Background: The physiological roles of NUAK1 are poorly understood because of embryonic lethality in NUAK1 null mice.
Results: Negative feedback regulation of insulin signaling was abrogated in skeletal muscle of muscle-specific NUAK1 knock-out mice.
Conclusion: NUAK1 controls glucose metabolism through negative regulation of insulin signal transduction in skeletal muscle.
Significance: This is the first report of a physiological role of NUAK1 in adult tissues.
Keywords: AMP-activated Kinase (AMPK), Glucose Metabolism, Insulin Resistance, Phosphoproteomics, Skeletal Muscle, NUAK1
Abstract
NUAK1 is a member of the AMP-activated protein kinase-related kinase family. Recent studies have shown that NUAK1 is involved in cellular senescence and motility in epithelial cells and fibroblasts. However, the physiological roles of NUAK1 are poorly understood because of embryonic lethality in NUAK1 null mice. The purpose of this study was to elucidate the roles of NUAK1 in adult tissues. We determined the tissue distribution of NUAK1 and generated muscle-specific NUAK1 knock-out (MNUAK1KO) mice. For phenotypic analysis, whole body glucose homeostasis and muscle glucose metabolism were examined. Quantitative phosphoproteome analysis of soleus muscle was performed to understand the molecular mechanisms underlying the knock-out phenotype. Nuak1 mRNA was preferentially expressed in highly oxidative tissues such as brain, heart, and soleus muscle. On a high fat diet, MNUAK1KO mice had a lower fasting blood glucose level, greater glucose tolerance, higher insulin sensitivity, and higher concentration of muscle glycogen than control mice. Phosphoproteome analysis revealed that phosphorylation of IRS1 Ser-1097 was markedly decreased in NUAK1-deficient muscle. Consistent with this, insulin signaling was enhanced in the soleus muscle of MNUAK1KO mice, as evidenced by increased phosphorylation of IRS1 Tyr-608, AKT Thr-308, and TBC1D4 Thr-649. These observations suggest that a physiological role of NUAK1 is to suppress glucose uptake through negative regulation of insulin signaling in oxidative muscle.
Introduction
NUAK1 is a serine/threonine kinase belonging to the AMP-activated protein kinase-related kinase (AMPK-RK)2 family. AMPK is a heterotrimer composed of an α catalytic subunit and two regulatory subunits: β and γ. The AMPK-RK family includes 12 proteins classified together on the basis of sequence similarity with AMPKαs (1, 2). The phosphorylation and activation of AMPKαs are up-regulated by AMP through binding to the γ regulatory subunit (3). On the other hand, AMPK-RKs do not have regulatory subunits; thus their activities are not directly regulated by the cellular AMP:ATP ratio (4). Like the AMPKαs and most other AMPK-RKs, NUAK1 can be phosphorylated by liver kinase B1 (LKB1) at a conserved threonine residue (corresponding to Thr-211 in NUAK1) (2). In addition to Thr-211, NUAK1 can be phosphorylated at Ser-600 by AKT (5). However, a recent study demonstrated that amino acid substitution at this site (S600A) has no influence on kinase activity (6). Thus, the functional significance of phosphorylation at Ser-600 is controversial.
In epithelial cells and fibroblasts, LKB1-NUAK1 phosphorylates myosin phosphatase target subunit 1 and large tumor suppressor homolog 1, which results in cell detachment and senescence, respectively (6, 7). NUAK1 also acts as a transcriptional coactivator in complex with LKB1 and tumor suppressor p53 to induce cell cycle G1 arrest in A549 lung adenocarcinoma cells (8). In mouse C2C12 myoblasts, NUAK1 is increasingly expressed with differentiation to myotubes (9). Apart from these in vitro studies, a study involving knock-out mice showed that the mouse homolog of NUAK1 (OMPHK1) is essential for closure of the ventral body wall in developing embryos (10). In a study with colorectal cancer clinical samples, increased NUAK1 mRNA has been observed (11). Overall, the functions and roles of NUAK1 have been investigated in the context of motility or proliferation of cultured cells, embryonic development, and cancer progression. However, little research has focused on the physiological roles of NUAK1 in adult tissues.
Skeletal muscle is the major tissue responsible for disposal of total body glucose (12). The two major physiological stimulators of skeletal muscle glucose uptake are insulin and muscle contraction (13). Contraction-stimulated glucose uptake has been shown to be mediated by LKB1 via AMPKα2 and/or NUAK2, an AMPK-RK with the highest homology to NUAK1 (14–20). In addition to the influences on contraction-stimulated glucose uptake, muscle-specific LKB1 knock-out mice display increased insulin sensitivity and improved whole body glucose homeostasis (21). In contrast, muscle-specific inhibition of AMPKα2 impairs insulin sensitivity and glucose tolerance (22). Other than AMPKα2 and NUAK2, little is known about the involvement of other AMPK-RKs in muscle glucose metabolism.
The purpose of this study was to elucidate the physiological roles of NUAK1 in adult tissues. For this purpose, we generated muscle-specific NUAK1 knock-out (MNUAK1KO) mice. To our knowledge, this is the first report of conditional knock-out of NUAK1. MNUAK1KO mice were apparently normal but exhibited improved glucose homeostasis under high fat diet (HFD) conditions. To understand the molecular mechanisms underlying the phenotype associated with the knock-out, we performed a quantitative phosphoproteome analysis of skeletal muscle. Our data suggest that one role of NUAK1 is suppression of insulin signal transduction in skeletal muscle.
EXPERIMENTAL PROCEDURES
Animal Protocols
All of the experimental protocols were approved by the Institutional Ethics Review Committee at the National Cancer Center. The mice were maintained on a 12-h light/dark cycle and housed in a temperature-controlled barrier facility with free access to water and a standard rodent chow composed of 20% calories from fat, 50% calories from carbohydrate, and 30% calories from protein (CMF; Oriental Yeast, Tokyo, Japan). Nuak1flox/flox mice were obtained from the Laboratory for Animal Resources and Genetic Engineering, Center for Developmental Biology, RIKEN Kobe (accession number CDB0555K). Prior to initiation of the present study, Nuak1flox/flox mice were backcrossed onto a C57BL/6J background using the speed cogenic method (Oriental Bioservice, Tokyo, Japan). To generate MNUAK1KO mice, Nuak1flox/flox mice were mated with muscle creatin kinase (Mck)-Cre mice (JAX, number 006475: B6.FVB (129S4)-Tg (Ckmm-cre) 5 Khn/J). As a control for MNUAK1KO mice, their Nuak1flox/flox littermate mice were used. For HFD-induced glucose intolerance, the mice were fed a HFD composed of 57% calories from fat, 23% calories from carbohydrate, and 20% calories from protein (HFD32; Clea Japan, Tokyo, Japan) starting from 5 weeks of age until the termination of the experiments. Male mice were used for all of the experiments.
Genotyping
Genomic DNA from various tissues was subjected to PCR involving 34 cycles of denaturation at 94 °C for 30 s, annealing at 60 °C for 30 s, and elongation at 72 °C for 90 s with the following primers: 5′-specific primer P1 (5′-GGTAGGTGGAGGTCGGCTGAGAAGG) and 3′-specific primer P2 (5′-TCGGATCCTAGTGAACCTCTTC).
Real Time RT-PCR
Total RNA was extracted using Isogen (Nippon Gene, Tokyo, Japan) and quantified using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA). Equal amounts (3.5 μg/15-μl reaction) of total RNA were subjected to first strand cDNA synthesis using a first strand cDNA synthesis kit (GE Healthcare Japan) according to the manufacturer's instructions. Real time PCR was carried out with TaqMan Universal PCR Master Mix (Invitrogen Japan) and TaqMan gene expression assays (Invitrogen Japan) according to the manufacturer's protocols. The TaqMan gene expression assays used in this study were Nuak1 (Mm01250701_m1), Nuak2 (Mm00546961_m1), PGC-1α (Mm_01208835_m1), TNNC1 (Mm00437111_m1), TNNC2 (Mm00437116_m1), and 18 S rRNA (4319413E). For relative quantification, Ct values for each gene were normalized to those for 18 S rRNA. For absolute quantification, synthesized oligonucleotide DNA fragments (Sigma-Aldrich Japan) containing the PCR amplicon regions were used to generate standard curves.
Immunoblotting
Following sacrifice, mouse tissues were rapidly dissected and frozen in liquid nitrogen. The frozen samples were homogenized in lysis buffer containing 1% SDS, 10 mm Tris (pH 7.5), 1 mm Na3VO4, and protease inhibitor mixture (Sigma-Aldrich Japan) and then subjected to SDS-PAGE. The proteins were transferred onto a polyvinylidene fluoride microporous membrane (Millipore, MA). The primary antibodies used were: anti-NUAK1 (4458), anti-phospho-LKB1 Ser-428 (3482), anti-LKB1 (3047), anti-IRS1 (2382), anti-phospho-AKT Thr-308 (9275), and anti-AKT (4685), all obtained from Cell Signaling Technologies (Beverly, MA), anti-phospho-TBC1D4 Thr-642 (ab65753), and anti-glucose transporter type 4 (GLUT4) (ab65976), obtained from Abcam (Cambridge, UK), anti-TBC1D4 (07-741) and anti-phospho-IRS1 Tyr-608 (09-432), obtained from Millipore (Billerica, MA), and anti-actin (sc-1615), obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Immunoblots were scanned using a Canoscan LiDE60 image scanner (Cannon, Tokyo, Japan), and the intensities of the protein bands were quantified using an image processing program ImageJ 1.44.
Measurements of Blood Glucose, Insulin, and Muscle Glycogen
Blood glucose levels were measured from tail blood using an Antsense II glucometer (Bayer Medical, Tokyo, Japan). Plasma insulin levels were determined from peripheral blood using mouse insulin ELISA kit S-Type (Shibayagi, Gunma, Japan). The glycogen concentration of the soleus muscle was determined using a glycogen assay kit (BioVision, Milpitas, CA). For oral glucose tolerance tests (OGTT), mice fasted overnight were administered glucose at a dose of 1 g/kg of body weight. Blood glucose levels were measured immediately before and 20, 40, 60, and 120 min after the administration. For insulin tolerance tests (ITT), mice fasted for 2 h were injected intraperitoneally with recombinant human insulin (Wako Pure Chemical Industries, Osaka, Japan) at a dose of 1 unit/kg of body weight. Blood glucose levels were measured immediately before and 15, 30, and 60 min after the injection.
Histological Analysis
Soleus muscles were fixed with 10% neutral buffered formalin, embedded in paraffin, and sectioned at 6-μm thickness. The sections were stained using Alexa Fluor 555-conjugated wheat germ agglutinin and Hoechst 33342 (Invitrogen Japan). The immunofluorescent images were visualized with a Zeiss LSM 710 confocal laser microscope (Carl Zeiss Japan). The myocyte cross-sectional areas obtained from 84 cells from six control mice and 83 cells from six MNUAK1KO mice were quantified by ZEN 2009 image viewer software (Carl Zeiss Japan).
Measurements of Glucose Uptake in Skeletal Muscles
All of the incubation media were pregassed with 95% O2, 5% CO2, and all incubation was performed at 30 °C. Ex vivo muscle incubation was performed as described previously (23). Soleus muscles were incubated for 30 min in Krebs-Henseleit buffer (KHB) (pH 7.4) containing 10 mm HEPES, 5.5 mm glucose, 2 mm pyruvate, and 0.05% free fatty acid free-bovine serum albumin without or with insulin (20 milliunits/ml). Subsequently, the muscles were washed with glucose-free KHB without or with insulin (20 milliunits/ml) three times. Glucose transport was then determined in KHB supplemented with 0.5 MBq/ml 2-deoxy-2-[18F]fluoro-d-glucose ([18F]FDG) (Nihon Medi-Physics, Tokyo, Japan), 0.75 mm FDG, and 9 μm mannitol in the absence or presence of insulin (20 milliunits/ml) for 20 min. [18F]FDG was washed out with ice-cold KHB without glucose and without insulin three times. Thereafter, the muscles were blotted and weighed. The remaining radioactivity derived from [18F]FDG was measured by a Wizard 2 automatic γ-counter (PerkinElmer Japan).
Phosphoproteome Analysis
Soleus muscles were frozen in liquid nitrogen and disrupted using a multibead shocker (MB400U; Yasui Kikai, Osaka, Japan). Homogenates of the muscles were reduced with dithiothreitol and then alkylated with iodoacetamide before being subjected to a dual enzymatic digestion (Lys-C followed by trypsin) (24). Digested samples were desalted using StageTips with SDB-XC Empore disk membranes (3M Company, St. Paul, MN) (25). Dimethyl labeling was performed according to the literature (26). Phosphopeptide enrichment based on hydroxy acid-modified metal oxide chromatography was performed using lactic acid-modified titania as described previously with a slight modification (27). The eluates from hydroxy acid-modified metal oxide chromatography were concentrated in a vacuum evaporator (CC-105; Tomy, Tokyo, Japan). NanoLC-MS/MS analysis was performed using a previously described setup (28). Peptides and proteins were identified by Mascot version 2.3 (Matrix Science, Tokyo, Japan) against the Uniprot/SwissProt data base release 2011_04 with a precursor mass tolerance of 3 ppm, a fragment ion mass tolerance of 0.8 Da, and strict trypsin specificity allowing for up to two missed cleavages. Cysteine carbamidomethylation was set as a fixed modification. Oxidation of methionine, phosphorylation of serine, threonine, and tyrosine, and [1H4,12C2/2H4,13C2]dimethylation of the N terminus and lysine were allowed as variable modifications. Peptides were considered identified if the Mascot score was over the 95% confidence limit based on the “identity” score of each peptide, and at least three successive y or b ions and two or more y, b, and/or precursor origin neutral loss ions were observed, based on the error-tolerant peptide sequence tag concept (29). False positive rates were estimated by searching against a randomized decoy data base created by the Mascot Perl program supplied by Matrix Science (averaged false positive rate = 1.02%). The details of experimental procedures are described in the supplemental information.
RESULTS
NUAK1 Is Preferentially Expressed in Highly Oxidative Tissues Such as Cerebrum, Heart, and Soleus Muscle
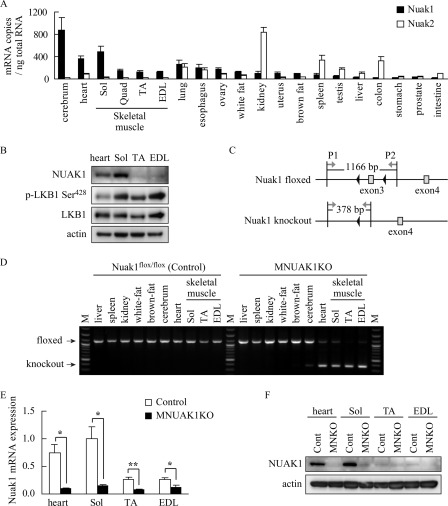
To determine the tissue distribution of NUAK1, the level of mouse Nuak1 mRNA in various organs and tissues was measured using quantitative RT-PCR. The distribution of Nuak2 mRNA was also examined because NUAK1 and NUAK2 may have redundant functions (7, 30). As shown in Fig. 1A, Nuak1 was most abundant in the cerebrum and heart, which is consistent with the distribution of human NUAK1 (31). We found that among the skeletal muscles examined, Nuak1 was selectively expressed in the soleus, at a level comparable with the heart. In contrast, Nuak2 was barely detectable in the skeletal muscles and was highly expressed in kidney.
FIGURE 1.
NUAK1 is preferentially expressed in highly oxidative tissues. A, quantitative RT-PCR analysis of Nuak1 and Nuak2. RNA was isolated from 10-week-old C57BL/6 mice. The data are the means ± S.E. (n = 3). B, immunoblotting analysis of NUAK1 and LKB1 in heart, Sol, TA, and EDL muscles. Actin was used as a loading control. Protein extracts were from three individual 10-week-old C57BL/6 mice. C, schematic representation of floxed and knock-out alleles of Nuak1. The black arrowheads represent loxP sites. Gray arrows indicate PCR primers P1 and P2 used for the genotyping. Amplification with the P1 and P2 primers yields the 1166-bp product from the floxed allele and the 378-bp product from the knock-out allele. D, genotyping of Nuak1 floxed mice without (control) or with (MNUAK1KO) the Mck-Cre transgene. M, molecular marker. E, real time RT-PCR for Nuak1 mRNA in muscles. RNA was isolated from 10-week-old control and MNUAK1KO mice. The mRNA levels are expressed relative to that in soleus muscle of control mice. The data are the means ± S.E. (n = 3). *, p < 0.05; **, p < 0.01 (Student's t test). F, immunoblot analysis of NUAK1 protein in muscles. Protein extracts of each type of muscle were prepared from three individual mice. Sol, soleus; Quad, quadriceps; Cont, control; MNKO, MNUAK1KO.
Expression of NUAK1 protein in the heart, soleus, tibialis anterior (TA), and extensor digitorum longus (EDL) muscles was analyzed using immunoblotting, which also showed muscle type-specific expression of NUAK1 (Fig. 1B). LKB1 was expressed in all of the muscles examined, and the phosphorylation level was lower in the heart than in the other muscles (Fig. 1B). Previous studies have shown that soleus muscle predominantly contains type I and type IIA fibers, which have a high mitochondrial density and oxidative capacity (32). Therefore, our data suggest that NUAK1 is preferentially expressed in highly oxidative tissues such as the cerebrum, heart, and soleus muscle.
Generation of MNUAK1KO Mice
To investigate the role of NUAK1 in skeletal muscle, we generated conditional knock-out (MNUAK1KO) mice. Mice carrying Nuak1 floxed allele (Nuak1flox/flox), in which exon 3 was flanked by loxP sequences as shown in Fig. 1C, were crossed with muscle creatin kinase (Mck)-Cre transgenic mice. The MNUAK1KO mice were born in a normal Mendelian ratio and had no gross abnormalities in appearance and behavior. Genomic PCR analysis confirmed deletion of the Nuak1 gene in the heart and skeletal muscles of MNUAK1KO mice. The deletion was not observed in any nonmuscle tissues in the MNUAK1KO mice or in tissues from their Nuak1flox/flox (control) littermates (Fig. 1D). Real time RT-PCR showed that the level of Nuak1 mRNA was substantially reduced in heart, soleus, TA, and EDL muscles of MNUAK1KO mice (Fig. 1E). The NUAK1 protein was barely detectable in those tissues of MNUAK1KO mice (Fig. 1F). These observations confirmed that muscle-specific knock-out of NUAK1 was achieved.
MNUAK1KO Mice Exhibit No Skeletal Muscle Morphological Abnormalities
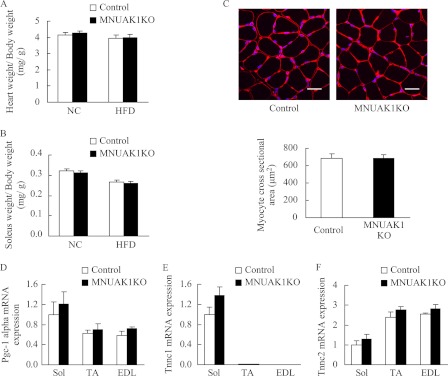
For phenotypic analysis, we first assessed the mass and fiber size of the soleus muscle, as well as the weight of heart and body, because the involvement of NUAK1 in myoblast differentiation has been implied (9). No difference was observed between MNUAK1KO and control mice with respect to body, heart, or soleus weights under both normal chow diet (NC) and HFD conditions at 13–15 weeks of age (Fig. 2, A and B, and supplemental Table S1). The myocyte cross-sectional area of the soleus muscle was almost identical in MNUAK1KO and control mice (Fig. 2C). The expression of NUAK1 in heart or soleus muscle of control mice was not affected by the HFD (data not shown).
FIGURE 2.
MNUAK1KO mice display no morphological abnormalities. A and B, weight of heart (A) and soleus muscle (B) of control and MNUAK1KO mice fed a NC or HFD. The indicated values are normalized to body weight. The data are the means ± S.E. (n = 10). C, cross-section of soleus muscle myocytes stained with wheat germ agglutinin. Scale bars, 20 μm. The graph below shows the average myocyte cross-sectional area for control and MNUAK1KO mice. The data are the means ± S.E. (n = 6). D–F, real time RT-PCR analysis of PGC-1α (D), TNNC1 (E), and TNNC2 (F). The mRNA levels are expressed relative to that of the soleus in control mice. The data are the means ± S.E. (n = 3). Mice at the age of 13–15 weeks were used for all experiments. Sol, soleus.
To determine whether NUAK1 is involved in the formation of type I fibers, we examined the expression level of peroxisome proliferator-activated receptor γ coactivator 1-α (PGC-1α), a transcriptional regulator that drives type I fiber formation (33). We also assessed myofiber composition by examining the expression levels of troponin C1 (TNNC1) and troponin C2 (TNNC2), which are specifically expressed in type I and type II fibers, respectively (32). Real time RT-PCR analysis revealed no significant difference between MNUAK1KO and control mice with respect to PGC-1α, TNNC1, or TNNC2 mRNA expression in the soleus, TA, and EDL muscles (Fig. 2, D–F). These observations suggest that NUAK1 is not involved in the determination of muscle mass, fiber size, or fiber type.
MNUAK1KO Mice Fed High Fat Diet Exhibit Improved Glucose Homeostasis
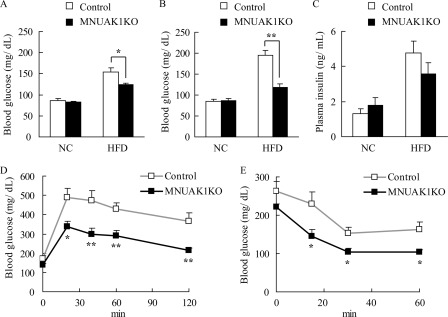
To assess the influence of muscle-specific knock-out of NUAK1 on whole body glucose homeostasis, MNUAK1KO and control mice were fed either a NC or a HFD, and the level of blood glucose was monitored. There was no difference in the fasting blood glucose concentration of MNUAK1KO and control mice fed a NC through age 19 weeks (Fig. 3, A and B). In contrast, on a HFD, the fasting blood glucose concentration at 13–15 weeks of age was significantly lower in MNUAK1KO mice than in control mice (Fig. 3A). This phenotype was even more prominent by age 18–19 weeks (Fig. 3B). At this age, MNUAK1KO mice fed a HFD exhibited slightly lower body weight compared with that of control mice (supplemental Table S1), which may reflect the improved glucose homeostasis in MNUAK1KO mice. In conjunction with these findings, the fasting plasma free fatty acid level was significantly lower in MNUAK1KO mice than in control mice under HFD conditions (supplemental Table S1). The HFD-induced hyperinsulinemia and hypertriglyceridemia also tended to be less pronounced in MNUAK1KO mice (Fig. 3C and supplemental Table S1). The level of food intake was unaltered between MNUAK1KO and control mice (supplemental Table S1).
FIGURE 3.
MNUAK1KO mice show improved glucose homeostasis under HFD conditions. A and B, fasting blood glucose levels of control and MNUAK1KO mice at 13–15 weeks (A) and 18–19 weeks of age (B). The data are the means ± S.E. (n = 8). C, plasma insulin levels of control and MNUAK1KO mice. The data are the means ± S.E. (n = 12). D, oral glucose tolerance of control and MNUAK1KO mice under HFD conditions. The data are the means ± S.E. (n = 12). E, insulin tolerance of control and NUAK1 KO mice under HFD conditions. The data are the means ± S.E. (n = 8). Mice at the age of 13–15 weeks were used for C–E. *, p < 0.05; **, p < 0.01 (Student's t test).
To further investigate the involvement of NUAK1 in glucose homeostasis, an oral glucose tolerance test (OGTT) and an insulin tolerance test (ITT) were performed. No differences in OGTT and ITT results were observed between MNUAK1KO and control mice fed a NC (data not shown). On a HFD, MNUAK1KO mice displayed significantly lower blood glucose levels than control mice as determined using both the OGTT (Fig. 3D) and the ITT (Fig. 3E). These results indicate that muscle-specific knock-out of NUAK1 increases the capacity for glucose disposal, at least in part, in response to insulin.
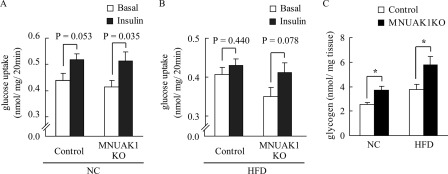
To determine whether the improved whole body glucose metabolism is attributed to NUAK1-deficient skeletal muscle, we measured glucose uptake into isolated soleus muscle without or with insulin. Insulin-stimulated glucose uptake was observed in the soleus muscle from both MNUAK1 and control mice fed a NC (Fig. 4A). Under HFD conditions, the effects of insulin were less pronounced in the soleus muscle from control mice, whereas NUAK1-deficient soleus muscle displayed insulin-stimulated glucose uptake comparable with that observed under NC conditions (Fig. 4B). We also measured the soleus muscle glycogen concentration in MNUAK1KO and control mice. The glycogen concentration was significantly higher in the soleus of MNUAK1KO mice under both NC and HFD conditions, indicating that NUAK1 plays a critical role in glucose storage in the soleus muscle (Fig. 4C). Taken together, our findings suggest that a muscle-specific knock-out of NUAK1 preserved insulin sensitivity under HFD conditions.
FIGURE 4.
MNUAK1KO mice show improved insulin sensitivity and increased glycogen storage in skeletal muscle. A and B, rate of glucose uptake in soleus muscles isolated from 18–20-week-old control and MNUAK1KO mice fed a NC (A) or a HFD (B) without or with insulin stimulation. The data are the means ± S.E. (n = 7 for NC and 8 for HFD). C, glycogen concentration in the soleus muscle of 13–15-week-old control and MNUAK1KO mice fed a NC or HFD. The data are the means ± S.E. (n = 8). *, p < 0.05 (Student's t test).
Phosphorylation of TBC1D4 Is Increased by Deletion of Nuak1 in Skeletal Muscle
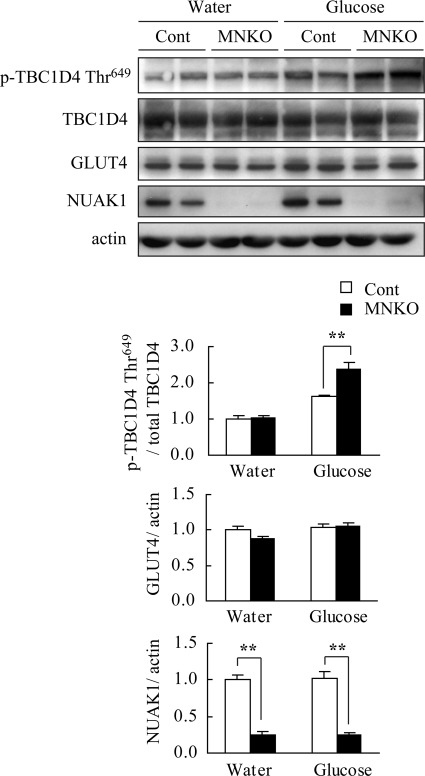
Glucose metabolism in skeletal muscle is highly sensitive to both the expression of glucose transporter type 4 (GLUT4) which is increased by exercise and decreased by a HFD (34, 35), and its translocation to the plasma membrane (36–38), which is facilitated by phosphorylated TBC1 domain family member 4 (TBC1D4) (39, 40). To investigate the molecular mechanism underlying improved glucose metabolism, the TBC1D4 phosphorylation in acute response to glucose in the soleus of HFD-fed MNUAK1KO and control mice was examined. The level of GLUT4 protein was also compared between MNUAK1KO and control mice. The phosphorylation level of TBC1D4 Thr-649 (corresponding to Thr-642 in the human isoform) was significantly higher in the soleus muscle from MNUAK1KO mice in the glucose-administered group (Fig. 5). This difference was not observed in the water-administered group, suggesting that postprandial glucose uptake is enhanced in the soleus of MNUAK1KO mice. No differences were observed in the GLUT4 protein level (Fig. 5). Our data indicate that the observed increase in glucose disposal in MNUAK1KO mice fed a HFD is associated with increased GLUT4 translocation rather than changes in GLUT4 expression in the soleus muscle.
FIGURE 5.
Phosphorylation of TBC1D4 is up-regulated in NUAK1-deficient muscle after glucose administration. Immunoblot analysis of TBC1D4, GLUT4, and NUAK1 in soleus muscle of HFD-fed control and MNUAK1KO mice. Mice at the age of 13–15 weeks were fasted overnight and sacrificed 40 min after oral administration of water (basal control) or glucose. The graphs show the intensities of phospho-TBC1D4, GLUT4, and NUAK1 bands. Phospho-TBC1D4 protein levels were normalized to total TBC1D4. GLUT4 and NUAK1 protein levels were normalized to actin. The protein levels are expressed relative to those in the water-administered control mice. The data are the means ± S.E. (n = 4 for NUAK1 and 5 for TBC1D4 and GLUT4). **, p < 0.01 (Student's t test). Cont, control; MNKO, MNUAK1KO.
Insulin Signal Transduction Is Enhanced in Soleus Muscle of MNUAK1KO Mice
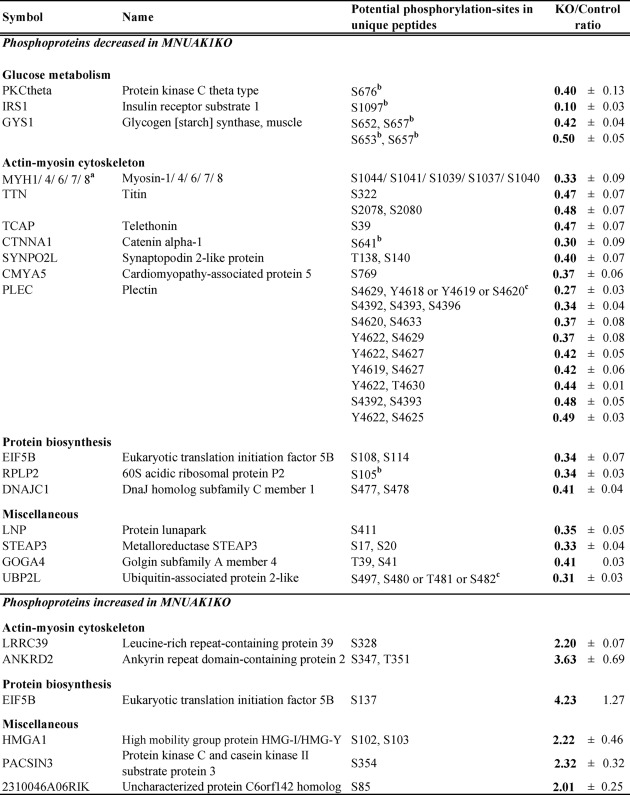
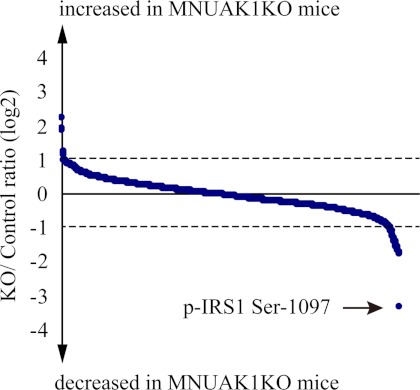
To obtain a comprehensive understanding of the molecular mechanism underlying the observed MNUAK1KO phenotype, a quantitative phosphoproteome analysis was performed on soleus muscle isolated from HFD-fed MNUAK1KO and control mice in random fed state. Total protein was enzymatically digested, differentially labeled with stable isotopes, and then subjected to simultaneous LC-MS/MS analysis, which allowed for precise comparison between the two samples. To ensure reproducibility, four independent experiments were performed. All of the mass spectra and a detailed list of all proteins identified in this study are provided in the supplemental mass spectra and supplemental Table 3, respectively. Table 1 shows proteins that were found to be differentially phosphorylated more than 1.5-fold in all four experiments and more than 2-fold in average between MNUAK1KO and control mice. Among the 1,229 phosphopeptides quantitatively detected, the abundance of 27 phosphopeptides, corresponding to 21 proteins, decreased as a result of the Nuak1 deletion, whereas the abundance of six phosphopeptides, corresponding to six proteins, increased. Most of the differences in phosphorylation status were also observed under NC conditions (supplemental Table S2). It should be noted that differential regulation of phosphoproteins involved in glucose metabolism was clearly shown. In particular, the phosphorylation of IRS1 at Ser-1097 was changed most drastically (KO/control ratio = 0.10 ± 0.03) (Fig. 6). This serine phosphorylation (Ser-1101 in the human isoform), together with the phosphorylation of its upstream regulator PKCθ at Ser-676 (the same residue in the human isoform), is known to mediate negative feedback regulation of insulin signaling through blocking of IRS1 tyrosine phosphorylation (41). Phosphorylation of glycogen synthase 1 (GYS1) at the C-terminal serine residues including Ser-653 and Ser-657 (both same in the human isoform) leads to its inactivation and decreases glycogen synthesis (42, 43). Therefore, the hypophosphorylation of these sites in NUAK1-deficient soleus muscle can enhance insulin sensitivity and glycogen synthesis, which is consistent with the observed decreased blood glucose, improved glucose tolerance and insulin sensitivity, and increased muscle glycogen content phenotype. Besides glucose metabolism, biological processes shown to be affected include cell motility. Components of the actin-myosin cytoskeleton, such as various myosin isoforms, titin, and plectin, were also hypophosphorylated in NUAK1-deficient soleus muscle. Although it is unclear how changes in the phosphorylation status of these cytoskeletal proteins affect glucose metabolism and muscle contraction, the data suggest that NUAK1 has a role in regulation of muscle cytoskeletal structure.
TABLE 1.
Phosphoproteome analysis in soleus muscles from MNUAK1KO and control mice fed a HFD
The listed phosphorylation sites were differentially regulated more than 2.0-fold in MNUAK1KO mice compared with control mice under HFD conditions. The data are means ± S.E. (n = 4).
a Protein isoforms that could not be distinguished by unique peptides.
b Phosphorylation sites previously identified using site-specific methods in reference to the PhosphoSitePlus database.
c Ambiguous phosphorylation sites (i.e., those that could not be determined from MS-MS spectra).
FIGURE 6.
Phosphoproteome of soleus muscles from MNUAK1KO and control mice. A total of 1,229 phosphopeptides were quantitatively detected in soleus muscle. KO/control ratio of phosphopeptides are shown by blue dots (mean, n = 4). Phosphopeptides with KO/control ratio of more than 2.0 or less than 0.5 are listed in Table 1. An IRS1 phosphopeptide containing Ser-1097 is indicated by an arrow.
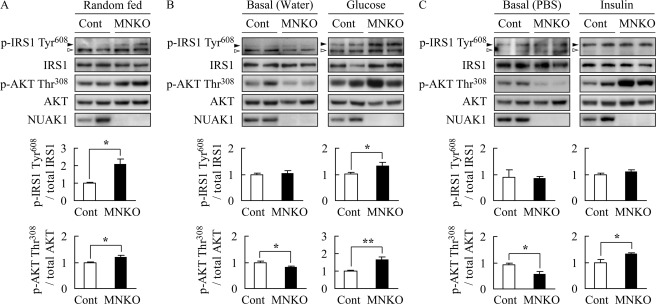
To validate the increased insulin signaling suggested by the phosphoproteome analysis, phosphorylation of IRS1 at Tyr-608 and AKT at Thr-308 in soleus muscle of mice fed a HFD were analyzed by immunoblotting. The phosphorylation levels of IRS1 and AKT were significantly higher in soleus muscle of MNUAK1KO mice than in that of control mice under fed conditions, confirming the enhancement of insulin signaling by a lack of NUAK1 (Fig. 7A). We also examined the phosphorylation of IRS1 and AKT in acute response to glucose and insulin in soleus muscle of mice fed a HFD. As shown in Fig. 7B, phosphorylation levels of IRS1 Tyr-608 and AKT Thr-308 in response to glucose administration were significantly higher in soleus muscle of MNUAK1KO mice than in that of control mice. The AKT Thr-308 phosphorylation in response to insulin injection was also significantly higher in the soleus muscle of MNUAK1KO mice than in that of control mice (Fig. 7C). No difference was observed in IRS1 phosphorylation in response to insulin injection, probably because of the time point for this analysis. Note that phosphorylation level of AKT under basal (fasted) conditions was significantly lower in soleus muscle of MNUAK1KO mice than in that of control mice (Fig. 7, B and C). An increase in the basal phosphorylation level of AKT is characteristic of insulin resistance induced by a HFD (44, 45). These observations strongly suggest that a HFD-induced insulin resistance was reduced in soleus muscle of MNUAK1KO mice. Therefore, we concluded that NUAK1 is involved in the negative regulation of insulin signal transduction in soleus muscle.
FIGURE 7.
Phosphorylation of insulin signaling proteins are up-regulated in NUAK1-deficient muscle. Immunoblot analysis of phosphorylation of IRS1 at Tyr-608 (black arrowhead) and AKT at Thr-308 in soleus muscle of HFD-fed control and MNUAK1KO mice. A white arrowhead denotes a nonspecific band. Graphs show the intensities of phospho-protein bands normalized to total protein levels in each case. The data are expressed relative to those from control mice. *, p < 0.05; **, p < 0.01 (Student's t test). A, mice under fed conditions. The data are the means ± S.E. (n = 3). B, mice were fasted overnight and administered water or glucose (1 g/kg of body weight). Soleus was excised 40 min after the administration. The data are the means ± S.E. (n = 6). C, mice were fasted overnight and intraperitoneally injected with PBS or insulin (1.5 units/kg of body weight). Soleus was excised 20 min after the injection. The data are the means ± S.E. (n = 3). Cont, control; MNKO, MNUAK1KO.
DISCUSSION
Skeletal muscle is the principle tissue for insulin-mediated glucose uptake. Oxidative muscle has higher insulin-stimulated glucose transport activity than does glycolytic muscle (46). Therefore, our finding that NUAK1 is highly expressed in soleus muscle as well as cerebrum and heart, which are also highly oxidative tissues, suggests the possibility that NUAK1 is involved in energy metabolism in oxidative tissues. In support of this hypothesis, muscle-specific LKB1 knock-out (MLKB1KO) mice show improved whole body glucose homeostasis (21). Thus, we examined whether muscle-specific deletion of Nuak1 affects whole body glucose homeostasis and found that MNUAK1KO mice fed a HFD exhibit decreased fasting blood glucose levels and improved glucose metabolism compared with control mice also fed a HFD. The phenotypic similarity, together with the results of previous in vitro studies (2, 6, 7), strongly suggests that LKB1 is an upstream kinase of NUAK1 in skeletal muscle. Note that the phenotype of MNUAK1KO mice was apparent only under HFD conditions. This can be explained by the contribution of skeletal muscle to whole body glucose disposal. Under euglycemic conditions, the majority of glucose disposal occurs in a non-insulin-mediated manner, and skeletal muscle accounts for only 20% of whole body glucose disposal (47, 48). On the other hand, skeletal muscle is responsible for virtually all glucose disposal increased in hyperglycemic conditions (47). We concluded that a lack of NUAK1 alters muscle glucose metabolism, which contributes significantly to whole body glucose disposal in mice with hyperglycemia.
Our data provide evidence suggesting that NUAK1 controls glucose metabolism through negative regulation of insulin signal transduction in skeletal muscle. Firstly, the lower blood glucose of MNUAK1KO mice in OGTT and ITT indicates increased insulin sensitivity. Secondly, the ex vivo experiment of muscle glucose uptake suggests that NUAK1-deficient muscle is protected from HFD-induced insulin resistance. Thirdly, decreased phosphorylation of PKCθ at Ser-676 and IRS1 at Ser-1097 in the soleus muscle of MNUAK1KO mice indicates that a negative feedback regulation of insulin signal transduction is down-regulated by a lack of NUAK1. Indeed, phosphorylation of IRS1 at Tyr-608, which is caused by insulin and leads to the activation of downstream molecules such as AKT and TBC1D4, was up-regulated in the soleus muscle of MNUAK1KO mice. Our observations in MNUAK1KO mice are consistent with a report that PKCθ knock-out mice are not susceptible to HFD-induced insulin resistance (49). The hypophosphorylation of PKCθ in NUAK1-deficient soleus muscle also explains our observation that basal glucose uptake in muscle tended to be decreased by a lack of NUAK1 (Fig. 4, A and B), because it is regulated by PKC isoforms (50, 51). Lastly, increased muscle glycogen concentration together with decreased phosphorylation of GYS1 at Ser-653 and Ser-657, which is involved in activation of GYS1, can be explained by enhancement of insulin signaling because GYS1 at Ser-653 is regulated by glycogen synthase kinase 3, a downstream effector of IRS1-AKT axis (42).
How does NUAK1 control the negative feedback loop of insulin signaling? Phosphoproteome analysis showed that phosphorylation of IRS1 at Ser-1097 was decreased remarkably among all phosphorylations in soleus muscle. In adipose tissues, IRS1 is regulated by salt-inducible kinase 2, another member of the AMPK-RK family (52), through phosphorylation of Ser-794. Therefore, although the amino acid sequence flanking Ser-1097 is not consistent with the optimal motif for phosphorylation by AMPK (53–55), it is plausible that IRS1 is a target of NUAK1. The hypophosphorylation of PKCθ, an upstream kinase of IRS1 Ser-1097, can be explained by the lower plasma free fatty acid levels in MNUAK1KO mice because the phosphorylation of PKCθ is stimulated by free fatty acid (56–58).
Our phosphoproteome analysis also revealed that deletion of Nuak1 leads to a decrease in phosphorylation of components of contraction apparatus such as myosin isoforms and titin, which is consistent with a recent report suggesting that myosin and paramyosin are potential substrates of UNC-82, a Caenorhabditis elegans ortholog of NUAK1 and NUAK2 (59). Current understanding of how changes in the phosphorylation status of cytoskeletal proteins affect glucose metabolism and muscle contraction is incomplete. Further phenotypic analyses of cytoskeletal organization and muscle contraction are required to fully understand the role of NUAK1 in skeletal muscle.
In summary, we found that NUAK1 is preferentially expressed in highly oxidative tissues such as the cerebrum, heart, and soleus muscle. We generated muscle-specific NUAK1 knock-out mice and found that they have improved glucose homeostasis compared with control mice when fed a hyperglycemia-inducing HFD. This phenotype is similar to that of muscle-specific LKB1 knock-out mice, suggesting that LKB1 is an upstream kinase for NUAK1 in skeletal muscle. A quantitative phosphoproteome analysis revealed that NUAK1 is involved in the negative feedback regulation of insulin signal transduction, possibly through the phosphorylation of IRS1. Our results strongly suggest that a physiological role of NUAK1 is to suppress insulin-mediated glucose uptake in skeletal muscle.
Supplementary Material
Acknowledgments
We thank Dr. Izumi O. Umeda and Dr. Hirofumi Fujii (Functional Imaging Division, Research Center for Innovative Oncology, National Cancer Center Hospital East, Kashiwa, Japan) for invaluable advice for the ex vivo glucose uptake experiment.
This work was supported in part by a Grant for the 3rd Term Comprehensive 10-Year Strategy for Cancer Control from the Ministry of Health, Labour and Welfare Japan.

This article contains supplemental text, references, mass spectra, and Tables S1–S3.
- AMPK
- AMP-activated protein kinase
- AMPK-RK
- AMPK-related kinase
- IRS1
- insulin receptor substrate 1
- NUAK1
- NUAK family SNF1-like kinase 1
- TBC1D4
- TBC1 domain family member 4
- PGC-1α
- peroxisome proliferator-activated receptor γ coactivator 1-α
- TNNC
- troponin C
- GLUT4
- glucose transporter type 4
- GYS1
- glycogen synthase 1
- TA
- tibialis anterior
- EDL
- extensor digitorum longus
- OGTT
- oral glucose tolerance test
- ITT
- insulin tolerance test
- NC
- normal chow diet
- HFD
- high fat diet
- LKB
- liver kinase B1
- KHB
- Krebs-Henseleit buffer
- FDG
- fluoro-d-glucose.
REFERENCES
- 1. Manning G., Whyte D. B., Martinez R., Hunter T., Sudarsanam S. (2002) The protein kinase complement of the human genome. Science 298, 1912–1934 [DOI] [PubMed] [Google Scholar]
- 2. Lizcano J. M., Göransson O., Toth R., Deak M., Morrice N. A., Boudeau J., Hawley S. A., Udd L., Mäkelä T. P., Hardie D. G., Alessi D. R. (2004) LKB1 is a master kinase that activates 13 kinases of the AMPK subfamily, including MARK/PAR-1. EMBO J. 23, 833–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Scott J. W., Ross F. A., Liu J. K., Hardie D. G. (2007) Regulation of AMP-activated protein kinase by a pseudosubstrate sequence on the γ subunit. EMBO J. 26, 806–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Al-Hakim A. K., Göransson O., Deak M., Toth R., Campbell D. G., Morrice N. A., Prescott A. R., Alessi D. R. (2005) 14-3-3 cooperates with LKB1 to regulate the activity and localization of QSK and SIK. J. Cell Sci. 118, 5661–5673 [DOI] [PubMed] [Google Scholar]
- 5. Suzuki A., Kusakai G., Kishimoto A., Lu J., Ogura T., Lavin M. F., Esumi H. (2003) Identification of a novel protein kinase mediating Akt survival signaling to the ATM protein. J. Biol. Chem. 278, 48–53 [DOI] [PubMed] [Google Scholar]
- 6. Humbert N., Navaratnam N., Augert A., Da Costa M., Martien S., Wang J., Martinez D., Abbadie C., Carling D., de Launoit Y., Gil J., Bernard D. (2010) Regulation of ploidy and senescence by the AMPK-related kinase NUAK1. EMBO J. 29, 376–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zagórska A., Deak M., Campbell D. G., Banerjee S., Hirano M., Aizawa S., Prescott A. R., Alessi D. R. (2010) New roles for the LKB1-NUAK pathway in controlling myosin phosphatase complexes and cell adhesion. Sci. Signal. 3, ra25. [DOI] [PubMed] [Google Scholar]
- 8. Hou X., Liu J. E., Liu W., Liu C. Y., Liu Z. Y., Sun Z. Y. (2011) A new role of NUAK1. Directly phosphorylating p53 and regulating cell proliferation. Oncogene 30, 2933–2942 [DOI] [PubMed] [Google Scholar]
- 9. Niesler C. U., Myburgh K. H., Moore F. (2007) The changing AMPK expression profile in differentiating mouse skeletal muscle myoblast cells helps confer increasing resistance to apoptosis. Exp. Physiol. 92, 207–217 [DOI] [PubMed] [Google Scholar]
- 10. Hirano M., Kiyonari H., Inoue A., Furushima K., Murata T., Suda Y., Aizawa S. (2006) A new serine/threonine protein kinase, Omphk1, essential to ventral body wall formation. Dev. Dyn. 235, 2229–2237 [DOI] [PubMed] [Google Scholar]
- 11. Kusakai G., Suzuki A., Ogura T., Kaminishi M., Esumi H. (2004) Strong association of ARK5 with tumor invasion and metastasis. J. Exp. Clin. Cancer Res. 23, 263–268 [PubMed] [Google Scholar]
- 12. DeFronzo R. A., Ferrannini E., Sato Y., Felig P., Wahren J. (1981) Synergistic interaction between exercise and insulin on peripheral glucose uptake. J. Clin. Invest. 68, 1468–1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Goodyear L. J., Kahn B. B. (1998) Exercise, glucose transport, and insulin sensitivity. Annu. Rev. Med. 49, 235–261 [DOI] [PubMed] [Google Scholar]
- 14. Mu J., Brozinick J. T., Jr., Valladares O., Bucan M., Birnbaum M. J. (2001) A role for AMP-activated protein kinase in contraction- and hypoxia-regulated glucose transport in skeletal muscle. Mol. Cell 7, 1085–1094 [DOI] [PubMed] [Google Scholar]
- 15. Jørgensen S. B., Viollet B., Andreelli F., Frøsig C., Birk J. B., Schjerling P., Vaulont S., Richter E. A., Wojtaszewski J. F. (2004) Knockout of the α2 but not α1 5′-AMP-activated protein kinase isoform abolishes 5-aminoimidazole-4-carboxamide-1-β-4-ribofuranoside but not contraction-induced glucose uptake in skeletal muscle. J. Biol. Chem. 279, 1070–1079 [DOI] [PubMed] [Google Scholar]
- 16. Sakamoto K., Göransson O., Hardie D. G., Alessi D. R. (2004) Activity of LKB1 and AMPK-related kinases in skeletal muscle. Effects of contraction, phenformin, and AICAR. Am. J. Physiol. Endocrinol. Metab. 287, E310–E317 [DOI] [PubMed] [Google Scholar]
- 17. Hayashi T., Hirshman M. F., Kurth E. J., Winder W. W., Goodyear L. J. (1998) Evidence for 5′ AMP-activated protein kinase mediation of the effect of muscle contraction on glucose transport. Diabetes 47, 1369–1373 [DOI] [PubMed] [Google Scholar]
- 18. Sakamoto K., McCarthy A., Smith D., Green K. A., Grahame Hardie D., Ashworth A., Alessi D. R. (2005) Deficiency of LKB1 in skeletal muscle prevents AMPK activation and glucose uptake during contraction. EMBO J. 24, 1810–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fujii N., Hirshman M. F., Kane E. M., Ho R. C., Peter L. E., Seifert M. M., Goodyear L. J. (2005) AMP-activated protein kinase α2 activity is not essential for contraction- and hyperosmolarity-induced glucose transport in skeletal muscle. J. Biol. Chem. 280, 39033–39041 [DOI] [PubMed] [Google Scholar]
- 20. Koh H. J., Toyoda T., Fujii N., Jung M. M., Rathod A., Middelbeek R. J., Lessard S. J., Treebak J. T., Tsuchihara K., Esumi H., Richter E. A., Wojtaszewski J. F., Hirshman M. F., Goodyear L. J. (2010) Sucrose nonfermenting AMPK-related kinase (SNARK) mediates contraction-stimulated glucose transport in mouse skeletal muscle. Proc. Natl. Acad. Sci. U.S.A. 107, 15541–15546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Koh H. J., Arnolds D. E., Fujii N., Tran T. T., Rogers M. J., Jessen N., Li Y., Liew C. W., Ho R. C., Hirshman M. F., Kulkarni R. N., Kahn C. R., Goodyear L. J. (2006) Skeletal muscle-selective knockout of LKB1 increases insulin sensitivity, improves glucose homeostasis, and decreases TRB3. Mol. Cell Biol. 26, 8217–8227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fujii N., Ho R. C., Manabe Y., Jessen N., Toyoda T., Holland W. L., Summers S. A., Hirshman M. F., Goodyear L. J. (2008) Ablation of AMP-activated protein kinase α2 activity exacerbates insulin resistance induced by high-fat feeding of mice. Diabetes 57, 2958–2966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Alkhateeb H., Chabowski A., Glatz J. F., Luiken J. F., Bonen A. (2007) Two phases of palmitate-induced insulin resistance in skeletal muscle. Impaired GLUT4 translocation is followed by a reduced GLUT4 intrinsic activity. Am. J. Physiol. Endocrinol. Metab. 293, E783–E793 [DOI] [PubMed] [Google Scholar]
- 24. Saito H., Oda Y., Sato T., Kuromitsu J., Ishihama Y. (2006) Multiplexed two-dimensional liquid chromatography for MALDI and nanoelectrospray ionization mass spectrometry in proteomics. J. Proteome Res. 5, 1803–1807 [DOI] [PubMed] [Google Scholar]
- 25. Rappsilber J., Ishihama Y., Mann M. (2003) Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 75, 663–670 [DOI] [PubMed] [Google Scholar]
- 26. Boersema P. J., Raijmakers R., Lemeer S., Mohammed S., Heck A. J. (2009) Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat. Protoc. 4, 484–494 [DOI] [PubMed] [Google Scholar]
- 27. Sugiyama N., Masuda T., Shinoda K., Nakamura A., Tomita M., Ishihama Y. (2007) Phosphopeptide enrichment by aliphatic hydroxy acid-modified metal oxide chromatography for nano-LC-MS/MS in proteomics applications. Mol. Cell. Proteomics 6, 1103–1109 [DOI] [PubMed] [Google Scholar]
- 28. Nakagami H., Sugiyama N., Mochida K., Daudi A., Yoshida Y., Toyoda T., Tomita M., Ishihama Y., Shirasu K. (2010) Large-scale comparative phosphoproteomics identifies conserved phosphorylation sites in plants. Plant Physiol. 153, 1161–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mann M., Wilm M. (1994) Error-tolerant identification of peptides in sequence databases by peptide sequence tags. Anal. Chem. 66, 4390–4399 [DOI] [PubMed] [Google Scholar]
- 30. Yamamoto H., Takashima S., Shintani Y., Yamazaki S., Seguchi O., Nakano A., Higo S., Kato H., Liao Y., Asano Y., Minamino T., Matsumura Y., Takeda H., Kitakaze M. (2008) Identification of a novel substrate for TNFα-induced kinase NUAK2. Biochem. Biophys. Res. Commun. 365, 541–547 [DOI] [PubMed] [Google Scholar]
- 31. Nagase T., Ishikawa K., Miyajima N., Tanaka A., Kotani H., Nomura N., Ohara O. (1998) Prediction of the coding sequences of unidentified human genes. IX. The complete sequences of 100 new cDNA clones from brain which can code for large proteins in vitro. DNA Res. 5, 31–39 [DOI] [PubMed] [Google Scholar]
- 32. Bassel-Duby R., Olson E. N. (2006) Signaling pathways in skeletal muscle remodeling. Annu. Rev. Biochem. 75, 19–37 [DOI] [PubMed] [Google Scholar]
- 33. Lin J., Wu H., Tarr P. T., Zhang C. Y., Wu Z., Boss O., Michael L. F., Puigserver P., Isotani E., Olson E. N., Lowell B. B., Bassel-Duby R., Spiegelman B. M. (2002) Transcriptional co-activator PGC-1α drives the formation of slow-twitch muscle fibres. Nature 418, 797–801 [DOI] [PubMed] [Google Scholar]
- 34. McGee S. L., Sparling D., Olson A. L., Hargreaves M. (2006) Exercise increases MEF2- and GEF DNA-binding activity in human skeletal muscle. FASEB J. 20, 348–349 [DOI] [PubMed] [Google Scholar]
- 35. Kim Y., Tamura T., Iwashita S., Tokuyama K., Suzuki M. (1994) Effect of high-fat diet on gene expression of GLUT4 and insulin receptor in soleus muscle. Biochem. Biophys. Res. Commun. 202, 519–526 [DOI] [PubMed] [Google Scholar]
- 36. Tsao T. S., Stenbit A. E., Factor S. M., Chen W., Rossetti L., Charron M. J. (1999) Prevention of insulin resistance and diabetes in mice heterozygous for GLUT4 ablation by transgenic complementation of GLUT4 in skeletal muscle. Diabetes 48, 775–782 [DOI] [PubMed] [Google Scholar]
- 37. Treadway J. L., Hargrove D. M., Nardone N. A., McPherson R. K., Russo J. F., Milici A. J., Stukenbrok H. A., Gibbs E. M., Stevenson R. W., Pessin J. E. (1994) Enhanced peripheral glucose utilization in transgenic mice expressing the human GLUT4 gene. J. Biol. Chem. 269, 29956–29961 [PubMed] [Google Scholar]
- 38. Garvey W. T., Maianu L., Zhu J. H., Brechtel-Hook G., Wallace P., Baron A. D. (1998) Evidence for defects in the trafficking and translocation of GLUT4 glucose transporters in skeletal muscle as a cause of human insulin resistance. J. Clin. Invest. 101, 2377–2386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kramer H. F., Witczak C. A., Taylor E. B., Fujii N., Hirshman M. F., Goodyear L. J. (2006) AS160 regulates insulin- and contraction-stimulated glucose uptake in mouse skeletal muscle. J. Biol. Chem. 281, 31478–31485 [DOI] [PubMed] [Google Scholar]
- 40. Thong F. S., Bilan P. J., Klip A. (2007) The Rab GTPase-activating protein AS160 integrates Akt, protein kinase C, and AMP-activated protein kinase signals regulating GLUT4 traffic. Diabetes 56, 414–423 [DOI] [PubMed] [Google Scholar]
- 41. Li Y., Soos T. J., Li X., Wu J., Degennaro M., Sun X., Littman D. R., Birnbaum M. J., Polakiewicz R. D. (2004) Protein kinase Cθ inhibits insulin signaling by phosphorylating IRS1 at Ser1101. J. Biol. Chem. 279, 45304–45307 [DOI] [PubMed] [Google Scholar]
- 42. Roach P. J. (1990) Control of glycogen synthase by hierarchal protein phosphorylation. FASEB J. 4, 2961–2968 [PubMed] [Google Scholar]
- 43. Fiol C. J., Mahrenholz A. M., Wang Y., Roeske R. W., Roach P. J. (1987) Formation of protein kinase recognition sites by covalent modification of the substrate. Molecular mechanism for the synergistic action of casein kinase II and glycogen synthase kinase 3. J. Biol. Chem. 262, 14042–14048 [PubMed] [Google Scholar]
- 44. Khamzina L., Veilleux A., Bergeron S., Marette A. (2005) Increased activation of the mammalian target of rapamycin pathway in liver and skeletal muscle of obese rats. Possible involvement in obesity-linked insulin resistance. Endocrinology 146, 1473–1481 [DOI] [PubMed] [Google Scholar]
- 45. Liu H. Y., Hong T., Wen G. B., Han J., Zuo D., Liu Z., Cao W. (2009) Increased basal level of Akt-dependent insulin signaling may be responsible for the development of insulin resistance. Am. J. Physiol. Endocrinol. Metab. 297, E898–E906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Richardson J. M., Balon T. W., Treadway J. L., Pessin J. E. (1991) Differential regulation of glucose transporter activity and expression in red and white skeletal muscle. J. Biol. Chem. 266, 12690–12694 [PubMed] [Google Scholar]
- 47. Baron A. D., Brechtel G., Wallace P., Edelman S. V. (1988) Rates and tissue sites of non-insulin- and insulin-mediated glucose uptake in humans. Am. J. Physiol. 255, E769–E774 [DOI] [PubMed] [Google Scholar]
- 48. Lang C. H. (1992) Rates and tissue sites of noninsulin- and insulin-mediated glucose uptake in diabetic rats. Proc. Soc. Exp. Biol. Med. 199, 81–87 [DOI] [PubMed] [Google Scholar]
- 49. Kim J. K., Fillmore J. J., Sunshine M. J., Albrecht B., Higashimori T., Kim D. W., Liu Z. X., Soos T. J., Cline G. W., O'Brien W. R., Littman D. R., Shulman G. I. (2004) PKC-θ knockout mice are protected from fat-induced insulin resistance. J. Clin. Invest. 114, 823–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Shillabeer G., Chamoun C., Hatch G., Lau D. C. (1995) Exogenous triacylglycerol inhibits insulin-stimulated glucose transport in L6 muscle cells in vitro. Biochem. Biophys. Res. Commun. 207, 768–774 [DOI] [PubMed] [Google Scholar]
- 51. Montell E., Turini M., Marotta M., Roberts M., Noé V., Ciudad C. J., Macé K., Gómez-Foix A. M. (2001) DAG accumulation from saturated fatty acids desensitizes insulin stimulation of glucose uptake in muscle cells. Am. J. Physiol. Endocrinol. Metab. 280, E229–E237 [DOI] [PubMed] [Google Scholar]
- 52. Horike N., Takemori H., Katoh Y., Doi J., Min L., Asano T., Sun X. J., Yamamoto H., Kasayama S., Muraoka M., Nonaka Y., Okamoto M. (2003) Adipose-specific expression, phosphorylation of Ser794 in insulin receptor substrate-1, and activation in diabetic animals of salt-inducible kinase-2. J. Biol. Chem. 278, 18440–18447 [DOI] [PubMed] [Google Scholar]
- 53. Dale S., Wilson W. A., Edelman A. M., Hardie D. G. (1995) Similar substrate recognition motifs for mammalian AMP-activated protein kinase, higher plant HMG-CoA reductase kinase-A, yeast SNF1, and mammalian calmodulin-dependent protein kinase I. FEBS Lett. 361, 191–195 [DOI] [PubMed] [Google Scholar]
- 54. Scott J. W., Norman D. G., Hawley S. A., Kontogiannis L., Hardie D. G. (2002) Protein kinase substrate recognition studied using the recombinant catalytic domain of AMP-activated protein kinase and a model substrate. J. Mol. Biol. 317, 309–323 [DOI] [PubMed] [Google Scholar]
- 55. Gwinn D. M., Shackelford D. B., Egan D. F., Mihaylova M. M., Mery A., Vasquez D. S., Turk B. E., Shaw R. J. (2008) AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol. Cell 30, 214–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Qu X., Seale J. P., Donnelly R. (1999) Tissue and isoform-selective activation of protein kinase C in insulin-resistant obese Zucker rats. Effects of feeding. J. Endocrinol. 162, 207–214 [DOI] [PubMed] [Google Scholar]
- 57. Itani S. I., Pories W. J., Macdonald K. G., Dohm G. L. (2001) Increased protein kinase Cθ in skeletal muscle of diabetic patients. Metabolism 50, 553–557 [DOI] [PubMed] [Google Scholar]
- 58. Griffin M. E., Marcucci M. J., Cline G. W., Bell K., Barucci N., Lee D., Goodyear L. J., Kraegen E. W., White M. F., Shulman G. I. (1999) Free fatty acid-induced insulin resistance is associated with activation of protein kinase Cθ and alterations in the insulin signaling cascade. Diabetes 48, 1270–1274 [DOI] [PubMed] [Google Scholar]
- 59. Hoppe P. E., Chau J., Flanagan K. A., Reedy A. R., Schriefer L. A. (2010) Caenorhabditis elegans unc-82 encodes a serine/threonine kinase important for myosin filament organization in muscle during growth. Genetics 184, 79–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.