Background: Stat5 transcription factors regulate genes required for cell growth, survival, and differentiation.
Results: A novel cytokine-induced Stat5b phospho-regulatory site was identified and found to be constitutively activated in hematopoietic cancers.
Conclusion: This newly defined phosphorylation site acts to regulate Stat5b DNA binding and transcriptional activity.
Significance: Therapeutic strategies that disrupt Stat5 serine phosphorylation may be important for controlling hematopoietic diseases such as cancer.
Keywords: Cell Signaling, Protein Kinases, Protein Phosphatase, Protein Phosphorylation, Signal Transduction, STAT Transcription Factor, Hematopoietic Malignancies
Abstract
Signal transducer and activator of transcription 5b (Stat5b) is a critical node in the signaling network downstream of external (cytokines or growth factors) or internal (oncogenic tyrosine kinases) stimuli. Maximum transcriptional activation of Stat5b requires both tyrosine and serine phosphorylation. Although the mechanisms governing tyrosine phosphorylation and activation of Stat5b have been extensively studied, the role of serine phosphorylation remains to be fully elucidated. Using mass spectrometry and phospho-specific antibodies, we identified Ser-193 as a novel site of cytokine-induced phosphorylation within human Stat5b. Stat5b Ser(P)-193 was detected in activated primary human peripheral blood mononuclear cells or lymphoid cell lines in response to several γ common (γc) cytokines, including interleukin (IL)-2, IL-7, IL-9, and IL-15. Kinetic and spatial analysis indicated that Stat5b Ser-193 phosphorylation was rapid and transient and occurred in the cytoplasmic compartment of the cell prior to Stat5b translocation to the nucleus. Moreover, inducible Stat5b Ser-193 phosphorylation was sensitive to inhibitors of mammalian target of rapamycin (mTOR), whereas inhibition of protein phosphatase 2A (PP2A) induced phosphorylation of Ser-193. Reconstitution assays in HEK293 cells in conjunction with site-directed mutagenesis, EMSA, and reporter assays indicated that Ser(P)-193 is required for maximal Stat5b transcriptional activity. Indeed, Stat5b Ser-193 was found constitutively phosphorylated in several lymphoid tumor cell lines as well as primary leukemia and lymphoma patient tumor cells. Taken together, IL-2 family cytokines tightly control Stat5b Ser-193 phosphorylation through a rapamycin-sensitive mechanism. Furthermore, constitutive Ser-193 phosphorylation is associated with Stat5b proto-oncogenic activity and therefore may serve as a novel therapeutic target for treating hematopoietic malignancies.
Introduction
Signal transducers and activators of transcription (Stats) are a family of latent transcription factors that upon activation mediate multiple cellular processes, including proliferation, differentiation, and survival (1). The Stat protein family is composed of seven members (Stat1–6), including the two closely related Stat5a and Stat5b molecules that share 96% amino acid sequence identity (2, 3). Stat5a and Stat5b are critical for lymphoid, myeloid, and erythroid cell development and function (4, 5). Indeed, Stat5 proteins are activated by multiple cytokines, including IL-2, IL-3, IL-5, IL-7, IL-9, IL-15, and erythropoietin (6–8). Following cytokine stimulation, human Stat5a and Stat5b are phosphorylated on the conserved tyrosine residues Tyr-694 and Tyr-699, respectively, which allows for their dissociation from the receptor complex, formation of hetero- or homodimers, and nuclear translocation to bind specific elements in the promoter of target genes and activate transcription (9). In addition to tyrosine phosphorylation, the activity of Stat5 has been shown to be modulated by serine phosphorylation. Previous studies from our group and others identified a novel phospho-serine site located in a conserved Pro-Ser-Pro (PSP)2 motif in the transactivation domain at amino acid positions 726 (Stat5a) and 731 (Stat5b) (10–12). In addition, human Stat5a harbors a unique serine phosphorylation site within a Ser-Pro (SP) motif (Ser-780) (13). These studies indicate that maximum transcriptional activation of Stat5 requires both tyrosine and serine phosphorylation. Most efforts to date have focused on determining the mechanisms that govern tyrosine phosphorylation and activation of Stat5; however, the role of serine phosphorylation remains to be fully elucidated.
In addition to the physiological role of Stat5 in hematopoietic cell development, dysregulation of the Stat5 signaling pathway plays a role in oncogenesis and leukemogenesis (14, 15). Specifically, Stat5 has been shown to be constitutively activated in several forms of lymphoid, myeloid, and erythroid leukemias (16–21). Indeed, the introduction of constitutively active Stat5 mutants into hematopoietic cells is sufficient to induce multilineage leukemia in mice (22, 23). Persistent tyrosine phosphorylation and activation of Stat5 was found to be a result of deregulated cytokine signaling (24) or the presence of oncogenic tyrosine kinases. Stat5 proteins are critical targets for many oncogenic tyrosine kinases, including Bcr-Abl (breakpoint cluster region-abelson), Tel-Jak2, mutated forms of Flt3 (fms-like tyrosine kinase receptor-3) and c-Kit, and the Jak2 V617F mutant (19, 23, 25–28). Although the aberrant regulation of Stat5 tyrosine phosphorylation is well established, the significance of Stat5 serine phosphorylation is less clear. The present study was initiated in an attempt to fully define the Stat5 serine phosphorylation sites and determine their role in normal and proto-oncogenic Stat5 activity. We provide novel evidence that Stat5b undergoes cytokine-induced phosphorylation at Ser-193 through a rapamycin-sensitive mechanism. Furthermore, Ser-193 phosphorylation was found to play an important role in Stat5b DNA binding and transcriptional activity. Indeed, Stat5b Ser-193 was constitutively phosphorylated in several lymphoid tumor cell lines as well as primary leukemia and lymphoma patient tumor cells. These insights provide further evidence regarding the importance of serine phosphorylation in Stat5 function and dysfunction.
EXPERIMENTAL PROCEDURES
Cell Culture and Patient Samples
The human YT, MT-2, HUT-78, HUT-102, and HEK293 cell lines were maintained in RPMI 1640 medium containing 10% fetal bovine serum (Atlanta Biologicals), 2 mm l-glutamine, and penicillin-streptomycin (50 IU/ml and 50 mg/ml, respectively) as reported previously (29). Human peripheral blood mononuclear cells (PBMCs) were obtained from healthy donors, purified by isocentrifugation, and activated with phytohemagglutinin (PHA) (1 μg/ml) for 72 h, as described previously (30). Quiescent PBMCs or YT cells were stimulated in the absence or presence of 10,000 IU human recombinant interleukin (IL)-2 (NCI Preclinical Repository, National Institutes of Health), IL-7, IL-9, or IL-15 (PeproTech) at 37 °C for the times indicated. Primary patient leukemia and lymphoma cells were obtained from de-identified excess diagnostic material (peripheral blood or lymph node biopsies) through a University of Texas at El Paso Institutional Review Board approved study. Additional information on the patient samples is shown in Table 1.
TABLE 1.
Clinical information and Stat5 activation status of primary human samples
Pre-existing, remnant primary human leukemia, and lymphoma cells were obtained from peripheral blood or lymph node biopsies at the time of diagnosis. Control samples were taken from healthy volunteer donors. The diagnoses were made according to French-American-British (FAB) criteria. ALL, acute lymphoblastic leukemia; AML, acute myeloid leukemia; AMML, acute myelomonocytic leukemia; AMoL, acute monocytic leukemia; n.a., not available; WBC, white blood cell count.
| No. | Diagnosis | Subtype | Proportion of malignant cells | Cytogenetics | WBC | Sample source | STAT5 Tyr(P)-694/699 | STAT5 Ser(P)-193 | STAT5 |
|---|---|---|---|---|---|---|---|---|---|
| 1 | ALL | Precursor T-ALL | 80% | 46, XX,t (5;7), inv (14) del (9p21) | 363,000 | Peripheral blood | + | + | + |
| 2 | AML | AML M0/M1 | >90% | 46, XY | 216,000 | Peripheral blood | + | + | + |
| 3 | B-cell lymphoma | Large follicular center B-cell | NA | Tetraploid,t (14;18) | NA | Lymph node biopsy | + | + | + |
| 4 | ALL | Precursor B-ALL | 70% | 54, XY; Gains in X, Y, 6, 10, 14, 18, 21 | 68,000 | Peripheral blood | + | + | + |
| 5 | Hairy cell leukemia | NA | 60% | ND | 6,900 | Peripheral blood | + | + | + |
| 6 | AMML | AML M4 | 50% | Monosomy 7, trisomies 12, 19, 21 | 11,000 | Peripheral blood | + | + | + |
| 7 | Hodgkin Lymphoma | Nodular sclerosing | NA | 46, XY | NA | Lymph node biopsy | − | − | + |
| 8 | Non-Hodgkin Lymphoma | Follicular center B-cell | 60% | 48, XY, Trisomy 2, add (6) (q27), add (9) (p24), dup (12) (q13-q22) | NA | Lymph node biopsy | − | − | + |
| 9 | AMoL | AML FAB M5b | 10% | 46, XX, inv (16) | 36,000 | Peripheral blood | − | − | + |
| 10 | Control Donor | NA | NA | NA | NA | Peripheral blood | − | − | + |
| 11 | Control Donor | NA | NA | NA | NA | Peripheral blood | − | − | + |
Antibodies and Chemical Reagents
The anti-phospho-serine 193 Stat5b (α-Ser(P)-193 Stat5) rabbit polyclonal antibody was custom-generated and affinity-purified by GenScript (Piscataway, NJ) using the immunogen CLAQL(pS)PQERL (where pS indicates phospho-serine) coupled to keyhole limpet hemocyanin. Anti-phospho-serine 726/731 Stat5 (α-Ser(P)-726/731 Stat5) polyclonal antibodies were applied as described previously (12). The anti-phospho-tyrosine 694/699 Stat5 (α-phospho-Tyr Stat5) and anti-phospho-tyrosine (α-phospho-Tyr) monoclonal antibodies were purchased from Millipore. The anti-Stat5 polyclonal antibodies (α-Stat5) were purchased from Santa Cruz Biotechnology. Kinase and phosphatase experiments employing PD98059 (New England Biolabs), wortmannin (Calbiochem), rapamycin (Calbiochem), calyculin A (CA) (Sigma-Aldrich), okadaic acid (OA) (Sigma-Aldrich), fostriecin (FOS) (Sigma-Aldrich), or tautomycin (TAU) (Sigma-Aldrich) were performed at 37 °C using the concentrations and time points indicated.
Cell Lysis, Immunoprecipitation, Western Blot, and Mass Spectrometry Analysis
Cells were pelleted, lysed, and subjected to immunoprecipitation and Western blot analysis as reported previously (32). For all samples, total protein was determined by the bicinchoninic acid method (Pierce). When reblotting, PVDF membranes were incubated with stripping buffer (100 mm β-mercaptoethanol, 2% SDS, 62.5 mm Tris-HCl, pH 6.7) at 55 °C for 30 min, blocked, and then reprobed. Liquid chromatography-tandem mass spectrometry analysis was performed by the Taplin Biological Mass Spectrometry Facility (Harvard University) or the Biomolecule Analysis Core Facility, a component of the Border Biomedical Research Center (University of Texas at El Paso) following their standard procedures.
Plasmids, Site-directed Mutagenesis, and Transfection of HEK293 Cells
The pcDNA3.1/GS human IL-2Rγ and IL-2Rβ expression plasmids were purchased from Invitrogen. The human Jak3 and β-casein-luciferase reporter plasmids were used as described previously (33). The pCMV-β-galactosidase (β-gal) expression plasmid was kindly provided by Dr. Hallgeir Rui and described in Ref. 34. Human Stat5b cDNA (OriGene) was PCR-amplified and subcloned into the pCMV-Tag2 (Stratagene). Mutant forms of Stat5b were prepared using the QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. The primers used for Stat5b mutation were as follows: S193A (5′-GCTGGCCCAGCTGGCCCCCC-3′); S193E (5′-CCGCTGGCCCAGCTGGAAC-3′). All subclones and mutations were verified by DNA sequencing. Transient transfections of HEK293 cells were performed with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions.
Luciferase and β-Galactosidase Assays
Subconfluent HEK293 cells in 10-cm dishes were transfected with the following plasmids: β-casein-luciferase reporter (3 μg), pCMV-β-gal (0.3 μg), Jak3 (0.5 μg), IL-2Rβ (6 μg), IL-2Rγ (6 μg), and wild-type or mutant Stat5b (3 μg). At 48 h after transfection, cells were stimulated with or without 1,000 IU/ml IL-2 for 6 h. The cells were then harvested, and luciferase/β-gal activities were measured with the Dual-Light luciferase assay reporter system (Applied Biosystems) according to the manufacturer's instructions. Statistical significance was determined by analysis of variance at the 0.05 confidence level.
Immunofluorescent Confocal Microscopy
Cells were cytocentrifuged onto glass slides, fixed, permeabilized, and immunostained using the indicated antibodies as described previously (30). Immunofluorescent confocal microscopy was performed at the Analytical Cytology Core Facility, a component of the Border Biomedical Research Center (University of Texas at El Paso). The cells were visualized using a Zeiss LSM 700 confocal microscope using a 40× oil immersion objective (Carl Zeiss) in the multitrack scanning mode with excitation wavelengths set at 488 (argon laser), 543, and 633 nm (He-Ne lasers); emission wavelengths were 505–530 nm and >560 nm for detection of the Cy2 and Cy3 coupled secondary antibodies, respectively. Images were captured using the ZEN 2009 (version 5.5) software and exported in a 12-bit TIFF RGB format to Adobe Photoshop and Illustrator CS4 for processing.
Electrophoretic Mobility Shift Assay (EMSA)
Nuclear extracts were prepared and assays were conducted as published previously (35). Oligonucleotides corresponding to the β-casein gene promoter element for Stat5 (5′-AGATTTCTAGGAATTCAATCC-3′) were obtained from Santa Cruz Biotechnology.
RESULTS
Identification of a Novel IL-2-induced Serine Phosphorylation Site in Human Stat5b
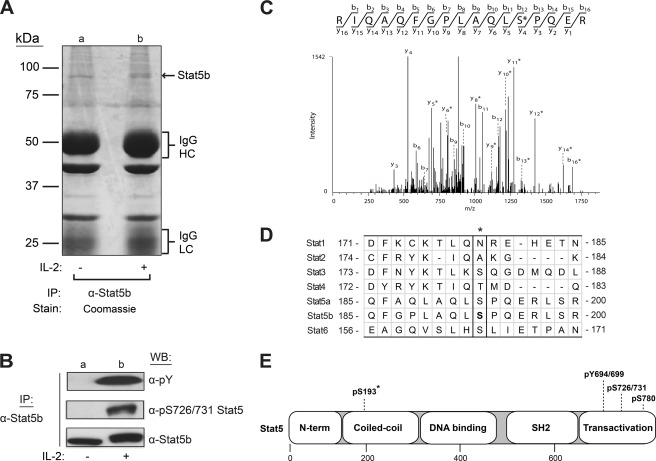
In an attempt to fully define the Stat5 phospho-regulatory sites, mass spectrometry analysis of activated Stat5 was performed. Stat5b was immunoprecipitated from YT cells stimulated without or with IL-2 for 10 min. The samples were separated by 7.5% SDS-PAGE and visualized by Coomassie Blue staining (Fig. 1A). Duplicate samples were prepared and analyzed by Western blot for α-phospho-Tyr, α-Ser(P)-726/731 Stat5, or α-Stat5b (Fig. 1B) to ensure IL-2-induced activation of Stat5b. Indeed, IL-2 stimulation induced both tyrosine and serine 731 phosphorylation of Stat5b (Fig. 1B, lane b). Consequently, the Stat5b-corresponding 89-kDa bands (Fig. 1A, lanes a and b) were excised and subjected to trypsin digestion, and peptides were analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS). Using the Sequest search algorithm, the resulting spectra covered 85% of the Stat5b protein and identified one novel IL-2-induced serine phosphorylation site. The tandem mass spectrum for the identified peptide R.IQAQFGPLAQLSPQE.R, which contains the phosphorylated Stat5b Ser-193 residue (underlined), is shown in Fig. 1C. To determine the extent of Ser-193 sequence conservation among other Stats, human Stat5a and Stat5b protein sequences were aligned with other human Stats (Stat1, Stat2, Stat3, Stat4, and Stat6) (Fig. 1D). Human Stat3 and Stat6 do possess a serine residue at the aligned Stat5 Ser-193 position; however, the surrounding amino acid sequence is not well conserved. Specifically, Stat5 Ser-193 is C-terminally flanked by a proline residue, which is not conserved in the other Stats. It is important to note that the Stat5a Ser-193 phospho-peptide was not detected by LC-MS/MS analysis; thus, the functional studies described herein focused on Stat5b. The location of Ser-193 in the coiled-coil domain of Stat5 is shown along with previously identified sites in the transactivation domain (Fig. 1E). Previously, large-scale phospho-proteomic analyses using HeLa (36–38) or Jurkat (39) cell extracts identified Ser-193 as a Stat5b phosphorylation site using discovery-mode mass spectrometry. Accordingly, the present work sought to functionally define Stat5b Ser-193 phosphorylation using site-specific methodologies.
FIGURE 1.
Identification of Ser-193 as a novel proline-flanked phosphorylation site in human Stat5b. YT cells were stimulated without (−) or with (+) IL-2 for 15 min, and Stat5b proteins were immunoprecipitated (IP) from soluble cell lysates with α-Stat5b antibodies. Two sets of immunoprecipitations were separated by SDS-PAGE. One set was Coomassie Blue-stained (A), and the other was Western blotted (WB) (B) with α-phospho-Tyr (α-pY), α-phospho-Ser (pS726/731), or α-Stat5b antibodies. HC, heavy chain; LC, light chain. C, tandem mass spectra of a monophosphorylated peptide showing site localization of Ser-193, as indicated by asterisks. D, amino acid sequence alignment of the region surrounding Ser-193 (asterisk) from each human Stat protein using the ClustalW program (progressive alignment) (31). E, domain architecture of human Stat5 with known and newly identified (asterisk) serine and tyrosine phosphorylation sites. Numbers indicate amino acid residues of human Stat5 (a/b).
Generation of α-Ser(P)-193 Stat5 Phospho-specific Antibody
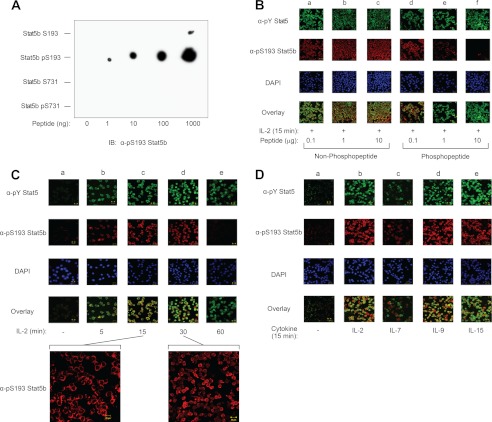
To verify that Stat5b is phosphorylated at serine 193 and to investigate the regulatory roles of this phosphorylation site, a phospho-specific polyclonal antibody was generated. Dot blot analysis was performed with the immunizing phospho-peptide and the corresponding nonphosphorylated peptide (see “Experimental Procedures” for sequences) to determine whether the Stat5 phospho-specific antibody cross-reacts with regions distal to the phosphorylated serine. Additionally, a Stat5b Ser-731-containing phospho-peptide and corresponding nonphosphorylated peptide (see “Experimental Procedures” for sequences) were used to determine whether the Stat5 Ser-193 phospho-specific antibody cross-reacts with the other known phosphorylated serines in Stat5b. Increasing amounts of Stat5b Ser-193, Ser(P)-193, Ser-731, or Ser(P)-731 peptides (Fig. 2A) were spotted onto PVDF membranes and immunoblotted with α-Ser(P)-193 Stat5b. The Ser(P)-193 Stat5b antibody primarily recognized the corresponding phosphorylated peptide but not its nonphosphorylated counterpart, nor the Stat5b Ser-731 peptides, indicating that the phospho-specific antibody does not cross-react significantly with the nonphosphorylated peptide nor other proline-flanked serine phosphorylation sites within Stat5b. Multiple studies to determine whether the Ser(P)-193 Stat5b antibody was amenable to SDS-PAGE-separated wild-type protein by Western blot analysis were unsuccessful (data not shown). Therefore, the Ser(P)-193 Stat5b antibody was tested for utilization in immunofluorescent confocal microscopy. For this analysis, YT cells were stimulated with IL-2 for 15 min and stained using α-phospho-Tyr Stat5 and α-Ser(P)-193 Stat5b in the presence of increasing amounts of Ser-193 non-phospho-peptide (lanes a–c) or phospho-peptide (lanes d–f) (Fig. 2B). The Ser(P)-193 Stat5b phospho-antibody was specifically blocked by the phospho-, but not the non-phospho-peptide, thus confirming antibody specificity, as well as its utility in immunofluorescent microscopy (Fig. 2B). Additionally, phospho-peptide inhibition of α-Ser(P)-193 Stat5b was dose-dependent relative to the non-phospho-peptide (Fig. 2B). Detection of phospho-Tyr-Stat5 was not affected by either peptide. Taken together, a novel Ser(P)-193 Stat5b antibody was generated that specifically recognizes Stat5b Ser-193 phosphorylation in vivo using immunofluorescent microscopy.
FIGURE 2.
Phosphorylation of Stat5b Ser-193 displays rapid kinetics and is inducible by multiple cytokines. A, a phospho-specific polyclonal Stat5b Ser(P)-193 antibody was generated and tested by dot blot analysis using increasing amounts of Stat5b Ser-193 (CLAQLSPQERL), Ser(P)-193 (CLAQL(pS)PQERL), Ser-731 (KDQAPSPAVCP), and Ser(P)-731 (KDQAP(pS)PAVCP) peptides spotted onto PVDF membrane (where pS indicates phospho-serine). IB, immunoblotting. B, YT cells stimulated with IL-2 for 15 min were fluorescently labeled using α-phospho-Tyr (α-pY) Stat5 (Cy2, green), α-Ser(P)-193 (α-pS193) Stat5b (Cy3, red), and DAPI (blue) and visualized by confocal microscopy. The overlay (bottom panel) shows co-localization of phospho-Tyr Stat5 and Ser(P)-193 Stat5b. For peptide competition analysis, the polyclonal Ser(P)-193 Stat5b antibody was preblocked with increasing amounts of non-phospho-peptide (lanes a–c) or phospho-peptide (lanes d–f) for 1 h at 4 °C followed by staining of YT cells treated with IL-2 for 15 min. C, YT cells were stimulated without (−) (lane a) or with IL-2 (+) from 0 to 60 min (lanes b–e) and fixed with cold methanol. Cells were then stained with α-phospho-Tyr Stat5 (Cy2, green), α-Ser(P)-193 Stat5b (Cy3, red), and DAPI (blue) and visualized by confocal microscopy. The overlay (bottom panel) shows co-localization of phospho-Tyr Stat5 and Ser(P)-193 Stat5b. Insets show a higher magnification view of α-Ser(P)-193 Stat5b (Cy3, red)-stained YT cells treated with IL-2 for the indicated time points. D, quiescent PHA-activated human PBMCs were stimulated with medium (−) (lane a) or IL-2 (lane b), IL-7 (lane c), IL-9 (lane d), or IL-15 (lane e) for 15 min and fixed with cold methanol. Confocal images using α-phospho-Tyr Stat5 (Cy2, green), α-Ser(P)-193 Stat5b (Cy3, red), and DAPI (blue) and overlay images are shown. Immunofluorescent images were captured using PASCAL software on a Zeiss LSM 510 Meta confocal microscope at 63× magnification. Representative data from three independent experiments are shown.
Phosphorylation of Stat5b Ser-193 Is Induced by γc Cytokines in Vivo
Cytokines belonging to the γc family, such as IL-2, IL-4, IL-7, and IL-9, are critical for the development and function of the immune system (40, 41). Binding of these cytokines to their receptors results in the rapid activation of Jaks, which in turn phosphorylate and activate Stats to control cellular function (41). To determine the kinetic and spatial distribution of Ser-193-phosphorylated Stat5b in vivo, YT cells were subjected to an IL-2 stimulation time course (0–60 min) and immunostained using α-phospho-Tyr Stat5 and α-Ser(P)-193 Stat5b. Stat5b Ser-193 was rapidly and transiently phosphorylated following IL-2 stimulation. Phosphorylation of Stat5 Ser-193 was detected at 5 min (lane b), reached maximal levels after 15–30 min (lanes c and d), and returned to the baseline by 60 min (lane e) (Fig. 2C). This was similar to the kinetics shown for IL-2-induced phospho-Tyr Stat5. Additionally, Ser-193-phosphorylated Stat5b was predominantly cytoplasmic localized after 15 min of IL-2 stimulation (Fig. 2C, inset). However, Ser(P)-193 Stat5b was distributed in both the cytoplasmic and the nuclear fractions after 30 min of IL-2 stimulation (Fig. 2C, inset). These data suggest that Stat5b Ser-193 phosphorylation occurs in the cytoplasm before translocation to the nucleus. To determine whether Stat5b is phosphorylated at Ser-193 in response to other γc-containing cytokines, PHA-activated primary human PBMCs were made quiescent and then stimulated with media (lane a), IL-2 (lane b), IL-7 (lane c), IL-9 (lane d), and IL-15 (lane e) for 15 min and analyzed by immunofluorescent microscopy (Fig. 2D). When compared with the unstimulated control cells (lane a), each cytokine was able to induce Stat5b Ser-193 phosphorylation (Fig. 2D). These data suggest that Stat5b Ser-193 phosphorylation may be important for diverse biological functions mediated by γc cytokines and Stat5. It is also important to note that IL-7 (lane c) consistently appeared as the weakest Ser(P)-193 activation signal in these cells but was similarly a poor phospho-Tyr Stat5 activator (Fig. 2D). Thus, the phosphorylation of Stat5b Ser-193 occurred in multiple cell types, including primary human PBMCs, with activation profiles indicating a common mechanism of Stat5b activation.
Phosphorylation of Stat5b Ser-193 Is Sensitive to Rapamycin Treatment
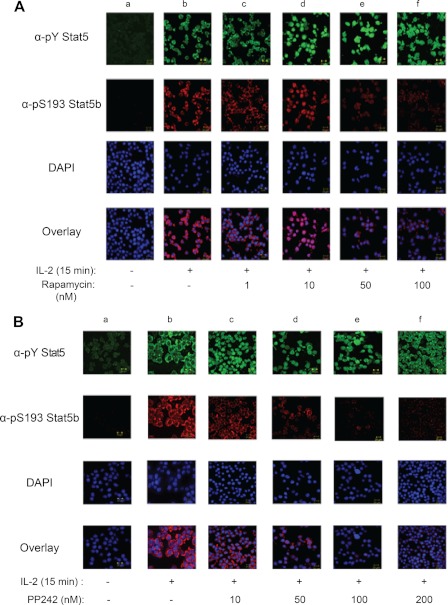
To identify the putative kinase(s) responsible for phosphorylation of Stat5b Ser-193, YT cells were incubated with inhibitors of candidate serine/threonine kinases prior to IL-2 stimulation. The candidate kinases were chosen based on previous studies that identified the RAS-RAF-MEK and PI3K-mTOR pathways as regulators of Stat serine phosphorylation (42–44). Inhibition of MEK (PD98059) had no visible effect on IL-2-induced phosphorylation of Stat5b Ser-193 (data not shown). However, when compared with the untreated controls (lane a), preincubation with increasing amounts of rapamycin (1–100 nm) (lanes c–f), a mammalian target of rapamycin (mTOR) inhibitor, significantly decreased IL-2-induced phosphorylation of Stat5b Ser-193 (Fig. 3A). Treatment of YT cells with rapamycin did not have an effect on IL-2-induced tyrosine phosphorylation of Stat5b. To corroborate these results, YT cells were treated with another mTOR inhibitor, PP242 hydrate, which resulted in a similar response at comparative concentrations (Fig. 3B). Taken together, these results suggest that Stat5b Ser-193 is dependent upon IL-2-induced activation of an mTOR-dependent pathway.
FIGURE 3.
Stat5b Ser-193 undergoes phosphorylation in an mTOR-dependent manner. A, YT cells were left untreated (lanes a and b) or pretreated with increasing concentrations of rapamycin (1–100 nm) (lanes c–f) for 1 h followed by stimulation with IL-2 (lanes b–f) for 15 min. α-pY, α-phospho-Tyr; α-pS193, α-Ser(P)-193. B, YT cells were left untreated (lanes a and b) or pretreated with 10–200 nm PP242 hydrate for 1 h followed by stimulation with IL-2 for 15 min (lanes b–f) as indicated. The cells were fixed and stained with α-phospho-Tyr Stat5 (Cy2, green), α-Ser(P)-193 Stat5b (Cy3, red), and DAPI (blue) and visualized by confocal microscopy. The overlay (bottom panel) shows co-localization of phospho-Tyr Stat5 and Ser(P)-193 Stat5b. Immunofluorescent images were captured using PASCAL software on a Zeiss LSM 510 Meta confocal microscope at 63× magnification. Representative data from three independent experiments are shown.
PP2A, but Not PP1, Negatively Regulates Phosphorylation of Stat5b Ser(P)-193
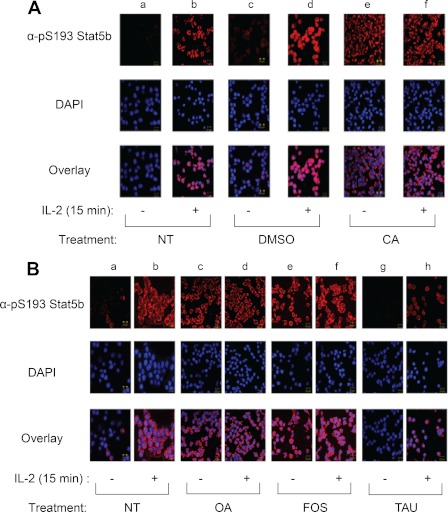
Our group and others have shown that inhibition of protein phosphatase types 1 (PP1) or 2A (PP2A) attenuates Stat3 (45), Stat5 (32), and Stat6 (45, 46) activity. To determine whether Stat5b Ser-193 phosphorylation is regulated by PP1 or PP2A activity, we tested the ability of the PP1 and PP2A inhibitor CA (47) to induce Stat5b Ser-193 phosphorylation (Fig. 4A). YT cells were left untreated (lanes a and b) or pretreated with either DMSO (lanes c and d) or 50 nm CA (lanes e and f) for 1 h before being stimulated without (−) or with (+) IL-2 for 15 min. Cells stained with α-Ser(P)-193 Stat5 displayed low basal levels of Ser-193 phosphorylation in the absence of IL-2 when left untreated (lane a) or pretreated with DMSO (lane c); however, treatment of YT cells with IL-2 (lanes b, d, and f) or calyculin A (lane e) resulted in a significant induction of Stat5b Ser-193 phosphorylation (Fig. 4A). To further define the phosphatase involved, PP1- or PP2A-specific inhibitors were administered, including OA and FOS, which selectively block PP2A, and TAU, which preferentially inhibits PP1 (32). For this analysis, YT cells were treated with inhibitory concentrations of 150 nm OA (lanes c and d), 25 nm FOS (lanes e and f), or 1 μm TAU (lanes g and h) for 60 min prior to stimulation without or with IL-2 for 15 min (Fig. 4B). When compared with untreated controls (lanes a and b), blockade of PP2A by OA and FOS showed constitutively phosphorylated Stat5b Ser-193 at levels comparable with that of IL-2-induced phosphorylation (lane b), whereas no change was observed following treatment with the PP1 inhibitor TAU (lanes g and h) (Fig. 4B). Collectively, these data suggest that PP2A negatively regulates Stat5b Ser-193 phosphorylation.
FIGURE 4.
PP2A, but not PP1, negatively regulates Stat5b Ser-193 phosphorylation in YT cells. A, cells were pretreated without (lanes a and b) or with DMSO (lanes c and d) or 50 nm CA (lanes e and f) for 1 h before stimulation without (lanes a, c, and e) or with IL-2 (lanes b, d, and f) for 15 min at 37 °C. The cells were fixed and stained with α-phospho-Tyr Stat5 (Cy2, green), α-Ser(P)-193 (α-pS193) Stat5b (Cy3, red), and DAPI (blue) and visualized by confocal microscopy. The overlay (bottom panel) shows co-localization of phospho-Tyr Stat5 and Ser(P)-193 Stat5b. NT, not treated. B, YT cells were left untreated (lanes a and b) or pretreated with 150 nm OA (lanes c and d), 25 nm FOS (lanes e and f), or 1 μm TAU (lanes g and h) for 1 h before stimulation without (−) or with (+) IL-2 for 15 min at 37 °C. The cells were fixed and analyzed using α-Ser(P)-193 Stat5b and DAPI as described above. Immunofluorescent images were captured using PASCAL software on a Zeiss LSM 510 Meta confocal microscope at 63× magnification. Representative data from three independent experiments are shown.
Phosphorylation of Ser-193 Is Required for Maximum Stat5b Transcriptional Activity
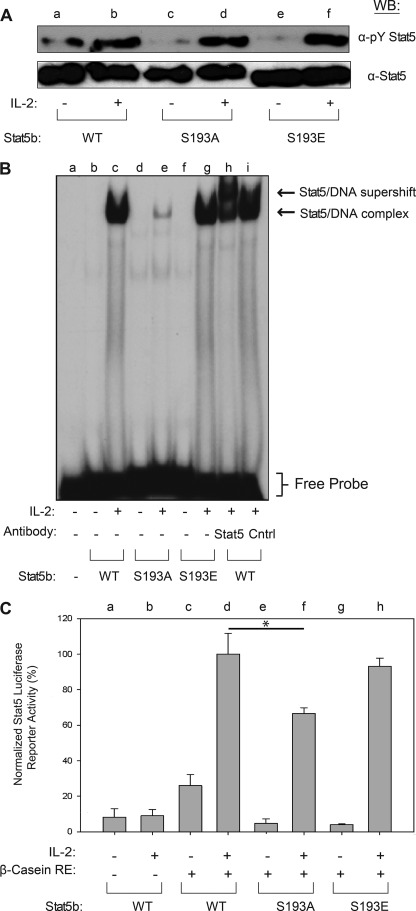
To determine the functional role of Stat5b Ser-193 phosphorylation, a HEK293 reconstitution system was employed using phospho-deletion (S193A) and phospho-mimetic (S193E) mutations of Stat5b. Plasmids encoding Jak3, IL-2Rβ, and IL-2Rγ were co-transfected into HEK293 cells with wild-type (WT) (lanes a and b), S193A (lanes c and d), or S193E (lanes e and f) forms of Stat5b. At 48 h after transfection, cells were stimulated in the absence (−) or presence (+) of IL-2 for 15 min. Western blot analysis of Stat5b immunoprecipitates using α-phospho-Tyr Stat5 indicated that when compared with WT, S193A and S193E mutations did not affect IL-2-induced tyrosine phosphorylation (Fig. 5A). Reprobing this blot with α-Stat5b confirmed that similar amounts of Stat5b protein were immunoprecipitated and measured (Fig. 5A). Upon tyrosine phosphorylation, Stat5 proteins dimerize, translocate to the nucleus, and bind specific promoter elements that stimulate transcription of target genes that control cell growth and differentiation (48, 49). Therefore, EMSA were utilized to determine whether Ser-193 phosphorylation was important for Stat5 DNA binding activity. For these assays, nuclear extracts were isolated from reconstituted HEK293 cells treated without (−) or with (+) IL-2 for 30 min and incubated with radiolabeled probe corresponding to the Stat5 binding element in the β-casein gene promoter. This complex was separated within a nondenaturing gel to determine its DNA binding activity. When compared with WT (lanes b and c), the S193A mutation (lanes d and e) displayed significantly decreased IL-2-induced Stat5b-DNA binding activity (Fig. 5B). In addition, the S193E mutation (lanes f and g) recovered IL-2-induced Stat5b DNA binding activity (Fig. 5B). Stat5b-DNA supershift assays were performed with an amino-terminal Stat5 antibody (lane h) to confirm the presence of Stat5 in the complex, whereas normal rabbit IgG (lane i) was used as a negative control. Thus, phosphorylation of Stat5b Ser-193 is required for optimal IL-2-induced DNA binding activity. To correlate the reduced DNA binding activity with decreased transcriptional activity, Stat5b luciferase reporter assays were performed in the HEK293 reconstitution system. Reconstituted HEK293 expressing WT (lanes c and d), S193A (lanes e and f), and S193E (lanes g and h) Stat5b proteins were co-transfected with a β-casein-firefly luciferase reporter construct harboring three Stat5 response elements in tandem (Fig. 5C). The cells were also transfected with a second β-gal reporter gene under the control of a constitutive CMV promoter to control for transfection efficiency. In these experiments, each luciferase activity was normalized first to the β-gal activity and second to WT Stat5b activity. Upon IL-2 stimulation, the S193A mutant Stat5b (lane f) showed a 40% decrease in Stat5b luciferase reporter activity relative to WT Stat5b (lane d) (Fig. 5C). Additionally, the Stat5b S193E (lane h) mutant displayed normal IL-2-induced transcriptional activity (Fig. 5C). As a control, cell lysates were immunoblotted with α-phospho-Tyr Stat5 to confirm equal expression (data not shown). These data suggest that Ser-193 phosphorylation is required for maximum Stat5b transcriptional activity.
FIGURE 5.
Stat5b Ser-193 phosphorylation is required for its maximal DNA binding and transcriptional activity. A, HEK293 cells were transfected with IL-2Rβ, IL-2Rγ, Jak3, and Stat5b (WT, S193A, or S193E) and incubated for 48 h. The cells were then stimulated without (−) or with IL-2 (+) for 15 min. Stat5b WT (lanes a and b), Stat5b S193A (lanes c and d), and Stat5b S193E (lanes e and f) were blotted with α-phospho-Tyr (α-pY) Stat5 (upper panel) or total Stat5 (lower panel). WB, Western blot. B, nuclear extracts (5 μg) isolated from transfected HEK293 described in A were incubated with a 32P-radiolabeled oligonucleotide probe corresponding to the Stat5 binding site in the β-casein gene promoter. The extracts indicated were co-incubated with N-terminal directed α-Stat5 (lane h) or normal rabbit IgG (Cntrl) (lane i). The bracket indicates the location of free probe, and the arrows indicate the location of non-supershifted and supershifted Stat5b-DNA complexes. Representative data from two independent experiments are shown. C, HEK293 cells transfected as described in A were treated without (−) or with (+) IL-2 for 6 h. Control cells were transfected with Stat5b alone (lanes a and b). At 48 h after transfection, the cells were lysed, and luciferase activities were measured and normalized to β-galactosidase activity. Statistical significance was determined using analysis of variance (*, p < 0.05). Representative data from three independent experiments are shown. Error bars indicate S.D.
Stat5 Ser-193 Is Constitutively Phosphorylated in HTLV-1-transformed T-cell Lines and Primary Hematopoietic Tumor Cells
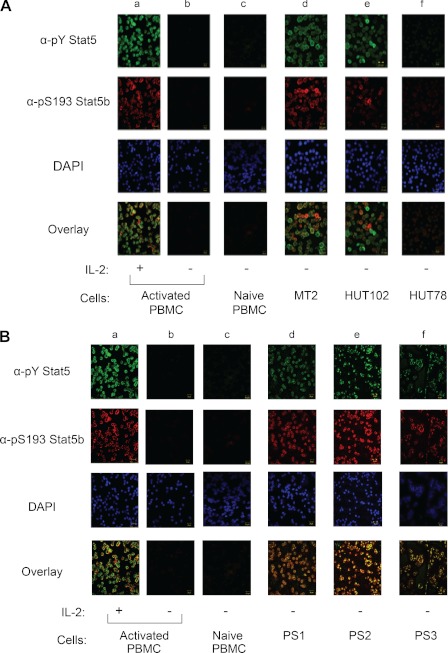
Elevated levels of Stat5 tyrosine phosphorylation and transcriptional activity have been observed in a number of primary tumors and tumor cell lines (50–52). However, the significance of serine phosphorylation for the proto-oncogenic function of Stat5 remains unclear. To correlate hyperactive Stat5 with constitutive Stat5 Ser-193 phosphorylation, human T lymphotropic virus type-1 (HTLV-1)-transformed cell lines and primary hematopoietic tumors were examined by α-phospho-Tyr Stat5- and α-Ser(P)-193 Stat5b-directed immunofluorescent confocal microscopy. In the absence of IL-2 stimulation, Stat5b was not tyrosine- or Ser-193-phosphorylated in naive (lane c) or quieted PHA-activated PBMCs (lane b) (Fig. 6A). However, similar to IL-2-stimulated PHA-activated PBMCs (lane a), MT-2 (lane d) and HUT-102 (lane e) cells displayed Stat5b tyrosine and Ser-193 phosphorylation in the absence of IL-2 stimulation (Fig. 6A). Interestingly, non-HTLV-1-transformed HUT-78 cells (lane f) did not display constitutive tyrosine or Ser-193 Stat5b phosphorylation (Fig. 6A). To substantiate these results, Stat5b Ser-193 and tyrosine phosphorylation was analyzed in primary patient tumor cells of hematopoietic origin. Indeed, immunofluorescent confocal microscopy analysis of tumor cells from patients diagnosed with T-cell acute lymphocytic leukemia (T-ALL) (lane d), acute myeloid leukemia (AML) (lane e), and B-cell lymphoma (lane f) showed constitutive phosphorylation of both Stat5 Tyr-694/699 and Ser-193 residues (Fig. 6B). The phosphorylation status of Stat5b Tyr-699 and Ser-193 was examined in a total of nine primary patient tumors and two control samples and is summarized in Table 1. Taken together, these data indicate that Stat5b is constitutively tyrosine-phosphorylated and Ser-193-phosphorylated in HTLV-1-transformed T-cell lines and primary hematopoietic tumor cells.
FIGURE 6.
Stat5b Ser-193 is constitutively phosphorylated in HTLV-1-transformed tumor T-cell lines and primary leukemia and lymphoma patient tumor cells. A, immunofluorescent confocal microscopy was utilized to detect phospho-Tyr Stat5 (α-pY Stat5) and Ser(P)-193 Stat5b (α-pS193 Stat5b) in normal PHA-activated quiescent human PBMCs stimulated with IL-2 or without for 15 min (lanes a and b), non-PHA-activated PBMCs cells (lane c), MT-2 (lane d), HUT-102 (lane e), and HUT-78 (lane f). B, immunofluorescent confocal microscopy was utilized to detect phospho-Tyr Stat5 and Ser(P)-193 Stat5b in primary tumor cells isolated from patients diagnosed with ALL (lane d), AML (lane e), and B-cell lymphoma (lane f). Normal PHA-activated quiescent human PBMCs stimulated with IL-2 or without IL-2 for 15 min (lanes a and b) and non-PHA-activated PBMCs cells (lane c) served as controls. Immunofluorescent images were captured using PASCAL software on a Zeiss LSM 510 Meta confocal microscope at 63× magnification. Representative data from three independent experiments are shown.
DISCUSSION
Stat5 activation is tightly controlled by a complex interplay between tyrosine and serine/threonine kinases and phosphatases. Although significant effort has established the importance of Stat5 tyrosine phosphorylation, the role of serine phosphorylation in Stat5 function and dysfunction remains to be fully elucidated. In the present study, a novel cytokine-induced serine phosphorylation site in Stat5b (Ser-193) was identified using mass spectrometry and further characterized with a site-specific phospho-specific antibody (Figs. 1 and 2). Stat5b Ser-193 was determined to be rapidly and transiently phosphorylated in response to IL-2 in YT and primary human PBMCs. Additionally, other γc cytokines, including IL-7, IL-9, and IL-15, similarly activated the phosphorylation of Stat5b Ser-193 in primary human PBMCs. Further evidence was provided that indicates phosphorylation of Stat5 Ser-193 is positively regulated by the serine/threonine kinase, mTOR, and negatively regulated by the protein phosphatase PP2A (Figs. 3 and 4). To evaluate the functional importance of Stat5b Ser-193 phosphorylation, WT, S193A, and S193E Stat5 variants were examined for their ability to bind DNA and activate transcription using EMSA and luciferase reporter assays, respectively. The S193A phospho-deletion mutation resulted in reduced ILb-2-stimulated Stat5b DNA binding and transcriptional activity, which could be rescued by the S193E phospho-mimetic mutation (Fig. 5). Importantly, multiple tumor cell lines and primary hematopoietic cancers were identified with constitutively phosphorylated Stat5b Ser-193, suggesting that it plays an important role in the proto-oncogenic activity of Stat5b in leukemias and lymphomas (Fig. 6). Taken together, these findings support the role of Ser-193 phosphorylation in normal and abnormal Stat5b activation, which represents a previously unrecognized positive regulatory mechanism that may guide novel therapeutic strategies to uncouple these critical regulators of lymphoid, myeloid, and erythroid tumor cell survival.
Previously identified Stat serine phospho-acceptor sites are represented by a positionally conserved serine residue that is located within one or more proline-rich motifs of the transactivation domain (reviewed in Ref. 10). For Stats 1, 3, and 4, serine phosphorylation sites are localized within a PMSP motif, whereas within Stat5a and Stat5b, they were mapped to a PSP motif. In addition, Stat5a has a unique phosphorylation site at Ser-779 that is flanked by a proline residue within an SP motif (34). Similarly, although Stat6 Ser-756 is not positionally conserved among other Stats, it does retain a juxtaposed proline residue localized within an SP motif (46). Extending the current model, our data indicate that Ser-193 represents a novel proline-flanked (SP motif) phosphorylation site within human Stat5b (Fig. 1D). Although Ser-193 is positionally conserved in Stat3 and Stat6, it is proline-flanked only in Stat5a and Stat5b. To further examine the extent of Ser-193 sequence conservation, phylogenetic analysis was performed using Stat5 homologous sequences from representative mammals and vertebrate outgroup taxa. Interestingly, Stat5 Ser-193 was found to be conserved in primates and marsupials, but not in bovine, murine, or rat Stat5 homologs, thus suggesting that Ser-193 was the result of an evolutionarily recent gain-of-function mutation. Additionally, Ser-193 is the first Stat5b phosphorylation site to be identified outside the transactivation domain (Fig. 1E). Ser-193 is located in the coiled-coil domain of Stat5b, which has been shown to be important for protein-protein interaction with co-regulators (53). Whether interaction of Stat5b with these coregulators involves phosphorylation of Ser-193 remains to be determined.
Current models hold that activation of Stat5 is mediated by two distinct kinases, a tyrosine kinase (such as Jak, Src, or Abl family members) and a proline-directed serine/threonine kinase(s). Although the identities of Stat5 tyrosine kinases are well established, the Stat5 serine/threonine kinase(s) have remained elusive. Several serine/threonine kinases have been demonstrated to phosphorylate the PMSP motif in Stat1, Stat3, and Stat4, including extracellular-regulated kinase (Erk)1/2 (42, 54). Indeed, the PMSP motif represents a consensus mitogen-activated protein kinase (MAPK) phosphorylation sequence (55). In contrast, the Stat5 PSP motif lacks the invariant methionine residue and thus is a weak MAPK target (54). Accordingly, phosphorylation of Stat5 Ser-726/731 has proved to be insensitive to inhibitors of MAPKs (11, 12, 46). A different picture has emerged after analysis of Stat6 (46), which displayed phosphorylation of Ser-756 localized in a conserved SP motif that is distinct from the PMSP and PSP motifs. Consequently, Ser-756 in the SP region of Stat6 was shown to be insensitive to MAPK inhibitors, but dependent upon mTOR activation (56). To identify the kinase(s) responsible for regulation of Stat5b Ser-193 phosphorylation, we investigated the role of MAPK and mTOR kinases using pharmacological inhibitors. MAPK inhibition had no visible effect on IL-2-induced phosphorylation of Stat5 Ser-193 (data not shown); however, pretreatment of YT cells with mTOR inhibitors significantly decreased IL-2-induced phosphorylation of Stat5b Ser-193 (Fig. 3). These finding are therefore in line with previous reports (10) of mTOR phosphorylation of Stat6, suggesting that mTOR preferentially phosphorylates Stat serine residues within SP motifs. Indeed, the mTOR phosphoproteome was recently characterized, and the primary consensus sequence was determined to include a proline residue at the +1 position (57).
Many studies have focused on the mechanisms driving Stat phosphorylation and activation; however, much less is known about its negative regulation. In addition to protein tyrosine phosphatases, serine/threonine phosphatases play an important role in the regulation of Stat activation. Recently, our group and others have reported that the serine/threonine phosphatase PP2A plays a role in regulating Stat3 (58), Stat5 (32), and Stat6 (45, 46) activity. To investigate PP2A regulation of Stat5 Ser(P)-193, attenuation of serine/threonine phosphatase activity by CA treatment of YT cells revealed that dephosphorylation of Stat5b Ser-193 occurred through a PP1- or PP2A-mediated process (Fig. 4A). To delineate the specific phosphatase, YT cells were treated with pharmacological inhibitors that preferentially target PP1 (TAU) or PP2A (OA and FOS). In accordance with our previous findings, phosphorylation of Stat5b Ser-193 was induced by OA and FOS, but not TAU, indicating that PP2A was the primary phosphatase responsible for regulating Stat5b Ser-193 phosphorylation (Fig. 4B).
Constitutive activation of Stat5 has been shown to be directly involved in oncogenic transformation (14). Moreover, Stat5 serine phosphorylation was recently shown to be a prerequisite for hematopoietic transformation (59, 60). To correlate hyperactive Stat5b with constitutive Stat5b Ser-193 phosphorylation, HTLV-1-transformed cell lines and primary hematopoietic tumors were examined. HTLV-1-transformed T-cells (MT-2, HUT-102) harbor constitutively tyrosine-phosphorylated Stat5b (61). Indeed, immunofluorescent confocal microscopy analysis showed that Stat5b is constitutively phosphorylated at both Tyr-699 and Ser-193 in MT-2 and Hut102 cells (Fig. 6A). To substantiate the clinical significance of Stat5b Ser-193 phosphorylation, primary patient tumor cells from multiple hematopoietic origins were analyzed (Fig. 6B). Stat5b was found to be constitutively tyrosine- and Ser-193 phosphorylated in various tumor cell types, including precursor T-ALL, AML-MO, AML-M4, large B-cell lymphoma, precursor B-cell ALL, and hairy cell leukemia (Table 1). Interestingly, in contrast to IL-2-stimulated Stat5b that translocates to the nucleus upon activation (Fig. 2), oncogenic Stat5b localizes primarily to the cytoplasm in primary hematopoietic tumor cells (Fig. 6). This suggests that the transforming capacity of Stat5b is not limited to its role as a transcription factor in the nucleus. Indeed, recent studies have demonstrated that oncogenic Stat5 localizes to the cytoplasm and plays an important role in the activation of the PI3K-Akt-mTOR signaling pathway via association with Gab2 (62–64). Extending this model, the results presented herein suggest that upon activation, mTOR serine phosphorylates Stat5b and allows for its maximal transcriptional activation. Therefore, the Stat5-mTor axis may play an important role in driving hematopoietic malignancy.
In summary, we have identified Stat5b Ser-193 as a novel site of cytokine-mediated phosphorylation. A phospho-antibody that specifically recognized this residue confirmed that Stat5b Ser-193 is rapidly and transiently induced in YT and primary human PBMCs. Using pharmacological inhibitors directed toward particular kinases and phosphatases, we show that phosphorylation of Stat5b Ser-193 is regulated by a mTOR/PP2A-dependent mechanism. Mechanistically, Ser-193 phosphorylation was shown to positively regulate the DNA binding and transcriptional activity of Stat5b. Lastly, Stat5b Ser-193 was found to be constitutively phosphorylated in several lymphoid tumor cell lines as well as primary leukemia and lymphoma patient tumor cells. Thus, drugs targeting the Stat5-mTOR axis could provide an alternative strategy for hematopoietic malignancies that display proto-oncogenic Stat5 activation.
Acknowledgments
We thank Dr. Max Shpak for the evolutionary divergence analysis, Derrick Oaxaca for assistance with Stat5b Ser(P)-193 antibody screening, and Dr. Armando Varela for technical assistance with immunofluorescent confocal microscopy.
This work was supported, in whole or in part, by grants from the Lizanell and Colbert Coldwell Foundation (to R. A. K.), the Edward N. and Margaret G. Marsh Foundation (to R. A. K. and J. A. R.), and the University Research Institute at the University of Texas at El Paso (to J. A. R.), and was made possible by Grant G12MD007592 from the National Institutes on Minority Health and Health Disparities (NIMHD), a component of the National Institutes of Health (NIH).
- PSP
- proline-serine-proline
- PMSP
- proline-methionine-serine-proline
- SP
- serine-proline
- CA
- calyculin A
- FOS
- fostriecin
- γc
- γ common chain
- mTOR
- mammalian target of rapamycin
- OA
- okadaic acid
- PBMC
- peripheral blood mononuclear cell
- PHA
- phytohemagglutinin
- PP1
- protein phosphatase 1
- PP2A
- protein phosphatase 2A
- TAU
- tautomycin
- ALL
- acute lymphocytic leukemia
- T-ALL
- T-cell ALL
- AML
- acute myeloid leukemia
- HTLV-1
- human T lymphotropic virus type-1
- IL-2R
- interleukin-2 receptor
- DMSO
- dimethyl sulfoxide.
REFERENCES
- 1. Darnell J. E., Jr. (1997) STATs and gene regulation. Science 277, 1630–1635 [DOI] [PubMed] [Google Scholar]
- 2. Liu X., Robinson G. W., Gouilleux F., Groner B., Hennighausen L. (1995) Cloning and expression of Stat5 and an additional homologue (Stat5b) involved in prolactin signal transduction in mouse mammary tissue. Proc. Natl. Acad. Sci. U.S.A. 92, 8831–8835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yamashita H., Xu J., Erwin R. A., Farrar W. L., Kirken R. A., Rui H. (1998) Differential control of the phosphorylation state of proline-juxtaposed serine residues Ser-725 of Stat5a and Ser-730 of Stat5b in prolactin-sensitive cells. J. Biol. Chem. 273, 30218–30224 [DOI] [PubMed] [Google Scholar]
- 4. Teglund S., McKay C., Schuetz E., van Deursen J. M., Stravopodis D., Wang D., Brown M., Bodner S., Grosveld G., Ihle J. N. (1998) Stat5a and Stat5b proteins have essential and nonessential, or redundant, roles in cytokine responses. Cell 93, 841–850 [DOI] [PubMed] [Google Scholar]
- 5. Cui Y., Riedlinger G., Miyoshi K., Tang W., Li C., Deng C. X., Robinson G. W., Hennighausen L. (2004) Inactivation of Stat5 in mouse mammary epithelium during pregnancy reveals distinct functions in cell proliferation, survival, and differentiation. Mol. Cell Biol. 24, 8037–8047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kirken R. A., Rui H., Malabarba M. G., Howard O. M., Kawamura M., O'Shea J. J., Farrar W. L. (1995) Activation of JAK3, but not JAK1, is critical for IL-2-induced proliferation and STAT5 recruitment by a COOH-terminal region of the IL-2 receptor β-chain. Cytokine 7, 689–700 [DOI] [PubMed] [Google Scholar]
- 7. Snow J. W., Abraham N., Ma M. C., Abbey N. W., Herndier B., Goldsmith M. A. (2002) STAT5 promotes multilineage hematolymphoid development in vivo through effects on early hematopoietic progenitor cells. Blood 99, 95–101 [DOI] [PubMed] [Google Scholar]
- 8. Smithgall T. E., Briggs S. D., Schreiner S., Lerner E. C., Cheng H., Wilson M. B. (2000) Control of myeloid differentiation and survival by Stats. Oncogene 19, 2612–2618 [DOI] [PubMed] [Google Scholar]
- 9. Leonard W. J. (2001) Role of Jak kinases and STATs in cytokine signal transduction. Int. J. Hematol. 73, 271–277 [DOI] [PubMed] [Google Scholar]
- 10. Decker T., Kovarik P. (2000) Serine phosphorylation of STATs. Oncogene 19, 2628–2637 [DOI] [PubMed] [Google Scholar]
- 11. Kirken R. A., Malabarba M. G., Xu J., DaSilva L., Erwin R. A., Liu X., Hennighausen L., Rui H., Farrar W. L. (1997) Two discrete regions of interleukin-2 (IL2) receptor β independently mediate IL2 activation of a PD98059/rapamycin/wortmannin-insensitive Stat5a/b serine kinase. J. Biol. Chem. 272, 15459–15465 [DOI] [PubMed] [Google Scholar]
- 12. Nagy Z. S., Wang Y., Erwin-Cohen R. A., Aradi J., Monia B., Wang L. H., Stepkowski S. M., Rui H., Kirken R. A. (2002) Interleukin-2 family cytokines stimulate phosphorylation of the Pro-Ser-Pro motif of Stat5 transcription factors in human T-cells: resistance to suppression of multiple serine kinase pathways. J. Leukoc. Biol. 72, 819–828 [PubMed] [Google Scholar]
- 13. Pircher T. J., Petersen H., Gustafsson J. A., Haldosén L. A. (1999) Extracellular signal-regulated kinase (ERK) interacts with signal transducer and activator of transcription (STAT) 5a. Mol. Endocrinol. 13, 555–565 [DOI] [PubMed] [Google Scholar]
- 14. Bunting K. D. (2007) STAT5 signaling in normal and pathologic hematopoiesis. Front. Biosci. 12, 2807–2820 [DOI] [PubMed] [Google Scholar]
- 15. Hennighausen L., Robinson G. W. (2008) Interpretation of cytokine signaling through the transcription factors STAT5A and STAT5B. Genes Dev. 22, 711–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chai S. K., Nichols G. L., Rothman P. (1997) Constitutive activation of JAKs and STATs in BCR-Abl-expressing cell lines and peripheral blood cells derived from leukemic patients. J. Immunol. 159, 4720–4728 [PubMed] [Google Scholar]
- 17. Ho J. M., Beattie B. K., Squire J. A., Frank D. A., Barber D. L. (1999) Fusion of the ets transcription factor TEL to Jak2 results in constitutive Jak-Stat signaling. Blood 93, 4354–4364 [PubMed] [Google Scholar]
- 18. Ilaria R. L., Jr., Van Etten R. A. (1996) P210 and P190BCR/ABL induce the tyrosine phosphorylation and DNA binding activity of multiple specific STAT family members. J. Biol. Chem. 271, 31704–31710 [DOI] [PubMed] [Google Scholar]
- 19. Schwaller J., Parganas E., Wang D., Cain D., Aster J. C., Williams I. R., Lee C. K., Gerthner R., Kitamura T., Frantsve J., Anastasiadou E., Loh M. L., Levy D. E., Ihle J. N., Gilliland D. G. (2000) Stat5 is essential for the myelo- and lymphoproliferative disease induced by TEL/JAK2. Mol. Cell 6, 693–704 [DOI] [PubMed] [Google Scholar]
- 20. Shuai K., Halpern J., ten Hoeve J., Rao X., Sawyers C. L. (1996) Constitutive activation of STAT5 by the BCR-ABL oncogene in chronic myelogenous leukemia. Oncogene 13, 247–254 [PubMed] [Google Scholar]
- 21. Spiekermann K., Pau M., Schwab R., Schmieja K., Franzrahe S., Hiddemann W. (2002) Constitutive activation of STAT3 and STAT5 is induced by leukemic fusion proteins with protein tyrosine kinase activity and is sufficient for transformation of hematopoietic precursor cells. Exp. Hematol. 30, 262–271 [DOI] [PubMed] [Google Scholar]
- 22. Kato Y., Iwama A., Tadokoro Y., Shimoda K., Minoguchi M., Akira S., Tanaka M., Miyajima A., Kitamura T., Nakauchi H. (2005) Selective activation of STAT5 unveils its role in stem cell self-renewal in normal and leukemic hematopoiesis. J. Exp. Med. 202, 169–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Moriggl R., Sexl V., Kenner L., Duntsch C., Stangl K., Gingras S., Hoffmeyer A., Bauer A., Piekorz R., Wang D., Bunting K. D., Wagner E. F., Sonneck K., Valent P., Ihle J. N., Beug H. (2005) Stat5 tetramer formation is associated with leukemogenesis. Cancer Cell 7, 87–99 [DOI] [PubMed] [Google Scholar]
- 24. Van Etten R. A. (2007) Aberrant cytokine signaling in leukemia. Oncogene 26, 6738–6749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Carron C., Cormier F., Janin A., Lacronique V., Giovannini M., Daniel M. T., Bernard O., Ghysdael J. (2000) TEL-JAK2 transgenic mice develop T-cell leukemia. Blood 95, 3891–3899 [PubMed] [Google Scholar]
- 26. Nieborowska-Skorska M., Wasik M. A., Slupianek A., Salomoni P., Kitamura T., Calabretta B., Skorski T. (1999) Signal transducer and activator of transcription (STAT)5 activation by BCR/ABL is dependent on intact Src homology (SH)3 and SH2 domains of BCR/ABL and is required for leukemogenesis. J. Exp. Med. 189, 1229–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mizuki M., Fenski R., Halfter H., Matsumura I., Schmidt R., Müller C., Grüning W., Kratz-Albers K., Serve S., Steur C., Büchner T., Kienast J., Kanakura Y., Berdel W. E., Serve H. (2000) Flt3 mutations from patients with acute myeloid leukemia induce transformation of 32D cells mediated by the Ras and STAT5 pathways. Blood 96, 3907–3914 [PubMed] [Google Scholar]
- 28. Levine R. L., Loriaux M., Huntly B. J., Loh M. L., Beran M., Stoffregen E., Berger R., Clark J. J., Willis S. G., Nguyen K. T., Flores N. J., Estey E., Gattermann N., Armstrong S., Look A. T., Griffin J. D., Bernard O. A., Heinrich M. C., Gilliland D. G., Druker B., Deininger M. W. (2005) The JAK2V617F activating mutation occurs in chronic myelomonocytic leukemia and acute myeloid leukemia, but not in acute lymphoblastic leukemia or chronic lymphocytic leukemia. Blood 106, 3377–3379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nagy Z. S., LeBaron M. J., Ross J. A., Mitra A., Rui H., Kirken R. A. (2009) STAT5 regulation of BCL10 parallels constitutive NFκB activation in lymphoid tumor cells. Mol. Cancer 8, 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ross J. A., Nagy Z. S., Kirken R. A. (2008) The PHB1/2 phosphocomplex is required for mitochondrial homeostasis and survival of human T-cells. J. Biol. Chem. 283, 4699–4713 [DOI] [PubMed] [Google Scholar]
- 31. Chenna R., Sugawara H., Koike T., Lopez R., Gibson T. J., Higgins D. G., Thompson J. D. (2003) Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31, 3497–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ross J. A., Cheng H., Nagy Z. S., Frost J. A., Kirken R. A. (2010) Protein phosphatase 2A regulates interleukin-2 receptor complex formation and JAK3/STAT5 activation. J. Biol. Chem. 285, 3582–3591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cheng H., Ross J. A., Frost J. A., Kirken R. A. (2008) Phosphorylation of human Jak3 at tyrosines 904 and 939 positively regulates its activity. Mol. Cell Biol. 28, 2271–2282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yamashita H., Nevalainen M. T., Xu J., LeBaron M. J., Wagner K. U., Erwin R. A., Harmon J. M., Hennighausen L., Kirken R. A., Rui H. (2001) Role of serine phosphorylation of Stat5a in prolactin-stimulated β-casein gene expression. Mol. Cell Endocrinol. 183, 151–163 [DOI] [PubMed] [Google Scholar]
- 35. Nagy Z. S., Rui H., Stepkowski S. M., Karras J., Kirken R. A. (2006) A preferential role for STAT5, not constitutively active STAT3, in promoting survival of a human lymphoid tumor. J. Immunol. 177, 5032–5040 [DOI] [PubMed] [Google Scholar]
- 36. Beausoleil S. A., Villén J., Gerber S. A., Rush J., Gygi S. P. (2006) A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat. Biotechnol. 24, 1285–1292 [DOI] [PubMed] [Google Scholar]
- 37. Daub H., Olsen J. V., Bairlein M., Gnad F., Oppermann F. S., Körner R., Greff Z., Kéri G., Stemmann O., Mann M. (2008) Kinase-selective enrichment enables quantitative phospho-proteomics of the kinome across the cell cycle. Mol. Cell 31, 438–448 [DOI] [PubMed] [Google Scholar]
- 38. Dephoure N., Zhou C., Villén J., Beausoleil S. A., Bakalarski C. E., Elledge S. J., Gygi S. P. (2008) A quantitative atlas of mitotic phosphorylation. Proc. Natl. Acad. Sci. U.S.A. 105, 10762–10767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mayya V., Lundgren D. H., Hwang S. I., Rezaul K., Wu L., Eng J. K., Rodionov V., Han D. K. (2009) Quantitative phospho-proteomic analysis of T-cell receptor signaling reveals system-wide modulation of protein-protein interactions. Sci. Signal. 2, ra46. [DOI] [PubMed] [Google Scholar]
- 40. Ihle J. N., Witthuhn B. A., Quelle F. W., Yamamoto K., Silvennoinen O. (1995) Signaling through the hematopoietic cytokine receptors. Annu. Rev. Immunol. 13, 369–398 [DOI] [PubMed] [Google Scholar]
- 41. Johnston J. A., Kawamura M., Kirken R. A., Chen Y. Q., Blake T. B., Shibuya K., Ortaldo J. R., McVicar D. W., O'Shea J. J. (1994) Phosphorylation and activation of the Jak-3 Janus kinase in response to interleukin-2. Nature 370, 151–153 [DOI] [PubMed] [Google Scholar]
- 42. Jain N., Zhang T., Kee W. H., Li W., Cao X. (1999) Protein kinase C δ associates with and phosphorylates Stat3 in an interleukin-6-dependent manner. J. Biol. Chem. 274, 24392–24400 [DOI] [PubMed] [Google Scholar]
- 43. Schuringa J. J., Wierenga A. T., Kruijer W., Vellenga E. (2000) Constitutive Stat3, Tyr-705, and Ser-727 phosphorylation in acute myeloid leukemia cells caused by the autocrine secretion of interleukin-6. Blood 95, 3765–3770 [PubMed] [Google Scholar]
- 44. Nguyen V. A., Chen J., Hong F., Ishac E. J., Gao B. (2000) Interferons activate the p42/44 mitogen-activated protein kinase and JAK-STAT (Janus kinase-signal transducer and activator transcription factor) signaling pathways in hepatocytes: differential regulation by acute ethanol via a protein kinase C-dependent mechanism. Biochem. J. 349, 427–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Woetmann A., Brockdorff J., Lovato P., Nielsen M., Leick V., Rieneck K., Svejgaard A., Geisler C., Ødum N. (2003) Protein phosphatase 2A (PP2A) regulates interleukin-4-mediated STAT6 signaling. J. Biol. Chem. 278, 2787–2791 [DOI] [PubMed] [Google Scholar]
- 46. Wang Y., Malabarba M. G., Nagy Z. S., Kirken R. A. (2004) Interleukin-4 regulates phosphorylation of serine 756 in the transactivation domain of Stat6: roles for multiple phosphorylation sites and Stat6 function. J. Biol. Chem. 279, 25196–25203 [DOI] [PubMed] [Google Scholar]
- 47. Ishihara H., Martin B. L., Brautigan D. L., Karaki H., Ozaki H., Kato Y., Fusetani N., Watabe S., Hashimoto K., Uemura D. (1989) Calyculin A and okadaic acid: inhibitors of protein phosphatase activity. Biochem. Biophys. Res. Commun. 159, 871–877 [DOI] [PubMed] [Google Scholar]
- 48. Lin J. X., Mietz J., Modi W. S., John S., Leonard W. J. (1996) Cloning of human Stat5B: reconstitution of interleukin-2-induced Stat5A and Stat5B DNA binding activity in COS-7 cells. J. Biol. Chem. 271, 10738–10744 [PubMed] [Google Scholar]
- 49. Lin J. X., Leonard W. J. (2000) The role of Stat5a and Stat5b in signaling by IL-2 family cytokines. Oncogene 19, 2566–2576 [DOI] [PubMed] [Google Scholar]
- 50. Funakoshi-Tago M., Tago K., Abe M., Sonoda Y., Kasahara T. (2010) STAT5 activation is critical for the transformation mediated by myeloproliferative disorder-associated JAK2 V617F mutant. J. Biol. Chem. 285, 5296–5307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gaipa G., Bugarin C., Longoni D., Cesana S., Molteni C., Faini A., Timeus F., Zecca M., Biondi A. (2009) Aberrant GM-CSF signal transduction pathway in juvenile myelomonocytic leukemia assayed by flow cytometric intracellular STAT5 phosphorylation measurement. Leukemia 23, 791–793 [DOI] [PubMed] [Google Scholar]
- 52. Kotecha N., Flores N. J., Irish J. M., Simonds E. F., Sakai D. S., Archambeault S., Diaz-Flores E., Coram M., Shannon K. M., Nolan G. P., Loh M. L. (2008) Single-cell profiling identifies aberrant STAT5 activation in myeloid malignancies with specific clinical and biologic correlates. Cancer Cell 14, 335–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Nakajima H., Brindle P. K., Handa M., Ihle J. N. (2001) Functional interaction of STAT5 and nuclear receptor co-repressor SMRT: implications in negative regulation of STAT5-dependent transcription. EMBO J. 20, 6836–6844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. David M., Petricoin E., 3rd, Benjamin C., Pine R., Weber M. J., Larner A. C. (1995) Requirement for MAP kinase (ERK2) activity in interferon α- and interferon β-stimulated gene expression through STAT proteins. Science 269, 1721–1723 [DOI] [PubMed] [Google Scholar]
- 55. Winston L. A., Hunter T. (1996) Intracellular signaling: putting JAKs on the kinase MAP. Curr. Biol. 6, 668–671 [DOI] [PubMed] [Google Scholar]
- 56. Yokogami K., Wakisaka S., Avruch J., Reeves S. A. (2000) Serine phosphorylation and maximal activation of STAT3 during CNTF signaling is mediated by the rapamycin target mTOR. Curr. Biol. 10, 47–50 [DOI] [PubMed] [Google Scholar]
- 57. Hsu P. P., Kang S. A., Rameseder J., Zhang Y., Ottina K. A., Lim D., Peterson T. R., Choi Y., Gray N. S., Yaffe M. B., Marto J. A., Sabatini D. M. (2011) The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science 332, 1317–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Woetmann A., Nielsen M., Christensen S. T., Brockdorff J., Kaltoft K., Engel A. M., Skov S., Brender C., Geisler C., Svejgaard A., Rygaard J., Leick V., Odum N. (1999) Inhibition of protein phosphatase 2A induces serine/threonine phosphorylation, subcellular redistribution, and functional inhibition of STAT3. Proc. Natl. Acad. Sci. U.S.A. 96, 10620–10625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Friedbichler K., Kerenyi M. A., Kovacic B., Li G., Hoelbl A., Yahiaoui S., Sexl V., Müllner E. W., Fajmann S., Cerny-Reiterer S., Valent P., Beug H., Gouilleux F., Bunting K. D., Moriggl R. (2010) Stat5a serine 725 and 779 phosphorylation is a prerequisite for hematopoietic transformation. Blood 116, 1548–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Friedbichler K., Hoelbl A., Li G., Bunting K. D., Sexl V., Gouilleux F., Moriggl R. (2012) Serine phosphorylation of the Stat5a C terminus is a driving force for transformation. Front Biosci. 17, 3043–3056 [DOI] [PubMed] [Google Scholar]
- 61. Kirken R. A., Erwin R. A., Wang L., Wang Y., Rui H., Farrar W. L. (2000) Functional uncoupling of the Janus kinase 3-Stat5 pathway in malignant growth of human T-cell leukemia virus type 1-transformed human T-cells. J. Immunol. 165, 5097–5104 [DOI] [PubMed] [Google Scholar]
- 62. Nyga R., Pecquet C., Harir N., Gu H., Dhennin-Duthille I., Régnier A., Gouilleux-Gruart V., Lassoued K., Gouilleux F. (2005) Activated STAT5 proteins induce activation of the PI3-kinase/Akt and Ras/MAPK pathways via the Gab2 scaffolding adapter. Biochem. J. 390, 359–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Harir N., Pecquet C., Kerenyi M., Sonneck K., Kovacic B., Nyga R., Brevet M., Dhennin I., Gouilleux-Gruart V., Beug H., Valent P., Lassoued K., Moriggl R., Gouilleux F. (2007) Constitutive activation of Stat5 promotes its cytoplasmic localization and association with PI3-kinase in myeloid leukemias. Blood 109, 1678–1686 [DOI] [PubMed] [Google Scholar]
- 64. Harir N., Boudot C., Friedbichler K., Sonneck K., Kondo R., Martin-Lannerée S., Kenner L., Kerenyi M., Yahiaoui S., Gouilleux-Gruart V., Gondry J., Bénit L., Dusanter-Fourt I., Lassoued K., Valent P., Moriggl R., Gouilleux F. (2008) Oncogenic Kit controls neoplastic mast cell growth through a Stat5/PI3-kinase signaling cascade. Blood 112, 2463–2473 [DOI] [PMC free article] [PubMed] [Google Scholar]