Background: Ail is a critical fibronectin (Fn)-binding protein in plague pathogenesis.
Results: Ail binds 9FNIII, unique among bacterial Fn-binding proteins.
Conclusion: Ail binding to 9FNIII facilitates binding to pure Fn and host cells.
Significance: Upon binding 9FNIII, Ail is adjacent to the RGD in 10FNIII and may alter integrin signaling events required for delivery of Yop cytotoxins and plague virulence.
Keywords: Bacterial Adhesion, Bacterial Pathogenesis, Epitope Mapping, Extracellular Matrix Proteins, Fibronectin, Plague
Abstract
The Yersinia pestis adhesin molecule Ail interacts with the extracellular matrix protein fibronectin (Fn) on host cells to facilitate efficient delivery of cytotoxic Yop proteins, a process essential for plague virulence. A number of bacterial pathogens are known to bind to the N-terminal region of Fn, comprising type I Fn (FNI) repeats. Using proteolytically generated Fn fragments and purified recombinant Fn fragments, we demonstrated that Ail binds the centrally located 120-kDa fragment containing type III Fn (FNIII) repeats. A panel of monoclonal antibodies (mAbs) that recognize specific epitopes within the 120-kDa fragment demonstrated that mAb binding to 9FNIII blocks Ail-mediated bacterial binding to Fn. Epitopes of three mAbs that blocked Ail binding to Fn were mapped to a similar face of 9FNIII. Antibodies directed against 9FNIII also inhibited Ail-dependent cell binding activity, thus demonstrating the biological relevance of this Ail binding region on Fn. Bacteria expressing Ail on their surface could also bind a minimal fragment of Fn containing repeats 9–10FNIII, and this binding was blocked by a mAb specific for 9FNIII. These data demonstrate that Ail binds to 9FNIII of Fn and presents Fn to host cells to facilitate cell binding and delivery of Yops (cytotoxins of Y. pestis), a novel interaction, distinct from other bacterial Fn-binding proteins.
Introduction
Binding of Yersinia pestis to host cells via the outer membrane protein, Ail, is critical for delivery of cytotoxic Yop proteins to host cells (1, 2). Yop delivery requires host cell contact to initiate type III secretion from the bacterial cell to the host cell (3, 4). The type III secretion process is hypothesized to occur by direct transfer of effector molecules from the bacterial cytoplasm to the host cell cytoplasm, without an extracellular intermediate (5). Consistent with a role for Ail in Yop delivery in vivo, an ail mutant is highly attenuated, with a >3000-fold increase in LD50 compared with the parental Y. pestis KIM5 strain in a mouse model of septicemic plague (1), and recent studies indicate that Ail plays an important role in bubonic and pneumonic plague in mice and rats, contributing to serum resistance and adhesion-dependent immunosuppression due to type III secretion of Yop proteins (6, 7). Thus, Ail is a major virulence factor for Y. pestis pathogenesis.
We previously reported that Ail from Y. pestis interacts with the host extracellular matrix protein, fibronectin (Fn)2 (2). This Ail-Fn interaction leads to efficient Yop delivery as inhibition of this interaction with anti-Fn antibody results in reduced levels of cytotoxicity. Thus, characterization of the mechanism of Ail-Fn interaction will contribute the understanding of virulence mechanisms of Y. pestis.
Fn is a critical extracellular matrix protein found throughout vertebrate hosts and is involved in many processes, including wound healing, tissue structure, cell migration, and embryonic development (8). Previous studies with cultured fibroblasts found that Fn mediates cell-cell interactions and cell-substratum contacts, thus imparting tissue architecture (9, 10). As part of this process, cells within tissues synthesize soluble Fn, which is arranged into insoluble fibrils that make up extracellular Fn matrices.
Fn is a large complex glycoprotein found in blood, on cell surfaces, and within tissues (11, 12) that exists as a dimer of ∼500 kDa, with the two monomers linked together by two disulfide bonds near the C terminus of the molecule. Fn is composed of 12 type I repeats (FNI), 2 type II repeats (FNII), and 15–17 type III repeats (FNIII) (13).
To facilitate cell attachment, Fn binds host cell integrins (14, 15) as well as many other host cell components, including collagen (16), heparin (17, 18), and fibrin (17, 19). The binding sites for these Fn substrates are depicted in Fig. 1A. The N terminus of Fn, including 1–5FNI, is termed the matrix assembly site because it is responsible for the assembly of soluble Fn into insoluble fibrils. Integrin α5β1 binds this N-terminal matrix assembly site (20, 21) in addition to the classical RGD binding motif in 10FNIII (22), but α5β1 binding to 1–5FNI initiates host cell signaling events distinct from those observed with interaction with the RGD domain (23).
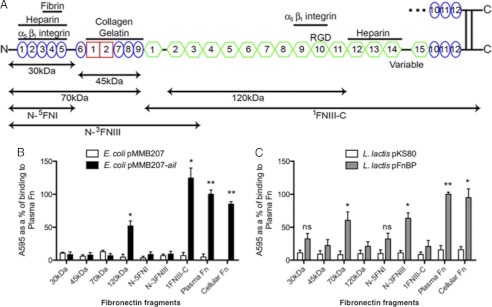
FIGURE 1.
Ail binds 120-kDa fragment of Fn. A, Fn fragments used in this study are illustrated. B, Fn, commercially available proteolytically generated Fn fragments, and purified recombinant Fn fragments were adsorbed onto plastic wells (10 μg/ml). E. coli AAEC185 derivatives expressing empty vector and Ail were added to the wells and allowed to bind at 37 °C. Bound bacteria were stained with 0.01% crystal violet, washed, and the cells were then solubilized, to read the A595. C, wells were coated with various forms of Fn (10 μg/ml), and L. lactis derivatives were added as described above. *, p < 0.01; **, p < 0.00005. FnBP binding to 30 kDa had a p = 0.055, FnBP binding to N-5FNI had a p = 0.071. Error bars, S.D.
Numerous bacteria encode Fn-binding proteins, taking advantage of Fn to facilitate host cell contact and, in some cases, cellular invasion (24). Protein F1 (Sfb1) of Streptococcus pyogenes is one example (25, 26). Protein F1 has multiple Fn binding repeats, each with the capability to interact with multiple FNI repeats in the N-terminal region of Fn, allowing high affinity, multivalent interactions (27, 28). Staphylococcus aureus also binds the N-terminal region of Fn (29–31), using FnBPA (Fn-binding protein A) (32). FnBPA interacts with Fn via multiple repeats, each of which binds to several FNI domains by β-strand addition, termed the β-zipper model (33, 34). S. aureus can also bind the 120-kDa fragments of Fn, indicating that there are multiple S. aureus binding sites along Fn (35–37). S. pyogenes and S. aureus protein F and FnBPA, respectively, use Fn to invade host cells by clustering of α5β1 integrins on the surface of host cells (26, 32). The Yersinia pseudotuberculosis autotransporter adhesion YadA also interacts with Fn (38) which then acts as a bridge to engage β1 integrins to mediate cell adhesion and invasion (39). Thus, the use of Fn as a bridge for bacteria to engage host cells is a common strategy of bacterial pathogens.
To gain insight into the mechanism of Ail interaction with Fn, an event that facilitates Yop delivery, we mapped the Ail binding site on Fn. We present evidence that Ail binds 9FNIII neighboring the RGD site in 10FNIII, a unique location relative to other bacterial Fn-binding proteins. The potential repercussions of such a binding mechanism on cell signaling and Yop delivery are discussed.
EXPERIMENTAL PROCEDURES
Strains and Culture Conditions
Escherichia coli AAEC185 strains were cultured in Luria-Bertani (LB) broth or LB agar at 28 °C or 37 °C. Lactococcus lactis strains were cultured in M-17 medium supplemented with 0.5% d-glucose (GM-17 medium) at 30 °C without shaking. Y. pestis strains were cultured in heart infusion broth (Difco) or heart infusion agar (Difco). Antibiotics were used at the following concentrations: chloramphenicol = 25 μg/ml and erythromycin = 10 μg/ml. Isopropyl-β-d-thiogalactopyranoside (IPTG) was used at a 100 μm concentration unless otherwise noted. Plasmids and strains used in this study are listed in supplemental Table S1.
Fn and Monoclonal Antibodies
The following Fn and Fn fragments were used: plasma Fn (Sigma, F2006), cellular Fn (Sigma, F2518), 30 kDa of Fn (Sigma F9911), 45 kDa of Fn (Sigma, F0162) 70 kDa of Fn (Sigma, F0287), 120 kDa of Fn (Chemicon, F1904). Recombinant N-5FNI, N-3FNIII, and 1FNIIII-C fragments were described previously (40). The following antibodies were used: polyclonal rabbit anti-human Fn antibody (Sigma, F3648), monoclonal mouse anti-Fn (anti-10FNIII, 3E3, Chemicon, MAB88916). Other mouse or rat anti-human Fn monoclonal antibodies used in this study have been described previously (28, 41, 42). Rat monoclonal antibodies used in this study were a kind gift from Dr. Kenneth Yamada, NIDCR, National Institutes of Health. Recombinant 9–10FNIII was overproduced in E. coli strain Rosetta (DE3) pLysS, following PCR from the vector pRSET-C′ (XhoI−) (43) and ligation into the His-tagging bacterial expression vector pET30b+ with a tobacco etch virus cleavage site in place of the enterokinase cleavage site primers 5′-gatatacatatgggtcttgattc-3′ and 5′-gatcggtacccatctgggatggtttgtc-3′. Protein was overexpressed in 1 liter of culture with 1 mm IPTG at room temperature overnight and purified over a nickel-agarose column as described by the manufacturer (Qiagen).
Preadsorbed Antibody Blocking Assay
20 μg/ml of full-length plasma Fn, 70-kDa, or 120-kDa fragments were immobilized onto a 96-well plastic plate overnight at 4 °C. Rabbit anti-Fn polyclonal antibody (Sigma, F3648), was added to each Fn derivative at a concentration of 1:25. After 2 h of incubation, the solution was transferred to a fresh well coated with plasma Fn, 70 kDa, or 120 kDa (as in the previous treatment) and incubated for another 2 h. This preadsorption was performed a total of four times. The resulting solution was then used in a subsequent bacterial binding assay as described below.
Bacterial Binding to Fn and Antibody Blocking Assays
These assays were performed as described previously (2). Briefly, polystyrene 96-well plates (BD Scientific) were coated with 10 μg/ml plasma Fn overnight at 4 °C. E. coli AAEC185 derivatives were cultured overnight at 28 °C in LB + 100 μm IPTG. Y. pestis derivatives were cultured overnight at 28 °C, followed by a 1:50 dilution of the overnight culture in fresh HIB + 100 μm IPTG. L. lactis was grown overnight in GM-17 medium at 30 °C without shaking. The following day, wells were washed with PBS before blocking with blocking buffer (PBS + 2% BSA). The bacterial cells were normalized to an OD600 (OD620 for Y. pestis) of 1.0 in PBS + 0.4% BSA. 50 μl of bacterial suspension was added to the appropriate wells. The plate was incubated at 37 °C for 2 h. The wells were washed with PBS before fixing with methanol. The bacteria bound to the wells were stained with crystal violet, washed, and solubilized with an 80% ethanol/20% acetone solution, and the absorbance was measured at ABS595.
Antibody blocking assays were performed similarly to the bacterial binding assays except antibodies were diluted to ∼40 μg/ml and were added to Fn-coated wells 1 h prior to the addition of 10 μl of 5-fold concentrated bacteria (i.e. OD600 = 5.0 for E. coli; OD620 = 5.0 for Y. pestis).
Cell Binding Assays
HEp-2 cells were cultured in 24-well tissue culture plates until reaching 80–90% confluence. Y. pestis KIM5 D27 strains were cultured overnight and then diluted 1:50 the next day in fresh medium. Strains were allowed to grow for an additional 4 h at 28 °C. Tissue culture cells were washed with PBS and polyclonal rabbit anti-human Fn antibody and monoclonal antibodies were diluted in tissue culture media without serum with 20 mm HEPES, pH 7.0, for a final concentration of 40 μg/ml. Bacteria were added at a multiplicity of infection of 100. Plates were incubated with bacteria at 37 °C in 5% CO2 for 105 min. Cells were then washed with PBS, and cell-associated bacteria were liberated by the addition of sterile H2O containing 0.1% Triton X-100 for 10 min. Percent adhesion was calculated by dividing bound cfu by total bacteria in the well at the end of a 105-min incubation and then multiplying by 100.
Fine Epitope Mapping
Monoclonal antibodies previously shown to react with 9FNIII or 10FNIII (42, 44, 45) were tested by direct enzyme-linked immunosorbent assay for recognition of a coating of purified Fn from human, cow, rat, or mouse or of diluted chicken serum. The method was as described previously (28) except that the secondary antibody was peroxidase-conjugated donkey anti-rat IgG (H+L) (Jackson ImmunoResearch Laboratories, West Grove, PA). Comparison of amino acid differences among species and reactivity of the antibody with Fn from the various species allowed identification of responsible residues. The residues were numbered in reference to human Fn (UniProtKB Entry P02751) in its nonprocessed pre-pro form (i.e. from the initiating methionine) that includes the alternatively splice ED-B FNIII module (i.e. the longest form) and located in the crystal structure of 7–10FNIII of human Fn (46).
RESULTS
Bacteria Expressing Ail Bind to 120-kDa Fragment of Fn
To characterize the mechanism of Ail interaction with Fn, we sought to map the Ail binding domain on Fn. Fn can be proteolytically cleaved into various sized fragments (Fig. 1A), and others have shown that some bacteria can bind to these smaller fragments with an efficiency similar to full-length Fn (47, 48). Thus, we used these fragments to identify the Ail binding site. Commercially available 30-, 45-, 70-, and 120-kDa Fn fragments were adsorbed to 96-well microtiter plates and used for bacterial binding assays. E. coli expressing Ail from the inducible plasmid pMMB207 (49) bound immobilized full-length Fn (plasma Fn) to high levels. This level of binding was set at 100%. Although Ail bound to the centrally located 120-kDa fragment of Fn, it did not mediate binding to the N-terminal 30-, 45-, or 70-kDa fragments (Fig. 1B). This indicates that Ail interacts with a region within the 120-kDa fragment, which includes most of the FNIII modules, including 2–11FNIII (Fig. 1A). E. coli expressing empty vector did not bind to any fragment tested, and binding to cellular Fn gave levels of binding similar to those seen with immobilized plasma Fn (Fig. 1B), indicating that upon immobilization on polystyrene plates, plasma Fn exposes domains inaccessible when present in its soluble globular conformation (supplemental Fig. S1).
Proteolytically generated Fn fragments can contain minor levels of contamination with other Fn fragments. Thus, we also tested purified recombinant Fn constructs with various N-terminal and C-terminal end points for Ail binding. These His6-tagged recombinant Fn proteins were generated previously from cultured insect SF9 or High 5 cells to allow appropriate eukaryotic modifications and purified via nickel-agarose (40).
Consistent with Ail binding to the 120-kDa proteolytically generated Fn Fragment, E. coli expressing Ail also bound to the 1FNIII-C fragment, containing Fn sequences from 1FNIII to the C terminus (Fig. 1, A and B). Furthermore, Ail did not facilitate E. coli binding to the most N-terminal fragment, N-5FNI, nor to the larger N-terminal fragment, N-3FNIII, containing sequences from the N terminus of Fn to 3FNIII (Fig. 1, A and B). 2FnIII-3FNIII are present in the 120-kDa fragment, thus, these data provide evidence that Ail does not bind this region of the 120-kDa domain. Again, these data support the position of the Ail binding site in the 120-kDa fragment of Fn.
To ensure that the N-terminal fragments of Fn were competent for bacterial binding, we demonstrated that L. lactis strains expressing FnBPA from S. aureus were able to mediate binding to proteolytically generated or recombinant Fn derivatives containing the N terminus of Fn (Fig. 1C). Thus, the N-terminal Fn derivatives were competent to support bacterial adhesion by bacterial proteins known to bind the N-terminal domain of Fn (50).
We also performed dose-response binding curves to full-length Fn and various Fn fragments and showed all fragments capable of supporting binding reached maximal binding at a coating concentration of 10–20 μg/ml (supplemental Fig. S2).
Antibodies to Regions in 120-kDa Fragment Block Ail Binding
As the Fn binding data indicated Ail binds to the 120-kDa fragment, we predicted that antibodies directed against this region would block Ail-mediated bacterial binding. To obtain polyclonal antibodies enriched for those reactive against the 120-kDa or 70-kDa Fn fragments, we preadsorbed a commercially available polyclonal anti-Fn antibody to the N-terminal 70-kDa Fn fragment, central 120-kDa fragment, or full-length Fn to remove antibodies that bind those fragments. Remaining unbound antibodies from each treatment were tested for inhibition of Ail-mediated bacterial binding to immobilized full-length Fn. Without any antibodies present, bacteria expressing Ail bound to immobilized Fn well, and preincubation with the nonadsorbed polyclonal anti-Fn antibody (at 20 μg/ml) led to ∼60% inhibition of Ail-mediated binding to immobilized full-length Fn (supplemental Fig. S3), as seen previously (2). Anti-Fn antibodies preadsorbed to the 70-kDa fragment (enriched for anti-120-kDa antibodies) were still able to block Ail-mediated binding, similar to nonadsorbed anti-Fn antibody, whereas antibodies preadsorbed to the 120-kDa Fn fragment (enriched for anti-70-kDa antibodies) did not block Ail-mediated binding (supplemental Fig. S3). Thus, antibodies capable of blocking Ail-mediated cell binding map to the 120-kDa fragment. Antibodies preadsorbed to full-length immobilized plasma Fn were unable to block Ail-mediated binding to Fn, as expected. This commercially available polyclonal anti-Fn antibody does contain antibodies that recognize various epitopes along the entire length of the Fn molecule including the N terminus (supplemental Fig. S4).
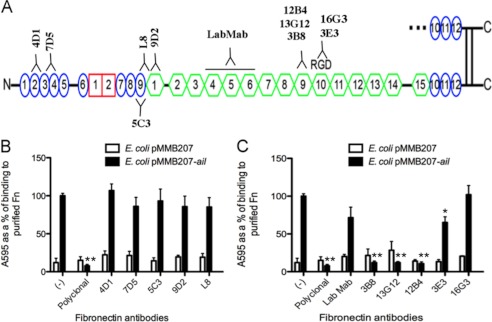
To define the Ail binding site on Fn further, we utilized monoclonal antibodies that bind previously mapped regions of Fn (Fig. 2A) (28, 41, 42, 44, 45). Plastic microtiter wells were coated with full-length plasma Fn and preincubated with 40 μg/ml (purified) or a 1:500 dilution (ascites) of the monoclonal antibodies before the addition of E. coli expressing Ail or empty vector. Without antibody addition, Ail mediated efficient binding to immobilized plasma Fn (set to 100%) whereas the polyclonal anti-Fn antibody completely blocked Ail binding (Fig. 2, B and C). Ail-mediated binding to Fn was completely inhibited by three monoclonal antibodies (3B8, 13G12, and 12B4; Fig. 2C). Based on their recognition of recombinant Fn fragments of variable length, these antibodies were deduced to bind to module 9FNIII (within the 120-kDa fragment of Fn) (42). The 3E3 and 16G3 antibodies, which map to module 10FNIII (42, 44, 45), and LabMab, which maps within 3–7FNIII (41), showed modest or no inhibiting activity (Fig. 2C). None of the antibodies directed against type I or early type III repeats blocked Ail-mediated binding (Fig. 2B) (28). These data indicate that Ail binds within module 9FNIII, a unique binding site for characterized bacterial Fn-binding proteins.
FIGURE 2.
Monoclonal antibodies to regions in 120-kDa fragment block Ail binding. A, binding sites for the antibodies used in this study are indicated. Plasma Fn was adsorbed onto plastic wells (10 μg/ml). Antibodies directed against the N terminus of FN (B) and more central region (C) were added to Fn-coated wells 1 h prior to the addition of E. coli AAEC185 derivatives expressing empty vector and Ail. Bacteria were allowed to bind for an additional 1 h at 37 °C. Bound bacteria were stained with 0.01% crystal violet. The cells and crystal violet were solubilized and washed, and the cells were solubilized and read at A595. E. coli expressing Ail without antibody treatment was set at 100% to normalize bacteria binding across multiple experiments (n = 6–9). *, p < 0.0005; **, p < 5 × 10−12 relative to Ail-mediated binding with no mAb treatment (−). Error bars, S.D.
Fine Epitope Mapping of Monoclonal Antibodies within Modules 9–10FNIII
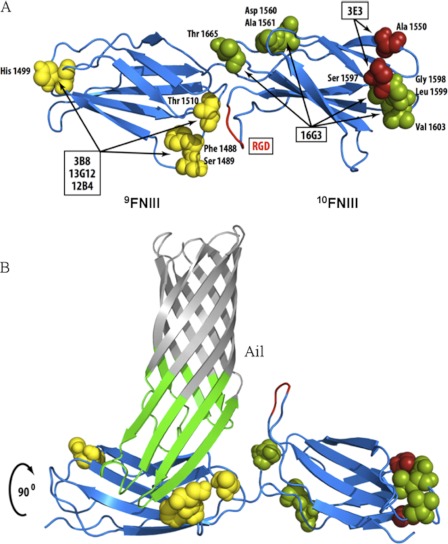
We next sought to map more finely the binding site for Ail on Fn. The three antibodies that inhibited Ail binding to Fn (3B8, 13G12, and 12B4) had previously been mapped to module 9FNIII, with 13G12 and 12B4 competing with one another for Fn binding (42). Using Fn molecules from five different species (human, mouse, bovine, rat, and chicken), we determined the reactivity of each antibody by direct ELISA (supplemental Fig. S5). The 13G12/12B4 and 3B8 antibodies that blocked Ail-mediated adhesion to Fn recognized only human Fn, narrowing their recognition epitopes to three potential residue clusters all on a similar face of 9FNIII in the crystal structure of 7–10FNIII (Fig. 3A and supplemental Fig. S5) (46). The noninhibitory antibody, 16G3, recognized human, bovine, and chicken Fn, defining epitopes that are either on the opposite face of 10FNIII from Ail or at the distal end of 10FNIII in the crystal structure (Fig. 3A and supplemental Fig. S4). The fact that antibody 3E3, previously mapped to 10FNIII (44, 45), recognized only human and bovine Fn indicates that binding by 3E3 requires residue Ala-1550 and/or Ser-1597, both of which are at the distal end of 10FNIII (Fig. 3A and supplemental Fig. S4). That both 16G3 and 3E3 block RGD-dependent cell adhesion, but showed only minimal inhibitory activity on Ail binding, further indicates that Ail binding to Fn is distinct from cell adhesion to 10FNIII. Thus, Ail appears to bind within 9FNIII on a similar face of the Fn molecule as the RGD in 10FNIII and the synergy region within 9FNIII that contributes to RGD-mediated cell adhesion by β1 integrins (Fig. 3B) (46, 51, 52).
FIGURE 3.
Defining binding sites of mAbs that map to 9–10FNIII. A, based on unique residues in Fn from five different species and the reactivity of each mAb, potential interaction residues in 9–10FNIII are proposed (see also supplemental Fig. S5). B, using OmpX of E. coli as an Ail homolog (62), a model of Ail binding to 9FNIII in a region overlapping the synergy region (51) but exposing the RGD domain is proposed. Repeats 7–10FNIII were structurally determined by Leahy et al. (46).
Antibodies That Interfere with Ail/Fn Interaction Inhibit Ail-mediated Cell Binding
Given that a major role for Ail bound to Fn is to facilitate cell binding and Yop delivery (2), we determined whether antibodies that block Ail-mediated Fn binding also affected binding to host cells. First, we assessed binding to HEp-2 cells in the presence of antibodies using Y. pestis strain KIM5 D27 Δpla. This strain lacks plasminogen activator (Pla), thus the remaining Fn binding activity is primarily Ail-dependent (2). Addition of mAbs 13G12 or 3B8 directed against 9FNIII blocked binding of KIM5 D27 Δpla to HEp-2 cells as expected, and they blocked binding with an efficiently similar to the polyclonal anti-Fn antibody (Fig. 4). Addition of both 13G12 and 3B8 in combination did not give any additional inhibition (Fig. 4). Antibodies to a more N-terminal region of Fn (5C3) did not inhibit Ail-mediated Y. pestis binding to HEp-2 cells, nor did the anti-10FNIII mAb, 3E3 (Fig. 4). Deletion of ail in the KIM5 D27 Δpla background reduced bacterial binding to 3% relative to the KIM5 D27 Δpla parent.
FIGURE 4.
Monoclonal antibodies can inhibit both Ail-mediated Y. pestis host cell binding and Yop delivery. Strain KIM5 D27 Δpla or ΔplaΔail was allowed to bind HEp-2 cells at a multiplicity of infection of 100 for 105 min at 37 °C 5% CO2. Bacterial binding was performed in the presence of 40 μg/ml antibody after allowing a 1-h preincubation with antibody. Unbound bacteria were washed off with PBS, and bound bacteria were released following cell lysis with 0.1% Triton X-100 and enumeration by cfu analysis. *, p = 0.056; **, p < 0.05. Error bars, S.D.
Ail Binds Minimal Fn Fragment Containing 9–10FNIII
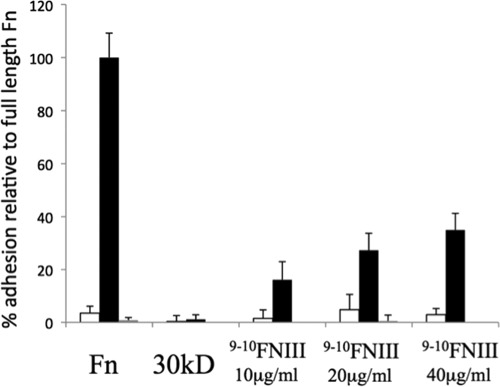
Because Ail binding to Fn is blocked by mAbs mapping to 9FNIII, we purified a derivative of human Fn harboring 9–10FNIII to determine whether it was sufficient to support Ail-mediated bacterial binding. Immobilization of this 193-amino acid fragment of Fn allowed E. coli expressing Ail to bind to microtiter wells, and binding was inhibited by the addition of mAb 13G12 (Fig. 5), which maps to 9FNIII. Thus, we have confirmed our antibody-based mapping of the Ail binding site to a minimal region of Fn, 9FNIII.
FIGURE 5.
Fn domains 9–10FNIII are sufficient to support Ail-mediated bacterial binding. Microtiter wells were coated with 20 μg/ml plasma Fn, a 30-kDa N-terminal fragment of Fn or increasing concentrations of 9–10FNIII. Binding was performed at 37 °C with E. coli AAEC185 expressing empty vector pMMB207 (open bars) or pMMB207-ail (black bars) for 90 min followed by washing with PBS, staining with crystal violet, and reading the bound bacterial by A595, after solubilization. Preincubation of the plates with the mAb 13G12 was performed for plasma Fn and 9–10FNIII at 20 μg/ml (gray bars) to block Ail-mediated binding. Error bars, S.D.
DISCUSSION
Ail interactions with Fn are important for Ail-mediated virulence functions such as host cell binding and Yop delivery (2). In this study, we sought to identify the Ail binding site on the Fn molecule to further understand this critical interaction. Proteolytically generated and recombinant Fn fragment binding studies and antibody inhibition assays indicated that Ail binds 9FNIII within the120-kDa fragment (Figs. 1–3 and 5). As most bacterial Fn-binding proteins characterized to date recognize FNI repeats near the N terminus of Fn (34, 53), binding within the 120-kDa domain represents an unusual, but not unprecedented (48, 54–56), mechanism of bacterial binding to Fn.
In the course of precisely defining the binding site of Ail on Fn, we epitope-mapped several anti-Fn monoclonal antibodies that inhibit Ail binding to Fn. Based on previous deletion mapping (42) and our epitope mapping of these mAbs, the three mAbs that inhibit Ail binding to Fn, 3B8, 13G12, and 12B4, bind to a similar face of 9FNIII (Fig. 3 and supplemental Fig. S4). Furthermore, they bind a similar face of Fn as the RGD cell adhesion motif in 10FNIII and the synergy region in 9FNIII (46, 51, 52). Two antibodies that map to the neighboring 10FNIII, 3E3 and 16G3, did not prevent Ail binding to Fn. Thus, the Ail binding site does not appear to extend into 10FNIII. Whereas inhibition of Ail-mediated binding to pure Fn was completely inhibited by anti-Fn antibodies (Fig. 2, B and C), binding to host cells was blocked less dramatically (40–50%, Fig. 4). This may be due to host cells synthesizing more Fn than is present in the purified Fn binding assays (thus substantially more Ab would be required for complete inhibition), the Fn deposited by cells being less accessible to anti-Fn antibodies and/or Ail having other substrates in addition to Fn on host cells. We have observed incomplete inhibition of Ail-mediated binding to HEp-2 cells previously (2). Laminin was recently shown to be a ligand for Ail, and laminin also contributes to Ail-host cell interactions (57). In this same study, the tip of extracellular loop 3 of Y. pestis Ail was disordered (57). This is of particular interest, as we have recently identified Phe-130 of Ail, present in the unstructured turn, as important for Ail-mediated binding to host cells.3 Thus, the third extracellular loop of Ail may obtain order upon interaction with Fn in a fashion similar to other β-zipper type Fn-binding proteins (33).
Given that Ail binds near the RGD of Fn and likely occludes the previously characterized synergy region in 9FNIII that is required for efficient RGD-dependent cell adhesion via β1 integrins (51, 52), Ail may present Fn to cells in a fashion that fails to generate the same cell signaling events associated with traditional Fn-β1 integrin interactions (58–60). Such altered signals may lead to activation of pathways required for efficient Ail-mediated type III secretion of Yop proteins, a process known to require specific cell signaling events (61). We demonstrate in this study that the RGD region of Fn does not contribute to Ail-mediated Fn binding (Fig. 4), but it may contribute to Ail-mediated Yop delivery. If the RGD region is required for Yop delivery, we hypothesize that either Ail utilizes the dimeric nature of Fn to bind one strand of Fn, leaving the other free to engage integrin α5β1, or RGD-mediated interaction with α5β1, even in the absence of contributions from the synergy region (which would be occluded by Ail binding to 9FNIII), generates sufficient signals for type III secretion of Yops. Differential signaling requirements for high affinity invasin-mediated cell invasion versus Yop delivery have been demonstrated previously (61). The contribution of the RGD region and specific integrin receptors in Ail-mediated Yop delivery will be the subject of future studies.
The findings presented here demonstrate that Ail binds a unique region of Fn, 9FNIII, for bacterial Fn-binding proteins. Upon binding this region, Ail then presents Fn to host cell receptors to facilitate Yop delivery (2), a process required for plague pathogenesis.
Supplementary Material
Acknowledgments
We thank Dr. Kenneth Yamada for providing anti-Fn monoclonal antibodies, 3B8, 13G12, 12B4, and 16G3, mapping to the 120-kDa domain and clone pRSET-C' (XhoI−) containing repeats 9–10FNIII of human Fn (43); Dr. Ruth Massey for the kind gifts of L. lactis strains SP9 and SP18; and Dr. Victor DiRita for providing helpful comments on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant R01HL021644 through the NHLBI (to D. F. M.) and Grants R21AI090194 and R03AI092318 through the NIAID (to E. S. K.). This work was also supported by the Office of the Vice President of Research (OVPR) at the University of Michigan (to E. S. K.) and the Frances Wang Chin Endowed Fellowship from the Department of Microbiology and Immunology at the University of Michigan (to T. M. T.).

This article contains supplemental Figs. S1–S5, Table S1, and additional references.
T. M. Tsang and E. S. Krukonis, unpublished data.
- Fn
- fibronectin
- FnBPA
- Fn-binding protein A
- FNI
- FNII, FNIII, types I, II, and III Fn repeats, respectively
- IPTG
- isopropyl-β-d-thiogalactopyranoside.
REFERENCES
- 1. Felek S., Krukonis E. S. (2009) The Yersinia pestis Ail protein mediates binding and Yop delivery to host cells required for plague virulence. Infect. Immun. 77, 825–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tsang T. M., Felek S., Krukonis E. S. (2010) Ail binding to fibronectin facilitates Yersinia pestis binding to host cells and Yop delivery. Infect. Immun. 78, 3358–3368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rosqvist R., Forsberg A., Rimpiläinen M., Bergman T., Wolf-Watz H. (1990) The cytotoxic protein YopE of Yersinia obstructs the primary host defence. Mol. Microbiol. 4, 657–667 [DOI] [PubMed] [Google Scholar]
- 4. Cornelis G. R., Boland A., Boyd A. P., Geuijen C., Iriarte M., Neyt C., Sory M. P., Stainier I. (1998) The virulence plasmid of Yersinia, an antihost genome. Microbiol. Mol. Biol. Rev. 62, 1315–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Galán J. E., Wolf-Watz H. (2006) Protein delivery into eukaryotic cells by type III secretion machines. Nature 444, 567–573 [DOI] [PubMed] [Google Scholar]
- 6. Kolodziejek A. M., Schnider D. R., Rohde H. N., Wojtowicz A. J., Bohach G. A., Minnich S. A., Hovde C. J. (2010) Outer membrane protein X (Ail) contributes to Yersinia pestis virulence in pneumonic plague and its activity is dependent on the lipopolysaccharide core length. Infect. Immun. 78, 5233–5243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hinnebusch B. J., Jarrett C. O., Callison J. A., Gardner D., Buchanan S. K., Plano G. V. (2011) Role of the Yersinia pestis Ail protein in preventing a protective polymorphonuclear leukocyte response during bubonic plague. Infect. Immun. 79, 4984–4989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mosher D. F. (1984) Physiology of fibronectin. Annu. Rev. Med. 35, 561–575 [DOI] [PubMed] [Google Scholar]
- 9. Vaheri A., Ruoslahti E., Linder E., Wartiovaara J., Keski-Oja J., Kuusela P., Saksela O. (1976) Fibroblast surface antigen (SF): molecular properties, distribution in vitro and in vivo, and altered expression in transformed cells. J. Supramol. Struct. 4, 63–70 [DOI] [PubMed] [Google Scholar]
- 10. Ruoslahti E., Vaheri A. (1974) Novel human-serum protein from fibroblast plasma-membrane. Nature 248, 789–791 [DOI] [PubMed] [Google Scholar]
- 11. Kuusela P., Ruoslahti E., Engvall E., Vaheri A. (1976) Immunological interspecies cross-reactions of fibroblast surface antigen (fibronectin). Immunochemistry 13, 639–642 [DOI] [PubMed] [Google Scholar]
- 12. Mosesson M. W., Chen A. B., Huseby R. M. (1975) The cold-insoluble globulin of human plasma: studies of its essential structural features. Biochim. Biophys. Acta 386, 509–524 [DOI] [PubMed] [Google Scholar]
- 13. Pankov R., Yamada K. M. (2002) Fibronectin at a glance. J. Cell Sci. 115, 3861–3863 [DOI] [PubMed] [Google Scholar]
- 14. Hynes R. O. (1987) Integrins: a family of cell surface receptors. Cell 48, 549–554 [DOI] [PubMed] [Google Scholar]
- 15. Ruoslahti E. (1988) Fibronectin and its receptors. Annu. Rev. Biochem. 57, 375–413 [DOI] [PubMed] [Google Scholar]
- 16. Engvall E., Ruoslahti E. (1977) Binding of soluble form of fibroblast surface protein, fibronectin, to collagen. Int. J. Cancer 20, 1–5 [DOI] [PubMed] [Google Scholar]
- 17. Stathakis N. E., Mosesson M. W. (1977) Interactions among heparin, cold insoluble globulin, and fibrinogen in formation of the heparin precipitable fraction of plasma. J. Clin. Invest. 60, 855–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ruoslahti E., Hayman E. G., Engvall E., Cothran W. C., Butler W. T. (1981) Alignment of biologically active domains in the fibronectin molecule. J. Biol. Chem. 256, 7277–7281 [PubMed] [Google Scholar]
- 19. Ruoslahti E., Vaheri A. (1975) Interaction of soluble fibroblast surface antigen with fibrinogen and fibrin: identity with cold insoluble globulin of human plasma. J. Exp. Med. 141, 497–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Christopher R. A., Kowalczyk A. P., McKeown-Longo P. J. (1997) Localization of fibronectin matrix assembly sites on fibroblasts and endothelial cells. J. Cell Sci. 110, 569–581 [DOI] [PubMed] [Google Scholar]
- 21. Dzamba B. J., Bultmann H., Akiyama S. K., Peters D. M. (1994) Substrate-specific binding of the amino terminus of fibronectin to an integrin complex in focal adhesions. J. Biol. Chem. 269, 19646–19652 [PubMed] [Google Scholar]
- 22. Pierschbacher M. D., Hayman E. G., Ruoslahti E. (1981) Location of the cell-attachment site in fibronectin with monoclonal antibodies and proteolytic fragments of the molecule. Cell 26, 259–267 [DOI] [PubMed] [Google Scholar]
- 23. Hocking D. C., Sottile J., McKeown-Longo P. J. (1998) Activation of distinct α5β1-mediated signaling pathways by fibronectin's cell adhesion and matrix assembly domains. J. Cell Biol. 141, 241–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Patti J. M., Allen B. L., McGavin M. J., Höök M. (1994) MSCRAMM-mediated adherence of microorganisms to host tissues. Annu. Rev. Microbiol. 48, 585–617 [DOI] [PubMed] [Google Scholar]
- 25. Hanski E., Caparon M. (1992) Protein F, a fibronectin-binding protein, is an adhesin of the group A streptococcus Streptococcus pyogenes. Proc. Natl. Acad. Sci. U.S.A. 89, 6172–6176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ozeri V., Rosenshine I., Mosher D. F., Fässler R., Hanski E. (1998) Roles of integrins and fibronectin in the entry of Streptococcus pyogenes into cells via protein F1. Mol. Microbiol. 30, 625–637 [DOI] [PubMed] [Google Scholar]
- 27. Ozeri V., Tovi A., Burstein I., Natanson-Yaron S., Caparon M. G., Yamada K. M., Akiyama S. K., Vlodavsky I., Hanski E. (1996) A two-domain mechanism for group A streptococcal adherence through protein F to the extracellular matrix. EMBO J. 15, 989–998 [PMC free article] [PubMed] [Google Scholar]
- 28. Maurer L. M., Tomasini-Johansson B. R., Ma W., Annis D. S., Eickstaedt N. L., Ensenberger M. G., Satyshur K. A., Mosher D. F. (2010) Extended binding site on fibronectin for the functional upstream domain (FUD) of protein F1 of Streptococcus pyogenes. J. Biol. Chem. 285, 41087–41099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mosher D. F., Proctor R. A. (1980) Binding and factor XIIIa-mediated cross-linking of a 27-kilodalton fragment of fibronectin to Staphylococcus aureus. Science 209, 927–929 [DOI] [PubMed] [Google Scholar]
- 30. Fröman G., Switalski L. M., Speziale P., Höök M. (1987) Isolation and characterization of a fibronectin receptor from Staphylococcus aureus. J. Biol. Chem. 262, 6564–6571 [PubMed] [Google Scholar]
- 31. Sottile J., Schwarzbauer J., Selegue J., Mosher D. F. (1991) Five type I modules of fibronectin form a functional unit that binds to fibroblasts and Staphylococcus aureus. J. Biol. Chem. 266, 12840–12843 [PubMed] [Google Scholar]
- 32. Massey R. C., Kantzanou M. N., Fowler T., Day N. P., Schofield K., Wann E. R., Berendt A. R., Höök M., Peacock S. J. (2001) Fibronectin-binding protein A of Staphylococcus aureus has multiple, substituting, binding regions that mediate adherence to fibronectin and invasion of endothelial cells. Cell. Microbiol. 3, 839–851 [DOI] [PubMed] [Google Scholar]
- 33. Schwarz-Linek U., Werner J. M., Pickford A. R., Gurusiddappa S., Kim J. H., Pilka E. S., Briggs J. A., Gough T. S., Höök M., Campbell I. D., Potts J. R. (2003) Pathogenic bacteria attach to human fibronectin through a tandem β-zipper. Nature 423, 177–181 [DOI] [PubMed] [Google Scholar]
- 34. Schwarz-Linek U., Höök M., Potts J. R. (2004) The molecular basis of fibronectin-mediated bacterial adherence to host cells. Mol. Microbiol. 52, 631–641 [DOI] [PubMed] [Google Scholar]
- 35. Kuusela P., Vartio T., Vuento M., Myhre E. B. (1985) Attachment of staphylococci and streptococci on fibronectin, fibronectin fragments, and fibrinogen bound to a solid phase. Infect. Immun. 50, 77–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sakata N., Jakab E., Wadström T. (1994) Human plasma fibronectin possesses second binding site(s) to Staphylococcus aureus on its C-terminal region. J. Biochem. 115, 843–848 [DOI] [PubMed] [Google Scholar]
- 37. Bozzini S., Visai L., Pignatti P., Petersen T. E., Speziale P. (1992) Multiple binding sites in fibronectin and the staphylococcal fibronectin receptor. Eur. J. Biochem. 207, 327–333 [DOI] [PubMed] [Google Scholar]
- 38. Tertti R., Skurnik M., Vartio T., Kuusela P. (1992) Adhesion protein YadA of Yersinia species mediates binding of bacteria to fibronectin. Infect. Immun. 60, 3021–3024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Eitel J., Dersch P. (2002) The YadA protein of Yersinia pseudotuberculosis mediates high-efficiency uptake into human cells under environmental conditions in which invasin is repressed. Infect. Immun. 70, 4880–4891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Xu J., Maurer L. M., Hoffmann B. R., Annis D. S., Mosher D. F. (2010) iso-DGR sequences do not mediate binding of fibronectin N-terminal modules to adherent fibronectin-null fibroblasts. J. Biol. Chem. 285, 8563–8571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chernousov M. A., Fogerty F. J., Koteliansky V. E., Mosher D. F. (1991) Role of the I-9 and III-1 modules of fibronectin in formation of an extracellular fibronectin matrix. J. Biol. Chem. 266, 10851–10858 [PubMed] [Google Scholar]
- 42. Nagai T., Yamakawa N., Aota S., Yamada S. S., Akiyama S. K., Olden K., Yamada K. M. (1991) Monoclonal antibody characterization of two distant sites required for function of the central cell binding domain of fibronectin in cell adhesion, cell migration, and matrix assembly. J. Cell Biol. 114, 1295–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Akiyama S. K., Aota S., Yamada K. M. (1995) Function and receptor specificity of a minimal 20-kilodalton cell-adhesive fragment of fibronectin. Cell Adhes. Commun. 3, 13–25 [DOI] [PubMed] [Google Scholar]
- 44. Pierschbacher M. D., Ruoslahti E., Sundelin J., Lind P., Peterson P. A. (1982) The cell attachment domain of fibronectin: determination of the primary structure. J. Biol. Chem. 257, 9593–9597 [PubMed] [Google Scholar]
- 45. Pierschbacher M. D., Ruoslahti E. (1984) Cell attachment activity of fibronectin can be duplicated by small synthetic fragments of the molecule. Nature 309, 30–33 [DOI] [PubMed] [Google Scholar]
- 46. Leahy D. J., Aukhil I., Erickson H. P. (1996) 2.0 Å crystal structure of a four-domain segment of human fibronectin encompassing the RGD loop and synergy region. Cell 84, 155–164 [DOI] [PubMed] [Google Scholar]
- 47. Kuusela P., Vartio T., Vuento M., Myhre E. B. (1984) Binding sites for streptococci and staphylococci in fibronectin. Infect. Immun. 45, 433–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Thomas D. D., Baseman J. B., Alderete J. F. (1985) Fibronectin mediates Treponema pallidum cytadherence through recognition of fibronectin cell binding domain. J. Exp. Med. 161, 514–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Morales V. M., Bäckman A., Bagdasarian M. (1991) A series of wide-host-range low-copy-number vectors that allow direct screening for recombinants. Gene 97, 39–47 [DOI] [PubMed] [Google Scholar]
- 50. Bingham R. J., Rudiño-Piñera E., Meenan N. A., Schwarz-Linek U., Turkenburg J. P., Höök M., Garman E. F., Potts J. R. (2008) Crystal structures of fibronectin binding sites from Staphylococcus aureus FnBPA in complex with fibronectin domains. Proc. Natl. Acad. Sci. U.S.A. 105, 12254–12258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Obara M., Kang M. S., Yamada K. M. (1988) Site-directed mutagenesis of the cell binding domain of human fibronectin: separable, synergistic sites mediate adhesive function. Cell 53, 649–657 [DOI] [PubMed] [Google Scholar]
- 52. Bowditch R. D., Halloran C. E., Aota S., Obara M., Plow E. F., Yamada K. M., Ginsberg M. H. (1991) Integrin αIIbβ3 (platelet GPIIb-IIIa) recognizes multiple sites in fibronectin. J. Biol. Chem. 266, 23323–23328 [PubMed] [Google Scholar]
- 53. Joh D., Wann E. R., Kreikemeyer B., Speziale P., Höök M. (1999) Role of fibronectin-binding MSCRAMMs in bacterial adherence and entry into mammalian cells. Matrix Biol. 18, 211–223 [DOI] [PubMed] [Google Scholar]
- 54. Dawson J. R., Ellen R. P. (1990) Tip-oriented adherence of Treponema denticola to fibronectin. Infect. Immun. 58, 3924–3928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Visai L., Bozzini S., Petersen T. E., Speciale L., Speziale P. (1991) Binding sites in fibronectin for an enterotoxigenic strain of E. coli B342289c. FEBS Lett. 290, 111–114 [DOI] [PubMed] [Google Scholar]
- 56. Westerlund B., Kuusela P., Vartio T., van Die I., Korhonen T. K. (1989) A novel lectin-independent interaction of P fimbriae of Escherichia coli with immobilized fibronectin. FEBS Lett. 243, 199–204 [DOI] [PubMed] [Google Scholar]
- 57. Yamashita S., Lukacik P., Barnard T. J., Noinaj N., Felek S., Tsang T. M., Krukonis E. S., Hinnebusch B. J., Buchanan S. K. (2011) Structural insights into Ail-mediated adhesion in Yersinia pestis. Structure 19, 1672–1682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Nojima Y., Morino N., Mimura T., Hamasaki K., Furuya H., Sakai R., Sato T., Tachibana K., Morimoto C., Yazaki Y. (1995) Integrin-mediated cell adhesion promotes tyrosine phosphorylation of p130Cas, a Src homology 3-containing molecule having multiple Src homology 2 binding motifs. J. Biol. Chem. 270, 15398–15402 [DOI] [PubMed] [Google Scholar]
- 59. Bellis S. L., Perrotta J. A., Curtis M. S., Turner C. E. (1997) Adhesion of fibroblasts to fibronectin stimulates both serine and tyrosine phosphorylation of paxillin. Biochem. J. 325, 375–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kornberg L., Earp H. S., Parsons J. T., Schaller M., Juliano R. L. (1992) Cell adhesion or integrin clustering increases phosphorylation of a focal adhesion-associated tyrosine kinase. J. Biol. Chem. 267, 23439–23442 [PubMed] [Google Scholar]
- 61. Mejía E., Bliska J. B., Viboud G. I. (2008) Yersinia controls type III effector delivery into host cells by modulating Rho activity. PLoS Pathog. 4, e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Vogt J., Schulz G. E. (1999) The structure of the outer membrane protein OmpX from Escherichia coli reveals possible mechanisms of virulence. Structure 7, 1301–1309 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.