Background: Both genetic and epigenetic factors are implicated in Type 1 diabetes (T1D).
Results: Variations in histone H3-lysine 9 acetylation are detected around the promoter/enhancer regions of key T1D susceptible genes in monocytes of T1D subjects versus normals.
Conclusion: The chromatin status of this key region is altered in T1D.
Significance: Epigenetic variations at T1D susceptible genes may be functionally important.
Keywords: Chromatin Regulation, Diabetes, Epigenetics, Genetic Diseases, Genome Structure, Histone Modification
Abstract
Both genetic and environmental factors are implicated in type 1 diabetes (T1D). Because environmental factors can trigger epigenetic changes, we hypothesized that variations in histone post-translational modifications (PTMs) at the promoter/enhancer regions of T1D susceptible genes may be associated with T1D. We therefore evaluated histone PTM variations at known T1D susceptible genes in blood cells from T1D patients versus healthy nondiabetic controls, and explored their connections to T1D. We used the chromatin immunoprecipitation-linked to microarray approach to profile key histone PTMs, including H3-lysine 4 trimethylation (H3K4me3), H3K27me3, H3K9me3, H3K9 acetylation (H3K9Ac), and H4K16Ac at genes within the T1D susceptible loci in lymphocytes, and H3K4me3, H3K9me2, H3K9Ac, and H4K16Ac at the insulin-dependent diabetes mellitus 1 region in monocytes of T1D patients and healthy controls separately. We screened for potential variations in histone PTMs using computational methods to compare datasets from T1D and controls. Interestingly, we observed marked variations in H3K9Ac levels at the upstream regions of HLA-DRB1 and HLA-DQB1 within the insulin-dependent diabetes mellitus 1 locus in T1D monocytes relative to controls. Additional experiments with THP-1 monocytes demonstrated increased expression of HLA-DRB1 and HLA-DQB1 in response to interferon-γ and TNF-α treatment that were accompanied by changes in H3K9Ac at the same promoter regions as that seen in the patient monocytes. These results suggest that the H3K9Ac status of HLA-DRB1 and HLA-DQB1, two genes highly associated with T1D, may be relevant to their regulation and transcriptional response toward external stimuli. Thus, the promoter/enhancer architecture and chromatin status of key susceptible loci could be important determinants in their functional association to T1D susceptibility.
Introduction
Type 1 diabetes (T1D)2 is an autoimmune disease that can occur when immune tolerance in genetically susceptible individuals is disrupted by environmental or other factors (1–3). Extensive studies have provided a wealth of evidence implicating both genetic and environment factors in the etiology of T1D (4). Multiple regions of the genome, known as T1D susceptible gene loci, have been clearly linked to the risk of T1D through extensive linkage and association studies (5–7). All susceptible loci are, however, not equally important. Insulin-dependent diabetes mellitus 1 (IDDM1) in the major histocompatibility complex (MHC) region in chromosome 6 is the prime susceptibility locus, providing up to 50% of the risk of inheritable incidence (8, 9). However, it appears that only a relatively small fraction of genetically susceptible individuals progress to T1D (10) and disease incidence continues to increase, which suggests a significant contribution from environmental factors. Indeed, many studies have uncovered key links between T1D and environmental factors such as infection, diet, perinatal and prenatal nutrition (10–13). Recent clinical trials, such as TEDDY (The Environmental Determinants in Diabetes of the Young), have aimed to identify the specific environmental triggers of T1D. It is plausible that such factors may regulate T1D in a gene-environment interaction or epigenetic manner and, based on our current understanding of the disease, T1D susceptible genes and genes involved in autoimmunity are logically the best candidate genes targeted by environmental factors.
Epigenetic mechanisms in chromatin are increasingly recognized to be a molecular link between genes and the environment. Notably, in eukaryotic cells, chromatin structure can be affected by environmental factors such as diet, chemicals, and pathogens (14). Nucleosome, the basic repeat unit of chromatin, consists of two copies each of histones H2A, H2B, H3, and H4 wrapped by 147 bp of DNA (15). Post-translational modifications (PTMs) at the N-terminal amino acids of histones play an essential role in modifying chromatin structure and, along with DNA methylation, form an epigenetic layer that can modulate gene transcription (16–19). Epigenetics usually refers to heritable changes, including those conferred mitotically or meiotically, which occur without changes in the DNA sequence. More recently the definition has been refined to include the structural adaptation of chromosomal regions (20). Thus, both DNA methylation and histone PTMs can work together to control epigenetic transmission (21). Importantly, the chromatin status of gene promoters plays a role in determining how the gene responds to various stimuli. Histone PTMs have varying effects on gene expression depending on the position, type of modification (acetylation, methylation, phosphorylation, or ubiquitylation), and also the degree of methylation (mono-, di-, or tri-) through recognition by key chromatin modifying proteins (16–19). In general, histone H3-lysine 4-trimethylation (H3K4me3) and H3K9-acetylation (H3K9Ac) are associated with promoters and active genes, whereas H3K9me2, H3K9me3, and H3K27me3 are generally associated with repressed genes, and H4K16Ac may control chromatin structure through interactions with key proteins (16–19, 22).
Recent studies have shown that diabetic stimuli like high glucose can alter the levels of key histone PTMs, including H3K9Ac, H3K9me2, H3K9me3, and H3K4me1 at the promoters of inflammatory and other genes related to the pathogenesis of diabetes as well as its vascular complications and metabolic memory (23–30). Genome-wide mapping of histone PTMs has greatly accelerated the field of epigenomics mainly due to the availability of high density microarrays and next generation sequencing technologies coupled with sophisticated bioinformatics analyses methods (31–33). We recently implemented the chromatin immunoprecipitation-linked to microarray (ChIP-array) epigenomic approach to detect variations in histone PTMs in a genome-wide scale in diabetes research (25, 28). In one study (28), we compared histone H3K9me2 patterns in peripheral blood lymphocytes and monocytes obtained from T1D patients versus healthy control subjects. The results revealed a significant increase in H3K9me2 in a subset of genes in lymphocytes of the T1D cohort. Pathway analyses showed that these methylated genes were part of key biological networks and canonical pathways relevant to the etiology of T1D and its complications. The study provided evidence of a link between T1D and altered histone methylation of key diabetes-related genes, including those associated with inflammation and autoimmunity. Interestingly, CTLA4, a known T1D susceptibility gene, was found to depict differential H3K9me2 methylation in T1D lymphocytes versus normal (28).
The massive datasets generated by ChIP-array and other high throughput epigenome profiling approaches have posed enormous challenges for subsequent data analyses, bioinformatics, and statistics as well as functional correlations. In the current study, we adopted a candidate gene-set focused approach because it is well established that a set of susceptible genes or loci are involved in T1D (5–7). A recent genome-wide association study (GWAS) confirmed and extended the results from previous linkage and association studies and defined 41 genome loci as T1D susceptible regions (34). We hypothesized that the chromatin near key T1D-susceptible genes might exhibit specific histone PTM variations, and that these PTM variations may be related to the pathogenesis of T1D. Such events could uncover a cross-talk between genetics and epigenetics to significantly enhance our understanding of T1D. Thus, to gain new insights into these T1D susceptible genes (34), we used the ChIP-array approach to profile and compare histone PTMs at T1D susceptible genes in lymphocyte and monocyte cells obtained from the peripheral blood of T1D individuals versus normal controls. To evaluate the changes in chromatin status at these T1D relevant regions of the genome, we first mapped key histone PTMs, including histone H3K4me3, H3K27me3, H3K9me3, H3K9Ac, and H4K16Ac in lymphocytes, and H3K4me3, H3K9me2, H3K9Ac, and H4K16Ac in monocytes from T1D patients versus healthy controls. We then examined potential variations in these histone PTMs around some 350 genes within these T1D susceptible regions, including the well characterized IDDM1 region. Noticeable, data analyses revealed the presence of significant variations in histone H3K9Ac at the promoter (enhancer) regions of HLA-DRB1 and HLA-DQB1 genes within IDDM1 in monocytes between T1D patients and healthy controls. Furthermore, experiments in THP-1 monocytes demonstrated increases in H3K9Ac at the same promoter regions as that seen in the patient monocytes when HLA-DRB1 and HLA-DQB1 expressions were stimulated by interferon-γ and TNF-α treatment of the THP-1 cells, suggesting that the H3K9Ac variations observed in vivo at these regions might be associated with gene expression.
EXPERIMENTAL PROCEDURES
Human Subject Enrollment
All studies were performed according to a protocol approved by the City of Hope Institutional Review Board. Written informed consents were obtained from all blood donors (T1D (n = 9) and healthy controls (n = 7)). Patient demographics have been previously described (28). There were no statistically significant differences in age or gender between the two groups. Blood monocyte and lymphocyte fractions were prepared as described (28).
ChIP and ChIP-array Experiments with Human Blood Cells
Peripheral blood monocytes and lymphocytes were freshly isolated from individual normal and T1D subjects and then cross-linked in 1% formaldehyde, sonicated to shear DNA, and immunoprecipitations (IP) performed with antibodies to histone PTMs. IP with no antibody was used as the input control. DNA precipitates were washed, eluted, and cross-links were reversed. Small aliquots of DNA before IP and the ChIP-enriched DNA were saved for follow-up validations of ChIP-chip data by ChIP-qPCRs. The remaining ChIP-enriched DNA and no antibody input control were amplified and purified. The amplified DNA samples were separately pooled from nine T1D or from seven normal control samples. These two groups were designated as T1D and normals in this study, and used for microarray hybridization as described (25, 28). ChIP-array experiments were performed with human 385K RefSeq promoter tiling arrays (for lymphocyte samples) at the Roche NimbleGen Core, and with chromosome 6 tiling arrays (NimbleGen human whole genome array 15 of 38) (for monocyte samples) at our Functional Genomics Core. The following antibodies were used for ChIPs: anti-histone H3K4me3 (Millipore 07-473), H3K9me3 (Abcam ab8898); H3K9me2 (Millipore 07-441), H3K27me3 (Millipore 07-449), H3K9Ac (Millipore 06-942), and H4K16Ac (Millipore 06-762).
Gene Expression Profiling
Human lymphocytes were prepared from four nondiabetic volunteers. RNAs were extracted using Qiagen RNeasy mini kits following the manufacturer's instructions. Biotinylated single strand cDNA was generated and subjected to expression profiling with Affymetrix Human Gene 1.0ST arrays followed by data analyses and conversion into expression measurements using Expression Console version 1.1.1 from Affymetrix.
T1D Susceptible Locus Gene Set Generation
The genes located in T1D susceptible human loci defined by Ensembl version 54 were from www.t1dbase.org. They were subsequently converted to NCBI Accession codes using BioMart. Transcripts not covered by either the Affymetrix GeneChip® Human Gene 1.0 ST Array or whose promoter regions (−2000 to +500 bp relative to TSS) were not covered by the Nimblegen 385K Refseq Promoter Array were filtered out. Redundant transcripts were finally removed by retaining only one transcript among those who shared the same gene symbol and shared the same transcription start site. The gene list is provided in supplemental Table S1.
Heatmap Generation
Gene promoter region (−2000 to 500 bp relative to transcript start site, TSS) of genes covered by both gene expression array and tiling array was divided into 25 nonoverlapping 100-bp regions (finest resolution available), each represented by a positional bin. For each mark, median-shifted log2-transformed enrichment ratios of the microarray probes falling in each promoter region of the transcript were retrieved, aligned, and assigned to the represented bin based on their positions relative to TSS. A data matrix with each row representing one transcript and 25 columns corresponding to the 25 bins was created. After removing redundant genes by retaining one transcript among those who share the same gene symbol and same TSS, the rows of the matrix are ordered by their corresponding genes' expression and visualized by Java TreeView version 1.1.3 with each row representing enrichment state of one promoter of the RefSeq gene.
Hyper- or Hypoenriched Peak Identification and False Discovery Rate (FDR) Calculation
For each histone PTM, the log2 ratio data from T1D patients or healthy controls' pooled lymphocytes were quantile normalized. The histone PTM-enriched regions (also called peaks, where log2 ratios of at least 5 continuous probes are ≥1.5 and peak p values are ≤0.05) in T1D patients and healthy individuals were then identified separately using TAMAL software (32). Hyper peaks (or T1D specific enriched regions) were subsequently selected from the peaks identified in the T1D sample by comparing the median log2 ratio of all the probes located inside the peak regions in the T1D sample with those in the healthy sample. FDR was calculated for each peak based on the difference median log2 ratio in T1D versus healthy samples, and the peaks with FDR < 5% were considered as hyper peaks, namely specific enriched regions that can be identified in T1D lymphocytes but not found at the same genomic location in normal lymphocytes. On the other hand, hypo peak refers to the peaks specific in healthy samples, and identified using similar criteria, with FDR of the difference median log2 ratio in healthy samples versus T1D less than 5%. The identified hyper/hypo peaks were further screened to identify those located on the promoters (−2 kb to +500 bp relative to TSS) of genes in T1D susceptible loci for lymphocyte samples. Monocyte data sets were similarly analyzed.
Because different PTMs have different enrichment levels, the tiling-array data distribution for each PTM mark is different. Thus, it is necessary to define different cutoffs of mean log2 ratio difference between T1D and normal individuals on PTM-enriched regions for each mark based on each dataset distribution. A null dataset was therefore generated for each mark using data from nondiabetic pooled sample. Specifically, the data were randomly shuffled and split into two equal size datasets. The average difference of probe signals of every five consecutive probes between these two datasets was then calculated. The average difference of the null dataset roughly follows a normal distribution with mean of 0 and its S.D. can be empirically calculated. Based on this distribution, the p values of the log2 ratio difference between T1D and the normal sample of each peak region were calculated. The p values were adjusted to obtain the corresponding FDR. FDR less than 5% was used to identify hyper/hypo PTM-enriched regions between T1D and normal for each mark.
Experiments with THP-1 Cells
THP1 cells were cultured in RPMI medium supplemented with 10% heat-inactivated fetal calf serum, HEPES (10 mm), glutamine (2 mm), streptomycin (50 g/ml), penicillin (50 units/ml), β-mercaptoethanol (50 μm), and glucose (5.5 mm) at 37 °C in a 5% CO2 incubator. Cells were treated with 20 μm IFNγ and 10 μm TNFα for 4, 16, and 24 h and total RNA was prepared from these samples and from untreated THP1 cells (0 h) using RNA-STAT-60 (Tel-Test). cDNAs were prepared according standard procedures and used for RT quantitative (q)PCR. For ChIP assays, THP1 cells (IFNγ and TNFα treated and untreated) were cross-linked in 1% formaldehyde, then sonicated to shear DNA, and IPs were performed with anti-histone H3K9Ac (Active Motif 39585) (24). A ChIP without antibody was used as control for enrichments and a small fraction of total chromatin saved as input control. The ChIP-enriched DNA and the controls were used in ChIP-qPCRs. ChIPs were also performed with untreated THP-1 cells using antibodies to H3K4me3 (Millipore 07-473), H3K4me1 (ab8895), and p300 (ab14984). These ChIP samples were amplified and used in ChIP-array experiments with human chromosome 6 tiling arrays at the Roche NimbleGen Core and the City of Hope Functional Genomics Core.
Real-time qPCR
ChIP-qPCRs were performed using the histone H3K9Ac ChIP-enriched DNA from each individual or from experiments with THP-1 cells and normalized to the input DNA as described (28). RT-qPCRs were performed as described (25). Student's t tests were used for statistical analyses of the data. Sequences of primers used in this study are provided in supplemental Table S2.
Data Deposition
All genomic data reported in this study have be deposited in the NCBI Gene Expression Omnibus (GEO) data base (accession number GSE36403).
RESULTS
Experimental Design
The T1D and control volunteer cohort used is the same as that previously reported (28). Primary lymphocytes and monocytes were isolated from the peripheral blood of these T1D patients (n = 9) and nondiabetic healthy controls (n = 7), respectively. ChIP assays with the indicated antibodies were performed and ChIP-enriched DNA samples were obtained from monocytes and lymphocytes of each volunteer separately, and then amplified. Here, amplified samples from each individual in T1D or healthy control cohorts were pooled and ChIP-array experiments were performed on each cell type. The study is summarized in Table 1. We used human promoter tiling arrays to map the histone PTMs H3K4me3, H3K27me3, H3K9me3, H3K9Ac, and H4K16Ac at promoters of genes located within the T1D susceptible loci in lymphocytes of T1D patients versus controls (Table 1). For monocytes from the same volunteers, we mapped H3K4me3, H3K9me2, H3K9Ac, and H4K16Ac marks at the IDDM1 locus within the MHC region using the chromosome 6 tiling arrays covering the MHC locus (Table 1).
TABLE 1.
Summary of the ChIP-array experiments in this study
Summary of Chip-array experiments performed in this study. Antibodies to histone PTM marks used are indicated. The lymphocyte experiments utilized human Ref- Seq promoter tiling arrays (Roche Nimblegen 385K RefSeq), while the monocyte experiments utilized chromosome 6 tiling arrays (Roche Nimblegen Human whole genome array 15 of 38). Details are described under “Experimental Procedures.”
| Lymphocytes | Monocytes | |
|---|---|---|
| Histone marks | H3K9Ac | H3K9Ac |
| H3K9me3 | H3K9me2 | |
| H3K4me3 | H3K4me3 | |
| H4K16Ac | H4K16Ac | |
| H3K27me3 | ||
| Microarray used | Human promoter tiling array | Human Ch. 6 tiling array |
| Genome regions | T1D susceptible locia | IDDM1 |
a T1D susceptible loci are from Ref. 34.
Profiling Histone PTMs in Blood Lymphocytes and Monocytes from Normal Healthy Volunteers
Increasing evidence links histone PTMs with disease states, including diabetes and its complications. However, very little is known about histone PTM profiles at key disease susceptible genetic regions. In our previous study, we observed that key diabetes-related genes displayed increased promoter histone H3K9me2 in T1D patient lymphocytes (28). Based on the fact that many T1D susceptible genes have been shown to play key roles in T-lymphocyte cell differentiation, proliferation, and signal transduction (35), we first mapped several key histone PTMs (H3K4me3, H3K27me3, H3K9me3, H3K9Ac, and H4K16Ac) at 24,659 gene promoter regions using promoter tiling arrays in lymphocytes obtained from the recruited seven healthy volunteers (28). To verify the functional association of these PTM marks to gene expression in general, we also performed expression profiling (with Affymetrix arrays) using RNA extracted from lymphocytes of another four separate normal healthy volunteers. These gene expression data were integrated into the ChIP-array data to evaluate the association between histone PTMs and gene expression.
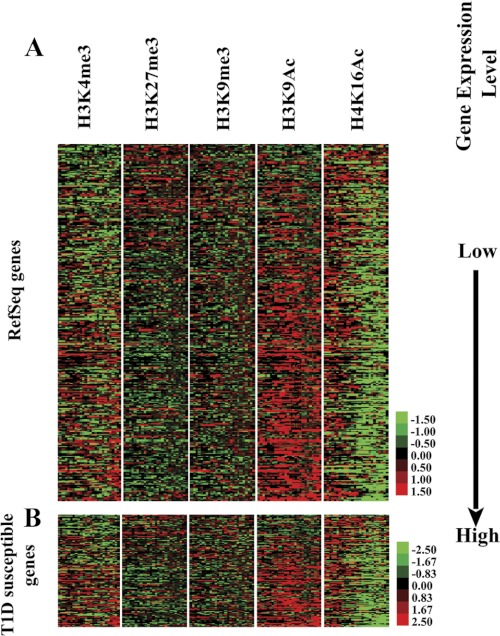
Heatmap results of the lymphocyte ChIP-array experiments are shown in Fig. 1A. Rows represent the enrichment status of H3K4me3, H3K27me3, H3K9me3, H3K9Ac, and H4K16Ac levels genome-wide at gene promoter regions from −2000 to 500 bp relative to TSS. Red indicates the relative enrichment of the corresponding mark, whereas green indicates depletion. Genes are ordered and sorted by expression level (low to high) from top to bottom based on the Affymetrix mRNA profiling data. As expected, genes depicting high expression levels in lymphocytes had higher enrichment of H3K4me3 and H3K9Ac, two chromatin marks generally associated with active genes. Reciprocally, RefSeq genes with low expression levels had the highest content of H3K27me3 and H3K9me3, marks usually associated with repressed genes (Fig. 1A). The composite files in lymphocytes also showed similar relationships between histone PTMs and the various gene expression groups from high, moderate, and low expression to silent (supplemental Fig. S1 and supplemental methods). These results confirmed the correlations between histone PTMs and gene expression as demonstrated in other cell types (32) and also verified the quality of the promoter array data sets generated in this study.
FIGURE 1.
Heatmap of various histone PTMs obtained from ChIP-promoter array profiling of human blood lymphocytes from normal healthy volunteers. A, heatmap of key histone PTMs at RefSeq gene promoters. Red represents enrichment, green represents depletion, and black represents no enrichment or missing data due to lack of probes. The enrichment level of each PTM at gene promoter regions is shown in the heatmap side-by-side with genes sorted by expression from the lowest (top) to highest (bottom). Details of heatmap generation are provided under “Experimental Procedures.” B, heatmap of histone PTMs at the promoter regions of 350 T1D genes within 41 T1D susceptibility loci. (The gene list is provided in supplemental Table S1).
Next, to gain insights into the potential role of histone modifications at T1D susceptible genes, we adopted a candidate gene-set survey approach by focusing on 41 T1D susceptible loci defined by recent GWAS. Fig. 1B shows the heatmap of the histone modification patterns at the promoter regions of some 350 T1D susceptible genes within these 41 loci in normal human lymphocytes (the gene list provided in supplemental Table S1). Notably, most of the expressed genes also appear to have high levels of histone H3K9Ac activity near their promoter regions (Fig. 1B) indicating that the H3K9Ac mark might be indispensable for the “open” chromatin status required for active or induced expression of these genes.
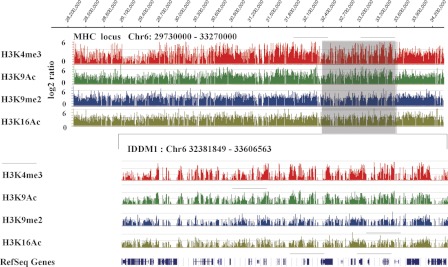
We next focused specifically on the human MHC locus, which contains the major T1D susceptible IDDM1 locus (5–7). MHC is a gene dense locus in the human genome that displays the highest density of single nucleotide polymorphism (SNP) variations (36). Its short form spans 3.6 Mb and contains ∼140 genes between flanking genetic marks MOG and COL11A2 on chromosome 6p21.3 (Fig. 2) and plays a vital role in immune system functions (37). MHC locus is tightly associated with many human autoimmune diseases including T1D, inflammatory disorders, and cancers (36). Innumerable reports have described the associations between MHC SNP variations and various human diseases. The IDDM1 region is located within MHC, and its SNPs are highly associated with T1D, providing up to 50% of the inheritable risk of incidence (9). Furthermore, compelling evidence shows that haplotypes of the human leukocyte antigen (HLA) class II genes HLA-DRB1 and HLA-DQB1 in IDDM1 within the MHC are among the major determinants of T1D risk (5–7). We hypothesized that there could be a correlation between the chromatin status (histone PTMs) of the IDDM1 locus in monocytes and T1D. Monocytes were assessed here because they are the source of macrophages and dendritic cells, potent antigen presenting cells (38), which express HLA class II genes and have well documented roles in inflammation and immune responses.
FIGURE 2.
Mapping the chromatin architecture of the human MHC locus and IDDM1 in blood monocytes from normal healthy volunteers. Histone H3K4me3, H3K9Ac, H3K9me2, and H3K16Ac maps of the genome region at the human MHC and IDDM1 locus. ChIP-chips were performed with chromosome 6 (Chr6) tiling arrays and the indicated antibodies are as described under “Experimental Procedures.” The enrichment level is presented in log2 ratio. Upper panel, the landscape of histone PTMs patterns (H3K4me3, H3K9me2, H3K9Ac, and H4K16Ac) of the MHC locus (Chr6: 29730000–33270000, human NCBI build 36) in normal human monocytes. Lower panel, IDDM1 (Chr6: 32381849–33606563), which contains about 43 genes including HLA genes located within the MHC locus. RefSeq genes are denoted in the lowest lane. The data shown is generated from SignalMap software (Roche Nimblegen).
We used the same ChIP-array approach to evaluate several key histone PTMs at the MHC locus and its chromatin status in blood monocytes of the normal controls but using chromosome 6 genomic tiling arrays, which allowed us to map the entire MHC locus. Fig. 2 illustrates the patterns of H3K4me3, H3K9me2, H3K9Ac, and H4K16Ac at the MHC locus (upper panel) and IDDM1 (lower panel) in monocytes from control volunteers. Similar to that observed in lymphocytes from these volunteers, composite files showed that active histone PTM marks (H3K4me3, H3K9Ac) in the normal monocytes correlated well with highly expressed genes, whereas inactive marks (H3K9me2) were associated with silenced or low expression genes (supplemental Fig. S2 and supplemental methods). These data also provide for the first time a snapshot of the epigenome of this important genomic region and a resource for studying the chromatin status of MHC locus in human monocytes. We used these data as a reference baseline for the next step to identify potential specific variations in these histone PTMs at the T1D susceptible regions in the blood lymphocytes and monocytes obtained from nondiabetic versus T1D volunteers.
Comparing Epigenomes of T1D Susceptible Genes between T1D and Nondiabetic Controls
In parallel, we profiled histone PTMs using ChIP-enriched DNA from lymphocytes and monocytes of a group of nine T1D patients. As discussed above, our approach was to map the histone PTMs at T1D susceptible genes in T1D patients and nondiabetic controls separately, and then scan for potential variations between these two cohorts through computational methods (25, 28). Five PTMs (histone H3K4me3, H3K27me3, H3K9me3, H3K9Ac, and H4K16Ac) were examined in lymphocytes, and four (H3K4me3, H3K9Ac, H2K9me2, and H4K16Ac) in monocytes. The H3K9me2 profile in lymphocytes was not evaluated in this study because we studied it earlier, albeit not specifically at T1D susceptible genes (28). The data obtained from profiling each PTM from the T1D or lymphocytes of monocytes from healthy individuals were quantile normalized and the TAMAL software (39) was used to identify promoter regions enriched by each PTM (peak) in T1D and control individuals separately (detailed analysis procedure is described under “Experimental Procedures”). Subsequently, we screened the data to identify those PTMs located specifically in the T1D susceptible gene loci for lymphocyte samples, or in the IDDMI locus (chromosome 6: 32381849–33606563, human NCBI build 36) for monocyte samples. As mentioned earlier, the reason for this different comparison is because many T1D susceptible genes have functions in T lymphocytes, whereas monocytes are more likely to show changes at the IDDM1 regions because they are precursors of potent antigen presenting cell cells.
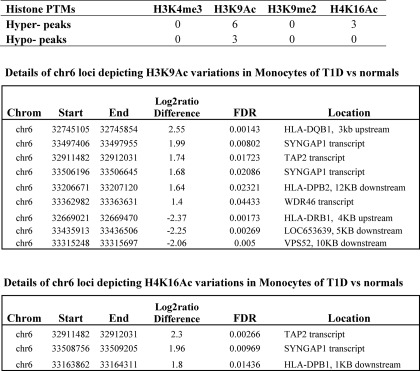
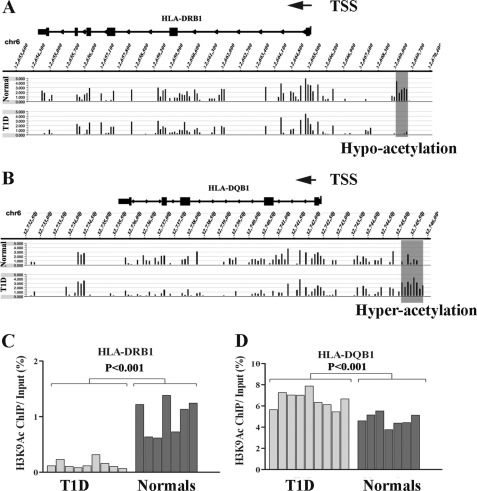
Tables 2 and 3 summarize the numbers of differentially methylated/acetylated regions between T1D versus controls in these specific regions as identified by computation in lymphocytes and monocytes, respectively. Our results revealed that most of these known T1D susceptible genes (34) show relatively few variations in their promoter regions of the key histone PTMs evaluated in this study in either lymphocytes or monocytes between T1D and healthy volunteers (Tables 2 and 3). However, the exception was that marked variations in histone H3K9Ac levels were detected at several regions in the IDDM1 locus in monocytes (Table 3, middle panel). Interestingly, two variations were located in the upstream region of HLA-DRB1 and HLA-DQB1 (Table 3, Fig. 3, A and B). Compared with controls, T1D patients had lower histone H3K9Ac levels at the HLA-DRB1 upstream promoter region (Fig. 3A), and higher H3K9Ac levels at the HLA-DQB1 upstream promoter region (Fig. 3B). This is noteworthy because HLA-DRB1 and HLA-DQB1 are known to be two top T1D susceptible genes and their SNPs have the highest association with T1D (5, 6). We also observed hyperacetylation at H4K16 at three regions within transcripts of TAP2, SYNGAP1, and downstream (1 kb) of HLA-DPB1 (Table 3, bottom panel).
TABLE 2.
Summary of histone PTM variations at T1D susceptible gene promoters between T1D patients and normals (lymphocytes)
The T1D susceptible genes are defined as genes located within 41 T1D susceptible loci. The gene list is provided in supplemental Table S1. Detailed methods for hyper- and hypo-enriched regions at T1D susceptible gene promoters are provided under “Experimental Procedures.”
| Histone PTMs | Hypo peaks | Hyper peaks |
|---|---|---|
| H3K4me3 | 0 | 0 |
| H3K9me3 | 1 | 1 |
| H3K9Ac | 0 | 0 |
| H3K27me3 | 0 | 0 |
| H4K16Ac | 0 | 0 |
TABLE 3.
Summary of histone PTMs variations at the IDDM1 locus between T1D patients and normals (monocytes)
The definitions of the enriched region, hyper-peak and hypo-peaks are provided under “Experimental Procedures.” IDDM1 locus spans from 32381849 to 33606563 on chromosome 6, based on human NCBI build 36. Computation indicates the key variations in chromatin marks histone H3K9Ac and H4K16Ac at key loci, but not other marks in monocytes of TID versus normals (in the IDDM1 locus).
FIGURE 3.
Histone H3K9Ac variations at HLA-DQB1 and HLA-DRB1 promoter (enhancer) regions in monocytes from T1D versus nondiabetic control volunteers. Snapshots of histone H3K9Ac status of HLA-DQB1 and HLA-DRB1 gene regions in T1D patients and normals. Pooled histone ChIP-enriched DNA samples from monocytes of T1D and normals were hybridized to human chromosome 6 (Chr6) tiling arrays and the data were extracted according to standard operating procedures of Nimblegen-Roche. Results were visualized by SignalMap software (Roche Nimblegen). Hyper- and hypo-histone H3K9Ac regions are indicated. A, HLA-DRB1; B, HLA-DQB1. For ChIP-qPCR validation of histone H3K9Ac variations at HLA-DQB1 and HLA-DRB1 promoter/enhancer regions in monocytes, histone H3K9Ac ChIPs were prepared from each individual volunteer (nine T1D and seven normal control samples). Standardized ChIP-qPCRs were used to quantify the amount of specific modified ChIP-enriched DNA in monocyte samples from individual T1D patients and normal controls. C, ChIP-qPCR validation of H3K9Ac at HLA-DRB1. Results shown are from one set of PCR primers (p < 0.001 T1D versus normal controls, by Student's t tests). Similar ChIP-qPCR results were obtained with two other sets of PCR primers amplifying nearby regions (data not shown). D, ChIP-qPCR validation of H3K9Ac at the HLA-DQB1 promoter region, which was done in triplicates (p < 0.001 T1D versus normal controls, by Student's t tests).
To validate the microarray data showing variations in H3K9Ac around the HLA-DRB1 and HLA-DQB1 promoters, we next performed follow-up ChIP-qPCR validations utilizing separate H3K9Ac antibody-enriched ChIP-DNA samples from each individual in the two cohorts. Fig. 3, C and D, respectively, confirm the decreases in H3K9Ac in T1D noted at the upstream promoter regions of HLA-DRB1 and the increases in H3K9Ac in T1D at the upstream promoter/enhancer region of HLA-DQB1 (p < 0.001).
It is interesting to determine whether the variations noted in the H3K9Ac levels are associated with any changes in gene expression. However, because all the human volunteer blood cells were used to isolate chromatin to perform multiple ChIP assays, there was insufficient cell material to isolate total RNA as well. Thus, further gene expression as well as additional ChIP and ChIP-array experiments were performed in THP-1 monocytes.
Histone H3K9Ac Variations within the Region Containing the Majority of Top T1D SNPs Are Associated with Up-regulation of HLA-DRB1 and HLA-DQB1
The transcription of MHC class II molecules is tightly regulated (40). The binding of key transcription factors, RFX, X2BP, and NF-Y, to the promoter region of MHC class II molecules is highly cooperative and results in the formation of a stable nucleoprotein complex, called the MHC-II enhanceosome. In addition, the master controller of MHC class II genes is class II transactivator (41). A more detailed picture emerged recently in which the insulator CCCTC binding factor was shown to interact with class II transactivator and the enhanceosome complex to form long-distance chromatin loop structures that co-regulate MHC-II genes (42, 43). Interferon-γ (IFN-γ) and TNF-α have been shown to increase the expression of class II molecules including HLA-DQB1 (44). Interestingly, the acetylation state of the upstream region of HLA-DRA has been linked to gene expression (45), suggesting that variations in histone acetylation observed in this study might be relevant to the expression of HLA-DRB1 and HLA-DQB1.
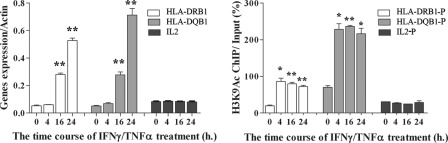
To further explore this in the context of the current results, we used THP1 cells, a human monocyte cell line, as a model because we noticed that HLA-DRB1 and HLA-DQB1 were up-regulated in response to IFNγ/TNFα treatment in these cells (Fig. 4left). Importantly, Fig. 4right shows for the first time that, along with this up-regulation of gene expression in these treatments, there was an increase in histone H3K9Ac levels at the same promoter regions of HLA-DRB1 and HLA-DQB1 that showed variations between T1D patients and normal controls in Fig. 3, C and D. H3K9Ac levels started to increase by 4 h after addition of IFNγ/TNFα, whereas gene expression was significantly increased by 16 h, indicating a time lag between an increase in gene expression and chromatin H3K9Ac changes, suggesting that chromatin remodeling is required for the gene stimulation. To ascertain specificity, we evaluated interleukin-2 (IL2), as a control gene that is not induced by IFNγ/TNFα. We observed that neither the expression of IL2 nor levels of histone H3K9Ac levels at its promoter region are altered upon IFNγ/TNFα treatment (Fig. 4, left and right). IL4 also behaved similarly (results not shown). These results suggest that specific changes in H3K9Ac levels at the genome region between HLA-DRB1 and HLA-DQB1 could reflect changes in the chromatin configuration that would alter accessibility to transcription factors, co-activators/repressors, which in turn could affect gene expression or the response to various stimuli relevant to T1D pathology. We speculate that the dysregulated expression of key T1D-associated genes can occur by at least two steps. First, the chromatin of these genes attains a poised state through alterations in the chromatin configuration marks such as histone PTMs, without affecting gene expression. Next, in response to a second “hit,” gene expression might be altered in response to an external triggering stimulus.
FIGURE 4.
Increased histone H3K9Ac at the promoter/enhancer regions of HLA-DRB1 and HLA-DQB1 correlates with gene expression in response to IFNγ/TNFα in THP1 cells. Left, time course of IFNγ/TNFα-induced HLA-DRB1 and HLA-DQB1 mRNA expression in THP1 cells. IL2 was included as a negative control gene, which was not induced under these conditions. THP1 cells were treated with 20 μm IFNγ and 10 μm TNFα and collected at 0 (no treatment), 4, 16, and 24 h. Total RNAs were prepared and RT-qPCRs were performed in triplicate. **, p < 0.0001 versus 0 h. Right, ChIP-qPCR data showing histone H3K9Ac enrichment levels at HLA-DRB1 and HLA-DQB1 promoter/enhancer regions in THP1 cells treated with IFNγ/TNFα. HLA-DRB1-P and HLA-DQB1-P refer to the same genomic regions where variations in acetylation were noted in the T1D patient monocytes (Fig. 3, C and D). The IL2 promoter (IL2-P) was examined as a negative control, which did not show any changes in H3K9Ac under these conditions. The ChIPs were performed as described under “Experimental Procedures.” ChIP-qPCRs were preformed in triplicates. Data shown are mean ± S.E. from triplicates. *, p < 0.002 versus 0 h; **, p < 0.0001 versus 0 h.
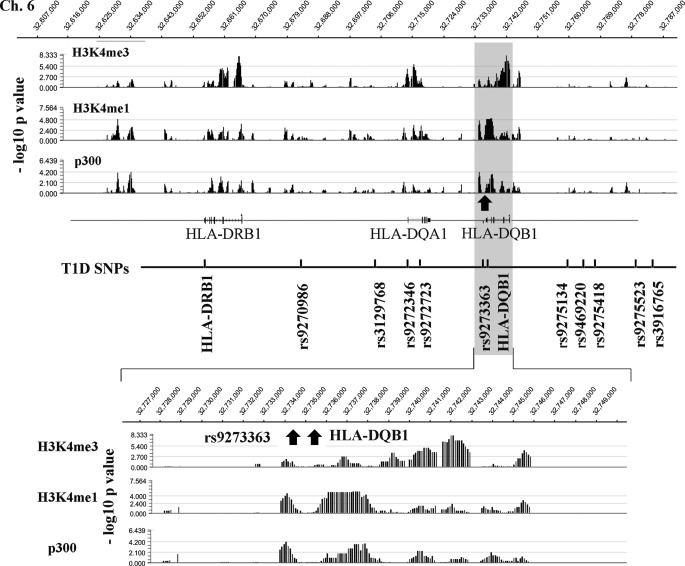
Recent studies show that active gene enhancer regions are typically marked by histone H3K4me1 and occupancy of the histone acetyltransferase p300, whereas promoter regions and TSS are marked by H3K4me3 (46, 47). To further explore such features of the chromatin structure in the HLA-DRB1 and HLA-DQB1 genome regions, we also mapped histone H3K4me3, H3K4me1, and p300 occupancy using the chromosome 6 tiling arrays in untreated THP1 cells. The top panel of Fig. 5 shows a snapshot of the profiles of these marks at this genome region, which reveals that histone H3K4me3 localizes around TSS of HLA-DRB1 and HLA-DQB1, whereas H3K4me1 co-localizes with p300 at regions that could potentially represent locations of enhancers within this key region. Notably, a cluster of top T1D SNPs are located within the loop and enhancer regions (Fig. 5). In particular, we found for the first time that two SNPs highly associated with T1D, rs9273363 and HLA-DQB1 (34), co-localize with H3K4me1 and p300 peaks (Fig. 5, lower panel, indicated by arrows), which suggests that both rs9273363 and HLA-DQB1 SNPs are located within potential enhancer regions of key T1D susceptibility genes.
FIGURE 5.
Mapping the chromatin conformation of the genome region between HLA-DRB1 and HLA-DQB1 in THP1 cells. Histone H3K4me3, H3K4me1, and p300 maps of the genome region between HLA-DRB1 and HLA-DQB1 in untreated THP1 cells. ChIP-chips were performed with chromosome 6 (Chr6) tiling arrays and the indicated antibodies as described under “Experimental Procedures.” Top panel, histone H3K4me3, H3K4me1, and p300 enrichment levels at genome region between HLA-DRB1 and HLA-DQB1 (Chr.6: 32607000–32787000). The enrichment level is presented in smoothed-data format −log10 (p value). p value at each probe was generated by Nimblescan using a nonparametric, one-sided Kolmogorov-Smirnov test of the probes located within the 750-bp sliding window centered by the probe. Genes and SNPs located in the regions are marked based on their genomic location in the region. Lower panel, zoom-in view of histone H3K4me3, H3K4me1, and p300 levels near rs9273363 and HLA-DQB1 with 100-bp resolution.
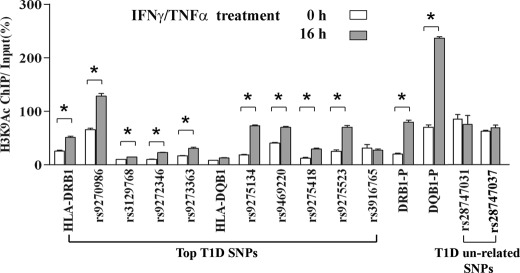
Because the majority of top T1D SNPs are located around the HLA-DRB1 and HLA-DQB1 genome regions (34), we next used ChIP assays to determine whether there are changes in histone H3K9Ac levels around these SNPs during IFNγ/TNFα-induced gene up-regulation in THP-1 cells. Results shown in Fig. 6 demonstrate that there is a significant increase in histone H3K9Ac levels at several of these T1D SNP loci at a time period when HLA-DRB1 and HLA-DQB1 are also up-regulated by IFNγ/TNFα, although to varying degrees. Interestingly, the highest increase of H3K9Ac was observed at the same promoter region of HLA-DQB1 (Fig. 6, DQB1-P) that displayed hyperacetylation in T1D patients, which might explain why acetylation at this region was detected in our study with T1D patients (Fig. 3D). As controls, we also examined two T1D un-related SNPs, rs28747031 and rs28747037, which are located about 20 kb upstream of the HLA-DQB1 locus. Results show that H3K9Ac levels at both of these unrelated SNPs are unchanged in response to IFNγ/TNFα treatment (Fig. 6, last four bars) unlike the increases seen at the T1D SNPs. Collectively, these results indicate an association between gene expression and the increased level of H3K9Ac at the genome region between HLA-DRB1 to HLA-DQB1, and reveal novel correlations between T1D-associated SNPs, histone PTMs, and the chromatin configuration of this key IDDM1 region. They further support our hypothesis that variations in histone PTMs can be found between T1D and normal controls at T1D susceptible loci, and may further enhance the risk of T1D susceptibility.
FIGURE 6.
ChIP-qPCR analysis of histone H3K9Ac-enrichment levels at key T1D SNPs loci in THP1 cells treated with and without IFNγ/TNFα. THP-1 cells were cultured in RPMI 1640 medium containing 5 mm glucose and treated with 20 μm IFNγ and 10 μm TNFα for 16 h. The ChIP was performed with histone H3K9Ac antibody, and ChIP-qPCRs with primers spanning the top T1D SNPs region shown, as well as HLA-DRB1-P and HLA-DQB1-P (same regions showing H3K9Ac changes in Figs. 3 and 4) were performed as described under “Experimental Procedures.” Two T1D unrelated SNPs, rs28747031 and rs28747037, were also included as controls and did not show any increases in H3K9Ac. Primer sequences are provided in supplemental Table S2. Data are shown as mean ± S.E. from triplicates. *, p < 0.001 treated versus untreated (0 h).
DISCUSSION
The recent GWAS in T1D have confirmed and extended the results from previous studies and defined 41 distinct genome locations as T1D susceptible regions (34). The huge datasets generated by GWAS have provided a rich resource of invaluable information, enormous challenges, and exciting opportunities for subsequent diabetes research. There is significant interest in exploring how the susceptible loci identified by GWAS are related to T1D. As such, in our current study, we focused on these 41 T1D susceptible regions to determine whether an epigenetic layer over these genetic loci can have additional correlations between these regions and T1D disease phenotype and pathogenesis. We profiled and mapped the key histone PTM status of genes located within these 41 genomic loci in lymphocytes, and more specifically, within the IDDM1 region in monocytes of patients with T1D versus healthy controls. Notably, we observed for the first time that there were marked variations in H3K9Ac levels at upstream regions of HLA-DRB1 and HLA-DQB1 in monocytes of this cohort (T1D versus control). On the other hand, the levels of the other tested histone PTMs at most of the T1D susceptible gene promoters were relatively quite similar between T1D and control volunteers in their lymphocytes or monocytes. It should be underscored that there were also variations noted in many of the histone PTMs beyond T1D susceptible regions in this cohort (data not shown). For reasons stated above, we focused only on T1D-related loci and adopted a T1D candidate gene-set profiling approach due to recent advances in GWAS of T1D (34).
It is well known that HLA-DRB1 and HLA-DQB1 are two major candidates associated with T1D incidence, with lesser contributions from the remaining T1D susceptible genes (5–6, 48, 49). This may explain why we detected variations at only HLA-DRB1 and HLA-DQB1 in the samples, although it could also be due to the level of sensitivity of detection in the ChIP-array approach, and that only limited genome regions and histone marks were assessed. It is possible that there are variations at T1D regions in other histone PTMs not examined in this study, or in DNA methylation, all of which are the focus of future endeavors. Interestingly, a recent study using next generation sequencing revealed that high glucose could induce genome-wide variations in histone H3K9Ac and DNA methylation in human aortic endothelial cells, and furthermore, the regions with histone H3K9Ac variations contained many common human disease SNPs including T1D (23).
Although GWAS have confirmed and cataloged a large number of SNP-associated T1D (34), most of these SNPs are intronic and intergenic and few SNPs have been linked to T1D at the functional or mechanistic level, a key step for a better understanding of T1D. Concerted efforts are ongoing to catalog additional SNPs with recent advances in sequencing technologies. Our results indicate the presence of marked variations in the levels of a key chromatin mark H3K9Ac at the upstream regions of two top T1D genes, HLA-DRB1 and HLA-DQB1, suggesting a link between the chromatin status of their promoter/enhancer region and T1D. We also observed that the epigenome or status of several histone PTMs at the MHC locus, when viewed globally, is quite similar between T1D and nondiabetic controls (Fig. 2), implying that, overall, most T1D-related SNPs do not affect the tested histone PTMs to a great extent. Hence, the variations we observed are most likely not due to SNPs but may be due to environmental factors that are known to affect epigenetic processes.
We are aware that our results are derived from a small number of T1D patients and controls, and therefore the exact similar type and intensity of variation might or might not be the same in all T1D patients. Disease duration and extent of complications may also play a role. However, based on the current data, we speculate that the chromatin architecture of the genome region near HLA-DRB1 and HLA-DQB1 within IDDM1 is quite sensitive to histone modifications and relevant to the expression of HLA-DRB1 and HLA-DQB1. This notion is further supported by our data with THP-1 cells showing increased gene expression along with increases in histone H3K9Ac in response to IFNγ/TNFα treatment at the same promoter/enhancer regions of HLA-DRB1 and HLA-DQB1 as that seen in the T1D patient monocytes. Moreover, we also noticed increases in H3K9Ac at several well documented T1D SNPs in the vicinity of HLA-DRB1/DQB1. Therefore, a combination of genetic and epigenetic changes at these regions could potentially further enhance the risk of disease incidence. Thus, the susceptibility to T1D might require not only the presence of disease-related SNPs, but also disease poised chromatin states such as hyperacetylation at the enhancer regions of these SNPs.
Finally, it is still an enigma why only a small fraction of genetically susceptible individuals appear to acquire T1D later in their lifetime. A general explanation is that there are protective SNPs that prevent T1D acquisition, whereas on the other hand, certain environmental factors might also influence T1D incidence. The role of epigenetics in this process is largely unknown. Based on various reports (4, 12, 23, 25, 28, 50), we are gradually gaining more details of epigenetic changes in T1D. Our current study links variations in histone H3K9Ac in monocytes at HLA-DQB1 and HLA-DRB1 promoter/enhancer regions to T1D, although it is limited to a small cohort and also cannot determine whether the variations are early triggers or late events. Parallel studies with THP-1 cells support the data with T1D patients and demonstrate novel connections between H3K9Ac at HLA-DQB1 and HLA-DRB1 especially at their enhancer regions and their expression. The pathological implications and significance of the findings need further investigation and reinforcement through similar studies in the future with larger well characterized cohorts as well as with T1D discordant twins and siblings. Notwithstanding, based on the current results, we speculate that the susceptibility to the development of T1D could require not only SNPs as documented by extensive genetic studies, but also potential “chromatin hits” from accompanying epigenetic events that could confer a functional context to the SNPs. Identifying such events and contributions will lead to a greater understanding of T1D susceptibility and, hopefully to better predictions or early detection of disease onset.
Supplementary Material
Acknowledgments
We are grateful to Dr. Wei Feng, City of Hope Medical Center, for recruiting T1D patients, Dr. Chih-Pin Liu, Beckman Research Institute of City of Hope, for valuable suggestions, Dr. Silvia DaCosta for help with the manuscript, to the nurses in the GCRC, City of Hope Medical Center, and to all the volunteers who generously donated blood. We also thank Dr. Parimal Majumder (Emory University, Atlanta, Georgia) for providing RT-PCR primer sequences for human HLA-DRB1 and HLA-DQB1.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 DK065073 and R01 HL087864 and the Juvenile Diabetes Research Foundation (to R. N.), and General Clinical Research Center Grant M01RR00043 from National Center for Research Resources (to City of Hope).

This article contains supplemental Methods, Tables S1 and S2, and Figs. S1 and S2.
All genomic and expression data can be accessed through NCBI Protein Database under NCBI accession number GSE36403.
- T1D
- type 1 diabetes
- PTM
- post-translational modification
- IP
- immunoprecipitation
- qPCR
- quantitative PCR
- TSS
- transcription start site
- MHC
- major histocompatibility complex
- SNP
- single nucleotide polymorphism
- HLA
- human leukocyte antigen
- IDDM1
- insulin-dependent diabetes mellitus 1
- H3K4me
- H3-lysine 4 methylation
- H3K9Ac
- H3-lysine 9 acetylation
- GWAS
- genome-wide association study
- FDR
- false discovery rate.
REFERENCES
- 1. Pociot F., Akolkar B., Concannon P., Erlich H. A., Julier C., Morahan G., Nierras C. R., Todd J. A., Rich S. S., Nerup J. (2010) Genetics of type 1 diabetes. What's next? Diabetes 59, 1561–1571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Eisenbarth G. S. (2010) Banting Lecture 2009, an unfinished journey. Molecular pathogenesis to prevention of type 1A diabetes. Diabetes 59, 759–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Concannon P., Rich S. S., Nepom G. T. (2009) Genetics of type 1A diabetes. N. Engl. J. Med. 360, 1646–1654 [DOI] [PubMed] [Google Scholar]
- 4. Todd J. A. (2010) Etiology of type 1 diabetes. Immunity 32, 457–467 [DOI] [PubMed] [Google Scholar]
- 5. Nerup J., Platz P., Andersen O. O., Christy M., Lyngsoe J., Poulsen J. E., Ryder L. P., Nielsen L. S., Thomsen M., Svejgaard A. (1974) HL-A antigens and diabetes melittis. Lancet 2, 864–866 [DOI] [PubMed] [Google Scholar]
- 6. Mein C. A., Esposito L., Dunn M. G., Johnson G. C., Timms A. E., Goy J. V., Smith A. N., Sebag-Montefiore L., Merriman M. E., Wilson A. J., Pritchard L. E., Cucca F., Barnett A. H., Bain S. C., Todd J. A. (1998) A search for type 1 diabetes susceptibility genes in families from the United Kingdom. Nat. Genet. 19, 297–300 [DOI] [PubMed] [Google Scholar]
- 7. Davies J. L., Kawaguchi Y., Bennett S. T., Copeman J. B., Cordell H. J., Pritchard L. E., Reed P. W., Gough S. C., Jenkins S. C., Palmer S. M. (1994) A genome-wide search for human type 1 diabetes susceptibility genes. Nature 371, 130–136 [DOI] [PubMed] [Google Scholar]
- 8. Aly T. A., Eller E., Ide A., Gowan K., Babu S. R., Erlich H. A., Rewers M. J., Eisenbarth G. S., Fain P. R. (2006) Multi-SNP analysis of MHC region. Remarkable conservation of HLA-A1-B8-DR3 haplotype. Diabetes 55, 1265–1269 [DOI] [PubMed] [Google Scholar]
- 9. Aly T. A., Ide A., Jahromi M. M., Barker J. M., Fernando M. S., Babu S. R., Yu L., Miao D., Erlich H. A., Fain P. R., Barriga K. J., Norris J. M., Rewers M. J., Eisenbarth G. S. (2006) Extreme genetic risk for type 1A diabetes. Proc. Natl. Acad. Sci. U.S.A. 103, 14074–14079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Peng H., Hagopian W. (2006) Environmental factors in the development of type 1 diabetes. Rev. Endocr. Metab. Dis. 7, 149–162 [DOI] [PubMed] [Google Scholar]
- 11. Villeneuve L. M., Natarajan R. (2010) The role of epigenetics in the pathology of diabetic complications. Am. J. Physiol. Renal Physiol 299, F14–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Litherland S. A. (2008) Immunopathogenic interaction of environmental triggers and genetic susceptibility in diabetes. Is epigenetics the missing link? Diabetes 57, 3184–3186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Harjutsalo V., Sjöberg L., Tuomilehto J. (2008) Time trends in the incidence of type 1 diabetes in Finnish children. A cohort study. Lancet 371, 1777–1782 [DOI] [PubMed] [Google Scholar]
- 14. Jirtle R. L., Skinner M. K. (2007) Environmental epigenomics and disease susceptibility. Nat. Rev. Genet. 8, 253–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Klug A., Rhodes D., Smith J., Finch J. T., Thomas J. O. (1980) A low resolution structure for the histone core of the nucleosome. Nature 287, 509–516 [DOI] [PubMed] [Google Scholar]
- 16. Strahl B. D., Allis C. D. (2000) The language of covalent histone modifications. Nature 403, 41–45 [DOI] [PubMed] [Google Scholar]
- 17. Zhang Y., Reinberg D. (2001) Transcription regulation by histone methylation. Interplay between different covalent modifications of the core histone tails. Genes Dev. 15, 2343–2360 [DOI] [PubMed] [Google Scholar]
- 18. Shilatifard A. (2006) Chromatin modifications by methylation and ubiquitination. Implications in the regulation of gene expression. Annu. Rev. Biochem. 75, 243–269 [DOI] [PubMed] [Google Scholar]
- 19. Campos E. I., Reinberg D. (2009) Histones, annotating chromatin. Annu. Rev. Genet. 43, 559–599 [DOI] [PubMed] [Google Scholar]
- 20. Bird A. (2007) Perceptions of epigenetics. Nature 447, 396–398 [DOI] [PubMed] [Google Scholar]
- 21. Berger S. L., Kouzarides T., Shiekhattar R., Shilatifard A. (2009) An operational definition of epigenetics. Genes Dev. 23, 781–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Smith E., Shilatifard A. (2010) The chromatin signaling pathway. Diverse mechanisms of recruitment of histone-modifying enzymes and varied biological outcomes. Mol. Cell 40, 689–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pirola L., Balcerczyk A., Tothill R. W., Haviv I., Kaspi A., Lunke S., Ziemann M., Karagiannis T., Tonna S., Kowalczyk A., Beresford-Smith B., Macintyre G., Kelong M., Hongyu Z., Zhu J., El-Osta A. (2011) Genome-wide analysis distinguishes hyperglycemia regulated epigenetic signatures of primary vascular cells. Genome Res. 21, 1601–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Miao F., Gonzalo I. G., Lanting L., Natarajan R. (2004) In vivo chromatin remodeling events leading to inflammatory gene transcription under diabetic conditions. J. Biol. Chem. 279, 18091–18097 [DOI] [PubMed] [Google Scholar]
- 25. Miao F., Wu X., Zhang L., Yuan Y. C., Riggs A. D., Natarajan R. (2007) Genome-wide analysis of histone lysine methylation variations caused by diabetic conditions in human monocytes. J. Biol. Chem. 282, 13854–13863 [DOI] [PubMed] [Google Scholar]
- 26. Villeneuve L. M., Reddy M. A., Lanting L. L., Wang M., Meng L., Natarajan R. (2008) Epigenetic histone H3 lysine 9 methylation in metabolic memory and inflammatory phenotype of vascular smooth muscle cells in diabetes. Proc. Natl. Acad. Sci. U.S.A. 105, 9047–9052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. El-Osta A., Brasacchio D., Yao D., Pocai A., Jones P. L., Roeder R. G., Cooper M. E., Brownlee M. (2008) Transient high glucose causes persistent epigenetic changes and altered gene expression during subsequent normoglycemia. J. Exp. Med. 205, 2409–2417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Miao F., Smith D. D., Zhang L., Min A., Feng W., Natarajan R. (2008) Lymphocytes from patients with type 1 diabetes display a distinct profile of chromatin histone H3 lysine 9 dimethylation. An epigenetic study in diabetes. Diabetes 57, 3189–3198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Brasacchio D., Okabe J., Tikellis C., Balcerczyk A., George P., Baker E. K., Calkin A. C., Brownlee M., Cooper M. E., El-Osta A. (2009) Hyperglycemia induces a dynamic cooperativity of histone methylase and demethylase enzymes associated with gene-activating epigenetic marks that coexist on the lysine tail. Diabetes 58, 1229–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sun G., Reddy M. A., Yuan H., Lanting L., Kato M., Natarajan R. (2010) Epigenetic histone methylation modulates fibrotic gene expression. J. Am. Soc. Nephrol. 12, 2069–2080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ren B., Robert F., Wyrick J. J., Aparicio O., Jennings E. G., Simon I., Zeitlinger J., Schreiber J., Hannett N., Kanin E., Volkert T. L., Wilson C. J., Bell S. P., Young R. A. (2000) Genome-wide location and function of DNA binding proteins. Science 290, 2306–2309 [DOI] [PubMed] [Google Scholar]
- 32. Barski A., Cuddapah S., Cui K., Roh T. Y., Schones D. E., Wang Z., Wei G., Chepelev I., Zhao K. (2007) High-resolution profiling of histone methylations in the human genome. Cell 129, 823–837 [DOI] [PubMed] [Google Scholar]
- 33. Hawkins R. D., Hon G. C., Ren B. (2010) Next-generation genomics. An integrative approach. Nat. Rev. Genet 11, 476–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Barrett J. C., Clayton D. G., Concannon P., Akolkar B., Cooper J. D., Erlich H. A., Julier C., Morahan G., Nerup J., Nierras C., Plagnol V., Pociot F., Schuilenburg H., Smyth D. J., Stevens H., Todd J. A., Walker N. M., Rich S. S. (2009) Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat. Genet. 41, 703–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bluestone J. A., Herold K., Eisenbarth G. (2010) Genetics, pathogenesis, and clinical interventions in type 1 diabetes. Nature 464, 1293–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Vandiedonck C., Knight J. C. (2009) The human major histocompatibility complex as a paradigm in genomics research. Brief. Funct. Genomics Proteomics 8, 379–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dausset J. (1981) The major histocompatibility complex in man. Science 213, 1469–1474 [DOI] [PubMed] [Google Scholar]
- 38. Cheong C., Matos I., Choi J. H., Dandamudi D. B., Shrestha E., Longhi M. P., Jeffrey K. L., Anthony R. M., Kluger C., Nchinda G., Koh H., Rodriguez A., Idoyaga J., Pack M., Velinzon K., Park C. G., Steinman R. M. (2010) Microbial stimulation fully differentiates monocytes to DC-SIGN/CD209(+) dendritic cells for immune T cell areas. Cell 143, 416–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bieda M., Xu X., Singer M. A., Green R., Farnham P. J. (2006) Unbiased location analysis of E2F1-binding sites suggests a widespread role for E2F1 in the human genome. Genome Res. 16, 595–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Choi N. M., Majumder P., Boss J. M. (2011) Regulation of major histocompatibility complex class II genes. Curr. Opin Immunol. 23, 81–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Masternak K., Muhlethaler-Mottet A., Villard J., Zufferey M., Steimle V., Reith W. (2000) CIITA is a transcriptional coactivator that is recruited to MHC class II promoters by multiple synergistic interactions with an enhanceosome complex. Genes Dev. 14, 1156–1166 [PMC free article] [PubMed] [Google Scholar]
- 42. Majumder P., Gomez J. A., Chadwick B. P., Boss J. M. (2008) The insulator factor CTCF controls MHC class II gene expression and is required for the formation of long-distance chromatin interactions. J. Exp. Med. 205, 785–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Majumder P., Boss J. M. (2010) CTCF controls expression and chromatin architecture of the human major histocompatibility complex class II locus. Mol. Cell. Biol. 30, 4211–4223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Arenzana-Seisdedos F., Mogensen S. C., Vuillier F., Fiers W., Virelizier J. L. (1988) Autocrine secretion of tumor necrosis factor under the influence of interferon-γ amplifies HLA-DR gene induction in human monocytes. Proc. Natl. Acad. Sci. U.S.A. 85, 6087–6091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Masternak K., Peyraud N., Krawczyk M., Barras E., Reith W. (2003) Chromatin remodeling and extragenic transcription at the MHC class II locus control region. Nat. Immunol. 4, 132–137 [DOI] [PubMed] [Google Scholar]
- 46. Heintzman N. D., Hon G. C., Hawkins R. D., Kheradpour P., Stark A., Harp L. F., Ye Z., Lee L. K., Stuart R. K., Ching C. W., Ching K. A., Antosiewicz-Bourget J. E., Liu H., Zhang X., Green R. D., Lobanenkov V. V., Stewart R., Thomson J. A., Crawford G. E., Kellis M., Ren B. (2009) Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459, 108–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Visel A., Blow M. J., Li Z., Zhang T., Akiyama J. A., Holt A., Plajzer-Frick I., Shoukry M., Wright C., Chen F., Afzal V., Ren B., Rubin E. M., Pennacchio L. A. (2009) ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature 457, 854–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Todd J. A., Bell J. I., McDevitt H. O. (1987) HLA-DQβ gene contributes to susceptibility and resistance to insulin-dependent diabetes mellitus. Nature 329, 599–604 [DOI] [PubMed] [Google Scholar]
- 49. Todd J. A., Walker N. M., Cooper J. D., Smyth D. J., Downes K., Plagnol V., Bailey R., Nejentsev S., Field S. F., Payne F., Lowe C. E., Szeszko J. S., Hafler J. P., Zeitels L., Yang J. H., Vella A., Nutland S., Stevens H. E., Schuilenburg H., Coleman G., Maisuria M., Meadows W., Smink L. J., Healy B., Burren O. S., Lam A. A., Ovington N. R., Allen J., Adlem E., Leung H. T., Wallace C., Howson J. M., Guja C., Ionescu-Tîrgovişte C., Simmonds M. J., Heward J. M., Gough S. C., Dunger D. B., Wicker L. S., Clayton D. G. (2007) Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat. Genet. 39, 857–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Seydel F., Garrigan E., Stutevoss B., Belkin N., Makadia B., Carter J., Shi J. D., Davoodi-Semiromi A., McDuffie M., Litherland S. A. (2008) GM-CSF induces STAT5 binding at epigenetic regulatory sites within the Csf2 promoter of nonobese diabetic (NOD) mouse myeloid cells. J. Autoimmun. 31, 377–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.