Background: Regiospecific installation of α-l-fucosyl residue into glycoconjugates is quite difficult.
Results: A glycosynthase mutant of 1,3-1,4-α-l-fucosidase specifically synthesized Lewis a/x trisaccharides using Galβ1–3/4GlcNAc as acceptors.
Conclusion: Structural studies provide a rationale to explain the unusually strict substrate specificity exhibited by the enzyme.
Significance: A new enzymatic route for specifically introducing Lewis a/x epitopes into type-1/2 chains becomes available.
Keywords: Carbohydrate Glycoconjugate, Enzyme Mechanisms, Glycobiology, Oligosaccharide, X-ray Crystallography
Abstract
α-l-Fucosyl residues attached at the non-reducing ends of glycoconjugates constitute histo-blood group antigens Lewis (Le) and ABO and play fundamental roles in various biological processes. Therefore, establishing a method for synthesizing the antigens is important for functional glycomics studies. However, regiospecific synthesis of glycosyl linkages, especially α-l-fucosyl linkages, is quite difficult to control both by chemists and enzymologists. Here, we generated an α-l-fucosynthase that specifically introduces Lea and Lex antigens into the type-1 and type-2 chains, respectively; i.e. the enzyme specifically accepts the disaccharide structures (Galβ1–3/4GlcNAc) at the non-reducing ends and attaches a Fuc residue via an α-(1,4/3)-linkage to the GlcNAc. X-ray crystallographic studies revealed the structural basis of this strict regio- and acceptor specificity, which includes the induced fit movement of the catalytically important residues, and the difference between the active site structures of 1,3-1,4-α-l-fucosidase (EC 3.2.1.111) and α-l-fucosidase (EC 3.2.1.51) in glycoside hydrolase family 29. The glycosynthase developed in this study should serve as a potentially powerful tool to specifically introduce the Lea/x epitopes onto labile glycoconjugates including glycoproteins. Mining glycosidases with strict specificity may represent the most efficient route to the specific synthesis of glycosidic bonds.
Introduction
α-l-Fucosyl residues attached at the non-reducing ends of glycoconjugates constitute histo-blood group antigens Lewis (Le)2 and ABO. The blood group antigens are involved in various important biological processes, and especially Lewis a and x epitopes (Galβ1–3/4(Fucα1–4/3)GlcNAc-; Lea/x) play fundamental roles in mammalian cell-to-cell communications at the developmental stages and at the sites of inflammation (1–5). In mouse embryogenesis, the stage-specific synthesis of Lex antigen has been observed, and this transient expression is thought to be involved in embryo compaction (2). Regulated expression of Lex epitope is also found in the brain and postulated to be important for the development of the central nervous system. In addition, recent studies show that Le antigens modulate host-pathogen interactions (6, 7). Helicobacter pylori shows phase-variable expression of Le antigens in its lipopolysaccharides (LPS), and the antigens act as adhesion pedestals between the organism and the host epithelial cells. The bacterium also utilizes the Lex epitope of LPS to suppress immune responses through the dendritic cell-specific intercellular adhesion molecule-3-grabbing non-integrin (8). The Lea/x antigen-containing oligosaccharides are therefore indispensable tools for functional glycomics studies and for pharmaceutical applications.
The synthesis of oligosaccharides with defined structures requires the precise control of regio- and stereospecificity at the glycosidic bond. In this context, enzymatic synthesis has several advantages over chemical synthesis because it enables the perfect control of anomeric configurations and provides relatively high regiospecificity without the requirement of laborious protection/deprotection steps. Enzymatic synthesis of oligosaccharides usually utilizes glycosyltransferases and glycosidases (9, 10). Glycosyltransferases generally have strict regio- and acceptor specificity and are therefore good catalysts for defined oligosaccharide syntheses. Efficient preparations of Lex and H antigen oligosaccharides using α-1,3- and α-1,2-fucosyltransferases from H. pylori have been reported (11–13). However, as described in the literature (11), the use of glycosyltransferases requires an expensive sugar nucleotide or a complex system for nucleotide recycling. Moreover, the expression of glycosyltransferases is generally difficult to handle. In contrast, glycosidase-catalyzed transglycosylation has been efficiently used as a method for oligosaccharide synthesis because of the simplicity and versatility of the reactions (9, 10). Notably, a new class of enzymes, glycosynthases, has been developed in the last decade (14–17). Glycosynthase is a mutant glycosidase that is devoid of hydrolysis but is able to perform the transglycosylation reaction when a suitably activated donor (generally glycosyl fluoride and in a few cases glycosyl azide) (18, 19) is used as a substrate. Use of glycosidases and their mutants has thus become a promising option for the synthesis of oligosaccharides (10, 20, 21). However, as the regioselectivity and acceptor specificity of glycosidase-catalyzed transglycosylation are usually not that high, the reaction products are frequently obtained as mixtures of several oligosaccharides with varied linkages and sometimes varied lengths. These drawbacks become apparent in the synthesis of fucosyl oligosaccharides using α-l-fucosidases (22–25) and α-l-fucosynthases (18) (details are described later) that belong to glycoside hydrolase family 29 (GH29) (26). Such a result prevents the use of these enzymes in the synthesis of oligosaccharides with defined structures. Thus, in view of the requirement of strict glycosidic bond formation, it is crucial to find glycosidases that provide strict specificity, both in linkage and in leaving groups that become acceptors for transglycosylation.
In previous studies, we have isolated two α-l-fucosidases from Bifidobacterium bifidum and revealed that the enzymes have strict substrate specificities. One is 1,2-α-l-fucosidase (BbAfcA) that belongs to GH95 (27, 28), and the other is 1,3-1,4-α-l-fucosidase (BbAfcB) belonging to GH29 (29). The high substrate specificities of the two α-l-fucosidases prompted us to examine the possible use of these enzymes in the defined synthesis of fucosyl oligosaccharides. In a recent study, we introduced the glycosynthase technology into BbAfcA (an inverting enzyme) and succeeded in generating 1,2-α-l-fucosynthase, which synthesized 2′-fucosyllactose (Fucα1–2Galβ1–4Glc; 2′-FL) exclusively when β-l-fucopyranosyl fluoride (FucF) and lactose (Lac) were used as a donor and an acceptor, respectively (30). No by-products were formed in that reaction.
In the present study, to expand the possibility of glycosynthase technology, we converted BbAfcB to a 1,3-1,4-α-l-fucosynthase and characterized the enzyme. The results indicated that the synthase should serve as a valuable tool to specifically introduce Lea and Lex epitopes into the type-1 (Galβ1-3GlcNAc; lacto-N-biose I (LNB)) and type-2 (Galβ1–4GlcNAc; N-acetyllactosamine (LacNAc)) chains, respectively. Moreover, the crystal structures of AfcB from Bifidobacterium longum subsp. infantis (BiAfcB; Blon_2336), a paralogue of BbAfcB, in complex with an inhibitor, deoxyfuconojirimycin (DFJ), and with lacto-N-fucopentaose II (Galβ1–3(Fucα1–4)GlcNAcβ1–3Galβ1–4Glc; LNFP II) (comprising the Lea antigen), were determined. The structures revealed how the enzyme exerts its strict regio- and acceptor specificity through an induced fit motion of catalytically important residues.
EXPERIMENTAL PROCEDURES
Chemicals
2′-FL, LacNAc, Fucα1–6GlcNAc disaccharide, and Lea and Lex trisaccharides were purchased from Dextra Laboratories (Reading, UK). Lacto-N-tetraose (Galβ1–3GlcNAcβ1-3Galβ1–4Glc; LNT) and lactodifucotetraose (Fucα1–2Galβ1–4(Fucα1–3)Glc) were from Isosep (Tullinge, Sweden). LNFP II and Fucα1–3/4GlcNAc disaccharides were from Carbosynth (Compton, UK). N,N′-Diacetylchitobiose, DFJ, and para-nitrophenyl-α-l-fucopyranoside (pNP-Fuc) were from Seikagaku Kogyo (Tokyo, Japan), Toronto Research Chemicals (North York, Ontario, Canada), and Sigma, respectively. LNB (31), galacto-N-biose (Galβ1–3GalNAc) (32), 2-acetamide-2-deoxy-4-O-(β-glucosyl)-glucose (Glcβ1–4GlcNAc; GlcGlcNAc) (33), and FucF (30) were synthesized as described previously. 3-Fucosyllactose (Galβ1–4(Fucα1–3)Glc; 3-FL) was purchased from Dextra Laboratories and further purified using Bio-Gel P2 gel filtration chromatography (Bio-Rad). l-Fucose dehydrogenase was purchased from Kikkoman (Noda, Japan). Other reagents of analytical grade were obtained from commercial sources.
Construction of BbAfcB Mutants
The mutants of BbAfcB (D703A, D703C, D703G, D703S, W742A, E746A, D763A, D766A, D778A, and D807A) were constructed using the QuikChange site-directed mutagenesis method (Stratagene) with the plasmid pET23b-afcB as the template (29). The following primers and their complementary primers were used: 5′-gaggtctggttcgcgggtgcccaaggc-3′ (D703A), 5′-aggtctggttctgcggtgcccaaggc-3′ (D703C), 5′-gaggtctggttcggcggtgcccaaggc-3′ (D703G), 5′-gaggtctggttcagcggtgcccaaggc-3′ (D703S), 5′-acgatgcccgagcggtgggcaacg-3′ (W742A), 5′-ggtgggcaacgcggacggctggg-3′ (E746A), 5′-ggcatacaacgccggcgtggaca-3′ (D763A), 5′cgacggcgtggccaaggtgtcgc-3′ (D766A), 5′-gatggcccccgccggtaagcttg-3′ (D778A), and 5′-ggccgaagtcgccgccaagaacc-3′ (D807A). The entire sequence used for later manipulation was sequenced to check that no base change other than those designed had occurred. The resulting plasmids were used to transform Escherichia coli BL21 ΔlacZ (DE3) (29).
Expression and Purification of BbAfcB Variants
The recombinant strains were cultured in Luria-Bertani medium containing 100 μg/ml ampicillin at 18 °C until the optical density at 600 nm reached 0.5. Isopropyl β-d-thiogalactopyranoside was then added to a final concentration of 0.1 mm to induce protein expression. Following further incubation for 15 h, the cells were harvested and disrupted by sonication. After centrifugation, the supernatant was applied to a nickel-nitrilotriacetic acid-agarose column (Qiagen, Hilden, Germany), and the protein was eluted according to the manufacturer's instructions. The fractions were combined, concentrated using Amicon Ultra 50K (Millipore), and loaded onto a Superdex 200 10/300 GL gel filtration column (GE Healthcare). The elution was carried out using 20 mm Tris-HCl (pH 8.0) containing 150 mm NaCl. The purified protein was extensively dialyzed against a 50 mm HEPES buffer (pH 7.0). Protein concentrations were determined using a BCA protein assay kit (Thermo Scientific) with bovine serum albumin as a standard.
Enzyme Assay
The hydrolytic activities of BbAfcB variants were determined using 3-FL and pNP-Fuc as the substrates. The reaction mixture contained 100 mm MOPS buffer (pH 6.5), 1 mm substrate, and the enzyme (for 3-FL hydrolysis: 14 nm wild-type (WT) and D766A, 12 μm Asp-703 mutants, 17 μm W742A and E746A, 44 nm D763A and D778A, and 890 nm D807A; for pNP-Fuc hydrolysis: 17 μm WT and mutants). The reaction was carried out for an appropriate time in which the linearity of the reaction rate was observed at 30 °C (i.e. in the 3-FL hydrolysis: ∼15 min for WT, W742A, D807A, D763A, D766A, and D778A mutants; ∼880 min for Asp-703 mutants; and ∼240 min for E746A mutant; in the pNP-Fuc hydrolysis: ∼150 min for WT and all mutants except for W742A and ∼500 min for W742A). The reaction was terminated by heating (for 3-FL hydrolysis) or by the addition of 1 m sodium carbonate (for pNP-Fuc hydrolysis). The amount of released Fuc was determined using a fucose dehydrogenase-coupled method (34). The amount of the liberated pNP was determined by measuring the absorbance at 405 nm. One unit of enzyme activity was determined as the amount of enzyme required to produce 1 μmol of Fuc or pNP under the specified conditions.
The glycosynthase activities of BbAfcB mutants were examined by incubating the reaction mixtures (50 μl) containing 100 mm MOPS buffer (pH 7.0), 20 mm FucF (donor), 100 mm LNB/LacNAc (acceptor), and a 17 μm concentration of the enzymes. The reaction was performed for 10 min at 30 °C and terminated by adding 5 μl of 30% trichloroacetic acid. The reaction products were analyzed by high-performance liquid chromatography (HPLC) equipped with a sugar-D column (4.6 × 250 mm; Nacalai Tesque, Kyoto, Japan). The elution was performed under a constant flow (1.0 ml/min) of 75 or 78% acetonitrile at 40 °C and monitored at 214 nm or using a charged aerosol detector (CAD) (Corona CAD, Thermo Scientific). The amounts of the reaction products were estimated by a standard curve created using known concentrations of Lea and Lex trisaccharides.
The optimal pH for the glycosynthase reaction was determined using the BbAfcB D703S mutant. The buffers (100 mm) used were as follows: citrate-NaOH (pH 4.0–5.0), MES (pH 5.0–6.5), and MOPS (pH 6.5–8.0). The acceptor specificity of the BbAfcB D703S mutant was examined using 13 different substrates. The reaction was carried out in 100 mm MES buffer (pH 5.0) containing 40 mm FucF and a 100 mm concentration of each acceptor for 40 min at 30 °C in the presence and absence of 17 μm D703S. The reaction products were analyzed by HPLC-CAD.
Synthesis, Purification, and Structural Analysis of Reaction Products
For Lea synthesis, the BbAfcB D703S mutant (17 μm) was incubated in the reaction mixture (300 μl) consisting of 100 mm MES buffer (pH 5.0), 40 mm FucF (donor), and 200 mm LNB (acceptor) at 30 °C for 40 min. Lex trisaccharide and LNFP II were synthesized in a similar manner except that 100 mm LacNAc and LNT were used as the acceptors, respectively. The reaction products were deionized with Amberlite MB-3, lyophilized, and purified using HPLC equipped with a sugar-D column. The elution was performed as described above. Peak fractions were collected, lyophilized, and further purified by a TSKgel ODS-80TS column (4.6 × 250 mm; Tosoh, Japan) to remove the remaining acceptors (LNB/LacNAc/LNT). The elution was carried out using water at a flow rate of 0.5 ml/min and monitored by a refractive index detector (RID-10A, Shimadzu, Kyoto, Japan). 1H and 13C nuclear magnetic resonance (NMR) spectra were recorded on a Bruker Avance 500 spectrometer. Electrospray ionization-mass spectrometry (MS) was carried out in positive mode using a Bruker APEX II 70e Fourier transformed ion-cyclotron resonance mass spectrometer.
X-ray Crystallography of BiAfcB
The gene (locus tag Blon_2336) was amplified by PCR using the genomic DNA of B. longum subsp. infantis ATCC15697 and a primer pair (5′-gcatatgaacaatcctgcagatgc-3′ and 5′-ctcgagtcagatgcgcacggcagcc-3′). The amplified fragment was inserted into the NdeI and XhoI sites of pET-30b to generate a non-tagged protein (Novagen). The resulting plasmid, pET30b-BiafcB, was used to transform E. coli BL21 ΔlacZ (DE3). The transformants were cultured in Luria-Bertani medium containing 30 μg/ml kanamycin at 18 °C to an optical density at 600 nm of 0.5. Isopropyl β-d-thiogalactopyranoside was added to a final concentration of 0.1 mm to induce protein expression. Following further incubation for 20 h, the harvested cells were disrupted by sonication. The protein was purified using Mono Q 5/50 GL column chromatography (GE Healthcare) followed by a Superdex 200 10/300 GL gel filtration chromatography step. The BiAfcB D172A/E217A double mutant was constructed using the QuikChange site-directed mutagenesis method with the plasmid pET30b-BiafcB as the template. The following primers and their complementary primers were used: 5′-ccgtctggcttgctggcgccaatgg-3′ (D172A) and 5′-tgggccgggaacgcagccgggcatgtg-3′ (E217A). Purification of the mutant protein was performed using a procedure similar to that described for the WT protein. The proteins were crystallized using the hanging drop-vapor diffusion method at 20 °C. The crystal of BiAfcB complexed with DFJ and ethylene glycol (WT-DFJ-EG) was obtained by mixing 1 μl of a protein solution (10 mg/ml in 10 mm HEPES buffer (pH 7.0) containing 40 mm DFJ) with 1 μl of a reservoir solution consisting of 0.1 m citrate buffer (pH 4.0) and 20% (w/v) polyethylene glycol (PEG) 6000. The crystal of BiAfcB D172A/E217A complexed with LNFP II (D172A/E217A-LNFP II) was obtained by mixing 1 μl of a protein solution (10 mg/ml in 10 mm HEPES buffer (pH 7.0) containing 100 mm LNFP II) with 1 μl of a reservoir solution consisting of 0.1 m sodium citrate buffer (pH 5.5) and 20% PEG 3000. After cryoprotection with 20% ethylene glycol, the crystals were flash cooled in a nitrogen stream at 100 K. Diffraction data were collected using beamline NW12A at the Photon Factory-Advanced Ring, KEK, Tsukuba (λ = 1.0 Å). Diffraction images were processed using HKL2000 (35). Molecular replacement was performed with MOLREP (36) using the ligand-free BiAfcB structure (Protein Data Bank code 3MO4) (37) as a search model. Model rebuilding and refinement were performed using Coot (38) and Refmac5 (39). Figures were prepared using PyMOL (Schrödinger, LLC).
Automated Docking
Automated docking analysis was performed as described previously (40). Ligand model preparation and automated docking were performed using the Sweet2 server (41) and AUTODOCK 4.0 (42), respectively. Rotatable ligand bonds (eight in Lex trisaccharide) were defined using the AutoDockTools interface. Water molecules in the D172A/E217A-LNFP II complex structure were removed. After adding polar hydrogens, Gasteiger charges were calculated for the ligand and protein. Grid maps were prepared with 40 × 40 × 40 points covering the substrate-binding pocket with a point spacing of 0.375 Å. For the Lamarckian genetic algorithm search, the size of the initial random population was 150 individuals, the maximal number of energy evaluations was 2.5 × 106, the maximal number of generations was 27,000, the number of top individuals that survived into the next generation was 1, the rate of mutation was 0.02, the rate of crossover was 0.80, and the average of the worst energy was calculated over a window of 10 generations. After 256 docking runs, all structures generated for a single compound were assigned to clusters based on a tolerance of 2.0 Å for all atom root mean square deviations from the lowest energy structure.
RESULTS
Conversion of 1,3-1,4-α-l-Fucosidase to 1,3-1,4-α-l-Fucosynthase
BbAfcB belongs to GH29 (26). The members of this family hydrolyze α-l-fucosidic bonds via a retaining mechanism (43, 44). We replaced Asp-703, which is assumed to be a nucleophile by sequence alignment with well studied GH29 members (discussed later) (45), with alanine, cysteine, glycine, and serine and examined the hydrolytic activity of these mutants. Note that only the specific activities were determined because the kinetic parameters could not be calculated (supplemental Table S1). The specific activities of D703A, D703C, D703G, and D703S were found to decrease by 160,000-, 46,000-, 17,000-, and 76,000-fold, respectively, as compared with that of the WT enzyme.
The glycosynthase activities of these mutants were then examined using 20 mm FucF and 100 mm LNB or LacNAc as the donor and the acceptor, respectively. Peaks corresponding to Lea and Lex trisaccharides appeared for the D703G/S mutants in the HPLC analysis of the reaction products, whereas no peak was observed for WT and the D703A/C mutants (supplemental Fig. S1). In either case, D703S gave a slightly larger peak area than D703G, and hence, D703S was further analyzed.
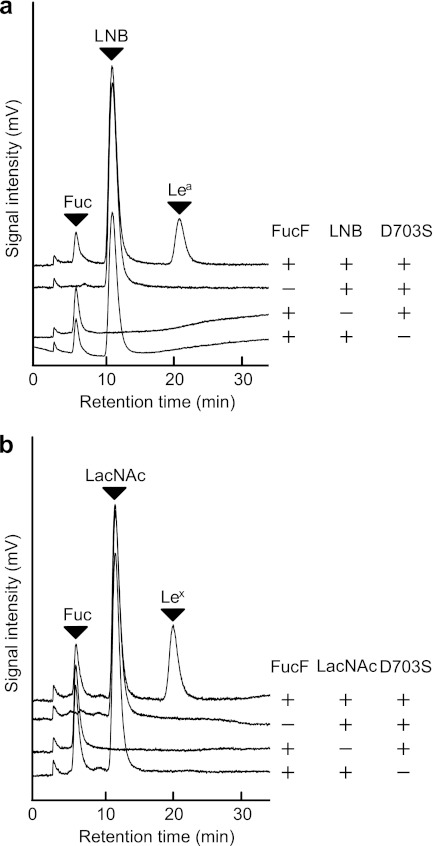
The optimal pH for the synthase reaction was found to be 5.0 (data not shown). The synthase activity was observed only when the reaction was carried out in the presence of FucF, LNB/LacNAc, and the enzyme. No peaks other than the added substrates were detected when the donor (FucF), the acceptor (LNB/LacNAc), or the enzyme was omitted (Fig. 1, a and b). Strikingly, no by-products other than the decomposed Fuc were detected in the reaction as revealed by HPLC-CAD. Identification of the reaction products is described later.
FIGURE 1.
Specific syntheses of Lea (a) and Lex (b) trisaccharides by the BbAfcB D703S mutant. a, the reaction was carried out for 40 min at 30 °C in 100 mm MES buffer (pH 5.0) containing 40 mm FucF (donor), 200 mm LNB (acceptor), and a 17 μm concentration of the BbAfcB D703S mutant. For the control experiments, the substrate or enzyme was omitted from the reaction. b, for the synthesis of Lex, 100 mm LacNAc was used as the acceptor. The reaction products were analyzed by HPLC-CAD. The peaks of l-fucose (Fuc), LNB, LacNAc, Lea, and Lex are shown. Note that FucF is decomposed when terminating the reaction by trichloroacetic acid.
The kinetic parameters of the reaction could not be calculated because of the rapid spontaneous decomposition of FucF. The half-life of FucF under the reaction conditions was determined to be 20 min (data not shown). In the time course of the optimized reaction, the concentrations of the products Lea and Lex reached a maximum at ∼40 and 60 min, respectively (supplemental Fig. S2). The reaction efficiency was estimated to be 56% against the added FucF when the reaction was carried out under the optimized conditions, i.e. in the presence of 40 mm FucF (donor), 200 mm LNB or 100 mm LacNAc (acceptor), and a 17 μm concentration of the enzyme. None of the BbAfcB Asp-703 variants used β-l-fucopyranosyl azide as a donor (18) or were rescued by sodium azide (data not shown).
Acceptor Specificity
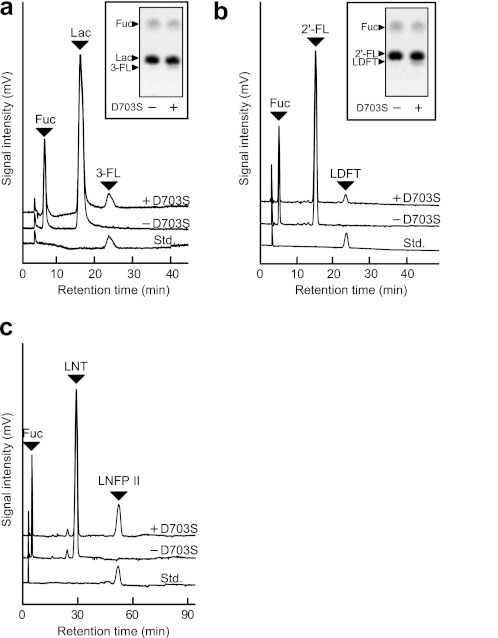
The acceptor specificity of BbAfcB D703S was examined using various mono- and oligosaccharides at a fixed concentration (100 mm) (Table 1, Fig. 2, and supplemental Fig. S3). In addition to LNB and LacNAc, the enzyme recognized Lac, 2′-FL, and LNT as acceptors and specifically produced 3-FL, lactodifucotetraose, and LNFP II in yields of 13, 5.5, and 41% against the added FucF, respectively. In contrast, the enzyme did not accept monosaccharides (Glc, Gal, GlcNAc, and GalNAc), cellobiose, GlcGlcNAc, N,N′-diacetylchitobiose, or galacto-N-biose as a substrate.
TABLE 1.
Acceptor specificity of 1,3-1,4-α-l-fucosynthase (BbAfcB D703S)
The reaction was carried out in 100 mm MES buffer (pH 5.0) containing 40 mm FucF (donor) and 100 mm of each acceptor for 40 min at 30 °C in the presence and absence of 17 μm D703S. The reaction products were analyzed by HPLC-CAD (Fig. 2 and supplemental Fig. S3).
| Substrates |
Synthesized compounds |
Yielda | ||
|---|---|---|---|---|
| Name | Structure | Name | Structure | |
| % | ||||
| Monosaccharides | ||||
| Glucose | 0 | |||
| Galactose | 0 | |||
| N-Acetylglucosamine | 0 | |||
| N-Acetylgalactosamine | 0 | |||
| Disaccharides | ||||
| Cellobiose | Glcβ1–4Glc | 0 | ||
| GlcGlcNAcb | Glcβ1–4GlcNAc | 0 | ||
| N,N′-Diacetylchitobiose | GlcNAcβ1–4GlcNAc | 0 | ||
| Lac | Galβ1–4Glc | 3-FL | Galβ1–4 (Fucα1–3)Glc | 13 |
| LNBb | Galβ1–3GlcNAc | Lea trisaccharide | Galβ1–3 (Fucα1–4)GlcNAc | 47 |
| LacNAc | Galβ1–4GlcNAc | Lex trisaccharide | Galβ1–4 (Fucα1–3)GlcNAc | 55 |
| Galacto-N-bioseb | Galβ1–3GalNAc | 0 | ||
| Tri- and tetrasaccharides | ||||
| 2′-FL | Fucα1–2Galβ1–4Glc | Lactodifucotetraose | Fucα1–2Galβ1–4 (Fucα1–3)Glc | 5.5 |
| LNT | Galβ1–3GlcNAcβ1–3Galβ1–4Glc | LNFP II | Galβ1–3 (Fucα1–4)GlcNAcβ1–3Galβ1–4Glc | 41 |
FIGURE 2.
Acceptor specificity of BbAfcB D703S glycosynthase. The reaction was carried out in 100 mm MES buffer (pH 5.0) containing 40 mm FucF and a 100 mm concentration of each acceptor for 40 min at 30 °C in the presence and absence of the D703S mutant (17 μm). The acceptors used were as follows: Lac (a), 2′-FL (b), and LNT (c). The reaction products were analyzed by HPLC-CAD. The peaks of l-fucose (Fuc), acceptor, and standard sugars (Std.) are indicated. Inset in a and b are the results of the thin-layer chromatography (TLC) analysis of the reaction products. See also Table 1 and supplemental Figs. S3–S7.
Regio- and Stereospecificity
We then analyzed the reaction products eluted from HPLC using electrospray ionization-MS and 1H and 13C NMR spectroscopy. The MS spectra indicated that one fucosyl residue was attached to LNB and LacNAc (calculated for the sodium adduct [M + Na]+, 552.19; observed, 552.18; supplemental Fig. S4). Both of the 1H and 13C NMR spectra were identical to the spectra of the authentic Lea and Lex trisaccharides (supplemental Figs. S5 and S6). In the case where LNT was used as an acceptor, only LNFP II, but not LNFP V (Galβ1–3GlcNAcβ1–3Galβ1–4(Fucα1–3)Glc)or lacto-N-difucohexaose II (Galβ1–3(Fucα1–4)GlcNAcβ1-3Galβ1–4(Fucα1–3)Glc), was synthesized as revealed by 1H NMR analysis (supplemental Fig. S7).
Crystallography of 1,3-1,4-α-l-Fucosidase
To elucidate the structural basis underlying the unique substrate specificity of this synthase, we tried to crystallize BbAfcB and its variants; however, no crystal was obtained. B. longum subsp. infantis possesses the paralogue (locus_tag 2336) of BbAfcB (46), and we found that this paralogue (referred to as BiAfcB) has a substrate specificity identical to that of BbAfcB (supplemental Fig. S8). The Pfam database suggested that BbAfcB is a multimodular enzyme, whereas BiAfcB possesses a simple domain organization. The structure of a ligand-free form of BiAfcB has been recently determined (Protein Data Bank code 3MO4) (37). We obtained the crystals of BiAfcB under different conditions from those used by Sela et al. (37) and determined two complex structures of BiAfcB: WT complexed with DFJ and EG (WT-DFJ-EG; 1.6-Å resolution) and an inactive mutant complexed with LNFP II (D172A/E217A-LNFP II; 2.1-Å resolution). Asp-172 and Glu-217 are a nucleophile and a general acid/base, respectively (described later). Data collection and refinement statistics are shown in Table 2. BiAfcB forms a dimer in solution (data not shown), and all three BiAfcB crystals contain homodimers in the asymmetric unit. Because the two chains (A and B) were almost identical in every case, we describe the chain A of each structure. The DFJ and ethylene glycol (cryoprotectant) molecules in the WT-DFJ-EG structure and the Fuc and Gal moieties in the D172A/E217A-LNFP II structure were clearly observed (supplemental Fig. S9). However, the GlcNAc moiety in LNFP II was partially ambiguous, and the β1,3-linked Lac moiety at the reducing end was not visible. Therefore, we included the Lea trisaccharide structure (Galβ1–3(Fucα1-4)GlcNAc) in the model. We also determined a complex structure of D172A/E217A with the Lea trisaccharide, and the resulting electron density map was virtually identical to that of the D172A/E217A-LNFP II structure (data not shown).
TABLE 2.
Crystallographic data collection and refinement statistics
r.m.s.d., root mean square deviation. Values in parentheses are for the highest-resolution shell.
| Data set | WT-DFJ-EG | D172A/E217A-LNFP II |
|---|---|---|
| Data collection | ||
| Protein Data Bank code | 3UES | 3UET |
| Beamline | NW12A | NW12A |
| Space group | P212121 | P212121 |
| Unit cell (Å) | a = 64.0, b = 120.6, c = 142.5 | a = 82.6, b = 106.0, c = 120.7 |
| Matthews coefficient (Å3/Da) | 2.59 | 2.49 |
| Solvent content (%) | 52.5 | 50.6 |
| Resolution (Å) | 50.00-1.60 (1.66-1.60) | 50.00-2.10 (2.14-2.10) |
| Total reflections | 1,029,177 | 358,658 |
| Unique reflections | 146,170 | 60,403 |
| Completeness (%) | 99.9 (100.0) | 96.4 (95.4) |
| Redundancy | 7.0 (6.7) | 5.9 (5.2) |
| Mean I/σ(I) | 36.2 (4.1) | 16.6 (2.6) |
| Rsym (%) | 5.9 (33.5) | 10.4 (47.4) |
| Refinement | ||
| Resolution (Å) | 25.91-1.60 | 32.05-2.10 |
| No. of reflections | 138,464 | 57,112 |
| R-factor/Rfree (%) | 16.0/19.1 | 17.3/22.3 |
| No. of atoms | 8,320 | 7,683 |
| r.m.s.d. from ideal values | ||
| Bond lengths (Å) | 0.029 | 0.020 |
| Bond angles (°) | 2.561 | 1.987 |
| Ramachandran plot (%) | ||
| Favored | 98.0 | 97.1 |
| Allowed | 1.8 | 2.7 |
| Outlier | 0.2 | 0.2 |
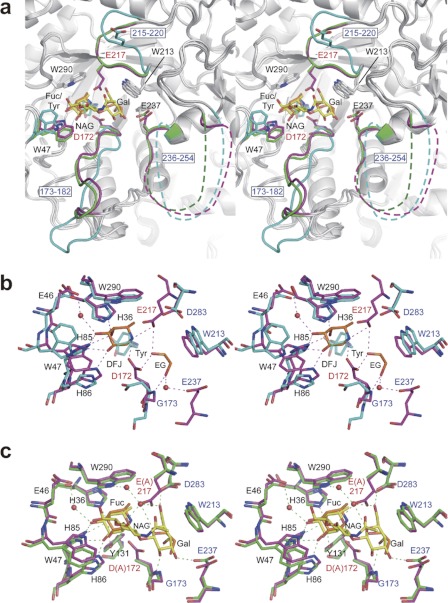
Fig. 3 shows the comparison between the three BiAfcB structures. Interestingly, upon ligand binding, BiAfcB undergoes a large conformational change at two loop regions (173–182 and 215–220) (Fig. 3a), and to our surprise, the acid/base catalyst Glu-217 is included in this induced fit motion. In the substrate-free structure, the catalytic residues (Asp-172 and Glu-217) are not appropriately poised (separated by 20.8 Å), and a tyrosine molecule occupies subsite −1 (Fuc-binding site) (Fig. 3b). In the WT-DFJ-EG structure, the loop (215–220) covers the catalytic pocket so that Glu-217 can be suitably poised to act as an acid/base. Consequently, the distance between the Oδ atom of Asp-172 and the Oϵ atom of Glu-217 becomes 5.8 Å, which is the typical length observed in retaining glycosidases (43). The Oδ atom of Asp-172 is located at a distance of 3.1 Å from C1 of the DFJ. Alanine replacement of each of these residues resulted in a ∼20,000-fold decrease of the hydrolytic activity (data not shown). The DFJ and ethylene glycol molecules are tightly bound by many hydrogen bonds with the protein. In the D172A/E217A-LNFP II structure, Fuc and Gal moieties of LNFP II are extensively recognized through the formation of hydrogen bonds, whereas the GlcNAc moiety does not make any notable interactions with the protein. The Fuc makes direct hydrogen bonds with His-36, Trp-47, His-85, His-86, and Tyr-131, and its C6 methyl group makes hydrophobic interactions with Phe-34, Trp-170, and Trp-290. The Gal moiety forms hydrogen bonds with the nitrogen atom of the main chain of Gly-173 and the side chains of Glu-237 and Asp-283, and its hydrophobic β-face is stacked by Trp-213 (Fig. 3c). The ethylene glycol in the WT-DFJ-EG complex occupies the Gal-binding site and interacts with the catalytic residues (Asp-172 and Glu-217) and the main chain of Gly-173. A water-mediated hydrogen bond is also formed with the side chain of Glu-237. However, its two hydroxyl groups do not overlap any of the Gal hydroxyl groups (Fig. 3, b and c).
FIGURE 3.
Induced fit movement of BiAfcB upon substrate binding (stereoviews). a, superimposed structures of the ligand-free form (cyan; Protein Data Bank code 3MO4) (37), WT-DFJ-EG complex (protein in magenta; ligand in orange), and D172A/E217A-LNFP II complex (protein in green; ligand in yellow). The ligand-free structure contains a tyrosine molecule at the Fuc-binding site (subsite −1). The side chain of Trp-47 and two loop regions (173–182 and 215–220) are significantly displaced. Disordered regions of the 236–254 loop are shown as dotted lines. b and c, the structure of the substrate-binding site. b, ligand-free (cyan) and WT-DFJ-EG complex (protein in magenta; ligand in orange). The tyrosine molecule bound to the ligand-free structure is shown transparently. Water molecules and hydrogen bonds in the WT-DFJ-EG complex are shown. c, WT-DFJ-EG complex (protein in magenta; ligand in orange) and D172A/E217A-LNFP II complex (protein in green; ligand in yellow). Water molecules and hydrogen bonds in the LNFP II complex are shown. Ethylene glycol and GlcNAc are labeled as EG and NAG, respectively.
Sequence identity between BbAfcB and BiAfcB is modest (34%) in the alignment (495–998 for BbAfcB and 4–472 for BiAfcB); however, the residues involved in substrate binding are essentially conserved to reflect the same substrate specificity (supplemental Fig. S10). Asp-172, Gly-173, Trp-213, Glu-217, and Asp-283 of BiAfcB correspond to Asp-703, Gly-704, Trp-742, Glu-746, and Asp-807 in BbAfcB, respectively. BbAfcB does not have a residue corresponding to Glu-237 of BiAfcB located in the disordered loop (236–254), but instead, three acid residues (Asp-763, Asp-766, and Asp-778) are present in the corresponding region. The specific activities of W742A, E746A, and D807A of BbAfcB for the hydrolysis of 3-FL decreased by 1,800-, 6,900-, and 80-fold, respectively, as compared with that of the WT enzyme (supplemental Table S1). Alanine substitution for Asp-763, Asp-766, and Asp-778 slightly lowered the enzyme activity.
DISCUSSION
A powerful tool to regiospecifically and stereospecifically install Fuc residues into glycoconjugates was developed. The BbAfcB D703S mutant can introduce Lea and Lex epitopes into type-1 and type-2 chains at the non-reducing ends, respectively, without producing by-products. The yields were ∼60% with respect to the added FucF. The reaction efficiency of this glycosynthase may be slightly lower than those of typical glycosynthases (80% or more), and this could be due to the spontaneous decomposition of FucF during the reaction as described above. Recently, Cobucci-Ponzano et al. (18, 19) demonstrated that some glycosynthases can accept glycosyl azide as a donor molecule. This finding is particularly valuable for oligosaccharide synthesis using α-glycosynthases because β-glycosyl azides are considerably more stable than β-glycosyl fluorides in aqueous solution. We examined the usage of β-l-fucopyranosyl azide in the synthase reaction of BbAfcB mutants, but no product was obtained in that reaction.
The glycosynthase exclusively recognized LNB, LacNAc, and Lac disaccharide structures (Galβ1–3/4GlcNAc(Glc)) at the non-reducing ends and did not recognize monosaccharides. Indeed, WT BbAfcB did not hydrolyze Fucα1–3/4GlcNAc disaccharides (supplemental Fig. S8), which had not been examined in our previous study (29). These results indicate that the presence of the branched Gal residue is critical for the catalytic activities of BbAfcB and the glycosynthase mutant.
Structural Basis for Substrate Specificity of 1,3-1,4-α-l-Fucosynthase
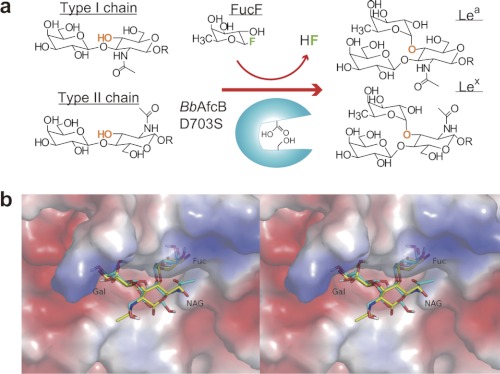
BiAfcB was identified as a paralogue of BbAfcB (46), and it showed the same substrate specificity as BbAfcB (supplemental Fig. S8). Structural analysis of BiAfcB enabled us to consider the mechanism of the unique enzymatic activity of 1,3-1,4-α-l-fucosidase(synthase). Binding of Fuc(F) and the Gal moiety of LNB/LacNAc would cause the induced fit movement at the catalytic site. The ethylene glycol molecule in the WT-DFJ-EG complex may stabilize the induced fit structure in the crystal. The Gal-binding site could be highly specific to Gal as the axial O4 hydroxyl group is recognized by the main chain (Gly-173 in BiAfcB and Gly-704 in BbAfcB), which is next to the nucleophile residue and is included in the mobile loop (Fig. 3c and supplemental Fig. S10). The glycosynthase neither accepted cellobiose nor GlcGlcNAc (Table 1). The Gal O2 hydroxyl group is not recognized by any protein residues and is exposed to the solvent (Fig. 3c), which rationalizes that the enzyme used 2′-FL as the acceptor. The GlcNAc-binding site appears to be rather promiscuous. It is interesting to note that the faces of the GlcNAc sugar rings are inverted between the LNB and LacNAc disaccharides, and consequently, the geometric positions of the respective O4 and O3 hydroxyl groups of the GlcNAc residues become identical (Fig. 4). When automated docking analysis was performed using Lex trisaccharide, the best docking result was found in the first ranked cluster containing the majority of the 256 docking run conformations. The number of conformations in this cluster, lowest binding energy, and mean binding energy were 81, −6.30, and −3.81 kcal/mol, respectively. In the predicted mode of Lex binding, the Fuc, Gal, and GlcNAc moieties of the molecule overlap well with those of the Lea trisaccharide observed in the D172A/E217A-LNFP II crystal except for the hydroxymethyl and N-acetyl groups of the GlcNAc (Fig. 4b). The O1 atoms of the inverted GlcNAc residues are exposed to the solvent in both cases and rather protrude from the catalytic pocket. These results explain why this synthase accepts LNB, LacNAc, and LNT almost equally (Table 1). The enzyme probably has no structural constraint at the reducing ends of the LNB/LacNAc disaccharides. Indeed, the Lac moiety is invisible in the D172A/E217A-LNFP II structure, and the rate of hydrolysis by BbAfcB is the same between LNFP II (type-1) and LNFP III (Galβ1–4(Fucα1–3)GlcNAcβ1–3Galβ1–4Glc) (type-2) (29). Galacto-N-biose cannot be used in the synthase reaction because the axial O4 hydroxyl group of the GalNAc moiety is not appropriately placed to make a glycosidic bond with Fuc. The GlcNAc-binding site can accept both GlcNAc and Glc, but the product yields in the synthetic reactions significantly differed between LNB and Lac (Table 1). The N-acetyl group of GlcNAc in the D172A/E217A-LNFP II structure does not form strong interactions with the protein, but the presence of the large N-acetyl group may stabilize the acceptor binding through a weak hydrophobic interaction with Trp-47 or Trp-213 (Fig. 3c). The enzyme synthesized LNFP II when LNT (Galβ1–3GlcNAcβ1–3Galβ1–4Glc) was included in the reaction mixture. LNT has two potential sites to be fucosylated by the enzyme, but in the analysis of the isolated product, we only detected LNFP II. This is because the substitution of the O3 hydroxyl group of the penultimate Gal impaired the accommodation and/or constrained the induced fit movement. Accordingly, when lacto-N-difucohexaose II (Galβ1–3(Fucα1–4)GlcNAcβ1–3Galβ1–4(Fucα1–3)Glc) was incubated with the WT AfcBs, the Fucα1–4GlcNAc linkage was hydrolyzed about 100-fold faster than the Fucα1–3Glc linkage (data not shown).
FIGURE 4.
a, regio- and stereospecific installation of a Fuc residue into type-1/2 chains catalyzed by the BbAfcB D703S glycosynthase. b, the predicted mode of binding of Lex trisaccharide (cyan) and its comparison with the Lea trisaccharide (yellow) observed in the D172A/E217A-LNFP II structure (electrostatic surface potential map) (stereoview). NAG, GlcNAc.
The mutational study of BbAfcB confirmed that Asp-703 and Glu-746 are the catalytic nucleophile and acid/base catalyst of the enzyme, respectively (supplemental Table S1). The amino acid replacement at the Gal-binding site (Trp-742 and Asp-807) resulted in a significant reduction (by 1800- and 80-fold, respectively) in the hydrolytic activity for 3-FL. It is interesting to note that the specific activities of the mutants decreased only by 2–4-fold for the pNP-Fuc hydrolysis (supplemental Table S1), which agrees with the finding that these residues are involved in the interaction with the Gal moiety and not in the Fuc binding. The presence of the branched Gal residue is important not only for the induced fit movement but also for fixing the substrate at the catalytic site. In the D172A/E217A-LNFP II structure, Glu-237 of BiAfcB interacts with the Gal O3 hydroxyl group. BbAfcB appears not to possess the corresponding residue as the alanine substitutions at Asp-763, Asp-766, and Asp-778 did not significantly affect hydrolytic activity. The sequence of the loop (236–254 in BiAfcB and 758–773 in BbAfcB) is not conserved between the two enzymes, and the loop is disordered in the BiAfcB structures (Fig. 3a and supplemental Fig. S10). BT_2192, an another close homologue of BiAfcB (see Fig. 5; described later), also does not possess the corresponding residue in its sequence, but the side chain of Glu-254 occupies the site in the structure (supplemental Figs. S11 and S12). These results suggest an auxiliary role for this loop region in the substrate binding.
FIGURE 5.
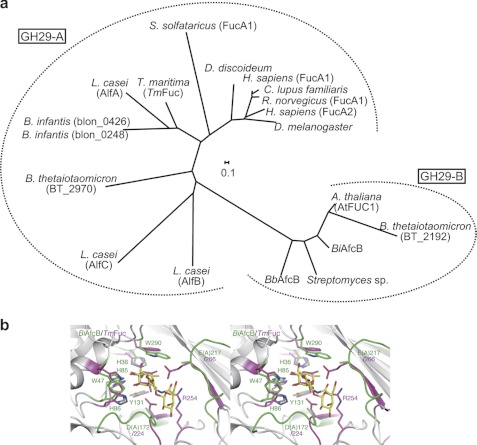
Phylogenetic and structural difference between two subfamilies of GH29. a, the phylogenetic analysis of enzymatically characterized GH29 α-l-fucosidases. The tree was constructed using the ClustalW program with a neighbor-joining method (55). b, comparison of the catalytic sites of BiAfcB (D172A/E217A-LNFP II complex; protein in green; ligand in yellow) and TmFuc (Fuc complex; magenta; Protein Data Bank code 1ODU) (stereoview) (44). The numbers of the residues of BiAfcB and TmFuc are labeled in green and magenta, respectively. See also supplemental Figs. S11 and S12. A., Arabidopsis; H., Homo; R., Rattus; D. discoideum, Dictyostelium discoideum; C., Canis; L., Lactobacillus; B. infantis, Bifidobacterium longum subsp. infantis; B. thetaiotaomicron, Bacteroides thetaiotaomicron; D. melanogaster, Drosophila melanogaster.
Comparison with Other α-l-Fucosidases
GH29 can be divided into two subfamilies by phylogenetic analysis (Fig. 5a), and this classification apparently correlates with the difference of the substrate specificities between the two groups (29). One subfamily (hereafter referred to as GH29-A) comprises enzymes that show relatively relaxed substrate specificity and act efficiently on a chromogenic substrate (e.g. pNP-Fuc) as do human and Thermotoga maritima (TmFuc) enzymes (α-l-fucosidases; EC 3.2.1.51) (44, 47, 48). In contrast, the members of the other subfamily specifically hydrolyze the terminal α-(1,3/4)-fucosidic linkages and hardly act on pNP-Fuc (referred to as GH29-B) (1,3-1,4-α-l-fucosidases; EC 3.2.1.111) (49, 50).3 BbAfcB and BiAfcB are thus categorized into the GH29-B subfamily (29). The overall structure of BiAfcB shows a resemblance to that of TmFuc (GH29-A) (Protein Data Bank code 1ODU) (root mean square deviation, 3.5 Å for 289 Cα atoms; Z score, 28.5; sequence identity, 24%). In particular, the structure of the Fuc-binding site (subsite −1) is similar between the two enzymes (Fig. 5b). The catalytic nucleophile Asp-224 and the acid/base catalyst Glu-266 of TmFuc overlap with Asp-172 and Glu-217 of BiAfcB (by induced fit movement), respectively (44, 45). TmFuc, however, does not have a Gal-binding site, and the side chain of Arg-254 occupies the corresponding site and forms a hydrogen bond with the endocyclic oxygen atom of Fuc. This Arg residue is highly conserved in GH29-A enzymes (13 of 14 members listed in Fig. 5a), whereas the Gal-binding residues (corresponding to Gly-173, Trp-213, and Asp-283 of BiAfcB) are invariable in the five characterized members of GH29-B. In addition, the constitution of the catalytic pocket is structurally conserved in the respective subfamilies (supplemental Figs. 11 and 12) (51). The structural difference observed at that site may therefore be the basis that differentiates the substrate specificity between GH29-A and GH29-B enzymes. But it is interesting to note that, in both TmFuc (GH29-A) and BiAfcB (GH29-B), two mobile loops (including or close to an acid/base residue) play critical roles in the catalytic processes (52) (supplemental Fig. S11).
Transglycosylation activity of α-l-fucosidases from Alcaligenes sp., Aspergillus niger, Penicillium multicolor, and T. maritima has been used to produce fucosyl oligosaccharides using pNP-Fuc as a donor (22–25). Recently, the glycosynthase methodology was introduced in α-l-fucosidase from Sulfolobus solfataricus (SsFuc D242S) and T. maritima (TmFuc D224G) (18). These enzymes/mutants are efficient catalysts that attach an α-l-fucosyl residue(s) to various acceptor molecules; however, because they belong to the GH29-A subfamily, they intrinsically fail to control the regioselectivity and acceptor specificity even though they have some preference. For example, TmFuc synthesized a mixture of Fucα1–3Fucα-pNP and Fucα1–2Galβ-pNP as a result of transglycosylation when incubated with pNP-Fuc (donor) and pNP-β-Gal (acceptor) (25). The SsFuc glycosynthase mutant produced Fucα1–6Galβ-pNP, Fucα1–3Galβ-pNP, Fucα1–4Galβ-pNP, and Fucα1–2(Fucα1–3)Galβ-pNP when incubated with β-l-fucopyranosyl azide and Galβ-pNP as the donor and the acceptor, respectively (18).
The strict specificity exhibited by the BbAfcB D703S mutant is thus a quite unusual feature. The synthase should therefore serve as a powerful tool for introducing the Lea and Lex antigens into the type-1 and type-2 chains at the non-reducing ends of various glycoconjugates including glycoprotein and glycolipids without attaching undesirable and unexpected α-l-fucosyl residues. Considering that 1,3-1,4-α-l-fucosynthase can recognize 2′-FL as the acceptor, the combination of this synthase with 1,2-α-l-fucosynthase, which we have already developed (30), may afford the one-pot tailor-made synthesis of all types of Le antigens including Leb and Ley. Mining glycosidases with strict specificity and exploiting those enzymes could be the most efficient route to the specific synthesis of glycosides. In this sense, intestinal microorganisms including bifidobacteria represent good enzyme resources as they are found to possess various glycosidases specific to host glycans (46, 53, 54).
Supplementary Material
Acknowledgments
We thank the staff of the Photon Factory for the x-ray data collection and Kaori Morii and Erika Tsutsumi for technical assistance.
This work was supported in part by Grant-in-aid for Scientific Research (C) 24580119 from the Ministry of Education, Culture, Sports, Science and Technology, Japan and a grant-in-aid from the Urakami Foundation.

This article contains supplemental Figs. S1–S12 and Table S1.
The atomic coordinates and structure factors (codes 3UES and 3UET) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
Unpublished results for BT_2192, H. Sakurama and T. Katayama.
- Le
- Lewis
- AfcA
- 1,2-α-l-fucosidase
- AfcB
- 1,3-1,4-α-l-fucosidase
- CAD
- charged aerosol detector
- DFJ
- deoxyfuconojirimycin
- FL
- fucosyllactose
- FucF
- β-l-fucopyranosyl fluoride
- GH
- glycoside hydrolase
- GlcGlcNAc
- 2-acetamide-2-deoxy-4-O-(β-glucosyl)-glucose
- LacNAc
- N-acetyllactosamine
- LNB
- lacto-N-biose I
- LNFP
- lacto-N-fucopentaose
- LNT
- lacto-N-tetraose;
- Bb
- B. bifidum
- Lac
- lactose
- Bi
- B. longum subsp. infantis
- pNP-Fuc
- para-nitrophenyl-α-l-fucopyranoside
- EG
- ethylene glycol
- Tm
- T. maritima
- Ss
- S. solfataricus.
REFERENCES
- 1. Becker D. J., Lowe J. B. (2003) Fucose: biosynthesis and biological function in mammals. Glycobiology 13, 41R–53R [DOI] [PubMed] [Google Scholar]
- 2. Gooi H. C., Feizi T., Kapadia A., Knowles B. B., Solter D., Evans M. J. (1981) Stage-specific embryonic antigen involves α1 goes to 3 fucosylated type 2 blood group chains. Nature 292, 156–158 [DOI] [PubMed] [Google Scholar]
- 3. Inoue M., Sasagawa T., Saito J., Shimizu H., Ueda G., Tanizawa O., Nakayama M. (1987) Expression of blood group antigens A, B, H, Lewis-a, and Lewis-b in fetal, normal, and malignant tissues of the uterine endometrium. Cancer 60, 2985–2993 [DOI] [PubMed] [Google Scholar]
- 4. Phillips M. L., Nudelman E., Gaeta F. C., Perez M., Singhal A. K., Hakomori S., Paulson J. C. (1990) ELAM-1 mediates cell adhesion by recognition of a carbohydrate ligand, sialyl-Lex. Science 250, 1130–1132 [DOI] [PubMed] [Google Scholar]
- 5. Walz G., Aruffo A., Kolanus W., Bevilacqua M., Seed B. (1990) Recognition by ELAM-1 of the sialyl-Lex determinant on myeloid and tumor cells. Science 250, 1132–1135 [DOI] [PubMed] [Google Scholar]
- 6. Nilsson C., Skoglund A., Moran A. P., Annuk H., Engstrand L., Normark S. (2006) An enzymatic ruler modulates Lewis antigen glycosylation of Helicobacter pylori LPS during persistent infection. Proc. Natl. Acad. Sci. U.S.A. 103, 2863–2868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Liu T. W., Ho C. W., Huang H. H., Chang S. M., Popat S. D., Wang Y. T., Wu M. S., Chen Y. J., Lin C. H. (2009) Role for α-l-fucosidase in the control of Helicobacter pylori-infected gastric cancer cells. Proc. Natl. Acad. Sci. U.S.A. 106, 14581–14586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bergman M. P., Engering A., Smits H. H., van Vliet S. J., van Bodegraven A. A., Wirth H. P., Kapsenberg M. L., Vandenbroucke-Grauls C. M., van Kooyk Y., Appelmelk B. J. (2004) Helicobacter pylori modulates the T helper cell 1/T helper cell 2 balance through phase-variable interaction between lipopolysaccharide and DC-SIGN. J. Exp. Med. 200, 979–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hancock S. M., Vaughan M. D., Withers S. G. (2006) Engineering of glycosidases and glycosyltransferases. Curr. Opin. Chem. Biol. 10, 509–519 [DOI] [PubMed] [Google Scholar]
- 10. Wang L. X., Huang W. (2009) Enzymatic transglycosylation for glycoconjugate synthesis. Curr. Opin. Chem. Biol. 13, 592–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang W., Hu T., Frantom P. A., Zheng T., Gerwe B., Del Amo D. S., Garret S., Seidel R. D., 3rd, Wu P. (2009) Chemoenzymatic synthesis of GDP-l-fucose and the Lewis X glycan derivatives. Proc. Natl. Acad. Sci. U.S.A. 106, 16096–16101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dumon C., Bosso C., Utille J. P., Heyraud A., Samain E. (2006) Production of Lewis x tetrasaccharides by metabolically engineered Escherichia coli. Chembiochem 7, 359–365 [DOI] [PubMed] [Google Scholar]
- 13. Drouillard S., Driguez H., Samain E. (2006) Large-scale synthesis of H-antigen oligosaccharides by expressing Helicobacter pylori α1,2-fucosyltransferase in metabolically engineered Escherichia coli cells. Angew. Chem. Int. Ed. Engl. 45, 1778–1780 [DOI] [PubMed] [Google Scholar]
- 14. Mackenzie L. F., Wang Q., Warren R. A. J., Withers S. G. (1998) Glycosynthases: mutant glycosidases for oligosaccharide synthesis. J. Am. Chem. Soc. 120, 5583–5584 [Google Scholar]
- 15. Malet C., Planas A. (1998) From β-glucanase to β-glucansynthase: glycosyl transfer to α-glycosyl fluorides catalyzed by a mutant endoglucanase lacking its catalytic nucleophile. FEBS Lett. 440, 208–212 [DOI] [PubMed] [Google Scholar]
- 16. Honda Y., Kitaoka M. (2006) The first glycosynthase derived from an inverting glycoside hydrolase. J. Biol. Chem. 281, 1426–1431 [DOI] [PubMed] [Google Scholar]
- 17. Cobucci-Ponzano B., Strazzulli A., Rossi M., Moracci M. (2011) Glycosynthases in biocatalysis. Adv. Synth. Catal. 353, 2284–2300 [Google Scholar]
- 18. Cobucci-Ponzano B., Conte F., Bedini E., Corsaro M. M., Parrilli M., Sulzenbacher G., Lipski A., Dal Piaz F., Lepore L., Rossi M., Moracci M. (2009) β-Glycosyl azides as substrates for α-glycosynthases: preparation of efficient α-l-fucosynthases. Chem. Biol. 16, 1097–1108 [DOI] [PubMed] [Google Scholar]
- 19. Cobucci-Ponzano B., Zorzetti C., Strazzulli A., Carillo S., Bedini E., Corsaro M. M., Comfort D. A., Kelly R. M., Rossi M., Moracci M. (2011) A novel α-d-galactosynthase from Thermotoga maritima converts β-d-galactopyranosyl azide to α-galacto-oligosaccharides. Glycobiology 21, 448–456 [DOI] [PubMed] [Google Scholar]
- 20. Umekawa M., Huang W., Li B., Fujita K., Ashida H., Wang L. X., Yamamoto K. (2008) Mutants of Mucor hiemalis endo-β-N-acetylglucosaminidase show enhanced transglycosylation and glycosynthase-like activities. J. Biol. Chem. 283, 4469–4479 [DOI] [PubMed] [Google Scholar]
- 21. Shaikh F. A., Withers S. G. (2008) Teaching old enzymes new tricks: engineering and evolution of glycosidases and glycosyl transferases for improved glycoside synthesis. Biochem. Cell Biol. 86, 169–177 [DOI] [PubMed] [Google Scholar]
- 22. Ajisaka K., Fujimoto H., Miyasato M. (1998) An α-l-fucosidase from Penicillium multicolor as a candidate enzyme for the synthesis of α(1→3)-linked fucosyl oligosaccharides by transglycosylation. Carbohydr. Res. 309, 125–129 [DOI] [PubMed] [Google Scholar]
- 23. Murata T., Morimoto S., Zeng X., Watanabe S., Usui T. (1999) Enzymatic synthesis of α-l-fucosyl-N-acetyllactosamines and 3′-O-α-l-fucosyllactose utilizing α-l-fucosidases. Carbohydr. Res. 320, 192–199 [DOI] [PubMed] [Google Scholar]
- 24. Farkas E., Thiem J., Ajisaka K. (2000) Enzymatic synthesis of fucose-containing disaccharides employing the partially purified α-l-fucosidase from Penicillium multicolor. Carbohydr. Res. 328, 293–299 [DOI] [PubMed] [Google Scholar]
- 25. Osanjo G., Dion M., Drone J., Solleux C., Tran V., Rabiller C., Tellier C. (2007) Directed evolution of the α-l-fucosidase from Thermotoga maritima into an α-l-transfucosidase. Biochemistry 46, 1022–1033 [DOI] [PubMed] [Google Scholar]
- 26. Cantarel B. L., Coutinho P. M., Rancurel C., Bernard T., Lombard V., Henrissat B. (2009) The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37, D233–D238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Katayama T., Sakuma A., Kimura T., Makimura Y., Hiratake J., Sakata K., Yamanoi T., Kumagai H., Yamamoto K. (2004) Molecular cloning and characterization of Bifidobacterium bifidum 1,2-α-l-fucosidase (AfcA), a novel inverting glycosidase (glycoside hydrolase family 95). J. Bacteriol. 186, 4885–4893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nagae M., Tsuchiya A., Katayama T., Yamamoto K., Wakatsuki S., Kato R. (2007) Structural basis of the catalytic reaction mechanism of novel 1,2-α-l-fucosidase from Bifidobacterium bifidum. J. Biol. Chem. 282, 18497–18509 [DOI] [PubMed] [Google Scholar]
- 29. Ashida H., Miyake A., Kiyohara M., Wada J., Yoshida E., Kumagai H., Katayama T., Yamamoto K. (2009) Two distinct α-l-fucosidases from Bifidobacterium bifidum are essential for the utilization of fucosylated milk oligosaccharides and glycoconjugates. Glycobiology 19, 1010–1017 [DOI] [PubMed] [Google Scholar]
- 30. Wada J., Honda Y., Nagae M., Kato R., Wakatsuki S., Katayama T., Taniguchi H., Kumagai H., Kitaoka M., Yamamoto K. (2008) 1,2-α-l-Fucosynthase: a glycosynthase derived from an inverting α-glycosidase with an unusual reaction mechanism. FEBS Lett. 582, 3739–3743 [DOI] [PubMed] [Google Scholar]
- 31. Nishimoto M., Kitaoka M. (2007) Practical preparation of lacto-N-biose I, a candidate for the bifidus factor in human milk. Biosci. Biotechnol. Biochem. 71, 2101–2104 [DOI] [PubMed] [Google Scholar]
- 32. Nishimoto M., Kitaoka M. (2009) One-pot enzymatic production of β-d-galactopyranosyl-(1→3)-2-acetamido-2-deoxy-f-galactose (galacto-N-biose) from sucrose and 2-acetamido-2-deoxy-d-galactose (N-acetylgalactosamine). Carbohydr. Res. 344, 2573–2576 [DOI] [PubMed] [Google Scholar]
- 33. Honda Y., Kitaoka M., Hayashi K. (2004) Reaction mechanism of chitobiose phosphorylase from Vibrio proteolyticus: identification of family 36 glycosyltransferase in Vibrio. Biochem. J. 377, 225–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cohenford M. A., Abraham A., Abraham J., Dain J. A. (1989) Colorimetric assay for free and bound l-fucose. Anal. Biochem. 177, 172–177 [DOI] [PubMed] [Google Scholar]
- 35. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 36. Vagin A., Teplyakov A. (1997) MOLREP: an automated program for molecular replacement. J. Appl. Crystallogr. 30, 1022–1025 [Google Scholar]
- 37. Sela D. A., Garrido D., Lerno L., Wu S., Tan K., Eom H. J., Joachimiak A., Lebrilla C. B., Mills D. A. (2012) Bifidobacterium longum subsp. infantis ATCC 15697 α-fucosidases are active on fucosylated human milk oligosaccharides. Appl. Environ. Microbiol. 78, 795–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 40. Suzuki R., Katayama T., Kitaoka M., Kumagai H., Wakagi T., Shoun H., Ashida H., Yamamoto K., Fushinobu S. (2009) Crystallographic and mutational analyses of substrate recognition of endo-α-N-acetylgalactosaminidase from Bifidobacterium longum. J. Biochem. 146, 389–398 [DOI] [PubMed] [Google Scholar]
- 41. Bohne A., Lang E., von der Lieth C. W. (1999) SWEET—WWW-based rapid 3D construction of oligo- and polysaccharides. Bioinformatics 15, 767–768 [DOI] [PubMed] [Google Scholar]
- 42. Morris G. M., Goodsell D. S., Halliday R. S., Huey R., Hart W. E., Belew R. K., Olson A. J. (1998) Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 19, 1639–1662 [Google Scholar]
- 43. McCarter J. D., Withers S. G. (1994) Mechanisms of enzymatic glycoside hydrolysis. Curr. Opin. Struct. Biol. 4, 885–892 [DOI] [PubMed] [Google Scholar]
- 44. Sulzenbacher G., Bignon C., Nishimura T., Tarling C. A., Withers S. G., Henrissat B., Bourne Y. (2004) Crystal structure of Thermotoga maritima α-l-fucosidase. Insights into the catalytic mechanism and the molecular basis for fucosidosis. J. Biol. Chem. 279, 13119–13128 [DOI] [PubMed] [Google Scholar]
- 45. Tarling C. A., He S., Sulzenbacher G., Bignon C., Bourne Y., Henrissat B., Withers S. G. (2003) Identification of the catalytic nucleophile of the family 29 α-l-fucosidase from Thermotoga maritima through trapping of a covalent glycosyl-enzyme intermediate and mutagenesis. J. Biol. Chem. 278, 47394–47399 [DOI] [PubMed] [Google Scholar]
- 46. Sela D. A., Chapman J., Adeuya A., Kim J. H., Chen F., Whitehead T. R., Lapidus A., Rokhsar D. S., Lebrilla C. B., German J. B., Price N. P., Richardson P. M., Mills D. A. (2008) The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proc. Natl. Acad. Sci. U.S.A. 105, 18964–18969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Liu S. W., Chen C. S., Chang S. S., Mong K. K., Lin C. H., Chang C. W., Tang C. Y., Li Y. K. (2009) Identification of essential residues of human α-l-fucosidase and tests of its mechanism. Biochemistry 48, 110–120 [DOI] [PubMed] [Google Scholar]
- 48. Rodríguez-Díaz J., Monedero V., Yebra M. J. (2011) Utilization of natural fucosylated oligosaccharides by three novel α-l-fucosidases from a probiotic Lactobacillus casei strain. Appl. Environ. Microbiol. 77, 703–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sano M., Hayakawa K., Kato I. (1992) Purification and characterization of α-l-fucosidase from Streptomyces species. J. Biol. Chem. 267, 1522–1527 [PubMed] [Google Scholar]
- 50. Zeleny R., Leonard R., Dorfner G., Dalik T., Kolarich D., Altmann F. (2006) Molecular cloning and characterization of a plant α1,3/4-fucosidase based on sequence tags from almond fucosidase I. Phytochemistry 67, 641–648 [DOI] [PubMed] [Google Scholar]
- 51. Fisher K. J., Aronson N. N., Jr. (1989) Isolation and sequence analysis of a cDNA encoding rat liver α-l-fucosidase. Biochem. J. 264, 695–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wu H. J., Ho C. W., Ko T. P., Popat S. D., Lin C. H., Wang A.H. (2010) Structural basis of α-fucosidase inhibition by iminocyclitols with Ki values in the micro- to picomolar range. Angew. Chem. Int. Ed. Engl. 49, 337–340 [DOI] [PubMed] [Google Scholar]
- 53. Yoshida E., Sakurama H., Kiyohara M., Nakajima M., Kitaoka M., Ashida H., Hirose J., Katayama T., Yamamoto K., Kumagai H. (2012) Bifidobacterium longum subsp. infantis uses two different β-galactosidases for selectively degrading type-1 and type-2 human milk oligosaccharides. Glycobiology 22, 361–368 [DOI] [PubMed] [Google Scholar]
- 54. Asakuma S., Hatakeyama E., Urashima T., Yoshida E., Katayama T., Yamamoto K., Kumagai H., Ashida H., Hirose J., Kitaoka M. (2011) Physiology of consumption of human milk oligosaccharides by infant gut-associated bifidobacteria. J. Biol. Chem. 286, 34583–34592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Saitou N., Nei M. (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.