Background: Increased translation of p53 mRNA is important for stress induction of p53 protein.
Results: RPL26 and nucleolin proteins interact with each other and with a double-stranded RNA structure in p53 mRNA to regulate p53 translation after stress.
Conclusion: Nucleolin represses basal p53 translation and recruits RPL26 after cellular stress to enhance p53 translation.
Significance: This study has increased our knowledge of the molecular details of the process regulating p53 induction.
Keywords: DNA Damage Response, p53, Protein-Nucleic Acid Interaction, Protein-Protein Interactions, Translational Control, RPL26, Nucleolin
Abstract
Ribosomal protein RPL26 enhances p53 translation after DNA damage, and this regulation depends upon interactions between the 5′- and 3′-UTRs of human p53 mRNA (Takagi, M., Absalon, M. J., McLure, K. G., and Kastan, M. B. (2005) Cell 123, 49–63; Chen, J., and Kastan, M. B. (2010) Genes Dev. 24, 2146–2156). In contrast, nucleolin (NCL) suppresses the translation of p53 mRNA and its induction after DNA damage. We confirmed reports that RPL26 and NCL interact with each other and then explored the potential role of this interaction in the translational control of p53 after stress. NCL repression of p53 translation utilizes both the 5′- and 3′-UTRs of p53 mRNA, and NCL binds to the same 5′-3′-UTR interaction region that is critical for the recruitment of RPL26 to p53 mRNA after DNA damage. We also found that NCL is able to oligomerize, consistent with a model in which NCL stabilizes this double-stranded RNA structure. We found that the RNA-binding domain of NCL participates in binding to p53 mRNA, is required for both NCL dimerization and NCL-mediated translational repression, and is the domain of NCL that interacts with RPL26. Excessive RPL26 disrupts NCL dimerization, and point mutations in the NCL-interacting region of RPL26 reduce NCL-RPL26 interactions and attenuate both RPL26 binding to human p53 mRNA and p53 induction by RPL26. These observations suggest a model in which the base pairings in the p53 UTR interaction regions are critical for both translational repression and stress induction of p53 by NCL and RPL26, respectively, and that disruption of a NCL-NCL homodimer by RPL26 may be the switch between translational repression and activation after stress.
Introduction
The p53 tumor suppressor is a critical mediator of cellular responses to DNA damage and other cellular stresses (3, 4). Although an increased half-life of p53 protein via modulation of the Mdm2 E3 ubiquitin ligase contributes to p53 induction after stress (5, 6), an important role for translational regulation of p53 mRNA is emerging (7–10). Our laboratory recently reported that increased translation of p53 mRNA by RPL26 is a requisite step for optimal induction of p53 protein following DNA damage (1, 11). We also demonstrated that RPL26 protein binds to a double-stranded RNA structure containing complementary sequences of the 5′- and 3′-UTRs of p53 mRNA after DNA damage (2). Disruption of the base pairing within this region abrogates the ability of RPL26 both to bind p53 mRNA and to stimulate p53 translation. We also identified nucleolin as an inhibitor of p53 translation (1). However, the mechanisms by which nucleolin represses p53 translation and its relationship to RPL26 in this regulation were not elucidated.
Nucleolin is a highly conserved, multifunctional protein implicated in transcription, translation, DNA replication, recombination, and repair (12, 13). As the most abundant non-ribosomal protein in the nucleolus, nucleolin affects rRNA synthesis, processing, and transportation (12). Among its activities, nucleolin binds a consensus “nucleolin-responsive element” in pre-rRNA and stabilizes a stem-loop structure of this sequence (14, 15). Nucleolin has also been reported to function as a stress-responsive mRNA-binding protein, binding to UTRs of mRNA in a sequence-nonspecific manner to either stabilize the mRNAs (16–18) or regulate their translation (19).
We report here that repression of the translation of p53 mRNA by nucleolin depends on both 5′- and 3′-UTR sequences. Nucleolin simultaneously and independently binds to both of the RNA sequences of the base pairing interaction region that is also critical for the translational regulation of p53 by RPL26 after DNA damage. Disruption of the base pairs in this region blunts nucleolin (NCL)3 repression. The RNA-binding domain (RBD) of nucleolin is indispensible for NCL binding to p53 mRNA, NCL self-interaction, NCL-RPL26 interaction, and NCL-dependent translational repression. Disruption of the NCL-RPL26 interaction attenuates both RPL26 binding to human p53 mRNA and p53 induction by RPL26. These observations provide new insights about the mechanisms of translational control of human p53 and suggest a collaborative relationship between NCL and RPL26 in this translational regulation.
EXPERIMENTAL PROCEDURES
Cell Culture, Stable Cell Line, and Transfection
MCF-7 cells were maintained in DMEM plus 10% FBS. MCF-7 cell lines stably expressing luciferase reporters were infected with a pBABE-based retrovirus carrying luciferase coding sequence, luciferase coding sequence after 145 nucleotides of the human p53 5′-UTR, or luciferase coding sequence flanked by 145 nucleotides of the human p53 5′-UTR and the full-length 3′-UTR, and the infected cells were selected and maintained in DMEM plus 10% FBS supplemented with 1 μg/ml puromycin. Plasmids were transfected into cells using Lipofectamine 2000 (Invitrogen), and nucleolin siRNA duplex (Invitrogen) was introduced using Lipofectamine RNAiMAX (Invitrogen).
Plasmids, Oligonucleotides, and Proteins
The plasmids used in this study include pCMV3-FLAG-nucleolin, pEGFP-C3-nucleolin, pCMV-FLAG-RPL26, and pLPCXp53mRNA (1). NCL, RPL26, and their deletion mutants were constructed in the pEGFP-C3 or pCMV-FLAG vector. The oligonucleotides used in this study were synthesized at the Hartwell Center of St. Jude Children's Research Hospital. Native nucleolin protein was purchased from Vaxron Corp. (Rockaway, NJ).
RNA Synthesis and in Vitro Transcription/Translation
Capped p53 mRNA was transcribed in vitro using the mMESSAGE mMACHINE kit (Ambion), followed by a poly(A) tailing kit (Ambion) to add a poly(A) tail modification. The synthesized mRNA was further purified with a MEGAclear column (Ambion) and quantified using NanoDrop spectrometer. RNA without modification was synthesized using a MEGAscript high yield transcription kit (Ambion). All synthesized RNA was purified using a MEGAclear kit (Ambion) according to the manufacturer's protocol. The in vitro transcription/translation reactions were performed using a TnT T7 Quick for PCR DNA kit (Promega, Madison, WI). Briefly, PCR primers for p53 were designed according to the manual and used to amplify the desired gene. Appropriate amounts of PCR products (usually a total of 400 ng of input PCR products/25-μl reaction) were then further used for in vitro transcription/translation in rabbit reticulocyte lysates provided with the kit at 30 °C for 90 min. 20 μCi of [35S]methionine (PerkinElmer Life Sciences) was added to each reaction to label newly synthesized protein. After the reaction, 3 μl of reaction mixture was separated on precast 4–12% SDS-polyacrylamide gel, transferred to nitrocellulose membrane, and subjected to autoradiography. The primers used in this study for in vitro transcription/translation reactions were as follows: human NCL, GGATCCTAATACGACTCACTATAGGAGCCATCATGGTGAAGCTCGCGAAGGCAGG (forward) and CTATTCAAACTTCGTCTTCTTTCCTTGTGGCTT (reverse); and luciferase, TAATACGACTCACTATAGGGAGACCCAAGC (forward) and GATATAGGCGCCAGCAACCGCAC (reverse), with the T7 luciferase plasmid as a template.
Dual-Luciferase Assay
Luciferase assays were performed using the Dual-Luciferase reporter assay system (Promega) according to the manual provided by the manufacturer. Briefly, MCF-7 cells were cotransfected with 2 μg of the indicated protein constructs, 100 ng of −145pGl3ctrl+3′-UTR (145 bases of the 5′-UTR, coding sequence, and the full-length 3′-UTR of human p53 mRNA), and 27 ng of pRL-TK. 24 h post-transfection, cell lysates were prepared and subjected to the reporter assay according to the manufacturer's instructions.
Immunoblotting, Immunoprecipitation, Co-immunoprecipitation, and Immunoprecipitation/RT-PCR
Cell lysates were prepared by freeze-thawing once, followed by incubation in radioimmune precipitation assay buffer for 30 min on ice, and supernatants were analyzed by immunoblotting or immunoprecipitation. For immunoblot analysis, 20-μg protein samples were denatured in an equal volume of SDS sample buffer, separated on 4–12% SDS-polyacrylamide gel, and transferred to nitrocellulose membrane. The blots were probed with primary antibody against p53 (DO-1; Santa Cruz Biotechnology, Santa Cruz, CA), nucleolin (MS-3; Santa Cruz Biotechnology), GFP (FL; Santa Cruz Biotechnology), FLAG (M2; Sigma or Cell Signaling), RPL26 (Bethyl Laboratories), or actin (Sigma). Primary antibody binding was detected by incubation with HRP-conjugated anti-rabbit or anti-mouse secondary antibody and visualized using an ECL system (Amersham Biosciences).
For immunoprecipitations, 1 mg of whole cell extract in radioimmune precipitation assay buffer (1) was cleared using protein A/G PLUS-agarose (Santa Cruz Biotechnology) and rabbit/mouse IgG (Sigma). Precleared lysates were incubated overnight with anti-GFP antibody (Abcam), anti-FLAG antibody M2, anti-p53 antibody (FL393; Santa Cruz Biotechnology), anti-RPL26 antibody, or anti-nucleolin antibody (H250; Santa Cruz Biotechnology). Immunoprecipitated proteins were then washed extensively with lysis buffer and subjected to Western blot analysis as described above. For co-immunoprecipitations, cells were lysed in TGN buffer (1, 30), and the rest of the procedure followed the immunoprecipitation protocol. For immunoprecipitation/RT-PCR, immunoprecipitated protein complexes were split in half: one half was used for immunoblot analysis, and the other half was used for real-time RT-PCR as described previously (1).
Northwestern Blot Analysis
The Northwestern blot analysis was adapted from a procedure described previously (16). Briefly, GFP or GFP-tagged proteins were immunoprecipitated from H1299 cells as described above, separated on 4–12% SDS-polyacrylamide gel, and transferred to nitrocellulose membrane. The blot was then renatured in RNA binding buffer and hybridized to a 5′-end-labeled RNA probe (∼106 cpm/5 ml of hybridization buffer) overnight at ambient temperature. RNA probes were first synthesized using the MEGAscript high yield transcription kit, labeled at the 5′-end using a KinaseMaxTM 5′-end labeling kit (Ambion), and later purified using the MEGAclear kit. After extensive washing with wash buffer at room temperature, the membrane was air-dried for autoradiography at −80 °C.
Real-time RT-PCR
1 μg of total RNA prepared using TRIzol reagent (Invitrogen) was treated with DNase I (Invitrogen) and then reverse-transcribed with a SuperScript first-strand synthesis system (Invitrogen) as recommended by the manufacturer using the random hexamer primer provided in the kit. Real-time RT-PCR was performed using an ABI PRISM 7900HT sequence detection system and an ABI TaqMan One-Step PCR Master Mix Reagents kit. The primer/TaqMan probe set for human p53 was Hs00153340_m1 (20× mixture; Applied Biosystems). Human 18 S rRNA (20×; Applied Biosystems) was used as an internal control. Total RNA extracted from MCF-7 cells was used for a standard curve. The reaction was performed with 20 ng of total RNA in triplicate reactions in a 30-μl volume containing 2× p53 primer/probe and 1× 18 S rRNA primer/probe. Cycling conditions were 25 °C for 10 min, 48 °C for 30 min, 95 °C for 10 min, and 40 cycles at 95 °C for 15 s and 60 °C for 1 min for amplification. The results were analyzed using SDS 2.2 software (Applied Biosystems). For comparison of total human endogenous p53 mRNA levels in cells, the p53 mRNA level was normalized to the internal 18 S rRNA level. For immunoprecipitation/RT-PCR, the absolute human p53 mRNA level was used for analysis.
RESULTS
Repression of p53 Translation by NCL Requires Both p53 5′- and 3′-UTR Sequences
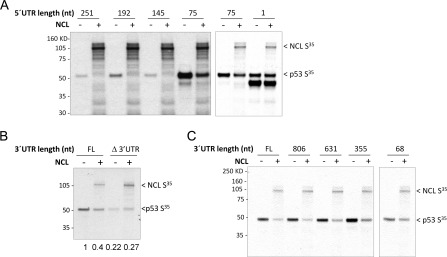
Although nucleolin binds to p53 mRNA and represses its translation (1), the mechanisms by which nucleolin affects p53 translation have not extensively characterized. We explored the mechanisms of p53 translational repression by NCL in both an in vitro translation system and a cell-based reporter system. A series of constructs were generated that contained various lengths of p53 mRNA 5′- and 3′-UTRs, and the ability of nucleolin to repress translation of these constructs was examined in an in vitro rabbit reticulocyte translation system. As noted previously both in cells and in vitro (1), adding a 75-base 5′-UTR sequence enhanced basal translation of p53, but UTR sequences longer than 75 bases then began to reduce basal p53 translation (Fig. 1A), suggesting the presence of basal inhibitory sequences in this 5′-region of p53 mRNA. Nucleolin failed to repress p53 translation in the absence of a 5′-UTR (Fig. 1A, right panel), but nucleolin repressed p53 translation of mRNAs containing 5′-UTR sequences of 75 bases or more, regardless of the basal level of translation (left and right panels). These effects were also seen when purified nucleolin protein was directly added to the system (supplemental Fig. S1A) and were not attributable to nonspecific inhibition of protein translation in this system (supplemental Fig. S1B) or changes in p53 RNA levels (data not shown). Furthermore, they were not altered in the presence of a 5′-m7GpppN cap or 3′-poly(A) tail structure (supplemental Fig. S1C). Translational repression was also evident in a cell-based system using a chimeric construct containing the 5′- and 3′-UTRs of p53 mRNA surrounding the luciferase coding sequence instead of the p53 coding sequence (data not shown). Thus, similar to the requirements of RPL26 to stimulate p53 translation, nucleolin repression of p53 translation requires the 5′-UTR, but not the p53 coding sequence, 5′-m7GpppN cap structure, or 3′-poly(A) tail.
FIGURE 1.
Both 5′- and 3′-UTR sequences of human p53 mRNA are required for translational repression by NCL. A, the 5′-UTR of human p53 mRNA is required for translational repression of p53 by NCL in vitro. NCL repression of translation was evaluated in rabbit reticulocyte lysates containing p53 cDNAs with variable lengths of 5′-UTR sequences (as indicated) plus the full-length p53 coding sequence and the 3′-UTR without (−) or with (+) the NCL coding sequence added. The expression of newly synthesized, [35S]methionine-labeled protein was assessed by autoradiography. The panels merge the results of two independent experiments with different exposure times of the film. B, the 3′-UTR of human p53 mRNA is required for translational repression by NCL in vitro. Translation of p53 mRNA containing 75 bases of the 5′-UTR and the p53 coding sequence without (Δ3′UTR) or with (FL) the full-length 3′-UTR was evaluated in rabbit reticulocyte lysates without (−) or with (+) the NCL coding sequence. The expression of newly synthesized, [35S]methionine-labeled protein and total p53 protein was assessed by autoradiography. The band intensity of 35S-labeled p53 protein in each lane was quantitated using NIH ImageJ software and compared with that in the first lane, and the -fold changes are shown under the panel. C, the first 355 nucleotides (nt) of the p53 3′-UTR are required for nucleolin repression in vitro. p53 cDNA (75 nucleotide of the 5′-UTR plus the full-length coding sequence) with variable lengths of the 3′-UTR sequence was coexpressed without (−) or with (+) the NCL coding sequence in rabbit reticulocyte lysates. The expression of newly synthesized, [35S]methionine-labeled protein was assessed by autoradiography.
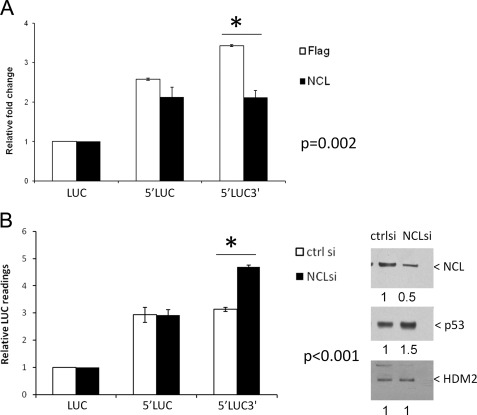
Deletion of the 3′-UTR of p53 decreased the basal level of p53 translation and unexpectedly also abrogated the ability of nucleolin to repress p53 translation in vitro (Fig. 1B). Serial deletion of the 3′-UTR region demonstrated that the first 355 nucleotides of the 3′-UTR were sufficient to mediate translational repression of p53 mRNA by nucleolin (Fig. 1C). Thus, nucleolin repression appears to require both the 5′- and 3′-UTR sequences of p53 mRNA. Similar results were obtained in the cell-based reporter system. Overexpression (Fig. 2A) or knockdown (Fig. 2B) of NCL affected luciferase reporter activity if the 5′- and 3′-UTR sequences of p53 mRNA were both present, but not if the reporter with the 5′-UTR (Fig. 2, A and B) or 3′-UTR (supplemental Fig. S2, A and B) was present alone. Because complete knockdown of NCL is detrimental to the cells, we chose a condition under which nucleolin is only partially depleted (Fig. 2B, right panels). Manipulation of NCL levels in these stable reporter lines did not affect the mRNA level of the luciferase reporter genes (supplemental Fig. S3).
FIGURE 2.
Both 5′- and 3′-UTR sequences of human p53 mRNA are required for translational repression by NCL in cells. A, overexpression of NCL represses the translation of human p53 mRNA in the presence of both 5′- and 3′-UTR sequences. MCF-7 stable cell lines were established to constitutively express a firefly luciferase reporter gene without any UTR sequences of human p53 mRNA (LUC) or containing the 145-nucleotide p53 5′-UTR with (5′LUC3′) or without (5′LUC) the full-length p53 3′-UTR. FLAG-tagged NCL (NCL) or empty vector (Flag) was cotransfected with a Renilla luciferase (19) expression vector (internal control; RL) into these stable cell lines. 24 h post-transfection, modulation of the reporter gene expression was assessed by comparing the LUC/RL ratio of each samples with that of LUC samples. The error bars represent the mean ± S.D. for three independent experiments. *, the p value was calculated using Student's t test. B, knock down of NCL in cells enhances the translation of human p53 mRNA in the presence of both 5′- and 3′-UTR sequences. 40 μm NCL siRNA duplex (NCLsi) (1) or non-target control siRNA (ctrl si) was introduced into the above MCF-7 stable reporter cell lines. 3 days post-transfection, the endogenous NCL levels was assessed by immunoblotting (right panels). Modulation of the reporter gene expression was assessed by comparing the firefly luciferase reading of each sample with that of LUC samples (left panel). The error bars represent the mean ± S.D. for three independent experiments. *, the p value was calculated using Student's t test. The band intensity of NCL (upper right panel), p53 (middle right panel) and HDM2 (lower right panel) proteins was quantitated using NIH ImageJ software, and -fold changes in the intensity compared with those in the NCL siRNA duplex sample are shown under the panels.
Nucleolin Independently Binds Both UTRs in Interaction Regions
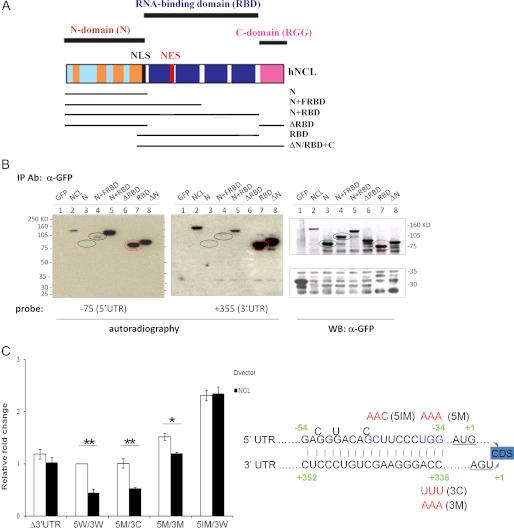
Nucleolin has been reported to bind ribosomal RNA in a sequence- and structure-specific manner, and its RBD has been characterized (14, 15, 20, 21). We initially identified nucleolin in a screen for proteins binding to the 5′-UTR of p53 mRNA (1), but had not demonstrated whether this was a direct interaction. Immunoprecipitation of nucleolin from cells brings down p53 mRNA in an immunoprecipitation/RT-PCR assay (1), but to explore whether nucleolin directly interacts with p53 mRNA, we made a series of epitope-tagged expression constructs coding for various domains of nucleolin protein (Fig. 3A). Using Northwestern blot analysis, we found that full-length nucleolin was able to directly and independently bind both the 5′-UTR sequence (−75 to +1) (Fig. 3B, left panel, lane 2) and the 3′-UTR sequence (first 355 bases) (middle panel, lane 2). Similar results were obtained with immunoprecipitated endogenous nucleolin (data not shown). Domain mapping revealed that the N terminus of nucleolin did not directly bind either of the UTR probes in vitro (Fig. 3B, lane 3) or p53 mRNA in vivo (supplemental Fig. S4, lane 5), and deletion of the N terminus did not affect the ability of nucleolin to bind these sequences (Fig. 3B, lane 8, and supplemental Fig. S4, lane 10). In contrast, the RBD was sufficient for the binding of p53 mRNA in vivo (supplemental Fig. S4, lane 9) and both the 5′- and 3′-UTR sequences in vitro (Fig. 3B, lane 7). Deletion of the RBD abrogated the binding of nucleolin to both sequences (Fig. 3B, lane 6, and supplemental Fig. S4, lane 8). We noted that a construct containing the N terminus plus the first half of the RBD appeared to weakly bind to the 5′-UTR sequence, but did not bind the 3′-UTR sequence (Fig. 3B, lane 4). These interactions were specific to these particular RNA sequences because full-length nucleolin failed to bind a control RNA sequence from the p53 coding sequence region (data not shown). We recently demonstrated that a region of double-stranded RNA containing complementary sequences from the 5′- and 3′-UTRs is critical for translational regulation of human p53 mRNA by RPL26 after stress (2). To investigate if NCL utilizes the same region for regulation, we performed Northwestern blotting with immunoprecipitated nucleolin and probed with labeled oligonucleotides representing the 21-base region of either the 5′- or 3′-UTR strands of the complementary region. Both the 5′- and 3′-UTR probes bound to the immunoprecipitated nucleolin protein (data not shown).
FIGURE 3.
NCL repression requires base pairings within UTR interaction region. A, schematic diagram of nucleolin functional domains adopted from Ref. 12 and nucleolin deletion mutants generated in this study. NLS, nuclear localization signal; NES, nuclear export signal; hNCL, human NCL; FRBD, first half of the RBD. B, NCL and its RBD directly and independently bind both the 5′- and 3′-UTRs of human p53 mRNA. GFP-tagged NCL deletion mutants were transiently transfected into H1299 cells and immunoprecipitated (IP) with anti-GFP antibody. The immunoprecipitated proteins were hybridized with a 32P-5′-end-labeled 5′-UTR (−75 to +1; left panel) or 3′-UTR (first 355 nucleotides; middle panel) RNA probe for Northwestern blot analysis. The immunoprecipitated proteins were also detected by Western blot (WB) analysis using anti-GFP antibody (right panel). C, disruption of the base pairings in UTR interaction region blunts NCL repression. Mutations were introduced into the last 3 bp of the 5′- and 3′-UTR interaction regions (5M/3M; depicted in the diagram on the right-hand side with mutations labeled in red), the middle 3 bases of the 5′-UTR in this region (5IM/3W), or a compensatory mutation (5M/3C) (2) that restores complementarity of the last 3 bp. The Dual-Luciferase reporter assay was performed as described under “Experimental Procedures.” Data shown are the mean ± S.D. for three independent experiments. p values were calculated using Student's t test. *, p < 0.01; **, p < 0.001.
One of the reported properties of nucleolin protein is that it binds both single- and double-stranded RNAs or DNAs and promotes base pairing of homologous or complementary nucleic acids (22–24). Because the 21-nucleotide regions of the 5′- and 3′-UTRs form a double-stranded RNA structure (2) and because nucleolin appeared to bind independently to each of the sequences involved in forming this double-stranded region, we speculated that the binding of NCL to these sequences secures/enhances the interaction between UTRs in this region. To investigate whether the UTR base pairs participate in the translational repression of human p53 mRNA by NCL, we introduced point mutations to disrupt the last 3 bp (5M/3M) (2) or the internal 3 bp (5IM/3W) of the UTR interaction region or to restore the last 3-bp interaction (5M/3C) (2). As long as the UTR interaction remained intact (5W/3W and 5M/3C), the reporter responded to NCL repression (Fig. 3C). Interestingly, the point mutations (5M/3M and 5IM/3W) that would partially disrupt the double-stranded structure in the UTR interaction region enhanced basal reporter expression levels, suggesting that the base pairs between UTRs are involved in regulating basal translation of human p53 mRNA. No alterations of reporter mRNA levels were detected in any samples (supplemental Fig. S5).
Interactions Occur between Nucleolin, RPL26, and p53 mRNA to Regulate p53 Translation
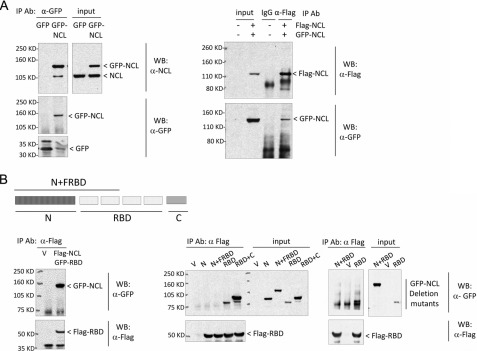
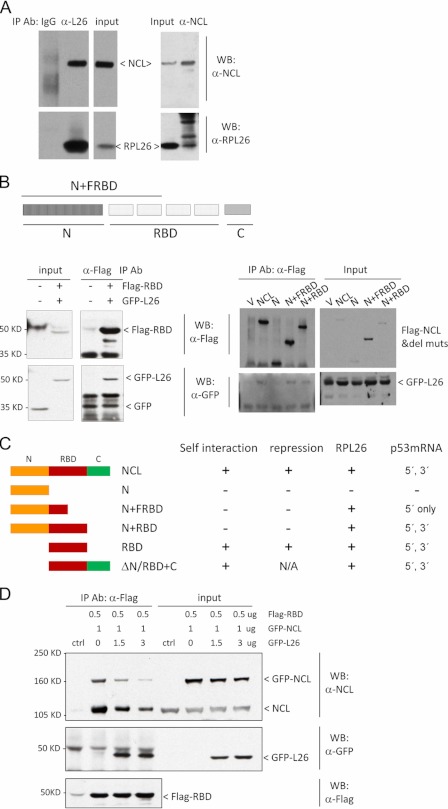
We noted that nucleolin protein appeared to be able to form a homodimer (or homo-oligomer) (Fig. 4A). This dimerization did not require p53 mRNA because it was observed in H1299 cells, which lack p53 genes, and because the NCL interaction was stable in the presence of nucleases that digest both DNA and RNA (data not shown). Domain mapping of nucleolin protein demonstrated that the RBD was sufficient for this self-interaction (Fig. 4B). Interestingly, the N terminus of nucleolin did not interact with the RBD, but its presence blunted nucleolin dimerization (Fig. 4B, middle and right panels). The C terminus of nucleolin did not appear to be involved in these interactions.
FIGURE 4.
NCL complexes with itself at RBD. A, NCL interacts with itself. GFP-tagged NCL was transfected (left panels) or cotransfected with FLAG-tagged NCL (right panels) into MCF-7 cells. GFP- or FLAG-tagged NCL was immunoprecipitated (IP) by anti-GFP or anti-FLAG antibody, respectively, and the bound protein was assessed by immunoblot analysis using anti-nucleolin (upper left panels) and anti-GFP (lower left panels) or anti-FLAG (upper right panel) and GFP (lower right panel) antibodies. 1–2 mg of total cell lysate was used for co-immunoprecipitation in each sample, and 10 μg of lysate was used as input. WB, Western blot. B, NCL interacts with itself at the RBD. The FLAG-tagged NCL RBD was cotransfected with GFP-NCL (left panels); with the GFP-tagged NCL N-terminal domain (N), the N-terminal domain plus the first half of the RBD (N+FRBD), the RBD, or the RBD plus the C-terminal domain (RBD+C) (middle panels); or with the N-terminal domain plus the full-length RBD (N+RBD) (right panels) into MCF-7 cells. FLAG-RBD was immunoprecipitated from cells using anti-FLAG antibody, and the bound proteins were detected by anti-FLAG (lower panels) or anti-GFP (upper panels) antibody. In these experiments, 3 mg of total cell lysate was used for co-immunoprecipitation in each sample, and 30 μg of lysate was used as input. V, empty vector.
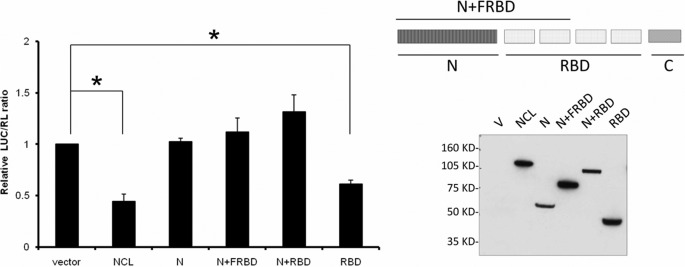
We then investigated whether NCL dimerization/oligomerization is required for its repression of p53 translation. Both full-length NCL and its RBD were able to repress the expression of a luciferase reporter containing both p53 5′-UTR (145 nucleotides) and 3′-UTR (full-length) sequences, whereas NCL deletion mutants (N-terminal domain plus the first half of or full-length RBD) that failed to self-interact were unable to repress the reporter (Fig. 5). No alterations of luciferase reporter mRNA levels were observed (supplemental Fig. S6). In both cases (self-interaction and repression), the nucleolin N-terminal domain seemed to counter the function of its RBD, and we speculate that this could be due to either the highly negatively charged nature of the N-terminal domain in cells or the conformational change in the protein when the N-terminal domain is included. On the basis of reported observations that nucleolin can facilitate annealing of single-stranded nucleic acids (23, 24) and our observations that nucleolin independently bound to each component of the UTR interaction regions (Fig. 3B) and that nucleolin could form a homodimer (Fig. 4), a property associated with translational repression of p53 mRNA (Fig. 5), we hypothesized that nucleolin homodimers facilitate the formation or stability of a repressive double-stranded RNA structure.
FIGURE 5.
NCL self-interaction is required for translational repression. MCF-7 cells were transiently transfected with empty vector (V), FLAG-NCL, or FLAG-tagged NCL deletion mutants together with a firefly luciferase construct (LUC) containing a 145-nucleotide p53 5′-UTR and the full-length p53 3′-UTR plus a control Renilla luciferase expression construct (RL). A simple diagram of the deletion mutants used is included (upper right panel). The relative LUC/RL ratio was calculated by normalizing the LUC/RL ratio of each sample to the ratio in cells transfected with the empty vector and both luciferase reporters. Data shown are the mean ± S.D. for three independent experiments. *, p < 0.0001 (Student's t test) (left panel). The expression level of FLAG-tagged proteins was assessed by immunoblotting with anti-FLAG antibody (lower right panel). FRBD, first half of the RBD.
Because nucleolin directly binds to the same interacting 5′- and 3′-UTR sequences (Fig. 3B) that are critical for RPL26 recruitment to p53 mRNA after damage (2), we explored the potential cooperation between nucleolin and RPL26 proteins in these interactions. Consistent with previous findings (25), we found that endogenous nucleolin and RPL26 proteins interacted with each other (Fig. 6A) and that RPL26 interacted with the RBD of nucleolin (Fig. 6B, left panel), the same domain involved in nucleolin homodimerization (Fig. 4B) and repression (Fig. 5). The N-terminal domain of nucleolin had no obvious effect on this interaction (Fig. 6B, right panel). The results of domain mapping experiments are summarized in Fig. 6C. Because RPL26 bound to the same domain of nucleolin protein as that involved in nucleolin homodimerization, we investigated if RPL26 could affect nucleolin homodimerization. RPL26 protein was able to compete with both full-length cotransfected GFP-nucleolin and endogenous nucleolin for binding to a FLAG-tagged RBD fragment (Fig. 6D), suggesting that RPL26 can inhibit nucleolin homodimerization.
FIGURE 6.
NCL interacts with RPL26. A, NCL interacts with RPL26 in cells. Endogenous NCL or RPL26 (L26) was immunoprecipitated (IP) with anti-nucleolin or anti-RPL26 antibody. The bound protein was assessed using appropriate antibodies. 1–2 mg of total cell lysate was used for co-immunoprecipitation in each sample, and 20 μg of lysate was used as input. WB, Western blot. B, the RBD of NCL is sufficient for NCL-RPL26 interaction. FLAG-tagged RBD (left panels) or FLAG-tagged NCL deletion mutants (del muts; right panels) were cotransfected with GFP-RPL26 into MCF-7 cells. FLAG-tagged proteins were immunoprecipitated with anti-FLAG antibody. The bound proteins were detected by anti-GFP antibody. 1–3 mg of total cell lysate was used for co-immunoprecipitation in each sample, and 30 μg of lysate was used as input. FRBD, first half of the RBD; V, empty vector. C, the schematic diagram summarizes the results from domain mapping experiments described in Figs. 3–6. D, RPL26 disrupts NCL self-interaction. The indicated amounts of FLAG-RBD, GFP-RPL26, and GFP-NCL were cotransfected into MCF-7 cells. FLAG-RBD was immunoprecipitated by anti-FLAG antibody, and the bound proteins were detected using the indicated antibodies. 6 mg of total cell lysate was used for co-immunoprecipitation in each sample, and 30 μg of lysate was used as input.
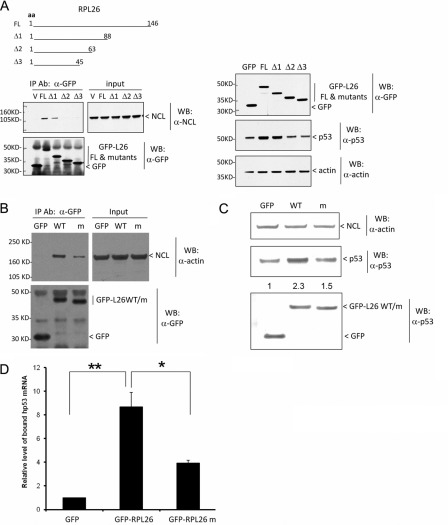
Finally, we explored the functional consequences of the interaction between RPL26 and nucleolin. Deletion analysis of RPL26 narrowed down the interaction region to 26 amino acids (positions 63–88) (Fig. 7A, left panels). These amino acids were also indispensible for p53 induction by RPL26 (Fig. 7A, right panels). Replacing amino acids 80–88 (the last 9 amino acids of the 26 amino acids) with alanine was sufficient to inhibit RPL26 and NCL interaction (Fig. 7B). This multiple-point mutant of RPL26 also reduced the ability to induce p53 protein when overexpressed (Fig. 7C) and the ability to bind endogenous p53 mRNA (Fig. 7D). We also observed that knocking down NCL attenuated the binding of nuclear RPL26 to biotin-labeled p53 mRNA in an in vitro RNA pulldown assay.4 These data suggest that nucleolin-RPL26 interaction at least partially contributes to the binding of RPL26 to p53 mRNA and its regulation of the translation of the message.
FIGURE 7.
NCL-RPL26 interaction affects human p53 induction by RPL26. A, a deletion mutant of RPL26 lost RPL26 binding to NCL (left panels) and the ability of RPL26 to induce p53 in cells (right panels). GFP-tagged RPL26 deletion mutants were transiently transfected into MCF-7 cells and immunoprecipitated (IP) using anti-GFP antibody. The bound NCL was detected using anti-nucleolin antibody (left panels). In this co-immunoprecipitation experiment, 30% of total cell lysate was used as input. The basal p53 protein level was also assessed in these samples before immunoprecipitation by immunoblotting (right panels). aa, amino acids; FL, full-length; V, empty vector; WB, Western blot. B, a point mutant of RPL26 lost RPL26 binding to NCL. GFP-tagged RPL26 point mutants (m; nine alanine replacements of amino acids 80–88) were transiently transfected into MCF-7 cells and immunoprecipitated using anti-GFP antibody 24 h post-transfection. The bound NCL was detected using anti-nucleolin antibody. 3 mg of total cell lysate was used for co-immunoprecipitation in each sample, and 20 μg of lysate was used as input. C, a point mutant of RPL26 lost RPL26 ability to induce p53 in cells. GFP-tagged RPL26 point mutants were transiently transfected into MCF-7 cells, and 24 h post-transfection, the basal p53 protein level was assessed by immunoblotting. D, a point mutant of RPL26 lost RPL26 binding to p53 mRNA in cells. GFP-tagged RPL26 point mutants were transiently transfected into MCF-7 cells and immunoprecipitated using anti-GFP antibody 24 h post-transfection. The bound p53 mRNA was measured by real-time RT-PCR. The bar graph shows the ratio of the bound p53 mRNA level compared with that seen in cells expressing GFP protein. The error bars represent the mean ± S.D. for three experiments. p values were calculated using Student's t test. *, p < 0.01; **, p < 0.001.
DISCUSSION
Expression of eukaryotic genes is tightly regulated at multiple levels, especially under stress conditions. Diverse RNA-binding proteins participate in the regulation of almost every aspect of the mRNA life cycle by binding to unique structures or sequences of the target RNA (15, 26). Protein-protein interactions mediated by specific domains in the RNA-binding proteins increase the complexity of the regulation of gene expression. Here, we have presented evidence for both positive and negative translational control of human p53 mRNA by the multifunctional RNA-binding protein NCL and ribosomal protein L26. We proposed a model (supplemental Fig. S7) in which NCL binds to an intrinsic double-stranded RNA region formed by base pairing between the UTRs of human p53 mRNA in unstressed cells, and based on the reported ability of NCL to stabilize nucleic acid hybrids, dimerization/oligomerization of NCL facilitates the formation/stability of the above repressive double-stranded RNA structure. Stress stimulation results in recruitment of RPL26 to this double-stranded RNA structure occupied by NCL, which disrupts the NCL homodimer by forming NCL-RPL26 heterodimers and facilitates the translation of p53 mRNA.
We suggest that this transition from NCL-NCL homodimer to NCL-RPL26 heterodimer occurs in the nucleus and represents an extraribosomal function of RPL26. At least two pieces of evidence support this suggestion. First, in vitro synthesized human p53 mRNA is able to bind RPL26 in nuclei in the absence of mature ribosomes, and this binding enrichment is enhanced by ionizing irradiation.4 Second, RPL26 is able to pull down endogenous human p53 mRNA from nuclear fractions, and the binding is increased in IR-treated cells (2). Because enhanced binding of RPL26 to p53 mRNA is a critical step for RPL26-dependent p53 induction and because it can occur in the nucleus after stress, RPL26 likely interacts with NCL and binds to p53 mRNA in the nucleus.
p53 induction acts as a double-edged sword. On the one hand, elevated p53 protein levels initiate apoptosis, cell cycle arrest, or senescence to restrain development of tumor cells. On the other hand, in certain settings, p53 induction is detrimental to normal tissue (27). Therefore, transiently blunting the potential detrimental effects of p53 induction could be beneficial in reducing p53-mediated cell death and tissue toxicity in settings of DNA damage or other stresses. Our study and previous investigations have demonstrated that translational regulation of p53 mRNA is a critical step in p53 induction under stress conditions. Several different aspects of this translational regulation could be potential targets for small molecule/drug interventions. The first is the UTR base pairing of p53 mRNA itself, which not only forms a closed structure to facilitate the repression of its own translation by nucleolin under unstressed conditions but also attracts RPL26 recruitment after stress stimuli. We have successfully designed targeting oligonucleotides to disrupt the UTR interaction region of human p53 mRNA, and these oligonucleotides blunt p53 induction and enhance cell survival after various stress stimuli by inhibiting RPL26 binding to p53 mRNA (2). The results reported here now support additional aspects of this regulatory mechanism that are potential targets for modulation of this response, including NCL-NCL, NCL-RPL26, and RPL26-p53 mRNA interactions. Although protein-protein interaction as a potential intervention point has been challenging due to large, featureless, or sporadic interaction surfaces, small molecules, peptides, or mimetics of protein structure have been developed as effective therapeutic reagents (28, 29). We have narrowed down the region of RPL26 that interacts with NCL to 9 continuous amino acids, and mutation of these amino acids interferes with the binding of RPL26 to p53 mRNA as well as p53 induction (Fig. 7). Detailed structural studies of the interaction between these two proteins and with p53 mRNA could help in the design of inhibitors for this interaction.
Supplementary Material
Acknowledgments
We thank Drs. Jacqueline Tait-Mulder and Frederick Derheimer for advice in retroviral packaging and Drs. Lyra Griffiths, Michael Goldstein, and John Crutchley for helpful comments on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants R37ES05777 and P50CA21765. This work was also supported by the American Lebanese Syrian Associated Charities (ALSAC) of St. Jude Children's Research Hospital.

This article contains supplemental Figs. S1–S7.
J. Chen and M. B. Kastan, unpublished data.
- NCL
- nucleolin
- RBD
- RNA-binding domain.
REFERENCES
- 1. Takagi M., Absalon M. J., McLure K. G., Kastan M. B. (2005) Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell 123, 49–63 [DOI] [PubMed] [Google Scholar]
- 2. Chen J., Kastan M. B. (2010) 5′-3′-UTR interactions regulate p53 mRNA translation and provide a target for modulating p53 induction after DNA damage. Genes Dev. 24, 2146–2156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kastan M. B., Bartek J. (2004) Cell cycle checkpoints and cancer. Nature 432, 316–323 [DOI] [PubMed] [Google Scholar]
- 4. Levine A. J., Oren M. (2009) The first 30 years of p53: growing ever more complex. Nat. Rev. Cancer 9, 749–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ashcroft M., Vousden K. H. (1999) Regulation of p53 stability. Oncogene 18, 7637–7643 [DOI] [PubMed] [Google Scholar]
- 6. Wade M., Wang Y. V., Wahl G. M. (2010) The p53 orchestra: Mdm2 and Mdmx set the tone. Trends Cell Biol. 20, 299–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mosner J., Mummenbrauer T., Bauer C., Sczakiel G., Grosse F., Deppert W. (1995) Negative feedback regulation of wild-type p53 biosynthesis. EMBO J. 14, 4442–4449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fu L., Benchimol S. (1997) Participation of the human p53 3′-UTR in translational repression and activation following γ-irradiation. EMBO J. 16, 4117–4125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mazan-Mamczarz K., Galbán S., López de Silanes I., Martindale J. L., Atasoy U., Keene J. D., Gorospe M. (2003) RNA-binding protein HuR enhances p53 translation in response to ultraviolet light irradiation. Proc. Natl. Acad. Sci. U.S.A. 100, 8354–8359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schumacher B., Hanazawa M., Lee M. H., Nayak S., Volkmann K., Hofmann E. R., Hofmann R., Hengartner M., Schedl T., Gartner A. (2005) Translational repression of C. elegans p53 by GLD-1 regulates DNA damage-induced apoptosis. Cell 120, 357–368 [DOI] [PubMed] [Google Scholar]
- 11. Ofir-Rosenfeld Y., Boggs K., Michael D., Kastan M. B., Oren M. (2008) Mdm2 regulates p53 mRNA translation through inhibitory interactions with ribosomal protein L26. Mol. Cell 32, 180–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ginisty H., Sicard H., Roger B., Bouvet P. (1999) Structure and functions of nucleolin. J. Cell Sci. 112, 761–772 [DOI] [PubMed] [Google Scholar]
- 13. Mongelard F., Bouvet P. (2007) Nucleolin: a multiFACeTed protein. Trends Cell Biol. 17, 80–86 [DOI] [PubMed] [Google Scholar]
- 14. Bouvet P., Allain F. H., Finger L. D., Dieckmann T., Feigon J. (2001) Recognition of preformed and flexible elements of an RNA stem-loop by nucleolin. J. Mol. Biol. 309, 763–775 [DOI] [PubMed] [Google Scholar]
- 15. Hall K. B. (2002) RNA-protein interactions. Curr. Opin. Struct. Biol. 12, 283–288 [DOI] [PubMed] [Google Scholar]
- 16. Yang C., Maiguel D. A., Carrier F. (2002) Identification of nucleolin and nucleophosmin as genotoxic stress-responsive RNA-binding proteins. Nucleic Acids Res. 30, 2251–2260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang J., Tsaprailis G., Bowden G. T. (2008) Nucleolin stabilizes Bcl-xL messenger RNA in response to UVA irradiation. Cancer Res. 68, 1046–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang Y., Bhatia D., Xia H., Castranova V., Shi X., Chen F. (2006) Nucleolin links to arsenic-induced stabilization of GADD45α mRNA. Nucleic Acids Res. 34, 485–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fähling M., Mrowka R., Steege A., Nebrich G., Perlewitz A., Persson P. B., Thiele B. J. (2006) Translational control of collagen prolyl 4-hydroxylase-α(I) gene expression under hypoxia. J. Biol. Chem. 281, 26089–26101 [DOI] [PubMed] [Google Scholar]
- 20. Allain F. H., Bouvet P., Dieckmann T., Feigon J. (2000) Molecular basis of sequence-specific recognition of pre-ribosomal RNA by nucleolin. EMBO J. 19, 6870–6881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bouvet P., Jain C., Belasco J. G., Amalric F., Erard M. (1997) RNA recognition by the joint action of two nucleolin RNA-binding domains: genetic analysis and structural modeling. EMBO J. 16, 5235–5246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dickinson L. A., Kohwi-Shigematsu T. (1995) Nucleolin is a matrix attachment region DNA-binding protein that specifically recognizes a region with high base-unpairing potential. Mol. Cell. Biol. 15, 456–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Thyagarajan B., Lundberg R., Rafferty M., Campbell C. (1998) Nucleolin promotes homologous DNA pairing in vitro. Somat. Cell Mol. Genet. 24, 263–272 [DOI] [PubMed] [Google Scholar]
- 24. Hanakahi L. A., Bu Z., Maizels N. (2000) The C-terminal domain of nucleolin accelerates nucleic acid annealing. Biochemistry 39, 15493–15499 [DOI] [PubMed] [Google Scholar]
- 25. Yanagida M., Shimamoto A., Nishikawa K., Furuichi Y., Isobe T., Takahashi N. (2001) Isolation and proteomic characterization of the major proteins of the nucleolin-binding ribonucleoprotein complexes. Proteomics 1, 1390–1404 [DOI] [PubMed] [Google Scholar]
- 26. Kishore S., Luber S., Zavolan M. (2010) Deciphering the role of RNA-binding proteins in the post-transcriptional control of gene expression. Brief. Funct. Genomics 9, 391–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Westphal C. H., Hoyes K. P., Canman C. E., Huang X., Kastan M. B., Hendry J. H., Leder P. (1998) Loss of atm radiosensitizes multiple p53 null tissues. Cancer Res. 58, 5637–5639 [PubMed] [Google Scholar]
- 28. Morelli X., Bourgeas R., Roche P. (2011) Chemical and structural lessons from recent successes in protein-protein interaction inhibition (2P2I). Curr. Opin. Chem. Biol. 15, 475–481 [DOI] [PubMed] [Google Scholar]
- 29. Mori M., Manetti F., Botta M. (2011) Targeting protein-protein and protein-nucleic acid interactions for anti-HIV therapy. Curr. Pharm. Des. 17, 3713–3728 [DOI] [PubMed] [Google Scholar]
- 30. Canman C. E., Lim D.-S., Cimprich K. A., Taya Y., Tamai K., Sakaguchi K., Appella E., Kastan M. B., Siliciano J. D. (1998) Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science 281, 1677–1679 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.