Background: Archaeal primase catalyzed the DNA-independent synthesis of oligonucleotide-like products from dNTPs.
Results: Mass spectrometry, NMR, and biochemical analyses identified these products as dNMP-glycerol and dNMP-Tris derivatives.
Conclusion: Archaeal primase catalyzes the formation of phosphodiester bonds between dNMPs and various acceptors.
Significance: Archaeal primase forms DNA-independent covalent adducts between deoxynucleotides and acceptors that mimic de novo aborted initiation reactions.
Keywords: DNA Enzymes, DNA Polymerase, DNA Primase, DNA Replication, DNA Synthesis, Initiation Reactions, Nucleotide Adducts, Template-independent
Abstract
In the presence of dATP, glycerol, and Tris buffer, the DNA primase isolated from Thermococcus kodakaraensis catalyzed the formation of dAMP and two products that were identified as dAMP-glycerol and dAMP-Tris. These products were formed by the T. kodakaraensis p41 catalytic subunit alone and the T. kodakaraensis p41-p46 complex in the absence of a DNA template. They were not formed with preparations containing the catalytically inactive p41 subunit. Similar glycerol and Tris derivatives as well as dNMPs were also formed with dGTP, dCTP, or dTTP. The mechanism contributing to the formation of these products and its implications in the initiation reaction catalyzed by the T. kodakaraensis primase are discussed.
Introduction
In the accompanying article (1), the isolation and characterization of the DNA primase from the archaeon Thermococcus kodakaraensis were described. Similar to eukaryotic DNA primases, the T. kodakaraensis primase is a two-subunit complex (p41-p46) in which the catalytic activity resides within the small subunit. The large subunit of both primases (T. kodakaraensis p46 and eukaryotic p58) contains a Fe-S cluster (2, 3), and sequence alignments between the two enzymes reveal homologies between both small and large subunits (4). In the small subunit, the region around the catalytic aspartate residues of the eukaryotic primase closely resemble the archaeal primase, suggesting that it is biologically important. The interaction of the small subunit with the large subunit stabilizes and modulates the catalytic activity of both primases (5–7).
In eukaryotes, the p48 catalytic subunit and the p48-p58 complex initiate short oligonucleotide synthesis only in the presence of rNTPs,2 and the size of these chains is influenced by the larger subunit (8). The eukaryotic complex and the catalytic subunit support dNTP incorporation only after de novo initiation with rNTPs. In archaea, the catalytic subunit initiates and extends DNA chains with dNTPs but fails to form or weakly forms RNA chains with rNTPs (1, 6, 7). Although the archaeal heterodimeric complex retains its ability to initiate and form DNA, the sizes of the DNA products synthesized are significantly shorter than those formed by the catalytic subunit (0.3–0.5 kb when compared with 3–7 kb). Furthermore, the archaeal complex synthesizes RNA chains with rNTPs (1, 6).
It has been reported that archaeal fragments formed in vivo contain oligoribonucleotides at their 5′ ends (9), suggesting that the archaeal primases initiate primer synthesis with rNTPs. In vitro studies with the Sulfolobus acidocaldarius DNA primase complex indicated that the apparent Km for rNTPs was 3 orders greater than the Km for dNTPs (10). However, studies with other archaeal primases isolated from Pyrococcus abyssi (7) and T. kodakaraensis (1) noted considerably less disparity in their Km values for rNTPs and dNTPs. Because archaea appears to lack DNA polymerase α activity, it has been suggested that the DNA synthetic activity of the archaeal primases may mimic the activity of this DNA polymerase (8). Thus, at present, the in vivo significance of dNTP initiation events observed with the archaeal DNA primase in vitro are unclear.
In the accompanying article (1), we noted the formation of two 32P-labeled products following incubation of the T. kodakaraensis primase complex or the T. kodakaraensis p41 catalytic subunit with [α-32P]dNTPs that migrated as 32P-labeled ∼16- and ∼25-nt bands following urea-PAGE separation. Their formation required a catalytically active p41 subunit but was independent of a DNA template. In this study, we identify these products as dAMP-glycerol (∼16-nt band) and dAMP-Tris (∼25-nt band). We show that each of the four common dNTPs support formation of these products, whereas rNTPs are considerably less active. In addition to these derivatives, dNMP is also produced in the reaction, indicating that H2O can replace glycerol and Tris in this reaction. Although the mechanism governing formation of these products is unclear, we discuss its possible significance in the DNA primase initiation and aborted initiation-elongation reactions.
EXPERIMENTAL PROCEDURES
Preparation of Proteins
The T. kodakaraensis primase proteins were isolated as described in the accompanying article (1). All enzymes were stored at −80 °C and stable to repeated freezing and thawing.
Commercial Enzymes
The enzymes and their sources included: spleen phosphodiesterase (46 units/mg, Amersham Biosciences); yeast inorganic pyrophosphatase and micrococcal nuclease (Worthington Biochemical); muscle myokinase, snake venom phosphodiesterase, yeast hexokinase, and calf intestinal phosphatase (CIP) (Sigma-Aldrich); and 3-phosphoglyceraldehyde dehydrogenase and 3-phosphoglycerate kinase (Roche Applied Science).
Assays for Production of dNMP Derivatives
Unless indicated, reaction mixtures (20 μl) containing 40 mm Tris-HCl, pH 8.0, 2 mm DTT, 10 mm magnesium acetate, 100 μm labeled dNTP or rNTP, 50 μg/ml BSA, and enzyme (as indicated) were incubated at 70 °C for a specified time. Unless noted, primase preparations were diluted with a solution containing 50% glycerol, 0.02 m Tris-HCl, pH 8.0, 1 mm DTT, 10 μm EDTA, and 50 μg/ml BSA (glycerol diluent). Aliquots of reactions were subjected to ascending thin layer chromatography (TLC) in the indicated solvents until the solvent front reached ∼1 cm from the top of the polyethyleneimine (PEI) cellular plate strips (Brinkmann Instruments); the strips were air-dried and subjected to autoradiography or phophorimaging. Aliquots of reactions were also subjected to 8 m urea-20% PAGE separation for 2–3 h at 300 V in 1× Tris-borate-EDTA, as described (1). Gels were usually subjected to autoradiography wet. Frozen gels were wrapped in paper towels to prevent moisture condensation.
Preparations of dAMP-glycerol and dAMP-Tris Products Using [13C,15N]dATP and Deuterated d-Tris for Analysis by High Mass Accuracy LC-MS and High Resolution NMR
Reaction mixtures (0.25 ml) containing 40 mm deuterated d-Tris-HCl (or light normal Tris-HCl), pH 8.0, 2 mm DTT, 10 mm magnesium acetate, 80 μm [13C,15N]dATP (or light ATP), 50 μg/ml BSA, and 5.84 μm p41 subunit (containing 0.64 m glycerol following dilution in the glycerol diluent described above) were incubated for 30 min at 70 °C. An identical reaction (25 μl) containing [α-32P]dATP revealed that following incubation with the p41 subunit and TLC analysis on PEI plates, >60% of the 32P was recovered in the fast migrating dAMP derivatives, >30% in dAMP, and <10% in ATP. This same distribution was observed in reactions containing d-Tris-HCl, pH 8.0, [13C,15N]dATP, or normally light reagents. Thus, under the conditions used, the use of heavy isotopes did not alter the utilization of dATP in the reaction. Samples from reaction mixtures were analyzed using a capillary reverse phase liquid chromatography system coupled to an Orbitrap Discovery mass spectrometer in negative mode (11). Negative mode nanospray mode was set to −1.5 kV, and chromatographic flow rate was <20 nl/min. Fragmentation was achieved using high collision dissociation at 45% energy. Isotope-labeled samples were run both individually and combined. By analyzing the mass difference between heavy and light fragmentation pattern, the identity of each fragmentation peak was established.
In parallel, isotope-labeled products of the reaction mixtures were analyzed directly using high resolution 31P NMR after the addition of 2H2O to the mixture, leading to a volume-to- volume percentage ratio of 90:10. Data were collected on a Bruker DRX 500-MHz spectrometer, equipped with a Broadband probe at 30 °C. Spectra were processed and analyzed using Topspin 1.3 on a Linux work station. One-dimensional 31P spectra were acquired using a carrier frequency of 5 ppm, sweep widths of 6000 Hz, 4097 complex points, and 4096 scans. 31P resonance assignments were made by comparison with one-dimensional 31P spectra of standard samples (dATP, dAMP, inorganic pyrophosphate (PPi), and inorganic phosphate (Pi)) and using two-dimensional 1H-31P hetero-COSY and 1H-13C HMQC correlated experiments applied to the isotope-labeled products.
Preparation of [γ-32P]dATP
The procedure used for this synthesis was a modified version of that described by Glynn and Chappell (12). Reaction mixtures (120 μl) containing 50 mm Tris-HCl, pH 8.0, 6 mm magnesium acetate, 2.5 mm cysteine, 10 mm 3-phosphoglycerate, 40 μm NAD, 0.2 mm dATP, 2 μg of 3-phosphoglycerate kinase, 40 μg of 3-phosphoglyceraldehyde dehydrogenase, and 32Pi (7.68 × 107 cpm) were incubated for 60 min at 37 °C. Mixtures were treated with 100 μl of 10% charcoal slurry (previously washed with 1 n HCl and H2O until neutral), 50 μl of 1 n HCl, and 50 μl of 10 mm sodium phosphate, pH 7.5. Charcoal was collected by centrifugation, washed three times with 0.3 ml of 10 mm sodium phosphate buffer, pH 7.5, and then washed twice with 0.3 ml of H2O. Charcoal adsorbed material was eluted with three washes of 100 μl of H2O-ethanol-concentrated NH4OH mixture (1:1, 0.08), which were pooled and concentrated in vacuo. The dried material was suspended in 0.2 ml of 1× TE and centrifuged at 20,000 rpm in an Eppendorf centrifuge at 4 °C. Approximately 45% of the input 32P was recovered in the eluted material. Assuming quantitative incorporation of 32Pi into dATP, [γ-32P]dATP would be expected to contain 3200 cpm/pmol. The 32P recovery (3.5 × 107 cpm) suggested that ∼11 nmol of dATP was isolated. TLC on PEI cellular plates in 0.5 m ammonium formate, pH 3.5, indicated that the preparation contained 94% [γ-32P]dATP and 6% 32Pi. Incubation with excess glucose and hexokinase resulted in the transfer of the 32P present in [γ-32P]dATP quantitatively to glucose 6-phosphate.
Preparation of [βγ-32P]dATP
Reaction mixtures (120 μl) containing 20 mm Tris-HCl, pH 8.0, 10 mm magnesium acetate, 0.4 mm cysteine, 2 mm 3-phosphoglycerate, 18 nm NAD, 32Pi (6 × 108 cpm), 0.25 mm dATP, 25 nm dAMP, 20 μg of 3-phosphoglycerate kinase, 30 μg of 3-phospho glyceraldehyde dehydrogenase, and 5 μg of muscle myokinase were incubated at 37 °C for 60 min. Following incubation, 100 μl of 20% charcoal and 220 μl of 1 n HCl were added. After 20 min on ice, the mixture was centrifuged, washed four times with 0.4 ml of 5 mm sodium phosphate buffer, pH 7.5, and then washed once with 0.5 ml of H2O. The adsorbed material was eluted with a freshly prepared mixture of H2O-ethanol-NH4OH (400 μl, six times). The washings were combined, vacuum-dried, and suspended in 0.2 ml of 1× TE. Approximately 40% of the 32Pi added to the reaction was recovered (2.36 × 108 cpm). Based on the level of dATP added and assuming complete utilization of the 32Pi, the isolated dATP had a specific activity of ∼20,000 cpm/pmol with 32P distributed equally between the β and γ phosphate residues of dATP. TLC on PEI plates in 0.5 m NH4 formate, pH 3.5, indicated that the preparation contained 92% [32P]dATP and 8% [32P]dADP; treatment with hexokinase and glucose resulted in the quantitative disappearance of [γ-32P]dATP and formation of equally labeled dADP and glucose 6-phosphate, in keeping with the equal distribution of 32P between the β and γ phosphate residues of dATP.
Preparation of [32P]dAMP
[32P]dAMP was synthesized from [α-32P]dATP by the combined action of hexokinase and myokinase. Reaction mixtures (20 μl) containing 40 mm Tris-HCl, pH 8.0, 10 mm magnesium acetate, 0.5 mm DTT, 50 μg/ml BSA, 50 mm glucose, 25 μm [α-32P]dATP (2 × 104 cpm/pmol), 2 μg of yeast hexokinase, and 6 μg of muscle myokinase were incubated at 37 °C. The mixture was extracted with phenol/CHCl3/isoamyl/alcohol mixture, the aqueous phase was adsorbed to activated charcoal (50 μl of 10% solution in 0.1 n HCl), which was collected by centrifugation and washed with 100 μl of H2O three times, and the bound nucleotide was eluted with H2O/ethanol/NH4OH mixture (twice). The pooled eluate were vacuum-concentrated and suspended in 30 μl of TE buffer. Based on the 32P recovered, approximately 50% of the input [α-32P]dATP was recovered as [32P]dAMP. TLC on PEI plates indicated that this material contained 92% [32P]dAMP, 7% dADP, and 1% dATP.
RESULTS
Requirements for Synthesis of [32P]dAMP Derivatives
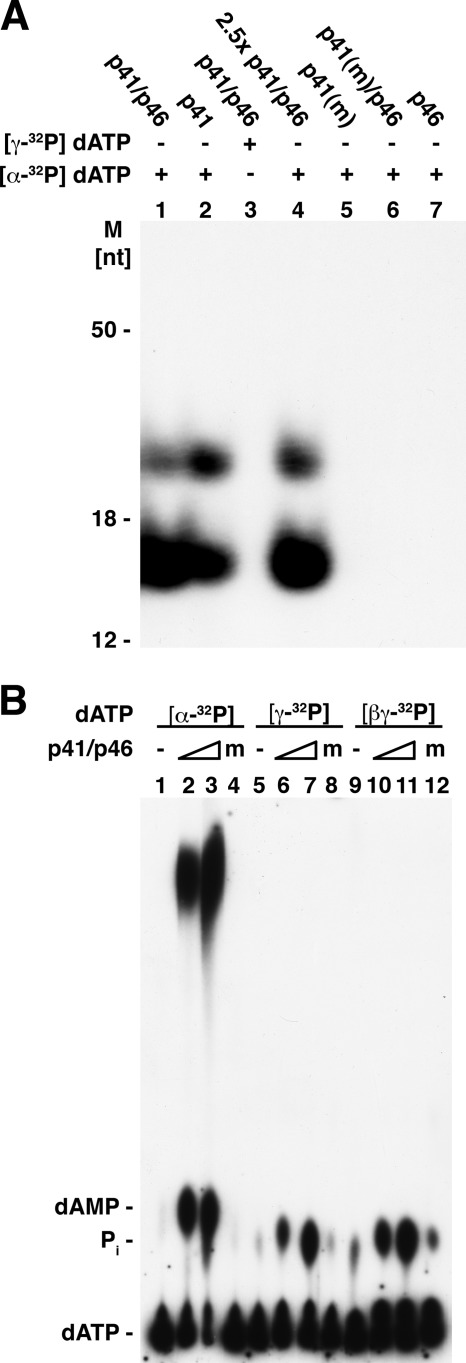
Both the T. kodakaraensis primase complex and the p41 subunit catalyzed the synthesis of two distinct labeled products from [α-32P]dATP that migrated as ∼16- and ∼25-nt bands following urea-PAGE separation (Fig. 1A). These derivatives were formed in the absence of a DNA template with any one of the [α-32P]dNTPs but were not readily detected with [α-32P]rNTPs (1). Their production required the catalytically active p41 subunit (compare lanes 1 and 2 with lanes 5 and 6) and were not observed when [α-32P]dATP was replaced by [γ-32P]dATP (Fig. 1A, lane 3) or following incubation with the p46 subunit alone (lane 7). Incubation of [α-32P]dATP with higher levels of the p41-p46 complex increased the amount of products formed (lane 4). These findings indicated that the products contained the α-phosphate moiety but not the γ-phosphate residue of dNTPs.
FIGURE 1.
A, urea-PAGE analysis of products formed by T. kodakaraensis primase preparations following incubation with [α-32P]dATP or [γ-32P]dATP. Reaction mixtures (20 μl) containing 40 mm Tris-HCl, pH 8.0, 10 mm magnesium acetate, 2 mm MnCl2, 50 μg/ml BSA, 2 mm DTT, 100 μm [α-32P]dATP (4250 cpm/pmol) or 100 μm [γ-32P]dATP (2430 cpm/pmol), where specified, and 0.5 μm (or 1.25 μm) of the indicated T. kodakaraensis primase preparations were incubated for 30 min at 70 °C. Aliquots (5 μl) from reactions were separated by urea-PAGE and subjected to autoradiography. m, mutant; M, molecular mass markers. B, TLC analysis of products formed following incubation of T. kodakaraensis primase preparations with labeled dATP. Reactions were as described in panel A with the exception that MnCl2 was omitted. Where indicated, mixtures contained [α-32P]dATP (4 × 103 cpm/pmol), [γ-32P]dATP (3.4 × 103 cpm/pmol), or [βγ-32P]dATP (5.1 × 103 cpm/pmol and 0.25 or 0.75 μm T. kodakaraensis primase complex or 0.75 μm mutant p41-p46 complex (indicated as m). Aliquots (1 μl) of each reaction were analyzed by TLC on a PEI plate developed in 0.3 m LiCl.
Products formed by the T. kodakaraensis primase complex with dATP were also analyzed by TLC on PEI plates (Fig. 1B). Incubation with [α-32P]dATP resulted in the production of a labeled fast migrating material and [32P]dAMP (Fig. 1B, lane 2). The amount of these products formed increased in the presence of increasing levels of the primase complex (Fig. 1B, lanes 2 and 3). They were not formed when the catalytically inactive primase complex (mutant p41-p46 complex) was used in place of the active complex (lane 4). When [γ-32P]dATP (lanes 5–8) or [βγ-32P]dATP (lanes 9–12) was added in lieu of [α-32P]dATP, the rapid migrating labeled product was not detected. However, 32Pi was formed, and its production required an active primase complex. As discussed in more detail below, PPi is produced in the primase reaction, which is partially hydrolyzed to Pi by the inorganic pyrophosphatase activity present in the primase preparations. We infer from these findings that T. kodakaraensis primase preparations convert [α-32P]dATP to multiple labeled products, including the 16- and 25-nt bands detected following urea-PAGE separation and dAMP and a rapid migrating material (which as described below includes the ∼16- and ∼25-nt products) following TLC separation.
Properties of Products Formed from [α-32P]dATP
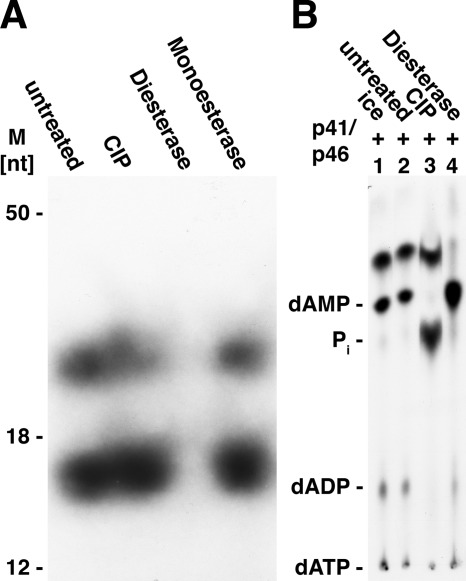
Products formed from [α-32P]dATP by the T. kodakaraensis primase complex were subjected, without isolation, to digestion with calf intestinal phosphatase (CIP), snake venom phosphodiesterase, or spleen phosphodiesterase. Urea-PAGE analysis indicated that both labeled products were hydrolyzed quantitatively by snake venom phosphodiesterase but unaffected by CIP or spleen phosphodiesterase (Fig. 2A). It should be noted that spleen phosphodiesterase requires the presence of a 5′-hydroxyl group for its action (13). The influence of CIP and venom phosphodiesterase was also examined by TLC separation using PEI plates (Fig. 2B). CIP treatment did not alter the level of the fast migrating dAMP derivatives, but hydrolyzed the residual dATP (and dADP) remaining after primase action to Pi (lane 3). Venom phosphodiesterase converted the fast migrating labeled derivatives to dAMP almost quantitatively (lane 4). Incubation of the 32P-labeled fast migrating labeled material with both venom phosphodiesterase and CIP converted the 32P present in this derivative to 32Pi. These findings indicated that the dAMP present in this product contained a phosphodiester linkage.
FIGURE 2.
A, treatment of products with CIP, venom phosphodiesterase, and spleen phosphodiesterase. Aliquots of reactions following incubation of [α-32P]dATP and the T. kodakaraensis primase complex (as described in the legend for Fig. 1) were subjected to the following treatment. Reactions (10 μl) contained 5 μl of the above reaction products, 40 mm Tris-HCl, pH 8.5, 2 mm magnesium acetate, CIP (1 unit), or snake venom phosphodiesterase (0.1 unit). Reactions with spleen phosphodiesterase (0.04 unit) contained 40 mm Tris-HCl, pH 7.0, and 1 mm magnesium acetate. All mixtures were incubated at 37 °C for 30 min and then subjected to urea-PAGE analysis and autoradiography. M, molecular mass markers. B, TLC analysis of products formed after treatment with CIP and snake venom phosphodiesterase. Reaction mixtures (5 μl) containing 1 μl of the products formed from [α-32P]dATP (as described in panel A), 40 mm Tris-HCl, pH 8.5, 2 mm magnesium acetate, and CIP (0.5 unit) or snake venom phosphodiesterase (0.1 unit) were incubated for 20 min at 37 °C. Mixtures devoid of these enzymes were incubated on ice (lane 1) or at 37 °C (untreated, lane 2). Following incubation, aliquots (1 μl) were plated on a PEI plate and developed in 0.5 m ammonium formate, pH 3.5. Markers indicating the migration of standards are shown on the left.
Analysis of Products Formed by Capillary HPLC-MS
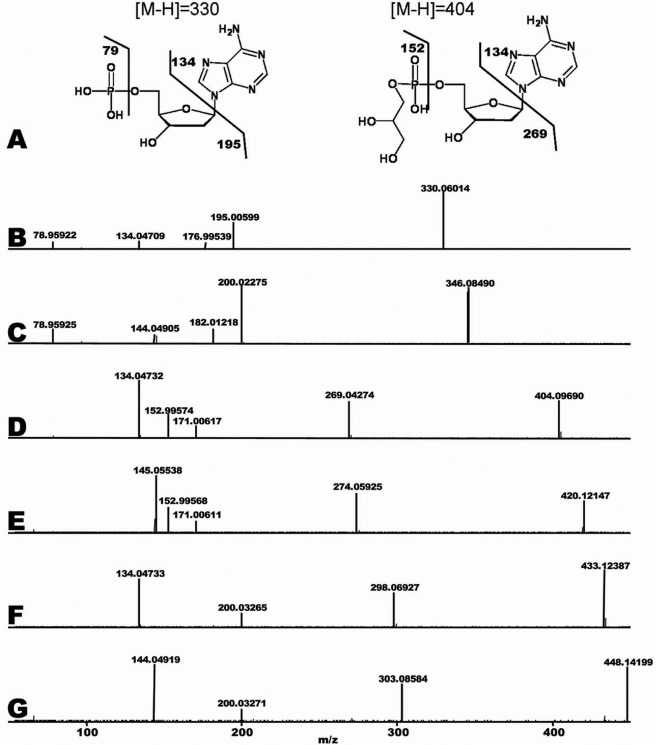
TLC and urea-PAGE separation indicated that the dAMP derivatives were not pdApdA, dApdA, pdApdApA, or dApdApdA (data not presented). We next turned to HPLC-MS analysis to identify the nature of the products. For this purpose, the dAMP derivatives formed from [C12,N14]dATP, [C13,N15]dATP, and d-Tris buffer were prepared as described under “Experimental Procedures,” analyzed by high mass accuracy LC-MS, and compared with dATP. Three chromatographic peaks were prominent in product samples with molecular weights of 330, 404, and 433 (the structure of the 330 and 404 peaks is shown in Fig. 3A). The peak with a molecular weight of 330 was identified as dAMP based on exact mass and fragmentation pattern matching the METLIN database. To further investigate the identity of peaks 404 and 433, 13C,15N (heavy) stable isotopes of dATP were used in conjunction with naturally occurring isotopes 12C,14N (light) of dATP. Heavy and light dATP were incubated in separate reactions with enzyme, and the products were analyzed for exact mass and fragmentation pattern. Subsequently, both heavy and light samples were mixed 1:1 to ensure proper correlation by chromatographic co-elution. As expected, peak 330 (light) yielded a corresponding peak 345 (heavy). Fragmentation of both these compounds and the products formed is shown in Fig. 3, B and C. The first major fragments formed (200/195) indicate the loss of the adenine base, which contains 10 heavy isotopes. A mass of 79 is indicative of a phosphate group. These findings establish the identity of the 330 m/z peaks as dAMP. To identify the unknown compound 404 m/z (structure shown in Fig. 3A), a similar analysis was undertaken. The 419/404 (heavy/light) m/z derivatives were thought to be a modified dAMP based on dATP being the sole substrate of the reaction and its mobility on TLC plate. The 419/404 m/z derivatives yielded fragmentation patterns similar to those of dAMP (345/330 m/z) with the exception of a 79 m/z peak. The difference between 419/404 and their major fragment peaks of 274/269 m/z was the exact mass and isotope composition of an adenine base. This confirms that 419/404 m/z is derived from dATP and that the unknown modification is not attached directly to the adenine base. The mass difference between dAMP and 404 m/z is 74.03676. The exact mass indicates that such a modification is C3H6O2. Fig. 3, D and E, show that peak 171 appears in both the 419 m/z and the 404 m/z samples. Subtracting a mass of 74.03676 from 171 m/z and accounting for water loss, which is common during MS fragmentation, suggests that 171.00615 m/z is phosphate + C3H6O2. This indicates that the modification is attached to the phosphate. As 171 m/z is found in both heavy and light samples, the 74.03676 m/z cannot be derived from the original dATP as all the dATP carbons in the heavy sample are labeled with 13C. Examination of the exact mass of 171 m/z and factoring in the dehydration reaction necessary to bond to phosphate identified the modification as glycerol, which is the sole match in the database. This modification is likely as the enzyme was diluted in 50% glycerol. Peak 448/433, examined in a similar manner, identified Tris base as the modifying conjugate (Fig. 3F, 433 m/z 12C,14N, and Fig. 3G, 448 m/z 13C,15N).
FIGURE 3.
Identification of dAMP modifications by LC-MS/MS. Reaction mixtures, as described under “Experimental Procedures” with dATP, resulted in products dAMP (330 m/z) and glycerol-dAMP (404 m/z) (structures consistent with the MS data are shown in A). B–G, reactions contained 0.75 μm T. kodakaraensis primase preparation diluted in 50% glycerol with [12C,14N]dATP (B, D, and F) or [13C,15N]dATP (C, E, and G) as substrates. Samples were analyzed by capillary LC-MS/MS. B, high resolution fragmentation of 330 m/z U-12C,14N; C, 345 m/z, the U-13C,15N counterpart to 330 m/z; D, 419 m/z U-12C,14N; E, 404 m/z U-13C,15N counterpart to 404 m/z; F, 433 m/z U-12C,14N; G, 448 m/z, the U-13C,15N counterpart to 433 m/z. See “Results” for analyses.
Analysis of Products Formed by 31P NMR
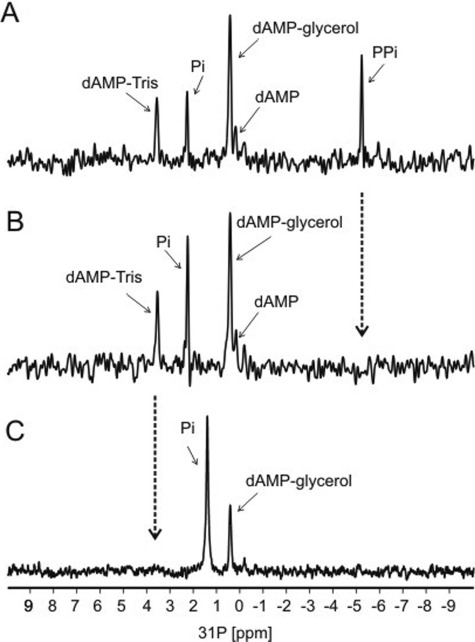
One-dimensional 31P NMR spectral analysis was used to confirm the nature of the phosphorylated products formed in the reaction. For this purpose, the dAMP derivatives prepared with [12C,14N]dATP, [13C,15N]dATP, and d-Tris buffer were again used. Initial one-dimensional 31P spectra of the reaction mixture (Fig. 4A) containing [13C,15N]ATP, glycerol, and d-Tris revealed four main 31P resonance peaks that could be assigned to phosphorus atoms contained in the dAMP-Tris and dAMP-glycerol adducts, as well as signals from Pi and PPi. In addition, a minor peak was also observed that could be assigned to dAMP. Note that in the 31P spectra, no signals were observed from the starting substrate dATP, indicating that the input dATP was converted quantitatively to products. After a few days at room temperature, the one-dimensional 31P spectra of the same reaction mixture (Fig. 4B) revealed only three main 31P peaks that were assigned to dAMP-Tris, dAMP-glycerol, dAMP, and Pi. In this spectrum, the PPi signal “disappeared” concomitant with an increased Pi peak, indicating that PPi was hydrolyzed to Pi. This finding is in keeping with the presence of inorganic pyrophosphatase in T. kodakaraensis primase preparations as described below. To confirm the formation of the dAMP-Tris adduct, a one-dimensional 31P spectrum of the reaction mixture containing [13C,15N]ATP, glycerol and glycine, substituted for d-Tris as the buffer, was examined. In this spectrum (Fig. 4C), 31P resonances assigned to dAMP-glycerol and Pi were observed, and as expected, the 31P resonance signal assigned to dAMP-Tris product was not observed. For both the dAMP-Tris and the dAMP-glycerol adducts, the 31P NMR confirms that these adducts formed via a 5′ a-phosphorus linkage to dAMP. In addition, 1H NMR indicated no other chemical modifications to the base or sugar of the dAMP moiety in these adducts.
FIGURE 4.
31P NMR analysis of reaction products. Reaction mixtures were as described in the legend for Fig. 3. A, initial one-dimensional 31P spectrum of the reaction mixture containing [13C,15N]ATP, glycerol, and d-Tris showing five 31P resonance peaks; 31P resonances in the spectra assigned to dAMP-Tris, dAMP-glycerol, dAMP, Pi, and PPi are labeled. Note that there is no observable signal from dATP, indicating the complete conversion of the input dATP to products. B, one-dimensional 31P spectrum of the reaction mixture (after a few days) showing four 31P peaks; 31P resonances in the spectrum assigned to dAMP-Tris, dAMP-glycerol, dAMP, and Pi are labeled. The PPi peak disappeared (noted by arrows) and the Pi peak increased, indicating a breakdown of PPi to Pi. C, one-dimensional 31P spectrum of the reaction mixture containing [13C,15N]ATP, glycerol, and glycine, substituted for d-Tris, as buffer showing two 31P peaks; 31P resonances in the spectrum assigned to dAMP-glycerol and Pi are labeled. The 31P resonances in the spectrum assigned to dAMP-Tris are not observed. Note that the resonance of Pi in this reaction was slightly shifted when compared with the Pi signal from the Tris-buffered reactions containing Tris buffer due to sensitivity of the inorganic phosphate to differences in the pH of the reactions.
Formation of dAMP-glycerol and dAMP-Tris Products
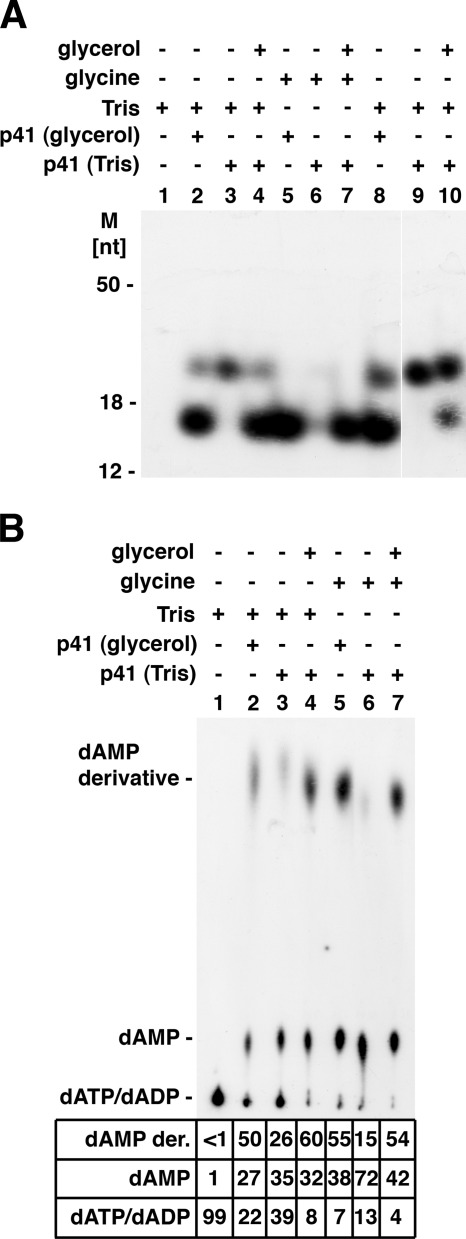
We next demonstrated that formation of the dAMP-Tris product (migrating as the ∼25-nt product) and the dAMP-glycerol product (migrating as an ∼16-nt product) required the addition of Tris buffer and glycerol, respectively, to primase reaction mixtures (Fig. 5). A glycerol diluent was added to primase reaction mixtures to dilute enzyme preparations (solutions containing 50% glycerol, equivalent to 5 m glycerol). For these reasons, reactions containing glycine buffer in place of Tris buffer were employed, and enzyme fractions were diluted with 10 mm Tris-HCl buffer, pH 8.0, and 1 mm EDTA (1× TE) rather than the 50% glycerol buffer diluent. As shown in Fig. 5A, primase reactions containing both 40 mm Tris buffer and enzyme diluted with the glycerol diluent (p41 (glycerol)) yielded the dAMP-Tris and dAMP-glycerol products following urea-PAGE analysis (Fig. 5A, lane 2). Reactions containing Tris buffer and primase fractions diluted in TE buffer (p41 (Tris)) rather than the glycerol diluent yielded the dAMP-Tris product but no dAMP glycerol derivative (lane 3); glycerol addition to such reactions (0.6 m glycerol, final concentration) led to the formation of both dAMP derivatives (lane 4). The omission of glycerol in reactions containing Tris buffer resulted in an increased formation of the dAMP-Tris derivative (compare lane 3 with lanes 2 and 4), suggesting that glycerol and Tris may compete in the reaction. When reactions contained enzyme diluted with the glycerol diluent and glycine buffer in place of Tris buffer, the dAMP-glycerol derivative was the only product formed (lane 5). When both Tris and glycerol were omitted, neither derivative was produced (lane 6). Identical results were obtained when primase reactions were carried out in 10-μl rather than 20-μl reaction mixtures (lanes 8–10). In the presence of Tris and glycerol, the catalytic p41 subunit generated both products (lane 8); omission of glycerol resulted in the formation of the dAMP-Tris derivative (lane 9), and the addition of low levels of glycerol (0.12 m rather than 0.6 m) resulted in the formation of low levels of the dAMP-glycerol product (compare lane 8 and lane 10).
FIGURE 5.
Influence of glycerol and Tris on formation of dAMP-glycerol and dAMP-Tris derivative. A, analysis of products by urea-PAGE separation. Reaction mixtures (lanes 1–7) were as described in the legend for Fig. 1 with [α-32P]dATP and (where indicated) 40 mm glycine buffer, pH 8.8 (glycine), 40 mm Tris-HCl buffer, pH 8.0 (Tris), 0.5 μm T. kodakaraensis p41 subunit diluted in 50% glycerol diluent (p41(glycerol)), or 0.5 μm T. kodakaraensis p41 subunit diluted in 1× TE (p41 (Tris)). MnCl2 was omitted in this experiment. Reactions described in lanes 8–10 were carried out in 10-μl reactions. Reaction described in lanes 4 and 7 (p41 glycerol) contained 0.6 m glycerol, whereas the reaction in lane 10 contained 0.2 m glycerol. M, molecular mass markers. B, analysis of products by thin layer chromatography. Reaction mixtures were as described in panel A; aliquots (1 μl) were analyzed on PEI plates developed in 0.3 m LiCl. The 32P distribution (in percentage) is presented. dAMP der., dAMP derivative.
The influence of glycerol and Tris on the formation of the dAMP-glycerol and dAMP-Tris derivative was also examined by TLC on PEI plates (Fig. 5B). As these derivatives are not separated by TLC and the dAMP-glycerol product represents the major dAMP derivative formed, omission of Tris had little quantitative effect on the level of the fast migrating products formed, whereas glycerol addition (compare lanes 3 and 4 with lanes 6 and 7) stimulated formation of dAMP-glycerol. The production of dAMP was also increased substantially in the absence of Tris and glycerol (lane 6). It should be noted that dATP hydrolysis to dAMP required the catalytically active p41 subunit. Collectively, these findings are in accord with the mass spectrometric and NMR analyses and indicate that the products formed are the dAMP-glycerol and dAMP-Tris derivatives.
We also examined whether RNA and DNA synthesis catalyzed by the T. kodakaraensis primase complex as well as the p41 subunit was affected by the presence or absence of glycerol and/or Tris. Their omission decreased DNA synthesis slightly (20–30%) with both primase preparations, whereas RNA synthesis catalyzed by the complex was unaffected (data not presented).
TLC Analysis of Products Synthesized in Presence of [α-32P]dATP or [α-32P]rATP
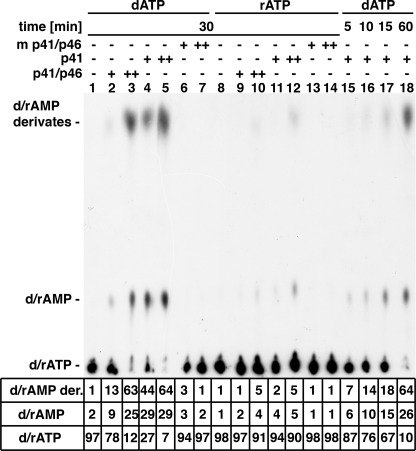
The synthesis of the fast migrating derivatives by the primase complex, the p41 subunit, and the mutant p41 catalytic subunit with [α-32P]dATP and [α-32P]rATP was examined by TLC analysis (Fig. 6). Labeled dAMP derivatives were formed more extensively with the p41 subunit than with the primase complex (∼3.5-fold, compare lane 2 and with lane 4). Their formation required the catalytically active p41 subunit as no derivatives were detected with the mutated p41 subunit (lanes 6 and 7). In addition to dAMP derivatives, primase preparations also generated dAMP from dATP. Identical levels of dAMP and the rapid migrating dAMP products were formed by the p41 subunit at early time points (5–15 min, lanes 15–17), but differed at later times. Approximately 90% of the input dATP was utilized in 30 min in the presence of 0.75 μm p41 subunit; 60% of this material was converted to the dAMP derivatives, and ∼30% was converted to dAMP (lane 5). When [α-32P]rATP was used in place of dATP, significantly lower levels of the putative AMP products were formed (lanes 9–12) than observed with dATP. Like dATP, rATP was not utilized by the mutant catalytic subunit (lanes 13 and 14). Thus, under the conditions used, the T. kodakaraensis DNA primase preparations utilized rATP less effectively than dATP (>10%).
FIGURE 6.
Analysis of products formed from [α-32P]dATP and [α-32P]rATP. Reaction mixtures (20 μl), as described in the legend for Fig. 1, containing 100 μm [α-32P]dATP (611 cpm/pmol) or 200 μm [α-32P]ATP (1090 cpm/pmol), 0.25 or 0.75 μm T. kodakaraensis DNA primase complex (lanes 2, 3, 9, and 10), and p41 subunit (lanes 4, 5, 11, and 12) or mutant (m) p41 subunit (lanes 6, 7, 13, and 14) were incubated for 30 min at 70 °C. Reactions containing 0.25 μm of the p41 subunit were incubated for 5 (lane 15), 10 (lane 16), 15 (lane 17), and 60 min (lane 18). Aliquots (1 μl) from reactions were separated by TLC on PEI plates in 0.3 m LiCl, after which plates were dried and quantified by phosphorimaging. d/rAMP der., dAMP/rAMP derivative; rAMP, 5′-riboadenosyl monophosphate.
Formation of Glycerol and Tris Derivatives with Other dNTPs
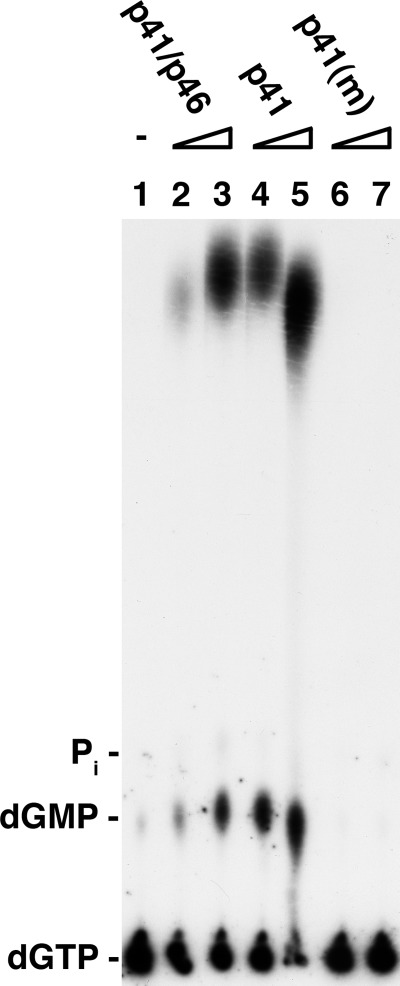
The T. kodakaraensis primase complex and p41 subunit converted the other [α-32P]dNTPs to the glycerol and Tris derivatives, and as observed with [α-32P]dATP, the mutant p41 subunit did not support these reactions. Products formed in reactions containing [α-32P]dGTP, analyzed by TLC on PEI plates, were dGMP and the dGMP derivatives (Fig. 7). Based on the 32P distribution, the level of the products formed equaled the amount of dGTP utilized. Maximally (with 0.75 μm p41 subunit), 90% of the input dGTP was utilized, leading to the production of ∼65% dGMP-glycerol and Tris derivatives and 20% dGMP. As observed with [α-32P]dATP, low levels of the catalytic subunit were more active than the primase complex (Fig. 7, compare lanes 2 and 3 with lanes 4 and 5). [α-32P]dTTP and [α-32P]dCTP were also utilized in a manner similar to that observed with dGTP and dATP (data not presented); thus, all four dNTPs are converted to the dNMP-glycerol and Tris derivatives and dNMP. The products formed with each of the four [α-32P]dNTPs, separated by urea-PAGE analysis, yielded the identical ∼16- and ∼25-nt species, which were hydrolyzed by venom diesterase and resistant to CIP and spleen phosphodiesterase (data not presented).
FIGURE 7.
Formation of dGMP-glycerol and dGMP-Tris products. Reactions (20 μl) were as described (Fig. 6) with [α-32P]dGTP (9400 cpm/pmol used in place of [α-32P]dATP. Aliquots of reactions (1 μl) were subjected to TLC on PEI plates in PEI thin layer chromatography in 0.3 m LiCl. m, mutants.
Formation of PPi and Pi from dNTPs
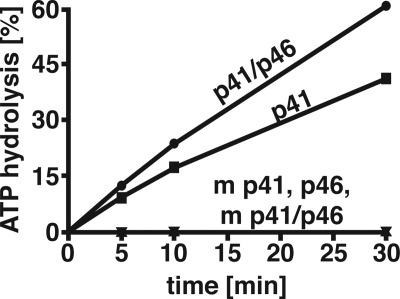
Formation of dAMP-glycerol, dAMP-Tris, and dAMP by T. kodakaraensis DNA primase preparations suggested that PPi or possibly Pi was produced as well. Incubation of [γ-32P]dATP with T. kodakaraensis DNA primase preparations resulted in the time-dependent formation of 32Pi (Fig. 8). After 30 min of incubation, ∼40 and 60% of the input [γ-32P]dATP was converted to 32Pi by the p41 subunit and primase complex, respectively, whereas 32Pi was not formed by mutant preparations or the p46 subunit alone. In the presence of [γ-32P]ATP, rather than [γ-32P]dATP, Pi formation was reduced ∼10-fold (data not presented). Examination of the different primase preparations for inorganic pyrophosphatase (PPase) activity revealed that all preparations contained this activity, but their levels varied. The p41, mutant p41, wild-type complex, mutant p41-p46 complex, and p46 subunit hydrolyzed PPi to Pi (pmol/min/pmol of preparation at 70 °C) 0.7, 0.9, 12.1, 18, and 12.0, respectively, suggesting the Pi could arise by hydrolysis of PPi generated by the action of primase. Preparations devoid of primase activity (mutant p41, mutant p41-p46, and p46 subunit), incapable of generating PPi from [γ-32P]dATP, did not form Pi (Fig. 8).
FIGURE 8.
Rate of [γ-32P]dATP hydrolysis by T. kodakaraensis primase preparations. Reaction mixtures (20 μl) containing 40 mm Tris-HCl, pH 8.0, 2.5 mm DTT, 10 mm magnesium acetate, 100 μm [γ-32P]dATP (1900 cpm/pmol), and 0.5 μm of T. kodakaraensis DNA primase preparation (as indicated) were incubated for 5, 10, and 30 min at 70 °C. Aliquots (1 μl) were subjected to TLC on PEI plates developed with 0.3 m LiCl. After drying, the plate was subjected to phosphorimaging, and the 32P that migrated as Pi (in percentage) was determined. The only other 32P material detected migrated in the dATP region and was not separated from PPi. Preparations labeled m p41 and m p41-p46 indicate the presence of mutated p41 subunit.
We next examined whether PPi was formed during the primase reaction. TLC on PEI plates, using a number of different solvents, failed to adequately resolve dATP (and other dNTPs) from PPi. To evaluate whether 32PPi was formed during [γ-32P]dATP hydrolysis, we used charcoal, which selectively binds nucleotides but not PPi or Pi, subjected the charcoal nonadsorbed material to yeast inorganic PPase treatment, and measured 32Pi formation following TLC separation (Table 1). Under the conditions used, 95% of the input 32P of the added [γ-32P]dATP was adsorbed to charcoal, whereas the low level of nonadsorbable material migrated as 32Pi. When [γ-32P]dATP was incubated with the p41 subunit, ∼20% of the [γ-32P]dATP was hydrolyzed to Pi. TLC separation of the charcoal nonadsorbable material showed that 80% of this material was PPi. When this material was incubated with yeast inorganic PPase, the 32P present in the fraction was converted quantitatively to Pi (Table 1). Similar analysis of the products formed by the T. kodakaraensis primase complex revealed that a major portion of the charcoal nonadsorbable material released from [α-32P]dATP was isolated at 32Pi; the 32PPi in this fraction was converted to 32Pi following yeast PPase digestion. These findings suggest that the PPi is most likely formed by hydrolysis of dATP, which is then converted to Pi by the PPase present in the enzyme fraction. A major source of PPase activity present in the primase complex appeared to be the p46 subunit. Because the PPase activity measurements were carried out at 70 °C, it is likely that this activity is associated with the archaeal proteins and not an Escherichia coli contaminant. The nature of this activity, however, remains to be explored.
TABLE 1.
Identification of PPi and Pi as products formed by T. kodakaraensis DNA primase preparations
Reaction mixtures, as described in the legend for Fig. 8, were incubated for 10 min at 70 °C. Charcoal (10 μl of 15% slurry) and 0.1 m HCl (10 μl) were added, and after 10 min on ice, the mixture was centrifuged. Aliquots of the supernatant (2 μl) were incubated in reactions (5 μl) containing 100 mm Tris-HCl, pH 7.5, and 2 mm magnesium acetate in the presence or absence of yeast inorganic PPase (1 μg) for 30 min at 37 °C. Aliquots (2 μl) were then subjected to TLC separation on PEI plates in 0.3 m LiCl and phosphorimaging.
| Primase preparation added and treatment | Charcoal nonadsorbable product (% of distribution) |
|
|---|---|---|
| PPi | Pi | |
| p41 | 80.4 | 19.6 |
| p41 + PPase | 4.0 | 96.0 |
| p41/p46 | 20.2 | 78.8 |
| p41/p46 + PPase | 4.2 | 95.8 |
DISCUSSION
In the accompanying article, we demonstrated that the T. kodakaraensis primase initiated primer synthesis in reactions containing only dNTPs (1). In this study, we show that the T. kodakaraensis p41-p46 complex and the T. kodakaraensis p41 catalytic subunit hydrolyzed dNTPs to dNMP and PPi. When these preparations were incubated in reactions containing high levels of Tris buffer and/or glycerol, dNMP-Tris and/or dNMP-glycerol derivatives were also formed. Surprisingly, these reactions were unaffected by DNA. We suggest that these findings may reflect the mechanism by which the archaeal T. kodakaraensis primase initiates the synthesis of oligonucleotide chains because they resemble an aborted initiation-elongation reaction. Where examined, all primases generate substantial levels of short oligonucleotides, presumably reflecting their instability and/or inability to be extended. In some cases, primases can form dinucleotides even in the absence of DNA (summarized in Ref. 14).
Frick and Richardson (14) proposed a model for the synthesis of primers by primases that involves two NTP binding sites on the primase protein. These include a site at which a NTP is to be incorporated at the 5′ end of the primer (initiation site) and a second site that binds the NTP added to the 3′ end of the primer (elongation site). During each elongation step of primer synthesis, the product (n + 1) oligonucleotide is transferred to the initiation site, leading to the binding of another NTP at the elongation site. To explain the formation of the dAMP derivatives described here, we propose that H2O, glycerol, or Tris occupy the initiation site and that dATP is loaded at the elongation site. Hydrolysis of dATP (to PPi) concomitant with dAMP transfer to the occupant at the initiation site could lead to the formation of dAMP, glycerol, or dAMP-Tris. In keeping with this notion, the binding of NTPs by primase appears to occur initially at the elongation site (15). The crystal structure of the Pyrococcus horikoshii DNA primase complex containing a single UTP molecule was determined (16). This complex was obtained by soaking the primase crystals with UTP. We suggest that this nucleotide is bound in the elongation site and that a similar complex would be formed if dNTPs rather than rNTPs were used.
Primers synthesized by prokaryotic and eukaryotic primases contain 5′-triphosphate termini (5), whereas only a small percentage (5–10%) of the products formed by the T. kodakaraensis primase complex retained triphosphate ends (1). We noted that the amount of dNMP derivatives formed in the T. kodakaraensis primase reaction was hardly altered by the presence of DNA template (see experiments with synthetic oligonucleotide templates (1)). We speculate that the transfer of dNTPs (or possibly rNTPs) from the elongation nucleotide binding site to the initiation site (occupied with H2O, glycerol, or Tris) in the presence of a DNA template could lead to the formation of polynucleotide chains deficient in triphosphate ends. This idea is in keeping with our findings that a substantial amount of chains formed contained five monophosphate ends. Because the dNMP-glycerol and dNMP-Tris derivatives were produced at rates significantly higher than that observed with the corresponding rNTP derivatives, our proposed model would be expected to lead to the formation of a higher percentage of triphosphate-terminated RNA chains. Our findings, however, indicated that the level of RNA and DNA chains containing triphosphate ends produced were similar (both ∼5–10%). Further studies on products synthesized with rNTPs in the T. kodakaraensis primase reaction may help explain this discrepancy.
In this study, we focused on the synthesis of the dNMP-glycerol and the dNMP-Tris derivatives because their production was more robust than those formed with rNTPs. However, we detected the production of various derivatives formed in reactions with rNTPs and the T. kodakaraensis primase complex that migrated as ∼15- and ∼5–8-nt bands (particularly with GTP) and small levels of fast migrating material with ATP following TLC on PEI plates. Similar to observations made with dNTPs, their formation did not require a DNA template and instead required an active p41 subunit or primase complex, glycerol, and Tris buffer. We also showed that other small molecule acceptors, including glucose, could be linked to dAMP, although they were not characterized in more detail. Because little is known about the physiology and metabolic activities of archaea, the biological role of these adducts remain to be explored.
As described here, T. kodakaraensis primase preparations formed oligonucleotides only in the presence of DNA templates, whereas production of the dNMP-glycerol and dNMP-Tris derivatives occurred in the absence of DNA. Because of their aberrant migration through polyacrylamide gels and cleavage by venom phosphodiesterase, these derivatives appeared to represent the de novo synthesis of oligonucleotides. A number of enzymes, including other archaeal primases, DNA polymerase λ, DNA polymerase μ, and terminal deoxynucleotidal transferase, have been reported to possess template-dependent and template-independent polymerase activities that synthesize oligodeoxynucleotides de novo (17, 18). Based on our findings, these conclusions warrant a more careful characterization.
Formation of the dNMP adducts, described above, suggests that the in vitro initiation reactions catalyzed by the purified T. kodakaraensis primase in vitro lack specificity. Initiation sites detected with purified eukaryotic DNA primases also appear to be nonspecific as they are altered by the level of NTPs added to reactions (8). These findings raise the possibility that other replication proteins (perhaps at the fork) interact with eukaryotic DNA primases and influence the specificity of the initiation reaction.
This work was supported, in whole or in part, by National Institutes of Health Grant GM034559 (to J. H.). This work was also supported by National Science Foundation Grant MCB-0815646 (to Z. K.).
- rNTP
- ribonucleoside 5′-triphosphate
- rATP
- riboadenosine 5′-triphosphate
- CIP
- calf intestinal phosphates
- PPase
- inorganic pyrophosphatase
- nt
- nucleotide
- TE
- Tris-EDTA
- dAp
- 3′-deoxyadenosine phosphate
- pdAp
- 5′,3′-deoxyadenosine diphosphate.
REFERENCES
- 1. Chemnitz Galal W., Pan M., Kelman Z., Hurwitz J. (February 17, 2012) Characterization of the DNA primase complex isolated from the archaeon, Thermococcus kodakaraensis. J. Biol. Chem. 287, 16209–16219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Klinge S., Hirst J., Maman J. D., Krude T., Pellegrini L. (2007) An iron-sulfur domain of the eukaryotic primase is essential for RNA primer synthesis. Nat. Struct. Mol. Biol. 14, 875–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Weiner B. E., Huang H., Dattilo B. M., Nilges M. J., Fanning E., Chazin W. J. (2007) An iron-sulfur cluster in the C-terminal domain of the p58 subunit of human DNA primase. J. Biol. Chem. 282, 33444–33451 [DOI] [PubMed] [Google Scholar]
- 4. Iyer L. M., Koonin E. V., Leipe D. D., Aravind L. (2005) Origin and evolution of the archaeo-eukaryotic primase superfamily and related palm-domain proteins: structural insights and new members. Nucleic Acids Res. 33, 3875–3896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zerbe L. K., Kuchta R. D. (2002) The p58 subunit of human DNA primase is important for primer initiation, elongation, and counting. Biochemistry 41, 4891–4900 [DOI] [PubMed] [Google Scholar]
- 6. Liu L., Komori K., Ishino S., Bocquier A. A., Cann I. K., Kohda D., Ishino Y. (2001) The archaeal DNA primase: biochemical characterization of the p41-p46 complex from Pyrococcus furiosus. J. Biol. Chem. 276, 45484–45490 [DOI] [PubMed] [Google Scholar]
- 7. Le Breton M., Henneke G., Norais C., Flament D., Myllykallio H., Querellou J., Raffin J. P. (2007) The heterodimeric primase from the euryarchaeon Pyrococcus abyssi: a multifunctional enzyme for initiation and repair? J. Mol. Biol. 374, 1172–1185 [DOI] [PubMed] [Google Scholar]
- 8. Kuchta R. D., Stengel G. (2010) Mechanism and evolution of DNA primases. Biochim. Biophys. Acta 1804, 1180–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Matsunaga F., Norais C., Forterre P., Myllykallio H. (2003) Identification of short “eukaryotic” Okazaki fragments synthesized from a prokaryotic replication origin. EMBO Rep. 4, 154–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lao-Sirieix S. H., Bell S. D. (2004) The heterodimeric primase of the hyperthermophilic archaeon Sulfolobus solfataricus possesses DNA and RNA primase, polymerase, and 3′-terminal nucleotidyl transferase activities. J. Mol. Biol. 344, 1251–1263 [DOI] [PubMed] [Google Scholar]
- 11. Yuan W., Edwards J. L. (2011) Thiol metabolomics of endothelial cells using capillary liquid chromatography mass spectrometry with isotope coded affinity tags. J. Chromatogr. A. 1218, 2561–2568 [DOI] [PubMed] [Google Scholar]
- 12. Glynn I. M., Chappell J. B. (1964) A simple method for the preparation of 32P-labelled adenosine triphosphate of high specific activity. Biochem. J. 90, 147–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kornberg A., Baker T. A. (1992) DNA Replication, 2nd Edition, p. 415, W. H. Freeman and Company, New York [Google Scholar]
- 14. Frick D. N., Richardson C. C. (2001) DNA primases. Annu. Rev. Biochem. 70, 39–80 [DOI] [PubMed] [Google Scholar]
- 15. Sheaff R. J., Kuchta R. D. (1993) Mechanism of calf thymus DNA primase: slow initiation, rapid polymerization, and intelligent termination. Biochemistry 32, 3027–3037 [DOI] [PubMed] [Google Scholar]
- 16. Ito N., Nureki O., Shirouzu M., Yokoyama S., Hanaoka F. (2003) Crystal structure of the Pyrococcus horikoshii DNA primase-UTP complex: implications for the mechanism of primer synthesis. Genes Cells 8, 913–923 [DOI] [PubMed] [Google Scholar]
- 17. Hubscher U., Maga G., Spadari S. (2002) Eukaryotic DNA polymerases. Annu. Rev. Biochem. 71, 133–163 [DOI] [PubMed] [Google Scholar]
- 18. Ramadan K., Shevelev I. V., Maga G., Hübscher U. (2004) De novo DNA synthesis by human DNA polymerase λ, DNA polymerase μ, and terminal deoxyribonucleotidyl transferase. J. Mol. Biol. 339, 395–404 [DOI] [PubMed] [Google Scholar]