Background: Drugs that target steroid receptors are notoriously promiscuous, causing an array of off-target side effects.
Results: Reversal of the historical mutation H853R in the mineralocorticoid receptor (MR) fully restores agonist activity by mometasone furoate, an MR antagonist.
Conclusion: A single residue outside of the ligand-binding pocket toggles agonism versus antagonism response by MR to synthetic ligands.
Significance: Ancestral proteins are ideal tools to elucidate the mechanisms of drug selectivity.
Keywords: Drug Design, Glucocorticoids, Molecular Evolution, Molecular Pharmacology, Steroid Hormone Receptor, Ancestral Gene Resurrection
Abstract
Steroid receptors (SRs) are the largest family of metazoan transcription factors and control genes involved in development, endocrine signaling, reproduction, immunity, and cancer. The entire hormone receptor system is driven by a molecular switch triggered by the binding of small lipophilic ligands. This makes the SRs ideal pharmaceutical targets, yet even the best clinically approved synthetic steroidal agonists are prone to cross-reactivity and off-target pharmacology. The mechanism underlying this promiscuity is derived from the fact that SRs share common structural features derived from their evolutionary relationship. More often than not, rational attempts to probe SR drug selectivity via mutagenesis fail even when high quality structural and functional data are available due to the fact that important mutations often result in nonfunctional receptors. This highlights the fact that SRs suffer from instability, preventing in-depth mutational analysis and hampering crystallization of key receptor-ligand complexes. We have taken a unique approach to address this problem by using a resurrected ancestral protein to determine the structure of a previously intractable complex and identified the structural mechanisms that confer activation and selectivity for a widely used glucocorticoid, mometasone furoate. Moreover, we have identified a single residue located outside of the ligand-binding pocket that controls mometasone furoate antagonism versus agonism in the human mineralocorticoid receptor.
Introduction
Complex life depends on intra- and intercellular communication, whereby secreted messengers are detected by specific receptors to regulate metabolism, reproduction, cell cycles, and more. This coordination tightly controls cellular activity within the higher organism. Poor coordination of these processes can result in many health concerns, including metabolic disorders, reproductive diseases, and cancer. Over time, a vast repertoire of receptors has evolved to respond to small chemical stimuli, making them attractive pharmacological targets. However, because most receptors belong to large classes of evolutionary-related proteins that show high structural similarity, targeting a single receptor subtype is a major challenge. Poor selectivity can cause serious off-target side effects, as seen in the treatments of major depression (3), heart disease (4, 5), asthma (4, 5), and allergies (6).
To fully understand the mechanisms supporting receptor-ligand recognition, selectivity, and activation, robust structure-function relationships must be built from extensive mutational analysis and ligand design. This analysis is hindered for several reasons. First, amino acid residues conferring protein function and ligand specificity between homologous receptors can be difficult to identify among the vastly more prevalent neutral mutations that accumulate over time (7). Second, restrictive mutations that are not directly related to the protein-ligand interaction can accumulate in extant proteins, preventing the tolerance of function-shifting mutations (8). Third, many mutations are destabilizing and result in loss of protein function, complicating the distinction between an effect that is specific to the protein-ligand interaction versus an effect that is globally inactivating to the protein (7). Although most conclusions are currently drawn from function-killing mutations, the insight needed to understand ligand selectivity among a class of homologous proteins would be better drawn from function-shifting mutations that preserve receptor activation.
These problems have hindered the design of selective drugs that target human steroid receptors (SRs).2 SRs are a family of ligand-regulated transcription factors that control genes involved in development, endocrine signaling, reproduction, immunity, and cancer (9). This makes them attractive pharmaceutical targets. Although SRs show exquisite selectivity for their endogenous hormones, SR-targeting drugs tend to be promiscuous and cause many off-target side effects (10, 11). This is because SRs, consisting of the estrogen receptor, progesterone receptor (PR), androgen receptor (AR), mineralocorticoid receptor (MR), and glucocorticoid receptor (GR), descended from a common ancestor >500 million years ago (see Fig. 1A) and show high structural similarity (9, 12). In the absence of ligand, SRs remain partially unfolded and associate with heat shock proteins (13, 14). This instability is necessary to permit the conformational changes that drive receptor activation upon ligand binding (1, 2, 7, 14–16), but it has limited our ability to probe receptor-ligand interactions via mutagenesis, as many mutations of interest disable the protein entirely.
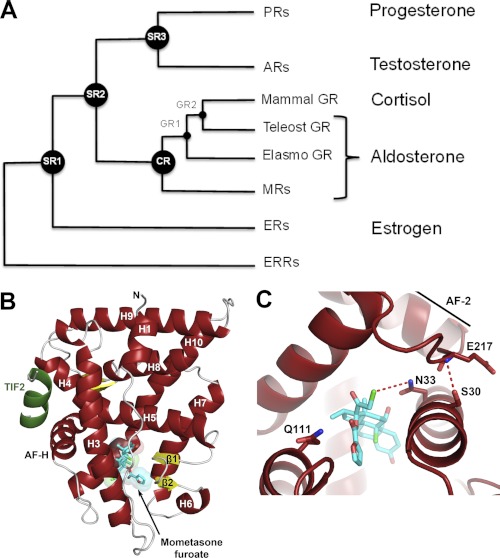
FIGURE 1.
Evolutionary history of corticosteroid receptors and structure of AncGR2-TIF2-MOF. A, simplified phylogenetic tree depicting the evolution of corticosteroid receptors. Activating hormones are listed on the right. ER, estrogen receptor; ERR, estrogen receptor-related receptor. B, structure of AncGR2 (red) in complex with human TIF2 (green) and MOF (cyan). C, AncGR2 LBP residues (red) with MOF shown (cyan).
With the advancement of whole-gene synthesis and pioneering efforts made in computational and evolutionary biology, it is now possible to predict and “resurrect” ancestral genes. Ancestral gene reconstruction is used to study the molecular evolution of a biological system (12, 17–19) but shows promising applications to the process of drug design. By comparing two ancestral proteins from nodes on an evolutionary tree, we are provided with a smaller subset of possible amino acid replacements to dissect between related proteins that have different ligand specificities. Our efforts can be focused on fewer residues when probing structure-function relationships than when looking only at extant proteins. This approach therefore allows us to avoid interference from neutral and restrictive mutations that have accumulated over time. Furthermore, unlike many extant proteins, ancestral proteins show remarkable tolerance toward changes in function-shifting residues, making them more stable under laboratory conditions.
We hypothesized that one could exploit these ancestral proteins to understand cross-pharmacology in human SRs. Ancestral SRs (AncSRs) are more tolerant to mutation than their extant descendants (20), and their molecular and structural evolution has already been characterized (8, 12, 20–23). AncSRs therefore make an effective model to study the structural mechanisms of SR pharmacology. To achieve this goal, we determined the structure of the ancestral glucocorticoid receptor 2 (AncGR2) ligand-binding domain in complex with a fragment of human transcription intermediary factor 2 (TIF2) and mometasone furoate (MOF). We draw upon functionally important historical amino acid substitutions to elucidate the mechanisms driving GR activation for this widely used glucocorticoid. Furthermore, we use a combination of structural analysis and functional assays to explain the selectivity of this drug against MR and AR and strong cross-reactivity with PR.
EXPERIMENTAL PROCEDURES
Chemicals and Reagents
Chemicals were purchased from Sigma or Fisher. The pMALCH10T and pMCSG7-based protein expression vectors were gifts from J. Tesmer (University of Texas, Austin) and J. Sondek (University of North Carolina, Chapel Hill), respectively. The empty pSG5-based mammalian expression vector and human MR and GR pSG5 constructs, pFRluc reporter, and phRLtk reporter were gifts from J. Thornton (University of Oregon, Eugene).
Protein Expression and Purification
AncGR2 (GenBankTM accession number EF631976.1) in a pMALCH10T vector was transformed into Escherichia coli strain BL21(DE3) and expressed as a maltose-binding protein-His fusion. Cultures (1.3 liters in Terrific Broth) were grown to an A600 of 0.6–0.7 and induced with 400 μm isopropyl β-d-thiogalactopyranoside and 50 μm MOF at 30 °C for 4 h. Cell mass was collected by centrifugation at 4000 × g for 15 min, lysed, and purified by nickel affinity chromatography. The maltose-binding protein-His tag was cleaved by tobacco etch virus protease at 4 °C overnight with simultaneous dialysis into buffer containing 300 mm NaCl, 20 mm Tris (pH 7.4), 5% glycerol and purified to homogeneity by nickel affinity, followed by gel filtration chromatography.
Crystallization, Data Collection, and Structural Refinement
Pure AncGR2 was concentrated to 3–5 mg/ml in buffer containing 300 mm NaCl, 20 mm Tris (pH 7.4), 5% glycerol, 50 μm CHAPS, and 50 μm MOF. Crystals were grown via hanging drop vapor diffusion at 4 °C from solutions containing 0.75 μl of AncGR2-TIF2-MOF solution, 0.75 μl of 1.5–3 m ammonium formate, and a dodecapeptide derived from the GR coactivator human TIF2 (+H3N-ENALLRYLLDKD-CO2−, SynBioSci Corp.). Crystals were cryoprotected by immersion in 1.5–3 m ammonium formate containing 25% glycerol and flash-frozen in liquid nitrogen. Data to a resolution of 2.5 Å were collected at the Southeast Regional Collaborative Access Team at the Advanced Photon Source (Argonne, IL) (Table 1). The structure of the AncGR2-MOF-TIF2 complex was solved by molecular replacement using PHASER in the CCP4 software suite. Model building and refinement were performed using Refmac and Coot. Cavity volumes were calculated using CASTp, and figures were generated in PyMOL. The refined AncGR2-MOF structure has been deposited in the Protein Data Bank (code 4E2J).
TABLE 1.
Data collection and refinement statistics
AU, asymmetric unit; r.m.s.d., root mean square deviation; PDB, Protein Data Bank.
| Resolution (highest shell; Å) | 2.50 (2.59–2.50) |
| Space group | P61 |
| Unit cell dimensions | a = 104.4, b = 104.4, c = 143.9 Å; α = β = 90.0°, γ = 120.0° |
| No. of reflections | 30,710 |
| Rsym (highest shell)a | 7.7% (42.2%) |
| Completeness (highest shell) | 99.90% (98.96%) |
| Average redundancy (highest shell) | 8.0 (7.9) |
| I/σ | 29.3 (5.3) |
| Monomers/AU | 2 |
| No. of protein atoms/AU | 4195 |
| No. of ligand atoms/AU | 85 |
| No. of waters/AU | 151 |
| Rworkingb (Rfree)c | 20.5 (25.5) |
| Average B-factors (Å2) | |
| Protein | 45.0 |
| Ligand | 53.5 |
| Water | 45.5 |
| r.m.s.d. | |
| Bond lengths (Å) | 0.005 |
| Bond angles | 1.078° |
| PDB code | 4E2J |
a Rsym = Σ|I − 〈I〉|/Σ|I|, where I is the observed intensity, and 〈I〉 is the average intensity of several symmetry-related observations.
b Rworking = Σ‖Fo| − |Fc‖/Σ|Fo, where Fo and Fc are the observed and calculated structure factors, respectively.
c Rfree = Σ‖Fo| − |Fc‖/Σ|Fo for 7% of the data not used at any stage of the structural refinement.
Mutagenesis
Wild-type AncGR1 (GenBankTM accession number JF896321.1) and AncGR2 were subcloned into a pMCSG7-maltose-binding protein-His expression vector, and the following mutations were created from these constructs: AncGR1-S106P, AncGR1-S106P/L111Q, AncGR1-S106P/L111A, AncGR2-P106S, AncGR2-P106S/Q111L, and AncGR2-P106S/Q111A. Mutagenesis was performed using a QuikChange II XL kit (Stratagene).
Ligand Binding Assays
Wild-type or mutant AncGR1 or AncGR2 was expressed as described above and assayed prior to tobacco etch virus protease cleavage as purified maltose-binding protein fusion proteins. All fluorescence polarization experiments were performed in buffer containing 150 mm NaCl, 10 mm HEPES (pH 7.4), 5 mm DTT, 3 mm EDTA, and 0.005% Tween 20. Binding affinity for dexamethasone-fluorescein was measured with a constant concentration of 12 nm dexamethasone and a variable protein concentration of 10−10–10−5 m. Competition assays were performed at a protein concentration 1.2 times its binding affinity for dexamethasone in the presence of 12 nm dexamethasone and 10−10–10−5 m competing ligand. Data were processed with GraphPad Prism 5. Statistical significance was determined by two-factor analysis of variance (ANOVA), and individual comparisons were made with Tukey's honestly significant difference (HSD) post hoc tests.
In-cell Activation Assays
All ancestral, and mutant ligand-binding domains were cloned into a pSG5 expression vector immediately following a Gal4 DNA-binding domain and a GR hinge sequence. CHO-K1 cells were grown and maintained in phenol red-free complete α-minimal essential medium (Invitrogen) supplemented with 10% charcoal/dextran-stripped FBS (Invitrogen) and penicillin/streptomycin. Cells grown in 96-well assay plates were transfected at 70–90% confluence with 1 ng of receptor, 100 ng of upstream activator sequence-driven firefly luciferase reporter (pFRluc), and 0.1 ng of constitutive Renilla luciferase reporter (phRLtk) for 4 h using Lipofectamine 2000 in Opti-MEM I (Invitrogen). Transfections were ended by replacement with complete α-minimal essential medium, and cells were allowed to recover overnight. After recovery, cells were treated in triplicate with 10−12–10−6 m ligand or vehicle (Me2SO) in complete α-minimal essential medium for 24 h (final working Me2SO of 1%) and then assayed with Dual-Glo luciferase substrate (Promega). Firefly activity was normalized to Renilla activity, and the -fold increase in activation was calculated relative to the vehicle control. Dose-response curves were generated in GraphPad Prism 5. Statistical significance was determined by two-factor ANOVA, and individual comparisons were made with Tukey's HSD post hoc tests.
RESULTS
AncGR2-TIF2-MOF Crystal Structure
MOF is a powerful topical anti-inflammatory drug for the skin and airways and is the active ingredient of Nasonex, Asmanex, and Elocon (24). Although MOF has been in clinical use for over 24 years, it suffers from severe cross-pharmacology, resulting in unwanted side effects and limiting its use to topical applications. MOF strongly activates GR, cross-reacts with PR, and is selective against AR and MR. The ternary AncGR2-TIF2-MOF crystal structure reveals the structural basis for MOF binding to vertebrate GRs (Fig. 1, B and C, and supplemental Fig. S1). The hydrogen bond network that is required for activation of corticoid receptors (25) is intact and is stabilized by a dipole-dipole interaction between MOF C21-Cl and AncGR2 Asn-33. MOF binding requires a rearrangement of the helix H6-H7 region of the receptor to accommodate the large 17α-furoate moiety, inducing a 200-Å3 (1.3-fold) increase in the volume of the ligand-binding pocket relative to dexamethasone; this highlights the ability of SRs to expand their ligand-binding pockets to accommodate exogenous ligands (26, 27). The AncGR2-TIF2-MOF structure also reveals that a strong H-bond is not possible between MOF and Gln-111 of GR (Fig. 1C), an interaction that plays a critical role in the specific recognition of 17α-OH-substituted ligands and is absolutely required for cortisol activation (8). Instead, hydrophobic interactions replace this interaction in a fashion analogous to the structure of the GR-fluticasone furoate complex (28).
Structural and Evolutionary Basis for PR Cross-reactivity
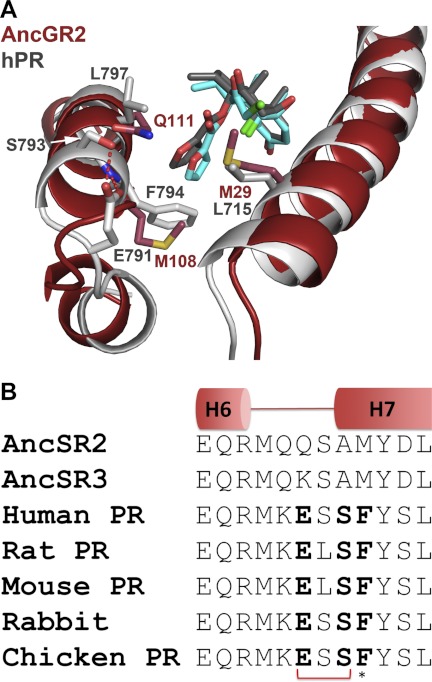
The AncGR2-TIF2-MOF structure allows for the direct structural comparison of GR-MOF and PR-MOF complexes and reveals how additional space in the H6-H7 loop region is created to accommodate the furoate moiety. PR residues 791ESSF794 on H7 appear to play a key role in allowing strong MOF binding by expanding the pocket via a conserved Glu-791–Ser-793 H-bond between the H6-H7 loop and H7 (Fig. 2A). Steric bulk provided by PR Phe-794 between H7 and H3 maintains space for the 17α-furoate moiety and contributes a hydrophobic interaction via the aromatized side chain (26). This motif is strictly conserved among PRs but is not present in AncSR2 (the common ancestor of all 3-keto-SRs) or AncSR3 (the common ancestor of PR and AR) (Figs. 1A and 2B). Therefore, PR response to MOF was probably a late evolutionary derivation resulting in this cross-reactivity.
FIGURE 2.
Structural basis for off-target activation of PR. A, the human PR (hPR)-MOF complex (white; Protein Data Bank code 1SR7) superimposed on the AncGR2-MOF complex (red). PR residue Phe-794 maintains space for the 17α-furoate moiety and contributes a hydrophobic interaction via the aromatized side chain (15). PR residues 791ESSF794 on H7 appear to play a key role in allowing strong MOF binding by positioning the H6-H7 loop and H7 via a conserved Glu-791–Ser-793 H-bond. B, this motif is strictly conserved among extant PRs but is not present in AncSR2.
Structural and Evolutionary Basis for Selectivity against MR
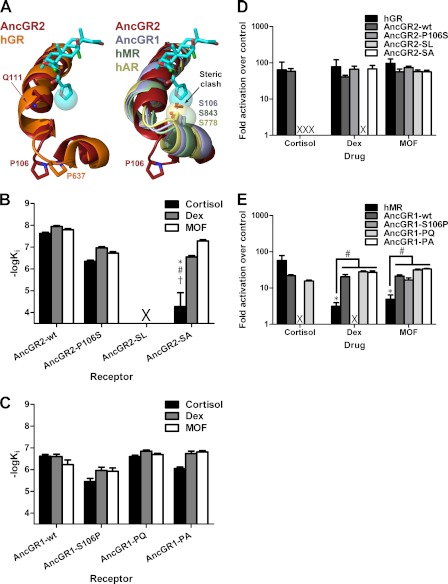
Our structure also suggests a mechanism for the selectivity of MOF against MR and AR. H6 and H7, which border the 17α-binding area, are partially unwound and stabilized by Pro-637/Pro-106 in GR/AncGR2, accommodating the furoate moiety (Fig. 3A, left); MR, AR, and AncGR1 have a serine at this site that caps H7, positioning the helix within 2.5 Å of where the furoate would rest, creating a steric incompatibility (Fig. 3A, right). We have shown in previous work that, during the evolution of GR, S106P and L111Q substitutions were critical in both reshaping the H6-H7 region of the receptor and generating a new H-bond with the 17-OH moiety of cortisol, the endogenous glucocorticoid (8, 22). To test the effect of reversing these critical substitutions with respect to MOF binding affinity, we generated two AncGR2 mutants, P106S and P106S/Q111L, and measured their binding affinities for cortisol, dexamethasone, and MOF using fluorescence polarization competition assays against dexamethasone-fluorescein. The P106S reversal reduced the affinity of AncGR2 for all three ligands by only an order of magnitude (Fig. 3B). This result was surprising because the P106S mutation was identified as the driving force behind the H6-H7 rearrangement required to open space in the receptor for specific recognition of hormones with C17 substituents (13, 22). Because MOF binding requires this structural rearrangement (Fig. 3A), Pro-106 likely plays a role in stabilizing the H6-H7 loop in a productive binding mode but is not absolutely required to induce this structural change. AncGR2-P106S/Q111L, which is known to be inactive to endogenous ligands (8), did not bind dexamethasone-fluorescein (supplemental Fig. S2). This prevented competition assays on this mutant but suggested that H7 indeed repositions to place Leu-111 in contact with C17 of the steroid. This generates a polar incompatibility with C17-OH-containing steroids, such as cortisol and dexamethasone, and introduces a steric clash with the bulky furoate substituent of MOF. To test this hypothesis, we generated a P106S/L111A mutant, designed to alleviate this steric clash in the Pro-106 background, which restored binding to MOF and dexamethasone (Fig. 3B). As expected, cortisol binding was only marginally restored because cortisol does not contain the additional bulky hydrophobic group present on MOF to stabilize the core of the receptor in the absence of the critical Gln-111–17-OH H-bond. Interestingly, dexamethasone binding was more fully restored than cortisol binding, presumably due to additional interactions on its modified backbone. Thus, the H6-H7 region of the receptor can adopt an expanded conformation in the absence of Pro-106, suggesting that the H6-H7 region of GRs is inherently flexible, allowing it to adapt to ligand-induced perturbation. This reshapes our understanding of the role of the H6-H7 region within the ligand-binding domain in the recognition of synthetic glucocorticoids.
FIGURE 3.
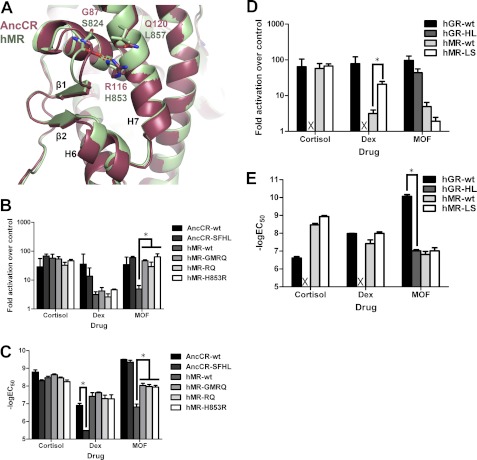
Binding of MOF by modern and ancestral SRs requires expansion of the LBP. A, structure of the 17α-binding pocket. Like PR and AncGR2, human GR (hGR) has an extended H6-H7 loop conformation (left); MR, AR, and AncGR1 have a tightened H6-H7 that would create a steric incompatibility with the furoate (right). B and C, the binding affinities of AncGR2 (B) and AncGR1 (C) mutants for the indicated ligand were measured by fluorescence polarization competition with dexamethasone-fluorescein (Dex). AncGR2-P106S/Q111L (AncGR2-SL) did not bind dexamethasone-fluorescein, and competition experiments could not be performed for this receptor. Statistical analyses were performed using two-factor ANOVA, with Tukey's HSD post hoc tests used for individual comparisons. Comparisons found to be statistically significant to p < 0.05 are marked. *, compared with the same ligand binding for the wild-type receptor; #, compared with dexamethasone binding for the same mutant; †, compared with MOF binding for the same mutant. AncGR1-SA, AncGR2-P106S/Q111A; AncGR1-PQ, AncGR1-S106P/L111Q; AncGR1-PA, AncGR1-S106P/L111A. D and E, receptor activation for GR-like (D) and MR-like (E) receptors was measured by Dual-Luciferase reporter gene activation in transiently transfected CHO-K1 cell cultures. The mean ± S.E. is shown (n = 3). Statistical analyses were performed using two-factor ANOVA, with Tukey's HSD post hoc tests used for individual comparisons. Comparisons found to be statistically significant to p < 0.05 are marked. *, compared with activation of the same receptor by cortisol; #, comparisons made as indicated on the figure. X, no binding or activation observed.
We have shown previously that AncGR1, which preceded the evolution of AncGR2, is a low sensitivity MR-like receptor with activation by both mineralocorticoids and glucocorticoids (22). Because MOF is selective against MR, we reasoned that MOF would display similar selectivity against AncGR1. Surprisingly, MOF bound AncGR1 with an affinity comparable with dexamethasone and cortisol (Fig. 3C), indicating that AncGR1 H7 had already acquired the plasticity needed to accommodate the bulky furoate moiety. The forward mutations AncGR1-S106P and AncGR1-S106P/L111Q had no effects specific to a particular ligand, but the AncGR1-S106P/L111A mutation selectively reduced cortisol binding while leaving dexamethasone and MOF binding unaffected. This is presumably due to the removal of a 17α-interaction. These data show that receptor-ligand interactions at the 17α-site are important for effective ligand binding, although poor interactions here can be surmounted by stronger interactions elsewhere along the ligand scaffold.
A Single Residue Controls MOF Selectivity and Transcriptional Activity
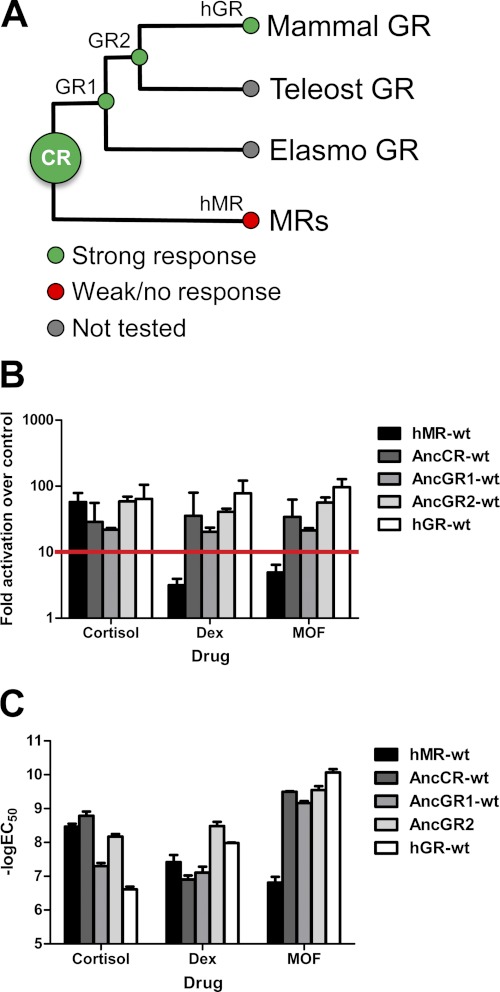
To determine the structural differences between GR and MR that govern MOF recognition, we characterized the ability of MOF to drive luciferase reporter gene activation across the entire ancestral corticosteroid phylogeny. Although MOF only very weakly activated MR (Fig. 4A), it strongly activated AncGR1 and the ancestral corticosteroid receptor (AncCR) with a subnanomolar potency, comparable with the strong activation seen in AncGR2 and GR (Fig. 4, A and B). This provides further evidence that the corticoid receptors from AncCR to AncGR2 are able to unwind H6-H7 to accommodate the MOF 17α-furoate moiety without requiring the S106P substitution.
FIGURE 4.
Activation of modern and ancestral corticosteroid receptors by synthetic glucocorticoids. A, corticoid receptor phylogeny with response to synthetic glucocorticoids (dexamethasone (Dex)/MOF) is shown. Full agonism is shown in green, and weak or no agonism is shown in red. Human GR (hGR) and MR (hMR) were used to represent extant mammalian GR and MR. The -fold activation (B) and potency (C) of corticosteroid receptor ligand-binding domains were measured via Dual-Luciferase reporter gene activation in transiently transfected CHO-K1 cells. The mean ± S.E. is shown (n = 3). For the purpose of this research, activation below 10-fold over the control (B, red line) was considered weak agonism/antagonism, whereas activation above this threshold was considered full agonism.
Furthermore, we have shown that MOF exhibits its selectivity for GR over MR not via a difference in potency but rather in efficacy: although MOF binds MR, with an ∼100 nm potency, it is unable to stabilize an active receptor conformation (25). Thus, MR must have accumulated epistatic changes that prohibit activation from this drug. We therefore examined the importance of residues that changed on the lineage leading to MR with respect to MOF activation. Mutation of residues in or near the ligand-binding pocket had no significant impact on MOF activation without affecting receptor activation toward cortisol and dexamethasone, consistent with our fluorescence polarization competition assays (Fig. 3, D and E) (data not shown). We therefore looked for changes outside of the ligand-binding pocket and activation function surface. Arg-116 and Gln-120 in AncCR, corresponding to His-853 and Leu-857 in MR, respectively, are located on the solvent-exposed face of H7 and interact with the main chain of the loop between H5 and β1, at AncCR residues Gly-87 and Met-89 (MR residues Ser-824 and Phe-826) (Fig. 5A). These residues are ∼14–18 Å from the furoate moiety of MOF and closest to the B ring of the steroid (10–16 Å), yet reversal of all four residues in MR to their ancestral states (MR-GMRQ) completely restored MOF activation (Fig. 5B). We narrowed down the cause of this effect, first to those residues on helix 7 (hMR-RQ), and further to the single residue at MR site 853. Reversal of this site via the substitution H853R conferred full MOF activation (Fig. 5B). In wild-type MR, His-853 interacts with the main chain atoms in the H5-β1 loop and appears to stabilize the MR-like configuration of H7, which must unwind to support activation by ligands with bulky C17α substituents. The stronger interaction provided by an arginine substitution at this site stabilized MR-H853R to enable MOF activation (Figs. 5A and 6). Importantly, these changes are neutral with respect to activation by cortisol: neither the EC50 nor activation of cortisol was affected by the MOF selectivity mutations (Fig. 5, B and C), indicating that the structural determinants of MOF activation are unique from those that support the endogenous ligand recognition. Introducing the equivalent forward substitutions in AncCR (AncCR-SFHL) failed to abrogate MOF response (Fig. 5B), which is in line with the more promiscuous phenotype of the ancestral protein. Intriguingly, making the equivalent site mutations horizontally between MR and GR (MR-H853L/L857S and GR-L647H/S651L) not only failed to enable MOF activation in MR but also abrogated activation by cortisol and dexamethasone in GR (Fig. 5, D and E). Mutations at these residues during the evolution of GRs were previously identified to be destabilizing to GRs, contributing to the low affinity but high selectivity of modern GRs for endogenous glucocorticoids (20). Here, disruption of this site in GR fully destabilized the active receptor during cortisol and dexamethasone binding. In contrast, MOF expanded the ligand-binding pocket (LBP) to make additional hydrophobic contacts offered by the furoate ring (Fig. 6) and was able to stabilize the active conformation, albeit at a much lower potency than in wild-type GR (Fig. 5, D and E).
FIGURE 5.
Distal residues control corticosteroid specificity. A, key residues preceding β-sheet 1 and H7 in AncCR and human MR (hMR) were cross-mutated. The -fold activation (B) and potency (C) were measured via Dual-Luciferase reporter gene activation in transiently transfected CHO-K1 cells. Dex, dexamethasone. The same residues in GR and MR were cross-mutated, and the -fold activation (D) and potency (E) were measured via Dual-Luciferase reporter gene activation in transiently transfected CHO-K1 cells. The mean ± S.E. is shown (n = 3). Statistical analyses were performed using two-factor ANOVA, with Tukey's HSD post hoc tests used for individual comparisons. Comparisons found to be statistically significant to p < 0.05 are marked (*). hGR-HL, human GR-L647H/S651L; hMR-LS, human MR-H853L/L857S. X, no binding or activation observed.
FIGURE 6.
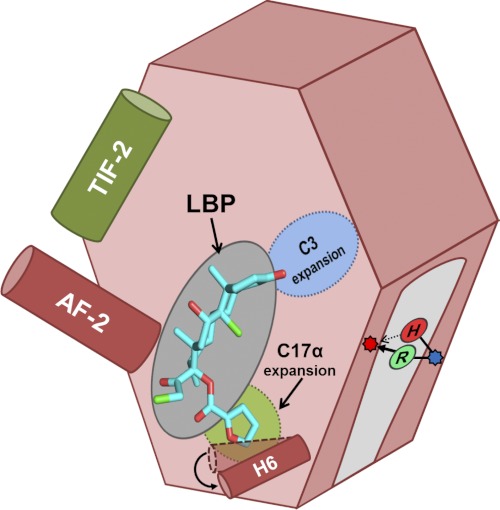
Schematic summarizing relevant features of SR LBP that dictate MOF activation. Activation occurs when a ligand (e.g. MOF (cyan)) binds to the LBP, stabilizing the AF-2 helix (dark red) to allow for coactivator binding (e.g. TIF2 (dark green)) and subsequent transcriptional control. In GRs and PRs, the LBP can expand to accommodate steroids that are substituted at C3 (blue) or C17α (light green). In MR, we identified a single site outside of the LBP that can toggle MOF agonism versus antagonism, ostensibly by forming a bridge between H7 (blue dot) and the H5-β1 loop (red dot). In wild-type MR, His-853 makes a weak hydrogen bond that cannot support MOF activation (red H); the historical substitution to a positively charged arginine (green R) strengthens this interaction, restoring activation.
We anticipate that the findings produced by this study will be applicable to ligands that protrude into extrasteroidal binding regions within the LBP. Furthermore, the finding that ligand specificity is strongly influenced by structural features that lie well outside of the LBP must be taken into consideration during the development of future drugs. The fact that these sites could not be identified using extant proteins highlights the power of using ancestral gene reconstruction to identify the obscure conserved structural mechanisms that support activation via endogenous versus synthetic ligands that may be exploited by selective therapeutics.
DISCUSSION
We have successfully adapted ancestral gene reconstruction to shed light on the structural mechanisms of drug selectivity for SRs. Our approach combines structural and evolutionary biology to overcome many of the obstacles that frequently hinder protein research using modern proteins. It is well known that function-shifting amino acid changes are not tolerated well in modern proteins because most proteins are only moderately stable (7, 15, 16). They display a narrow thermal window of activity dictated by the effects of natural selection on both thermal and kinetic stability (15) and by the accumulation of neutral mutations over evolutionary time (7). A fine balance is necessary to allow small perturbations or signals, such as ligand binding, to functionally alter protein structure: although too little stability prevents proper protein folding, too much stability prevents a receptor from adopting an active conformation in response to stimuli within the host organism. We are therefore limited by the effects of both natural selection and neutral drift, as we are left with mesophilic proteins to use for structure-function analysis. This is exemplified in the SR family and, in particular, with modern GRs, which are notoriously difficult to manipulate under laboratory conditions (23, 29). Furthermore, modern proteins have accumulated millions of years of neutral mutations that make it difficult to identify functionally important amino acid residues, as well as restrictive mutations that can further prohibit mutational analysis.
Workarounds to these problems are limited and frequently involve the incorporation of stabilizing mutations. Although this approach does improve the stability of modern proteins, including GR (23, 29), mutations such as these may alter the way ligands interact with their target receptors. As a result, the behavior of these mutants may not accurately mirror the behavior of the wild-type proteins. In contrast, ancestral proteins are subjected to rigorous testing during the reconstruction process to ensure their behavior is consistent with the behavior of other proteins within their phylogeny (e.g. that the structural mechanisms for activation are conserved). Ancestral proteins are inherently more tolerant to mutation and may serve as ideal models in which to study structure-activity relationships for moderately stable eukaryotic proteins (8, 22, 23). Even when the resurrection of an entire protein is not feasible, the insertion of ancestral residues in modern proteins can increase stability and enhance adaptability and tolerance to mutations (30). In addition, we have found that ancestral proteins tend to be more promiscuous to synthetic ligands or drug activation, especially in cases in which the ancestral proteins display a more promiscuous phenotype than the extant proteins for endogenous ligands. Thus, resurrected proteins may permit the crystallization and functional analysis of previously intractable complexes due to their enhanced stability and promiscuity.
We have shown that, by mirroring what has been done in evolutionary studies aimed at discovering the structural mechanism that conferred hormone selectivity, ancestral proteins may be used to examine cross-pharmacology among homologous proteins. The advantages of using ancestral proteins to study the structural mechanisms of drug promiscuity lie not only in their enhanced stability but also in locating the structural features that contribute to differences in ligand recognition. Ancestral gene reconstruction therefore provides an elegant solution to some of the troubling problems that currently interfere with the process of drug design.
Supplementary Material

This article contains supplemental Figs. S1 and S2.
The atomic coordinates and structure factors (code 4E2J) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- SR
- steroid receptor
- PR
- progesterone receptor
- AR
- androgen receptor
- MR
- mineralocorticoid receptor
- GR
- glucocorticoid receptor
- AncSR
- ancestral SR
- AncGR
- ancestral GR
- TIF2
- transcription intermediary factor 2
- MOF
- mometasone furoate
- ANOVA
- analysis of variance
- HSD
- honestly significant difference
- AncCR
- ancestral corticosteroid receptor
- LBP
- ligand-binding pocket.
REFERENCES
- 1. Nagy L., Schwabe J. W. (2004) Mechanism of the nuclear receptor molecular switch. Trends Biochem. Sci. 29, 317–324 [DOI] [PubMed] [Google Scholar]
- 2. Kallenberger B. C., Love J. D., Chatterjee V. K., Schwabe J. W. (2003) A dynamic mechanism of nuclear receptor activation and its perturbation in a human disease. Nat. Struct. Biol. 10, 136–140 [DOI] [PubMed] [Google Scholar]
- 3. Remick R. A. (1988) Anticholinergic side effects of tricyclic antidepressants and their management. Prog. Neuropsychopharmacol. Biol. Psychiatry 12, 225–231 [DOI] [PubMed] [Google Scholar]
- 4. Decalmer P. B., Chatterjee S. S., Cruickshank J. M., Benson M. K., Sterling G. M. (1978) Beta-blockers and asthma. Br. Heart J. 40, 184–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hanania N. A., Singh S., El-Wali R., Flashner M., Franklin A. E., Garner W. J., Dickey B. F., Parra S., Ruoss S., Shardonofsky F., O'Connor B. J., Page C., Bond R. A. (2008) The safety and effects of the beta-blocker, nadolol, in mild asthma: an open-label pilot study. Pulm. Pharmacol. Ther. 21, 134–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Roy M., Dumaine R., Brown A. M. (1996) HERG, a primary human ventricular target of the nonsedating antihistamine terfenadine. Circulation 94, 817–823 [DOI] [PubMed] [Google Scholar]
- 7. Taverna D. M., Goldstein R. A. (2002) Why are proteins marginally stable? Proteins 46, 105–109 [DOI] [PubMed] [Google Scholar]
- 8. Bridgham J. T., Ortlund E. A., Thornton J. W. (2009) An epistatic ratchet constrains the direction of glucocorticoid receptor evolution. Nature 461, 515–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Carson-Jurica M. A., Schrader W. T., O'Malley B. W. (1990) Steroid receptor family: structure and functions. Endocr. Rev. 11, 201–220 [DOI] [PubMed] [Google Scholar]
- 10. Stahn C., Löwenberg M., Hommes D. W., Buttgereit F. (2007) Molecular mechanisms of glucocorticoid action and selective glucocorticoid receptor agonists. Mol. Cell. Endocrinol. 275, 71–78 [DOI] [PubMed] [Google Scholar]
- 11. Madauss K. P., Stewart E. L., Williams S. P. (2007) The evolution of progesterone receptor ligands. Med. Res. Rev. 27, 374–400 [DOI] [PubMed] [Google Scholar]
- 12. Thornton J. W. (2001) Evolution of vertebrate steroid receptors from an ancestral estrogen receptor by ligand exploitation and serial genome expansions. Proc. Natl. Acad. Sci. U.S.A. 98, 5671–5676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bledsoe R. K., Montana V. G., Stanley T. B., Delves C. J., Apolito C. J., McKee D. D., Consler T. G., Parks D. J., Stewart E. L., Willson T. M., Lambert M. H., Moore J. T., Pearce K. H., Xu H. E. (2002) Crystal structure of the glucocorticoid receptor ligand-binding domain reveals a novel mode of receptor dimerization and coactivator recognition. Cell 110, 93–105 [DOI] [PubMed] [Google Scholar]
- 14. Gee A. C., Katzenellenbogen J. A. (2001) Probing conformational changes in the estrogen receptor: evidence for a partially unfolded intermediate facilitating ligand binding and release. Mol. Endocrinol. 15, 421–428 [DOI] [PubMed] [Google Scholar]
- 15. Godoy-Ruiz R., Ariza F., Rodriguez-Larrea D., Perez-Jimenez R., Ibarra-Molero B., Sanchez-Ruiz J. M. (2006) Natural selection for kinetic stability is a likely origin of correlations between mutational effects on protein energetics and frequencies of amino acid occurrences in sequence alignments. J. Mol. Biol. 362, 966–978 [DOI] [PubMed] [Google Scholar]
- 16. DePristo M. A., Weinreich D. M., Hartl D. L. (2005) Missense meanderings in sequence space: a biophysical view of protein evolution. Nat. Rev. Genet 6, 678–687 [DOI] [PubMed] [Google Scholar]
- 17. Gaucher E. A., Thomson J. M., Burgan M. F., Benner S. A. (2003) Inferring the paleoenvironment of ancient bacteria on the basis of resurrected proteins. Nature 425, 285–288 [DOI] [PubMed] [Google Scholar]
- 18. Jermann T. M., Opitz J. G., Stackhouse J., Benner S. A. (1995) Reconstructing the evolutionary history of the artiodactyl ribonuclease superfamily. Nature 374, 57–59 [DOI] [PubMed] [Google Scholar]
- 19. Wouters M. A., Liu K., Riek P., Husain A. (2003) A despecialization step underlying evolution of a family of serine proteases. Mol. Cell 12, 343–354 [DOI] [PubMed] [Google Scholar]
- 20. Carroll S. M., Ortlund E. A., Thornton J. W. (2011) Mechanisms for the evolution of a derived function in the ancestral glucocorticoid receptor. PLoS Genet. 7, e1002117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Carroll S. M., Bridgham J. T., Thornton J. W. (2008) Evolution of hormone signaling in elasmobranchs by exploitation of promiscuous receptors. Mol. Biol. Evol. 25, 2643–2652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ortlund E. A., Bridgham J. T., Redinbo M. R., Thornton J. W. (2007) Crystal structure of an ancient protein: evolution by conformational epistasis. Science 317, 1544–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bridgham J. T., Carroll S. M., Thornton J. W. (2006) Evolution of hormone-receptor complexity by molecular exploitation. Science 312, 97–101 [DOI] [PubMed] [Google Scholar]
- 24. Tan R. A., Corren J. (2008) Mometasone furoate in the management of asthma: a review. Ther. Clin. Risk Manag. 4, 1201–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bledsoe R. K., Madauss K. P., Holt J. A., Apolito C. J., Lambert M. H., Pearce K. H., Stanley T. B., Stewart E. L., Trump R. P., Willson T. M., Williams S. P. (2005) A ligand-mediated hydrogen bond network required for the activation of the mineralocorticoid receptor. J. Biol. Chem. 280, 31283–31293 [DOI] [PubMed] [Google Scholar]
- 26. Madauss K. P., Deng S. J., Austin R. J., Lambert M. H., McLay I., Pritchard J., Short S. A., Stewart E. L., Uings I. J., Williams S. P. (2004) Progesterone receptor ligand-binding pocket flexibility: crystal structures of the norethindrone and mometasone furoate complexes. J. Med. Chem. 47, 3381–3387 [DOI] [PubMed] [Google Scholar]
- 27. Nettles K. W., Bruning J. B., Gil G., O'Neill E. E., Nowak J., Guo Y., Hughs A., Kim Y., DeSombre E. R., Dilis R., Hanson R. N., Joachimiak A., Greene G. L. (2007) Structural plasticity in the estrogen receptor ligand-binding domain. EMBO Rep. 8, 563–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Biggadike K., Bledsoe R. K., Hassell A. M., Kirk B. E., McLay I. M., Shewchuk L. M., Stewart E. L. (2008) X-ray crystal structure of the novel enhanced affinity glucocorticoid agonist fluticasone furoate in the glucocorticoid receptor ligand-binding domain. J. Med. Chem. 51, 3349–3352 [DOI] [PubMed] [Google Scholar]
- 29. Seitz T., Thoma R., Schoch G. A., Stihle M., Benz J., D'Arcy B., Wiget A., Ruf A., Hennig M., Sterner R. (2010) Enhancing the stability and solubility of the glucocorticoid receptor ligand-binding domain by high throughput library screening. J. Mol. Biol. 403, 562–577 [DOI] [PubMed] [Google Scholar]
- 30. Yamashiro K., Yokobori S., Koikeda S., Yamagishi A. (2010) Improvement of Bacillus circulans β-amylase activity attained using the ancestral mutation method. Protein Eng. Des. Sel. 23, 519–528 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.