Background: Release of microvesicles, including exosomes, is a novel mechanism of intercellular communication. At Drosophila synapses, the transmembrane Wnt-binding protein Evi/Wls is released in vesicles.
Results: Evi-exosome release requires Rab11, Syntaxin 1A, and Myosin5.
Conclusion: We established an in vivo system to elucidate the mechanisms of exosomal release.
Significance: This is the first in vivo characterization of exosomal communication in the nervous system.
Keywords: Drosophila Genetics, Exosomes, Neuroscience, Synapses, Wnt Signaling, Myosin 5, Rab11, Syntaxin1A, Neuromuscular Junction, Vesicular Release
Abstract
Wnt signaling plays critical roles during synaptic development and plasticity. However, the mechanisms by which Wnts are released and travel to target cells are unresolved. During synaptic development, the secretion of Drosophila Wnt1, Wingless, requires the function of Evenness Interrupted (Evi)/Wls, a Wingless-binding protein that is secreted along with Wingless at the neuromuscular junction. Given that Evi is a transmembrane protein, these studies suggested the presence of a novel vesicular mechanism of trans-synaptic communication, potentially in the form of exosomes. To establish the mechanisms for the release of Evi vesicles, we used a dsRNA assay in cultured cells to screen for genes that when down-regulated prevent the release of Evi vesicles. We identified two proteins, Rab11 and Syntaxin 1A (Syx1A), that were required for Evi vesicle release. To determine whether the same mechanisms were used in vivo at the neuromuscular junction, we altered the activity of Rab11 and Syx1A in motoneurons and determined the impact on Evi release. We found that Syx1A, Rab11, and its effector Myosin5 were required for proper Evi vesicle release. Furthermore, ultrastructural analysis of synaptic boutons demonstrated the presence of multivesicular bodies, organelles involved in the production and release of exosomes, and these multivesicular bodies contained Evi. We also used mass spectrometry, electron microscopy, and biochemical techniques to characterize the exosome fraction from cultured cells. Our studies revealed that secreted Evi vesicles show remarkable conservation with exosomes in other systems. In summary, our observations unravel some of the in vivo mechanisms required for Evi vesicle release.
Introduction
Synaptic transmission through chemical and electrical synapses is the major means by which signals are propagated within neuronal circuits. However, recent studies suggest alternative pathways to convey signals between neurons. In particular, the release of entire vesicles containing membrane receptors and other cargo has been documented to transmit signals between pre- and postsynaptic compartments (1). One mechanism of intercellular vesicular transmission is through exosomes, small vesicles released by many different cell types. They can serve to purge obsolete cellular components (2), to mediate the presentation of major histocompatibility complex (MHC) class II molecules (3), to transfer receptor ligands (4, 5), soluble proteins, and lipids (6–8), and to transport genetic information in the form of mRNAs and microRNAs (9–12). For example, in the immune system, antigen-presenting cells secrete exosomes bearing MHC bound to pathogen antigens. These exosomes activate humoral and T helper responses that can protect against acute infection (13, 14).
Although exosomes have been shown to have potent biological effects in the recipient cells, their function in the nervous system is virtually unknown. We recently demonstrated that a Wnt/Wingless (Wg)3 signal is transmitted through the release of intact vesicles, containing the Wg-binding protein Evenness Interrupted (Evi)/Wntless (Wls), from perisynaptic regions of synaptic boutons at the Drosophila larval neuromuscular junction (NMJ). The function of this vesicular release is to carry hydrophobic Wg from its site of release to distant areas of the postsynaptic muscle membrane (the subsynaptic reticulum (SSR)), where Drosophila Frizzled-2 (DFz2) receptors are localized (15). We proposed that Evi vesicles could represent exosomes (1), but the exact nature of the vesicles or the mechanisms required for their release were not known. A role for exosomes in the nervous system is beginning to emerge (16). However, virtually nothing is known about their function, their biosynthesis, or the signaling pathways that utilize this type of intercellular communication.
The finding that Wnt signals could potentially use an exosomal release pathway offered a unique opportunity to examine this mode of transmission in an in vivo system. Because such systems used to study the function of exosomes are sorely lacking (17), the sophisticated genetic tools available in Drosophila and the critical role of Wnt signaling during synapse development offer a highly amenable approach to study mechanisms of exosome function.
We took advantage of the observation that Evi-GFP is sorted to and released from vesicular structures both in cultured Drosophila cells as well as at the larval NMJ. First, we developed a cell culture assay to begin the identification of factors required for the Evi vesicle release. Using this system, we found that the small GTPase Rab11 and Syntaxin 1A (Syx1A), known for its role in neurotransmitter release, were required for the release of Evi-containing vesicles. Neuron-specific interference with Rab11 and Syx1A in motoneurons in vivo demonstrated that the same molecules were required for the release of Evi vesicles at the larval NMJ. In addition, we showed that the Rab11 effector and Syx1A-binding protein Myosin-5 (Myo5), an unconventional myosin, was also required for Evi vesicle release. We also detected Evi-containing multivesicular bodies (MVBs), endosomal organelles known to release exosomes at the NMJ. Having provided a proof of principle for the use of cultured cells as an assay to study the mechanisms of Evi vesicle release, we isolated the exosome fraction from S2 cells. Using mass spectrometry, Western blot analysis, and electron microscopy, we provide evidence that the Evi-containing vesicles are exosomes and that the composition of these exosomes is remarkably conserved across species (14, 18). These studies represent an important effort to elucidate the function of exosomes in vivo in the nervous system.
EXPERIMENTAL PROCEDURES
Molecular Biology
pUASt-attB was from Dr. Konrad Basler, Institute of Molecular Life Sciences, Zurich, Switzerland. pUASt-Syx1A(T254I) was prepared by inserting a genomic fragment containing the mutant syntaxin 1A, syx(3–69), into 5′ AgeI and 3′ EagI sites of a pUASt-DBB-stinger green vector. The genomic fragment was obtained by PCR amplification using genomic DNA isolated from syx(3–69) mutant flies (19) using the following set of primers: forward primer, ATACCGGTCCCGCGCGAGGTGCTGTGTTTCGTT; reverse primer, AATCGGCCGTTTACATGAAATAACTGCTAACATA. Sequences were verified by sequencing. pUASt-attB-Evi-HA encoding the long isoform of Evi tagged with three HA tags on the C terminus was made by subcloning the Evi sequence from pAc5.1-Evi-EGFP (20) to pUASt-attB-HA. pUASt-attB-HA was made by inserting a synthetic oligonucleotide coding for three tandem HA tags (YPYDVPDYASGYPYDVPDYAGSYPYDVPDYAS, where GS are linker amino acids) followed by a stop codon into the KpnI-XbaI sites of the multiple cloning site of pUASt-attB. For generating stable cell lines expressing copper-inducible Evi-EGFP or mCherry, the Evi-EGFP or mCherry coding region from pAc5.1-Evi-EGFP or pAc5.1-mCherry (1) was cloned into pMK33, containing a hygromycin selection cassette. pMK33-BiP-ProteinA-dSiaT was described in Ref. 21. The number of samples analyzed is specified in figure legends.
Transgenic Strains
pUASt-Evi-HA carrying flies were prepared by targeted transgenesis at the attP2 docking site (3L; 68A4; GenetiVision). pUASt-Syx1A(T254I) was injected for random P-element insertion (Rainbow Transgenic Flies).
Immunocytochemistry
The 3rd instar larval body wall muscles were dissected in Ca2+-free saline (22) and fixed in 4% paraformaldehyde in 0.1 m phosphate buffer for 10 min, unless otherwise indicated. Fixed larvae were washed and permeabilized in PBT (0.1 m phosphate buffer; 0.2% (v/v) Triton X-100) and incubated in primary antibody overnight. Following secondary antibody incubations, samples were washed with PBT and mounted in Vectashield (Vector Laboratories). The following antibodies were used: anti-Late bloomer (monoclonal antibody (mAb) 10C9, Developmental Studies Hybridoma Bank, 1:1000 for Western blots (WB)); anti-Wg (mAb 4D4, Developmental Studies Hybridoma Bank, 1:10 for WB); anti-Bruchpilot (mAb nc82, Developmental Studies Hybridoma Bank, 1:1000 for WB, and 1:100 for immunocytochemistry (ICC)); anti-GFP (Abcam, 6556, preabsorbed against S2 cell lysate, 1:8000 for WB); anti-Wg (15) (1:300 for ICC); anti-GFP (Invitrogen, mAb 3E6, 1:300 for ICC); anti-GFP (MBL International, 598, 1:500 for immuno-EM); anti-Rab11 (BD Biosciences, 1:50 for ICC after nonalcoholic Bouin's (71.4% (v/v) picric acid, 23.8% (v/v) paraformaldehyde (using 37% w/v methanol-free stock prepared from powder), 4.76% glacial acetic acid) fixation for 10 min and 1:500 for WB); anti-dPak (23) (1:2000 for ICC, 20-min fixation); anti-Alix (24) (1:500 for WB); mouse anti-tubulin (Sigma, 1:5000 for WB); affinity-purified N-terminal anti-Evi (1) (1:400 for ICC, 20 min fixation in Bouin's, 1:300 for immuno-EM); anti-HA (Roche Applied Science, mAb 3F10, 1:500 for ICC). DyLight-conjugated secondary antibodies were from Jackson ImmunoResearch and used at the following dilutions: DyLight-594- or DyLight-649-conjugated goat anti-HRP at 1:200 for ICC; DyLight-405-, DyLight-488-, or DyLight-594-conjugated anti-rat, anti-mouse, or anti-rabbit at 1:400 for tissue ICC and 1:800 for cultured cells. For immuno-EM, 18 nm of gold-conjugated donkey anti-rabbit secondary antibodies (Jackson ImmunoResearch) were used at 1:70.
Fly Stocks
pUASp-YFP-Rab11DNS25N (23261) and pUASt-YFP-Rab35S22N (9820) were from Bloomington Stock Center. pUASt-Syx1A-RNAi was from the Vienna Drosophila RNAi Center (VDRC; transformant ID 33112) (25). pWiz-Rab11-RNAi, pUASt-Rab11-DNN124I, and pUASt-GFP-Myo5(CT) were from Dr. Donald Ready (Purdue University). yw; syx4 ry506; yw; syxΔ229 ry506, and yw; pUASt-Syx1A were from Dr. Hugo Bellen (Baylor College of Medicine, Houston, TX). evi2 was from Dr. Michael Boutros (German Cancer Research Center, Heidelberg, Germany) (20). C380-Gal4 is a neuron-specific Gal4 driver, which has no detectable Gal4 expression in muscle (26). Neuron-specific expression of transgenes and RNAi was performed by using the Gal4 system (27).
Confocal Microscopy and Signal Intensity Measurements
Images were acquired on Zeiss Pascal or LSM 700 scanning confocal microscope equipped with a Zeiss ×63 Plan-Apochromat 1.4 NA DIC oil immersion objective at xyz pixel settings of 0.05 × 0.05 × 0.3 μm unless otherwise indicated. For quantification of signal intensity, NMJs were imaged at identical settings for control and experimental groups on a Zeiss Axioplan microscope equipped with a Yokogawa CSU10 spinning disk confocal scanning unit and a Hamamatsu 9100 EM-CCD camera (512 × 512) and a ×40 EC-Plan-NeoFluar 1.3 NA objective. Briefly, after image acquisition, the bouton volume bounded by HRP staining was selected, and fluorescence intensity inside (presynaptic) and outside (postsynaptic) the boutons was determined as the sum of total pixel intensity and normalized to bouton volume, as described previously (1). Deconvolution was performed using Volocity 5.0 deconvolution software using reference beads (Molecular Probes PS-Speck beads (505/515, 540/560, and 633/660)) dried onto coverslips that were mounted in Vectashield on microscope slides). To determine the point-spread function for use in subsequent deconvolution, the reference beads were imaged on a Zeiss LSM 700 confocal microscope at xyz pixel settings of 0.05 × 0.05 × 0.16 μm and 1 Airy unit pinhole size for each fluorophore used. Confocal Z-stacks of larval specimens for deconvolution were acquired on the same confocal microscope under identical settings except for adjusting the laser intensity to avoid pixel saturation.
Electron and Immunoelectron Microscopy of Purified Exosomes
Exosomes were examined using negative staining as described previously (28). Briefly, the purified exosome pellet was fixed in 2% paraformaldehyde at 4 °C, and 5 μl were deposited on Formvar-coated EM grids. After 20 min of absorption, grids were rinsed in PBS and fixed with 1% glutaraldehyde for 5 min. After washing in distilled water, grids were contrasted with uranyl oxalate (75 mm oxalic acid in 2% (w/v) uranyl acetate, final pH adjusted to 7.0 with NH4OH) and embedded in 4% (w/v) uranyl acetate and 2% (w/v) methylcellulose for transmission electron microscopy. Immunoelectron microscopy of exosomes was performed without permeabilization, as described previously (28). Briefly, 5 μl of purified exosomes were deposited on Formvar-coated EM grids, washed with PBS, then with PBS containing 50 mm glycine, and subsequently blocked with PBS containing 0.5% (w/v) BSA. Next, the grids were incubated with antibodies diluted with blocking solution for 30 min, then washed, and incubated with gold-conjugated secondary antibodies. The immunoreaction was stabilized by postfixing the grids with 1% glutaraldehyde in 0.1 m sodium phosphate buffer. After washing in distilled water, they were contrasted and embedded as above for EM.
Electron and Immunoelectron Microscopy of Cultured Cells and Larval NMJs
Cell culture dishes containing S2 cells or dissected larval body wall muscles were fixed overnight in modified Trump's fixative (0.2% glutaraldehyde, 4% paraformaldehyde, 1 μm MgCl2 in 0.1 m sodium cacodylate buffer (pH 7.2) at 4 °C). Samples were then rinsed in 0.1 m cacodylate buffer (pH 7.2). For ultrastructural analysis of cultured cells, S2 cells were removed from culture dishes with a cell scraper and pelleted in a 1.5-ml microcentrifuge tube by spinning at 6000 × g for 15 min. Dehydration was performed over crushed ice with N,N-dimethylformamide (Sigma) in a graduated series (N,N-dimethylformamide/double distilled water = 30, 50, and 70% for 5 min each; 85 and 95% for 15 min each; 100% for 1 h). Infiltration was performed using LR White resin (Electron Microscopy Sciences, LR White resin/N,N-dimethylformamide = 1:2 for 30 min and 2:1 for 30 min, pure LR White resin overnight). Resin was then replaced with fresh LR White resin and cured in the microcentrifuge tube for 48 h at 60 °C. For ultrastructural analysis of body wall muscles, fixed preparations were cut longitudinally along the midline, and each half was placed in a clear polypropylene plastic capsule (Ted Pella Inc., 21460) for embedding. For immunolabeling, sections were collected on nickel or gold grids, and subsequent steps were done by floating the grids on 100 μl drops. Sections were conditioned with 0.1 m Tris-HCl (pH 7.2) buffer for 5 min and then treated with 35 mm glycine for 10 min to block unbound aldehydes. Nonspecific binding was blocked with 0.5% BSA in Tris-HCl buffer (pH 7.2) for 1–2 h and then rinsed in buffer for 30 min. Incubation with primary antibody ranged from 2 h to overnight at 4 °C. After washing in buffer for 30 min, grids were incubated with 18-nm gold-conjugated secondary antibodies for 2–3 h and then rinsed with Tris-HCl buffer for 15 min. Staining was done with saturated uranyl acetate and lead citrate. In some cases, uranyl acetate was omitted to allow better contrast of the gold labeling. Some grids were exposed to 1% osmium tetroxide vapors for 1 h to enhance membrane electron density.
S2 Cell Culture
S2 cells were cultured at 26 °C in SFX insect medium (HyClone) supplemented with 10% fetal bovine serum (Hyclone), 50 unit/ml penicillin, and 0.05 mg/ml streptomycin solution (Sigma) in NunclonTMΔT-flasks (Thermo Scientific). Stable S2 cells lines with copper-inducible expression of Evi-GFP, mCherry, or BiP-ProteinA-dSiaT were prepared by transfecting the cells with the pMK33 vector containing the corresponding tagged cDNAs using Effectene transfection reagent (Qiagen) and maintained under continuous hygromycin (Invitrogen) selection at 0.5 mg/ml.
Western Blotting
S2 cell lysates were prepared by sonication of cell pellets resuspended in PBS containing protease inhibitors (Complete Protease Inhibitor Mixture; Roche Applied Science). Larval body wall muscles (typically from 20 larvae) attached to the central nervous system (CNS) were homogenized in 200 μl of ES2 buffer (20 mm HEPES (pH 7.5), 100 mm KCl, 0.05% Triton X-100, 2.5 mm EDTA, 5 mm DTT, 5% glycerol and protease inhibitors) with 0.5-mm glass beads at 4 °C using a BBX24B Bullet Blender Blue homogenizer (Next Advance Inc.). Larval homogenates were centrifuged at 16,000 × g for 10 min at 4 °C, and then the supernatant was centrifuged at 16,000 × g for 10 min at 4 °C. The resulting pellet was used as the source of larval lysate. Appropriate volumes were mixed with 5× sample buffer (250 mm Tris (pH 6.8), 10% SDS, 30% glycerol, 5% β-mercaptoethanol, 0.02% bromphenol blue), incubated at 95 °C for 5 min, and resolved by 10% SDS-PAGE under reducing and denaturing conditions. For exosomes, the 5× loading buffer also contained 6 m urea. Proteins were transferred onto pure nitrocellulose membranes (Bio-Rad) and blocked in 5% instant nonfat dry milk (SACO Foods, Inc.) in TBST (50 mm Tris (pH 7.4), 150 mm NaCl, 0.05% Tween 20) or in 5% BSA in TBST for the anti-Wg 4D4 antibody for 1 h at room temperature. Blots were incubated with primary antibodies (diluted to working concentration in blocking solution) overnight at 4 °C. After washing in TBST, blots were incubated with HRP-conjugated secondary antibodies diluted 1:3000 in blocking solution for 1 h at room temperature. Western blots were visualized using the ECL Plus chemiluminescent detection kit (GE Healthcare). Quantification of signal intensity was carried out after scanning the blots on a Hewlett-Packard Photosmart C5180 scanner at 400 dpi resolution and using the ImageJ 1.44p (imagej.nih.gov) software gel quantification module.
Exosome Preparation
S2 cells were cultured in serum-free medium to eliminate bovine serum exosome contamination. Large scale cultures were grown in spinner flasks (BellCo Glass Inc.) at 26 °C and harvested at 1–1.5 × 106 cells/ml density. Exosome preparation using the additional sucrose/D2O density gradient purification step was as described previously (28). Briefly, cells were pelleted at 300 × g, and the supernatant was cleared of dead cells at 2000 × g for 10 min. The cleared supernatant was then centrifuged at 10,000 × g for 30 min to remove cell debris. Finally, the supernatant was centrifuged at 100,000 × g for 70 min to pellet the exosome fraction. This exosome fraction was further purified by layering on top of a Tris/sucrose/D2O (200 mm, 30% (w/v), 50% (v/v)) solution. Samples were then centrifuged in an SW28 ultracentrifuge rotor at 100,000 × g for 75 min. Approximately 3.5 ml of the Tris/sucrose/PBS interface was collected with a syringe needle through the side of the centrifuge tube. Sucrose and D2O were removed by resuspending the fraction in 60 ml of PBS, and the exosomes were collected after 100,000 × g centrifugation for 70 min. The final pellet, representing the purified exosome fraction, was resuspended in a small volume of PBS and stored at −80 °C.
Preparation of dsRNA for S2 Cell Treatment
Primers for dsRNA templates were designed either by using the E-RNAi software (German Cancer Research Center) or based on the known amplicon primers published by the Drosophila RNAi Screening Center website (supplemental Table 2). When needed, T7 recognition sequences (TAATACGACTCACTATAGGG) were added to the 5′ of gene-specific primers via primer synthesis. dsRNA templates were first amplified via PCR from cDNAs (if available) or from genomic DNA using specific primers. Using the purified PCR products another round of touchdown PCR was performed with the T7-tagged primers to generate the T7 flanked templates used for transcription. dsRNA synthesis was performed using the MEGAscript RNAi kit (Ambion).
For dsRNA treatment, 10 μg/ml dsRNA was added to S2 cells seeded at 60% confluency and incubated for 4 days before the co-culture assay. For Evi-GFP-S2 or ProtA-dSiaT-S2 cells, on day 3, 0.7 mm CuSO4 was added to induce protein expression for 24 h. Most experiments were performed in 12-well plates using 0.4 ml of total cell culture volume per well. To eliminate evaporation during dsRNA treatment, plates were covered in aluminum foil, stored in humidified sterile Tupperware boxes, and cultured in humidified incubators.
Exosome Uptake by S2 Cells
Donor S2 cells were pelleted by centrifugation at 800 × g for 3 min, and the supernatant (medium containing the Evi-GFP exosomes) was then cleared of floating cells and larger debris by centrifugation at 2500 × g for 3 min. 100–200 μl of cleared supernatant was added to 400 μl of 90–100% confluent recipient S2 cells, and the cells were incubated for 2 h, unless otherwise indicated. The supernatant was then removed, and cells were rinsed twice with 1 ml of PBS immediately before fixation in 2% paraformaldehyde in 0.1 m phosphate buffer for 10 min. After mounting, internalized Evi-GFP positive puncta were counted under a Zeiss Axioscope 2 plus fluorescent microscope using a Zeiss Plan NeoFluar 63 × 1.25 NA oil immersion objective. All experiments were performed in triplicate.
Real Time Quantitative PCR
Cells were harvested and resuspended in PBS, and 5 volumes of TRIzol reagent (Invitrogen) was added. After 5 min at room temperature, 0.2 volume of chloroform was added, and the mixture was vigorously agitated by hand for 30 s and incubated at room temperature for 10 min. Samples were then centrifuged at 12,000 × g for 10 min, and the aqueous phase was precipitated with 0.7 volume of isopropyl alcohol by incubating at room temperature for 10 min. RNA was spun down at 12,000 × g for 10 min at 4 °C, and the pellet was rinsed in 70% ethanol and dried for 5 min at room temperature. Samples were resuspended in RNase-free water. Concentration and purity of total RNA samples were determined using a NanoDrop 2000 spectrophotometer (Thermo Scientific). For cDNA synthesis, 1 μg of RNA was used per sample, using the SuperScript III first strand synthesis system for RT-quantitative PCR (Invitrogen). The RT-quantitative PCRs were performed in triplicate in a 96-well plate (Applied Biosystems) using an ABI PRISM 7000 sequence detection system. For the reactions, a TaqMan® gene expression master mix (Applied Biosystems) was used with the following TaqMan® gene expression assay primers: Rab35 (Dm01845617_m1 (6-carboxyfluorescein-conjugated TaqMan probe)) or Rab27 (Dm01820985_m1 (6-carboxyfluorescein)). GAPDH (Dm01841185_m1 (6-carboxyfluorescein)) was used as a reference gene. All reactions were run at 50 °C for 2 min and 95 °C for 10 min and 40 cycles of 95 °C for 15 s and 60 °C for 1 min. To check amplification efficiency, calibration curves for both target and reference genes were performed. Reactions without template were run in parallel for all plates as internal controls. The relative quantification (ΔΔCT) method was used to compare the mRNA expression between samples. All transcript levels were normalized to GAPDH transcript level, using the same cDNA template. Data were expressed in graphs as 2−ΔΔCT (29).
Statistical Analysis
Statistical analysis was performed using a Student's t test when a single experimental sample was compared with control. For comparison of multiple experimental groups with a single control, a one-way analysis of variance was used followed by Dunnett's post hoc test. *, p < 0.05; **, p < 0.001; ***, p < 0.0001.
Mass Spectrometry
Purified exosome fractions were resuspended in 5× protein sample buffer containing 6 m urea, incubated at 95 °C for 5 min, and loaded onto 10% denaturing polyacrylamide protein gels (10 μg of total protein per lane). Single gel lanes containing the purified exosome fraction were excised into 15 slices. Each gel slice was cut into 1 × 1-mm fragments, and the gel fragments were extensively washed with water prior to addition of 1 ml of 50 mm ammonium bicarbonate and 50% acetonitrile and incubated at room temperature for 1 h. The supernatant was discarded, and gel fragments were then suspended in 200 μl of acetonitrile for 20 min. Excess acetonitrile was discarded, and the resulting white opaque gel fragments were dried in a SpeedVac. For tryptic digestion, dried gel fragments were rehydrated with 25 μl of 50 mm ammonium bicarbonate solution containing 20 ng of trypsin (Sigma Proteomics grade). Once the trypsin solution was absorbed, 30 μl of 50 mm ammonium bicarbonate was added to fully submerge the gel fragments. Samples were then incubated in a heat block at 37 °C overnight. After digestion, the peptide fraction in the supernatant of each sample was removed and placed in a separate vial. The gel fragments were suspended in 50 μl of 1% formic acid and 80% acetonitrile for 30 min at room temperature. The supernatant was removed and combined with the previous supernatant fraction. Gel fragments were then discarded, and the supernatant fractions were further dried down to about 10 μl in a SpeedVac. Samples were then brought to a 40-μl volume with 0.1% TFA prior to injection on the LC/MS system.
For liquid chromatography tandem mass spectrometry (LC/MS/MS), eluted peptides were separated on a LC Packings ultimate nanoflow HPLC system as follows. Ten μl of the peptide digest solution (∼0.25 of the total digest) was manually injected onto a micro trap column (LC Packings precolumn cartridge 0.3 × 5 mm C18 PM), and the trap column was manually washed with 10 μl of 0.1% formic acid prior to switching in line with the reverse phase separating column (LC Packings 100 μm × 15 cm C18 PepMap). A gradient was developed from 100% solvent A (0.1% formic acid) to 60% solvent B (0.1% formic acid in acetonitrile/water 70:30) over 60 min at a flow rate of 500 nl/min. The outlet of the column was connected to an electrospray needle (New Objective Taper Tip 20 μm). Electrospray mass spectrometry was performed on a ThermoElectron Finnigan LCQ DeCA ion trap mass spectrometer. Data-dependent acquisitions were set up according to a triple play experiment program where full MS scans from 400 to 2000 Da were ongoing until an MS signal grew above a specified threshold upon which a high resolution scan (Zoom Scan) was performed to determine monoisotopic mass and charge state followed by a single MS/MS scan. Dynamic exclusion was applied to prevent repeat scans of the same peptide masses. The raw data files were converted into mass peak lists using the LCQ_DTA program and then searched against the Drosophila taxonomy of the SwissProt protein data base using the Mascot search engine using 1.5-Da mass tolerances for the parent and 1.0 Da for the fragment masses. Cutoff criteria for selection of proteins in supplemental Table 1 were set at a minimum protein identity of 99.9%, minimum peptide identity of 95%, and minimum number of two peptides as determined by Scaffold 3 (Proteome Software, Inc.).
RESULTS
Development of a Cell Culture Assay to Study Exosome Release
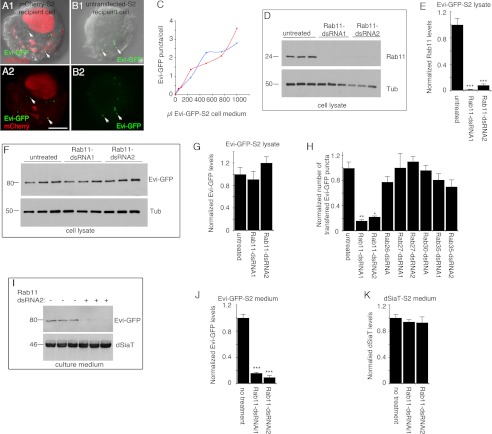
We have previously demonstrated that Evi-containing vesicles are transferred from the presynaptic compartment to postsynaptic muscles (1). In addition, we have shown that these Evi vesicles are specifically transferred between Schneider-2 (S2) cells (1). In those experiments, S2 cells expressing Evi-GFP were mixed with untransfected mCherry-expressing S2 cells, which led to the transfer of Evi-GFP puncta to the untransfected mCherry cells in contact with the Evi-GFP-expressing cells (1). This transfer was specific to Evi-GFP, and it was not simply due to an unspecific phagocytic activity of S2 cells, as expressing membrane proteins, such as DFrizzled-2 (DFz2) and rCD2-mRFP instead of Evi-GFP, did not result in such transfer across cells (1). To establish if cell contact was required for transfer and to develop an assay to screen for genes required for the release of Evi vesicles, mCherry-expressing cells were co-cultured with the conditioned medium of Evi-GFP-S2 cells that was cleared of cells and cellular debris. Consistent with the idea that cell contact is not required for the uptake of Evi vesicles, we found Evi-GFP puncta within the mCherry-expressing recipient cells (Fig. 1, A1 and A2). Importantly, cells incubated with a purified exosome fraction from Evi-GFP-S2 cells obtained by differential centrifugation and sucrose density gradient separation (28) showed a similar uptake of Evi-GFP (Fig. 1, B1 and B2), indicating that the Evi-GFP containing vesicles are present within the exosomal fraction. This uptake was dependent on the amount of Evi-GFP-containing medium (Fig. 1C), which allowed this assay to be used to understand the mechanisms of exosome release described below.
FIGURE 1.
Exosome uptake assay and role of Rab11 in exosome release from Evi-GFP-S2 cells. A, mCherry-S2 recipient cell incubated with the cleared culture medium from Evi-GFP-S2 donor cells showing Evi-GFP puncta (arrows) in the cytoplasm. Image is shown after deconvolution. B, untransfected S2 recipient cell incubated with purified exosome fraction isolated from Evi-GFP-S2 donor cells. C, number of Evi-GFP puncta per cell inside mCherry-S2 recipient cells after incubation with increasing amounts of culture medium from Evi-GFP-S2 donor cells. Two independent experiments are shown. n = 100 for each point. D, Western blots showing that endogenous Rab11 protein levels are significantly decreased after Rab11-dsRNA treatment. E, quantification of Rab11 protein levels in Evi-GFP-S2 cell lysate after Rab11-dsRNA treatment. n = 3. F, Western blots of Evi-GFP-S2 cell lysate, showing that Evi-GFP levels are not significantly affected by Rab11-dsRNA treatment. Tub, tubulin. G, quantification of Evi-GFP levels in Evi-GFP-S2 cell lysate after Rab11-dsRNA treatment. n = 3. H, number of Evi-GFP puncta in untransfected recipient cells incubated with dsRNA-treated Evi-GFP-S2 donor cells normalized to control, showing that Rab11 dsRNA treatment drastically reduces the number of internalized Evi vesicles. n = 100 cells per condition, per experiment. Three independent experiments were performed for each dsRNA. I, Western blot of Evi-GFP-S2 or dSiaT-S2 cell culture medium showing that Evi-GFP, but not dSiaT levels, are significantly reduced by Rab11-dsRNA treatment. Quantification of Evi-GFP in the culture medium of Evi-GFP-S2 (J) and, dSiaT in the culture medium of dSiaT-S2 cells after treatment with Rab11-dsRNA (K). n = 3. Calibration bar is 8 μm in A and B.
Rab11 and Syx1A Are Required for the Exosomal Release of Evi-GFP in Cultured Cells
To begin understanding the mechanisms of Evi vesicle trafficking and release, we conducted a small scale dsRNA screen in Evi-GFP-expressing S2 cells. In particular, we centered on the role of Rab proteins, key regulators of intracellular vesicular trafficking that are highly specific to subcellular organelles (30). In addition, we examined a number of candidate molecules previously implicated in vesicle trafficking and/or release. In these experiments, Evi-GFP-S2 donor cells were treated with dsRNA for 4 days; Evi-GFP expression was induced on day 3, and on day 4 the culture medium containing the secreted Evi-GFP exosomes was cleared of cell debris and added to recipient S2 cells for transfer assays.
Notably, treating Evi-GFP exosome donor cells with Rab11-dsRNA, which dramatically reduced Rab11 levels in the donor cells (Fig. 1, D and E), but not Evi-GFP (Fig. 1, F and G) in these cells, resulted in a drastic decrease in Evi-GFP transfer to S2 recipient cells (Fig. 1H), suggesting that Rab11 is required for the release of Evi vesicles. In contrast, down-regulating Rab27 or Rab35, Rabs previously implicated in exosome release in HeLa and oligodendroglial cells (31, 32), were without effect (Fig. 1H and supplemental Fig. S1, A and B). The requirement for Rab11 in Evi vesicle release was supported by Western blot analysis of the Evi-GFP-S2 cell media ruling out the possibility that their uptake, but not their release, was affected. Rab11-dsRNA treatment was accompanied by a substantial decrease of Evi-GFP from the culture medium (Fig. 1, I and J). Although eliminating Rab11 from Evi-GFP-S2 cells suppressed the release of Evi-GFP containing exosomes, it did not affect the classical secretory pathway, as secretion of Drosophila sialyltransferase (dSiaT), which is secreted via the classical pathway in this cell line (21), was unaffected by Rab11-dsRNA treatment (Fig. 1, I and K).
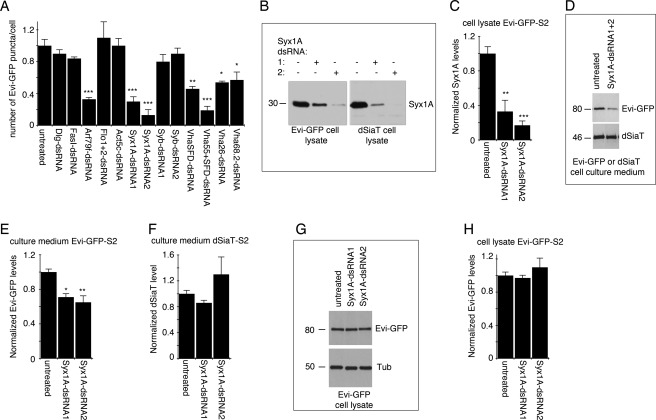
We also examined a number of candidate exosomal proteins that could be involved in exosome formation, trafficking, and/or release, including the following: Arf79f, a protein involved in the formation and targeting of vesicles; Actin 5c; the lipid raft and exosome-associated proteins Flotillin1 and Flotillin2; V-type ATPase subunits Vha68.2, Vha26, Vha55, and VhaSFD, which are involved in the acidification of endosomal compartments but have also been implicated in secretion (33, 34) and are present in exosomes; Syntaxin 1A (Syx1A), a protein well characterized for its role in synaptic vesicle exocytosis (35, 36); the cell adhesion molecule Fasciclin I (FasI); and the scaffolding protein Discs-Large (DLG). We found that several of the above proteins showed reduced Evi vesicle release, including Arf79f, the V-type ATPase subunits Vha68.2, Vha26, Vha55, and VhaSFD, and Syx1A (Fig. 2A). In contrast, other proteins tested, such as FasI, DLG, Flotillin1 and Flotillin2, Actin 5c, and synaptobrevin, had no significant effect on exosome release/transfer (Fig. 2A).
FIGURE 2.
dsRNA screen of exosomal candidates and role of Syx1A in Evi vesicle release from Evi-GFP-S2 cells. A, number of exosomes internalized by untransfected S2 cells after treatment of Evi-GFP-S2 donor cells with the indicated dsRNA, showing that Syx1A down-regulation significantly decreases the number of internalized exosomes. Data are normalized to untreated control cells. n = 100 cells per experiment, per condition. Three independent experiments were performed for each dsRNA. B, Western blots of Evi-GFP and dSiaT-S2 cell lysates treated with Syx1A-dsRNA constructs showing that Syx1A protein levels are significantly reduced by this treatment. C, quantification of Syx1A levels in Evi-GFP-S2 cell lysates after dsRNA treatment. n = 3. D, Western blots of Evi-GFP and dSiaT-S2 cell culture medium showing that Syx1A-dsRNA treatment decreases the levels of secreted Evi-GFP but not dSiaT. E, quantification of Evi-GFP in the cleared culture medium of Evi-GFP-S2 cells after dsRNA treatment. n = 3. F, quantification of dSiaT levels in the culture medium of dSiaT-S2 cells after dsRNA treatment. n = 3. G, Western blots of Evi-GFP-S2 cell lysate, showing that Syx1A-dsRNA treatment does not affect Evi-GFP levels in these cells. Tub, tubulin. H, quantification of Evi-GFP levels in cell lysates of Evi-GFP-S2 cells after dsRNA treatment. n = 3.
We were particularly interested in the function of Syx1A, because together with Rab3a, this protein targets the exocytosis of synaptic vesicles at the presynaptic active zone (37, 38). The finding that Rab11 is required for Evi vesicle release raised the possibility that Rab11 and Syx1A might target the fusion of multivesicular bodies to the perisynaptic region of the NMJ. We used two Syx1A-dsRNA constructs targeting two different regions of Syx1A transcript, which were effective in down-regulating Syx1A levels (Fig. 2, B and C). Western blot analysis of donor cell culture medium showed that levels of Evi vesicle release were substantially reduced upon treatment of these cells with Syx1A-dsRNA (Fig. 2, D and E) without any change in the secretion of dSiaT (Fig. 2, D and F). This was likely due to a defect in the secretion of Evi-GFP, as the levels of Evi-GFP in the cell lysate were not changed (Fig. 2, G and H).
Rab11 Is Also Required for Release of Evi Vesicles at the NMJ
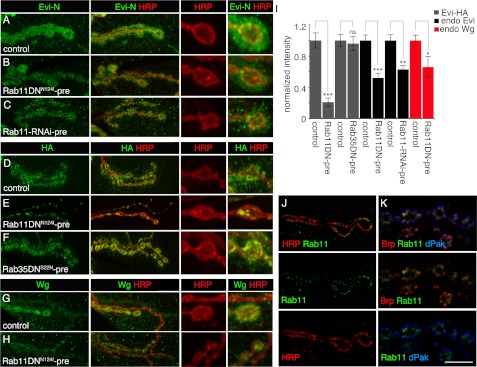
We next tested whether the findings in S2 cell culture could be applied to the NMJ. Although many processes are expected to occur in a cell-specific manner, many others appear conserved in different cell types, and S2 cells have been widely used to elucidate such conserved signal transduction pathways (39–41). To determine whether Rab11 was also required for the release of Evi vesicles at the NMJ, we determined the pre- and postsynaptic intensities of endogenous Evi staining after interfering with Rab11 function. rab11 is an essential gene required in all cell types, and thus, rab11 null alleles are early embryonic lethal (42). In addition, we wanted to determine the function of Rab11 specifically in motoneurons. Therefore, we examined NMJs in larvae expressing Rab11 dominant negative transgenes (Rab11S25N (30) and Rab11N124I (43), referred here as Rab11DNS25N and Rab11DNN124I, respectively) in motoneurons. Consistent with the requirement of Rab11 for Evi release from synaptic boutons, endogenous postsynaptic Evi levels were reduced when Rab11DNN124I was expressed in motoneurons (Fig. 3, A, B, and I). Further support for the above results was obtained by expressing Rab11-RNAi in motoneurons and examining the levels of endogenous Evi at the postsynaptic compartment. Again, a substantial decrease in postsynaptic Evi levels was observed (Fig. 3C).
FIGURE 3.
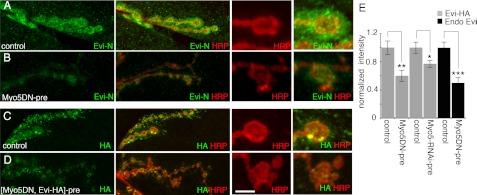
Rab11 is required for Evi-HA release at the NMJ and is localized at synaptic boutons. A–C, endogenous Evi localization at the NMJ of the following genotypes, in preparations labeled with anti-Evi (N-terminal epitope) and anti HRP. Insets are high magnification views of synaptic boutons. A, control (C380-Gal4/+) animal; B, larva expressing Rab11DNN124I in motoneurons (C380-Gal4 > Rab11DNN124I), showing a decrease in endogenous Evi levels; C, larva expressing Rab11-RNAi in motoneurons (C380-Gal4 > Rab11-RNAi), showing a similar decrease in endogenous Evi levels. D–F, localization of Evi-HA at the NMJ in larvae expressing Evi-HA in motoneurons in the following genotypes: D, control larva expressing Evi-HA in motoneurons (C380-Gal4 > Evi-HA); E, larva expressing Rab11DNN124I and Evi-HA in motoneurons (C380-Gal4 > Evi-HA, Rab11DNN124I), showing the localization of Evi-HA in large aggregates within the boutons and absent from the postsynaptic region; F, larva expressing Rab35DNS22N and Evi-HA in motoneurons (C380-Gal4 > Evi-HA, Rab35DNS22N) showing the normal presence of Evi-HA at the postsynaptic region. G and H, endogenous Wg levels at the NMJ of a control larva (C380-Gal4/+), showing the presence of Wg at the postsynaptic region (G), and larva expressing Rab11DNN124I in motoneurons (C380-Gal4 > Rab11DNN124I), showing a decrease in Wg levels at the postsynaptic region (H). J and K, deconvolved images showing the localization of endogenous Rab11 at synaptic boutons of a wild type larva in relationship to HRP (J) and Bruchpilot (Brp) and dPak (K). I, quantification of Evi-HA, endogenous Evi and Wg immunoreactivity levels, shown as ratios of postsynaptic to presynaptic intensities, normalized to controls. At least five larvae and 10 NMJs were analyzed for each genotype, and the following number of boutons were quantified for Evi-HA: 48 boutons for control C380-Gal4 > Evi-HA and 72 boutons for C380-Gal4 > Evi-HA, Rab11DNN124I; 46 boutons for control C380-Gal4 > EviHA and 50 boutons for C380-Gal4 > EviHA, Rab35DN; endogenous Evi, 68 boutons for control (C380-Gal4/+) and 45 boutons for C380-Gal4 > Rab11DNN124I; endogenous Evi, 75 boutons for C380-Gal4/+ and 56 boutons for C380-Gal4 > Rab11-RNAi; endogenous Wg, 47 boutons for C380-Gal4/+ and 38 boutons for C380-Gal4 > Rab11DNN124I. Calibration bar is 5 μm for J–K, 15 μm for A–H, and 5 μm for insets on A–H. Images are confocal Z-stack projections of NMJs on muscles 6/7 at segment A3. NS, not statistically significant.
Our previous studies show that Evi protein is synthesized both by motoneurons and muscles (1). Therefore, the total pool of Evi protein at the postsynaptic region is composed both of a pool derived from muscle plus a pool transferred from presynaptic boutons (1). The above experiments demonstrated that interfering with Rab11 function in motoneurons resulted in a decrease in postsynaptic Evi levels. However, given that muscles also synthesize Evi, the full extent of Rab11 function in transferring Evi from the presynaptic bouton cannot be completely resolved by measuring endogenous Evi levels in the postsynaptic region. To isolate the effect of interfering with Rab11 function on the pool of Evi protein that is transferred from the presynaptic compartment independently of postsynaptic Evi, we expressed Evi-HA in motoneurons (the expression pattern of which is indistinguishable from the previously reported (1) secretion of Evi-GFP by synaptic boutons (supplemental Fig. S1C)). We then examined the transfer of presynaptic Evi-HA to the postsynaptic muscle region in the presence of Rab11DNN124I in motoneurons. Expressing Evi-HA in neurons resulted in the presence of the HA signal at the postsynaptic region (Fig. 3D), showing that Evi-HA is secreted from synaptic boutons as reported previously (1). In contrast, expressing both Evi-HA and either of the Rab11DN transgenes blocked Evi-HA transfer to the postsynaptic region (Fig. 3, E and I, and supplemental Fig. S1, D and E). Instead, Evi-HA or Evi-GFP became distributed in aggregates inside the presynaptic boutons (Fig. 3E and supplemental Fig. S1E). This effect was specific to Rab11, as interfering with Rab35 by expressing the dominant negative transgene Rab35DNS22N (30) in motoneurons did not alter the transfer of Evi to the postsynaptic muscle cell (Fig. 3, F and I). Thus, interfering with Rab11 function does not prevent the trafficking of Evi to presynaptic boutons, but it does prevent its release.
Previously, we reported that Evi was required for Wg release from synaptic boutons (1). Therefore, we investigated whether interfering with Rab11 function had any effect on Wg release. As expected, expressing Rab11DNN124I in motoneurons resulted in a significant decrease in the levels of Wg at the synaptic junctional region (Fig. 3, G–I).
If Rab11 functions in the release of Evi from presynaptic boutons as our results suggest, then the Rab11 protein should be found at the NMJ. Indeed, previous reports indicated the presence of Rab11 at synaptic boutons, and we found that Rab11 was localized to the synaptic bouton boundary in a tubulo-vesicular-like pattern (Fig. 3, J and K, and supplemental Fig. S1, F and G for specificity control) (44, 45). Colabeling preparations with antibodies against the active zone component Bruchpilot and the postsynaptic marker dPak demonstrated that Rab11 did not localize to active zones but rather to the perisynaptic region (Fig. 3K) (44), the same region where Evi was proposed to be released (1). Together, these observations suggest that Rab11 is specifically required for the release of Evi from synaptic boutons and that this function is likely to be carried out locally at synaptic boutons.
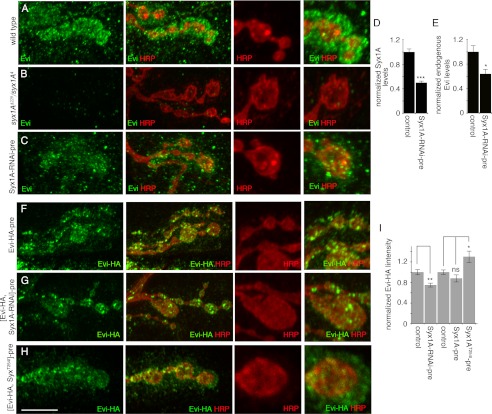
Requirement for Syx1A in Evi Vesicle Release
Our S2 cell assays demonstrated that Syx1A is also required for the release of Evi vesicles. Therefore, we next determined if this was the case in vivo, at the NMJ. Notably, transheterozygous syx1A4/syx1AΔ229 mutant animals did not show any endogenous Evi staining either pre- or postsynaptically (Fig. 4, A and B), suggesting that at the NMJ Syx1A is required for proper Evi synthesis and/or transport. To address the potential function of Syx1A in Evi release at synaptic termini, we reduced Syx1A by about 50% by expressing Syx1A-RNAi in motoneurons (Fig. 4D). Under these conditions, endogenous Evi is detected at synapses, indicating that the residual Syx1A protein level in motoneurons is sufficient for the transport of Evi to synaptic termini. Quantification of endogenous Evi in the pre- and postsynaptic compartments upon expressing Evi-RNAi in motoneurons revealed a significant decrease in postsynaptic Evi levels (Fig. 4, C and E). Similarly, when Evi-HA was expressed together with Syx1A-RNAi in motoneurons, a significant decrease in postsynaptic Evi-HA levels was observed (Fig. 4, F, G and I), suggesting that synaptic Syx1A is involved in the release and transfer of Evi (or Evi-HA) to postsynaptic sites.
FIGURE 4.
Syntaxin 1A is involved in Evi vesicle release at the larval NMJ. A–C, endogenous Evi localization at the larval NMJ of a y w control, showing endogenous Evi localization in both the pre- and postsynaptic region (A) and a y w/w; syx1A4/syx1AΔ229 animal, showing virtual absence of endogenous Evi (B). C, larva expressing Syx1A-RNAi in motoneurons (C380-Gal4 > Syx1A-RNAi), showing a decrease in postsynaptic Evi. D, quantification of endogenous Syx1A levels in synaptic boutons of larvae expressing Syx1A-RNAi in motoneurons (C380-Gal4 > Syx1A-RNAi). Four larvae (eight NMJs) were quantified for control C380-Gal4/+ (38 boutons) and C380-Gal4 > Syx1A-RNAi (38 boutons). E, quantification of endogenous Evi levels in the indicated genotypes, shown as ratios of postsynaptic to presynaptic intensities, normalized to controls. 75 boutons were quantified for control C380-Gal4/+ and 56 boutons for C380-Gal4 > Syx1A-RNAi. F–H, Evi-HA localization at the larval NMJ of a control larva expressing Evi-HA in motoneurons (C380-Gal4 > Evi-HA), showing transfer of Evi-HA to the postsynaptic region (F); larva expressing both Evi-HA and Syx1A-RNAi in motoneurons (C380-Gal4 > Evi-HA, Syx1A-RNAi), showing a decrease in postsynaptic Evi-HA levels (G); larva expressing overactive Syx1A(T254I) in motoneurons (C380-Gal4 > Evi-HA, Syx1A(T254I)), showing an increase in postsynaptic Evi-HA levels (H). I, Evi-HA immunoreactivity levels shown as ratios of postsynaptic to presynaptic intensities, normalized to controls. At least five larvae and 10 NMJs were analyzed for each genotype, and the following number of boutons were quantified: 55 boutons for control C380-Gal4 > Evi-HA; 83 boutons for C380-Gal4 > Evi-HA, Syx1A-RNAi; 61 boutons for control C380-Gal4 > Evi-HA; 76 for C380-Gal4 > Syx1A, Evi-HA; 66 for C380-Gal4 > Syx1A(T254I), Evi-HA. NS, not statistically significant. Calibration bar is 8 μm in A–G (left and middle panel), 3 μm in A–E, insets. Images are confocal Z-stack projections of NMJs on muscles 6/7 at segment A3.
To further test the idea that Syx1A is specifically involved in Evi vesicle release, we took advantage of a point mutation in syx1A, found in the syx(3–69) mutant, that dominantly enhances both spontaneous and evoked vesicle fusion (46). To specifically overactivate Syx1A in motoneurons, we expressed Evi-HA together with Syx1A(T254I), which mimics the syx(3–69) mutation (46) and fully recapitulates the syx(3–69) phenotype. This resulted in an increase in the amount of Evi-HA at the postsynaptic region (Fig. 4, H and I). This phenotype was not just due to the overexpression of Syx1A, as overexpressing wild type Syx1A did not result in an increase in Evi-HA (Fig. 4I). Thus, Syx1A overactivation stimulates Evi vesicle release.
Rab11 Effector and Syx1A Interactor Myosin 5 Is Required for Evi Vesicle Release at the NMJ
Previously, Syx1A has been shown to interact with Myo5 (47). Myo5 is an unconventional myosin involved in anterograde transport pathways that are required for the movement and/or anchoring of cargo to the cell periphery (48). In addition, Myo5 has been shown to interact with Rab11 in a number of contexts, and it is a well established Rab11 effector (49, 50). The requirement of both Rab11 and Syx1A in the release of Evi vesicles raised the possibility that Myo5 may also be involved in this process.
Interestingly, expressing a dominant negative C-terminal fragment of GFP-tagged Myo5 (Myo5DN) phenocopied the alterations in Evi release that were observed both when interfering with Rab11 or Syx1A. In particular, expressing Myo5DN resulted in a reduction of endogenous Evi (Fig. 5, A, B, and E) as well as transgenic Evi-HA (Fig. 5, C, D, and E) transfer to the postsynaptic region. This phenotype was also observed when Myo5-RNAi was expressed together with Evi-HA in motoneurons (Fig. 5E). In addition, Evi-HA accumulated in bright puncta in the presynaptic compartment (Fig. 5D), similar to our observations with Rab11DNN124I expression.
FIGURE 5.
Myo5 is required for Evi secretion at the NMJ. A and B, endogenous Evi localization at the larval NMJ in a control (C380-Gal4/+) larva (A); larva expressing both Myo5DN in motoneurons (C380-Gal4 > Myo5DN) showing a marked reduction of synaptic Evi levels at the postsynaptic region (B). C, control (C380-Gal4 > Evi-HA) larva expressing Evi-HA in motoneurons; D, larva expressing both Evi-HA and Myo5DN in motoneurons (C380-Gal4 > Myo5DN, Evi-HA), showing a decrease in postsynaptic Evi-HA. E, Evi-HA or endogenous Evi immunoreactivity levels shown as ratios of postsynaptic to presynaptic intensities that were normalized to controls. At least five larvae and 10 NMJs were analyzed for each genotype, and the following number of boutons were quantified: 41 boutons for control C380-Gal4 > Evi-HA and 19 for C380-Gal4 > Evi-HA, Myo5DN; 35 boutons for control C380-Gal4 > Evi-HA and 46 for C380-Gal4 > Evi-HA, Myo5DN; 43 boutons for control C380-Gal4/+ and 54 boutons for C380-Gal4 > Myo5DN. Calibration bar is 8 μm for A–E (left and middle panels), 3 μm for insets. Images are confocal Z-stack projections of NMJs on muscles 6/7 at segment A3.
Evi Vesicles Are Exosomes
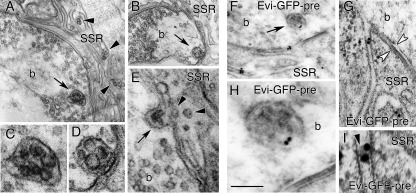
Exosomes are released from cells by the exocytosis of multivesicular bodies (2). Therefore, if Evi vesicles represent exosomes, then multivesicular bodies should be found within synaptic boutons, and these multivesicular bodies should contain Evi. We found that many synaptic boutons contained multivesicular bodies (Fig. 6, A–E), and that these multivesicular bodies were sometimes observed in close contact with the presynaptic membrane (Fig. 6, B and D). Immunoelectron microscopic labeling of Evi at the NMJ demonstrated that these multivesicular bodies contained Evi (Fig. 6, F and H) and that when Evi-GFP was expressed in motoneurons, Evi-labeled vesicles (black arrowhead in Fig. 6I) could be observed in cisternae of the postsynaptic SSR, a system of muscle-derived convoluted membranes surrounding the boutons (Fig. 6, G and I) (see supplemental Fig. S2B for no primary antibody controls). Evi was also found within multivesicular bodies in S2 cells (Fig. 7A and supplemental Fig. S2C for no primary antibody controls).
FIGURE 6.
Electron microscopy of multivesicular bodies and exosomes at larval NMJs. A–E, transmission electron micrographs of type I synaptic boutons (b) from a third instar larva, showing the presence of a multivesicular body (arrow) inside the bouton (A), shown at high magnification in C. Arrowheads point to small vesicular structures at the SSR. B, multivesicular body (arrow) in the presynaptic bouton appearing closely attached to the presynaptic membrane, shown at high magnification in D. E, a multivesicular body (arrow) appearing to fuse with the presynaptic membrane of a synaptic bouton (b). Arrowheads point to vesicular structures in the SSR that might have been released from the presynaptic terminal. F–I, immunoelectron micrographs of body wall muscles from larvae expressing Evi-GFP in motoneurons labeled with GFP antibodies showing a type I synaptic bouton (b) with gold particles representing Evi-GFP immunoreactivity at a presynaptic multivesicular body (arrow) (F), shown at high magnification in H, and at the postsynaptic SSR. G, Evi-GFP localization in the postsynaptic SSR; white arrowheads point to the synaptic region, recognized by the size of the synaptic cleft and its increased electron density. I, high magnification view of the Evi-GFP immunogold label in G, showing that in the crevices of SSR Evi-GFP appears to be present on a membrane encapsulated vesicle (black arrowhead), consistent with the view that Evi-GFP is released via exosomes. Calibration bars: 450 nm in A and B; 150 nm in C and D; 170 in E; 400 nm in F; 130 nm in H; 250 nm in G; and 85 nm in I.
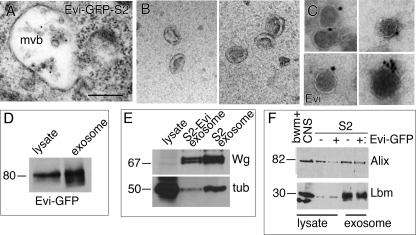
FIGURE 7.
Electron microscopic analysis of multivesicular bodies and exosomes in S2 cells. A, immunoelectron micrograph showing an Evi-GFP immunoreactive multivesicular body (mvb) within an S2 cell. B, transmission electron micrographs of negatively stained exosome preparations from S2 cells showing the characteristic cup-shaped appearance of exosomes. C, immunoelectron micrograph of exosomes from Evi-GFP-S2 cells stained with an antibody against the N terminus of Evi, showing the presence of gold particles decorating the exosomes. D, Western blot of Evi-GFP-S2 cell lysate and the purified exosome fraction showing enrichment of Evi-GFP in the exosomal fraction. 30 ng of Evi-GFP cell lysate and 0.3 ng of Evi-GFP exosome fraction were loaded. E, Western blot of S2 cell lysate and purified exosomes from Evi-GFP-S2 or untransfected S2 cells showing Wg enrichment in the exosome fraction. tub, tubulin. F, Western blot of larval body walls containing the central nervous system (CNS), as well as of untransfected S2 and Evi-GFP-S2 cell lysates and their corresponding exosome fractions. Note the enrichment of Lbm in the exosome fraction. Equal amounts of protein were loaded for each sample. Calibration bar: 300 nm in A; 200 nm in B; and 130 nm in C.
The results described above suggest that Rab11 and Syx1A are required for normal Evi vesicle release both at the NMJ and in S2 cells. Furthermore, Evi is present in similar intracellular organelles both at the NMJ and in S2 cells, and both cell types are able to release Evi vesicles. To examine the nature of the Evi vesicles, the supernatant from a stable Evi-GFP-S2 cell line was subjected to differential centrifugation followed by density separation on a sucrose gradient, following the same protocol as used previously to isolate exosomes (28). GFP fluorescence was enriched in this fraction (see below) suggesting that Evi-GFP vesicles were concentrated in this fraction. Negative staining and visualization by transmission electron microscopy showed that this Evi-GFP fraction was enriched in 50–100-nm cup-shaped vesicles (Fig. 7B), which is consistent with the reported morphology of exosomes (14). Immunostaining this fraction with an antibody against the N-terminal region of Evi, expected to be located on the outer surface of Evi vesicles (1), followed by electron microscopy, demonstrated the presence of gold particles decorating the outer edge of the vesicles (Fig. 7C). On exosomes of plain S2 cells that do not express Evi-GFP, Evi immunogold labeling was also observed albeit at much lower frequencies (supplemental Fig. S2A), indicating that endogenous Evi is normally targeted to exosomes and that Evi-GFP localization to exosomes is not a side effect of Evi-GFP protein overexpression.
By Western blot analysis, Evi-GFP was over 100-fold enriched in this fraction (Fig. 7D). Unfortunately, we could not assess the enrichment of endogenous Evi levels in this fraction because our Evi antibodies are not suitable for Western blots. Similarly, the Evi-interacting protein, Wg, whereas at nearly undetectable levels in the lysate, was also enriched in this fraction (Fig. 7E). The Evi-GFP-S2 exosome fraction also contained components typically associated with exosomes, such as the Drosophila homolog of Alix (24) and a Drosophila member of the tetraspanin family, Late bloomer (Lbm) (Fig. 7F) (51). Lbm was highly enriched in this fraction compared with total cell lysate (Fig. 7F), similar to the enrichment of this family of proteins in human exosomes (52). In summary, our observations in S2 cells strongly support the conclusion that Evi is released in exosomes.
To elucidate the exosomal proteome, the S2 cell exosome fraction in both untransfected and Evi-GFP-S2 cells was analyzed by liquid chromatography-tandem mass spectrometry (LC/MS/MS). In agreement with findings from vertebrate systems (14) and insect cells (53), all key classes of proteins previously found to be associated with exosomes were found in the S2 exosome fraction (supplemental Table 1 and supplemental Fig. S3). Most of these could be classified as cytoskeletal, cytoplasmic, plasma membrane, and nuclear proteins (supplemental Fig. S3), and among others, they included cytoskeletal core components (Actin, Tubulin, Cofilin, and Myosin); membrane trafficking components (Annexins, Rho proteins, and Ras-related proteins); adhesion molecules (Integrins, Fasciclin-1, and Tetraspanin); V-ATPase subunits; signaling proteins (G-protein subunits, 14-3-3); lipid raft proteins (Flotillin 1); cytoplasmic proteins (RNA elongation factors and ADP-ribosylation factor); chaperones (heat-shock proteins), and molecules thought to be involved in MVB formation (Alix and Clathrin). From the cellular component of the gene ontology chart (supplemental Fig. S3A), the exosomal proteins were dominated by membrane, cytoskeletal, and cytosolic proteins, indicating that little contamination is present from mitochondria, endoplasmic reticulum, Golgi, or other cellular organelles. Molecular function gene ontology categories were mostly represented by enzymatic activities of a hydrolytic nature, followed by transporter and transferase activities (supplemental Fig. S3B). Finally, biological process gene ontology was dominated by processes involved in cellular component organization and localization, as well as metabolic processes and cell communication (supplemental Fig. S3C). These findings further support the model that Evi is specifically sorted to and released in exosomes and that the core components of exosomes are remarkably conserved across species. Notably, some of the proteins identified by Western blot in the exosome fraction, including Alix, Lbm, and Wg, were not identified by mass spectrometry. This is likely due to their relatively lower abundance, low molecular weight, or hydrophobicity, which might make their detection and analysis by mass spectrometry challenging (54).
DISCUSSION
Role of Rab11 in Exosome Release in S2 Cells and at the NMJ
Here, we provide important insights into the mechanisms of Evi-exosome release and transfer across cells, and we establish the Drosophila NMJ as an in vivo model system to study the function of exosomes in the nervous system. The use of a cell culture-based assay allowed us to identify components of the Evi vesicle release machinery, Rab11 and Syx1A, which were also required for the release of Evi vesicles at the NMJ.
Interfering with Rab11 function in S2 cells through dsRNA resulted in a marked reduction of Evi-exosome secretion without affecting the classical secretory pathway. Similarly, interfering with Rab11 function specifically in motoneurons in vivo reduced the secretion of Evi vesicles by synaptic boutons at the NMJ. The localization of Rab11 at perisynaptic regions of presynaptic boutons further substantiates a role for Rab11 in the local release of Evi vesicles. The periactive zone has been suggested to function as a region for local endosome recycling (44), with Cdc42 and Nervous Wreck (Nwk) modulating actin-polymerization-dependent trafficking from this compartment. Both Cdc42 and Nwk have been shown to be required for the transmission of a muscle-derived BMP retrograde signal that regulates the growth and proliferation of synaptic boutons as the muscle fibers grow in size (44, 55). It has been suggested that in the absence of Nwk, Cdc42, or Rab11, the activated BMP receptor, Thickveins (Tkv), fails to be silenced via endocytosis after binding its ligand, Glass bottom boat, resulting in persistent BMP receptor activation and unregulated synaptic growth (44, 56). The involvement of Rab11 in both BMP endocytosis and Evi vesicle release indicates that it may represent a central hub for coordinating the endo- and exocytic processes at the presynaptic terminal and that these two activities are tightly coordinated. Indeed, a recent report showing the surprising co-localization of the majority of neuronal Rab proteins (RabX4, Rab26, Rab19, Rab32, RabX1, and Rab21) with Rab11 at synapses in the fly also points to a central role of Rab11 in locally regulating synaptic membrane traffic (57).
Role of Syx1A in Exosome Release at the NMJ
Fusion of vesicles with target organelles largely depends on the interaction of a vesicular v-SNARE and of a cognate target t-SNARE present in the membrane of the target organelle (58). Our studies suggested that Syx1A, which is present at synaptic boutons and is critically required for fusion of synaptic vesicles (35), is also required for the release of Evi exosomes in vivo at the NMJ and in S2 cells. This model was supported by partial Syx1A loss of function experiments, which lead to a significant decrease in the secretion of Evi-HA exosomes without affecting presynaptic Evi-HA levels, and by gain of function experiments, which increased the levels of secreted Evi-HA at the NMJ. Syx1A(T254I) contains a point mutation that markedly increases the rate of both evoked and constitutive vesicle fusion, an effect that is not observed when wild type Syx1A is overexpressed (46). In addition to this synaptic function, Syx1A also functions upstream of this release in regulating the stability or trafficking of Evi at the NMJ, as larger decrease in Syx1A levels by loss of function mutants prevented endogenous Evi from reaching the NMJ.
It is interesting to note that although the release of neurotransmitter-containing vesicles requires both Syx1A and Rab3, our result suggests that the release of exosomes requires both Syx1A and Rab11. This is consistent with the notion that Rabs serve to target and tether different vesicle types to alternative membrane targets (59). Thus, through the use of different Rabs, synaptic vesicles and multivesicular bodies could be targeted to different membrane domains (the active zone and the perisynaptic zone) within the presynaptic bouton.
A recent study has reported the presence of Syx1A within the exosome fraction (53). Although we also found Syx1A within the exosome fraction through mass spectrometry, its score fell below the confidence level. At this point, we cannot rule out the possibility that Syx1A in this fraction might be present as a contaminant. In this regard, it would be interesting to elucidate the identity of the v-SNARE on MVBs that interact with Syx1A. Our studies did reveal the presence of a v-SNARE, synaptobrevin, in the exosome fraction. However, interfering with its function did not prevent the release of exosomes.
Rab Effector and Syx1A Interactor, Myo5, Is Also Required for Release of Evi-Exosomes at the NMJ
Myo5 is one of the proteins known to function as a Rab11 effector (50). In addition, vertebrate Myo5a interacts with Syx1A in a calcium-dependent manner (47). Our data linking Myo5 to the release of exosomes point to a possible molecular convergence at the synapse involving Syx1A, Myo5, and Rab11. Because Myo5 is an actin-binding processing motor that mediates short range transport of diverse axonal or dendritic vesicles (60) in actin-rich synaptic regions, it is conceivable that Myo5 functions in the transport and/or localization of MVBs to the synaptic membrane. At the synaptic membrane, Myo5 may contribute to the tethering of MVBs via its potential interaction with the SNARE domain of Syx1A (47).
Evi-containing Vesicles Are Exosomes
Based on size, morphology (cup shape), the marked enrichment of a tetraspanin, Lbm, and their composition elucidated by mass spectrometry, in particular the presence of Flotillin, Alix, V-type ATPase subunits, and heat-shock proteins (14, 61), we suggest that Evi is sorted to exosomes. The unusually high enrichment of Evi-GFP in exosomes also indicates an active sorting mechanism to exosomes.
The composition of Evi-GFP exosomes was in excellent agreement with other published exosome data (53, 62). Contamination from other organelles was minimal, which is also demonstrated by comparison with the proteomic analysis of specific cellular organelles (63). For example, the Golgi proteome is characterized by the presence of 45% endoplasmic reticulum and Golgi resident proteins (64). The presence of similar core protein components (with the exception for cell type-specific molecules, such as the MHC class molecules) in exosomes across vertebrate and invertebrate species suggests a conserved mechanism for their biosynthesis. This notion is supported by the observation that the cytosolic components of exosomes do not randomly include all cytoplasmic proteins, but many of these are selectively enriched and sorted to exosomes (65).
Diversity of Exosomal Pathways
The study of exosomes is still in its infancy, and the variety of pathways that involve the use of exosomes is not fully understood. The best characterized exosomal pathway functions during the degradation of obsolete or malfunctioning cellular material in reticulocytes (66). In this case, multivesicular bodies containing exosomes either fuse with lysosomes to induce cargo degradation or are discarded by fusion of multivesicular bodies with the plasma membrane, thus releasing cellular “debris.” However, many studies, particularly in the immune system, suggest that certain exosome classes serve a signaling function (14). Given this diverse function of exosomes, either as degradation pathways or signaling entities, it is likely that a number of different molecules are required for their formation and trafficking. For example, three Rab proteins have been implicated in exosome release so far: Rab11 in a K562 myelogenous leukemia cell line (67); Rab35 in oligodendrocytes (32), and Rab27a as well as Rab27b but not Rab11 in HeLa cells (31). In our studies, we found that interfering with Rab11, but not Rab35 or Rab27 (there is a single Rab27 in flies), prevented exosome release. Interestingly, this finding was supported both in cultured S2 cells as well as in vivo at the NMJ. Although our studies do not rule out the use of alternative Rabs in different cellular contexts, it will be important to verify the potential roles of Rab35 and Rab27 in the release of exosomes in their normal physiological context in vivo.
Another prominent example of emerging differential pathways used for the formation and trafficking of exosomes regards the use of the endosomal sorting complex required for transport (ESCRT) complex and Alix. ESCRT is composed of four protein complexes involved in the sequential sorting of cargo to MVBs and lysosomes, and Alix plays a role in the inward budding of the limiting membrane of endosomes (68). Exosomes are thought to form through the inward budding of the MVB limiting membrane. Interfering with the ESCRT-0 protein complex component hepatocyte growth factor-regulated tyrosine kinase substrate (Hrs) leads to reduced exosome release (69). However, recent studies reveal ESCRT- and Alix-independent mechanisms of exosome formation (70–72). Similarly, our unpublished results suggest that components of the ESCRT complex (Hrs, Vsp28, and Vps4), Alix, and neutral sphingomyelinase (71) are not required for Evi-exosome release in S2 cells. These dissonant results may reflect the variety of exosome populations and pathways used during exosome formation within a cell and in diverse cell types.
In summary, our results reveal the existence of an in vivo exosomal pathway in the nervous system, and they identify some of the molecular players regulating this novel communication pathway.
Supplementary Material
Acknowledgments
We thank Drs. Hong Bao and Rudi Bohm (B. Z. laboratory) for assistance in generating the pUASt-Syx1A(T254I) line. We thank Dr. James Ashley for comments on the manuscript and the Budnik laboratory members for critical discussions. We also thank Dr. Xi-Xi Jia for the earlier ultrastructural studies carried out in serial sections (73), which allowed us to gather material documenting the presence of multivesicular bodies at the NMJ. We thank Drs. Donald Ready (Purdue University), Hugo Bellen (Baylor College of Medicine), Matthew P. Scott (Stanford School of Medicine), Nicholas Harden (Simon Fraser University, Canada), and Michael Boutros (German Cancer Research Center, Heidelberg, Germany) for antibodies, reagents, and fly stocks. We additionally thank the Electron Microscopy Facility at the University of Massachusetts Medical School for facilitating our ultrastructural studies.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 MH0700000 (to V. B.) and Grant R01 NS06878 (to the B. Z. laboratory). This work was also supported by Mizutani Foundation Grant 090054 (to K. K.).

This article contains supplemental Figs. S1–S3 and Tables S1 and S2.
- Wg
- Wingless
- ESCRT
- endosomal sorting complex required for transport
- Evi
- Evenness Interrupted
- HRP
- horseradish peroxidase
- Lbm
- Late bloomer
- MVB
- multivesicular body
- NMJ
- neuromuscular junction
- SSR
- subsynaptic reticulum
- WB
- Western blot
- ICC
- immunocytochemistry
- BMP
- bone morphogenetic protein.
REFERENCES
- 1. Korkut C., Ataman B., Ramachandran P., Ashley J., Barria R., Gherbesi N., Budnik V. (2009) Trans-synaptic transmission of vesicular Wnt signals through Evi/Wntless. Cell 139, 393–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Johnstone R. M. (2005) Revisiting the road to the discovery of exosomes. Blood Cells Mol. Dis. 34, 214–219 [DOI] [PubMed] [Google Scholar]
- 3. Raposo G., Nijman H. W., Stoorvogel W., Liejendekker R., Harding C. V., Melief C. J., Geuze H. J. (1996) B lymphocytes secrete antigen-presenting vesicles. J. Exp. Med. 183, 1161–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nolte-'t Hoen E. N., Buschow S. I., Anderton S. M., Stoorvogel W., Wauben M. H. (2009) Activated T cells recruit exosomes secreted by dendritic cells via LFA-1. Blood 113, 1977–1981 [DOI] [PubMed] [Google Scholar]
- 5. Hwang I., Ki D. (2011) Receptor-mediated T cell absorption of antigen presenting cell-derived molecules. Front. Biosci. 16, 411–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hawari F. I., Rouhani F. N., Cui X., Yu Z. X., Buckley C., Kaler M., Levine S. J. (2004) Release of full-length 55-kDa TNF receptor 1 in exosome-like vesicles. A mechanism for generation of soluble cytokine receptors. Proc. Natl. Acad. Sci. U.S.A. 101, 1297–1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Al-Nedawi K., Meehan B., Micallef J., Lhotak V., May L., Guha A., Rak J. (2008) Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumor cells. Nat. Cell Biol. 10, 619–624 [DOI] [PubMed] [Google Scholar]
- 8. Subra C., Grand D., Laulagnier K., Stella A., Lambeau G., Paillasse M., De Medina P., Monsarrat B., Perret B., Silvente-Poirot S., Poirot M., Record M. (2010) Exosomes account for vesicle-mediated transcellular transport of activatable phospholipases and prostaglandins. J. Lipid Res. 51, 2105–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mittelbrunn M., Gutiérrez-Vázquez C., Villarroya-Beltri C., González S., Sánchez-Cabo F., González M. Á., Bernad A., Sánchez-Madrid F. (2011) Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat. Commun. 2, 282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Skog J., Würdinger T., van Rijn S., Meijer D. H., Gainche L., Sena-Esteves M., Curry W. T., Jr., Carter B. S., Krichevsky A. M., Breakefield X. O. (2008) Glioblastoma microvesicles transport RNA and proteins that promote tumor growth and provide diagnostic biomarkers. Nat. Cell Biol. 10, 1470–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Deregibus M. C., Cantaluppi V., Calogero R., Lo Iacono M., Tetta C., Biancone L., Bruno S., Bussolati B., Camussi G. (2007) Endothelial progenitor cell-derived microvesicles activate an angiogenic program in endothelial cells by a horizontal transfer of mRNA. Blood 110, 2440–2448 [DOI] [PubMed] [Google Scholar]
- 12. Valadi H., Ekström K., Bossios A., Sjöstrand M., Lee J. J., Lötvall J. O. (2007) Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 9, 654–659 [DOI] [PubMed] [Google Scholar]
- 13. Aline F., Bout D., Amigorena S., Roingeard P., Dimier-Poisson I. (2004) Toxoplasma gondii antigen-pulsed dendritic cell-derived exosomes induce a protective immune response against T. gondii infection. Infect. Immun. 72, 4127–4137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Théry C., Ostrowski M., Segura E. (2009) Membrane vesicles as conveyors of immune responses. Nat. Rev. Immunol. 9, 581–593 [DOI] [PubMed] [Google Scholar]
- 15. Packard M., Koo E. S., Gorczyca M., Sharpe J., Cumberledge S., Budnik V. (2002) The Drosophila Wnt, wingless, provides an essential signal for pre- and postsynaptic differentiation. Cell 111, 319–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lachenal G., Pernet-Gallay K., Chivet M., Hemming F. J., Belly A., Bodon G., Blot B., Haase G., Goldberg Y., Sadoul R. (2011) Release of exosomes from differentiated neurons and its regulation by synaptic glutamatergic activity. Mol. Cell. Neurosci. 46, 409–418 [DOI] [PubMed] [Google Scholar]
- 17. Théry C. (2011) Exosomes. Secreted vesicles and intercellular communications. F1000 Biol. Rep. 3, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Simpson R. J., Jensen S. S., Lim J. W. (2008) Proteomic profiling of exosomes. Current perspectives. Proteomics 8, 4083–4099 [DOI] [PubMed] [Google Scholar]
- 19. Littleton J. T., Chapman E. R., Kreber R., Garment M. B., Carlson S. D., Ganetzky B. (1998) Temperature-sensitive paralytic mutations demonstrate that synaptic exocytosis requires SNARE complex assembly and disassembly. Neuron 21, 401–413 [DOI] [PubMed] [Google Scholar]
- 20. Bartscherer K., Pelte N., Ingelfinger D., Boutros M. (2006) Secretion of Wnt ligands requires Evi, a conserved transmembrane protein. Cell 125, 523–533 [DOI] [PubMed] [Google Scholar]
- 21. Koles K., Irvine K. D., Panin V. M. (2004) Functional characterization of Drosophila sialyltransferase. J. Biol. Chem. 279, 4346–4357 [DOI] [PubMed] [Google Scholar]
- 22. Stewart B. A., Atwood H. L., Renger J. J., Wang J., Wu C. F. (1994) Improved stability of Drosophila larval neuromuscular preparations in hemolymph-like physiological solutions. J. Comp. Physiol. A 175, 179–191 [DOI] [PubMed] [Google Scholar]
- 23. Harden N., Lee J., Loh H. Y., Ong Y. M., Tan I., Leung T., Manser E., Lim L. (1996) A Drosophila homolog of the Rac- and Cdc42-activated serine/threonine kinase PAK is a potential focal adhesion and focal complex protein that colocalizes with dynamic actin structures. Mol. Cell. Biol. 16, 1896–1908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tsuda M., Seong K. H., Aigaki T. (2006) POSH, a scaffold protein for JNK signaling, binds to ALG-2 and ALIX in Drosophila. FEBS Lett. 580, 3296–3300 [DOI] [PubMed] [Google Scholar]
- 25. Dietzl G., Chen D., Schnorrer F., Su K. C., Barinova Y., Fellner M., Gasser B., Kinsey K., Oppel S., Scheiblauer S., Couto A., Marra V., Keleman K., Dickson B. J. (2007) A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448, 151–156 [DOI] [PubMed] [Google Scholar]
- 26. Budnik V., Koh Y. H., Guan B., Hartmann B., Hough C., Woods D., Gorczyca M. (1996) Regulation of synapse structure and function by the Drosophila tumor suppressor gene dlg. Neuron 17, 627–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Brand A. H., Perrimon N. (1993) Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401–415 [DOI] [PubMed] [Google Scholar]
- 28. Théry C., Amigorena S., Raposo G., Clayton A. (2006) Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. Chapter 3, Unit 3.22 [DOI] [PubMed] [Google Scholar]
- 29. Livak K. J., Schmittgen T. D. (2001) Analysis of relative gene expression data using real time quantitative PCR and the 2−ΔΔC(T) Method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 30. Zhang J., Schulze K. L., Hiesinger P. R., Suyama K., Wang S., Fish M., Acar M., Hoskins R. A., Bellen H. J., Scott M. P. (2007) Thirty one flavors of Drosophila rab proteins. Genetics 176, 1307–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ostrowski M., Carmo N. B., Krumeich S., Fanget I., Raposo G., Savina A., Moita C. F., Schauer K., Hume A. N., Freitas R. P., Goud B., Benaroch P., Hacohen N., Fukuda M., Desnos C., Seabra M. C., Darchen F., Amigorena S., Moita L. F., Thery C. (2010) Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat. Cell Biol. 12, 19–30 [DOI] [PubMed] [Google Scholar]
- 32. Hsu C., Morohashi Y., Yoshimura S., Manrique-Hoyos N., Jung S., Lauterbach M. A., Bakhti M., Grønborg M., Möbius W., Rhee J., Barr F. A., Simons M. (2010) Regulation of exosome secretion by Rab35 and its GTPase-activating proteins TBC1D10A–C. J. Cell Biol. 189, 223–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Coombs G. S., Yu J., Canning C. A., Veltri C. A., Covey T. M., Cheong J. K., Utomo V., Banerjee N., Zhang Z. H., Jadulco R. C., Concepcion G. P., Bugni T. S., Harper M. K., Mihalek I., Jones C. M., Ireland C. M., Virshup D. M. (2010) WLS-dependent secretion of WNT3A requires Ser-209 acylation and vacuolar acidification. J. Cell Sci. 123, 3357–3367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kolotuev I., Apaydin A., Labouesse M. (2009) Secretion of Hedgehog-related peptides and WNT during Caenorhabditis elegans development. Traffic 10, 803–810 [DOI] [PubMed] [Google Scholar]
- 35. Schulze K. L., Broadie K., Perin M. S., Bellen H. J. (1995) Genetic and electrophysiological studies of Drosophila syntaxin-1A demonstrate its role in non-neuronal secretion and neurotransmission. Cell 80, 311–320 [DOI] [PubMed] [Google Scholar]
- 36. Bhattacharya S., Stewart B. A., Niemeyer B. A., Burgess R. W., McCabe B. D., Lin P., Boulianne G., O'Kane C. J., Schwarz T. L. (2002) Members of the synaptobrevin/vesicle-associated membrane protein (VAMP) family in Drosophila are functionally interchangeable in vivo for neurotransmitter release and cell viability. Proc. Natl. Acad. Sci. U.S.A. 99, 13867–13872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schlüter O. M., Basu J., Südhof T. C., Rosenmund C. (2006) Rab3 superprimes synaptic vesicles for release: implications for short term synaptic plasticity. J. Neurosci. 26, 1239–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Graf E. R., Daniels R. W., Burgess R. W., Schwarz T. L., DiAntonio A. (2009) Rab3 dynamically controls protein composition at active zones. Neuron 64, 663–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rohn J. L., Sims D., Liu T., Fedorova M., Schöck F., Dopie J., Vartiainen M. K., Kiger A. A., Perrimon N., Baum B. (2011) Comparative RNAi screening identifies a conserved core metazoan actinome by phenotype. J. Cell Biol. 194, 789–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wendler F., Gillingham A. K., Sinka R., Rosa-Ferreira C., Gordon D. E., Franch-Marro X., Peden A. A., Vincent J. P., Munro S. (2010) A genome-wide RNA interference screen identifies two novel components of the metazoan secretory pathway. EMBO J. 29, 304–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sathyanarayanan S., Zheng X., Kumar S., Chen C. H., Chen D., Hay B., Sehgal A. (2008) Identification of novel genes involved in light-dependent CRY degradation through a genome-wide RNAi screen. Genes Dev. 22, 1522–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Perrimon N., Lanjuin A., Arnold C., Noll E. (1996) Zygotic lethal mutations with maternal effect phenotypes in Drosophila melanogaster. II. Loci on the second and third chromosomes identified by P-element-induced mutations. Genetics 144, 1681–1692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Satoh A. K., O'Tousa J. E., Ozaki K., Ready D. F. (2005) Rab11 mediates post-Golgi trafficking of rhodopsin to the photosensitive apical membrane of Drosophila photoreceptors. Development 132, 1487–1497 [DOI] [PubMed] [Google Scholar]
- 44. Rodal A. A., Motola-Barnes R. N., Littleton J. T. (2008) Nervous wreck and Cdc42 cooperate to regulate endocytic actin assembly during synaptic growth. J. Neurosci. 28, 8316–8325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Khodosh R., Augsburger A., Schwarz T. L., Garrity P. A. (2006) Bchs, a BEACH domain protein, antagonizes Rab11 in synapse morphogenesis and other developmental events. Development 133, 4655–4665 [DOI] [PubMed] [Google Scholar]
- 46. Lagow R. D., Bao H., Cohen E. N., Daniels R. W., Zuzek A., Williams W. H., Macleod G. T., Sutton R. B., Zhang B. (2007) Modification of a hydrophobic layer by a point mutation in syntaxin 1A regulates the rate of synaptic vesicle fusion. PLoS Biol. 5, e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Watanabe M., Nomura K., Ohyama A., Ishikawa R., Komiya Y., Hosaka K., Yamauchi E., Taniguchi H., Sasakawa N., Kumakura K., Ushiki T., Sato O., Ikebe M., Igarashi M. (2005) Myosin-Va regulates exocytosis through the submicromolar Ca2+-dependent binding of syntaxin-1A. Mol. Biol. Cell 16, 4519–4530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Trybus K. M. (2008) Myosin V from head to tail. Cell. Mol. Life Sci. 65, 1378–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Roland J. T., Bryant D. M., Datta A., Itzen A., Mostov K. E., Goldenring J. R. (2011) Rab GTPase-Myo5B complexes control membrane recycling and epithelial polarization. Proc. Natl. Acad. Sci. U.S.A. 108, 2789–2794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Li B. X., Satoh A. K., Ready D. F. (2007) Myosin V, Rab11, and dRip11 direct apical secretion and cellular morphogenesis in developing Drosophila photoreceptors. J. Cell Biol. 177, 659–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kopczynski C. C., Davis G. W., Goodman C. S. (1996) A neural tetraspanin, encoded by late bloomer, that facilitates synapse formation. Science 271, 1867–1870 [DOI] [PubMed] [Google Scholar]
- 52. Escola J. M., Kleijmeer M. J., Stoorvogel W., Griffith J. M., Yoshie O., Geuze H. J. (1998) Selective enrichment of tetraspan proteins on the internal vesicles of multivesicular endosomes and on exosomes secreted by human B-lymphocytes. J. Biol. Chem. 273, 20121–20127 [DOI] [PubMed] [Google Scholar]
- 53. Koppen T., Weckmann A., Muller S., Staubach S., Bloch W., Dohmen R. J., Schwientek T. (2011) Proteomics analyses of microvesicles released by Drosophila Kc167 and S2 cells. 11, 4397–4410 [DOI] [PubMed] [Google Scholar]
- 54. Lubec G., Afjehi-Sadat L. (2007) Limitations and pitfalls in protein identification by mass spectrometry. Chem. Rev. 107, 3568–3584 [DOI] [PubMed] [Google Scholar]
- 55. O'Connor-Giles K. M., Ho L. L., Ganetzky B. (2008) Nervous wreck interacts with thick veins and the endocytic machinery to attenuate retrograde BMP signaling during synaptic growth. Neuron 58, 507–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. O'Connor-Giles K. M., Ganetzky B. (2008) Satellite signaling at synapses. Fly 2, 259–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Chan C. C., Scoggin S., Wang D., Cherry S., Dembo T., Greenberg B., Jin E. J., Kuey C., Lopez A., Mehta S. Q., Perkins T. J., Brankatschk M., Rothenfluh A., Buszczak M., Hiesinger P. R. (2011) Systematic discovery of Rab GTPases with synaptic functions in Drosophila. Curr. Biol. 21, 1704–1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wickner W., Schekman R. (2008) Membrane fusion. Nat. Struct. Mol. Biol. 15, 658–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hutagalung A. H., Novick P. J. (2011) Role of Rab GTPases in membrane traffic and cell physiology. Physiol. Rev. 91, 119–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Desnos C., Huet S., Darchen F. (2007) “Should I stay or should I go?” Myosin V function in organelle trafficking. Biol. Cell 99, 411–423 [DOI] [PubMed] [Google Scholar]
- 61. Lee T. H., D'Asti E., Magnus N., Al-Nedawi K., Meehan B., Rak J. (2011) Microvesicles as mediators of intercellular communication in cancer. The emerging science of cellular “debris.” Semin. Immunopathol. 33, 455–467 [DOI] [PubMed] [Google Scholar]
- 62. Simpson R. J., Lim J. W., Moritz R. L., Mathivanan S. (2009) Exosomes. Proteomic insights and diagnostic potential. Expert Rev. Proteomics 6, 267–283 [DOI] [PubMed] [Google Scholar]
- 63. Yates J. R., 3rd, Gilchrist A., Howell K. E., Bergeron J. J. (2005) Proteomics of organelles and large cellular structures. Nat. Rev. Mol. Cell Biol. 6, 702–714 [DOI] [PubMed] [Google Scholar]
- 64. Wu C. C., MacCoss M. J., Mardones G., Finnigan C., Mogelsvang S., Yates J. R., 3rd, Howell K. E. (2004) Organellar proteomics reveals Golgi arginine dimethylation. Mol. Biol. Cell 15, 2907–2919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Sahu R., Kaushik S., Clement C. C., Cannizzo E. S., Scharf B., Follenzi A., Potolicchio I., Nieves E., Cuervo A. M., Santambrogio L. (2011) Microautophagy of cytosolic proteins by late endosomes. Dev. Cell 20, 131–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Johnstone R. M. (2006) Exosomes biological significance. A concise review. Blood Cells Mol. Dis. 36, 315–321 [DOI] [PubMed] [Google Scholar]
- 67. Savina A., Fader C. M., Damiani M. T., Colombo M. I. (2005) Rab11 promotes docking and fusion of multivesicular bodies in a calcium-dependent manner. Traffic 6, 131–143 [DOI] [PubMed] [Google Scholar]
- 68. Matsuo H., Chevallier J., Mayran N., Le Blanc I., Ferguson C., Fauré J., Blanc N. S., Matile S., Dubochet J., Sadoul R., Parton R. G., Vilbois F., Gruenberg J. (2004) Role of LBPA and Alix in multivesicular liposome formation and endosome organization. Science 303, 531–534 [DOI] [PubMed] [Google Scholar]
- 69. Tamai K., Tanaka N., Nakano T., Kakazu E., Kondo Y., Inoue J., Shiina M., Fukushima K., Hoshino T., Sano K., Ueno Y., Shimosegawa T., Sugamura K. (2010) Exosome secretion of dendritic cells is regulated by Hrs, an ESCRT-0 protein. Biochem. Biophys. Res. Commun. 399, 384–390 [DOI] [PubMed] [Google Scholar]
- 70. Fang Y., Wu N., Gan X., Yan W., Morrell J. C., Gould S. J. (2007) Higher order oligomerization targets plasma membrane proteins and HIV gag to exosomes. PLoS Biol. 5, e158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Trajkovic K., Hsu C., Chiantia S., Rajendran L., Wenzel D., Wieland F., Schwille P., Brügger B., Simons M. (2008) Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science 319, 1244–1247 [DOI] [PubMed] [Google Scholar]
- 72. Babst M. (2011) MVB vesicle formation: ESCRT-dependent, ESCRT-independent. and everything inbetween. Curr. Opin. Cell Biol. 23, 452–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Jia X. X., Gorczyca M., Budnik V. (1993) Ultrastructure of neuromuscular junctions in Drosophila. Comparison of wild type and mutants with increased excitability. J. Neurobiol. 24, 1025–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.