Background: Light harvesting genes associated with photosystem II (LHCBM) form a conserved multigene family involved in light absorption and photoprotection.
Results: Silencing of LHCBM1 and LHCBM2/7 genes resulted, respectively, in reduced capacity for energy dissipation and state transitions.
Conclusion: Despite the high homology, each LHCB protein exerts a specific role.
Significance: Comprehension of LHCBM protein specific functions paves the way for the manipulation of the Chlamydomonas photosynthetic antennas.
Keywords: Algae, Chlamydomonas, Gene Silencing, Photosynthesis, Photosystem II, Lhc, Lhcbm, Light Harvesting, Photoprotection, Solar Conversion Efficiency
Abstract
The photosystem II antenna of Chlamydomonas reinhardtii is composed of monomeric and trimeric complexes, the latter encoded by LHCBM genes. We employed artificial microRNA technology to specifically silence the LHCBM2 and LHCBM7 genes, encoding identical mature polypeptides, and the LHCBM1 gene. As a control, we studied the npq5 mutant, deficient in the LHCBM1 protein. The organization of LHCII complexes, functional antenna size, capacity for photoprotection, thermal energy dissipation and state transitions, and resistance to reactive oxygen species was studied in the various genotypes. Silencing of the LHCBM2/7 genes resulted in a decrease of an LHCII protein with an apparent molecular mass of 22 kDa, whereas silencing/lack of LHCBM1 caused the decrease/disappearance of a 23-kDa protein. A decrease in the abundance of trimeric LHCII complexes and in functional antenna size was observed in both LHCBM2/7 and LHCBM1 knockouts. In agreement with previous data, depletion of LHCBM1 decreased the capacity for excess energy dissipation but not the ability to perform state transitions. The opposite was true for LHCBM2/7, implying that this polypeptide has a different functional role from LHCBM1. The abundance of LHCBM1 and LHCBM2/7 is in both cases correlated with resistance to superoxide anion, whereas only LHCBM1 is also involved in singlet oxygen scavenging. These results suggest that different LHCBM components have well defined, non-redundant functions despite their high homology, implying that engineering of LHCBM proteins can be an effective strategy for manipulating the light harvesting system of Chlamydomonas reinhardtii.
Introduction
Chlamydomonas reinhardtii is a unicellular green alga currently used in production of biofuels such as bio-hydrogen (1–4), biodiesel (5, 6), and bioethanol (7) as well as molecular farming (8). As a photosynthetic organism, Chlamydomonas converts solar energy into biomass using as inputs just CO2, light, and H2O. Light is absorbed by large multisubunit chlorophyll-carotenoid-protein complexes (light harvesting complexes (LHC)2) embedded in the thylakoid membranes of chloroplasts, which are tightly packed around photosystems (PS) I and II (9). Light absorbed by these protein complexes is thus transferred as excitation energy to the PSII and PSI reaction centers and drives photosynthesis.
Like many unicellular green algae, Chlamydomonas assembles large arrays of light-absorbing chlorophyll (Chl) molecules in its photosynthetic antenna complexes, which confer a competitive advantage in natural environments where light is often limiting. However, under typical mass culture conditions, the high optical density causes the superficial layers to absorb light in strong excess with respect to what they can use for photoelectron transport (10). As a consequence, most energy is dissipated as heat, and when this photoprotective mechanism is saturated, cells are photoinhibited. Conversely, light is limited in layers located deeper in the culture, and cell growth is slow or negative due to dark respiration. Thus, the large size of photosynthetic antenna complexes reduces the solar conversion efficiency of Chlamydomonas mass cultures to unacceptably low levels, resulting in disappointingly low biomass yield. Reduction of the functional antenna size is a possible way to overcome this bottleneck (11, 12). This can be achieved through inhibition of biosynthesis of Chl b (13) of the xanthophylls lutein, violaxanthin, and neoxanthin (14) through mutation of regulatory genes involved in photosystem biogenesis (15) or through RNAi inhibition of the entire LHC gene family (16). Nevertheless, besides light harvesting, LHC proteins are involved in photoprotection mechanisms such as 3Chl* triplet quenching (17) (18, 19) and reactive oxygen species (ROS) scavenging (20–22), thus making photosynthetic organisms depleted in antenna proteins prone to photoinhibition (23, 24). Due to the functional specialization of different LHC subunits, some of which are more active in light harvesting while others are more active in heat dissipation, ROS scavenging, and/or 3Chl* quenching (25–27), selective down-regulation of individual components according to their specific function appears more suitable than a general reduction of antenna size for an improvement of biomass yield in mass cultures. To this aim, a detailed knowledge of the physiological role of individual light harvesting components is needed. LHC genes form a conserved multigene family composed by at least 20 genes (9), and based on their predominant association with PSI or PSII, LHC proteins are classified, respectively, as LHCA (or LHCI) and LHCB (or LHCII).
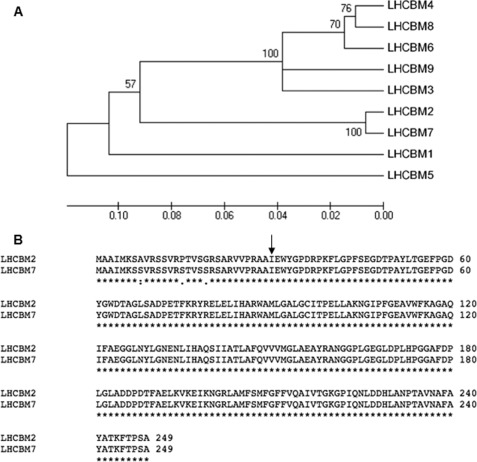
The antenna of PSII is formed by three major domains as follows. The first is the core antenna complex, made up by plastid-encoded proteins including D1, D2, CP47, and CP43. These subunits together bind ∼35 Chl a and 11 β-carotene molecules (28, 29), and the resulting core complex is dimeric in each PSII supercomplex (30). Next is the inner antenna layer, made by the monomeric LHCB4 (CP29) and LHCB5 (CP26) subunits. These are nuclear-encoded proteins, each binding 9–13 Chl a and b and three xanthophyll chromophores and are, respectively, bound to the CP47 and CP43 subunits of the core complex (31). A trimeric LHCII complex is peripherally bound to LHCB4 and LHCB5 to form the so-called C2S2 supercomplex (32). This LHCII trimer is called LHCII-S, being strictly bound to the PSII core complex. Several other LHCII trimers are organized and functionally connected to the PSII core complex, forming an outer antenna layer (iii) whose organization is not yet defined. In higher plants, high Mr complexes have been isolated where a third monomeric complex, LHCB6, bridges LHCB4 to LHCII-M trimers, including C2S2M2 (33, 34). However, LHCB6 is not present in Chlamydomonas, thus making the location of additional LHCII trimers unclear (35). The polypeptides composing LHCBM trimers are encoded by nine genes sharing high homology (see the phylogenetic tree in Fig. 1A), eight of which have been identified at the protein level by mass spectrometry (36, 37). Reverse genetic approaches have been extensively used to elucidate the function of individual LHC genes, particularly in Arabidopsis (33, 38–40). In Chlamydomonas, the LHCBM1 protein has been inactivated by mutation (41), and the monomeric LHCB4 and LHCB5 proteins have been down-regulated by RNAi (42). LHCBM1 inactivation causes a decrease in thermal dissipation of excess light energy and a reduction of LHCII trimers but does not affect state transitions. To the opposite, down-regulation of LHCB4 causes reduced state transitions (42). Additional evidence for differential functions of different LHCB polypeptides includes the preferential expression of LHCBM9 under sulfur starvation and anaerobiosis (43). In this work, we have focused on the effect of depleting the two most highly expressed polypeptides in C. reinhardtii, namely LHCBM1 and LHCBM2/7 (9). Although LHCBM1 is encoded by a single gene, LHCBM2/7 is encoded by two highly homologous genes (LHCBM2 and LHCBM7), which encode identical mature polypeptides (9, 36). To this aim, we have used the artificial microRNA (amiRNA) silencing technology (44), which allows to discrimination precisely between different genes and coordinately silence genes sharing identical regions while keeping unaltered the level of expression of other genes in the family. In addition, we analyzed the npq5 mutant (41) in which LHCBM1 is inactive.
FIGURE 1.
Phylogenetic tree of LHCBM CDSs (panel A) and alignment of the LHCBM2 and LHCBM7 polypeptide sequences (panel B). For LHCBM nomenclature and accession numbers, see supplemental Table 1. The phylogenetic tree and molecular evolutionary analyses were conducted using MEGA Version 4 (72). In panel B, the arrow indicates the start site of the mature LHCBM2 and LHCBM7 polypeptides. The two mature polypeptides are identical as the three amino acid substitutions are localized in the transit peptide.
The genotypes obtained were analyzed for the organization of their pigment-protein complexes and for the major physiological functions associated to the PSII antenna system, namely functional antenna size (45), capacity for photoprotection in high light conditions (46), non-photochemical quenching (NPQ) (47, 48), capacity for state 1-state 2 transitions, which regulates PSI versus PSII antenna size under limiting light conditions (49), and resistance to ROS (46).
EXPERIMENTAL PROCEDURES
Strains and Culture Conditions
Unless indicated differently, cells were grown at 25 °C with fluorescent white light (60 μE·m−2s−1). A 16-h light:8-h dark photoperiod was used for growth in TAP medium and continuous irradiation for growth in HS medium. The cell wall less cw15 strain was transformed with the pChlamyRNA3 vectors (44) containing the LHCBM1a, LHCBM1b, LHCBM2a, LHCBM2b, LHCBM2/7 amiRNAs. Nuclear transformation was performed as described (50). Plasmids were digested with ScaI, and 1 μg of DNA was used for each transformation. Transformants were selected on TAP agar plates containing paromomycin (10 μg/ml). To screen the silenced LHC1, LHC2, and LHC2/7 transformants based on Chl a/b ratios, cells were grown in 96-well microtiter plates in 200 μl of TAP at 25 °C until the stationary phase (2 × 107 cells/ml). 90 transformants were analyzed for each construct. Chl a/b ratios were determined on pigment extracts using a SAFAS spectrophotometer (see below). To perform quantitative real-time PCR, transformants showing increased Chl a/b ratios were grown in 4 ml in 24-well microtiter plates until the late-log phase, and cells were harvested for RNA extraction.
Physiological and biochemical experiments were performed on cells cultured at about 60 μE·m−2s−1 in liquid high salt medium (74) and harvested in their exponential growth phase. NPQ was measured on cells acclimated for about 10 cell divisions at 500 μE·m−2s−1. In experiments involving CC425 (an Arg auxotrophic strain (51)), the medium was supplemented with 50 μg/ml arginine.
Plasmid Construction
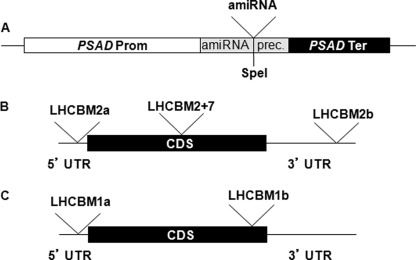
amiRNAs used to silence LHCBM genes were designed using the WMD3 software (Web micro RNA designer Version3) and verified using the EST data base. The LHCBM1a and LHCBM1b amiRNAs anneal, respectively, on the 5′-UTR and CDS sequence of the LHCBM1 gene. The LHCBM2a and LHCBM2b amiRNAs are predicted to silence only the LHCBM2 gene and anneal, respectively, in the 5′-UTR and 3′-UTR. The LHCBM2/7 amiRNA is predicted to silence both LHCBM2 and LHCBM7 genes and targets the CDS (Fig. 2, Table 1). The amiRNAs were cloned in pChlamyRNA3 vector at the unique SpeI site located in the amiRNA precursor as 90-bp oligonucleotides. Primers AmiRNAprec-for (GGTGTTGGGTCGGTGTTTTTG) and Spacer-rev (TAGCGCTGATCACCACCACCC) were used to screen the right orientation of the amiRNA cloned using the following PCR conditions: 1 cycle at 95 °C for 4 min + 35 cycles at 95 °C for 30 s, 65 °C for 30 s, and 72 °C for 30 s. The clones with correct orientation were confirmed by sequencing using the AmiRNAprec-for primer. Because the amiRNA precursor has a strong secondary structure, the following protocol was used for sequencing; 4 ng of fragment amplified with AmiRNAprec-for and Spacer-rev, purified through Sephadex G-100 resin (GE Healthcare, catalog no. 17-0060-01), or 300 ng of recombinant pChlamyRNA3 vector were mixed with 1.6 μl of 2 μm primer solution, 2 μl of Big Dye buffer 5×, 1 μl of Big Dye v3.0 (Applied Biosystems), and 0.5 μl of ultra-pure DMSO in a final volume of 10 μl. Cycling conditions were 1 cycle at 96 °C for 45 s + 30 cycles at 96 °C for 10 s + 60 °C for 4 min.
FIGURE 2.
Schematic maps of the constructs used for silencing. Panel A, the silencing cassette in the pChlamyRNA3 vector is shown. This vector was engineered to express a 21-nucleotide silencing amiRNA. The PSAD promoter and terminator control the rate of amiRNA transcription. Panel B, shown are the target regions of the amiRNAs on LHCBM2 and LHCBM7 mRNAs. To silence the LHCBM2 gene, two different amiRNAs were chosen (LHCBM2a and LHCBM2b) targeting, respectively, the 5′-UTR and the 3′-UTR. To silence both LHCBM2 and LHCBM7 genes, one amiRNA was chosen (LHCBM2/7), targeting the CDS. Panel C, shown are the target regions of the amiRNAs on LHCBM1 mRNA. For details on the amiRNAs sequence and features, see Table 1.
TABLE 1.
Nucleotide sequence of the amiRNAs used and position on LHCBM1, LHCBM2, and LHCBM7 mRNAs
Hybridization energy between the amiRNA and its target is expressed as kcal/mol. The position of the mismatches is indicated in parentheses and is relative to the reverse complement sequence of the amiRNA starting from the 5′ nucleotide. The schematic position of the amiRNAs on the LHCBM mRNAs is shown in Fig. 2.
| Target | amiRNA | amiRNA sequence | Position on mRNA | Hybridization energy | Mismatches |
|---|---|---|---|---|---|
| LHCBM1 | LHCBM1a | TTCTGTTGAAATCACTGTCGG | 5′-UTR 11–31 | −38.05 | 1 (2) |
| LHCBM1 | LHCBM1b | TATTTACTGGGGGGTGAACGT | CDS 800–820 | −42.50 | 2 (2,21) |
| LHCBM2 | LHCBM2a | TTTGATATGAGTGTAGTGGAC | 5′-UTR 20–40 | −41.6 | 0 |
| LHCBM2 | LHCBM2b | TTGTCGAGAACCCATGTCGCT | 3′-UTR 1165–1185 | −42.38 | 1 (3) |
| LHCBM2 and LHCBM7 | LHCBM2/7 | TACAGCCTCACCGAAAGGGAT | CDS LHCBM2, 359–379; LHCBM7, 363–383 | −42.17 | 2 (6, 21) |
Quantitative Real-time RT-PCR
Total RNA was extracted using the RNeasy Plant Mini Kit (Qiagen, catalog no. 74904). cDNA was transcribed using the Omniscript RT kit (Qiagen, catalog no. 205111) according to the manufacturer's instructions. 1 μg of total RNA was reverse-transcribed using random nonamers (Sigma, catalog no. R7647) for 2 h at 37 °C, and 2 μl of the reaction was used to perform real-time PCR. Amplification conditions were: 95 °C for 10 min; 50 cycles of 95 °C for 15 s, and 60 °C for 1 min. SYBR Green PCR Master Mix (Applied, catalog no. 4309155) was used in a final volume of 15 μl.
The oligonucleotides used to amplify the LHCBM genes are as described by Nguyen et al. (43). Ubiquitin ligase (Ubiq ligase, accession number BE237718) and ribulose-1,5-bisphosphate carboxylase/oxygenase small subunit 1 (RBCS1, accession number XM_001702357) were used as “housekeeping” genes. The oligonucleotides used to amplify the Ubiquitin ligase mRNA are ubiq-ligase-for (TTTGCGCACTTCTTCGTGG) and ubiq-ligase-rev (TTCACCATGAGCAGGTTCTTGT), and those for RBCS1 are rbcs1-for (CCTGCCTGGAGTTCGCTG) and rbcs1-rev (CCAGTAGCGGTTGTCGTAGTACAG). Both pairs of oligonucleotides amplify a fragment of 101 bp of the ubiquitin ligase and RBCS1 mRNAs.
Pigment Content Determination
Pigments were extracted from cell pellets; samples were frozen in liquid nitrogen and resuspended in 80% acetone buffered with Na2CO3, then the supernatant of each sample was recovered after centrifugation (15 min at 15,000 g, 4 °C); separation and quantification of pigments were performed by HPLC (52). Chl a/b and Chl/cars ratio were corrected through fitting analysis of the absorption spectrum (53).
Thylakoid Preparation from C. reinhardtii Cells
C. reinhardtii-stacked thylakoids were purified after a modified version of the protocol described in Dainese and Bassi (54). Briefly, cells were harvested and resuspended in 20 mm Tricine, pH 8, 0.4 m sorbitol, 10 mm MgCl2, 0.2% BSA. After 2 cycles of sonication (5 s) broken cells were centrifuged at 6000 × g and resuspended in 20 mm Tricine, pH 8, 0.1 m NaCl, 5 mm MgCl2, 0.2% BSA. After 15 min of ice, incubation membranes were centrifuged at 8000 × g and resuspended in 20 mm Hepes, pH 7.6, 15 mm NaCl, 5 mm MgCl2. After a last washing step, membranes were stored in 20 mm Hepes, pH 7.6, 5 mm MgCl2, 10 mm NaCl, 20% sorbitol. Stroma and grana membranes were isolated from thylakoids purified as described in Bassi and Wollman (55) but with the thylakoid preparation described in this section.
SDS-PAGE Electrophoresis and Immunoblotting
Denaturing SDS-PAGE was performed in the presence of 6 m urea with the Tris-Tricine buffer systems (56). The gel was transblotted to a nitrocellulose filter, decorated with homemade anti-LHCII, anti-CP47, and anti-LHCSR sera (57), and developed by means of the alkaline phosphatase detection system.
Native Electrophoresis
Thylakoid membranes were solubilized in the presence of 1.2% α-dodecyl-maltoside, incubated in ice for 15 min, and spun in a bench centrifuge, and the supernatant was loaded on native electrophoresis as previously reported (58).
PSII Functional Antenna Size
Relative PSII antenna size has been estimated from Fm saturation kinetics (1/t⅔) in the presence of 10−5 m DCMU (59). Kinetics were measured with a home-built apparatus. Fluorescence was excited using a green LED with a peak emission at 520 nm (intensity 20 μmol·m−2s−1) and detected in the near infrared.
State Transitions
The extent of state 1-state 2 transition capacity was obtained according to a protocol proposed by Fleischmann et al. (60); state 1 was induced by vigorous shaking in the dark and by adding 10−5 m 3-(3,4-dichlorophenyl)-1,1-dimethylurea 1 min before the measurement, whereas state 2 was induced through a 20-min-long dark incubation in the presence of 250 μm NaN3. State 1-state 2 transition was calculated as (Fm in state1 − Fm in state 2/Fm in state 1) × 100.
NPQ Measurements
Cells acclimated to high light conditions were harvested in the exponential growth phase (∼2 × 106 cells/ml), pelleted, and resuspended at a concentration of ∼108 cells/ml). Cells were preilluminated for 2 min with a weak (3 μE·m−2s−1) far-red LED before NPQ analysis with a PAM-101 (Waltz, Effeltrich, Germany); actinic light was 1600 μE·m−2s−1, and saturating light was 4080 μE·m−2s−1. The far-red LED was kept on during dark recovery.
PSI Functional Antenna Size
Relative PSI antenna size was estimated measuring the kinetics of P700 oxidation using a BioLogic Joliot-type spectrophotometer (JTS-10). State 1 and State 2 were chemically induced as described in the previous section, and the stacked thylakoids was isolated as reported in Iwai (61) and Takahashi (62). P700 oxidation kinetics were measured under limiting light (12 μmol·m−2s−1) upon dark adaptation and treatment with 10−5 m 3-(3,4-dichlorophenyl)-1,1-dimethylurea and 250 μm methyl viologen as previously described (63).
Spot Test with ROS
To test the susceptibility of the strains to reactive oxygen species, cells were grown mixotrophically in TAP liquid medium in continuous light (60 μE·m−2s−1) up to mid-exponential phase (2–3 × 106 cells/ml). Cells were collected by centrifugation at 1500 × g for 5 min at 21 °C and spotted at different dilutions on TAP (6 × 104, 6 × 103 cells) and HS (6 × 105, 6 × 104 cells) agar (1%) media containing 0.5 mm H2O2, 7.5 μm rose bengal or 0.17 μm methyl viologen. The plates were grown for 8 days in continuous light (60 μE·m−2s−1) before growth was scored.
RESULTS
Selective Silencing of LHCBM Genes
We employed amiRNA technology (44) to silence three genes (LHCBM1, LHCBM2, and LHCBM7) of the major light-harvesting complex of photosystem II. The nomenclature of LHCBM genes used in this paper, the correspondence to the sequences published in (9), and accession numbers of mRNA and protein sequences are reported is supplemental Table 1. In the newest version of the C. reinhardtii genome (Version 4.0), LHCBM1 is localized on scaffold_23: 78554–80401 (+), ProteinID 185533, LHCBM2 is reported on chromosome 12: 7646897–7648838, ProteinID: 184067, and LHCBM7 is reported on chromosome12: 7698723–7700417, ProteinID: 184071.
LHCBM2 and LHCBM7 are paralogous genes showing high levels of identity to each other (Fig. 1A). The two genes encode identical mature polypeptides, as the three amino acid substitutions map in the transit peptide domain that is cleaved upon import in the chloroplast (Fig. 1B). Two different amiRNAs were designed for silencing LHCBM2, targeting, respectively, the 5′- and 3′-UTR of the gene, and one for the simultaneous silencing of LHCBM2 and LHCBM7, targeting the coding sequence of the genes (Fig. 2B). We also designed two different amiRNAs for LHCBM1, targeting the 5′-UTR and the CDS (Fig. 2C). The amiRNAs were cloned in pChlamyRNA3 (44) under the control of the PSAD constitutive promoter (64) (Fig. 2A). The sequence of all the amiRNAs employed in this study, the position on the LHCBM mRNA, the hybridization energy, and the number and the position of the mismatches are reported in Table 1. The silencing vectors containing the different amiRNAs were used to transform C. reinhardtii strain cw15.
Pigment Analysis of Silenced Transformants
90 transformants for each construct were grown in liquid TAP culture in microtiter wells, pelleted, and resuspended in 80% (v/v) acetone. The acetone extracts were then analyzed in the chlorophyll a and b spectral regions using a microtiter spectrophotometer to discriminate lines with an increased Chl a/b ratio with respect to wild type, diagnostic of a reduced LHCII antenna (11).
The results (supplemental Fig. 1) show that ∼30% of the LHCBM1b-, LHCBM2b-, and LHCBM2/7-silenced strains show increased Chl a/b ratios, with the phenotype being more evident in LHCBM1b and LHCBM2/7 strains. One LHCBM1-silenced strain (termed LHC1), two LHCBM2-silenced strains (termed LHC2a and LHC2b), and two LHCBM2/7-silenced strains (termed LHC2/7a and LHC2/7b), showing the highest Chl a/b values in the initial screening, were chosen for further analysis.
Because PSII antennas bind Chl a, Chl b and xanthophylls, whereas PSII reaction centers bind Chl a and β-carotene, it is expected that transformants with a reduced antenna size will have increased Chl a/b as well as β-carotene/xanthophyll ratios. These ratios are shown in Table 2 for the selected silenced lines and the wild type cw15 strain. The LHCBM1 null mutant, npq5, and its wild type line, CC425, grown in the same conditions, are shown for comparison. The results are consistent with a reduced antenna size in npq5 and the LHC1, LHC2, and LHC2/7 transformants. The phenotype observed in the LHC2/7 transformants, both in terms of Chl a/b and of β-carotene/xanthophyll ratios, is more severe than in the npq5 mutant, which lacks completely the LHCBM1 polypeptide, encoded by the most highly expressed LHCBM gene (41).
TABLE 2.
Pigment ratios in wild type and LHCBM mutated and silenced strains
Data shown are the means ± S.D. (n = 3). The npq5 mutant (41) and its wild type progenitor CC425 are shown for a comparison. The values were determined with high accuracy using HPLC by measuring the absorption spectra of acetone-extracted pigments and spectral fitting analysis (53). Xanth, xanthophyll. B-Car, β-carotenoid.
| CC425 | npq5 | cw15 | LHC1 | LHC2a | LHC2b | LHC2/7a | LHC2/7b | |
|---|---|---|---|---|---|---|---|---|
| Chl a/b | 2.32 ± 0.04 | 2.43 ± 0.01 | 2.39 ± 0.01 | 2.56 ± 0.02 | 2.47 ± 0.01 | 2.53 ± 0.01 | 2.58 ± 0.01 | 2.55 ± 0.02 |
| β-Car/xanth | 0.33 ± 0.00 | 0.37 ± 0.06 | 0.35 ± 0.01 | 0.39 ± 0.03 | 0.36 ± 0.02 | 0.40 ± 0.00 | 0.40 ± 0.01 | 0.43 ± 0.01 |
Quantification of LHCBM mRNA Levels
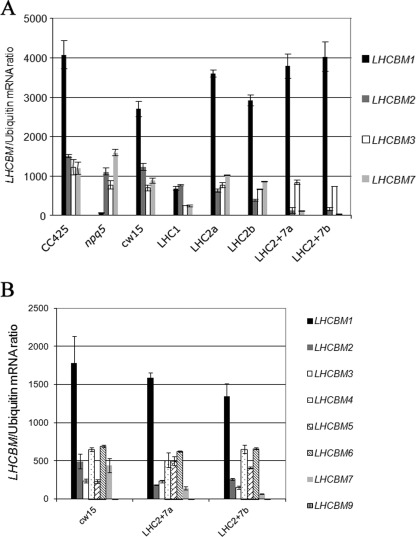
To correlate the reduction in antenna size to the level of silencing, the abundance of LHCBM mRNAs was measured by quantitative real-time PCR (Fig. 3A). The LHCBM1 mRNA is the most abundantly expressed and was greatly reduced or absent in the npq5 mutant. A 75% reduction of the LHCBM1 transcript was observed in the LHC1 transformant, confirming the silencing of this gene. However, the levels of expression of non-target genes (LHCBM2/3/7) are also decreased (respectively a reduction of 39, 69, and 73%). Similar levels of reduction of LHCBM3 were observed in the npq5 mutant and the LHC1 transformant, suggesting that a regulatory mechanism, sensing the levels of the LHCBM1 polypeptide, regulates LHCBM3 mRNA levels. However, LHCBM2/7 mRNA levels are selectively reduced in the LHC1 transformant but not in the npq5 mutant, suggesting that the LHCBM1b amiRNA performs off-target silencing of the LHCBM2 and LHCBM7 transcripts. The same analysis was also performed on transformants containing the LHCBM1a amiRNA, which targets the 5′-UTR (Fig. 2C), but a significant reduction in LHCBM2/7 mRNA was also observed (data not shown).
FIGURE 3.
Quantification of LHCBM mRNA levels by real-time RT PCR. Panel A, the genotypes shown in Table 2 were analyzed through quantitative real-time RT-PCR to determine the amount of the LHCBM1/2/3/7 mRNAs in TAP medium. LHCBM1 is the most abundant mRNA, is absent in the npq5 mutant, and has reduced levels in the LHC1 transformant. The LHC1 transformant also shows reduced levels of LHCBM2/3/7. LHC2 transformants (LHC2a and LHC2b) show decreased levels of LHCBM2 mRNA and unaltered levels of LHCBM3/7 mRNAs with respect to cw15. LHC2/7 transformants show decreased levels of both LHCBM2 and LHCBM7 mRNAs. The levels of LHCBM3 transcript are constant in LHC2 and LHC2/7 transformants. Panel B, the LHC2/7 transformants were further analyzed to determine the amount of the LHCBM1/2/3/4/5/6/7/9 mRNAs in HS medium. They show decreased levels of LHCBM2 and LHCBM7 mRNAs, whereas all the other mRNAs are unaffected.
In contrast, silencing of LHCBM2 and LHCBM2/7 was specific. The LHCBM2b amiRNA caused a 50–70% reduction of LHCBM2 mRNA, whereas the level of expression of LHCBM3 and LHCBM7 genes was unaffected. The LHCBM2/7 amiRNA caused a 75–95% reduction of both LHCBM2 and LHCBM7, whereas LHCBM3 expression was unaffected. A compensatory increase of LHCBM1 was observed in the LHC2 and LHC2/7 transformants. To confirm the lack of off-target silencing in the LHC2/7 transformants, we studied the abundance of other LHCBM mRNAs (1, 2, 3, 4, 5, 6, 7, and 9) in HS medium (Fig. 3B), which was not down-regulated, confirming the high specificity of silencing of the LHCBM2/7 mRNAs.
Taken together, these data indicate that the LHCBM1 amiRNA is not very specific, performing off-target silencing of other LHCBM transcripts. However, the LHCBM2/7 amiRNA is both efficient (resulting in 75–95% silencing of both genes) and highly specific, with no off-target effects detected in both TAP and HS media. Thus, all phenotypes described in the LHC2/7 transformants (see below) can be exclusively attributed to the reduction in the level of the LHCBM2/7 polypeptide.
Biochemical Characterization of LHCBM2 and LHC2/7 Transformants
Cells grown in control conditions (60 μE·m−2s−1) were dark-adapted, and the PSII quantum efficiency was estimated by measuring the Fv/Fm ratio (Table 3). This value was similar in the silenced LHC1, LHC2, and LHC2/7 transformants with respect to cw15 and in the npq5 mutant with respect to CC425. These results indicate that inactivation of selected LHCII genes by insertional mutagenesis or silencing did not affect PSII efficiency.
TABLE 3.
Fv/Fm in wild type and LHCBM-mutated and -silenced strains
Data shown are the means ± S.D. (n = 4). The npq5 mutant (41) and its wild type progenitor CC425 are shown for a comparison.
| CC425 | npq5 | cw15 | LHC1 | LHC2a | LHC2b | LHC2/7a | LHC2/7b |
|---|---|---|---|---|---|---|---|
| 0.741 ± 0.022 | 0.757 ± 0.010 | 0.746 ± 0.047 | 0.763 ± 0.010 | 0.717 ± 0.010 | 0.726 ± 0.049 | 0.709 ± 0.004 | 0.707 ± 0.011 |
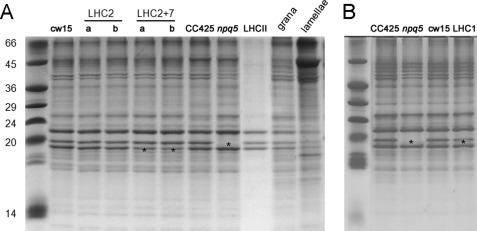
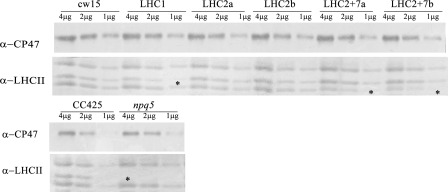
The LHCBM polypeptides isolated from thylakoid membranes migrate as three distinct bands, with apparent masses of 26, 23, and 22 kDa, in denaturing Tris-Tricine-urea-PAGE gels (Fig. 4A, lane LHCII). The three bands react with an anti-LHCII antibody that recognizes all nine LHCBM polypeptides (Fig. 5). The band with intermediate mobility (23 kDa, indicated by an asterisk) is absent in the npq5 mutant and reduced in the LHC1 transformant, suggesting that it corresponds to the LHCBM1 polypeptide (Figs. 4, A and B, and 5). Densitometric analysis (supplemental Table 2) shows a 40% reduction of the 23-kDa band in the LHC1 transformant as well as minor reductions of the 22- and 26-kDa bands, consistent with the off-target silencing observed by quantitative RT-PCR (Fig. 3A). The LHC2 transformants do not show reduced levels of the three LHCII bands in protein gels (Fig. 4, supplemental Table 2) and Western blotting (Fig. 5).
FIGURE 4.
Tris-Tricine-urea-PAGE of denatured thylakoid proteins. Panel A, SDS-PAGE separation of thylakoidal proteins extracted from all genotypes except LHC1 transformant is shown. In panel A the protein separation pattern of LHCII trimers, grana membranes, and stroma lamellae is also shown. The LHCII sample was obtained after mild thylakoid solubilization and purification of trimeric complex upon ultracentrifugation (73). Granal and stroma lamellae-enriched fractions were obtained following a modified version of the protocol by (55). Panel B, shown is SDS-PAGE separation of thylakoidal protein extracted from the LHC1 transformant and appropriate control strains. Asterisks highlight bands showing differential expression with respect to wild type strains.
FIGURE 5.
Western blot of thylakoid proteins. The lower panel was probed with an anti-LHCII antibody that recognizes all the nine LHCBM polypeptides. The upper panel was probed with an anti-CP47 antibody, used as loading control. Gels are loaded on a protein basis (4, 2, and 1 μg of thylakoidal proteins). Asterisks highlight bands showing differential accumulation with respect to wild type strains.
In contrast, the 22-kDa band is decreased in the LHC2/7 transformants, suggesting that this band corresponds to the LHCBM2/7 polypeptide (and, possibly, to polypeptides encoded by additional genes). This hypothesis is consistent with the real-time data (Fig. 3A), showing that only LHC2/7 transformants show significant silencing of both genes.
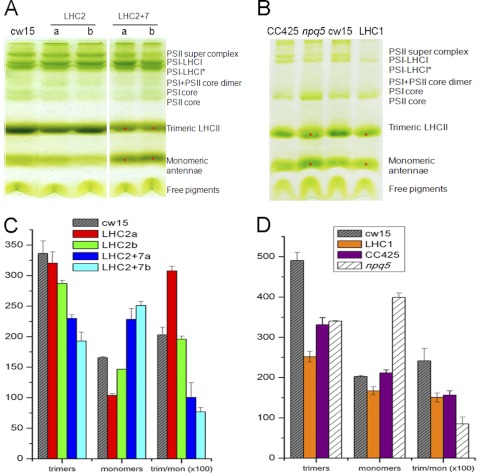
Pigment-protein complexes isolated from thylakoid membranes were separated according to their molecular mass through non-denaturing electrophoresis (Fig. 6). PSI-LHCI components migrate as a high Mr supercomplex, forming two bands in the upper part of the gel. PSII components migrate either as a high Mr supercomplex forming a low intensity, high Mr band, or as multiple bands representing the PSII core and the LHCII components in either a monomeric or a trimeric aggregation state.
FIGURE 6.
Deriphat-PAGE of native thylakoid proteins. Thylakoid proteins were extracted from cw15-, LHC2-, and LHC2/7-silenced transformants (panel A) and CC425-, npq5-, cw15-, and LHC1-silenced transformant (panel B). Proteins were solubilized with 1.2% α-DM. Bands showing an altered intensity are marked with a red asterisk. Densitometric analysis of the gels is shown, respectively, in panels C and D.
Silencing of LHCBM2 did not alter the trimer/monomer ratio (Fig. 6, A and C), whereas silencing of LHCBM2/7 led to a strong decrease in the intensity of the trimeric LHCII band and a corresponding increase of the monomeric band, suggesting a role of the LHCBM2/7 polypeptide in stabilizing the trimeric organization of LHCII. This result also suggests that LHCII monomerization requires reduction of the LHCBM2/7 polypeptide below a threshold level, which is reached only via silencing of both genes.
In the npq5 mutant and in the LHC1 transformant, a decrease in the trimer/monomer ratio (Fig. 6, B and D) are observed. This decrease was more evident in the LHC1 transformant than in the npq5 mutant, compared with their respective wild type strains, suggesting that it is partially due to off-target silencing of other LHCBM genes (Fig. 3A). Interestingly, in LHC2/7 transformants compared with cw15, an increase of the PSI-LHCI complex was evident in the upper part of the gel, suggesting an involvement of the LHCBM2/7 polypeptide in photosystem balance in thylakoids.
Roles of LHCBM1 and LHCBM2/7 in Light Harvesting and Photoprotection
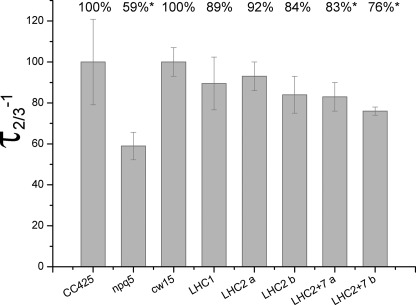
To investigate the role of the LHCBM1 and LHCBM2/7 polypeptides in light harvesting, the functional antenna size of PSII was measured (45) (Fig. 7). The highest reduction was observed in the npq5 mutant, with its antenna size reduced to 59% compared with CC425. The reduction observed in the LHC1 and LHC2 transformants is not significant considering the standard deviation associated with the measurement; in contrast, in the LHC2/7 transformants, a significant (17–24%) reduction in functional antenna size was observed. These data indicate that both the LHCBM1 and LHCBM2/7 polypeptides contribute to the building of the PSII antenna system.
FIGURE 7.
PSII functional antenna size. PSII functional antenna size was calculated as t⅔−1 of the kinetics of reduction of QA in the presence of 3-(3,4-dichlorophenyl)-1,1-dimethylurea. Differences with respect to wild type CC425 (only in the case of npq5 mutant) or cw15 are reported as a percent. Significant differences are highlighted with an asterisk. Data are means with S.D. (n = 6).
To assess the role of the LHCBM1 and LHCBM2/7 polypeptides in photoprotection, we measured the kinetics of the Fv/Fm decay during exposure to high light (1500 μE·m−2s−1). The results yielded only minor differences between different strains and transformants (data not shown), indicating that photoprotection in high light is a property shared redundantly with other LHCBM gene products.
Involvement of LHCBM2/7 in NPQ
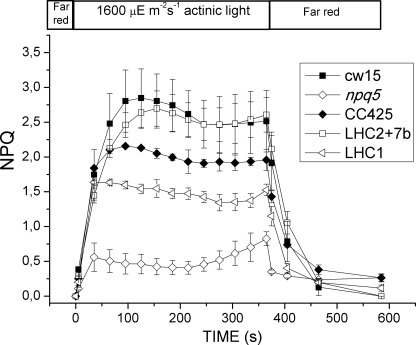
NPQ is a mechanism evolved by photosynthetic organisms to respond to varying light environments. It consists of several components, including energy-dependent quenching (qE) and state transition-dependent quenching (qT). In Chlamydomonas, LHCBM1 has been reported to be involved in thermal dissipation of excess light energy via energy-dependent quenching (41). Moreover, NPQ is induced upon high light acclimation with the concomitant accumulation of the LHCSR3 protein, directly involved in the quenching mechanism (48, 57). To assess the role of the LHCBM1 and LHCBM2/7 polypeptides in NPQ, we measured the kinetics of NPQ induction during exposure to high light (1600 μE·m−2s−1). The results (Fig. 8) indicate that both npq5 and the LHC1 transformant showed a significant reduction in NPQ with respect to their parental strains, respectively, CC425 and cw15. The LHC2/7 transformant showed amplitude of NPQ comparable with that of its parental strain, cw15. This observation indicates that the LHCBM2/7 polypeptide is not involved in NPQ and/or that the residual levels present in the silenced line still allow efficient NPQ. Protein extracts from cells grown in high light (500 μE·m−2s−1) were subjected to Western blot to check for accumulation of LHCSR3, the polypeptide responsible of NPQ in C. reinhardtii (48) (57). No significant differences with respect to wild type strains were found (supplemental Fig. 2). This result demonstrates that the NPQ phenotype of the npq5 mutant is directly due to the absence of LHCBM1 rather than to a modulation of LHCSR3 levels.
FIGURE 8.
NPQ analysis. CC425, cw15, the npq5 mutant, and the LHC2/7b and LHC1 transformants were acclimated in high light (500 μE·m−2s−1) for at least 10 generations. NPQ capacity under 1600 μE·m−2s−1 light was analyzed. Cells were preilluminated for 2 min with a weak (3 μE·m−2s−1) far-red LED before NPQ analysis. The far-red LED was kept on during dark recovery. Only the npq5 mutant and LHC1 transformant show reduced NPQ capacity respect to their wild type (CC425 and cw15, respectively), whereas the LHC2/7 transformant behaves as the cw15 strain.
LHCBM1 and LHCBM2/7 Are Involved in Protection against ROS
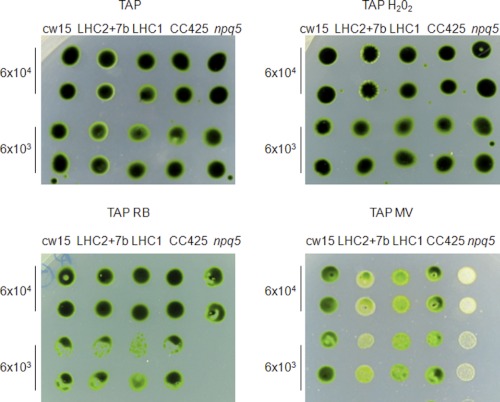
ROS are produced normally under any light conditions (21), and their production rate is enhanced when the input of incoming excitation energy and the efficiency of downstream electron acceptors are imbalanced (22). To investigate if LHCBM1 and LHCBM2/7 polypeptides are involved in protection against ROS, a spot test was performed on wild type (CC425 and cw15) cells, the npq5 mutant, and one LHC1 and one LHC2/7 transformant. Cells were grown mixotrophically until mid-exponential phase in TAP medium and then spotted at several dilutions on HS or TAP agar medium containing oxygen peroxide, rose bengal (generating singlet oxygen), or methyl viologen (generating superoxide anion) (46). The growth rate of the strains was scored after 8 days in continuous light (60 μE·m−2s−1). Mutant npq5 had a reduced growth rate in mixotrophic conditions in the presence of both rose bengal and methyl viologen, whereas the LHC1 and LHC2/7 transformants exhibited a reduced growth only in the presence of methyl viologen (Fig. 9). These results suggest a different role of LHCBM1 and LHCBM2/7 polypeptides in ROS scavenging, as npq5 is sensitive to both singlet oxygen (released from rose bengal) and oxygen superoxide (released from methyl viologen), whereas the growth of the LHC2/7 transformant was impaired in the medium supplemented with methyl viologen only. We did not observe significant differences in the growth rate of the different strains/transformants in photoautotrophic conditions (supplemental Fig. 3).
FIGURE 9.
Response of wild type, LHC2/7 and LHC1-silenced transformants, and npq5 mutant to different ROS in TAP medium. Cells were cultured in TAP medium until mid-log phase. Then 6 × 104 and 6 × 103 cells were spotted on TAP agar (1%) containing 0.5 mm H2O2, 7.5 μm rose bengal (RB), and 0.17 μm methyl viologen (MV) and on plain TAP as control. TAP medium was supplemented with arginine 50 μg/ml as CC425 is an Arg auxotrophic strain. Plates were grown in continuous light at 60 μE·m−2s−1 at 25 °C for 8 days, and finally growth was scored. The silenced LHC2/7 and LHC1 transformants show a slight sensitivity to methyl viologen and are not affected by H2O2 and rose bengal. The npq5 strain is highly sensitive to rose bengal and methyl viologen but not to H2O2. The experiment was repeated three times.
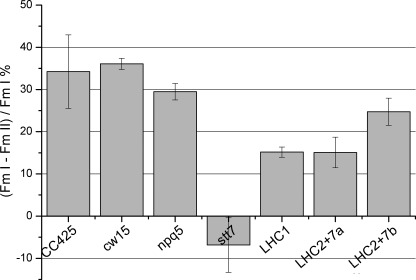
Involvement of LHCBM2/7 Polypeptide in State Transitions
Photosynthetic eukaryotes balance the light excitation energy between PSI and PSII through a process called state 1-state 2 transition. Over-excitation of PSII leads to changes in the redox state of the plastoquinone pool, which activate the Stt7 kinase and lead to phosphorylation of LHCII followed by the release of LHCII from PSII and its migration to PSI (49, 65). LHCB4 (CP29), LHCB5 (CP26), and LHCBM5 have been shown to be phosphorylated and move from PSII to PSI during state transitions in Chlamydomonas (62, 66), although only LHCB4 is crucial for association with PSI (42). Fig. 10 shows the state transition capacity of npq5, of the silenced LHC1 and LHC2/7 transformants, and of the respective wild type strains. The stt7 mutant, which is locked in state 1, was included as a control. This mutant has a lesion in a gene encoding a chloroplast thylakoid-associated serine-threonine protein kinase that is required for the phosphorylation of the major LHCII and for state transitions (49). Fig. 10 shows that, in contrast to stt7, npq5 shows normal state transition activity, considering the standard deviation associated with the measurement, suggesting that the LHCBM1 polypeptide is not involved in this process. In contrast, both LHC2/7 transformants show a significant reduction in the amplitude of state transitions, indicating that the LHCBM2/7 polypeptide is involved in this mechanism. The LHC1 transformant showed a significant reduction in state transitions, which can be attributed to silencing of non-target genes, including LHCBM2 and LHCBM7, as shown in Fig. 3A. To confirm the LHCBM2/7-dependent increase of PSI antenna size upon induction of state transition, we measured the PSI antenna size in state 1 and 2 in CC425 and cw15 wild type cells, in the npq5 mutant, and in the LHC1, zLHC2/7a, and LHC2/7b transformants by measuring the kinetics of P700 oxidation (supplemental Fig. 4). Faster kinetics of P700 oxidation are evident in both wild type strains and in the npq5 mutant but not in the LHC1, LHC2/7a, and LHC2/7b transformants. The LHC2/7 transformants show a reduced PSI antenna size in state 2 as compared with cw15, indicating that the LHCBM2/7 polypeptide plays a direct role in state transitions. The reduction of PSI antenna size in the LHC1 transformant can be attributed to nonspecific silencing of other LHCBM genes (see Fig. 3A), as PSI antenna size in state 2 of the npq5 mutant is not affected compared with its wild type (CC425).
FIGURE 10.
State transition analysis. CC425, cw15, npq5, stt7, LHC1, LHC2/7a, and LHC2/7b transformants were analyzed for the capacity of switching the LHCII antenna from PSII to PSI in conditions promoting plastoquinone pool over-reduction. State transition capacity is calculated as the % difference between maximum PSII fluorescence in state 1 and state 2.
DISCUSSION
Although photosynthetic core complexes (PSI and PSII) show a high level of conservation in higher plants, algae, and cyanobacteria (32), LHC complexes are encoded by a very diverse group of genes, allowing the colonization of very different habitats by photosynthetic organisms (67). The light harvesting antenna of C. reinhardtii is composed by at least 20 different proteins, associated with PSI (LHCA or LHCI) or with PSII (LHCB or LHCII). LHCB polypeptides are mainly associated with the phenomena of photo-acclimation and with adaptation to light quality and quantity. This is achieved through transcriptional and post-transcriptional modulation of the LHCBM polypeptides and through migration of selected complexes from PSII to PSI. Even if the nine LHCBM genes are highly conserved, it is likely that each gene or distinct group of genes exerts a specific role in light harvesting and/or in light dissipation. Except for LHCBM1 (41), the specific function of each LHCBM polypeptide is unknown. amiRNA technology can be a powerful technology to investigate the specific function of different LHCBM polypeptides, as it allows specific silencing of paralogs showing a high degree of identity. Conversely, RNAi allows the simultaneous silencing of many different LHC genes (16), thus allowing generating strains with an reduced antenna size but not allowing the discrimination between different paralogs, whereas amiRNA technology allows the simultaneous and specific silencing of smaller groups of genes (this work).
Here, we aimed to study the function of the LHCBM2/7 polypeptide and to compare it with the function of LHCBM1. Efficient and specific silencing was achieved for the LHCBM2/7 genes but not for LHCBM1. Analysis of pigment composition of the single- and double-silenced transformants is compatible with a reduced antenna size phenotype (Table 2). Silencing of LHCBM2 alone does not result in remarkable differences with respect to the wild type cw15 strain in terms of PSII-LHCII complex organization, amount of monomers and trimers, and intensity of the three LHCII bands resolved in SDS-PAGE. On the other hand, simultaneous silencing of both LHCBM2 and LHCBM7 results in a higher percentage of LHCII monomers over trimers (Fig. 6) and a significant reduction of the functional and biochemical antenna size (Fig. 7). Similar results were obtained in the case of the transformants silenced in the LHCBM1 gene and in the npq5 mutant. These results show that LHCBM1, LHCBM2, and LHCBM7 are important in maintaining the trimeric state of the antenna and the association of the LHCII antenna to the PSII complex. Silencing of LHCBM2 alone does not produce a significant phenotype, partly because LHCBM7 is unaffected and partly because LHCBM2 in less affected than in LHC2/7 transformants (Fig. 3A), resulting in unaltered levels of the LHCBM2/7 polypeptide (Figs. 4 and 5).
To investigate the functional role of the LHCBM2/7 polypeptide with respect to LHCBM1, NPQ and state transition analysis were carried out on the LHC1, LHC2, and LHC2/7 transformants and in the npq5 mutant. The results (Fig. 8) showed that the LHCBM2/7 polypeptide is not central to NPQ, whereas a strong effect on NPQ is produced both in the npq5 mutant, in agreement with a previous report (41), and in the LHC1-silenced transformant. The reduction in NPQ amplitude observed in npq5 and LHC1 could be explained with a direct role of this gene product in quenching excitation energy or with its specific interaction with a subunit carrying quenching activity (41). In C. reinhardtii, NPQ is fully dependent on the presence of the LHCSR3 polypeptide (48) that exhibits a very short fluorescence lifetime, making it a good quencher for interacting pigment-protein complexes (57). Because this polypeptide is correctly accumulated in npq5 and in LHC2/7 silenced cells, we suggest that LHCBM1, but not LHCBM2/7, has a role in NPQ by acting as a partner of LHCSR3 in the mechanism of heat dissipation. Interestingly, we observed a linear relationship between NPQ induction and LHCBM1 amounts in the CC425 and cw15 strains, in the npq5 mutant, and in two LHCBM1 transformants (LHC1 and LHC1b), suggesting that the amount of LHCBM1 per PSII core is determinant for NPQ activation (supplemental Fig. 5).
Conversely, the LHC2/7 silenced transformants, but not the npq5 mutant, display an altered state transition phenotype. Previous work showed that the LHCB5 (CP26), LHCB4 (CP29), and LHCBM5 LHCII proteins are found to be attached to PSI in state 2-promoting conditions (61, 62). We cannot state whether LHCBM2/7 is also phosphorylated and migrates toward PSI. However, part of the LHCII that remains connected to the PSII core and favors disconnection of the “mobile” LHCII fraction by repulsion of the negatively charged phosphate groups added to both the mobile and non-mobile fractions by the Stt7 kinase (68) (69). LHCBM2/7 could be involved in this repulsion mechanism, if it is not directly involved in the migration. On the contrary, LHCBM1 is not involved in state transitions, as already reported for the npq5 mutant (41). The increase of PSI-LHCI in thylakoid membrane in LHC2/7 transformants can be inferred to be a counteraction to the limitation in state transition, to increase the electron sink downstream of PSII and maintain the excitation pressure balance between the two photosystems. Alternatively, LHCBM2/7 proteins, which are resident in grana partitions unless phosphorylated, might be more involved than LHCBM1 to maintain the segregation of PSII core complexes in this compartment. In their absence, PSII might partition more largely in stroma membranes where it undergoes turnover, leading to increased degradation of PSII, with consequent apparent increase of PSI content per chlorophyll. Our data suggest that LHCBM1 and LHCBM2/7 not only have different functions but also have different localizations within the PSII supercomplex, with LHCBM2/7 being part or interacting with the peripheral mobile fraction of LHCII and LHCBM1 being more strictly connected to the PSII core, as suggested by the similar state transition induction in the npq5 mutant as compared with CC425. The hypothetic localization of LHCBM1 in between peripheral LHCBM proteins and PSII reaction center can also be relevant for its proposed role as a partner of the LHCSR3 protein in the process of excitation energy quenching. The energy absorbed by peripheral antenna proteins and directed to the reaction center can thus transit through LHCBM1 where it can be spilled-over for quenching by LHCSR3 before reaching the PSII core (57). Interestingly, in higher plants, depletion of the major subunits of the LHCII trimers, LHCB1 and LHCB2, yields impairment of state transitions but not of NPQ (38). It is worth noting that we also observed a reduced state transition in the LHC1 transformant that could be due to off-target silencing of other LHCBM polypeptides (Fig. 3A). Off-target silencing is a well known phenomenon and has been attributed to partial sequence complementarity between the small RNA and its off-target sequences (70). The differences in specificity observed here between LHCBM1, LHCBM2, and LHCBM2/7 amiRNAs, all designed using the same software and criteria, suggest that additional, transcript-specific mechanisms may influence off-target silencing. LHCBM1 and LHCBM2/7 have also a differential role in ROS scavenging, with LHCBM1 being specifically involved in singlet oxygen scavenging (Fig. 9), whereas both LHCBM1 and LHCBM2/7 are involved in resistance to superoxide anion. The peculiar role of LHCBM1 against singlet oxygen could be derived from its location in the inner shell of LHC subunits around the PSII reaction center with specifically enhanced scavenging function toward singlet oxygen produced by charge recombination in PSII RC; interestingly, even a small amount of residual LHCBM1, as in LHC1 mutant, is sufficient to restore resistance to single oxygen. The enhanced sensitivity of transformants toward superoxide can be ascribed to the role of neoxanthin. In fact both LHCBM1 and LHCBM2/7, from their high homology to plant LHCII subunits, are likely to be enriched in neoxanthin that has a specific role in superoxide anion scavenging (20).
The work presented here clearly demonstrates that, despite high sequence identity, different LHCBM polypeptides play different roles in Chlamydomonas. To investigate the functional properties of the other LHCBM genes whose roles are still unknown, we plan to silence unique and specific combinations of LHCBM genes using amiRNA technology. The transformants showing the highest silencing of the target gene(s) and unchanged levels of the other LHCBM transcripts will be then phenotypically analyzed to infer the function(s) specific to the LHCBM gene(s) silenced.
CONCLUSIONS
One of the major factors limiting the productivity of unicellular algae in photobioreactors is the high optical density of cells, causing strong light gradients with over-excitation of cells located near the surface and subsaturating light intensity for cells located further inside (10, 71), causing, respectively, excess energy dissipation and respiratory loss of biomass. Selective silencing of genes encoding polypeptides of the antenna system could allow for improved optical properties while preserving any specific photoprotective functions important for biomass accumulation. In this work we show that two antenna polypeptides are specifically involved in two regulatory mechanisms, active in optimizing efficiency in low light (state 1-state 2 transitions) and dissipating energy in excess light (NPQ). Moreover, we show that other LHC polypeptides are unaffected in the npq5 mutant and in LHC2/7-silenced cells, which still maintain the capacity for light stress resistance, known to be catalyzed through ROS scavenging and triplet quenching reactions. These results indicate that experimental manipulation of individual polypeptides of the light-harvesting antenna is a promising approach for gaining knowledge on their function and increasing efficiency of light energy conversion in mass culture.
Supplementary Material
Acknowledgment
We thank A. Molnar for providing the pChlamyRNA3 vector and for helpful suggestions about amiRNA design and use.
This work was supported by the Italian Ministry of Agriculture, Hydrobio project, and by Euopean Union Project 245070 FP7-KBBE-2009-3 SUNBIOPATH.

This article contains supplemental Tables 1 and 2 and Figs. 1–5.
- LHC
- light harvesting complex
- LHCA (b)
- LHC of photosystem I (II)
- LHCBM
- major subunits of photosystem II LHC in C. reinhardtii
- LHCII
- major LHC of photosystem II
- NPQ
- non-photochemical quenching
- PSI (II)
- photosystem I (II)
- ROS
- reactive oxygen species
- LHC1
- C. reinhardtii strain transformed with construct designed for silencing the LHCBM1 gene
- LHC2
- C. reinhardtii strain transformed with construct designed for silencing the LHCBM2 gene
- LHC2/7
- C. reinhardtii strain transformed with construct designed for silencing the LHCBM2 and LHCBM7 genes
- Chl
- chlorophyll
- μE
- microeinsteins
- amiRNA
- artificial microRNA
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine.
REFERENCES
- 1. Gaffron H. (1939) Reduction of CO2 with H2 in green plants. Nature 143, 204–205 [Google Scholar]
- 2. Gaffron H. (1940) Carbon dioxide reduction with molecular hydrogen in green algae. Am. J. Bot. 27, 273–283 [Google Scholar]
- 3. Melis A., Zhang L., Forestier M., Ghirardi M. L., Seibert M. (2000) Sustained photobiological hydrogen gas production upon reversible inactivation of oxygen evolution in the green alga Chlamydomonas reinhardtii. Plant. Physiol. 122, 127–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Melis A., Happe T. (2004) Trails of green alga hydrogen research. From hans gaffron to new frontiers. Photosynth. Res. 80, 401–409 [DOI] [PubMed] [Google Scholar]
- 5. Wang Z. T., Ullrich N., Joo S., Waffenschmidt S., Goodenough U. (2009) Algal lipid bodies. Stress induction, purification, and biochemical characterization in wild type and starchless Chlamydomonas reinhardtii. Eukaryot. Cell 8, 1856–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hu Q., Sommerfeld M., Jarvis E., Ghirardi M., Posewitz M., Seibert M., Darzins A. (2008) Microalgal triacylglycerols as feedstocks for biofuel production. Perspectives and advances. Plant. J. 54, 621–639 [DOI] [PubMed] [Google Scholar]
- 7. Choi S. P., Nguyen M. T., Sim S. J. (2010) Enzymatic pretreatment of Chlamydomonas reinhardtii biomass for ethanol production. Bioresour. Technol. 101, 5330–5336 [DOI] [PubMed] [Google Scholar]
- 8. Rasala B. A., Muto M., Lee P. A., Jager M., Cardoso R. M., Behnke C. A., Kirk P., Hokanson C. A., Crea R., Mendez M., Mayfield S. P. (2010) Production of therapeutic proteins in algae. Analysis of expression of seven human proteins in the chloroplast of Chlamydomonas reinhardtii. Plant. Biotechnol. J. 8, 719–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Elrad D., Grossman A. R. (2004) A genome's-eye view of the light-harvesting polypeptides of Chlamydomonas reinhardtii. Curr. Genet. 45, 61–75 [DOI] [PubMed] [Google Scholar]
- 10. Mitra M., Melis A. (2008) Optical properties of microalgae for enhanced biofuels production. Opt. Express 16, 21807–21820 [DOI] [PubMed] [Google Scholar]
- 11. Polle J. E., Kanakagiri S., Jin E. S., Masuda T., Melis A. (2002) Truncated chlorophyll antenna size of the photosystems. A practical method to improve microalgal productivity and hydrogen production in mass culture. Int. J. Hydrogen Energy 27, 1257–1264 [Google Scholar]
- 12. Melis A. (2009) Solar energy conversion efficiencies in photosynthesis. Minimizing the chlorophyll antennae to maximize efficiency. Plant Science 177, 272–280 [Google Scholar]
- 13. Polle J. E., Benemann J. R., Tanaka A., Melis A. (2000) Photosynthetic apparatus organization and function in the wild type and a chlorophyll b-less mutant of Chlamydomonas reinhardtii. Dependence on carbon source. Planta 211, 335–344 [DOI] [PubMed] [Google Scholar]
- 14. Polle J. E., Niyogi K. K., Melis A. (2001) Absence of lutein, violaxanthin, and neoxanthin affects the functional chlorophyll antenna size of photosystem-II but not that of photosystem-I in the green alga Chlamydomonas reinhardtii. Plant. Cell Physiol. 42, 482–491 [DOI] [PubMed] [Google Scholar]
- 15. Tetali S. D., Mitra M., Melis A. (2007) Development of the light-harvesting chlorophyll antenna in the green alga Chlamydomonas reinhardtii is regulated by the novel Tla1 gene. Planta. 225, 813–829 [DOI] [PubMed] [Google Scholar]
- 16. Mussgnug J. H., Thomas-Hall S., Rupprecht J., Foo A., Klassen V., McDowall A., Schenk P. M., Kruse O., Hankamer B. (2007) Engineering photosynthetic light capture. Impacts on improved solar energy to biomass conversion. Plant. Biotechnol. J. 5, 802–814 [DOI] [PubMed] [Google Scholar]
- 17. Peterman E. J., Gradinaru C. C., Calkoen F., Borst J. C., van Grondelle R., van Amerongen H. (1997) Xanthophylls in light harvesting complex II of higher plants. Light harvesting and triplet quenching. Biochemistry 36, 12208–12215 [DOI] [PubMed] [Google Scholar]
- 18. Dall'Osto L., Lico C., Alric J., Giuliano G., Havaux M., Bassi R. (2006) Lutein is needed for efficient chlorophyll triplet quenching in the major LHCII antenna complex of higher plants and effective photoprotection in vivo under strong light. BMC Plant. Biol. 6, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mozzo M., Dall'Osto L., Hienerwadel R., Bassi R., Croce R. (2008) Photoprotection in the antenna complexes of photosystem II. Role of individual xanthophylls in chlorophyll triplet quenching. J. Biol. Chem. 283, 6184–6192 [DOI] [PubMed] [Google Scholar]
- 20. Dall'Osto L., Cazzaniga S., North H., Marion-Poll A., Bassi R. (2007) The Arabidopsis aba4-1 mutant reveals a specific function for neoxanthin in protection against photooxidative stress. Plant Cell 19, 1048–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dall'Osto L., Fiore A., Cazzaniga S., Giuliano G., Bassi R. (2007) Different roles of α- and β-branch xanthophylls in photosystem assembly and photoprotection. J. Biol. Chem. 282, 35056–35068 [DOI] [PubMed] [Google Scholar]
- 22. Niyogi K. K. (1999) Photoprotection Revisited. Genetic and Molecular Approaches. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 333–359 [DOI] [PubMed] [Google Scholar]
- 23. Dall'Osto L., Cazzaniga S., Havaux M., Bassi R. (2010) Enhanced photoprotection by protein-bound vs free xanthophyll pools. A comparative analysis of chlorophyll b and xanthophyll biosynthesis mutants. Mol. Plant. 3, 576–593 [DOI] [PubMed] [Google Scholar]
- 24. Kim E. H., Li X. P., Razeghifard R., Anderson J. M., Niyogi K. K., Pogson B. J., Chow W. S. (2009) The multiple roles of light-harvesting chlorophyll a/b-protein complexes define structure and optimize function of Arabidopsis chloroplasts. A study using two chlorophyll b-less mutants. Biochim. Biophys. Acta 1787, 973–984 [DOI] [PubMed] [Google Scholar]
- 25. Morosinotto T., Castelletti S., Breton J., Bassi R., Croce R. (2002) Mutation analysis of Lhca1 antenna complex. Low energy absorption forms originate from pigment-pigment interactions. J. Biol. Chem. 277, 36253–36261 [DOI] [PubMed] [Google Scholar]
- 26. Dall'Osto L., Caffarri S., Bassi R. (2005) A mechanism of nonphotochemical energy dissipation, independent from PsbS, revealed by a conformational change in the antenna protein CP26. Plant Cell 17, 1217–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Avenson T. J., Ahn T. K., Zigmantas D., Niyogi K. K., Li Z., Ballottari M., Bassi R., Fleming G. R. (2008) Zeaxanthin radical cation formation in minor light-harvesting complexes of higher plant antenna. J. Biol. Chem. 283, 3550–3558 [DOI] [PubMed] [Google Scholar]
- 28. Ferreira K. N., Iverson T. M., Maghlaoui K., Barber J., Iwata S. (2004) Architecture of the photosynthetic oxygen-evolving center. Science 303, 1831–1838 [DOI] [PubMed] [Google Scholar]
- 29. Umena Y., Kawakami K., Shen J. R., Kamiya N. (2011) Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 Å. Nature 473, 55–60 [DOI] [PubMed] [Google Scholar]
- 30. Morris E. P., Hankamer B., Zheleva D., Friso G., Barber J. (1997) The three-dimensional structure of a photosystem II core complex determined by electron crystallography. Structure 5, 837–849 [DOI] [PubMed] [Google Scholar]
- 31. Harrer R., Bassi R., Testi M. G., Schäfer C. (1998) Nearest-neighbor analysis of a photosystem II complex from Marchantia polymorpha L. (liverwort), which contains reaction center and antenna proteins. Eur. J. Biochem. 255, 196–205 [DOI] [PubMed] [Google Scholar]
- 32. Dekker J. P., Boekema E. J. (2005) Supramolecular organization of thylakoid membrane proteins in green plants. Biochim. Biophys. Acta 1706, 12–39 [DOI] [PubMed] [Google Scholar]
- 33. Kovács L., Damkjaer J., Kereïche S., Ilioaia C., Ruban A. V., Boekema E. J., Jansson S., Horton P. (2006) Lack of the light-harvesting complex CP24 affects the structure and function of the grana membranes of higher plant chloroplasts. Plant Cell 18, 3106–3120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Caffarri S., Kouril R., Kereïche S., Boekema E. J., Croce R. (2009) Functional architecture of higher plant photosystem II supercomplexes. EMBO J. 28, 3052–3063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Minagawa J., Takahashi Y. (2004) Structure, function, and assembly of Photosystem II and its light-harvesting proteins. Photosynth. Res. 82, 241–263 [DOI] [PubMed] [Google Scholar]
- 36. Stauber E. J., Fink A., Markert C., Kruse O., Johanningmeier U., Hippler M. (2003) Proteomics of Chlamydomonas reinhardtii light-harvesting proteins. Eukaryot. Cell 2, 978–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Turkina M. V., Vener A. V. (2007) Identification of phosphorylated proteins. Methods Mol. Biol. 355, 305–316 [DOI] [PubMed] [Google Scholar]
- 38. Andersson J., Wentworth M., Walters R. G., Howard C. A., Ruban A. V., Horton P., Jansson S. (2003) Absence of the Lhcb1 and Lhcb2 proteins of the light-harvesting complex of photosystem II. Effects on photosynthesis, grana stacking, and fitness. Plant J. 35, 350–361 [DOI] [PubMed] [Google Scholar]
- 39. de Bianchi S., Dall'Osto L., Tognon G., Morosinotto T., Bassi R. (2008) Minor antenna proteins CP24 and CP26 affect the interactions between photosystem II subunits and the electron transport rate in grana membranes of Arabidopsis. Plant Cell 20, 1012–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Damkjaer J. T., Kereïche S., Johnson M. P., Kovacs L., Kiss A. Z., Boekema E. J., Ruban A. V., Horton P., Jansson S. (2009) The photosystem II light-harvesting protein Lhcb3 affects the macrostructure of photosystem II and the rate of state transitions in Arabidopsis. Plant Cell 21, 3245–3256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Elrad D., Niyogi K. K., Grossman A. R. (2002) A major light-harvesting polypeptide of photosystem II functions in thermal dissipation. Plant Cell 14, 1801–1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tokutsu R., Iwai M., Minagawa J. (2009) CP29, a monomeric light-harvesting complex II protein, is essential for state transitions in Chlamydomonas reinhardtii. J. Biol. Chem. 284, 7777–7782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nguyen A. V., Thomas-Hall S. R., Malnoë A., Timmins M., Mussgnug J. H., Rupprecht J., Kruse O., Hankamer B., Schenk P. M. (2008) Transcriptome for photobiological hydrogen production induced by sulfur deprivation in the green alga Chlamydomonas reinhardtii. Eukaryot. Cell 7, 1965–1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Molnar A., Bassett A., Thuenemann E., Schwach F., Karkare S., Ossowski S., Weigel D., Baulcombe D. (2009) Highly specific gene silencing by artificial microRNAs in the unicellular alga Chlamydomonas reinhardtii. Plant J. 58, 165–174 [DOI] [PubMed] [Google Scholar]
- 45. Sundby C., Melis A., Maenpaa P., Andersson B. (1986) Temperature-dependent changes in the antenna size of photosystem II. Reversible conversion of photosystem II-α to photosystem II-β. Biochim. Biophys. Acta 851, 475–483 [Google Scholar]
- 46. Baroli I., Gutman B. L., Ledford H. K., Shin J. W., Chin B. L., Havaux M., Niyogi K. K. (2004) Photo-oxidative stress in a xanthophyll-deficient mutant of Chlamydomonas. J. Biol. Chem. 279, 6337–6344 [DOI] [PubMed] [Google Scholar]
- 47. Demmig-Adams B., Adams W. W., III, Barker D. H., Logan B. A., Bowling D. R., Verhoeven A. S. (1996) Using chlorophyll fluorescence to assess the fraction of absorbed light allocated to thermal dissipation of excess excitation. Physiologia Plantarum 98, 253–264 [Google Scholar]
- 48. Peers G., Truong T. B., Ostendorf E., Busch A., Elrad D., Grossman A. R., Hippler M., Niyogi K. K. (2009) An ancient light-harvesting protein is critical for the regulation of algal photosynthesis. Nature 462, 518–521 [DOI] [PubMed] [Google Scholar]
- 49. Depège N., Bellafiore S., Rochaix J. D. (2003) Role of chloroplast protein kinase Stt7 in LHCII phosphorylation and state transition in Chlamydomonas. Science 299, 1572–1575 [DOI] [PubMed] [Google Scholar]
- 50. Kindle K. L. (1990) High frequency nuclear transformation of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. U.S.A. 87, 1228–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Niyogi K. K., Bjorkman O., Grossman A. R. (1997) Chlamydomonas Xanthophyll cycle mutants identified by video imaging of chlorophyll fluorescence quenching. Plant Cell 9, 1369–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lagarde D., Beuf L., Vermaas W. (2000) Increased production of zeaxanthin and other pigments by application of genetic engineering techniques to Synechocystis sp. strain PCC 6803. Appl. Environ. Microbiol. 66, 64–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Croce R., Morosinotto T., Castelletti S., Breton J., Bassi R. (2002) The Lhca antenna complexes of higher plants photosystem I. Biochim. Biophys. Acta 1556, 29–40 [DOI] [PubMed] [Google Scholar]
- 54. Dainese P., Bassi R. (1991) Subunit stoichiometry of the chloroplast photosystem II antenna system and aggregation state of the component chlorophyll a/b binding proteins. J. Biol. Chem. 266, 8136–8142 [PubMed] [Google Scholar]
- 55. Bassi R., Wollman F. A. (1991) The chlorophyll a/b proteins of photosystem II in Chlamydomonas reinhardtii. Isolation, characterization, and immunological cross-reactivity to higher plants polypeptides. Planta 183, 423–433 [DOI] [PubMed] [Google Scholar]
- 56. Schägger H., von Jagow G. (1987) Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166, 368–379 [DOI] [PubMed] [Google Scholar]
- 57. Bonente G., Ballottari M., Truong T. B., Morosinotto T., Ahn T. K., Fleming G. R., Niyogi K. K., Bassi R. (2011) Analysis of LhcSR3, a protein essential for feedback de-excitation in the green alga Chlamydomonas reinhardtii. PLoS Biol. 9, e1000577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Morosinotto T., Ballottari M., Klimmek F., Jansson S., Bassi R. (2005) The association of the antenna system to photosystem I in higher plants. Cooperative interactions stabilize the supramolecular complex and enhance red-shifted spectral forms. J. Biol. Chem. 280, 31050–31058 [DOI] [PubMed] [Google Scholar]
- 59. Cardol P., Bailleul B., Rappaport F., Derelle E., Béal D., Breyton C., Bailey S., Wollman F. A., Grossman A., Moreau H., Finazzi G. (2008) An original adaptation of photosynthesis in the marine green alga Ostreococcus. Proc. Natl. Acad. Sci. U.S.A. 105, 7881–7886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Fleischmann M. M., Ravanel S., Delosme R., Olive J., Zito F., Wollman F. A., Rochaix J. D. (1999) Isolation and characterization of photoautotrophic mutants of Chlamydomonas reinhardtii deficient in state transition. J. Biol. Chem. 274, 30987–30994 [DOI] [PubMed] [Google Scholar]
- 61. Iwai M., Takizawa K., Tokutsu R., Okamuro A., Takahashi Y., Minagawa J. (2010) Isolation of the elusive supercomplex that drives cyclic electron flow in photosynthesis. Nature 464, 1210–1213 [DOI] [PubMed] [Google Scholar]
- 62. Takahashi H., Iwai M., Takahashi Y., Minagawa J. (2006) Identification of the mobile light-harvesting complex II polypeptides for state transitions in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. U.S.A. 103, 477–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Bonente G., Pippa S., Castellano S., Bassi R., Ballottari M. (2012) Acclimation of Chlamydomonas reinhardtii to different growth irradiances. J. Biol. Chem. 287, 5833–5847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Fischer N., Rochaix J. D. (2001) The flanking regions of PsaD drive efficient gene expression in the nucleus of the green alga Chlamydomonas reinhardtii. Mol. Genet. Genomics 265, 888–894 [DOI] [PubMed] [Google Scholar]
- 65. Lemeille S., Rochaix J. D. (2010) State transitions at the crossroad of thylakoid signalling pathways. Photosynth. Res. 106, 33–46 [DOI] [PubMed] [Google Scholar]
- 66. Lemeille S., Willig A., Depège-Fargeix N., Delessert C., Bassi R., Rochaix J. D. (2009) Analysis of the chloroplast protein kinase Stt7 during state transitions. PLoS Biol. 7, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ballottari M., Girardon J., Dall'osto L., Bassi R. (2012) Evolution and functional properties of Photosystem II light harvesting complexes in eukaryotes. Biochim. Biophys. Acta 1817, 143–157 [DOI] [PubMed] [Google Scholar]
- 68. Bassi R., Ghiretti Magaldi A., Tognon G., Giacometti G. M., Miller K. R. (1989) Two-dimensional crystals of the photosystem II reaction center complex from higher plants. Eur. J. Cell Biol. 50, 84–93 [PubMed] [Google Scholar]
- 69. Fristedt R., Granath P., Vener A. V. (2010) A protein phosphorylation threshold for functional stacking of plant photosynthetic membranes. PLoS One 5, e10963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Jackson A. L., Burchard J., Schelter J., Chau B. N., Cleary M., Lim L., Linsley P. S. (2006) Widespread siRNA “off-target” transcript silencing mediated by seed region sequence complementarity. RNA. 12, 1179–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Bonente G., Formighieri C., Mantelli M., Catalanotti C., Giuliano G., Morosinotto T., Bassi R. (2011) Mutagenesis and phenotypic selection as a strategy toward domestication of Chlamydomonas reinhardtii strains for improved performance in photobioreactors. Photosynth. Res. 108, 107–120 [DOI] [PubMed] [Google Scholar]
- 72. Tamura K., Dudley J., Nei M., Kumar S. (2007) MEGA4. Molecular Evolutionary Genetics Analysis (MEGA) software Version 4.0. Mol. Biol. Evol. 24, 1596–1599 [DOI] [PubMed] [Google Scholar]
- 73. Caffarri S., Frigerio S., Olivieri E., Righetti P. G., Bassi R. (2005) Differential accumulation of Lhcb gene products in thylakoid membranes of Zea mays plants grown under contrasting light and temperature conditions. Proteomics. 5, 758–768 [DOI] [PubMed] [Google Scholar]
- 74. Sueoka N. (1960) Mitotic replication of deoxyribonucleic acid in Chlamydomonas reinhardi. Proc. Natl. Acid. Sci. USA 46, 83–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.