurofins MWG Operon (Ebersberg, Germany).
Expression in Vitro
Constructs cloned in pGEM1 were transcribed and translated in the TnT® Quick-coupled transcription/translation system, except for the kinetic studies, where uncoupled transcription and translation reactions were performed (see below). One microgram of DNA template, 1 μl of [35S]Met (10 μCi; 1 Ci = 37 GBq), and 1 μl of dog pancreas RMs were mixed with 10 μl of TnT lysate mixture, and samples were incubated for 90 min at 30 °C. The sample was mixed with SDS sample buffer (10) and incubated at 90 °C for 5 min before loading on a 12% SDS-polyacrylamide gel.
Proteins bands were visualized in a Fuji FLA-3000 phosphorimager (Fujifilm, Tokyo, Japan). Image Gauge version 4.23 software (Fujifilm) was used to generate a two-dimensional intensity profile of each gel lane and the multi-Gaussian fit program from the QtiPlot software package was used to calculate the peak areas of the protein bands.
Glycosylation Mapping Assay
Two glycan acceptor sites were engineered into the Lep fusion proteins, one serving as a marker for lumenal localization at position 97 and another upstream of the test segment (at a distance of 10 amino acid or less), which can only be glycosylated if the H segment reaches the ER lumen (11). After expression and SDS-PAGE, an apparent equilibrium constant between the membrane-integrated and nonintegrated forms was calculated as: Kapp = f1g/f2g, where f1g is the fraction of singly glycosylated Lep molecules and f2g is the fraction of doubly glycosylated Lep molecules. The results were then converted to apparent free energies, ΔGapp = −RT ln(Kapp).
PI-PLC and EndoH Treatment
After in vitro expression, Lep-GPI fusions were subjected to Triton X-114 extraction and EndoH and/or PI-PLC treatments as described in Doering et al. (12) with some modifications. Briefly, the reaction mixture was solubilized with ice-cold lysis buffer (1% Triton X-114, 10 mm Tris-Cl, pH 7, 150 mm NaCl, 1 mm EDTA) and incubated at 37 °C to induce phase separation. The detergent-rich phase was re-extracted with 10 mm Tris-Cl, pH 7, 150 mm NaCl, 1 mm EDTA and resuspended in 100 mm Tris-Cl, pH 7, 50 mm NaCl, 1 mm EDTA. 4–8 milliunits of PI-PLC and/or 70 milliunits of EndoH per μl of translation mixture were added and incubated 60 min at 37 °C with shaking.
GPI-anchored bands were identified on SDS-polyacrylamide gels by a shift to higher apparent molecular weight after treatment with PI-PLC. EndoH treatment removed the N-linked glycans, after which the proteins migrated faster, at a size similar to the protein synthesized in the absence of microsomes (which is nonglycosylated).
Quantification of GPI Anchoring
Two glycan acceptor sites were engineered into the Lep-GPI fusions (positions 97 and 215) that were glycosylated in the ER lumen irrespective of the transmembrane or lumenal localization of the C terminus. The added glycans decreased the electrophoretic mobility enough for a GPI-anchored band to be distinguished from the nonglycosylated band.
In case the GPI-anchored band migrated differently from the nonanchored double-glycosylated form (generally for C termini with predicted ΔGapp > −1.5 kcal mol−1), GPI anchoring efficiency was expressed as IGPI/(IGPI + Ifl), where IGPI represents the intensity of the band of the GPI-anchored form and Ifl the full-length nonanchored protein. For hydrophobic C termini, anchored and nonanchored forms exhibit very similar electrophoretic mobilities. In these cases, IGPI was calculated from the PI-PLC modified band. Comparison of both quantification methods reveals that, due to possibly incomplete hydrolysis by PI-PLC, the latter method underestimates the GPI-anchoring efficiency by maximally 15%.
Measurement of Glycosylation and GPI-anchoring Kinetics
For kinetic studies, constructs were transcribed for 60 min at 37 °C using a standard SP6 polymerase transcription protocol (13). Translation was initiated by adding the resulting mRNA to a pre-warmed reaction mixture containing rabbit reticulocyte lysate (75 ng of mRNA/μl of translation mixture) [35S]Met (0.85 μCi/μl of translation mixture), dog pancreas RMs (40 nl/μl of translation mixture), an amino acid mixture (each 80 μm/μl of translation mixture) and RNasin (1.5 units/μl of translation mixture). After 4 min, 1.8 μl of aurintricarboxylic acid (3.5 mm) was added to inhibit further translation initiation. 4-μl samples were transferred to tubes containing 1 μl of 5% Triton X-100 at specific time points as indicated in Fig. 7 and incubation was continued at 30 °C until 60 min after addition of aurintricarboxylic acid. Control samples without aurintricarboxylic acid and Triton X-100 were run in parallel, and all translation products were analyzed as described above.
FIGURE 7.
Kinetics of glycosylation and GPI attachment to Lep-CD24 (top) and Lep-EFNA5 (bottom). Four minutes after the addition of mRNA to the translation reaction, translation initiation was blocked by the addition of aurintricarboxylic acid (t = 0). Aliquots from the reaction mixture were solubilized with Triton X-100 at the indicated time points to inhibit further glycosylation and GPI attachment, whereas allowing nascent chains to be elongated to full-length size (see “Experimental Procedures”). A control sample (C) was synthesized in the absence of the initiation inhibitor and detergent. Single and double glycosylated species are indicated by one and two white dots, respectively, and GPI-modified forms are indicated by an arrowhead. The G2 acceptor site in these constructs is located so close to the hydrophobic C-terminal segment that it can be glycosylated only if the hydrophobic segment is released from the membrane, as seen for CD24.
RESULTS
Natural GPI-anchoring Signals Have Marginal Hydrophobicity
We first used the ΔG predictor software to analyze a sequence dataset comprising 87 GPI-anchored proteins (14). The prediction program was fed the C-terminal 40 amino acids of each sequence and yielded the predicted ΔGapp for the most hydrophobic stretch. In most cases (77%), the proprotein ended with the hydrophobic stretch, that is, there were no hydrophilic flanking residues at the C-terminal end, and 96% of the sequences had less than three hydrophilic residues following the hydrophobic segment.
Strikingly, the distribution of the predicted ΔGapp values (Fig. 1B) is rather narrow and centered around ΔGapp = 0 kcal mol−1 (a sequence with ΔGapp = 0 kcal mol−1 has equal probabilities for membrane insertion and translocation). The distribution is distinct from the ΔGapp distributions found for transmembrane α-helices in single-spanning transmembrane proteins and for secreted proteins, with only small overlaps. Thus, the hydrophobicity of GPI signals is intermediate between that of transmembrane helices that are efficiently integrated into the ER membrane via the Sec61 translocon and that of secreted proteins that are translocated across the ER membrane by the same translocon. This observation raises two issues: does GPI addition depend on the ΔGapp value in the way suggested by the data in Fig. 1B, and is the degree of membrane anchoring of a GPI signal accurately reflected in its ΔGapp value? To address these questions, we studied membrane anchoring and GPI addition for a series of natural and designed GPI signals by in vitro translation in the presence of dog pancreas RMs.
GPI Attachment and Membrane Insertion Assays
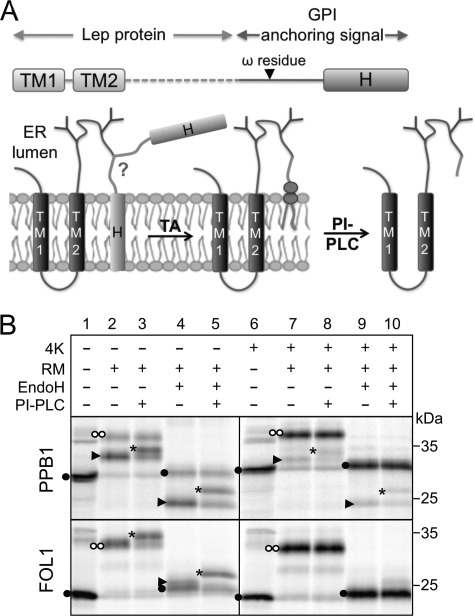
To analyze different GPI signals in a common sequence context that allows studies both of membrane insertion and GPI attachment, we used the well characterized model protein leader peptidase (Lep) as the host protein (Fig. 2A). Lep has two N-terminal transmembrane helices (TM1, TM2) and a large C-terminal domain that is translocated into the lumen of the RMs (15). In most of the constructs used here, two acceptor sites for N-linked glycosylation are present in the C-terminal domain (Asn-Ser-Thr at position 97 and Asn-Glu-Thr at position 215); both can be efficiently glycosylated by the lumenal oligosaccharyl transferase and serve as markers for lumenal localization of the C-terminal domain (16). Different GPI signals followed by a stop codon were introduced at position 229, replacing the C-terminal 95 residues in Lep.
FIGURE 2.
A, experimental setup used in this study. Leader peptidase (Lep) fusion proteins bearing C-terminal GPI-anchoring signals are transcribed and translated in vitro in the presence of RM, where the proprotein becomes cleaved by the transamidase (TA) at the ω residue and anchored to a GPI lipid. PI-PLC can be used to remove the GPI lipid after solubilization of the microsomes. B, fusions of Lep with PPB1 and FOL1 signal sequences are efficiently GPI anchored. The GPI-anchored protein can migrate faster (PPB1, arrowhead, lane 2, upper panel) or similarly (FOL1) to the full-length protein (two white dots, lane 2, lower panel). GPI anchoring can in both cases be assayed for by treatment with PI-PLC, which results in a slower electrophoretic mobility (asterisk, lane 3). Because all constructs had two N-glycosylatable motifs, protein that has not been translocated into the microsomes can be identified as a low molecular weight band (black dot). Addition of 4 lysines (4K) at the C terminus greatly inhibits GPI anchoring of both constructs (lanes 6–10).
Most GPI-anchored proteins have a cleavable N-terminal signal peptide targeting them to the ER translocon. Lep, in contrast, has no cleavable signal peptide, which is an advantage here because it reduces the complexity of the products obtained in the in vitro translation reaction, as signal peptides often are not removed from the protein with 100% efficiency in the RM system, giving rise to multiple protein forms.
Results for two constructs, Lep-PPB1 (predicted ΔGapp = 1.0 kcal mol−1) and Lep-FOL1 (predicted ΔGapp = −1.9 kcal mol−1), are shown in Fig. 2B. Both proteins undergo GPI modification, as seen in lane 3 by their sensitivity to PI-PLC, an enzyme that cleaves the ester bond between phosphoinositol and diacylglycerol, thus releasing the membrane anchor from the C terminus of the protein (12). Because the GPI anchor binds large amounts of SDS, PI-PLC treatment generally leads to a decreased mobility during SDS-PAGE (17) (Fig. 2, lanes 3 and 5).
GPI attachment by the transamidase has only a small net effect on the mass of the protein: loss of the residues downstream of the ω position (corresponding to 2–3 kDa) is counteracted by addition of the ∼1.5-kDa GPI moiety (18). Whether different bands can be detected upon SDS-PAGE for the nonprocessed and the GPI-anchored proteins depends largely on the respective amounts of SDS bound by the GPI-lipid and the removed amino acid tail. Whereas GPI-anchored PPB1 (arrowhead, Fig. 2B, lane 2, upper panel) can readily be distinguished from the doubly glycosylated uncleaved form (two white dots), identification of the GPI-anchored protein is difficult for more hydrophobic GPI signals such as FOL1, which bind higher amounts of SDS (19). In this case, the uncleaved and GPI-anchored proteins have similar electrophoretic mobilities (lane 2, lower panel) and PI-PLC digestion is required for positive identification of the GPI-anchored species (lane 3, lower panel). Deglycosylation by treatment with EndoH sometimes can improve the separation between the GPI-anchored and PI-PLC-cleaved forms (lanes 4 and 5).
As a control, we made two constructs with four lysine residues added to the C-terminal end of the GPI signals of Lep-PPB1 and Lep-FOL1 (Fig. 2B, lanes 6–10). A positively charged cytoplasmic tail is almost invariably present in transmembrane helices from single-spanning membrane proteins (20) but is rarely seen in GPI signals, as noted above. Only ∼10% of Lep-PPB1(4K) and Lep-FOL1(4K) are sensitive to PI-PLC (lanes 8 and 10), and hence these mutant GPI signals are essentially resistant to GPI addition, as expected.
Because, as noted above, most GPI-anchored proteins have a cleaved N-terminal signal peptide targeting them to the Sec61 translocon in the ER, we further validated the Lep-GPI signal chimeras by introducing a cleavage cassette (CC) of 7 amino acids at the end of TM2 (Fig. 3A). This sequence has been shown to render TM2 susceptible to cleavage by signal peptidase both in Escherichia coli (21) and in RMs (22). Translation of a set of Lep-GPI and Lep(CC)-GPI constructs in the presence of RMs gave similar results with and without the cleavage cassette (Fig. 3B). In both cases, two bands could be distinguished in addition to the protein that was not targeted to the RMs. As shown above for Lep-PPB1 (Fig. 2B), the faster-migrating bands (Fig. 3B, arrowheads) correspond to GPI-anchored proteins. This was further confirmed by PI-PLC treatment (see supplemental Fig. S1). We conclude that Lep-GPI signal chimeras with or without the N-terminal transmembrane segments are efficiently converted by the transamidase in the ER to their GPI-anchored forms. In line with these results, it was previously shown that a type II membrane protein that is attached to the membrane via an uncleaved N-terminal signal-anchor sequence can be GPI anchored (23, 24).
FIGURE 3.
The TM1 and TM2 transmembrane segments in Lep do not affect GPI anchoring. A, schematic representation of the Lep-GPI fusions and the processing of the CC+ constructs, where the cleavage cassette SPSAQA↓A (cleavage site marked by the arrow) was introduced at the end of TM2. After cleavage of the preproprotein by signal peptidase (SP), the proprotein is further processed into its GPI-anchored form by the transamidase (TA). B, in vitro synthesis of the indicated constructs (see Table 1 for the amino acid sequences) with or without the cleavage cassette, which leads to efficient cleavage by signal peptidase as inferred from the higher electrophoretic mobility. GPI-anchored proteins can be distinguished by increased electrophoretic mobility (arrowhead) compared with the full-length protein (two white dots).
ΔG Predictor Is Accurate for C-terminal Hydrophobic Segments
Because the ΔG predictor is based on data generated using hydrophobic segments located in the middle of the Lep C-terminal domain (7, 25), it was necessary to ascertain whether it could be reliably applied to GPI signals located at the extreme C terminus of the protein. To test this, we engineered a series of Lep-GPI fusions (Fig. 4A). In these constructs, the ω site and linker region were omitted; the hydrophobic part (H segment) of the GPI signal was fused directly to Lep and a glycosylation acceptor site (Asn-Ser-Thr) was introduced just upstream of the H segment. The Asn residue is located only 2 to 3 residues away from the H segment (supplemental Fig. S2) and can therefore be glycosylated only if the H segment is released into the lumen of the RMs (11). The glycosylation site at position 215 was not present in these constructs. Hence, constructs in which the H segment is retained in the membrane are modified only on one acceptor site (G1), whereas constructs in which the H segment is released to the lumen become doubly glycosylated. Two artificial H segments composed only of Ala and Leu residues, derived from constructs used in earlier studies by Hessa et al. (25) were also included (supplemental Fig. S2) to allow for a direct comparison to the results obtained with H segments located in the middle of the lumenal domain.
FIGURE 4.
The ΔG predictor is accurate for C-terminal protein segments. A, artificial hydrophobic Leu/Ala sequences or hydrophobic C-terminal segments from GPI-anchored proteins (H segments; see supplemental Fig. S2) were fused to Lep. We chose sequences with predicted |ΔGapp| < 2 kcal mol−1 because the glycosylation mapping method is maximally sensitive in the ΔGapp range (−1.2, 1.2) kcal mol−1 (25). Besides the glycosylation site at position 97 (G1), a glycosylation acceptor site (G2) was introduced immediately upstream of the H segments. No glycan acceptor site was present at position 215 in these constructs. Single and double glycosylated proteins are indicated with one or two white dots, respectively. Nonglycosylated protein is marked with a black dot. Experimental ΔGapp values were determined as described in the text. B, the experimental ΔGapp values were plotted against the theoretical values calculated by the ΔG predictor. Error bars represent ± 1 S.D. of the experimental values (n = 2–5) and a dotted line of slope = 1 and intersect = 0 is included as a reference.
We verified that these constructs did not undergo GPI modification (Fig. 4A and supplemental Fig. S2) and determined the extent of insertion into the ER membrane after in vitro transcription and translation in the presence of RMs by quantifying the amount of singly glycosylated (one white dot) versus doubly glycosylated protein (two white dots). We express the membrane insertion efficiency as the apparent free energy difference ΔGapp between the inserted and noninserted forms, ΔGapp = −RT ln(f1g/f2g), where f1g and f2g denote the fractions of singly and doubly glycosylated protein (see “Experimental Procedures”). As shown in Fig. 4B, the experimental and predicted ΔGapp values agree within the error margins determined previously (±0.45 kcal mol−1 for 90% of the tested sequences (7)). These results confirm that the ΔG predictor provides a faithful description of the membrane insertion behavior of C-terminal located segments, including GPI-anchoring signals, at least in the context of the Lep fusion proteins.
GPI Anchoring Is Efficient for C-terminal Signals with ΔGapp values in the Interval |ΔGapp| <2.5 kcal mol−1
To study the relationship between GPI addition and the hydrophobicity of the GPI signal, we chose a representative set of 15 putative or verified GPI signals (14) with ΔGapp values spanning from −4 to 4 kcal mol−1 (Table 1), and engineered Lep-GPI fusions as in Fig. 2A. As shown in Fig. 5 for a subset of the sequences, all Lep-GPI proteins studied except those with extreme ΔGapp values (MMP19 and EFNA4) were found to be efficiently GPI anchored. For six of the proteins (see Table 1), this constitutes, to the best of our knowledge, the first experimental evidence of their GPI anchoring.
TABLE 1.
The C-terminal GPI signals studied in this work
The indicated sequences were fused to residue 228 in the Lep protein. The C-terminal hydrophobic segments, as determined by the ΔG predictor, are shown in bold. The predicted or confirmed ω residue is depicted in bold italics. Asterisks represent stop codons. The presence of a GPI anchor was inferred as described in the legend to Fig. 2. ΔGapp values are in kcal mol−1.
a To the best of our knowledge, this is the first experimental evidence that this sequence is GPI anchored.
FIGURE 5.
GPI addition is efficient for GPI signals with |ΔGapp| < 2.5 kcal mol−1 (see Table 1 for amino acid sequences). GPI-anchored proteins (arrowhead) can be distinguished by increased electrophoretic mobility compared with the full-length protein (two white dots) and modification by PI-PLC (asterisk). See supplemental Fig. S3 for replicates of Lep-EFNA3 PI-PLC treatment.
The extent of the GPI modification observed ranged between 50 and 70% of the total protein targeted to the RMs (see “Experimental Procedures” for details of the quantitation procedure). In line with these results, previous studies carried out in vitro with RMs obtained from HeLa or CHO cells show maximal GPI-modification efficiencies of 60 and 70%, respectively (2, 26).
No GPI addition was observed for MMP19 (ΔGapp = 4.2 kcal mol−1) and very little for EFNA4 (ΔGapp = −4.1 kcal mol−1). The matrix metalloproteinase MMP19 was initially annotated as a GPI-modified protein by similarity to other GPI-anchored members of the family, notably MMP17 and 25 (27), but was later reclassified as a secreted protein (28), and our data confirms this. The EFNA4 protein belongs to the Ephrin A family of receptor tyrosine kinase ligands. We analyzed the four other members of this family present in the human genome (with ΔGapp values ranging from −2.4 to 1.6 kcal mol−1, cf. Table 1) and found them to be efficiently GPI anchored (shown for EFNA1 and -3 in Fig. 5). Cell-surface located EFNA4 has been shown to be sensitive to PI-PLC treatment (29), but the fraction released was not determined. We suggest that EFNA4 in human cells might be at least partially present as a nonprocessed transmembrane protein, anchored by the C-terminal polypeptide tail in the ER membrane. The presence of transmembrane members in a family of mostly GPI-anchored proteins has been observed previously, for example, in the COBRA protein family in plants (30).
Among the proteins found to be GPI-anchored, we do not see any significant correlation between predicted ΔGapp values and the observed GPI-anchoring efficiency. Our results therefore suggest that C-terminal sequences with intermediate ΔGapp values can be efficiently modified by the transamidase. However, processing efficiency appears to drop beyond a threshold value, such that only sequences with |ΔGapp| < 2.5 kcal mol−1, can be efficiently modified (Table 1), broadly consistent with the shape of the ΔGapp distribution for GPI-anchored proteins shown in Fig. 1B.
The Lep-GPI signal constructs differ in sequence not only in the hydrophobic C-terminal segment (accounting for the different ΔGapp values), but also in the upstream ω site and linker regions. Because the sequence environment around the ω site influences the efficiency of GPI anchoring (26, 31), our results probably reflect more than the contribution of the ΔGapp values alone. We therefore constructed Lep-GPI chimeras where the ω site and linker segment were kept constant but the hydrophobic C-terminal segment was replaced with 19-residue long model segments composed only of Ala, Leu, Val, and Phe residues. The membrane insertion properties of such model segments have been extensively studied (25), and 19 residues is a reasonably good approximation to the length of the hydrophobic segments found in native GPI signals (the average length and S.D. of the hydrophobic segments in the data base compiled by Poisson et al. (14) is 20 ± 3 residues).
Four sets of constructs were made, using the ω site/linker segment of FOL1, PPB1, CNTN1, and COBL9 (Table 2). Representative results for the Lep-COBL9 and Lep-PPB1 chimeric proteins are shown in Fig. 6A. Replacement of the native C-terminal hydrophobic segment of these proteins with a stretch of 19 Ala (ΔGapp = 1.5 kcal/mol) inhibited the GPI addition reaction to a great extent. Gradual introduction of more hydrophobic residues (Leu, Val, and Phe) increased GPI-anchoring efficiency, which again dropped at more negative ΔGapp values (Fig. 6B).
TABLE 2.
Chimeric constructs prepared by fusing poly-Ala sequences containing increasing number of Leu, Val, and Phe residues to the ω environment of FOL1, COBL9, CNTN1, and PPB1
The predicted or confirmed ω residue is depicted in bold italics. The C-terminal hydrophobic segments, as determined by the ΔG predictor, are shown in bold. Asterisks represent stop codons. ΔGapp values are in kcal mol−1.
FIGURE 6.
Differential effects of the ω site/linker region and the hydrophobic C-terminal tail on GPI attachment. A, in vitro synthesis and GPI anchoring of Lep-COBL9 (top) and Lep-PPB1 (bottom) chimeras bearing C-terminal model Leu/Ala (0L to 9L) and Val/Phe/Ala segments (3V to 4F7V) (see Table 2 for sequences). The leftmost lanes show the results for Lep fused to the native GPI signals. The presence of a GPI anchor was confirmed by treatment with PI-PLC. Symbols are as described in the legend to Fig. 5. B, GPI-attachment efficiencies of GPI chimeras with C-terminal model Leu/Ala (left panel) and Val/Phe/Ala (right panel) segments (see Table 2 for sequences). Anchoring efficiencies were estimated by quantitation of the bands corresponding to the full-length unprocessed protein and the GPI-anchored form (see Fig. 6A for some of the gels used for quantitation). In case these two forms could not be distinguished, the PI-PLC sensitive and -insensitive bands were quantitated. Average values ±1 S.D. are plotted (n = 3) and data obtained for the native sequences are shown in both graphs as a reference (gray dashed trace).
The constructs display different extents of GPI modification for a given C-terminal hydrophobic tail; efficiencies increase in the order CNTN1 < FOL1 < PPB1 < COBL9. This observation underscores the impact of the ω site/linker segment on the GPI-addition reaction. Because the native sequences, in contrast to some of the corresponding model constructs, are all efficiently GPI modified (60–70%, Fig. 5), there seems to be a certain amount of co-adaptation between the ω site, linker region and hydrophobic segment in the natural sequences.
With few exceptions, the curves in Fig. 6B are bell-shaped, reminiscent of the shape of the ΔGapp distribution for GPI-anchored proteins shown in Fig. 1B. The large error bars in constructs with ΔGapp ≤ −1.5 kcal mol−1 arise from difficulties in the quantification of the PI-PLC-modified bands as explained above.
For ΔGapp > 0, the efficiency of GPI attachment drops with increasing ΔGapp values in all cases and is essentially zero at ΔGapp = 1.5 kcal mol−1. This observation is in line with a mutagenesis study performed on the folate receptor by Yan et al. (32). The results of this work, when expressed in terms of ΔGapp values (supplemental Fig. S5), show a decrease in functional GPI-anchored receptor with mutations that increase the predicted ΔGapp of the GPI signal. The degree of GPI attachment is very low for mutants with ΔGapp > 2.5 kcal mol−1, consistent with the ΔGapp distribution for natural GPI-anchored proteins in Fig. 1B.
Other sequence preferences in the ω site/linker region can be inferred from the results shown in Fig. 6B. For a given ΔGapp value (for instance, 1 kcal mol−1) and the same presequence (PPB1), anchoring efficiencies are lowest for the segment composed of 1L/18A, intermediate for 3V/16A, and highest for the PPB1 native sequence. A similar trend is seen at ΔGapp ≈ 0 kcal mol−1 for the series 7V/12A, 3L/16A, and the CNTN native C terminus. It is noteworthy that all native sequences assessed here are equally or more efficiently GPI-anchored than any of the Leu/Val/Phe/Ala chimeras with the same predicted ΔGapp value. This observation again points to an evolutionary pressure exerted on the C-terminal sequences toward an optimal processing efficiency.
GPI Anchoring Is a Slow Reaction and Works Both with a Membrane-anchored and Lumenal GPI Signal
N-Linked glycosylation is known to occur in a co-translational manner, whereas GPI attachment by necessity is a post-translational reaction (2). To more precisely define the kinetics of glycosylation relative to GPI addition of the Lep-GPI chimeras, we utilized a pulse-chase assay (33) in which translation initiation was blocked 4 min after mRNA addition to the in vitro translation system, followed by the addition of the detergent Triton X-100 to aliquots extracted at subsequent time points to solubilize the RM membrane and inhibit any further N-glycosylation and GPI addition. After the addition of detergent, the translation reaction was allowed to proceed to completion, such that only full-length protein was produced.
Results for Lep-CD24 (ΔGapp = 1.8 kcal mol−1) and Lep-EFNA5 (ΔGapp = −2.4 kcal mol−1) are shown in Fig. 7. The two constructs were chosen on the basis of their different hydrophobicities and the presence of a distinct band for the GPI-anchored form even without PI-PLC treatment. Two glycosylation acceptor sites (G1, G2) were engineered into the two constructs similarly to the constructs described in Fig. 4, with the G2 site placed close enough to the hydrophobic part of the GPI signal to allow glycosylation only if the hydrophobic segment was released from the membrane (34). For both proteins, glycan addition to the G1 site was complete ∼4 min after blocking translation initiation. For Lep-CD24, the G2 site was fully modified after ∼6 min, whereas GPI attachment to the doubly glycosylated form became apparent only after ∼20 min. Thus, the GPI signal in Lep-CD24 is released from the membrane before the GPI addition. In contrast, only small amounts of Lep-EFNA5 are ever glycosylated on the G2 site, and GPI addition of the singly glycosylated, nonreleased form, becomes apparent after ∼20 min. The identity of the bands seen in Fig. 7 was confirmed by PI-PLC and EndoH treatment (supplemental Fig. S4). Because the synthesis rate in our in vitro system is on the order of 0.5 residues s−1 (35), translation of the full-length Lep-GPI chimeras (both ∼260 amino acids long) takes only ∼10 min. GPI addition to these chimeras is therefore a slow, post-translational modification, whereas glycosylation is co-translational. Although the absolute rate of GPI addition is dependent on the amino acids at the ω to ω + 2 positions (31), these results are in broad agreement with previous data by Chen et al. (2).
The two proteins thus appear to undergo the GPI-attachment reaction from different locations. For Lep-EFNA5, the GPI-anchored form derives from the singly glycosylated, membrane-integrated species (because the C-terminal segment has a high hydrophobicity in this case, the mobility shift caused by the GPI modification is small). In contrast, for Lep-CD24, it is the doubly glycosylated form in which the hydrophobic segment has been released into the lumen of the RMs that receives the GPI anchor (the mobility shift is larger in this case, because the C-terminal polypeptide segment is less hydrophobic). We therefore conclude that the transamidase can act on substrate GPI signals irrespective of whether the C-terminal hydrophobic segment is retained in or is released from the membrane prior to the reaction.
GPI-anchoring Reaction Tolerates Extensive Sequence Variation around the ω Site
Because many of the Lep-GPI chimeras with artificial C termini undergo extensive GPI-anchoring (Fig. 6), we asked whether an artificial ω environment could also be processed by the transamidase. To this end, we aligned the sequences in the dataset of Poisson et al. (14) at the first residue (n) of their hydrophobic segments, and the most frequent amino acid was chosen for positions n-7 to n-1, resulting in the consensus sequence GSGSSGS. An Asn-Ser-Thr glycosylation acceptor site was further engineered at positions n-10 to n-8. The consensus ω environment was then fused to the Leu/Ala or Val/Phe/Ala C-terminal hydrophobic segments. As shown in Fig. 8A, all constructs with ΔGapp < 0 kcal mol−1 became GPI-anchored, indicating that the consensus peptide could indeed be recognized and modified by the transamidase. The efficiency of GPI attachment was highest for the segments composed of 6L/13A (ΔGapp = −1.5 kcal mol−1) and 7V/12A (ΔGapp = −0.2 kcal mol−1).
FIGURE 8.
GPI modification tolerates extensive sequence variation in the ω site/linker region. A, artificial GPI signals consisting of the flexible ω site/linker consensus sequence NSTGSGSSGS followed by Ala/Leu or Ala/Val/Phe hydrophobic stretches as shown were fused to Lep and tested in the in vitro translation system. B, GPI anchoring persists upon deletion of the ω environment. The C termini of CD24, COBL9, and EFNA1 were fused directly to the Lep protein replacing the ω site/linker by an Asn-Ser-Thr glycosylation acceptor site. C, addition of four lysines (4K) to the C terminus of Lep-GPI signal fusions efficiently blocks GPI anchoring, shown here for EFNA5, EFNA2, and CD24, and in Fig. 2B for PPB1 and FOL1. GPI signals are shown in italics and glycan acceptor sites are underlined. In panels B and C, GPI-anchored proteins that can be distinguished because of modified electrophoretic mobility are depicted with one or two arrowheads (single and double glycosylated, respectively). Other symbols are as in previous figures.
Surprisingly, a segment derived by chance from the C-terminal domain of Lep also contained a cryptic ω site as it served as a substrate for the transamidation reaction (Fig. 8B). Removal of the ω site and linker segment in the GPI signal (that is, attaching the C-terminal hydrophobic segment directly to the Lep protein) was compatible with GPI anchoring in several cases, shown in Fig. 8B. Nevertheless, it is unlikely that this cryptic ω site in Lep is processed by the transamidase in the constructs studied earlier, because in those it is far away (at least 10 amino acids, see Tables 1 and 2) from the hydrophobic segment, so that the native ω sites are probably modified instead. Interestingly, both glycosylation of the Asn residue at position n-3 and GPI attachment were observed for Lep-CD24 (double arrowhead, Fig. 8B). This indicates that the added N-glycan did not inhibit GPI addition, although the ω residue is presumably very close to the glycosylated Asn because no linker residues are present besides the Asn-Ser-Thr triad.
The only sequence modification that we found invariably to prevent the GPI-modification reaction was the addition of four lysines to the C terminus of the GPI signal (Figs. 2B and 8C). We observed extensive double glycosylation of Lep-EFNA2–4K (Fig. 8C), implying that the C-terminal segments of this protein are released from the membrane despite the positively charged C-terminal flank. Thus, the inhibition of the transamidation reaction by the positively charged flank does not rely on the retention of the C-terminal segment in the ER membrane.
DISCUSSION
What is the role of the C-terminal hydrophobic segment that is invariably found as part of the GPI signal present in all GPI-anchored proteins? Its length is similar to that of typical type I transmembrane anchor segments, yet it lacks the positively charged C-terminal flanking residues normally found in the latter. This suggests that the unprocessed precursors of GPI-anchored proteins may be less firmly anchored in the ER membrane than bona fide transmembrane proteins, or may even be released to the ER lumen before the GPI-addition reaction.
To better define the role of the hydrophobic part of the GPI signal, we have combined theoretical and experimental analyses of a large number of known or predicted GPI signals, also including “synthetic” signals constructed from simplified hydrophobic and upstream hydrophilic segments. The initial observation that piqued our interest was the peculiar distribution of predicted ΔGapp values for GPI signals obtained with the ΔG predictor software (7) (Fig. 1B). The distribution is centered around ΔGapp = 0 kcal mol−1 (corresponding to marginally hydrophobic segments that insert into the ER membrane with ∼50% efficiency) and is distinct from ΔGapp distributions of both secreted and single-span transmembrane proteins, showing that the GPI signal is intermediate between secreted and transmembrane segments in terms of hydrophobicity.
In fact, we find that GPI-anchored proteins can be rather well predicted by applying the following simple rules. 1) The protein should have a signal peptide predicted by SignalP 4.0 (36). 2) After removal of the predicted signal peptide, the mature part of the protein should contain no internal transmembrane helix with ΔGapp < 0 kcal mol−1 predicted by the ΔG predictor (7). 3) There should be one segment with |ΔGapp| < 2.0 kcal mol−1 at the C terminus of the protein. 4) There should be no more than 5 residues downstream of the predicted hydrophobic region, and no more than one of these should be Arg or Lys. Among 22,151 reviewed human protein sequences present in the UniProt data bank on October 21, 2011, 3,614 pass rule 1. Of these, 121 are annotated as GPI anchored. Applying rules 2–4 on these 3,614 proteins predicts 158 proteins to be GPI anchored, 95 of which are annotated as such (accuracy = 0.60, coverage 0.79). We also tested three dedicated GPI-anchoring predictors on the same dataset of 3,614 proteins and obtained the following accuracy/coverage values: BigPI (37) 0.66/0.73, PredGPI (38) 0.68/0.83, and FragAnchor (14) 0.73/0.75. Not surprisingly, the sophisticated machine-learning methods perform somewhat better than our simple rule-based scheme, but it is nevertheless striking how far one can get by just requiring the presence of a marginally hydrophobic C-terminal segment while making no attempt to identify an ω site.
To study the effect of the C-terminal hydrophobic segment on the GPI-attachment reaction in a common sequence context, we have developed a new model protein for the study of GPI anchoring based on the well characterized Lep protein, which is efficiently targeted to RMs in a standard in vitro translation system (15). The model protein is composed of a truncated version of Lep fused to C-terminal GPI-anchoring signals and can be GPI modified with high efficiency in the in vitro RM system, comparable with, for example, the mini-PLAP substrate that has long been used as a model GPI-anchored protein (26). Lep thus provides a new scaffold for addressing questions related to GPI modification.
Systematic variation of the GPI signal has allowed us to identify a clear influence of hydrophobicity on the GPI-processing reaction catalyzed by the transamidase. For a given ω environment, the extent of processing mirrors the ΔGapp distribution obtained for the GPI-signal dataset: GPI attachment is most efficient around ΔGapp = 0 kcal mol−1 and drops for both higher and lower values of ΔGapp. However, for a couple of the simplified GPI signals that we have tested the efficiencies of GPI modification are not fully symmetric around ΔGapp = 0 kcal mol−1 (Fig. 6); whether this lack of symmetry depends on, for example, the precise sequence of the ω site or the linker length remains to be addressed.
The study of natural GPI signals (Fig. 5, Table 1) reveals a hydrophobicity threshold of |ΔGapp| ∼2.5 kcal mol−1 for GPI addition that mirrors the ΔGapp distribution of the GPI signal dataset in Fig. 1B. The ΔGapp interval compatible with GPI addition found for artificial hydrophobic segments is somewhat broader (Fig. 6), especially in the negative range. This difference suggests that the efficiency of GPI addition by the transamidase is not the only driving force acting on the sequence evolution of GPI anchor proteins. Also, whereas the absence of a C-terminal flank is a general feature of GPI signals, GPI addition has been observed in a couple of cases when GPI signals were placed internally in protein sequences (39, 40); the sequence requirements for this to be possible are not clear at present.
Our results provide new insights into the kinetics of GPI anchoring and show that this modification occurs in a post-translational manner (Fig. 7). Because N-glycosylation is carried out co-translationally, this difference in time scales argue against a scenario where the transamidase and the glycosyltransferase compete for their substrate. The relatively long time lapse between the two modification reactions allows us to ascertain whether a given GPI signal is released into the ER-lumen before undergoing GPI anchoring. Strikingly, the examples presented in Fig. 7 show that the C-terminal hydrophobic segment in the GPI signal can either be released into the ER lumen (CD24) or retained in the ER membrane (EFNA5) before being processed by the transamidase.
Further support for this notion is provided by the observation that efficient GPI anchoring (60–70%) is seen for GPI signals with a 3 kcal mol−1 difference in ΔGapp, for instance, FOL1 (−1.9 kcal mol−1) and PPB1 (1 kcal mol−1), shown in Figs. 2 and 5. These values correspond to 96% probability of membrane insertion (FOL1) and 84% probability of lumenal location (PPB1). The most plausible scenario is therefore that the transamidase substrates can be recruited from both lumenal and transmembrane locations, thereby reconciling earlier data (3–6).
Our results also suggest that there is some degree of interdependence between the ω site environment and the C-terminal hydrophobic segment. On the one hand, for a given set of simplified C-terminal hydrophobic segments (for instance, those composed of Leu and Ala, in Fig. 6), the four different ω environments assessed here show consistent differences in the extent of GPI anchoring. On the other hand, for a given ω environment, different C-terminal segments (native, Leu/Ala, and Val/Phe/Ala sequences) with the same predicted ΔGapp value show different efficiencies of GPI attachment. This result is especially noteworthy because sequence conservation among GPI-anchored proteins drops dramatically after the ω residue and it has proven difficult to identify any conserved sequence motifs in the C-terminal segment beyond the hydrophobicity constraint (41–44).
Although only low-resolution structural data are available for the transamidase (45), it seems clear from previous studies that the size and polarity requirements on the ω environment (26, 31, 44) reflect the active site architecture of the catalytic subunit, Gpi8 in yeast and PIG-K in human (46). Whether there is a separate binding site for the C-terminal hydrophobic region is unknown. It has been proposed that Gaa1, a subunit of the transamidase with multiple predicted transmembrane helices (47), could funnel substrates to the catalytic subunit. Indeed, co-precipitation of this subunit by a substrate protein is lost when the C-terminal hydrophobic segment is deleted (48). It is thus tempting to propose a role for the Gaa1 subunit in the recognition and/or release from the membrane of the hydrophobic C-terminal segment. The sequence preferences at the C terminus observed for a given ΔGapp value (see above) could also fit into such a molecular-recognition scenario.
Regardless, our analysis shows that GPI can be efficiently added to sequences that are either released into the ER lumen or retained in the ER membrane. Whether this means that the transamidase recruits its substrates from either location, or whether a transient repartitioning of membrane-embedded proteins into the ER lumen (or of lumenal proteins into the ER membrane) is required for the reaction remains an open question. In any case, the fact that the ΔGapp distribution and GPI-anchoring efficiency both peak at 0 kcal mol−1, which corresponds to a situation of 50% membrane and 50% lumenal location, indicates that GPI signals are able to sample both the ER membrane and lumen prior to recognition by the transamidase.
Supplementary Material
This work was supported by grants from the Swedish Cancer Foundation, Swedish Research Council, Swedish Foundation for Strategic Research, and European Research Council Grant ERC-2008-AdG 232648 (to G. v. H.).

This article contains supplemental Figs. S1–S5.
- GPI
- glycosylphosphatidylinositol
- CC
- cleavage cassette
- ER
- endoplasmic reticulum
- Lep
- leader peptidase
- PI-PLC
- phosphatidylinositol-specific phospholipase C
- RMs
- dog pancreas rough microsomes
- TM
- transmembrane
- EndoH
- endoglycosidase H
- MMP
- matrix metalloproteinase.
REFERENCES
- 1. Orlean P., Menon A. K. (2007) Thematic review series. Lipid post-translational modifications. GPI anchoring of protein in yeast and mammalian cells, or how we learned to stop worrying and love glycophospholipids. J. Lipid Res. 48, 993–1011 [DOI] [PubMed] [Google Scholar]
- 2. Chen R., Knez J. J., Merrick W. C., Medof M. E. (2001) Comparative efficiencies of C-terminal signals of native glycophosphatidylinositol (GPI)-anchored proproteins in conferring GPI-anchoring. J. Cell. Biochem. 84, 68–83 [DOI] [PubMed] [Google Scholar]
- 3. Berger J., Howard A. D., Brink L., Gerber L., Hauber J., Cullen B. R., Udenfriend S. (1988) COOH-terminal requirements for the correct processing of a phosphatidylinositol glycan-anchored membrane protein. J. Biol. Chem. 263, 10016–10021 [PubMed] [Google Scholar]
- 4. Caras I. W., Weddell G. N. (1989) Signal peptide for protein secretion directing glycophospholipid membrane anchor attachment. Science 243, 1196–1198 [DOI] [PubMed] [Google Scholar]
- 5. Wang J., Maziarz K., Ratnam M. (1999) Recognition of the carboxyl-terminal signal for GPI modification requires translocation of its hydrophobic domain across the ER membrane. J. Mol. Biol. 286, 1303–1310 [DOI] [PubMed] [Google Scholar]
- 6. Dalley J. A., Bulleid N. J. (2003) The endoplasmic reticulum (ER) translocon can differentiate between hydrophobic sequences allowing signals for glycosylphosphatidylinositol anchor addition to be fully translocated into the ER lumen. J. Biol. Chem. 278, 51749–51757 [DOI] [PubMed] [Google Scholar]
- 7. Hessa T., Meindl-Beinker N. M., Bernsel A., Kim H., Sato Y., Lerch-Bader M., Nilsson I., White S. H., von Heijne G. (2007) Molecular code for transmembrane-helix recognition by the Sec61 translocon. Nature 450, 1026–1030 [DOI] [PubMed] [Google Scholar]
- 8. Nørholm M. H. (2010) A mutant Pfu DNA polymerase designed for advanced uracil-excision DNA engineering. BMC Biotechnol. 10, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bitinaite J., Nichols N. M. (2009) DNA cloning and engineering by uracil excision. Curr. Protoc. Mol. Biol. 86, 3.21.1–3.21.16 [DOI] [PubMed] [Google Scholar]
- 10. Schagger H. (2006) Tricine-SDS-PAGE. Nat. Protocols 1, 16–22 [DOI] [PubMed] [Google Scholar]
- 11. Nilsson I., Sääf A., Whitley P., Gafvelin G., Waller C., von Heijne G. (1998) Proline-induced disruption of a transmembrane α-helix in its natural environment. J. Mol. Biol. 284, 1165–1175 [DOI] [PubMed] [Google Scholar]
- 12. Doering T. L., Englund P. T., Hart G. W. (2001) Detection of glycophospholipid anchors on proteins. Curr. Protoc. Mol. Biol. 22, 17.8.1–17.8.13 [DOI] [PubMed] [Google Scholar]
- 13. Monné M., Gafvelin G., Nilsson R., von Heijne G. (1999) N-tail translocation in a eukaryotic polytopic membrane protein. Synergy between neighboring transmembrane segments. Eur. J. Biochem. 263, 264–269 [DOI] [PubMed] [Google Scholar]
- 14. Poisson G., Chauve C., Chen X., Bergeron A. (2007) FragAnchor, a large-scale predictor of glycosylphosphatidylinositol anchors in eukaryote protein sequences by qualitative scoring. Genomics Proteomics Bioinformatics 5, 121–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Johansson M., Nilsson I., von Heijne G. (1993) Positively charged amino acids placed next to a signal sequence block protein translocation more efficiently in Escherichia coli than in mammalian microsomes. Mol. Gen. Genet. 239, 251–256 [DOI] [PubMed] [Google Scholar]
- 16. Nilsson I. M., von Heijne G. (1993) Determination of the distance between the oligosaccharyltransferase active site and the endoplasmic reticulum membrane. J. Biol. Chem. 268, 5798–5801 [PubMed] [Google Scholar]
- 17. Walmsley A. R., Hooper N. M. (2003) Glycosylation efficiency of Asn-Xaa-Thr sequons is independent of distance from the C terminus in membrane dipeptidase. Glycobiology 13, 641–646 [DOI] [PubMed] [Google Scholar]
- 18. Meri S., Lehto T., Sutton C. W., Tyynelä J., Baumann M. (1996) Structural composition and functional characterization of soluble CD59. Heterogeneity of the oligosaccharide and glycophosphoinositol (GPI) anchor revealed by laser-desorption mass spectrometric analysis. Biochem. J. 316, 923–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Newstead S., Kim H., von Heijne G., Iwata S., Drew D. (2007) High-throughput fluorescent-based optimization of eukaryotic membrane protein overexpression and purification in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 104, 13936–13941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. von Heijne G., Gavel Y. (1988) Topogenic signals in integral membrane proteins. Eur. J. Biochem. 174, 671–678 [DOI] [PubMed] [Google Scholar]
- 21. Nilsson I., von Heijne G. (1991) A de novo designed signal peptide cleavage cassette functions in vivo. J. Biol. Chem. 266, 3408–3410 [PubMed] [Google Scholar]
- 22. Nilsson I., Whitley P., von Heijne G. (1994) The COOH-terminal ends of internal signal and signal-anchor sequences are positioned differently in the ER translocase. J. Cell Biol. 126, 1127–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Howell S., Lanctôt C., Boileau G., Crine P. (1994) A cleavable N-terminal signal peptide is not a prerequisite for the biosynthesis of glycosylphosphatidylinositol-anchored proteins. J. Biol. Chem. 269, 16993–16996 [PubMed] [Google Scholar]
- 24. Walmsley A. R., Zeng F., Hooper N. M. (2001) Membrane topology influences N-glycosylation of the prion protein. EMBO J. 20, 703–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hessa T., Kim H., Bihlmaier K., Lundin C., Boekel J., Andersson H., Nilsson I., White S. H., von Heijne G. (2005) Recognition of transmembrane helices by the endoplasmic reticulum translocon. Nature 433, 377–381 [DOI] [PubMed] [Google Scholar]
- 26. Kodukula K., Micanovic R., Gerber L., Tamburrini M., Brink L., Udenfriend S. (1991) Biosynthesis of phosphatidylinositol glycan-anchored membrane proteins. Design of a simple protein substrate to characterize the enzyme that cleaves the COOH-terminal signal peptide. J. Biol. Chem. 266, 4464–4470 [PubMed] [Google Scholar]
- 27. Sohail A., Sun Q., Zhao H., Bernardo M. M., Cho J. A., Fridman R. (2008) MT4-(MMP17) and MT6-MMP (MMP25), A unique set of membrane-anchored matrix metalloproteinases. Properties and expression in cancer. Cancer Metastasis Rev. 27, 289–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Brinckerhoff C. E., Matrisian L. M. (2002) Matrix metalloproteinases. A tail of a frog that became a prince. Nat. Rev. Mol. Cell Biol. 3, 207–214 [DOI] [PubMed] [Google Scholar]
- 29. Kozlosky C. J., Maraskovsky E., McGrew J. T., VandenBos T., Teepe M., Lyman S. D., Srinivasan S., Fletcher F. A., Gayle R. B., 3rd, Cerretti D. P. (1995) Ligands for the receptor tyrosine kinases hek and elk. Isolation of cDNAs encoding a family of proteins. Oncogene 10, 299–306 [PubMed] [Google Scholar]
- 30. Cruz-García F., Gómez A., Zúñiga J. J., Plasencia J., Vázquez-Ramos J. M. (2003) Cloning and characterization of a COBRA-like gene expressed de novo during maize germination. Seed Sci. Res. 13, 209–217 [Google Scholar]
- 31. Gerber L. D., Kodukula K., Udenfriend S. (1992) Phosphatidylinositol glycan (PI-G) anchored membrane proteins. Amino acid requirements adjacent to the site of cleavage and PI-G attachment in the COOH-terminal signal peptide. J. Biol. Chem. 267, 12168–12173 [PubMed] [Google Scholar]
- 32. Yan W., Shen F., Dillon B., Ratnam M. (1998) The hydrophobic domains in the carboxyl-terminal signal for GPI modification and in the amino-terminal leader peptide have similar structural requirements. J. Mol. Biol. 275, 25–33 [DOI] [PubMed] [Google Scholar]
- 33. Rothman J. E., Lodish H. F. (1977) Synchronized transmembrane insertion and glycosylation of a nascent membrane protein. Nature 269, 775–780 [DOI] [PubMed] [Google Scholar]
- 34. Lundin C., Johansson S., Johnson A. E., Näslund J., von Heijne G., Nilsson I. (2007) Stable insertion of Alzheimer Aβ peptide into the ER membrane strongly correlates with its length. FEBS Lett. 581, 3809–3813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nilsson I., von Heijne G. (2000) Glycosylation efficiency of Asn-Xaa-Thr sequons depends both on the distance from the C terminus and on the presence of a downstream transmembrane segment. J. Biol. Chem. 275, 17338–17343 [DOI] [PubMed] [Google Scholar]
- 36. Petersen T. N., Brunak S., von Heijne G., Nielsen H. (2011) SignalP 4.0, discriminating signal peptides from transmembrane regions. Nat. Meth. 8, 785–786 [DOI] [PubMed] [Google Scholar]
- 37. Eisenhaber B., Bork P., Eisenhaber F. (1999) Prediction of potential GPI-modification sites in proprotein sequences. J. Mol. Biol. 292, 741–758 [DOI] [PubMed] [Google Scholar]
- 38. Pierleoni A., Martelli P. L., Casadio R. (2008) PredGPI, a GPI-anchor predictor. BMC Bioinformatics 9, 392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Caras I. W. (1991) An internally positioned signal can direct attachment of a glycophospholipid membrane anchor. J. Cell Biol. 113, 77–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Vidugiriene J., Vainauskas S., Johnson A. E., Menon A. K. (2001) Endoplasmic reticulum proteins involved in glycosylphosphatidylinositol-anchor attachment. Photocross-linking studies in a cell-free system. Eur. J. Biochem. 268, 2290–2300 [DOI] [PubMed] [Google Scholar]
- 41. Yan W., Ratnam M. (1995) Preferred sites of glycosylphosphatidylinositol modification in folate receptors and constraints in the primary structure of the hydrophobic portion of the signal. Biochemistry 34, 14594–14600 [DOI] [PubMed] [Google Scholar]
- 42. Moran P., Caras I. W. (1991) Fusion of sequence elements from nonanchored proteins to generate a fully functional signal for glycophosphatidylinositol membrane anchor attachment. J. Cell Biol. 115, 1595–1600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Udenfriend S., Kodukula K. (1995) How glycosylphosphatidylinositol-anchored membrane proteins are made. Annu. Rev. Biochem. 64, 563–591 [DOI] [PubMed] [Google Scholar]
- 44. Eisenhaber B., Bork P., Eisenhaber F. (1998) Sequence properties of GPI-anchored proteins near the ω-site. Constraints for the polypeptide binding site of the putative transamidase. Protein Eng. 11, 1155–1161 [DOI] [PubMed] [Google Scholar]
- 45. Toh Y. K., Kamariah N., Maurer-Stroh S., Roessle M., Eisenhaber F., Adhikari S., Eisenhaber B., Grüber G. (2011) Structural insight into the glycosylphosphatidylinositol transamidase subunits PIG-K and PIG-S from yeast. J. Struct. Biol. 173, 271–281 [DOI] [PubMed] [Google Scholar]
- 46. Ohishi K., Inoue N., Maeda Y., Takeda J., Riezman H., Kinoshita T. (2000) Gaa1p and gpi8p are components of a glycosylphosphatidylinositol (GPI) transamidase that mediates attachment of GPI to proteins. Mol. Biol. Cell 11, 1523–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Vainauskas S., Maeda Y., Kurniawan H., Kinoshita T., Menon A. K. (2002) Structural requirements for the recruitment of Gaa1 into a functional glycosylphosphatidylinositol transamidase complex. J. Biol. Chem. 277, 30535–30542 [DOI] [PubMed] [Google Scholar]
- 48. Chen R., Anderson V., Hiroi Y., Medof M. E. (2003) Proprotein interaction with the GPI transamidase. J. Cell. Biochem. 88, 1025–1037 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.