Background: Binding of the vertebrate protease plasminogen is critical during the infectious cycle of Borrelia burgdorferi, although the mechanism of immobilization is unknown.
Results: Only OspC-expressing spirochetes immobilize plasminogen.
Conclusion: OspC, a dominant surface protein during the tick-to-host transition, is a potent plasminogen receptor.
Significance: Determining the timing and location of specific protein partnerships is crucial to understanding the infectious cycle.
Keywords: Bacteria, Host-Pathogen Interactions, Microscopic Imaging, Plasminogen, Protease
Abstract
Host-derived proteases are crucial for the successful infection of vertebrates by several pathogens, including the Lyme disease spirochete bacterium, Borrelia burgdorferi. B. burgdorferi must traverse tissue barriers in the tick vector during transmission to the host and during dissemination within the host, and it must disrupt immune challenges to successfully complete its infectious cycle. It has been proposed that B. burgdorferi can accomplish these tasks without an endogenous extra-cytoplasmic protease by commandeering plasminogen, the highly abundant precursor of the vertebrate protease plasmin. However, the molecular mechanism by which B. burgdorferi immobilizes plasminogen to its surface remains obscure. The data presented here demonstrate that the outer surface protein C (OspC) of B. burgdorferi is a potent plasminogen receptor on the outer membrane of the bacterium. OspC-expressing spirochetes readily bind plasminogen, whereas only background levels of plasminogen are detectable on OspC-deficient strains. Furthermore, plasminogen binding by OspC-expressing spirochetes can be significantly reduced using anti-OspC antibodies. Co-immunofluorescence staining assays demonstrate that wild-type bacteria immobilize plasminogen only if they are actively expressing OspC regardless of the expression of other surface proteins. The co-localization of plasminogen and OspC on OspC-expressing spirochetes further implicates OspC as a biologically relevant plasminogen receptor on the surface of live B. burgdorferi.
Introduction
Membrane-associated or secreted bacterial proteases are essential to many bacterial species during vertebrate infections. Bacterial proteases can target host proteins to provide essential nutrients for the pathogen, to disarm components of the host immune response, and to remove obstructions to efficient dissemination within the host (1, 2). Many bacterial pathogens that do not encode or produce endogenous membrane-associated or secreted proteases appropriate host proteases to increase the probability of successful host colonization (3, 4). Borrelia burgdorferi, the spirochete bacterium that causes Lyme disease in North America, traverses many membranes en route from the bite of a tick to secondary infection sites deep within hosts, although it does not produce an extra-cytoplasmic protease (5). To overcome the absence of endogenous extra-cytoplasmic proteases necessary for efficient migration across host tissue barriers, B. burgdorferi is hypothesized to immobilize host-derived plasminogen to its outer membrane (6–8). However, no B. burgdorferi protein has yet been demonstrated to immobilize plasminogen to the surface of the bacterium.
The vertebrate plasminogen system, which has a central role in the extracellular matrix degradation essential for eukaryotic cell migration (9–11), is used by numerous pathogenic bacterial species during vertebrate infections (3–5, 12). Immobilization of plasmin, the enzymatically active form of plasminogen, to the surface of Borrelia species promotes invasiveness in tick vectors (7) and in laboratory animals (7, 13, 14) to enhance transmission and dissemination. Plasmin localized to the surface of Borrelia species turns the bacterium into a proteolytically active organism capable of degrading tissue barriers such as basement membranes and extracellular matrices in vitro (6, 8, 15–17) and enhances dissemination in ticks and vertebrates in vivo (6, 7, 14, 18). Bacteremia is significantly greater in animals with intact plasminogen systems than in plasminogen-deficient animals suggesting that plasminogen may enhance the capacity for invasion of the cardiovascular system (7).
The molecular mechanism by which B. burgdorferi immobilizes plasminogen to its surface has been elusive due in part to the inherent difficulties in genetically manipulating B. burgdorferi (19). Furthermore, experimentally eliminating a B. burgdorferi plasminogen receptor may be problematic as all bacterial plasminogen-binding proteins identified to date in other bacterial species perform other essential functions (3, 12). In vitro binding assays employing soluble recombinant proteins have identified several proteins with plasminogen-binding potential (20–24). However, the in vivo relevance of in vitro protein-protein assays must be cautiously interpreted as membrane-associated proteins produced natively differ considerably from their soluble recombinant forms in structure and availability of binding sites (25, 26). A direct demonstration of physical interactions between plasminogen and native proteins embedded in the bacterial membrane is necessary to identify only the specific physiological associations between proteins that occur naturally. Deciphering where and when specific protein partnerships occur in or on living cells is critical to assess the biological relevance of membrane-associated proteins to plasminogen immobilization and utilization during infections.
Although active plasmin may be needed during multiple phases of the infectious cycle, plasmin use by B. burgdorferi has been best demonstrated during the transmission from ticks and during the early stages of vertebrate infections (7, 8). The expression of several proteins such as the outer surface protein C (OspC) are dramatically up-regulated during this period leading to the hypothesis that these proteins may be plasminogen receptors. OspC is a 21-kDa membrane-associated lipoprotein (27, 28) that is expressed on the surface of the bacterium as it migrates from the tick midgut to the vertebrate and remains expressed during the first weeks of infection (29–31), the time frame in which plasminogen is utilized by infecting B. burgdorferi (7, 8). OspC expression is also tightly correlated with the invasion of the tick salivary glands and host tissues suggesting a mechanistic role for OspC during these processes (32–36). However, the function of OspC remains the subject of considerable debate (37), and a native OspC-plasminogen interaction remains to be demonstrated.
Unraveling the molecular mechanism by which plasminogen is immobilized to the surface of B. burgdorferi requires a detailed understanding of plasminogen binding in the context of the membrane. This study focuses on the plasminogen-binding potential of the native OspC protein while embedded in the surface of live bacteria. Here, we employ genetically manipulated B. burgdorferi strains to test the hypothesis that OspC is a plasminogen receptor on the surface of B. burgdorferi. The results of quantitative proteome analyses, co-immunofluorescence microscopy, and whole-cell binding assays are all in agreement and strongly suggest that OspC is a potent plasminogen receptor on the surface of the Lyme disease bacterium. These analyses represent a significant methodological and conceptual step forward in dissecting the molecular mechanism of plasminogen utilization in a human pathogen of major public health importance.
EXPERIMENTAL PROCEDURES
Reagents
Polyclonal anti-OspC antibodies were produced at AnaSpec, Inc. (Fremont, CA), by subcutaneously injecting rabbits with 250 μg of recombinant OspC protein in Complete Freund's adjuvant on four occasions. Recombinant OspC was produced by cloning a 560-bp gene fragment, amplified using forward primer OCF-BsrGI (5′-GCGGCATGTACACGAATAATTCAGGGAAAGATGGG-3′) and reverse primer OCR-XhoI (5′-GCCGCACTCGAGTTAAGGTTTTTTTGGACTTTC-3′) from B. burgdorferi strain B31, into a pET45(b) vector using the BsrGI and XhoI restriction sites. The plasmid insert was sequenced to confirm identity with previously published ospC sequences. Recombinant OspC proteins were expressed in Escherichia coli BL21 (DE3) and purified using nickel-nitrilotriacetic acid Superflow Resin for His6-tagged proteins (Qiagen, 30410) and by ion exchange chromatography using Q-Sepharose Fast Flow (GE Healthcare). Anti-OspC antibodies were detected using goat anti-rabbit antibodies conjugated to AlexaFluor594 (Alexa, A11012). Native human plasminogen was purchased from Calbiochem (528178) and was detected using polyclonal FITC-conjugated sheep anti-plasminogen antibodies (Abcam Inc., Cambridge). α2-Antiplasmin and uPA4 were obtained from Calbiochem. Plasmin from human plasma was purchased from Sigma. Plasmin activity was assayed using the fluorometric plasmin substrate H-d-Val-Leu-Lys-7-amino-4-methylcoumarin (Bachem, Germany).
Bacterial Strains and Growth Conditions
B. burgdorferi strains were generously provided by P. Rosa (35). Briefly, strain ospCK1 (hereafter ospC-ko) is an ospC-deficient mutant derived from the wild-type B. burgdorferi clone B31–A3. ospCK1/pBSV2G-ospC (hereafter ospC-comp) is the ospCK1 strain complemented by homologous recombination using a shuttle vector carrying ospC. Assays used cultures grown to mid-log phase at 34 °C in BSK-II complete medium (38). In additional experiments, presented in the supplemental material, coumermycin (20 ng/ml), an antibiotic previously shown to up-regulate OspC (39), was added to cultures 24 h prior to experimental assays. B. burgdorferi densities were estimated using dark field microscopy with a Petroff-Hauser cell counter. Cultures were maintained at 34 °C and pH 7 prior to harvesting, conditions demonstrated to promote the expression of host-associated outer surface proteins (31, 40–43). For each culture, we harvested 5 × 108 bacteria by centrifugation at 2,000 × g for 20 min. Cells were washed three times in 1 ml of Hanks' balanced salt solution (HBSS), and 108 cells were resuspended in 215 μl of HBSS. The remaining cells were stored at −20 °C to assess plasmid content, as described previously (44), and protein expression by mass spectrometry (45).
Mass Spectrometry
The level of expression of all putative B. burgdorferi plasminogen receptors in the B31–A3, ospC-ko, and ospC-comp strains was assessed by mass spectrometry. Mass spectrometry is a high throughput technique that provides accurate quantification of all proteins in a sample. The mass spectrometric data were used to control for potential differences among the experimental strains that may affect plasminogen immobilization. To perform mass spectrometry, cells were first lysed by mechanical disruption using 0.1-mm diameter silica beads (Biospec Products, Inc.). Protein extracts were digested in-solution using trypsin (Promega, Madison, WI) at 37 °C overnight after DTT reduction and iodoacetamide alkylation. The resulting peptide mixtures were analyzed using an LCQ Deca XP Plus mass spectrometer (Thermo Fisher Scientific Inc., San Jose, CA) coupled to a nano-HPLC system (DIONEX) as described previously (45). Bioinformatics data analysis was accomplished in the BioWorks 3.3.1 SP1 platform using the SEQUEST algorithm (Thermo Scientific) and the B. burgdorferi proteome database. Peptide identity was considered confident if ΔCn was greater than 0.1 and the cross correlation score (Xcorr) was greater than 1.5, 2.0, or 2.5 for peptides with charges (z) of +1, +2, or +3, respectively (45). Relative protein abundance was determined by a label-free mass spectral method based on precursor peak intensity measurement of individual peptides (i.e. the maximum ion count observed for a precursor ion) (46, 47). The precursor peak intensity was obtained directly from the raw data acquisition file. The mass-to-charge ratios (m/z), chromatographic retention times, and patterns of peaks near the focal peak were used to confirm that the chromatogram peak intensities compared among samples corresponded to identical peptides. Protein concentrations were determined by Bradford assay using BSA as standard.
Immunofluorescence Localization
Each strain was incubated on an uncoated cover glass for 1 h at 34 °C with 0.167 μm human plasminogen, one-tenth the physiological concentration of plasminogen in vertebrate blood (48). Coverslips were washed five times in 1 ml of HBSS to remove weakly bound plasminogen and fixed using 16% paraformaldehyde. The coverslips were washed in PBS and incubated for 1 h in a 1:100 dilution of rabbit anti-OspC antibodies, followed by a 1:200 dilution of goat anti-rabbit antibodies conjugated to AlexaFluor594 (red), and finally a 1:100 dilution of FITC-conjugated sheep anti-plasminogen polyclonal antibodies (green) with the appropriate wash steps between. Coverslips were mounted using Mowiol with 2.5% 1,4-diazabicyclo-[2,2,2]-octane and examined at ×1,000 magnification by epifluorescence microscopy (Eclipse TE2000; Nikon, Tokyo, Japan).
Plasmin Binding and Activity
Each strain (108 cells) was incubated with gentle agitation for 1 h at 34 °C with 1.67 μm human plasmin or human plasminogen. Cells were then washed five times in 1 ml of HBSS to remove weakly bound plasmin or plasminogen. Cells were incubated for an additional 20 min with gentle agitation in 250 μl of HBSS with 15 units of uPA (Calbiochem 672081) as determined by the manufacturer. Urokinase-type plasminogen activator was included as no plasminogen activator endogenous B. burgdorferi has been detected to date (8). Cells were washed three times, and the residual plasmin activity was determined. The number of cells was confirmed by measuring the absorbance at 210 nm after the final wash. Each well was supplemented with 50 μl of the fluorogenic plasmin substrate d-Val-Leu-Lys-7-amino-4-methylcoumarin (1.2 mm in HBSS). Fluorescence intensity generated by the enzymatic degradation of the fluorogenic substrate was measured every 30 s for 30 min (excitation, 370 nm; emission, 430 nm read on a Analyst GT microplate reader, Molecular Devices). The number of active plasmin molecules in each well was determined by comparing fluorescence intensities to standards with known amounts of active plasmin. The retention of active plasmin bound by each strain was normalized by the density of spirochetes in each microplate well. To test for specific associations, α2-antiplasmin (100 μg/ml) was added to the plasmin-loaded bacteria and allowed to react for 20 min at room temperature prior to measuring plasmin activity levels in some assays. Anti-OspC antibodies (1:100) or the lysine analog ϵ-aminocaproic acid (ϵ-ACA; 10 mm) was added to the bacteria during the plasminogen or plasmin incubation in some assays to assess the role of OspC or lysine residues in plasminogen binding. Each strain was assayed in triplicate from six independent cultures per strain.
RESULTS
Expression and Relative Quantification of B. burgdorferi Surface Proteins
Surface protein expression was detected and quantified from whole-cell lysates of each strain using tandem mass spectrometry (Table 1). As expected, OspC peptides were observed only in wild-type (B31–A3) and ospC-complement (ospC-comp) strains at similar levels, but the knock-out strain, ospC-ko, did not have detectable OspC expression (Table 1 and supplemental Figs. S1 and S2). Borrelia plasminogen-binding protein, OspE-, and OspF-related proteins, and BbCRASP-1, each of which has been hypothesized as a potential plasminogen receptor (20, 22, 24), are expressed in all strains under the conditions studied (Table 1). Importantly, the expression levels of these proteins were similar among strains thus allowing us to isolate the effect of OspC on plasminogen immobilization from the effects caused by the other putative plasminogen receptors, i.e. no hypothesized plasminogen receptor aside from OspC can account for the observed differences in plasminogen binding among these strains. Additionally, any residual plasminogen detected in cultures of ospC-deficient strains may implicate one or more of the other surface proteins as alternative plasminogen receptors.
TABLE 1.
Expression and relative quantification of putative plasminogen receptors in strains B31–A3, ospC-ko, and ospC-comp
Borrelia plasminogen-binding protein (BPBP), BbCRASP-1 (cspA), and members of the OspE- and OspF-related proteins, all of which have been proposed as potential plasminogen receptors, are expressed at similar levels in all experimental strains as determined by the precursor peak intensities of the identified peptides in mass spectrometry analyses. As expected, OspC was detected only in strains B31–A3 and ospC-comp. Thus, no hypothesized plasminogen receptor aside from OspC can account for observed differences in plasminogen binding among these strains. Protein description, accession numbers, peptide sequences, precursor ion m/z, and the charge states (z) are listed. ND means not detected.
| Protein peptidea | GenBankTM accession no. | Precursor m/z, z | Precursor peak intensitiesb |
||
|---|---|---|---|---|---|
| B31–A3 | ospC-ko | ospC-comp | |||
| Outer surface protein C (OspC) | NP_047005 | ||||
| GPNLTEISK | 480.2, 2 | 1.25 × 108 | ND | 1.40 × 108 | |
| EVEALLSSIDEIAAK | 794.8, 2 | 1.25 × 107 | ND | 1.79 × 107 | |
| ELTSPVVAESPK | 629.3, 2 | 8.44 × 107 | ND | 9.63 × 107 | |
| BPBP | NP_212462 | ||||
| NAQEYFDETVPESELGIK | 1035.5, 2 | 8.36 × 108 | 7.03 × 108 | 4.60 × 108 | |
| KLLAEAGYPDGK | 631.7, 2 | 1.23 × 107 | 5.36 × 107 | 8.88 × 107 | |
| IRDDYYSGLK | 410.9, 3 | 2.84 × 108 | 1.37 × 108 | 5.28 × 108 | |
| BbCRASP-1 (CspA) | NP_045741 | ||||
| TLYSSLDYK | 545.6, 2 | 7.52 × 107 | 6.05 × 107 | 8.31 × 107 | |
| DFDTLKPAFY | 609.2, 2 | 7.99 × 107 | 8.89 × 107 | 6.45 × 107 | |
| KITNPGENTQNFEDK | 579.5, 3 | 3.35 × 107 | 4.58 × 107 | 5.02 × 107 | |
| OspE group | |||||
| ErpA (BbCRASP-5) | NP_051199 | ||||
| IHTSYDEQSNGEVKVKK | 981.7, 2 | 4.68 × 107 | 4.79 × 107 | 2.59 × 107 | |
| ErpP (BbCRASP-3) | NP_051449 | ||||
| TAEYAIPLEVLKK | 737.1, 2 | 3.45 × 106 | 2.14 × 106 | 8.41 × 106 | |
| ErpC (BbCRASP-4) | AAC34910 | ||||
| KIEFSEFTVK | 614.9, 2 | 1.28 × 107 | 1.20 × 107 | 1.31 × 107 | |
| OspF group | |||||
| ErpG | NP_051244 | ||||
| QNVKEKVEGFLDK | 512.0, 3 | 5.94 × 107 | 4.32 × 107 | 9.46 × 107 | |
| EKEIQELK | 1016.8, 1 | 1.01 × 108 | 1.41 × 108 | 1.34 × 108 | |
| ErpL | NP_051372 | ||||
| NSEQNLESSEQNVK | 803.2, 2 | 1.14 × 108 | 1.44 × 108 | 8.95 × 107 | |
| NSEQNLESSEQNVKK | 867.3, 2 | 1.40 × 108 | 1.54 × 108 | 2.98 × 108 | |
| Elp | |||||
| ErpX | NP_051509 | ||||
| LNKDNK | 732.5, 1 | 2.52 × 107 | 1.54 × 107 | 3.40 × 107 | |
| ENVDVSEIKEDLEK | 549.5, 3 | 1.13 × 108 | 6.37 × 107 | 5.39 × 107 | |
a The peptides listed from each protein were detected with high confidence by mass spectrometry (ΔCn >0.1 and the cross-correlation score (Xcorr) is greater than 1.5, 2.0, or 2.5 for peptides with charges (z) of +1, +2, or +3, respectively) and checked visually to confirm correct peptide identification.
b Precursor peak intensities of each peptide are listed for wild-type (B31–A3), OspC knockout (ospC-ko), and complement (ospC-comp) strains. Assays of 0.50 pm/μl bovine cytochrome c digest (Dionex) under similar conditions yielded peak intensities of (5–10) × 108 for major peptides.
OspC and Plasminogen Co-localize on Surface of B. burgdorferi
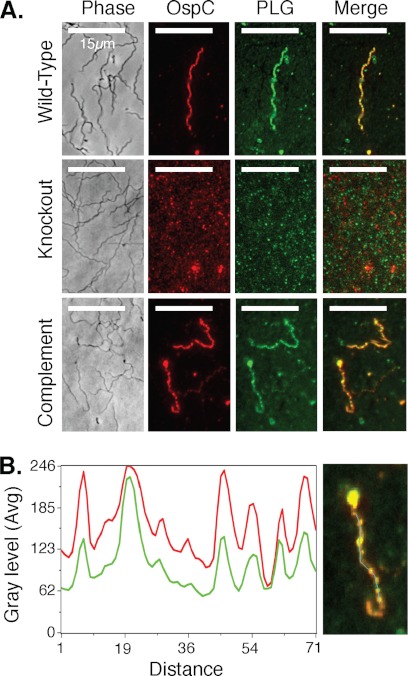
Immunofluorescence microscopy revealed that OspC was expressed and accessible to anti-OspC antibodies on the surface of a subset of the spirochetes from B31–A3 (wild type) and ospC-comp (complement) cultures, but no spirochetes from the ospC-ko (knock-out) culture (Fig. 1A and supplemental Figs. S3–S8). OspC was distributed along the surface of the bacteria in a nonhomogeneous punctate manner (Fig. 1A, OspC). Immunofluorescence microscopy of cultures incubated with plasminogen and then anti-plasminogen antibodies revealed that plasminogen was only associated with spirochetes from the OspC expression strains B31–A3 and ospC-comp (Fig. 1A, PLG). Anti-plasminogen antibody staining of ospC-ko spirochetes that were incubated with plasminogen did not reveal plasminogen bound to any spirochetes (Fig. 1A, PLG). Co-immunofluorescence staining using rabbit anti-OspC antibodies, AlexaFluor594-conjugated goat anti-rabbit antibodies, and FITC-conjugated anti-plasminogen antibodies demonstrated that the plasminogen bound to spirochetes perfectly co-localized with OspC (Fig. 1A, Merge). Epifluorescence microscopy images taken of spirochetes at both 594 nm (Fig. 1A, OspC) and 488 nm (Fig. 1A, PLG) are overlaid on the phase contrast microscopy images (Fig. 1A, Phase) to estimate the proximity of OspC and bound plasminogen (Fig. 1B). The fluorescence intensity profiles of the double-stained spirochetes suggest that areas with high plasminogen staining have correspondingly high levels of OspC staining (Fig. 1B). The perfect co-localization of OspC and plasminogen suggests that OspC may be responsible for the immobilization of plasminogen to the surface of the bacterium. The OspC-deficient strain showed only background fluorescence on both channels.
FIGURE 1.
OspC and plasminogen co-localize on the surface of OspC-expressing spirochetes. A, OspC is expressed and distributed along the surface of a fraction of the spirochetes (OspC) from strains B31–A3 (wild-type) and ospC-comp (Complement) in a nonhomogeneous pattern but is not expressed in the OspC-deficient strain (Knockout). Spirochetes that express OspC (wild-type and complement) are associated with plasminogen (PLG), again in a nonhomogeneous pattern, whereas spirochetes that do not express OspC (knock-out) are not associated with plasminogen. Anti-plasminogen antibody staining of ospC-ko spirochetes that were incubated with PLG result in only background staining, similar to the pattern found after anti-OspC staining (OspC) of this strain. Each row (Wild-type, Knock-out, and Complement) represents ×1000 images of paraformaldehyde-fixed slides taken with either phase contrast (Phase) or epifluorescence filters (594 nm, OspC; 488 nm, plasminogen). These images were overlaid (Merge) to estimate the proximity of OspC and bound plasminogen. B, importantly, bound plasminogen co-localizes with OspC on B31–A3 and ospC-comp spirochetes that were incubated with plasminogen. Fluorescence intensities of individual image pixels along the length of the double-stained spirochete (cyan line in right panel) were measured at both 594 nm (OspC) and 488 nm (plasminogen) and plotted against the distance in pixels (left panel). The fluorescence intensity profiles of double-stained spirochetes (red, OspC; green, plasminogen) demonstrate that areas with high plasminogen staining have correspondingly high levels of OspC staining, i.e. OspC and plasminogen are located in the exact same positions on the surface of the spirochetes (right panel, manifested by yellow overlap regions in the final merged image) suggesting that OspC is a potent plasminogen receptor.
OspC expression, and thus plasminogen-binding, was variable among spirochetes of strains B31–A3 and ospC-comp in cultures not induced with coumermycin. OspC could be detected by immunofluorescence on a fraction of individual spirochetes from strains B31–A3 and ospC-comp, similar to what has been observed in previous OspC expression experiments (31). Interestingly, OspC-expressing spirochetes had similar quantitative levels of OspC detectable by immunofluorescence, i.e. it appeared that variation in OspC expression among spirochetes is not continuous but that OspC expression is either on or off in each spirochete. By contrast, nearly all spirochetes of strains B31–A3 and ospC-comp expressed OspC at the maximally distinguishable level in cultures induced with coumermycin (supplemental Figs. S6–S8). The proportion of spirochetes expressing OspC decreases steadily as strains were continuously maintained in culture as noted previously (31). Regardless of the age of the culture, plasminogen binding was always restricted to OspC-expressing spirochetes in cultures of B31–A3 and ospC-comp, suggesting that OspC is a potent plasminogen receptor on the surface of the bacterium.
OspC Enhances the Ability of B. burgdorferi to Immobilize Active Plasmin and Plasminogen
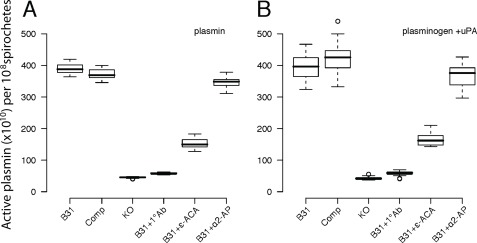
The binding capacity and ability to use the enzymatic activity of plasmin was measured for each of the B. burgdorferi isolates by two methods. First, each strain was incubated with active plasmin and washed to remove weakly bound plasmin, and the activity of the remaining plasmin bound to the bacteria was measured using the fluorogenic plasmin substrate, d-Val-Leu-Lys-7-amino-4-methylcoumarin. Cultures of the OspC-expressing strains B31–A3 (B31) and ospC-comp (Comp) each retain over 4 × 1010 molecules of active plasmin per culture (108 spirochetes), while cultures of the ospC-ko (KO) strain were associated with fewer than 0.25 × 1010 (108 spirochetes) (Fig. 2A). The plasminogen binding capacity of the OspC-expressing strains was even greater when OspC expression was induced with coumermycin, whereas plasminogen binding by the OspC-deficient strain remained constantly low across culture conditions (supplemental Fig. S9). Preincubating B31–A3 with polyclonal anti-OspC antibodies (B31 + 1°Ab) prevented the spirochetes from binding active plasmin, suggesting that the reduction in binding observed in the OspC-deficient strain was not caused by a reduction of total surface protein or membrane integrity but was caused by the loss of available OspC (Fig. 2A). Incubating B31–A3 with anti-OspA serum did not affect plasminogen binding (data not shown). Spirochetes did not show any increase in mortality or decrease in activity after a 3-h incubation with either anti-OspC or anti-OspA antibodies compared with spirochetes that were not incubated with antibodies. Binding of plasminogen to strain B31–A3 is reduced in the presence of ϵ-ACA, suggesting lysine residues are important in the OspC-plasminogen interaction, similar to what was previously observed in a protein-protein binding assay (23). The surface-bound plasmin was protected against inactivation by the serum-derived inhibitor α2-antiplasmin (Fig. 2, A and B).
FIGURE 2.
OspC-expressing strains bind significantly more plasmin and plasminogen than OspC-deficient strains. Cultures of the OspC-expressing strains B31–A3 (B31) and ospC-comp (Comp) each retain over 4 × 1010 molecules of active plasmin (A) or plasminogen (B) per culture containing 108 spirochetes, whereas the OspC-deficient strain, ospC-ko (KO) was associated with fewer than 0.25 × 1010 plasmin molecules per culture containing 108 spirochetes. Preincubating strain B31–A3 with polyclonal anti-OspC antibodies (B31 + 1°Ab) inhibits the spirochetes from binding active plasmin or plasminogen. The lysine analog ϵ-ACA inhibits binding of plasmin and plasminogen to the OspC-expressing spirochetes (B31+ϵ-ACA) confirming the role of lysine molecules in this interaction. Active plasmin bound to the spirochetes is protected from inhibition by α2-antiplasmin (α2-AP), a potent serum-derived inactivator of unbound plasmin. The number of active plasmin molecules that remained in spirochete cultures after incubation and extensive washing was estimated by comparing plasmin activity levels associated with each culture to experimentally prepared standards. Plasmin activity data were normalized by the number of spirochetes remaining in each experimental culture after all washing steps.
The OspC-expressing strains are also significantly better at capturing plasminogen than the ospC-ko strain or the B31–A3 parent strain preincubated with anti-OspC antibodies (Fig. 2B and supplemental Fig. S9B). The bound plasminogen could subsequently be converted to active plasmin by the addition of the exogenous activator uPA. Each strain was incubated with plasminogen, washed to remove weakly bound plasminogen, and then incubated with uPA before assessing the amount of active plasmin that remained bound to the spirochetes. No proteolytic activity was detected with any strain when the bacteria were incubated with plasminogen or uPA alone.
DISCUSSION
B. burgdorferi, the invasive bacterial pathogen that causes human Lyme disease in North America (49), completes its natural infectious cycle without expressing an endogenous extra-cytoplasmic protease (5). B. burgdorferi rapidly traverses many basement membranes and extracellular matrices as it migrates from the tick to the host dermis and subsequently enters and exits the vascular system en route to disseminated infection sites. Experimental infections of laboratory animals have shown that the plasminogen system is required for efficient transmission of B. burgdorferi from ticks to vertebrate hosts (7). However, the molecular mechanism by which B. burgdorferi immobilizes plasminogen to enhance dissemination was previously unknown. We used wild-type and genetically manipulated B. burgdorferi in immunofluorescence microscopy and live cell binding assays to test the hypothesis that the OspC is a plasminogen receptor on the surface of the bacterium. Immunofluorescence microscopy images demonstrate that only OspC-expressing spirochetes immobilize plasminogen to their surface, while plasminogen does not associate with OspC-deficient spirochetes (Fig. 1A). Furthermore, wild-type spirochetes that were not actively expressing OspC did not show measurable plasminogen binding (Fig. 1A). Importantly, plasminogen co-localizes with OspC on the cell surface, strongly implicating OspC as a biologically relevant plasminogen receptor (Fig. 1B). Plasminogen immobilization by live spirochetes was also much greater in cultures of OspC-expressing strains than in cultures of OspC-deficient strains (Fig. 2 and supplemental Fig. S9). Plasminogen binding is severely diminished in the presence of anti-OspC antibodies but not in the presence of anti-OspA antibodies suggesting a specific protein partnership between OspC and plasminogen. Plasmin bound to OspC is protected from inhibition by serum-derived α2-antiplasmin further indicating that this is a specific interaction that can provide the bacteria with active protease activity within a vertebrate host (Fig. 2, A and B). The data presented here demonstrate that OspC, a surface-exposed protein expressed during the critical tick-to-host transition, is a potent plasminogen receptor on the surface of living bacteria. These results are consistent with the hypothesis that OspC can enhance the efficacy of the infectious cycle in B. burgdorferi by immobilizing plasminogen.
Several lines of in vitro and in vivo evidence support the hypothesis that plasminogen enhances the efficacy of the infectious cycle of B. burgdorferi and related species. Specifically, extensive studies have demonstrated that the use of plasminogen by B. burgdorferi enhances invasiveness in in vitro assays through degradation of extracellular matrix components, penetration of biological barriers, and dissemination within ticks and into the vasculature of vertebrates (28–34). Furthermore, plasminogen increases the efficacy of internal organ invasion by relapsing fever Borrelia (6, 14). We propose that immobilization of plasminogen by the OspC surface receptor of B. burgdorferi increases the probability of a successful vertebrate infection from a tick bite at one or more of the following points in the infectious cycle. First, the probability of infection per tick bite may increase in the presence of plasminogen if more spirochetes can successfully migrate to the salivary glands and thus a greater number enter a host during tick feeding. Second, plasmin immobilized to the bacterial surface may increase the probability and efficacy of a bacterium entering the host vasculature. Finally, plasmin-coated bacteria in the host may be less likely to be destroyed by the host immune response as plasmin actively cleaves immunoglobulins and damages components of the complement pathway (50).
Although the data presented suggest that OspC-bound plasmin can enhance the infectivity of B. burgdorferi during the transmission from ticks to hosts, neither plasmin nor OspC is strictly required for successful infection (7, 51, 52). Overexpression of alternate surface-bound proteins can rescue the infectious phenotype of OspC-deficient strains, albeit with severely reduced infectiousness (51). The reduction of infectivity in wild-type B. burgdorferi observed in plasminogen-deficient laboratory animals is much less severe than the reduction in infectiousness observed in OspC-deficient and some OspC mutant strains infecting wild-type laboratory animals (33) suggesting that OspC has functions other than plasminogen immobilization. The pleiotropic nature of OspC is similar to the plasminogen receptors identified from all other plasminogen-utilizing bacteria that have alternative functions ranging from glycolytic enzymes to cellular adhesion molecules, many of which are essential even in in vitro culture (3). Indeed, a recent study suggests that OspC functions as an immune modulator and is also required for efficient dissemination, functions that are accomplished by different parts of the molecule (53). Thus, it is likely that OspC functions as a plasminogen receptor facilitating efficient dissemination in addition to other molecular functions. OspC, like most proteins, is likely pleiotropic and assumes several functions during the tick-to-host transition (reviewed in Ref. 37).
Although overexpression of alternative outer membrane proteins can partially rescue the infectious phenotype of OspC-deficient B. burgdorferi (51), the data presented here suggest that these surface proteins cannot completely rescue the plasminogen immobilization capacity provided by OspC during the tick-to-host transmission. Indeed, OspC-deficient strains express the other putative plasminogen receptors (Table 1) yet bound very little plasminogen in these assays (Fig. 2, A and B). This is unlikely the result of simultaneous down-regulation of OspC and other putative plasminogen receptors (54–57) as all strains expressed the other surface proteins of interest at similar levels (Table 1). Nevertheless, the other putative plasminogen receptors may be responsible for the low level plasminogen binding observed by ospC-ko, and thus genetic modification to dramatically overexpress these genes (51) may partially rescue plasminogen immobilization. Furthermore, the expression of alternative surface proteins may be much greater in different in vitro and in vivo conditions than the culture conditions used here (however, see Refs. 56, 57), which could potentially restore plasminogen immobilization in spirochetes not actively expressing OspC. Further research into the binding capabilities of alternative surface proteins embedded in the bacterial membrane in alternative experimental conditions is needed to assess their roles in plasminogen binding during the infectious cycle. We expect that plasminogen interacts with several other B. burgdorferi proteins embedded in the membrane of living cells at different stages of the infectious cycle that could potentially be detected using different culture conditions (i.e. conditions that mimic the divergent microenvironments that B. burgdorferi may inhabit during its life cycle). Regardless of the role that proteins may play in plasminogen immobilization, the data presented here demonstrate that OspC, a dominant surface protein during the critical tick-to-host transition (51), readily immobilizes plasminogen to the bacterial surface thus providing the bacteria with proteolytic activity in the presence of host tissues.
The results presented are consistent with the hypothesis that the sequence differences among ospC alleles affect the dissemination potential of B. burgdorferi due to the effect on OspC-plasminogen binding affinity. Several previous studies demonstrate that B. burgdorferi lineages that differ at the ospC locus have significantly different probabilities of disseminating in human patients (58–60). Furthermore, a protein-protein binding assay demonstrated that OspC sequence variation affects binding affinity to human plasminogen (23). Interestingly, recombinant OspC variants from two B. burgdorferi lineages that regularly cause disseminated human infections bind to human plasminogen with significantly higher affinity than two variants from lineages that rarely infect humans (23). Future studies using B. burgdorferi strains with genetically modified ospC alleles in whole-cell assays are essential to determine the relevance of OspC sequence differences to plasminogen immobilization on the surface of live cells and to the probability of human infection.
The distribution of OspC and plasminogen along the length of the bacteria was not homogeneous in this study (Fig. 1). OspC exists naturally as a multimer (61, 62) that could cause the nonhomogeneous pattern observed and thus could concentrate the proteolytic activity of plasmin to promote degradation of intercellular membranes. However, the nonhomogeneous distribution of OspC may be a methodological artifact resulting from the fixation process. Previous reports demonstrate that the outer surface protein A (OspA) forms highly localized aggregates when the bacteria are antibody labeled prior to fixation but show a more even distribution when the bacteria are fixed prior to exposure to anti-OspA antibodies (63). Regardless of the microvariation in the distribution of OspC, the co-localization of OspC and plasminogen along the length of the bacteria can serve to focus proteolytic activity to the extracellular matrices during the stationary adhesions to intercellular junctions (64). Thus, the OspC-plasminogen association could act to increase the ability of spirochetes to traverse membranes in ticks or hosts and thus increase the efficacy of the tick-to-host transition.
Many infectious bacteria including Yersinia pestis, Helicobacter pylori, Staphylococcus aureus, and Streptococcus pyogenes use host-derived plasmin to eliminate barriers to migration across extracellular matrices and basement membranes (3, 12), closely resembling the process of eukaryotic cell migration during repair of damaged tissues and cancer metastasis (65). The use of a host-derived proteolytic enzyme, as opposed to an endogenous bacterial enzyme, may allow the bacteria to efficiently migrate within a host without invoking an acute inflammatory response during early infections (16). In this study, we have identified OspC as a potent plasminogen receptor that is abundant on the surface of B. burgdorferi during the tick-to-host transition, a point in the enzootic cycle when proteolytic activity is critical. The approaches employed here, coupled with the developing genetic tool kit in B. burgdorferi, will allow further dissection of the quantitative effects of the OspC-plasminogen interaction on infectiousness and can be used to explore the specific regions or residues responsible for this interaction.
Supplementary Material
Acknowledgments
We thank P. Rosa for generously donating the genetically modified isolates used in this study and M. Pohlschröder and D. E. Dykhuizen for comments on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant AI076342.

This article contains supplemental Figs. S1–S9.
- uPA
- urokinase-type plasminogen activator
- HBSS
- Hanks' balanced salt solution
- ϵ-ACA
- ϵ-aminocaproic acid.
REFERENCES
- 1. Travis J., Potempa J., Maeda H. (1995) Are bacterial proteinases pathogenic factors? Trends Microbiol. 3, 405–407 [DOI] [PubMed] [Google Scholar]
- 2. Goguen J. D., Hoe N. P., Subrahmanyam Y. V. (1995) Proteases and bacterial virulence. A view from the trenches. Infect. Agents Dis. 4, 47–54 [PubMed] [Google Scholar]
- 3. Lähteenmäki K., Kuusela P., Korhonen T. K. (2001) Bacterial plasminogen activators and receptors. FEMS Microbiol. Rev. 25, 531–552 [DOI] [PubMed] [Google Scholar]
- 4. Lähteenmäki K., Edelman S., Korhonen T. K. (2005) Bacterial metastasis. The host plasminogen system in bacterial invasion. Trends Microbiol. 13, 79–85 [DOI] [PubMed] [Google Scholar]
- 5. Coleman J. L., Benach J. L. (1999) Use of the plasminogen activation system by microorganisms. J. Lab. Clin. Med. 134, 567–576 [DOI] [PubMed] [Google Scholar]
- 6. Grab D. J., Perides G., Dumler J. S., Kim K. J., Park J., Kim Y. V., Nikolskaia O., Choi K. S., Stins M. F., Kim K. S. (2005) Borrelia burgdorferi, host-derived proteases, and the blood-brain barrier. Infect. Immun. 73, 1014–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Coleman J. L., Gebbia J. A., Piesman J., Degen J. L., Bugge T. H., Benach J. L. (1997) Plasminogen is required for efficient dissemination of B. burgdorferi in ticks and for enhancement of spirochetemia in mice. Cell 89, 1111–1119 [DOI] [PubMed] [Google Scholar]
- 8. Coleman J. L., Sellati T. J., Testa J. E., Kew R. R., Furie M. B., Benach J. L. (1995) Borrelia burgdorferi binds plasminogen, resulting in enhanced penetration of endothelial monolayers. Infect. Immun. 63, 2478–2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mignatti P., Rifkin D. B. (1993) Biology and biochemistry of proteinases in tumor invasion. Physiol. Rev. 73, 161–195 [DOI] [PubMed] [Google Scholar]
- 10. Plow E. F., Herren T., Redlitz A., Miles L. A., Hoover-Plow J. L. (1995) The cell biology of the plasminogen system. FASEB J. 9, 939–945 [DOI] [PubMed] [Google Scholar]
- 11. Plow E. F., Ploplis V. A., Carmeliet P., Collen D. (1999) Plasminogen and cell migration in vivo. Fibrinolysis & Proteolysis 13, 49–53 [Google Scholar]
- 12. Sun H., Ringdahl U., Homeister J. W., Fay W. P., Engleberg N. C., Yang A. Y., Rozek L. S., Wang X., Sjöbring U., Ginsburg D. (2004) Plasminogen is a critical host pathogenicity factor for group A streptococcal infection. Science 305, 1283–1286 [DOI] [PubMed] [Google Scholar]
- 13. Klempner M. S., Noring R., Epstein M. P., McCloud B., Hu R., Limentani S. A., Rogers R. A. (1995) Binding of human plasminogen and urokinase-type plasminogen activator to the Lyme disease spirochete, Borrelia burgdorferi. J. Infect. Dis. 171, 1258–1265 [DOI] [PubMed] [Google Scholar]
- 14. Gebbia J. A., Monco J. C., Degen J. L., Bugge T. H., Benach J. L. (1999) The plasminogen activation system enhances brain and heart invasion in murine relapsing fever borreliosis. J. Clin. Invest. 103, 81–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Coleman J. L., Roemer E. J., Benach J. L. (1999) Plasmin-coated Borrelia burgdorferi degrades soluble and insoluble components of the mammalian extracellular matrix. Infect. Immun. 67, 3929–3936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hu L. T., Perides G., Noring R., Klempner M. S. (1995) Binding of human plasminogen to Borrelia burgdorferi. Infect. Immun. 63, 3491–3496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Klempner M. S., Noring R., Epstein M. P., McCloud B., Rogers R. A. (1996) Binding of human urokinase type plasminogen activator and plasminogen to Borrelia species. J. Infect. Dis. 174, 97–104 [DOI] [PubMed] [Google Scholar]
- 18. Nordstrand A., Shamaei-Tousi A., Ny A., Bergström S. (2001) Delayed invasion of the kidney and brain by Borrelia crocidurae in plasminogen-deficient mice. Infect. Immun. 69, 5832–5839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Blevins J. S., Revel A. T., Smith A. H., Bachlani G. N., Norgard M. V. (2007) Adaptation of a luciferase gene reporter and lac expression system to Borrelia burgdorferi. Appl. Environ. Microbiol. 73, 1501–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hallström T., Haupt K., Kraiczy P., Hortschansky P., Wallich R., Skerka C., Zipfel P. F. (2010) Complement regulator-acquiring surface protein 1 of Borrelia burgdorferi binds to human bone morphogenic protein 2, several extracellular matrix proteins, and plasminogen. J. Infect. Dis. 202, 490–498 [DOI] [PubMed] [Google Scholar]
- 21. Fuchs H., Wallich R., Simon M. M., Kramer M. D. (1994) The outer surface protein A of the spirochete Borrelia burgdorferi is a plasmin(ogen) receptor. Proc. Natl. Acad. Sci. U.S.A. 91, 12594–12598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Brissette C. A., Haupt K., Barthel D., Cooley A. E., Bowman A., Skerka C., Wallich R., Zipfel P. F., Kraiczy P., Stevenson B. (2009) Borrelia burgdorferi infection-associated proteins ErpP, ErpA, and ErpC bind human plasminogen. Infect. Immun. 77, 300–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lagal V., Portnoï D., Faure G., Postic D., Baranton G. (2006) Borrelia burgdorferi sensu stricto invasiveness is correlated with OspC-plasminogen affinity. Microbes Infect. 8, 645–652 [DOI] [PubMed] [Google Scholar]
- 24. Hu L. T., Pratt S. D., Perides G., Katz L., Rogers R. A., Klempner M. S. (1997) Isolation, cloning, and expression of a 70-kilodalton plasminogen binding protein of Borrelia burgdorferi. Infect. Immun. 65, 4989–4995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Inaba K. (2009) Disulfide bond formation system in Escherichia coli. J. Biochem. 146, 591–597 [DOI] [PubMed] [Google Scholar]
- 26. Demain A. L., Vaishnav P. (2009) Production of recombinant proteins by microbes and higher organisms. Biotechnol. Adv. 27, 297–306 [DOI] [PubMed] [Google Scholar]
- 27. Fuchs R., Jauris S., Lottspeich F., Preac-Mursic V., Wilske B., Soutschek E. (1992) Molecular analysis and expression of a Borrelia burgdorferi gene encoding a 22-kDa protein (pC) in Escherichia coli. Mol. Microbiol. 6, 503–509 [DOI] [PubMed] [Google Scholar]
- 28. Schwan T. G., Piesman J., Golde W. T., Dolan M. C., Rosa P. A. (1995) Induction of an outer surface protein on Borrelia burgdorferi during tick feeding. Proc. Natl. Acad. Sci. U.S.A. 92, 2909–2913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Masuzawa T., Kurita T., Kawabata H., Yanagihara Y. (1994) Relationship between infectivity and OspC expression in Lyme disease Borrelia. FEMS Microbiol. Lett. 123, 319–324 [DOI] [PubMed] [Google Scholar]
- 30. Fingerle V., Hauser U., Liegl G., Petko B., Preac-Mursic V., Wilske B. (1995) Expression of outer surface proteins A and C of Borrelia burgdorferi in Ixodes ricinus. J. Clin. Microbiol. 33, 1867–1869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schwan T. G., Piesman J. (2000) Temporal changes in outer surface proteins A and C of the lyme disease-associated spirochete, Borrelia burgdorferi, during the chain of infection in ticks and mice. J. Clin. Microbiol. 38, 382–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gilmore R. D., Jr., Piesman J. (2000) Inhibition of Borrelia burgdorferi migration from the midgut to the salivary glands following feeding by ticks on OspC-immunized mice. Infect. Immun. 68, 411–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Grimm D., Tilly K., Byram R., Stewart P. E., Krum J. G., Bueschel D. M., Schwan T. G., Policastro P. F., Elias A. F., Rosa P. A. (2004) Outer surface protein C of the Lyme disease spirochete. A protein induced in ticks for infection of mammals. Proc. Natl. Acad. Sci. U.S.A. 101, 3142–3147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pal U., Yang X., Chen M., Bockenstedt L. K., Anderson J. F., Flavell R. A., Norgard M. V., Fikrig E. (2004) OspC facilitates Borrelia burgdorferi invasion of Ixodes scapularis salivary glands. J. Clin. Invest. 113, 220–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tilly K., Krum J. G., Bestor A., Jewett M. W., Grimm D., Bueschel D., Byram R., Dorward D., Vanraden M. J., Stewart P., Rosa P. (2006) Borrelia burgdorferi OspC protein required exclusively in a crucial early stage of mammalian infection. Infect. Immun. 74, 3554–3564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fingerle V., Goettner G., Gern L., Wilske B., Schulte-Spechtel U. (2007) Complementation of a Borrelia afzelii OspC mutant highlights the crucial role of OspC for dissemination of Borrelia afzelii in Ixodes ricinus. Int. J. Med. Microbiol. 297, 97–107 [DOI] [PubMed] [Google Scholar]
- 37. Radolf J. D., Caimano M. J. (2008) The long strange trip of Borrelia burgdorferi outer-surface protein C. Mol. Microbiol. 69, 1–4 [DOI] [PubMed] [Google Scholar]
- 38. Zuckert W. R. (2007) Laboratory maintenance of Borrelia burgdorferi. Curr. Protoc. Microbiol. Chapter 12, Unit 12C.1 [DOI] [PubMed] [Google Scholar]
- 39. Alverson J., Bundle S. F., Sohaskey C. D., Lybecker M. C., Samuels D. S. (2003) Transcriptional regulation of the ospAB and ospC promoters from Borrelia burgdorferi. Mol. Microbiol. 48, 1665–1677 [DOI] [PubMed] [Google Scholar]
- 40. Carroll J. A., Garon C. F., Schwan T. G. (1999) Effects of environmental pH on membrane proteins in Borrelia burgdorferi. Infect. Immun. 67, 3181–3187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. El-Hage N., Stevenson B. (2002) Simultaneous coexpression of Borrelia burgdorferi Erp proteins occurs through a specific, erp locus-directed regulatory mechanism. J. Bacteriol. 184, 4536–4543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kumru O. S., Bunikis I., Sorokina I., Bergström S., Zückert W. R. (2011) Specificity and role of the Borrelia burgdorferi CtpA protease in outer membrane protein processing. J. Bacteriol. 193, 5759–5765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sarkar A., Hayes B. M., Dulebohn D. P., Rosa P. A. (2011) Regulation of the virulence determinant OspC by bbd18 on linear plasmid lp17 of Borrelia burgdorferi. J. Bacteriol. 193, 5365–5373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Elias A. F., Stewart P. E., Grimm D., Caimano M. J., Eggers C. H., Tilly K., Bono J. L., Akins D. R., Radolf J. D., Schwan T. G., Rosa P. (2002) Clonal polymorphism of Borrelia burgdorferi strain B31 MI. Implications for mutagenesis in an infectious strain background. Infect. Immun. 70, 2139–2150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Onder O., Turkarslan S., Sun D., Daldal F. (2008) Overproduction or absence of the periplasmic protease DegP severely compromises bacterial growth in the absence of the dithiol:disulfide oxidoreductase DsbA. Mol. Cell. Proteomics 7, 875–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wiener M. C., Sachs J. R., Deyanova E. G., Yates N. A. (2004) Differential mass spectrometry. A label-free LC-MS method for finding significant differences in complex peptide and protein mixtures. Anal. Chem. 76, 6085–6096 [DOI] [PubMed] [Google Scholar]
- 47. Xie F., Liu T., Qian W. J., Petyuk V. A., Smith R. D. (2011) Liquid chromatography-mass spectrometry-based quantitative proteomics. J. Biol. Chem. 286, 25443–25449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cederholm-Williams S. A. (1981) Concentration of plasminogen and antiplasmin in plasma and serum. J. Clin. Pathol. 34, 979–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Burgdorfer W., Barbour A. G., Hayes S. F., Benach J. L., Grunwaldt E., Davis J. P. (1982) Lyme disease. A tick-borne spirochetosis? Science 216, 1317–1319 [DOI] [PubMed] [Google Scholar]
- 50. Rooijakkers S. H., van Wamel W. J., Ruyken M., van Kessel K. P., van Strijp J. A. (2005) Anti-opsonic properties of staphylokinase. Microbes Infect. 7, 476–484 [DOI] [PubMed] [Google Scholar]
- 51. Xu Q., McShan K., Liang F. (2008) Essential protective role attributed to the surface lipoproteins of Borrelia burgdorferi against innate defences. Mol. Microbiol. 69, 15–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Earnhart C. G., Leblanc D. V., Alix K. E., Desrosiers D. C., Radolf J. D., Marconi R. T. (2010) Identification of residues within ligand-binding domain 1 (LBD1) of the Borrelia burgdorferi OspC protein required for function in the mammalian environment. Mol. Microbiol. 76, 393–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Seemanapalli S. V., Xu Q., McShan K., Liang F. T. (2010) Outer surface protein C is a dissemination-facilitating factor of Borrelia burgdorferi during mammalian infection. PLoS ONE 5, e15830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Babb K., McAlister J. D., Miller J. C., Stevenson B. (2004) Molecular characterization of Borrelia burgdorferi erp promoter/operator elements. J. Bacteriol. 186, 2745–2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Burns L. H., Adams C. A., Riley S. P., Jutras B. L., Bowman A., Chenail A. M., Cooley A. E., Haselhorst L. A., Moore A. M., Babb K., Fried M. G., Stevenson B. (2010) BpaB, a novel protein encoded by the Lyme disease spirochete's cp32 prophages, binds to erp Operator 2 DNA. Nucleic Acids Res. 38, 5443–5455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Narasimhan S., Santiago F., Koski R. A., Brei B., Anderson J. F., Fish D., Fikrig E. (2002) Examination of the Borrelia burgdorferi transcriptome in Ixodes scapularis during feeding. J. Bacteriol. 184, 3122–3125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Tokarz R., Anderton J. M., Katona L. I., Benach J. L. (2004) Combined effects of blood and temperature shift on Borrelia burgdorferi gene expression as determined by whole genome DNA array. Infect. Immun. 72, 5419–5432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wormser G. P., Brisson D., Liveris D., Hanincová K., Sandigursky S., Nowakowski J., Nadelman R. B., Ludin S., Schwartz I. (2008) Borrelia burgdorferi genotype predicts the capacity for hematogenous dissemination during early Lyme disease. J. Infect. Dis. 198, 1358–1364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Dykhuizen D. E., Brisson D., Sandigursky S., Wormser G. P., Nowakowski J., Nadelman R. B., Schwartz I. (2008) The propensity of different Borrelia burgdorferi sensu stricto genotypes to cause disseminated infections in humans. Am. J. Trop. Med. Hyg. 78, 806–810 [PMC free article] [PubMed] [Google Scholar]
- 60. Seinost G., Dykhuizen D. E., Dattwyler R. J., Golde W. T., Dunn J. J., Wang I. N., Wormser G. P., Schriefer M. E., Luft B. J. (1999) Four clones of Borrelia burgdorferi sensu stricto cause invasive infection in humans. Infect. Immun. 67, 3518–3524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Eicken C., Sharma V., Klabunde T., Owens R. T., Pikas D. S., Höök M., Sacchettini J. C. (2001) Crystal structure of Lyme disease antigen outer surface protein C from Borrelia burgdorferi. J. Biol. Chem. 276, 10010–10015 [DOI] [PubMed] [Google Scholar]
- 62. Kumaran D., Eswaramoorthy S., Luft B. J., Koide S., Dunn J. J., Lawson C. L., Swaminathan S. (2001) Crystal structure of outer surface protein C (OspC) from the Lyme disease spirochete, Borrelia burgdorferi. EMBO J. 20, 971–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Barbour A. G., Tessier S. L., Todd W. J. (1983) Lyme disease spirochetes and ixodid tick spirochetes share a common surface antigenic determinant defined by a monoclonal antibody. Infect. Immun. 41, 795–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Moriarty T. J., Norman M. U., Colarusso P., Bankhead T., Kubes P., Chaconas G. (2008) Real time high resolution three-dimensional imaging of the lyme disease spirochete adhering to and escaping from the vasculature of a living host. PLoS Pathog. 4, e1000090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Boyle M. D., Lottenberg R. (1997) Plasminogen activation by invasive human pathogens. Thromb. Haemost. 77, 1–10 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.