Background: Endoproteolytic α-cleavage of cellular prion protein (PrPC) regulates PrPC toxicity and functions; the responsible protease(s) is uncertain.
Results: ADAM8 performs α-cleavage of PrPC and PrPC overexpression up-regulates ADAM8 in muscle.
Conclusion: ADAM8 is the primary protease for the α-cleavage of PrPC that appears self-regulated through ADAM8 in muscle.
Significance: This advances our understandings on physiological processing and functions of PrPC.
Keywords: Metalloprotease, Muscle, Prions, Protein Processing, Transgenic Mice, ADAM8, Prion Protein
Abstract
The ubiquitously expressed cellular prion protein (PrPC) is subjected to the physiological α-cleavage at a region critical for both PrP toxicity and the conversion of PrPC to its pathogenic prion form (PrPSc), generating the C1 and N1 fragments. The C1 fragment can activate caspase 3 while the N1 fragment is neuroprotective. Recent articles indicate that ADAM10, ADAM17, and ADAM9 may not play a prominent role in the α-cleavage of PrPC as previously thought, raising questions on the identity of the responsible protease(s). Here we show that, ADAM8 can directly cleave PrP to generate C1 in vitro and PrP C1/full-length ratio is greatly decreased in the skeletal muscles of ADAM8 knock-out mice; in addition, the PrP C1/full-length ratio is linearly correlated with ADAM8 protein level in myoblast cell line C2C12 and in skeletal muscle tissues of transgenic mice. These results indicate that ADAM8 is the primary protease responsible for the α-cleavage of PrPC in muscle cells. Moreover, we found that overexpression of PrPC led to up-regulation of ADAM8, suggesting that PrPC may regulate its own α-cleavage through modulating ADAM8 activity.
Introduction
The cellular prion protein (PrPC)3 is a ubiquitous glycosylphophatidylinositol (GPI)-anchored glycoprotein that is highly expressed in the nervous system (1). PrP is the central factor in prion diseases, a group of fatal neurodegenerative disorders characterized by severe neuronal dysfunction and loss, spongiosis and accumulation of pathogenic prion protein (PrPSc) that is converted from PrPC (2). PrP is also implicated in Alzheimer disease (3–5) and cancer (6). The physiological roles of PrPC remain elusive, but many normal functions have been proposed for PrPC (6–9). These include signal transduction (10), resistance to oxidative stress, anti-apoptosis, neuroprotection (11), cell adhesion, neurogenesis, axonal growth, neurite outgrowth and neuritogenesis, neuronal differentiation, hematopoietic stem cell self renewal, lymphocyte activation, and metal ion trafficking (12–13).
PrP is differentially cleaved in normal and prion-affected brains. In the brains of Creutzfeldt-Jakob disease subjects (14–15) and prion-affected animals (16–17) and in prion-infected cells (18), PrP is cleaved around the end of the octapeptide repeats (termed β-cleavage) to generate the C2 and N2 fragments (14, 19–21). β-Cleavage preserves the cytotoxic and fibrillogenic PrP106–126 core that is also critical for the conversion of PrPC to PrPSc (22–25). β-Cleavage of PrP is mediated by reactive oxygen species in CHO cells (26) and human neuroblastoma SH-SY5Y cells (27). The identity of cellular proteases involved in C2 production remains unclear. Calpain was shown to be critical for C2 production in scrapie-infected mouse brain cells (18), and the contributions of cysteine proteases seem to depend on the cell models (28–30). Both C2 and N2 fragments appear to be biologically inert (31–32). Nevertheless, the β-cleavage of PrPC was reported to be critical for the anti-oxidative and neuroprotective effect of PrPC (27, 33). PrPC can also be cleaved and shedded directly by ADAM10, but not ADAM9, at a site near the GPI anchor (34).
In addition, PrPC undergoes a physiological endoproteolytic cleavage at the 110/111 or 111/112 peptide bond (termed α-cleavage) (15, 21), yielding the C-terminal C1 fragment tethered to the plasma membrane (14–15, 19, 35–36) and releasing the corresponding N-terminal N1 fragment (37–39). The α-cleavage of PrPC is stimulated by protein kinase C agonists (37) and takes place mostly in a late compartment of the secretory pathway (40). The α-cleavage disrupts the PrP106–126 region critical for both PrP toxicity and PrPC to PrPSc conversion (22–25). In addition, both products of PrPC α-cleavage are biologically active: the N1 fragment is neuroprotective in vitro and in vivo by modulating the p53 pathway (31) while the C1 fragment potentiates staurosporine-induced caspase 3 activation in the HEK293 cell line (32).
ADAMs (A Disintegrin And Metalloproteinase) is a family of transmembrane peptidases with a unique multidomain organization, including a prodomain, a proteolytic domain (metalloprotease) that sheds ectodomains of membrane-anchored cell surface proteins and cleaves extracellular matrix proteins (ECMs), adhesive domains (including a disintegrin domain that binds to integrin and a cysteine-rich domain that binds to heparin sulfate proteoglycans) that interact with ECMs, an EGF-like domain, a transmembrane domain, and a cytoplasmic tail that modulates the sheddase activity (41–42). The substrates for the ADAM sheddases include Notch, growth factors (such as EGF), cytokines (such as TNF-α, TRANCE) and their receptors (such as TNF receptors I and II, NGF receptor, IL-1 receptor, and IL-6 receptor), implicating a critical role for ADAMs in extracellular signaling events (41–43). ADAMs can also cleave adhering molecules (such as cadherins) and ECMs (such as fibronectin and laminin), thereby promoting cell migration and releasing ECM-bound growth factors for signaling (41–42).
Three ADAMs have been implicated in the α-cleavage of PrPC. In HEK293 cells, ADAM10 appears to participate in the constitutive formation of C1 (37–38) while ADAM17 seems responsible for protein kinase C-dependent formation of C1 (38, 44). ADAM9 was also reported to indirectly participate in C1 formation by modulating ADAM10 activity in HEK293 cells, mouse fibroblasts and TSM1 neurons (38–39). In addition, one article associates high levels of C1 with the presence of active ADAM10 in the human brain, but other ADAMs were not examined (45). However, more recent articles showed that overexpression of ADAMs 9, 10, and 17 and depletion of ADAMs 9 and 10 failed to change the levels of C1 in HEK cell lysates (34) and neuronal overexpression of ADAM10 influenced the amount of PrPC instead of its processing in vivo (46). Furthermore, Altmeppen et al. (47) reported that ADAM10 is not responsible for the α-cleavage of PrPC in neurons using neuron-specific ADAM10 knock-out mice. These reports suggest the involvement of an unidentified protease(s) in the α-cleavage of PrPC. PrPC is expressed at significant levels (48–49) and implicated in physiological and pathological processes in skeletal muscles.
On the one hand, skeletal muscles in PrP-null mice exhibited enhanced oxidative damage (50) and diminished tolerance for physical exercise (51). In addition, fast muscle fibers, which during exercise undergo very active oxidative phosphorylation and produce more reactive oxygen species, present a higher level of PrPC than slow fibers (52). This evidence suggests a protective role for PrPC. Moreover, PrPC is up-regulated when primary or immortalized myoblasts differentiate into myotubes (52–53), and it promotes regeneration of adult muscle tissues through the stress-activated p38 pathway (54). These data associate PrPC with muscle differentiation and regeneration.
On the other hand, skeletal muscles showed elevated levels of PrP in patients with sporadic and hereditary inclusion body myositis (55–56), polymyositis, dermatomyositis, and neurogenic muscle atrophy (57). In addition, transgenic (Tg) mice constitutively overexpressing wild type PrPs from hamster, sheep, or mice developed myopathy in aged animals (58). We also found that induced overexpression of wild type human PrP in the skeletal muscles of Tg(HQK) mice led to a primary myopathy that is correlated with preferential accumulation of C1 (59) and accompanied by activation of the p53-dependent apoptosis pathway (60), suggesting the involvement of C1 and p53 in PrP-mediated myopathy. However, the detailed pathogenic mechanism of the muscle diseases induced by overexpressed wild type PrPC is still unclear. What protease(s) performs the α-cleavage of PrPC and how it is regulated in the skeletal muscles are also unknown.
Here we present evidence to show that (1) ADAM8 can directly perform α-cleavage of PrP in vitro, (2) ADAM8 plays a major role in the α-cleavage of PrPC in muscle cells, and (3) overexpression of PrPC leads to elevated ADAM8 levels in muscle, suggesting a feedback loop where PrPC regulates its own α-cleavage through up-regulation of ADAM8.
EXPERIMENTAL PROCEDURES
Transgenic and Knock-out Mice
The Tg(HQK) mice that show skeletal muscle-specific, doxycycline (Dox)-inducible expression of human PrPC were described previously (59). The Tg43, Tg4, Tg21, and Tg17 mice constitutively expressing human PrP were created essentially as described for the Tg40 mice (61). The Tga20 mice overexpressing mouse PrP at ∼8-fold were kindly provided by Charles Weissmann at Scripps Florida. The wild type FVB and C57BL6 mice were from the Jackson Laboratory. The ADAM8 knock-out mice (62) were kindly provided by Carl Blobel at Hospital for Special Surgery and Weill Medical College of Cornell University with permission from Andy Docherty from UCB Celltech.
Animal Treatment and Specimen Collection
For Tg(HQK) and wild type FVB (Wt) mice, 8-week-old females were fed food pellets either lacking or containing 6g Dox/kg food (Bio-Serv, Frenchtown, NJ) to induce PrPC expression, and skeletal muscles from the quadriceps of hind legs were removed after 0–60 days of Dox treatment. For Tg43, Tg4, Tg21, and Tg17 mice, skeletal muscles from quadriceps of hind legs of 2-month-old female mice were directly removed. For ADAM8 knock-out mice and control C57BL6 mice, skeletal muscles from quadriceps of hind legs of 3–5-month-old mice were directly removed. The collected muscle tissues were immediately frozen on dry ice and stored at −80 °C before analysis.
Cell Culture and Transfection
Proliferating mouse C2C12 myoblasts were maintained in growth medium (Dulbecco's modified Eagle's medium (Invitrogen, Carlsbad, CA), supplemented with 10% fetal bovine serum (Atlanta Biologicals), 100 μg/ml penicillin (Invitrogen, Carlsbad, CA), 100 units/ml streptomycin (Invitrogen)) in 5% CO2 in a humid incubator at 37 °C. To minimize spontaneous differentiation, cells were always kept in subconfluent (<70%) conditions. Transfection was performed using Effectene (Qiagen, Valencia, CA) according to the manufacturer's instructions.
PrP Expression in C2C12 Cells
Mouse PrP (MoPrP) ORF sequence was amplified by PCR from mouse genomic DNA with primers ENS-PRPO-F (GAGAATTCGCGGCCGCGGTCATYATGGCGAACCTTGG, Y = C+T) and PRPO-Bam-R (CGGGATCCTCATCCCACKATCAGGAAG, K = T+G) and cloned into pCEP4 vector (Invitrogen) to obtain pCEP-MoPrP. C2C12 cells were transfected with pCEP-MoPrP and selected with 300 μg/μl hygromycin B (Invitrogen) to obtain several stable cell clones expressing MoPrP.
RNAi Knockdown of ADAM8 in C2C12 Cells
The Invitrogen siRNA design algorithm (BLOCK-IT RNAi Designer) was utilized to design two 64-mer miR RNAi sequences targeting the ORF region of mouse ADAM8: NM-1241 (TGCTGTCTCCATGCTCCACAAACAGGGTTTTGGCCACTGACTGACCCTGTTTGGAGCATGGAGA) and NM-1741 (TGCTGCAATGTTGCTGCCTGTGCCAAGTTTTGGCCACTGACTGACGTTTGTGGAGCATGGAGAC). These 64-mer sequences were cloned into the pcDNA 6.2-GW/EmGFP-miR vector (Invitrogen) to obtain the ADAM8 miR RNAi constructs, which were then transfected into the MoPrP-expressing C2C12 clones. The transfected cells were selected with blasticidin S (Invitrogen) and multiple cell clones were obtained. The ADAM8 protein levels in the RNAi clones were then examined by Western blot (see below).
Western Blot Analysis
Mouse skeletal muscle tissues were homogenized in a tissue lysis buffer containing 50 mm Tris/HCl (pH 7.5), 200 mm sodium chloride, 0.5% sodium deoxycholate, and 5 mm EDTA supplemented with a protease inhibitor mixture (Roche, Indianapolis, IN). C2C12 cells grown in 35-mm dishes were washed once with phosphate-buffered saline (PBS) and lysed with 500 μl of a cell lysis buffer containing 10 mm Tris/HCl (pH 7.5), 150 mm NaCl, 0.5% Triton X-100, 0.5% deoxycholate, and 5 mm EDTA supplemented with a protease inhibitor mixture (Roche, Indianapolis, IN). Total protein concentrations in the tissue homogenate and cell lysate were determined by the BCA protein assay (Pierce). For C1 and full-length PrP detection, the samples were first deglycosylated with 10,000 units/ml PNGase F (New England Biolabs, Ipswich, MA) following the manufacturer's instructions except for incubation at 37 °C overnight. Then 20 μg of total proteins for each sample were separated by SDS-PAGE on a 10–20% Criterion Tris-Triton Precast gel (Bio-Rad), transferred onto PVDF membranes under 360 mA for 90 min, incubated with the 8H4 antibody (for residues 176–186 of human PrP or residues 175–185 of mouse PrP) (63) (1:5000 diluted in 0.5% normal goat serum, 1× TBS and 0.05% Tween) at 4 °C with gentle shaking overnight, followed by incubation with sheep anti-mouse IgG (Amersham Biosciences, Buckinghamshire, UK). For ADAM8 protein detection, 25 μg of total proteins were separated by SDS-polyacrylamide gel electrophoresis on a 10% Criterion Tris-HCl Precast gel (Bio-Rad), transferred onto a PVDF membrane for 90 min at 380 mA, incubated with the M-80 polyclonal rabbit anti-ADAM8 antibody raised against residues 1–80 of mouse ADAM8 (Santa Cruz Biotechnology) (1:1000 diluted in 1% nonfat milk, 1× TBS and 0.05% Tween), followed by incubation with a donkey anti-rabbit IgG (Amersham Biosciences). After stripping the blots with a stripping buffer containing 1.4% 2-mercaptomethanol, 2% SDS, and 62.5 mm Tris (pH 6.8), actin was probed with a rabbit polyclonal anti-skeletal muscle actin antibody (Abcam, Cambridge, MA) (1:5000 diluted in 1% milk, 1× TBS, and 0.05% Tween) for skeletal muscle samples or with a rabbit polyclonal anti-β-actin antibody (Cell Signaling, Boston, MA) (1:2500 diluted in 1% milk, 1× TBS and 0.05% Tween) for C2C12 cell samples. The blots were developed with ECL Western blotting Detection Reagents (Amersham Biosciences) according to the manufacturer's instructions. X-ray films were exposed to the blots, developed, scanned, and the bands quantified with the UN-SCAN-IT gel 6.1 software (Silk Scientific, Orem, Utah).
In Vitro ADAM8 Protease Activity Assay
Purified recombinant human ADAM8 (rhADAM8) (aa158–497), which contains a partial pro-peptide domain (aa158–199), the metalloprotease domain (aa200–400), and the disintegrin-like domain (aa408–494), was purchased from R&D Systems (Minneapolis, MN). First, the rhADAM8 was activated by incubating 2.0 μg of rhADAM8 with 7.5 ng of thermolysin (Sigma) at 37 °C for 30 min in 10 μl of an assay buffer containing 50 mm Tris, 10 mm CaCl2, 150 mm NaCl, pH 7.5; then 1.0 μl of 0.5 mm phosphoramidon (Sigma) was added and incubated at 25 °C for 15 min to stop the reaction. To assay for ADAM8 protease activity, 2.0 μg of recombinant human PrP (aa23–231 with an extra Gly-Ser at the N terminus) expressed and purified from Escherichia coli (64) was incubated with the activated rhADAM8 at 37 °C overnight in 50 μl of buffer containing 20 mm Tris-Cl, 5 mm CaCl2, 100 μm ZnCl2, 200 mm NaCl, pH 7.4. A fraction (1/10) of the cleavage products were separated by SDS-PAGE on a 10–20% Criterion Tris-Triton Precast gel (Bio-Rad), and subjected to Western blot analysis with the 8H4 antibody or 3F4 antibody. The rest of the human PrP cleavage products were separated on 10–20% Criterion Tris-Triton Precast SDS-PAGE gels for Coomassie Blue staining with Imperial Protein Stain (Thermo Scientific, Rockford, IL).
Mass Spectrometry Analysis
All protein samples were loaded onto a PepMap reverse-phase trapping column (C18, 100 μm × 2 cm) in a nano UltiMate-3000 Rapid Separation LC (Thermo Fisher Scientific, Bremen, Germany) to pre-concentrate and wash away excess salts. The samples were desalted for 10 min with water and 0.1% formic acid (FA) at the flow rate of 15 μl/min. Reverse-phase separation was then performed on an analytical C18 PepMap column (75 μm × 25 cm) using an 4%-to-95% acetonitrile gradient (containing water and 0.1% FA) in 90 min. Proteins eluting from the column at a flow rate of 400 nl/min were directed to a LTQ-FT mass spectrometer (Thermo Fisher Scientific) equipped with a nanospray ion source and with the needle voltage of 2.4 kV. Mass spectra were acquired in the positive ion mode in data-dependent experiments such as MS and tandem MS with the following acquisition cycle: a full scan recorded in the FT-ICR (Fourier transform ion cyclotron resonance) analyzer followed by MS/MS of the first most-intense peptide ions recorded in the FT-ICR mass analyzer at resolution R of 100,000. Sequences of all protein fragments were manually verified using a series of b- and y-fragment ions generated from FT-ICR tandem MS spectra.
Quantitative Real-time PCR
Total RNA was isolated from frozen skeletal muscle using the RNeasy skeletal muscle RNA isolation kit (Qiagen, Valencia, CA) following the manufacturer's specifications. The total RNA preparations were further treated with Turbo DNA-Free DNase (Ambion, Austin, TX) to remove residual genomic DNA contamination, and examined with a Bioanalyzer 2100 (Agilent Technologies, Santa Clara CA) for purity and quantity. Quantitative real-time PCR of ADAM gene expression was performed using probe specific TaqMan gene expression assays on the Applied Biosystems 7500 Fast Real-Time PCR System. 100 ng of total RNA was reverse transcribed using the High Capacity cDNA Reverse Transcription kit (Applied Biosystems, Foster City, CA). Subsequently, 1 μl from each reverse transcription reaction was assayed in a 20 μl single-plex reaction for real-time quantification with the 7500 Fast PCR System using probes specific to those genes of interest. Each sample was run in biological triplicate, of which three technical replicates were performed. GAPDH was used as the endogenous control, and the expression of target genes in Tg(HQK) mouse samples was quantitatively measured relative to the wild type FVB mouse samples. Relative quantification values were determined using the 2−ΔΔct method, and expressed as fold-change over the wild type FVB samples.
RESULTS
ADAM8 mRNA Level Is Up-regulated in Skeletal Muscle of Doxycycline-treated Tg(HQK) Mice
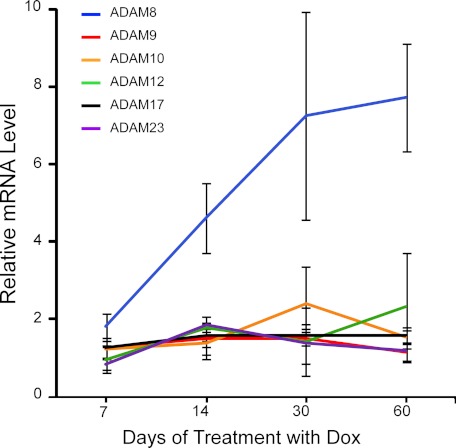
To investigate the protease(s) responsible for the α-cleavage of PrPC in skeletal muscles, Tg(HQK) mice with strictly Dox-dependent, skeletal muscle-specific expression of human PrP (59) were fed with Dox-laced food for 7, 14, 30, or 60 days; three animals were taken at each time point. Total RNA samples were isolated from skeletal muscle tissues (quadriceps) and subjected to quantitative real time PCR analysis for the three ADAMs already implicated in the α-cleavage of PrPC (ADAM10, ADAM17, and ADAM9) as well as three other ADAMs (ADAM8, ADAM12, and ADAM23). In Dox-treated wild type FVB mice, no significant changes were found for any of the six ADAMs (data not shown); in Dox-treated Tg(HQK) mice, ADAM8 mRNA level was significantly up-regulated whereas the other five ADAMs were largely unchanged (Fig. 1). These data indicate that, instead of the three previously implicated ADAMs (ADAM10, ADAM9 and ADAM17), ADAM8 may be the candidate protease for C1 production in skeletal muscles.
FIGURE 1.
Quantitative real-time PCR analysis of ADAMs gene expression. Hind leg skeletal muscles (quadriceps) were taken from Tg(HQK) mice treated with 6 g of Dox/kg food for 7, 14, 30, and 60 days. Three animals were used for each time point. Total RNA was extracted and subjected to quantitative real-time PCR analysis for ADAM8, ADAM9, ADAM10, ADAM12, ADAM17, and ADAM23 gene expression. Wild type FVB mice were similarly treated and analyzed. No significant changes were observed in the wild type FVB mice over the time course. Results are relative to the average expression of the corresponding ADAM genes in the skeletal muscles of similarly treated wild-type FVB control mice and represent the mean ± S.E. of triplicate measurements performed.
ADAM8 Protein Level Correlates with PrP C1 Production in the Skeletal Muscles of Transgenic and Knock-out Mice
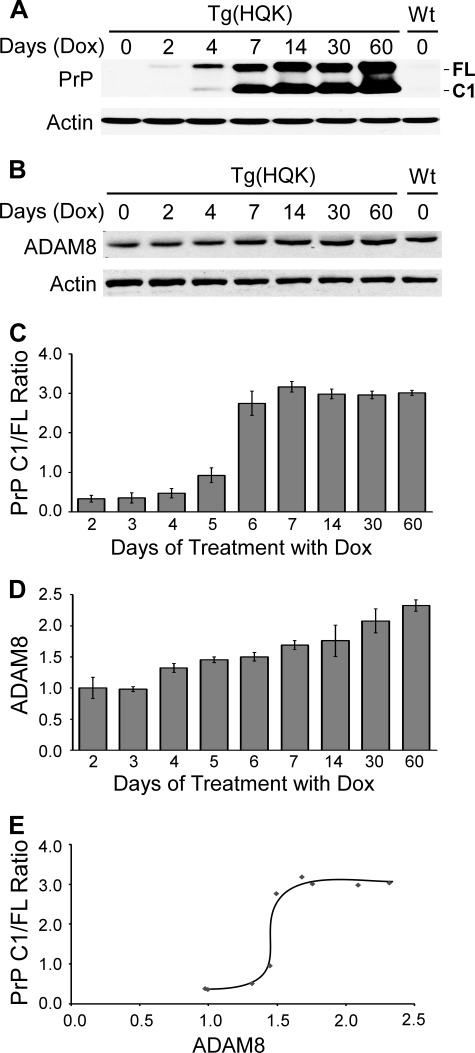
Another time course experiment was performed to examine the relationship between ADAM8 protein level and PrP C1/full-length ratio, the latter serves as a marker for C1 production. Young female adult Tg(HQK) mice were fed with Dox-laced food for 0, 2, 3, 4, 5, 6, 7, 14, 30, and 60 days; 3–6 animals were sacrificed at each time point and hind leg (quadriceps) muscles were collected and homogenized. The muscle homogenates were either treated with PNGase F to remove N-linked glycans and immunoblotted with 8H4 (a monoclonal antibody against the human PrP 176–186 epitope) or were directly subjected to Western blot with a rabbit polyclonal antibody that detects ADAM8 protein.
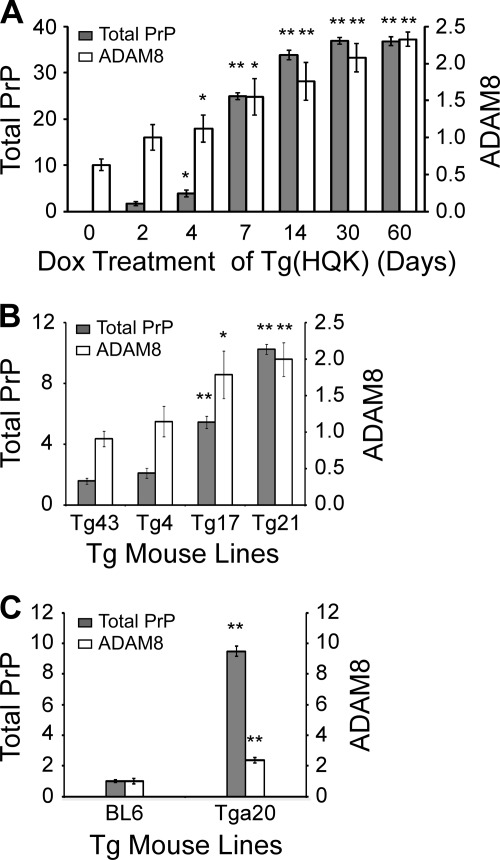
The PrP C1/full-length ratio in the skeletal muscle of Dox-treated Tg(HQK) mice started to rise from day 4, quickly reached a peak at day 7, and stayed at a plateau of ∼3.0 afterward (Fig. 2, A and C), confirming our previous report (59). The ADAM8 protein level in the skeletal muscle showed statistically significant increase from day 4 and kept rising slowly with time (Fig. 2, B and D). Plotting the PrP C1/full-length ratio against ADAM8 protein level revealed that the PrP C1/full-length ratio rose dramatically from 0.5 to ∼3.0 when the ADAM8 level increased from 1.4 to 1.6 during days 4–7, but it stayed at ∼3.0 despite further increase of the ADAM8 level after day 7 (Fig. 2E).
FIGURE 2.
ADAM8 protein level correlates with PrP C1/full-length ratio in skeletal muscle tissue of Dox-induced Tg(HQK) mice. Tg(HQK) mice were treated with 6 g of Dox/kg food for 0–60 days as indicated and 3–6 animals were taken at each time point. Skeletal muscles (quadriceps) from the hind legs of these animals were homogenized and subjected to SDS-PAGE and immunoblot analysis in three Western blots. The homogenates were either treated (for PrP probing) or not treated (for ADAM8 probing) with PNGase F before SDS-PAGE. A, representative immunoblot probed with anti-PrP antibody 8H4 followed by probing with an anti-actin antibody after stripping; samples for day 3, 5, and 6 are not shown. B, representative immunoblot probed with an antibody against ADAM8 followed by probing with an anti-actin antibody after stripping; samples for day 3, 5, and 6 are not shown. C and D, diagrams of PrP C1/full-length ratios (C) and ADAM8 protein level (D) over increasing duration of Dox treatment. The ADAM8 protein level for each sample was normalized against the actin level for the sample in each blot and expressed as the ratio against the normalized ADAM8 protein level in the untreated wild type FVB mouse on the same blot. The error bars denote standard deviations calculated from the duplicate blots. E, plot of PrP C1/full-length ratio against ADAM8 protein level over the time course of Dox treatment.
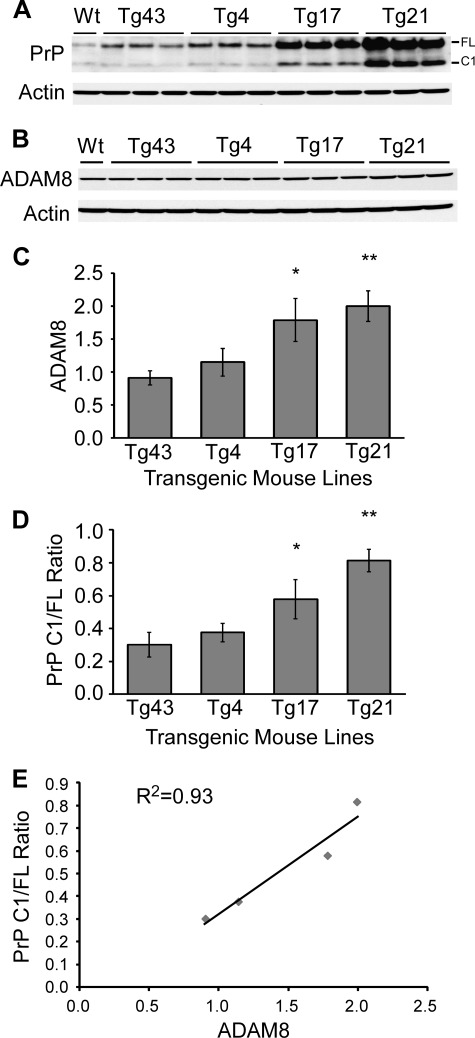
In Tg(HQK) mice treated with Dox, both PrP and ADAM8 expressions were in a dynamic state (rising with time) and the PrP levels were extremely high from day 14, which complicate data interpretation. To address this issue, the relationship between ADAM8 protein level and C1 production in the skeletal muscles was assessed in 4 transgenic (Tg) mouse lines, Tg43, Tg4, Tg17, and Tg21, which all constitutively express human PrP in the skeletal muscles albeit at different levels (Fig. 3A). Skeletal muscles (quadriceps) from three young female adult animals for each transgenic line were examined by Western blot for ADAM8 level and PrP C1/full-length ratio as described above for Tg(HQK) mice; skeletal muscle from a wild type FVB mouse was used as control. Tg lines with higher ADAM8 protein levels in skeletal muscles also showed higher PrP C1/full-length ratios in skeletal muscles, and the two exhibited an excellent linear correlation with a R2 value of 0.93 (Fig. 3).
FIGURE 3.
ADAM8 protein level correlates with PrP C1/full-length ratio in skeletal muscle tissue of Tg mice constitutively expressing human PrP at different levels. Skeletal muscles (quadriceps) from the hind legs of wild type FVB mice (Wt) and four different Tg mice lines (Tg43, Tg4, Tg17, and Tg21) constitutively expressing human PrP at different levels were collected. Muscle homogenates were either treated (for PrP probing) or not treated (for ADAM8 probing) with PNGase F, subjected to SDS-PAGE, and probed with an anti-PrP antibody (8H4) or an antibody against ADAM8. A, representative immunoblot probed with anti-PrP antibody 8H4 followed by probing with an anti-actin antibody after stripping. B, representative immunoblot probed with an antibody against ADAM8 followed by probing with an anti-actin antibody after stripping. C and D, diagrams of ADAM8 protein level (C) and PrP C1/full-length ratios (D) in the skeletal muscles of the four Tg mouse lines. The ADAM8 protein level for each sample was normalized against the actin level and expressed as the ratio against the normalized ADAM8 protein level in the untreated sex and age-matched wild type FVB mouse on the same blot. The error bars denote standard errors calculated from the three animals for each Tg line. The bars with asterisk(s) indicate statistical significance when compared with the Tg43 mouse samples; *, p < 0.05; **, p < 0.01. E, plot of PrP C1/full-length ratios against ADAM8 protein levels in the four Tg lines. Linear regression analysis revealed an R2 value of 0.93, highly indicative of a linear correlation between ADAM8 protein level and the PrP C1/full-length ratio.
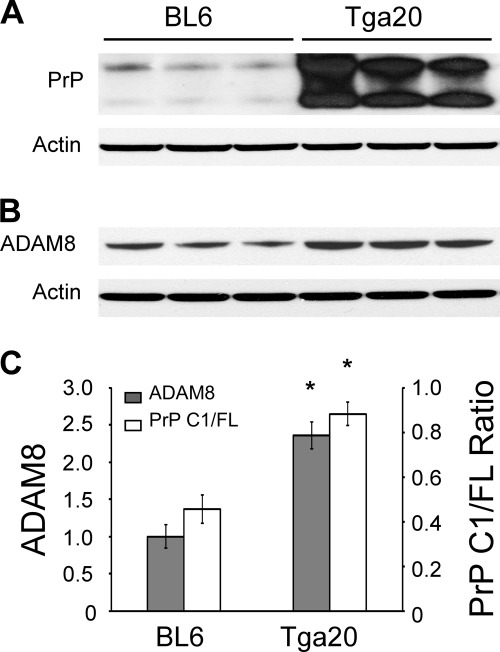
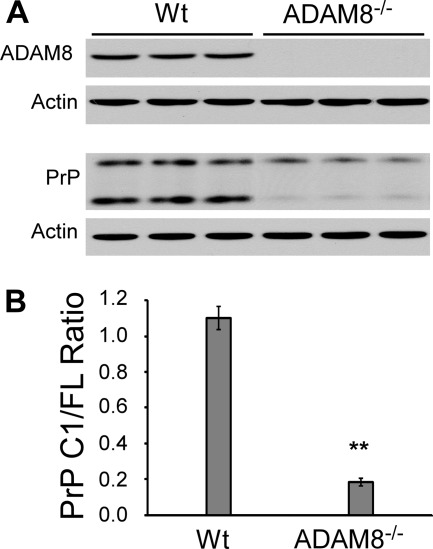
ADAM8 levels and PrP C1/full-length ratios were also examined in the skeletal muscles of wild type C57BL6 mice and Tga20 mice that overexpress mouse PrP. When compared with those of C57BL6 mice, the ADAM8 protein level and PrP C1/full-length ratio in the Tga20 mice were both significantly higher (2.3-fold and 1.8-fold, respectively) (Fig. 4), confirming that ADAM8 protein level is positively correlated with C1/full-length ratio irrespective of the species origin of the PrP. Furthermore, the PrP C1/full-length ratio is reduced by over 80% in skeletal muscles of ADAM8 knock-out mice compared with that of control wild type mice (Fig. 5). In addition, the total PrP level is also significantly decreased in ADAM8 knock-out mice (Fig. 5), which is reminiscent of the effect of ADAM10 knock-out (46) and suggests a prominent role for ADAM-mediated PrP processing in apparent PrP expression levels from the endogenous PrP gene.
FIGURE 4.
ADAM8 protein level correlates with PrP C1/full-length ratio in skeletal muscle tissue of mice constitutively expressing mouse PrP at different levels. Skeletal muscles (quadriceps) from the hind legs of wild type C57BL6 mice (BL6) and the Tga20 line constitutively expressing mouse PrP at high levels were collected. Muscle homogenates were either treated (for PrP probing) or not treated (for ADAM8 probing) with PNGase F, and subjected to SDS-PAGE and immunoblotting with an anti-PrP antibody (8H4) or an antibody against ADAM8. A, immunoblot probed with anti-PrP antibody 8H4 followed by probing with an anti-actin antibody after stripping. B, immunoblot probed with an antibody against ADAM8 followed by probing with an anti-actin antibody after stripping. C, plot of PrP C1/full-length ratios against relative ADAM8 protein levels. The error bars denote standard errors calculated from the three animals for each mouse line. The bars with an asterisk indicate statistical significance (p < 0.05) when compared with the wild type C57BL6 (BL6) samples.
FIGURE 5.
The PrP C1/full-length fragment ratio is significantly reduced in skeletal muscles of ADAM8 knock-out mice. Skeletal muscles (quadriceps) from the hind legs of three each of wild type C57BL/6 mice (Wt) and ADAM8−/− mice were collected. Muscle homogenates were either treated (for PrP probing) or not treated (for ADAM8 probing) with PNGase F, subjected to SDS-PAGE, and probed with an anti-PrP antibody (8H4) or an antibody against ADAM8. A, immunoblots probed with an antibody against ADAM8 or 8H4 followed by probing with an anti-actin antibody after stripping. B, diagram of PrP C1/full-length ratio in the skeletal muscles of Wt and ADAM8−/− mice. The error bars denote standard errors calculated from the three animals for each mouse line. The bars with asterisk(s) indicate statistical significance when compared with the Wt mouse samples. **, p < 0.005.
ADAM8 Protein Level Shows Linear Correlation with PrP C1/Full-length Ratio in the Myoblast Cell Line C2C12
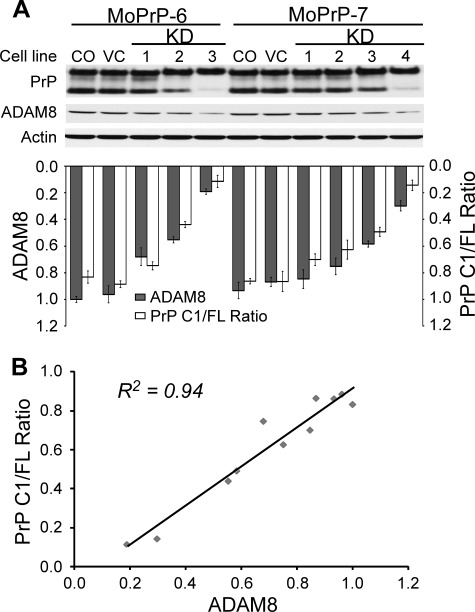
C2C12, a murine myoblast cell line, was utilized to examine the role of ADAM8 in C1 production in pure muscle cells. C2C12 cells normally express very little PrP, but PrP expression is activated during myoblast differentiation (52–53). To study the α-cleavage of PrPC in the C2C12 cell line, C2C12 cells were first transfected with pCEP4 carrying full-length mouse PrP (MoPrP) ORF and many C2C12 clones expressing MoPrP at various levels were established (Fig. 6A). Two of such C2C12 clones (MoPrP-6 and MoPrP-7) that express comparable levels of MoPrP were selected for ADAM8 knockdown (KD) with miR RNAi sequences cloned in the pcDNA vector. Three stable ADAM8-KD cell lines derived from MoPrP-6 and four stable ADAM8-KD cell lines derived from MoPrP-7 were obtained. Cell lysates from these stable ADAM8-KD cell lines were immunoblotted for PrP or ADAM8, and the PrP C1/full-length ratios were plotted against the levels of active ADAM protein in these cell lines (Fig. 7). The result shows that decreasing ADAM8 protein level in the ADAM8-KD cells is accompanied by a proportional drop in the PrP C1/full-length ratio; linear regression analysis reveals an excellent linear correlation between the ADAM8 level and the C1/full-length ratio with an R2 value of 0.94 (Fig. 7). Together with the data from the Dox-treated Tg(HQK) mice, the constitutive Tg mouse lines and the ADAM8 knock-out mice, this result strongly argues for a primary role for ADAM8 in the α-cleavage of PrPC for C1 production in skeletal muscle cells.
FIGURE 6.
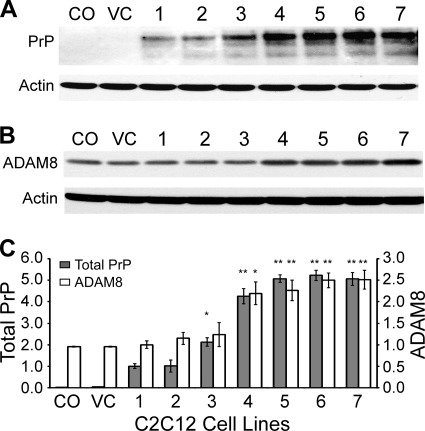
Overexpression of mouse PrP leads to up-regulation of ADAM8 in C2C12 cells. C2C12 cells were transfected with a vector expressing mouse PrP and selected with hygromycin to establish seven stable cell clones (numbered 1–7). The original C2C12 cells (CO) and blank vector-transfected C2C12 cells (VC) serve as controls. Lysates from these clones were directly subjected to Western blotting and the experiments repeated three times. A, representative immunoblot probed with 8H4 followed by probing with an anti-actin antibody after stripping. B, representative immunoblot probed with an antibody against ADAM8 followed by probing with an anti-actin antibody after stripping. C, diagram of total PrP levels and ADAM8 levels in the C2C12 clones. The total PrP and ADAM8 protein levels for each sample were normalized against the actin level and expressed as the ratio against the normalized protein levels in the #1 clone. The error bars denote standard deviations calculated from the three blots. The bars with asterisk(s) indicate statistical significance when compared with the #1 clone; *, p < 0.05; **, p < 0.01.
FIGURE 7.
The PrP C1/full-length ratio is proportional to ADAM8 protein level in C2C12 myoblast cells. Two stable C2C12 clones expressing mouse PrP (MoPrP-6 and MoPrP-7) were transfected with miR RNAi sequence targeting the ORF region of ADAM8 and several stable cell lines were obtained. Lysates from these stable cell lines were either treated (for PrP probing) or not treated (for ADAM8 probing) with PNGase F, then subjected to electrophoresis and immunoblotting with an anti-PrP antibody (8H4) or an antibody against ADAM8. The immunoblotting was repeated three times. A, representative immunoblots probed with anti-PrP antibody 8H4 or with an antibody against ADAM8 followed by probing with an anti-actin antibody after stripping; the relative ADAM8 protein level and PrP C1/full-length ratio for each cell line were quantified and diagramed underneath the Western blot. B, plot of PrP C1/full-length ratios against ADAM8 protein levels in ADAM8-knockdown C2C12 cell lines. The ADAM8 protein level for each C2C12 cell line was normalized against the actin level and expressed as the ratio against the normalized ADAM8 protein level in the original MoPrP-6 control cell line (CO) on the same blot. Data from the average of three duplicate blots are plotted. CO, the original MoPrP6/MoPrP7 cell line; VC, blank vector-transfected MoPrP6/MoPrP7 cell line; KD, knockdown.
ADAM8 Directly Cleaves PrP to Generate C1 in Vitro
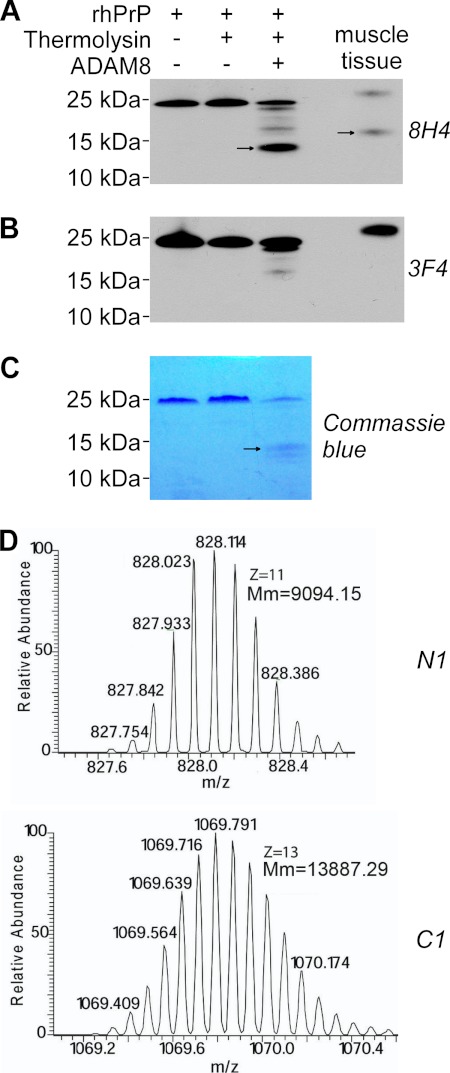
A recombinant human ADAM8 peptide (rhADAM8, aa158–497), which contains part of the propeptide and the entire metalloprotease and disintegrin-like domains, was incubated with recombinant human PrP (rhPrP, aa23–231 plus a Gly-Ser extension at the N terminus) before or after activation with thermolysin (Fig. 8). Electrophoretic analysis shows that only thermolysin-activated ADAM8 generated a prominent ∼14 kDa fragment from the rhPrP substrate. The size of this fragment is consistent with the expected molecular mass (13.9 kDa) of the C-terminal α-cleavage product (rhPrP111–231). The ∼14 kDa fragment was detected by the 8H4 antibody which recognizes residues 176–186 of hPrP, but not by the 3F4 antibody which recognizes residues 106–112 of hPrP (65–66), further indicating that this fragment corresponds to C1. The identity of hPrP fragments generated by treatment with thermolysin-activated ADAM8 was further verified by mass spectrometry (MS). Protein fragments with the monoisotopic molecular weights corresponding precisely to C1 with native Cys179-Cys214 disulfide bond (13,887.29 Da) and N1 (9,094.15 Da) were detected with a mass accuracy of 10 ppm (Fig. 8D). Furthermore, the expected amino acid sequences of the C1 and N1 fragments were confirmed by FT-ICR tandem MS using a series of b- and y-fragment ions (data not shown). C1 and N1 fragments were not detected when rhPrP was incubated with either unactivated ADAM8 or inactivated thermolysin alone. Together, these results prove that ADAM8 can directly perform α-cleavage of PrP.
FIGURE 8.
ADAM8 directly cleaves PrP to generate the C1 fragment. Recombinant human PrP (rhPrP) was cleaved at 37 °C for 14 h with a recombinant human ADAM8 that had been treated or untreated with thermolysin for activation, followed by SDS-PAGE and subsequent immunoblot analysis (A and B) or direct staining with Coomassie Blue (C), or by direct mass spectrometry analysis (D). A, Western blot probed with antibodies 8H4 (against human PrP176–186). B, Western blot probed with 3F4 (against human PrP 106–112). C, SDS-PAGE gel directly stained with Coomassie Blue. D, mass spectrometry analysis. An in vitro reaction sample with thermolysin-activated ADAM8 and rhPrP was analyzed. Monoisotopic distributions of protonated (M+11H)11+ N1 fragment ion (top) and protonated (M+13H)13+ C1 fragment ion (bottom) are shown; Mm: molecular mass. In panels A-B, skeletal muscle tissue homogenate from a Tg(HQK) mouse treated with Dox for 60 days was loaded as a control. In panels A–C, the arrows point to the C1 band. The rhPrP sample contained an extension of Gly-Ser at the N terminus as a leftover from the His tag; accordingly, the resulting N1 fragment also contained the extra Gly-Ser residues at the N terminus.
ADAM8 Is Up-regulated by PrP
Further examination of Western blots of total PrP and ADAM8 levels in the skeletal muscles of Dox-treated Tg(HQK) mice (Fig. 2) revealed that, starting from day 4 when total PrP level reached four times that of wild type mice, the ADAM8 protein level rose significantly (Fig. 9A). Quantification of Western blots for the four Tg lines constitutively expressing human PrP (Fig. 3) and the Tga20 mice overexpressing mouse PrP (Fig. 4) also indicates that PrP overexpression resulted in elevated ADAM8 protein levels (Fig. 9, B and C). Similarly, when compared with the C2C12 clone #1, the four C2C12 clones (#4, #5, #6, and #7) that expressed MoPrP at >4-fold levels showed >2-fold ADAM8 protein levels as well (Fig. 6). Together these in vivo and in vitro data demonstrate that overexpression of PrP leads to elevated ADAM8 activities in muscle cells. Since ADAM8 mRNA was also up-regulated in the muscles of Dox-treated Tg(HQK) mice (Fig. 1), PrP overexpression-induced up-regulation of ADAM8 is likely to be at the transcription level.
FIGURE 9.
Overexpression of human PrPC leads to up-regulation of ADAM8 in transgenic mice. Western blots for PrP and ADAM8 in homogenates of skeletal muscles from Dox-treated Tg(HQK) mice (shown in Fig. 1, A and B), Tg lines constitutively expressing human PrP at various levels (shown in Fig. 2, A and B), and C57BL6 (BL6) and Tga20 mice (shown in Fig. 4, A and B) were quantified after scanning the x-ray films. The ADAM8 protein level and total PrP protein level for each sample were normalized against the actin level and expressed as the ratio against the corresponding normalized protein level in the untreated sex and age matched wild type FVB mouse (for panels A and B) or wild type C57BL6 mice (for panel C) on the same blot. A, diagram of total PrP levels and ADAM8 levels in Tg(HQK) mice over the time course of Dox treatment. The statistical difference was calculated based on comparison with day 0 Tg(HQK) samples for ADAM8 or with day 2 Tg(HQK) samples for total PrP. B, diagram of the total PrP levels and ADAM8 levels in the four constitutive Tg mouse lines. The statistical difference was calculated based on comparison with Tg43 samples. C, diagram of the total PrP levels and ADAM8 levels in C57BL6 and Tga20 mice. The statistical difference was calculated based on comparison with C57BL6 (BL6) samples. For all panels, the error bars denote standard deviations calculated from the three animals for each Tg line; *, p < 0.05; **, p < 0.01.
DISCUSSION
The α-cleavage of PrPC disrupts the protein in a region critical for both toxicity and prion conversion and generates the biologically active C1 and N1 fragments. Our current understanding of the α-cleavage of PrPC is based almost exclusively on studies in vitro using cultured HEK and mouse fibroblasts or neuronal cells. However, the earlier reports implicating the direct involvement of ADAM10 and ADAM17 and an indirect role for ADAM9 (38–39, 44) were challenged by more recent articles (34, 46–47), putting a question mark on the identity of the protease(s) primarily responsible for the α-cleavage of PrPC.
We have addressed this issue using Tg mouse models, the C2C12 myoblast cell line, and in vitro protease assays. We found that in the skeletal muscles of Dox-treated Tg(HQK) mice, only the ADAM8 mRNA was significantly up-regulated (Fig. 1) and ADAM8 protein level increased starting from day 4 of Dox treatment, preceding the dramatic preferential accumulation of C1 that began at day 5 of Dox treatment (Fig. 2). The ADAM8 protein level was also found to be linearly correlated with the PrP C1/full-length ratio in the skeletal muscles of the Tg mouse lines constitutively expressing human PrP (Fig. 3). When compared with the wild type C57BL6 mice, the PrP C1/full-length ratio in skeletal muscles was increased in the Tga20 mice that expressed more ADAM8 (Fig. 4) and decreased in the ADAM8 knock-out mice (Fig. 5), confirming that the positive correlation between ADAM8 level and C1/full-length ratio is not specific to human PrP. Moreover, a linear correlation between ADAM8 protein level and PrP C1/full-length ratio was observed in C2C12 myoblast clones expressing the same level of mouse PrP but with different levels of ADAM8 due to RNAi knockdown (Fig. 7). Furthermore, efficient in vitro α-cleavage of recombinant human PrPC by recombinant ADAM8 was observed (Fig. 8). Together, these in vivo and in vitro results strongly argue for a primary role for ADAM8 in C1 production in skeletal muscles. ADAM10, the proposed protease for C1 production in the brain (45), was not up-regulated in the skeletal muscles of Dox-treated Tg(HQK) mice (Fig. 1). We found that the ADAM10 protein level in skeletal muscle is only about 2% of the brain level in wild type FVB mice (data not shown). These data indicate that ADAM10 is unlikely to play a major role in C1 production in the skeletal muscles, which is consistent with the recent reports refuting the involvement of ADAM10 in the α-cleavage of PrPC (46–47). Nevertheless, the presence of low level of C1 in the muscles of ADAM8 knock-out mice (Fig. 5) indicates that another protease(s) plays a minor role in the α-cleavage of PrPC in the skeletal muscles. It remains to be investigated whether ADAM8 also participates in the α-cleavage of PrPC in brain and other tissues.
We also discovered that overexpression of wild type PrP up-regulates ADAM8 in the skeletal muscles of Dox-treated Tg(HQK) mice (Fig. 9A) and Tg lines constitutively expressing different levels of human or mouse PrP (Fig. 9, B and C), and in C2C12 cells expressing different levels of mouse PrP (Fig. 6). Given that the ADAM8 mRNA level was significantly increased in the skeletal muscles of Dox-treated Tg(HQK) mice (Fig. 1), it is very likely that PrPC regulates ADAM8 at the transcriptional level.
The molecular mechanism underlying the over-expressed PrP-induced ADAM8 expression is unclear. PrPC was recently reported to promote regeneration of adult muscle tissue through affecting the generation of TNF-α (54, 67), a cytokine actively involved in myogenesis and muscle repair (68–72). In addition, the expression of ADAM8 was induced by TNF-α in primary cerebellar neurons and in motoneuron-like NSC19 cells in a dose-dependent manner (73). Moreover, TNF-α regulates the shedding of TNF-α receptor 1 through elevating ADAM8 activity in the mouse brain and primary neurons and microglia (74). Furthermore, antibody-mediated PrPC ligation was reported to cause TNF-α shedding in cultured neurons (75). It is conceivable that, in the muscles, the overexpressed PrPC somehow activates TNF-α to induce ADAM8 transcription, and the increased ADAM8 activity results in augmented α-cleavage of PrPC.
It is not clear why the PrP C1/full length ratio remained unchanged from day 7 to day 60 despite the continued rise in the ADAM8 level in the skeletal muscle of Dox-treated Tg(HQK) mice (Fig. 2). The very high levels of PrP at these later time points could counter the increase of ADAM8 activity to maintain a stable PrP C1/full-length ratio. Alternatively, an unknown inhibitor(s) of ADAM8 may be activated at very high PrP levels.
Our findings that ADAM8 can perform α-cleavage of PrPC and that PrPC can regulate ADAM8 expression have significant implications. In particular, they point to a self-regulatory loop where overexpressed PrPC modulates its own α-cleavage through upregulating ADAM8, leading to accumulation of cytotoxic C1 when PrPC is overexpressed. This self-regulatory loop may not only explain the myopathy incurred by over-expressed wild type PrPC in aged Tg mice (58) and Dox-induced Tg(HQK) mice (59–60), but also provide a plausible mechanism for PrP toxicity observed in human HEK293 cells, mouse neurons, and erythroleukemia cells (76–80). When PrP is overexpressed for an extended period of time, the up-regulation of ADAM8 will lead to accumulation of the cytotoxic and more stable C1 fragment (35) at a higher molar ratio than full-length PrPC. The concurrently released neuroprotective N1 fragment has a shorter half-life (31) and may provide only transient and possibly insufficient protection from the toxicity of the accumulated C1. However, a very recent report suggests that C1 itself is nontoxic when expressed in transgenic mice (81), making it uncertain how elevated C1 level may play a direct role in PrP-mediated myopathy. It is possible that the elevated ADAM8 activity may induce further effects stemming from the enhanced cleavage of its many other substrates (42), some of which may participate in the cytotoxic effect of overexpressed PrP.
Acknowledgments
We thank Meiling Wang and Laure Farnbauch for help with animal care. We would also like to thank Dr. Andy Docherty at UCB Celltech for giving us permission to use the ADAM8 knock-out mice and Dr. Carl Blobel at Hospital for Special Surgery and Weill Medical College of Cornell University for sending us the ADAM8 knock-out mice.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 NS052319 (to Q. K.).
- PrPC
- cellular prion protein
- ADAM
- A Disintegrin And Metalloproteinase
- Dox
- doxycycline
- ECMs
- extracellular matrix proteins
- MS
- mass spectrometry
- PrP
- prion protein
- Tg
- transgenic
- TRANCE
- TNF-related activation-induced cytokine.
REFERENCES
- 1. Miele G., Alejo Blanco A. R., Baybutt H., Horvat S., Manson J., Clinton M. (2003) Embryonic activation and developmental expression of the murine prion protein gene. Gene Expr. 11, 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Prusiner S. B. (1998) Prions. Proc. Natl. Acad. Sci. U.S.A. 95, 13363–13383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Laurén J., Gimbel D. A., Nygaard H. B., Gilbert J. W., Strittmatter S. M. (2009) Cellular prion protein mediates impairment of synaptic plasticity by amyloid-β oligomers. Nature 457, 1128–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gimbel D. A., Nygaard H. B., Coffey E. E., Gunther E. C., Laurén J., Gimbel Z. A., Strittmatter S. M. (2010) Memory impairment in transgenic Alzheimer mice requires cellular prion protein. J. Neurosci. 30, 6367–6374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kessels H. W., Nguyen L. N., Nabavi S., Malinow R. (2010) The prion protein as a receptor for amyloid-β. Nature 466, E3–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mehrpour M., Codogno P. (2010) Prion protein: From physiology to cancer biology. Cancer. Lett. 290, 1–23 [DOI] [PubMed] [Google Scholar]
- 7. Aguzzi A., Baumann F., Bremer J. (2008) The prion's elusive reason for being. Annu. Rev. Neurosci. 31, 439–477 [DOI] [PubMed] [Google Scholar]
- 8. Linden R., Martins V. R., Prado M. A., Cammarota M., Izquierdo I., Brentani R. R. (2008) Physiology of the prion protein. Physiol. Rev. 88, 673–728 [DOI] [PubMed] [Google Scholar]
- 9. Westergard L., Christensen H. M., Harris D. A. (2007) The cellular prion protein (PrP(C)): its physiological function and role in disease. Biochim. Biophys. Acta 1772, 629–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sorgato M. C., Peggion C., Bertoli A. (2009) Is, indeed, the prion protein a Harlequin servant of “many” masters? Prion 3, 202–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Steinacker P., Hawlik A., Lehnert S., Jahn O., Meier S., Görz E., Braunstein K. E., Krzovska M., Schwalenstöcker B., Jesse S., Pröpper C., Böckers T., Ludolph A., Otto M. (2010) Neuroprotective function of cellular prion protein in a mouse model of amyotrophic lateral sclerosis. Am. J. Pathol. 176, 1409–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pauly P. C., Harris D. A. (1998) Copper stimulates endocytosis of the prion protein. J. Biol. Chem. 273, 33107–33110 [DOI] [PubMed] [Google Scholar]
- 13. Singh A., Kong Q., Luo X., Petersen R. B., Meyerson H., Singh N. (2009) Prion protein (PrP) knock-out mice show altered iron metabolism: a functional role for PrP in iron uptake and transport. PLoS One 4, e6115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jiménez-Huete A., Lievens P. M., Vidal R., Piccardo P., Ghetti B., Tagliavini F., Frangione B., Prelli F. (1998) Endogenous proteolytic cleavage of normal and disease-associated isoforms of the human prion protein in neural and non-neural tissues. Am. J. Pathol. 153, 1561–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen S. G., Teplow D. B., Parchi P., Teller J. K., Gambetti P., Autilio-Gambetti L. (1995) Truncated forms of the human prion protein in normal brain and in prion diseases. J. Biol. Chem. 270, 19173–19180 [DOI] [PubMed] [Google Scholar]
- 16. Owen J. P., Rees H. C., Maddison B. C., Terry L. A., Thorne L., Jackman R., Whitelam G. C., Gough K. C. (2007) Molecular profiling of ovine prion diseases by using thermolysin-resistant PrPSc and endogenous C2 PrP fragments. J. Virol. 81, 10532–10539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dron M., Moudjou M., Chapuis J., Salamat M. K., Bernard J., Cronier S., Langevin C., Laude H. (2010) Endogenous proteolytic cleavage of disease-associated prion protein to produce C2 fragments is strongly cell- and tissue-dependent. J. Biol. Chem. 285, 10252–10264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yadavalli R., Guttmann R. P., Seward T., Centers A. P., Williamson R. A., Telling G. C. (2004) Calpain-dependent endoproteolytic cleavage of PrPSc modulates scrapie prion propagation. J. Biol. Chem. 279, 21948–21956 [DOI] [PubMed] [Google Scholar]
- 19. Pan K. M., Stahl N., Prusiner S. B. (1992) Purification and properties of the cellular prion protein from Syrian hamster brain. Protein Sci. 1, 1343–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Taraboulos A., Raeber A. J., Borchelt D. R., Serban D., Prusiner S. B. (1992) Synthesis and trafficking of prion proteins in cultured cells. Mol. Biol. Cell 3, 851–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mangé A., Béranger F., Peoc'h K., Onodera T., Frobert Y., Lehmann S. (2004) α- and β-cleavages of the amino-terminus of the cellular prion protein. Biol. Cell 96, 125–132 [DOI] [PubMed] [Google Scholar]
- 22. Gasset M., Baldwin M. A., Lloyd D. H., Gabriel J. M., Holtzman D. M., Cohen F., Fletterick R., Prusiner S. B. (1992) Predicted α-helical regions of the prion protein when synthesized as peptides form amyloid. Proc. Natl. Acad. Sci. U.S.A. 89, 10940–10944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Forloni G., Angeretti N., Chiesa R., Monzani E., Salmona M., Bugiani O., Tagliavini F. (1993) Neurotoxicity of a prion protein fragment. Nature 362, 543–546 [DOI] [PubMed] [Google Scholar]
- 24. Jobling M. F., Stewart L. R., White A. R., McLean C., Friedhuber A., Maher F., Beyreuther K., Masters C. L., Barrow C. J., Collins S. J., Cappai R. (1999) The hydrophobic core sequence modulates the neurotoxic and secondary structure properties of the prion peptide 106–126. J. Neurochem. 73, 1557–1565 [DOI] [PubMed] [Google Scholar]
- 25. Norstrom E. M., Mastrianni J. A. (2005) The AGAAAAGA palindrome in PrP is required to generate a productive PrPSc-PrPC complex that leads to prion propagation. J. Biol. Chem. 280, 27236–27243 [DOI] [PubMed] [Google Scholar]
- 26. McMahon H. E., Mangé A., Nishida N., Créminon C., Casanova D., Lehmann S. (2001) Cleavage of the amino terminus of the prion protein by reactive oxygen species. J. Biol. Chem. 276, 2286–2291 [DOI] [PubMed] [Google Scholar]
- 27. Watt N. T., Taylor D. R., Gillott A., Thomas D. A., Perera W. S., Hooper N. M. (2005) Reactive oxygen species-mediated β-cleavage of the prion protein in the cellular response to oxidative stress. J. Biol. Chem. 280, 35914–35921 [DOI] [PubMed] [Google Scholar]
- 28. Doh-Ura K., Iwaki T., Caughey B. (2000) Lysosomotropic agents and cysteine protease inhibitors inhibit scrapie-associated prion protein accumulation. J. Virol. 74, 4894–4897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Luhr K. M., Nordström E. K., Löw P., Kristensson K. (2004) Cathepsin B and L are involved in degradation of prions in GT1–1 neuronal cells. Neuroreport 15, 1663–1667 [DOI] [PubMed] [Google Scholar]
- 30. Taguchi Y., Shi Z. D., Ruddy B., Dorward D. W., Greene L., Baron G. S. (2009) Specific biarsenical labeling of cell surface proteins allows fluorescent- and biotin-tagging of amyloid precursor protein and prion proteins. Mol. Biol. Cell 20, 233–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Guillot-Sestier M. V., Sunyach C., Druon C., Scarzello S., Checler F. (2009) The alpha-secretase-derived N-terminal product of cellular prion, N1, displays neuroprotective function in vitro and in vivo. J. Biol. Chem. 284, 35973–35986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sunyach C., Cisse M. A., da Costa C. A., Vincent B., Checler F. (2007) The C-terminal products of cellular prion protein processing, C1 and C2, exert distinct influence on p53-dependent staurosporine-induced caspase-3 activation. J. Biol. Chem. 282, 1956–1963 [DOI] [PubMed] [Google Scholar]
- 33. Mitteregger G., Vosko M., Krebs B., Xiang W., Kohlmannsperger V., Nölting S., Hamann G. F., Kretzschmar H. A. (2007) The role of the octarepeat region in neuroprotective function of the cellular prion protein. Brain Pathol. 17, 174–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Taylor D. R., Parkin E. T., Cocklin S. L., Ault J. R., Ashcroft A. E., Turner A. J., Hooper N. M. (2009) Role of ADAMs in the ectodomain shedding and conformational conversion of the prion protein. J. Biol. Chem. 284, 22590–22600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shyng S. L., Huber M. T., Harris D. A. (1993) A prion protein cycles between the cell surface and an endocytic compartment in cultured neuroblastoma cells. J. Biol. Chem. 268, 15922–15928 [PubMed] [Google Scholar]
- 36. Harris D. A., Huber M. T., van Dijken P., Shyng S. L., Chait B. T., Wang R. (1993) Processing of a cellular prion protein: identification of N- and C-terminal cleavage sites. Biochemistry 32, 1009–1016 [DOI] [PubMed] [Google Scholar]
- 37. Vincent B., Paitel E., Frobert Y., Lehmann S., Grassi J., Checler F. (2000) Phorbol ester-regulated cleavage of normal prion protein in HEK293 human cells and murine neurons. J. Biol. Chem. 275, 35612–35616 [DOI] [PubMed] [Google Scholar]
- 38. Vincent B., Paitel E., Saftig P., Frobert Y., Hartmann D., De Strooper B., Grassi J., Lopez-Perez E., Checler F. (2001) The disintegrins ADAM10 and TACE contribute to the constitutive and phorbol ester-regulated normal cleavage of the cellular prion protein. J. Biol. Chem. 276, 37743–37746 [DOI] [PubMed] [Google Scholar]
- 39. Cissé M. A., Sunyach C., Lefranc-Jullien S., Postina R., Vincent B., Checler F. (2005) The disintegrin ADAM9 indirectly contributes to the physiological processing of cellular prion by modulating ADAM10 activity. J. Biol. Chem. 280, 40624–40631 [DOI] [PubMed] [Google Scholar]
- 40. Walmsley A. R., Watt N. T., Taylor D. R., Perera W. S., Hooper N. M. (2009) α-cleavage of the prion protein occurs in a late compartment of the secretory pathway and is independent of lipid rafts. Mol. Cell. Neurosci. 40, 242–248 [DOI] [PubMed] [Google Scholar]
- 41. White J. M. (2003) ADAMs: modulators of cell-cell and cell-matrix interactions. Curr. Opin. Cell Biol. 15, 598–606 [DOI] [PubMed] [Google Scholar]
- 42. Edwards D. R., Handsley M. M., Pennington C. J. (2008) The ADAM metalloproteinases. Mol. Aspects. Med. 29, 258–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Moss M. L., Lambert M. H. (2002) Shedding of membrane proteins by ADAM family proteases. Essays Biochem. 38, 141–153 [DOI] [PubMed] [Google Scholar]
- 44. Alfa Cissé M., Sunyach C., Slack B. E., Fisher A., Vincent B., Checler F. (2007) M1 and M3 muscarinic receptors control physiological processing of cellular prion by modulating ADAM17 phosphorylation and activity. J. Neurosci. 27, 4083–4092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Laffont-Proust I., Faucheux B. A., Hässig R., Sazdovitch V., Simon S., Grassi J., Hauw J. J., Moya K. L., Haïk S. (2005) The N-terminal cleavage of cellular prion protein in the human brain. FEBS Lett. 579, 6333–6337 [DOI] [PubMed] [Google Scholar]
- 46. Endres K., Mitteregger G., Kojro E., Kretzschmar H., Fahrenholz F. (2009) Influence of ADAM10 on prion protein processing and scrapie infectiosity in vivo. Neurobiol. Dis. 36, 233–241 [DOI] [PubMed] [Google Scholar]
- 47. Altmeppen H. C., Prox J., Puig B., Kluth M. A., Bernreuther C., Thurm D., Jorissen E., Petrowitz B., Bartsch U., De Strooper B., Saftig P., Glatzel M. (2011) Lack of a-disintegrin-and-metalloproteinase ADAM10 leads to intracellular accumulation and loss of shedding of the cellular prion protein in vivo. Mol. Neurodegener. 6, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kaneko K., Zulianello L., Scott M., Cooper C. M., Wallace A. C., James T. L., Cohen F. E., Prusiner S. B. (1997) Evidence for protein X binding to a discontinuous epitope on the cellular prion protein during scrapie prion propagation. Proc. Natl. Acad. Sci. U.S.A. 94, 10069–10074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Telling G. C., Scott M., Mastrianni J., Gabizon R., Torchia M., Cohen F. E., DeArmond S. J., Prusiner S. B. (1995) Prion propagation in mice expressing human and chimeric PrP transgenes implicates the interaction of cellular PrP with another protein. Cell 83, 79–90 [DOI] [PubMed] [Google Scholar]
- 50. Petropoulos C. J., Rosenberg M. P., Jenkins N. A., Copeland N. G., Hughes S. H. (1989) The chicken skeletal muscle α-actin promoter is tissue specific in transgenic mice. Mol. Cell. Biol. 9, 3785–3792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nico P. B., Lobão-Soares B., Landemberger M. C., Marques W., Jr., Tasca C. I., de Mello C. F., Walz R., Carlotti C. G., Jr., Brentani R. R., Sakamoto A. C., Bianchin M. M. (2005) Impaired exercise capacity, but unaltered mitochondrial respiration in skeletal or cardiac muscle of mice lacking cellular prion protein. Neurosci. Lett. 388, 21–26 [DOI] [PubMed] [Google Scholar]
- 52. Massimino M. L., Ferrari J., Sorgato M. C., Bertoli A. (2006) Heterogeneous PrPC metabolism in skeletal muscle cells. FEBS Lett. 580, 878–884 [DOI] [PubMed] [Google Scholar]
- 53. Brown D. R., Schmidt B., Groschup M. H., Kretzschmar H. A. (1998) Prion protein expression in muscle cells and toxicity of a prion protein fragment. Eur. J. Cell Biol. 75, 29–37 [DOI] [PubMed] [Google Scholar]
- 54. Stella R., Massimino M. L., Sandri M., Sorgato M. C., Bertoli A. (2010) Cellular prion protein promotes regeneration of adult muscle tissue. Mol. Cell. Biol. 30, 4864–4876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Askanas V., Bilak M., Engel W. K., Alvarez R. B., Tomé F., Leclerc A. (1993) Prion protein is abnormally accumulated in inclusion-body myositis. Neuroreport 5, 25–28 [DOI] [PubMed] [Google Scholar]
- 56. Sarkozi E., Askanas V., Engel W. K. (1994) Abnormal accumulation of prion protein mRNA in muscle fibers of patients with sporadic inclusion-body myositis and hereditary inclusion-body myopathy. Am. J. Pathol. 145, 1280–1284 [PMC free article] [PubMed] [Google Scholar]
- 57. Zanusso G., Vattemi G., Ferrari S., Tabaton M., Pecini E., Cavallaro T., Tomelleri G., Filosto M., Tonin P., Nardelli E., Rizzuto N., Monaco S. (2001) Increased expression of the normal cellular isoform of prion protein in inclusion-body myositis, inflammatory myopathies, and denervation atrophy. Brain Pathol. 11, 182–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Westaway D., DeArmond S. J., Cayetano-Canlas J., Groth D., Foster D., Yang S. L., Torchia M., Carlson G. A., Prusiner S. B. (1994) Degeneration of skeletal muscle, peripheral nerves, and the central nervous system in transgenic mice overexpressing wild-type prion proteins. Cell 76, 117–129 [DOI] [PubMed] [Google Scholar]
- 59. Huang S., Liang J., Zheng M., Li X., Wang M., Wang P., Vanegas D., Wu D., Chakraborty B., Hays A. P., Chen K., Chen S. G., Booth S., Cohen M., Gambetti P., Kong Q. (2007) Inducible overexpression of wild-type prion protein in the muscles leads to a primary myopathy in transgenic mice. Proc. Natl. Acad. Sci. U.S.A. 104, 6800–6805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Liang J., Parchaliuk D., Medina S., Sorensen G., Landry L., Huang S., Wang M., Kong Q., Booth S. A. (2009) Activation of p53-regulated pro-apoptotic signaling pathways in PrP-mediated myopathy. BMC Genomics 10, 201–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kong Q., Huang S., Zou W., Vanegas D., Wang M., Wu D., Yuan J., Zheng M., Bai H., Deng H., Chen K., Jenny A. L., O'Rourke K., Belay E. D., Schonberger L. B., Petersen R. B., Sy M. S., Chen S. G., Gambetti P. (2005) Chronic wasting disease of elk: transmissibility to humans examined by transgenic mouse models. J. Neurosci. 25, 7944–7949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kelly K., Hutchinson G., Nebenius-Oosthuizen D., Smith A. J., Bartsch J. W., Horiuchi K., Rittger A., Manova K., Docherty A. J., Blobel C. P. (2005) Metalloprotease-disintegrin ADAM8: expression analysis and targeted deletion in mice. Dev. Dyn. 232, 221–231 [DOI] [PubMed] [Google Scholar]
- 63. Pan T., Li R., Kang S. C., Wong B. S., Wisniewski T., Sy M. S. (2004) Epitope scanning reveals gain and loss of strain specific antibody binding epitopes associated with the conversion of normal cellular prion to scrapie prion. J. Neurochem. 90, 1205–1217 [DOI] [PubMed] [Google Scholar]
- 64. Morillas M., Swietnicki W., Gambetti P., Surewicz W. K. (1999) Membrane environment alters the conformational structure of the recombinant human prion protein. J. Biol. Chem. 274, 36859–36865 [DOI] [PubMed] [Google Scholar]
- 65. Lund C., Olsen C. M., Tveit H., Tranulis M. A. (2007) Characterization of the prion protein 3F4 epitope and its use as a molecular tag. J. Neurosci. Methods 165, 183–190 [DOI] [PubMed] [Google Scholar]
- 66. Zou W. Q., Langeveld J., Xiao X., Chen S., McGeer P. L., Yuan J., Payne M. C., Kang H. E., McGeehan J., Sy M. S., Greenspan N. S., Kaplan D., Wang G. X., Parchi P., Hoover E., Kneale G., Telling G., Surewicz W. K., Kong Q., Guo J. P. (2010) PrP conformational transitions alter species preference of a PrP-specific antibody. J. Biol. Chem. 285, 13874–13884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Stella R., Massimino M. L., Sorgato M. C., Bertoli A. (2010) Prion and TNFα: TAC(E)it agreement between the prion protein and cell signaling. Cell Cycle 9, 4616–4621 [DOI] [PubMed] [Google Scholar]
- 68. Chen S. E., Gerken E., Zhang Y., Zhan M., Mohan R. K., Li A. S., Reid M. B., Li Y. P. (2005) Role of TNF-{α} signaling in regeneration of cardiotoxin-injured muscle. Am. J. Physiol. Cell. Physiol. 289, C1179–C1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Chen S. E., Jin B., Li Y. P. (2007) TNF-α regulates myogenesis and muscle regeneration by activating p38 MAPK. Am. J. Physiol. Cell. Physiol. 292, C1660–C1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Zhan M., Jin B., Chen S. E., Reecy J. M., Li Y. P. (2007) TACE release of TNF-alpha mediates mechanotransduction-induced activation of p38 MAPK and myogenesis. J. Cell Sci. 120, 692–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Keren A., Tamir Y., Bengal E. (2006) The p38 MAPK signaling pathway: a major regulator of skeletal muscle development. Mol. Cell. Endocrinol. 252, 224–230 [DOI] [PubMed] [Google Scholar]
- 72. Zetser A., Gredinger E., Bengal E. (1999) p38 mitogen-activated protein kinase pathway promotes skeletal muscle differentiation. Participation of the Mef2c transcription factor. J. Biol. Chem. 274, 5193–5200 [DOI] [PubMed] [Google Scholar]
- 73. Schlomann U., Rathke-Hartlieb S., Yamamoto S., Jockusch H., Bartsch J. W. (2000) Tumor necrosis factor α induces a metalloprotease-disintegrin, ADAM8 (CD 156): implications for neuron-glia interactions during neurodegeneration. J. Neurosci. 20, 7964–7971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Bartsch J. W., Wildeboer D., Koller G., Naus S., Rittger A., Moss M. L., Minai Y., Jockusch H. (2010) Tumor necrosis factor-α (TNF-α) regulates shedding of TNF-α receptor 1 by the metalloprotease-disintegrin ADAM8: evidence for a protease-regulated feedback loop in neuroprotection. J. Neurosci. 30, 12210–12218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Pradines E., Loubet D., Mouillet-Richard S., Manivet P., Launay J. M., Kellermann O., Schneider B. (2009) Cellular prion protein coupling to TACE-dependent TNF-α shedding controls neurotransmitter catabolism in neuronal cells. J. Neurochem. 110, 912–923 [DOI] [PubMed] [Google Scholar]
- 76. Paitel E., Alves da Costa C., Vilette D., Grassi J., Checler F. (2002) Overexpression of PrPc triggers caspase 3 activation: potentiation by proteasome inhibitors and blockade by anti-PrP antibodies. J. Neurochem. 83, 1208–1214 [DOI] [PubMed] [Google Scholar]
- 77. Paitel E., Fahraeus R., Checler F. (2003) Cellular prion protein sensitizes neurons to apoptotic stimuli through Mdm2-regulated and p53-dependent caspase 3-like activation. J. Biol. Chem. 278, 10061–10066 [DOI] [PubMed] [Google Scholar]
- 78. Paitel E., Sunyach C., Alves da Costa C., Bourdon J. C., Vincent B., Checler F. (2004) Primary cultured neurons devoid of cellular prion display lower responsiveness to staurosporine through the control of p53 at both transcriptional and post-transcriptional levels. J. Biol. Chem. 279, 612–618 [DOI] [PubMed] [Google Scholar]
- 79. Zhang Y., Qin K., Wang J., Hung T., Zhao R. Y. (2006) Dividing roles of prion protein in staurosporine-mediated apoptosis. Biochem. Biophys. Res. Commun. 349, 759–768 [DOI] [PubMed] [Google Scholar]
- 80. Gougoumas D. D., Vizirianakis I. S., Triviai I. N., Tsiftsoglou A. S. (2007) Activation of Prn-p gene and stable transfection of Prn-p cDNA in leukemia MEL and neuroblastoma N2a cells increased production of PrP(C) but not prevented DNA fragmentation initiated by serum deprivation. J. Cell. Physiol. 211, 551–559 [DOI] [PubMed] [Google Scholar]
- 81. Westergard L., Turnbaugh J. A., Harris D. A. (2011) A naturally occurring C-terminal fragment of the prion protein (PrP) delays disease and acts as a dominant-negative inhibitor of PrPSc formation. J. Biol. Chem. 286, 44234–44242 [DOI] [PMC free article] [PubMed] [Google Scholar]