Background: Glutathione peroxidase 3 was identified as a new tumor suppressor. However, the mechanism is not clear.
Results: Our analysis indicates that glutathione peroxidase 3 activates p53-induced gene 3 both in vivo and in vitro.
Conclusion: p53-induced gene 3 is a major mediator of GPx3-induced cell death.
Significance: Glutathione peroxidase 3-p53-induced gene 3 signaling represents a novel signaling pathway for cell death.
Keywords: Cell Death, Prostate Cancer, Reactive Oxygen Species (ROS), Tumor Marker, Tumor Suppressor Gene
Abstract
Expression of glutathione peroxidase 3 (GPx3) is down-regulated in a variety of human malignancies. Both methylation and deletion of GPx3 gene underlie the alterations of GPx3 expression in prostate cancer. A strong correlation between the down-regulation of GPx3 expression and progression of prostate cancer and the suppression of prostate cancer xenografts in SCID mice by forced expression of GPx3 suggests a tumor suppression role of GPx3 in prostate cancer. However, the mechanism of GPx3-mediated tumor suppression remains unclear. In this report, GPx3 was found to interact directly with p53-induced gene 3 (PIG3). Forced overexpression of GPx3 in prostate cancer cell lines DU145 and PC3 as well as immortalized prostate epithelial cells RWPE-1 increased apoptotic cell death. Expression of GPx3x73c, a peroxidase-negative OPAL codon mutant, in DU145 and PC3 cells also increased cell death. The induced expression of GPx3 in DU145 and PC3 cells resulted in an increase in reactive oxygen species and caspase-3 activity. These activities were abrogated by either knocking down PIG3 or mutating the PIG3 binding motif in GPx3 or binding interference from a peptide corresponding to PIG3 binding motif in GPx3. In addition, UV-treated RWPE-1 cells underwent apoptotic death, which was partially prevented by knocking down GPx3 or PIG3, suggesting that GPx3-PIG3 signaling is critical for UV-induced apoptosis. Taken together, these results reveal a novel signaling pathway of GPx3-PIG3 in the regulation of cell death in prostate cancer.
Introduction
Glutathione peroxidase 3 (GPx3)3 belongs to the family of glutathione peroxidases that catalyze the reduction of hydrogen peroxide, organic hydrogen peroxide, and lipid peroxides by oxidizing glutathione (1). GPx3 is thought to protect cells from the oxidative damage generated by these reactive oxygen species (ROS) (2–7). It was recently found that the GPx3 gene is widely methylated or deleted in prostate cancer, resulting in low expression (8–11). In addition, the down-regulation of GPx3 was found closely associated with cancer progression (12). Forced expressions of GPx3 in prostate cancer cell lines PC3 (androgen-insensitive), DU145 (androgen-insensitive), and LNCaP (androgen-sensitive) in the absence of oxidative stress were shown to be associated with a reduction of tumor growth and a decrease in tumor metastasis in SCID mice (9). GPx3 contains a selenocysteine encoded by a UGA opal codon (13), which requires selenium to translate GPx3 to a full-length and enzymatically active protein. Coincidently, a population with higher selenium levels was found to be associated with a lower incidence of prostate cancer (14), which suggests that selenoproteins, such as GPx3, may have a protective role in the development of prostate cancer.
p53-induced gene 3 (PIG3), a NADPH-dependent reductase, is transactivated by p53 to generate ROS (15) and is essential for p53-mediated apoptosis (16–18). PIG3 is a critical signaling molecule that functions in response to DNA damage, which possibly occurs through an enhancement of Chk1 and Chk2 phosphorylation and the recruitment of DNA repair components (16). In addition, microsatellite instability has been associated with pentanucleotide repeats present in the promoter region of the PIG3 gene in a subset of lymphoblastic leukemia cases (19). In this study, we show that PIG3 is an important factor in GPx3-mediated cell death and is essential for UV-mediated cell death.
EXPERIMENTAL PROCEDURES
Cloning and Plasmid DNA Construction
Full-length cDNA of wild-type or a point mutant (GPx3x73c) of GPx3 was obtained by PCR amplification of GPx3 from Human Prostate Marathon-Ready cDNA (Clontech) and ligated into TOPO-TA cloning vectors. Deletion mutants of GPx3 were created using PCR assays containing full-length cDNAs of GPx3x73c as a template and the primers listed in Table 1. PCR products were agarose gel-purified, digested with BamHI and SalI, and then ligated with pGEX5-X-3 linearized with the same restriction enzymes. The constructs were transformed into Escherichia coli BL21 cells, and ampicillin-resistant clones were selected and sequenced to confirm the absence of mutations or frameshifts. For the His-PIG3 construct, full-length PIG3 was also obtained by PCR from Human Prostate Marathon-Ready cDNA using primers 5′-GGGAGCCGGGCCAGGGAATTCATGTTAGCCGTGC-3′ and 5′-CTGCCCCATCCTCCTCTCGAGGGGCAGTTCCAGG-3′. Amplified PIG3 was ligated into pET28a for fusion with a His tag. Using the same strategy, wild-type, full-length GPx3 was cloned into pSG5 using the GPx3 cDNA template and primers 5′-CACCTCTCTGCCCAGATTTGCTTA-3′ and 5′-AGAAAGGCTTTTACTGGGCAGACG-3′. Tetracycline-inducible clones of pCDNA4-GPx3 were then obtained by PCR using primers 5′-CCACGTCCACAGGTACCTGACTTCCACCTCTCTGC-3′ and 5′-TTCAGTTACTTCCTCTCGAGCCCCAGGGCTGCCTG-3′. pCDNA4-ΔGPx3, which represents a motif deletion mutant of GPx3, was obtained using mutagenesis PCR and the following pairs of primers: 5′-CCACGTCCACAGGTACCTGACTTCCACCTCTCTGC-3′, 5′-TTCAATGTACTGGCCCGTCAGCTTCGACTTCTCTTGTCCCCG-3′, 5′-CGGGGACAAGAGAAGTCGAAGCTGACGGGCCAGTACATTGAA-3′, and 5′-TTCAGTTACTTCCTCTCGAGCCCCAGGGCTGCCTG-3′. Additional PCR was performed by using the products of these two PCRs as templates with primers CCACGTCCACAGGTACCTGACTTCCACCTCTCTGC and TTCAGTTACTTCCTCTCGAGCCCCAGGGCTGCCTG to create a full-length GPx3 deletion mutant (ΔGPx3) ligated into pCDNA4. The pCDNA4 expression vectors containing wild-type or mutated GPx3 were co-transfected with pCDNA6 into PC3 or DU145 cells, respectively. The pCDNA4-GPx3x73c was created by mutagenic PCR to create a GPx3 mutant with a cysteine replacing the opal codon in codon 73. Transformants were selected with zeocin (250 μg/ml) and blasticidin (10 μg/ml) and designated PDG (pCDNA4-GPx3/pCDNA6 in PC3), PΔG (pCDNA4-ΔGPx3/pCDNA6 in PC3), DDG (pCDNA4-GPx3/pCDNA6 in DU145), DΔG (pCDNA4-ΔGPx3/pCDNA6 in DU145), PGPxx73c (pCDNA4-GPx3x73c/pCDNA6 in PC3), and DGPxx73c (pCDNA4-GPx3x73c/pCDNA6 in DU145).
TABLE 1.
Primers used for making GST-GPx3 deletion constructs
| Primers | Forward/reverse (5′ to 3′) |
|---|---|
| GPx3-full length | CCCACCCCGCCAGGATCCGGCTGCTGCAGGCGTCCTG/AGTTACTTCCTCTTGTCGACCAGGGCTGCCTGCCGCCT |
| GPx3-N-deletion | GGAGAGAACTCAGGGATCCTTCCTACCCTCAAGTAT/AGTTACTTCCTCTTGTCGACCAGGGCTGCCTGCCGCCT |
| GPx3-C-deletion | CCCACCCCGCCAGGATCCGGCTGCTGCAGGCGTCCTG/ATACTTGAGGGTAGGGTCGACCTCTGAGTTCTCTCC |
| GPx3_B1 | CCCACCCCGCCAGGATCCGGCTGCTGCAGGCGTCCTG/CCACTTATGCCACCATGGCAGTCGACCTTCGACTTCTC |
| GPx3_B2 | CCCACCCCGCCAGGATCCGGCTGCTGCAGGCGTCCTG/TTCAGTTCAATGTACTGGCGTCGACGGCCTCAGTAGC |
| GPx3_B3 | CCCACCCCGCCAGGATCCGGCTGCTGCAGGCGTCCTG/CGGACATACTTGAGGGTAGGTCGACTCTCTGAGTTCT |
| GPx3_B4 | CCCACCCCGCCAGGATCCGGCTGCTGCAGGCGTCCTG/AGGAGCTCCGAGGTGGGTCGACAGGAGTTCTTTAGGA |
| GPx3_B5 | CCCACCCCGCCAGGATCCGGCTGCTGCAGGCGTCCTG/GTGGTCCGGTGGTGCCAGCGTCGACTGGGTATACCAT |
| GPx3_D1 | CACCCCGCCAGGATCCGGCTGCTGCAGTCGAAGATGGACTGCCATGGT/AGTTACTTCCTCTTGTCGACCAGGGCTGCCTGCCGCCT |
| GPx3_D2 | CACCCCGCCAGGATCCGGCTGCTGCAGAACGTGGCCAGCTACTGAGGC/AGTTACTTCCTCTTGTCGACCAGGGCTGCCTGCCGCCT |
| GPx3_D3 | CACCCCGCCAGGATCCGGCTGCTGCAGGAACCAGGAGAGAACTCAGAG/AGTTACTTCCTCTTGTCGACCAGGGCTGCCTGCCGCCT |
| GPx3_D4 | CACCCCGCCAGGATCCGGCTGCTGCAGTTCTACACTTTCCTAAAGAAC/AGTTACTTCCTCTTGTCGACCAGGGCTGCCTGCCGCCT |
| GPx3_D5 | CACCCCGCCAGGATCCGGCTGCTGCAGGTGGGGCCAGATGGTATACCC/AGTTACTTCCTCTTGTCGACCAGGGCTGCCTGCCGCCT |
Cell Lines and Cell Culture
Prostate cancer cell lines DU145 and PC3, as well as the immortalized prostate epithelial cell line RWPE-1 were purchased from American Type Culture Collection (ATCC, Manassas, VA) in 2007. These cell lines underwent one cycle of growth before being stored in liquid nitrogen until needed. The cells were used for transfections within 2 weeks of thawing. DU145 cells were cultured in modified Eagle's medium (Invitrogen), and PC3 cells were cultured in F12K (Invitrogen) supplemented with 10% fetal bovine serum (FBS) (Cell Gro, Manassas, VA). Both cell lines were incubated at 37 °C and 5% CO2. RWPE1 cells were cultured in keratinocyte serum-free medium supplemented with 0.05 mg/ml bovine pituitary extract and 5 ng/ml recombinant epidermal growth factor (Invitrogen). PDG and PΔG were PC3-based clones and were cultured in F12K medium. DDG and DΔG were DU145-transformed clones and were cultured in modified Eagle's medium. Medium was supplemented with sodium selenium (1 nm final concentration; Sigma-Aldrich) for GPx3 expression and was changed every 3–4 days.
Fluorescence-activated Cell Sorting Analysis (FACS) of Cell Death
Cultures of PDG1 (one of the clones of pCDNA4-GPx3/pCNDA6-transfected PC3 cells) or DDG3 (one of the clones of pCDNA4-GPx3/pCNDA6-transfected DU145 cells) at 60–80% confluence were treated with or without tetracycline (5 μg/ml) for 3 days. These cells were then transfected with PIG3-targeted siRNA (siPig3) or a scrambled siRNA control (siScr) using Lipofectamine 2000TM (Invitrogen). Tetracycline induction was maintained for an additional 72 h. The cells were then harvested, and cell suspensions were incubated with 5 μl of Alexa Fluor 488-conjugated annexin V and 1 μl of propidium iodide (PI, 100 μg/ml) in binding buffer for 20 min (Molecular Probes). FACS analysis was performed using a LSR II flow cytometer (BD Bioscience). Unstained cells were used as negative controls. Cells irradiated with UV light and stained with either Alexa Fluor 488 or PI were used to calibrate or adjust for compensation. All samples were assessed in triplicate. For UV-induced cell death analysis, RWPE-1 cells were transfected with siRNA (siPig3 or siGPx3) or a scrambled siRNA control (siScr) using Lipofectamine 2000TM and irradiated with 300 mJ of UV light 48 h later. The cells were then harvested 6 h later for analysis in an annexin V binding assay.
Yeast Two-hybrid Analysis
Yeast competent cells were prepared as described previously (20). Briefly, 100 μl (2.5 million cells) of freshly prepared AH109 competent cells were mixed with pBD-GPx3 DNA, a prostate cDNA library, and herring testes carrier DNA in 0.6 ml of PEG/LiAc. The mixture was incubated at 30 °C for 30 min. Dimethyl sulfoxide (70 μl) was then added, and the cells were heat-shocked at 42 °C for 15 min. Cells were then pelleted, resuspended in 1 ml of YPD medium, and shaken at 30 °C for 40 min. Transformed cells were pelleted, resuspended in 0.5 ml of 0.9% NaCl, and plated onto low stringency SD−Leu/−Trp plates or medium stringency SD−Leu/−Trp/−His plates, respectively. Colony lift assays were performed as described previously (20), and blue colonies were transferred to high stringency SD−Ade/−His/−Leu/−Trp plates containing an indicator for galactosidase activity.
Validation of Protein-Protein Interactions in Yeast
Plasmid DNA from positive clones was isolated from yeast and sequenced. For validation, an isolated clone of pACT2-PIG3 was co-transformed with pBD-GPx3 into AH109 yeast cells and plated on SD−Ade/−His/−Leu/−Trp high stringency plates. Co-transformation of pGADT7T and pGBKT7–53 was used as a positive control for protein-protein interactions, and pGADT7T and pGBKT7-Lam were used as negative controls.
Immunoprecipitation
Antibodies raised against GPx3 or PIG3 (Santa Cruz Biotechnology) were incubated with ExactaCruzTM A or B immunoprecipitation (IP) matrix (Santa Cruz Biotechnology), respectively, for 1 h at 4 °C. The IP matrices were pelleted, washed with PBS, and further incubated with precleared cell lysates of GPx3 expression clones for 16 h at 4 °C. The IP matrix was then pelleted and washed four times with radioimmuneprecipitation assay buffer, and the protein complex was eluted with SDS-PAGE sample buffer. Samples were then electrophoresed on 7% SDS-polyacrylamide gels and immunoblotted with anti-PIG3 or anti-GPx3 antibodies.
GST Fusion Protein Pulldown Assays
Cells containing GST-GPx3 constructs were grown overnight at 37 °C in 10 ml of LB medium supplemented with ampicillin (100 μg/ml), diluted to 100 ml in LB the next morning, and shaken continuously at 37 °C until the cell density at 600 nm reached 0.5. IPTG was then added, and the cells were grown for an additional 4 h. Cells were then pelleted and sonicated at 50% of the maximal amplitude with a 10-s pulse 12 times. Total protein was solubilized in 1% Triton X-100 and separated from insoluble proteins by centrifugation at 15,000 × g for 5 min. For purification of GST and GST fusion proteins, protein extracts were applied to the glutathione-Sepharose 4B column for 1 h at 4 °C (GE Bioscience) and then incubated with precleared cell lysates prepared from prostate cancer cells at 4 °C for 2 h. The column was then centrifuged at 3000 × g at room temperature for 1 min and washed three times with PBS. Proteins were eluted with 40 μl of SDS-PAGE loading buffer, resolved by 7% SDS-PAGE, and subjected to Western blot analyses.
Immunofluorescence Staining
RWPE1 cells (1000 cells) were plated on chamber slides at 20% confluence and grown for 24 h. The slides were then washed twice with PBS, fixed with 4% paraformaldehyde for 1 h at room temperature, then washed two more times with PBS. Slides were blocked with PBS containing 10% donkey serum and 0.4% Triton X-100 for 30 min and then incubated with anti-GPx3 rabbit and anti-PIG3 goat antibodies (Santa Cruz Biotechnology) at 4 °C for 16 h. After washing the slides two times with PBS, the cells were incubated with rhodamine-conjugated donkey anti-rabbit and FITC-conjugated donkey anti-goat antibodies, respectively, for 1 h at room temperature. Slides were also incubated with 4′-6-diamidino-2-phenylindole (DAPI) for 5 min, washed with PBS, and mounted. Immunofluorescence staining was examined using a confocal microscope.
ROS Activity Assays
PDG1 or DDG3 cells (1 × 106) were treated with tetracycline (5 μg/ml) for 2 days and then transfected with siPig3 or siScr using Lipofectamine 2000TM. After 2 days, ROS activity for these cells was quantified using an Oxidelect ROS assay kit (Cell Biolabs, San Diego, CA). Briefly, cells were washed twice with PBS and then incubated with 100 μl of 2′,7′-dichlorfluorescein-diacetate (DCFH-DA) in culture medium at 37 °C for 1 h. Cells were washed twice with DPBS buffer, harvested, and analyzed by flow cytometry. For in vitro assays, the purified fusion proteins GST-GPx3C, GST-GPx3N, or GST-GPx3F in combination with His-PIG3 or controls, respectively, were incubated with PC3 cell lysates at room temperature for 1 h followed by a 1-h incubation with 100 μl of DCFH-DA at 37 °C. The reactions were terminated with the addition of 100 μl of 2× cell lysis buffer. Fluorescence was measured at 480 nm/530 nm and each clone, and conditions were assayed in triplicate. DCFH-DA-treated cells incubated with or without H2O2 were used as controls. For UV-induced cell death analysis, RWPE-1 cells were transfected with PIG3-targeted siRNA (siPig3), siRNA (siGPx3), or a scrambled siRNA control (siScr) using Lipofectamine 2000TM. After 48 h of incubation, the cells were irradiated with 300 mJ of UV light, incubated for an additional 6 h, and then harvested for ROS activity analysis.
Assays of Caspase-3 Activity
DDG3 cells (2 × 106) were treated with or without tetracycline and transfected with siPig3 or siScr. Two days later, the cell lysates were collected and transferred to black 96-well plates. Caspase-3 substrate (DEVD-AFC) (5 μl) and reaction buffer (50 μl) were then added to each well. After a 1-h incubation at 37 °C, caspase-3 activity was quantified in a fluoroplate reader at 400 nm/505 nm. Experiments with caspase-3 inhibitor or without caspase-3 substrate were used as controls.
Data Analysis
Flow cytometry of annexin V and PI staining was analyzed using WinMDI2.9 software. The intensity reading from cells without annexin V and PI was used as a background control. The output data were then imported to an Excel spreadsheet for statistical analysis.
RESULTS
GPx3 Interacts with PIG3
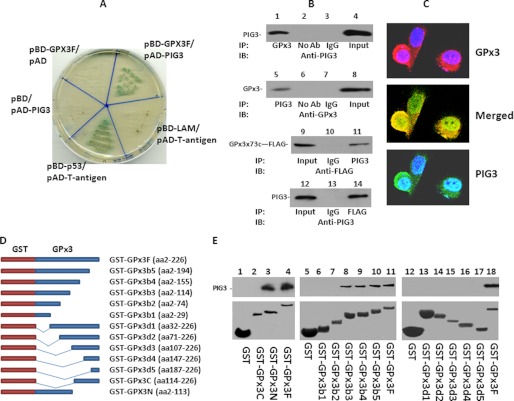
GPx3 was previously shown to exhibit tumor suppressor activity both in vitro and in vivo (12). To investigate signaling molecules that mediate the tumor suppression activity of GPx3, a yeast two-hybrid analysis was performed using a GAL4 bait domain (BD)-GPx3 fusion protein to screen a prostate cDNA library. In this assay, 26 colonies were found to grow on high stringency nutrient agar plates (i.e. SD−Trp−Leu−Ade−His), which were also positive for β-galactosidase activity. The pAD-cDNAs of these colonies were purified and sequenced. The sequence of one of these cDNAs was identified as the PIG3 coding region in-frame with vector pACT2. To validate this interaction, pAD-PIG3 was co-transformed with pBD-GPx3 into yeast AH109 cells. These transformants grew on high stringency nutrient agar plates and formed blue colonies in the presence of X-α-Gal together with the positive control pBD-p53/pAD-T-antigen (Fig. 1A). In contrast, yeast cells transformed with pBD-Lam/pAD-T-antigen did not survive on the high stringency agar plate, neither did pBD-GPx3/pAD nor pBD/pAD-PIG3 (negative control; Fig. 1A).
FIGURE 1.
GPx3 interacts with PIG3 in vivo and in vitro. A, GPx3-bound PIG3 in yeast two-hybrid analyses. GPx3 was ligated in-frame with the DNA binding domain (BD) of GAL4 in the pGBKT7 vector and co-transformed into yeast AH109 cells with pAD-PIG3. Growth and α-galactosidase activity of yeast AH109 cells after co-transformation of the indicated vectors in SD−4 (−Leu/−Trp/−Ade/−His) medium are shown. pBD-p53/pAD-T-antigen denotes the positive control, and pBD-Lam/pAD-T-antigen denotes the negative control. pBD-GPx3/pAD and pBD/pAD-PIG3 denote additional negative controls. B, co-IP of PIG3 and GPx3 from RWPE-1 cells. Protein extracts from RWPE-1 cells were immunoprecipitated with the indicated antibodies, electrophoresed on 7% SDS-PAGE, and immunoblotted (IB) with either anti-PIG3 (lanes 1–4) or anti-GPx3 (lanes 5–8) antibodies. Similar co-immunoprecipitations were also performed in PC3 cells transformed with pCDNA4-GPx3x73c-FLAG/pCDNA6 and induced with tetracycline (lanes 9–14). C, co-localization of GPx3 and PIG3 in RWPE-1 cells. RWPE-1 cells were immunostained with antibodies specific for GPx3 (rabbit) and PIG3 (goat). Immunofluorescence staining was then performed using FITC-conjugated antibodies against goat (PIG3) or rhodamine-conjugated antibodies against rabbit (GPx3). Cells incubated with second antibodies only were used as negative controls. D, schematic diagram of a series of GPx3 deletion constructs with GST. E, in vitro binding analysis of a series of GST-GPx3 fusion proteins with PIG3. GST-GPx3 fusion proteins were purified through a glutathione-Sepharose 4B column. The purified fusion proteins were incubated with His-tagged PIG3, which was purified from E. coli and eluted from a His-tagged column. The GST-GPx3 fusion protein-bound column was then washed vigorously, and the bound protein complex was resolved by 7% SDS-PAGE. Immunoblot analyses were performed with antibodies specific for PIG3. Upper panel, immunoblots of bound His-tagged PIG3. Lower panel, Coomassie staining of GST-GPx3 fusion proteins as indicated.
To investigate the binding of GPx3 and PIG3 in prostate cells, co-immunoprecipitation (co-IP) assays with GPx3 or PIG3 antibodies were performed using cell lysates of RWPE-1, which is an immortalized prostate epithelial cell line. Immunoblot analysis indicated that PIG3 was present in GPx3 co-IP complexes, and GPx3 was detected in PIG3 co-IP complexes (Fig. 1B). Similar results were obtained in co-IP assays of PC3 clones overexpressing GPx3. Interestingly, GPx3 × 73c, a mutant with cysteine replacing selenocysteine was also co-immunoprecipitated with PIG3. Immunofluorescence staining of RWPE-1 cells using anti-GPx3 and anti-PIG3 antibodies also detected the co-localization of these proteins in the cytoplasm (Fig. 1C).
We next constructed N-terminal, C-terminal, and full-length coding regions of GPx3 into pGEX-5T vector to express GST-GPx3 fusion proteins in E. coli. In vitro binding assays were then performed using these truncated GPx3 fusion proteins with His-tagged PIG3 purified from E. coli. The binding assays indicated that PIG3 only bound N-terminal and full-length versions of GPx3 (Fig. 1D). Because only purified recombinant GPx3 and PIG3 were used in these binding assays, the bindings suggest that no bridge proteins are required for the interactions between GPx3 and PIG3. To determine whether a PIG3 binding motif is present in GPx3, a series of deletion mutants of GPx3 was constructed (Fig. 1D). The results of the binding assays indicated that PIG3 was able to bind all GST-GPx3 fusion proteins containing amino acids 2–114, whereas in the absence of this region, no binding was observed (Fig. 1D). These results indicated that this sequence of GPx3 is critical for PIG3 binding.
GPx3 Induces Cell Death in Prostate Cancer Cell Lines
To study GPx3-mediated tumor suppression activity, the role of GPx3 in cell death was investigated. PC3, DU145, and RWPE-1 cells were transfected with pSG5-GPx3 to transiently express GPx3 in these cells, and cell death was quantified 48 h later using annexin V and PI staining. As shown in Fig. 2, transient expression of GPx3 in PC3, DU145, and RWPE-1 cells induced significant cell death involving apoptosis and necrosis (p < 0.001). The expression of GPx3 in PC3 cells resulted in a 2.7-fold increase (p < 0.001) in apoptosis and a 9.1-fold (p < 0.001) increase in necrotic cell death (Fig. 2B), whereas expression of GPx3 in DU145 cells induced a 2.3-fold increase in apoptosis (p < 0.001; Fig. 2C). Forced overexpression of GPx3 in RWPE-1 cells resulted in a 1.8-fold increase of apoptotic cell death (p = 0.005; Fig. 2D) and minimal increase of necrotic death. The patterns of GPx3-mediated cell death in these three cell lines varied, which probably reflected the sensitivities or the variation of cell death signaling among these cells.
FIGURE 2.
GPx3 induces cell death of prostate cancer cell lines. A, immunoblot analyses of GPx3 expression in PC3, DU145, and RWPE-1 cells. PC3, DU145, and RWPE-1 cells were transiently transfected with pSG5-GPx3 or pSG5script control. Forty-eight hours after transfection, the cells were harvested for annexin V binding analysis. B–D, representative FACS analyses and summary of annexin V and PI staining of PC3 cells (B), DU145 (C), and RWPE-1 (D) transfected with pSG5-GPx3 or its control pSG5 as in A. Triplicate experiments were performed for each condition. *, between the groups in necrotic cell death (p < 0.05); **, significant difference between the groups in apoptotic cell death (p < 0.05). Error bars, S.D.
Knockdown of PIG3 or Loss of PIG3-GPx3 Interaction Decreases GPx3-mediated Cell Death
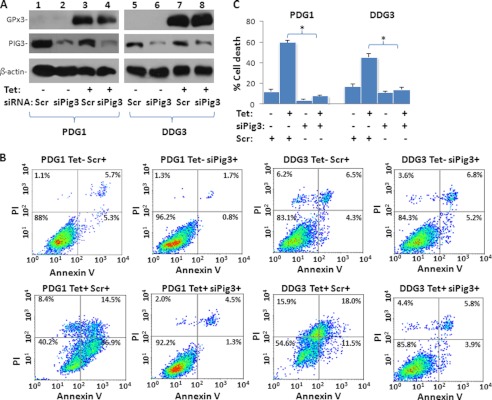
To investigate the functional significance of PIG3 in GPx3-mediated tumor suppression or cell death, PDG1 (pCDNA4-GPx3/pCDNA6 transformed PC3 cells) and DDG3 (pCDNA4-GPx3/pCDNA6 transformed DU145 cells) were induced with 5 μg/ml tetracycline for 4 days, and significant cell death was observed in PDG1 and DDG3 cells (Fig. 3). However, knockdown of PIG3 reversed cell death by >90% (p < 0.001) in PDG1 cells and by 63% (p = 0.003) in DDG3 cells (Fig. 3). The reversal of cell death by PIG3 knockdown occurred at both the apoptotic and necrotic levels, suggesting that PIG3 is broadly involved in the mechanism of GPx3-induced cell death. To evaluate whether the selenocysteine at amino acid residue 73, which is a crucial residue of GPx3 for peroxidase activity, was critical for GPx3-induced cell death, OPAL codon TGA of GPx3 was mutated to the TGC codon that codes for cysteine. This mutant was constructed into the pCDNA4 vector to create pCDNA4-GPx3x73c and co-transfected with pCDNA6 into PC3 and DU145 cells to create clones PGPx3x73c (PC3 cells transfected with pCDNA4-PGPx3x73c) and DGPx3x73c (DU145 cells transfected with pCDNA4-PGPx3x73c). When GPx3x73c was induced with tetracycline, cell death was similarly induced, albeit to a lesser extent than cells with wild-type GPx3. The knockdown of PIG3 in these cells also improved cell survival (Fig. 4), suggesting that selenocysteine is not essential for GPx3-induced cell death or GPx3-PIG3 signaling.
FIGURE 3.
Knockdown of PIG3 decreases GPx3-mediated cell death. A, immunoblots of GPx3 and PIG3 expression in PDG1 and DDG3 cells. PDG1 (PC3 transformed with pCDNA4-GPx3/pCDNA6) and DDG3 (DU145 transformed with pCNDA4-GPx3/pCDNA6) cells were induced with or without tetracycline (Tet) and transfected with siRNA specific for PIG3 (siPig3) or a scramble control (Scr). Cells were harvested for analyses 48 h after transfection. B, representative images of FACS analysis of annexin V and PI staining of PDG1 and DDG3 cells transfected with a scrambled siRNA or PIG3-siRNA induced with (lower panel) or without (upper panel) tetracycline. C, summary of FACS analysis of cell death in B. Triplicate experiments of each condition were averaged in the analyses. *, statistically significant difference in cell death between the PDG1 or DDG3 cell groups treated with Scr and siPIG3 (p < 0.01). Error bars, S.D.
FIGURE 4.
Knockdown of PIG3 decreases GPx3x73c-mediated cell death. A, immunoblots of GPx3x73c and PIG3 expression in PGPx3x73c and DGPx3x73c cells. The PGPx3x73c (PC3 cells transfected with pCDNA4-PGPx3x73c) and DGPx3x73c (DU145 cells transfected with pCDNA4-PGPx3x73c) cells treated with or without tetracycline and transfected with siPIG3 or Scr. Cells were harvested for analyses 48 h after transfection. B, representative images of FACS analysis of annexin V and PI staining of PGPx3x73c and DGPx3x73c transfected with a scrambled siRNA (Scr) or PIG3-siRNA (siPig3) induced with (lower panel) or without (upper panel) tetracycline. C, summary of the FACS analysis of cell death in B. Triplicate experiments of each condition were averaged in the analyses. *, statistically significant difference in cell death between groups treated with Scr and siPIG3 (p < 0.01). Error bars, S.D.
To investigate whether the PIG3-GPx3 interaction was crucial for GPx3-induced cell death, a GPx3 mutant lacking the PIG3 binding capability (amino acids 75–114) was created. This mutant was constructed into pCDNA4 expression vector to create pCDNA4-ΔGPx3 and transfected into PC3 and DU145 cells. Clones of mutant cell lines (PΔG and DΔG from PC3 and DU145 cells, respectively) were created. Induction of PΔG and DΔG with tetracycline did not induce significant cell death compared with wild-type controls (Fig. 5; p = 0.64), indicating that both the expression of GPx3 and the GPx3-PIG3 interaction are essential for GPx3-mediated cell death. Interestingly, when GPx3 was knocked down in RWPE-1 cells, cell death was minimized, which suggested that GPx3 plays a role in normal cell death (Fig. 5, A and B; p = 0.02).
FIGURE 5.
Loss of the PIG3 binding motif in GPx3 abrogates GPx3 cell death induction. A, immunoblots of GPx3 and β-actin expression in PDG1, DDG3, PΔG (PC3 transformed with pCDNA4-GPx3Δ75–114/pCDNA6), DΔG (DU145 transformed with pCDNA4-GPx3Δ75–114/pCDNA6), and RWPE-1 cells. PDG1, DDG3, PΔG, and DΔG cells were treated with or without tetracycline (Tet) for 4 days, and RWPE-1 cells transfected with siRNA specific for GPx3 (siGPx3) or scramble control (Scr) were treated for 3 days. B, representative images of the FACS analysis of annexin V and PI staining of cells listed in A. C, summary of FACS analysis of cell death in B. Triplicate experiments of each treatment were analyzed. *, statistically significant difference in cell death between Scr and siGPx3 groups (p < 0.05); **, statistically significant difference in cell death between the groups treated with or without tetracycline (p < 0.001). Error bars, S.D.
To investigate whether the interference of the GPx3-PIG3 interaction would impact on GPx3-induced cell death, the PIG3 binding motif of GPx3 (amino acids 1–114) was constructed into pCMV-FLAG to create pCMV-ΔGPx31–114-FLAG. This vector was transfected into PDG1 and DDG3 cells. As shown in Fig. 6, this mutant competed for the binding of PIG3 with wild-type GPx3 and abrogated virtually all GPx3-induced cell death activities. These results suggest that cell death induced by wild-type GPx3 is dependent on its interaction with PIG3.
FIGURE 6.
GPx3-PIG3 binding interference abrogates GPx3 cell death induction. A, immunoblots of GPx3, ΔGPx31–114-FLAG, and β-actin expression in PDG1 and DDG3. PDG1 and DDG3 cells were transfected with pCMV-FLAG (pCMV) or pCMV-ΔGPx31–114-FLAG and treated with or without tetracycline (Tet) for 4 days. B, representative images of the FACS analysis of annexin V and PI staining of cells listed in A. C, summary of FACS analysis of cell death in B. Triplicate experiments of each treatment were analyzed. **, statistically significant difference in cell death between the groups treated with or without tetracycline (p < 0.001). Error bars, S.D.
Increased Protease Activity of Caspase-3 by GPx3
Members of the cysteine aspartic acid-specific protease (caspase) family play key roles as effectors of apoptosis in mammalian cells (21). Activated caspases participate in a cascade of cleavage events that disable key homeostatic and repair enzymes and facilitate the systematic structural disassembly of dying cells. Initiation of the caspase cascade leads to activation of caspases-3, -6, and -7. To investigate the impact of GPx3 and PIG3 on caspase activity, the protease activity of caspase-3 was studied. As shown in Fig. 7, induction of GPx3 dramatically increased the protease activity of caspase-3 (3.3-fold, p < 0.001). Addition of a caspase-3-specific inhibitor completely abrogated caspase-3 protease activity, confirming the specificity of the caspase-3 protease assays performed. Knockdown of PIG3 or expression of a GPx3 mutant lacking PIG3 binding activity or of a PIG3 binding competition peptide was able to reverse GPx3-induced caspase-3 activity completely (Fig. 7). Taken together, these results suggest that the GPx3-PIG3 interaction increases the activity of the caspase cascade, which leads cells to disintegration and death.
FIGURE 7.
Increase of protease activity of caspase-3 by GPx3. PDG1 and DDG3 cells were induced with or without tetracycline (Tet) and transfected with siPig3 or Scr or pCMV-FLAG or pCMV-ΔGPx31–114-FLAG. PDG1 and DDG3 and their corresponding GPx3 mutants (PΔG and DΔG) were induced with or without tetracycline to express wild-type or mutant GPx3. Cell lysates of these cells were incubated with caspase-3 substrate (DEVD-AFC) as indicated. The protease activity of caspase-3 was then quantified in a fluororeader. Triplicate experiments of each treatment were analyzed. The reactions without DEVD-AFC, which is the caspase-3 substrate, were used as negative controls. Assay specificity was evaluated using a caspase-3 inhibitor, DEVD-CHO. Quintuplicate assays were performed. Tetracycline-induced PDG1 or DDG3 had a significant increase in caspase-3 activity (p < 0.001). PDG1 or DDG3 cells induced with tetracycline and treated with siPig3 had lower caspase-3 activity compared with cells treated with Scr (p < 0.001). Error bars, S.D.
Generation of ROS by PIG3 Is Enhanced by GPx3
PIG3 expression is associated with an increase in ROS levels in cells (15). To investigate whether GPx3 expression has an impact on the generation of ROS, PDG1 and DDG3 cells were induced to express GPx3. ROS were subsequently quantified using flow cytometry and a fluorometric reader. As shown in Fig. 8, a >100-fold increase (p < 0.001) in ROS levels was detected in GPx3-induced cells compared with uninduced controls. In contrast, knockdown of PIG3 by siRNA completely reversed the ROS generation in both PDG1 and DDG3 cells (Fig. 8, A and E). To evaluate the significance of the selenocysteine of GPx3 in ROS generation, PGPx3x73c and DGPx3x73c cells expressing GPx3x73c were induced, and the ROS generations in these cells were then analyzed. As shown in Fig. 8, B and E, expression of GPx3x73c significantly increased the level of ROS in these cells compared with the uninduced control. When PIG3 was knocked down with siPIG3, similar reversals of ROS generation were found. These results suggest that the selenocysteine residue in GPx3 has a negligible impact on ROS generation, even though this residue is located within the PIG3 binding motif of GPx3. In contrast, when PΔG and DΔG cells were induced to express the GPx3 mutant lacking the PIG3 binding domain, no increase in ROS levels in these cells was found (Fig. 8, C and E). The expression of PIG3 binding competing peptide through pCMV-ΔGPx31–114-FLAG similarly abrogated the ROS generation activity of GPx3 (Fig. 8, D and E). These results suggest that interaction with PIG3, rather than the intrinsic peroxidase activity, is critical for GPx3-mediated generation of ROS. When recombinant PIG3 and GPx3 were expressed in E. coli (Fig. 8F), full-length GPx3 (p < 0.001) construct was able to enhance the generation of ROS by >1000-fold compared with PC3 cell lysate alone, whereas no individual GPx3 fragment generated a similar level of ROS. Based on these observations, GPx3 appears to function as an activator of PIG3 for ROS generation.
FIGURE 8.
ROS generation is enhanced by GPx3-PIG3 interaction. A, representative FACS analyses of ROS generation in PDG1 and DDG3 cells treated with or without tetracycline (T) and transfected with siPIG3 or Scr. B, representative FACS analyses of ROS generation in PGPx3x73c (PC3 cells transfected with pCDNA4-PGPx3x73c) and DGPx3x73c (DU145 cells transfected with pCDNA4-PGPx3x73c) cells treated with or without tetracycline and transfected with siPIG3 or Scr. C, representative FACS analyses of ROS generation in PDG1, DDG3, PΔG, or DΔG cells treated with or without tetracycline. D, representative FACS analyses of ROS generation in PDG1 and DDG3 cells treated with or without tetracycline and transfected with pCMV-FLAG or pCMV-ΔGPx31–114-FLAG. E, summary of ROS generation from samples in A, B, and C. Triplicate experiments of each condition were performed. *, significant difference in ROS generation between tetracycline-treated and untreated, Scr- and siPIG3-transfected cells (p < 0.001). F, recombinant GPx3 enhances ROS generation in vitro. Purified GST-GPx3 or its N- or C-terminal fragments or His-tagged PIG3 was incubated with DCFH-DA and the proteins indicated. ROS was then quantified by measuring dichlorfluorescein fluorescence intensity in a fluorometric plate reader. The ROS from the incubation of PC3 cell lysate with GST and pET28 was treated as a background control. Quintuplicate assays were performed. *, significant difference between groups as indicated (p < 0.01). Error bars, S.D.
Knockdown of GPx3 Inhibits Cell Death in UV-treated Cells
To investigate whether GPx3-PIG3 signaling is involved in stress-induced cell death, such as UV irradiation, GPx3 was knocked down in the normal epithelial cell line RWPE-1 by siRNA specific for GPx3. These cells were then UV-irradiated (300 mJ), and cell death was analyzed 6 h later. As shown in Fig. 9, A and B, UV treatment induced significant cell death in RWPE-1. However, the UV-induced cell death was largely prevented by knocking down PIG3 or GPx3, and a 5-fold decrease in cell death in cells treated with siPIG3 and 4-fold decrease in death in cells treated with siGPx3 was observed. Consistently, the UV-treated cells had >100-fold higher levels of ROS (p < 0.001) (Fig. 9C). In addition, knocking down either PIG3 or GPx3 significantly lowered the level of ROS in the cells treated with UV light (p < 0.001). These analyses suggest that the GPx3-PIG3 signaling pathway critically contributes to UV-induced ROS generation and cell death.
FIGURE 9.
GPx3 or PIG3 is required in UV-induced cell death. A, immunoblots of GPx3, PIG3, and β-actin expression in RWPE1 cells transfected with siPIG3, siGPx3, or scramble siRNA for 48 h and irradiated with 300 mJ of UV light. B, representative images of FACS analyses of annexin V and PI staining of cells listed in A. Right, summary of FACS analysis of cell death in B, left. Triplicate experiments of each treatment were analyzed. *, statistically significant difference in cell death between the cells treated with Scr versus siPIG3 or Scr versus siGPx3 (p < 0.01). C, representative FACS analyses of ROS generation in UV-irradiated RWPE1 cells treated with siPIG3, siGPx3, or Scr transfected as in A. Right, summary of ROS generation of samples in C, left. Triplicate experiments of each condition were performed. *, statistically significant difference in ROS generation between cells treated with Scr versus siPIG3 or Scr versus siGPx3 (p < 0.001). Error bars, S.D.
DISCUSSION
GPx3 is a glutathione peroxidase that oxidizes reduced glutathione to reduce hydrogen peroxide. However, increasing evidence suggests that GPx3 also possesses tumor suppressor activity (5, 9, 22–29). In this study, GPx3 carries out tumor suppressor signaling at least partly through the activation of PIG3, which is a protein associated with the generation of ROS that induces cell death. Several lines of evidence in this study indicate that GPx3 interacts with PIG3. First, GPx3 and PIG3 were co-immunoprecipitated from either immortalized prostate epithelial cells or prostate cancer cell lines that express GPx3. Second, GPx3 and PIG3 have been shown to bind each other in cell-free systems in the absence of other potential bridge proteins. Third, the co-localization of GPx3 and PIG3 has been observed in the cytoplasm of RWPE-1 cells. Yeast two-hybrid analyses also indicated that the binding of GPx3 and PIG3 brings the AD and BD of these proteins together, thereby facilitating transcription of nutrient-synthesizing enzymes to ensure yeast survival and growth in nutritionally restricted medium. The binding of GPx3 and PIG3 appears to be independent of post-translational modifications.
PIG3 is an essential component for the generation of ROS and the induction of apoptosis (15, 18, 30). In the present study, the expression of PIG3 was found to be moderately abundant in both DU145 and PC3 cells as well as in the immortalized cell line RWPE-1. We hypothesize that the expression of GPx3 dramatically increases intracellular levels of ROS due to its interaction with PIG3. Support for this hypothesis is based on the finding that induced expression of GPx3 in the absence of PIG3 did not generate significant levels of ROS. A GPx3 mutant that does not interact with PIG3 also failed to generate significant levels of ROS in these cells. The dependence of GPx3 on PIG3 in generating ROS suggests that GPx3 may be an activator of PIG3 or act as a subunit of the PIG3-ROS enzyme. Alternatively, binding of GPx3 to PIG3 may activate a currently unknown protein to produce ROS. Overall, the mechanistic details of the ROS enzyme complex remain unclear, although it is clear that GPx3 induces PIG3 to mediate cell death.
The GPx3 and GPx families of proteins are known to be antioxidative enzymes of detoxification and can protect cells from oxidative stress (31–33). However, the results of the present study suggest that GPx3 acts as an essential mediator of ROS generation, suggesting that a complex pathway for reduction-oxygenation metabolism exists. GPx3 is known to be a secreted peroxidase and is found abundantly in the plasma. Nevertheless, a significant amount of GPx3 is also detected in the cytoplasm, which thereby raises the possibility that two different forms of GPx3 exist: the extracellular form of GPx3 is devoid of PIG3 or other co-enzymes in plasma and functions as an antioxidant enzyme for the reduction of ROS. In contrast, an intracellular version of GPx3 readily forms a complex with PIG3 or other proteins and activates these enzymes to produce ROS and cell death.
GPx3-mediated cell death occurs in both cancer cells as well as normal epithelium. Our experiments suggest that UV stress generates high levels of ROS and induces cell death. GPx3-PIG3 signaling is critical for UV-induced ROS generation and cell death because the removal of either GPx3 or PIG3 dramatically negates the effects of UV. This was a surprising observation in light of only a mild increase of GPx3 and PIG3 expression in normal epithelial cells (RWPE-1) in response to UV treatment (Fig. 8A), which suggests that GPx3 and PIG3 are constitutive components for ROS generation and cell death. Additional factor(s) induced by UV may activate this cell death machinery and lead to apoptosis. However, increased expression of GPx3 without UV induction also results in dramatic cell death. These results suggest that a critical level of intracellular GPx3 determines the fate of a cell: a high level of GPx3 may promote apoptosis, whereas a low level of the enzyme prevents cell death even under stress. Our finding may also explain the uneven expression level of GPx3 across prostate tissues, where GPx3 is abundant in maturing prostate acinar cells that have a high turnover but only sparse expression in proliferating basal stem cells (9). As a result, the likely function of GPx3 in the prostate gland is to prevent the overaccumulation and hyperproliferation of acinar cells by hastening cell death.
The current study also supports the previous finding that PIG3 alone, although transactivated by TP53, is insufficient to induce cell death (15), possibly due to the lack of a co-enzyme to generate ROS. This study may provide a new link for potential therapeutic intervention against prostate cancer through the molecular activation of PIG3. Expression of GPx3 is selenium-dependent due to the presence of an opal stop codon in the mid segment of the GPx3 open reading frame, and therefore selenium supplements in cancer treatment may enhance GPx3 expression and could hold promise in the suppression of tumor growth. In addition, gene targeting therapy of GPx3 and PIG3 signaling may be warranted due to the highly efficient induction of ROS generation and tumor cell death that has been associated with GPx3 expression.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 CA098249 through the NCI (to J.-H. L.). This work was also supported by American Cancer Society Grant RSG-08-137-01-CNE (to Y. P. Y.).
- GPx3
- glutathione peroxidase 3
- BD
- bait domain
- DCFH-DA
- 2′,7′-dichlorfluorescein-diacetate
- IP
- immunoprecipitation
- PI
- propidium iodide
- PIG3
- p53-induced gene 3
- ROS
- reactive oxygen species
- AD
- activation domain.
REFERENCES
- 1. Takahashi K., Avissar N., Whitin J., Cohen H. (1987) Purification and characterization of human plasma glutathione peroxidase: a selenoglycoprotein distinct from the known cellular enzyme. Arch. Biochem. Biophys. 256, 677–686 [DOI] [PubMed] [Google Scholar]
- 2. Brigelius-Flohé R. (1999) Tissue-specific functions of individual glutathione peroxidases. Free Rad. Biol. Med. 27, 951–965 [DOI] [PubMed] [Google Scholar]
- 3. Brigelius-Flohé R. (2006) Glutathione peroxidases and redox-regulated transcription factors. Biol. Chem. 387, 1329–1335 [DOI] [PubMed] [Google Scholar]
- 4. Brigelius-Flohé R., Flohé L. (2003) Is there a role of glutathione peroxidases in signaling and differentiation? BioFactors 17, 93–102 [DOI] [PubMed] [Google Scholar]
- 5. Brigelius-Flohé R., Kipp A. (2009) Glutathione peroxidases in different stages of carcinogenesis. Biochim. Biophys. Acta 1790, 1555–1568 [DOI] [PubMed] [Google Scholar]
- 6. Brigelius-Flohé R., Wingler K., Müller C. (2002) Estimation of individual types of glutathione peroxidases. Methods Enzymol. 347, 101–112 [DOI] [PubMed] [Google Scholar]
- 7. Müller C., Wingler K., Brigelius-Flohé R. (2003) 3′-UTRs of glutathione peroxidases differentially affect selenium-dependent mRNA stability and selenocysteine incorporation efficiency. Biol. Chem. 384, 11–18 [DOI] [PubMed] [Google Scholar]
- 8. Lodygin D., Epanchintsev A., Menssen A., Diebold J., Hermeking H. (2005) Functional epigenomics identifies genes frequently silenced in prostate cancer. Cancer Res. 65, 4218–4227 [DOI] [PubMed] [Google Scholar]
- 9. Yu Y. P., Yu G., Tseng G., Cieply K., Nelson J., Defrances M., Zarnegar R., Michalopoulos G., Luo J. H. (2007) Glutathione peroxidase 3, deleted or methylated in prostate cancer, suppresses prostate cancer growth and metastasis. Cancer Res. 67, 8043–8050 [DOI] [PubMed] [Google Scholar]
- 10. Falck E., Karlsson S., Carlsson J., Helenius G., Karlsson M., Klinga-Levan K. (2010) Loss of glutathione peroxidase 3 expression is correlated with epigenetic mechanisms in endometrial adenocarcinoma. Cancer Cell Int. 10, 46–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang X., Yang J. J., Kim Y. S., Kim K. Y., Ahn W. S., Yang S. (2010) An 8-gene signature, including methylated and down-regulated glutathione peroxidase 3, of gastric cancer. Int. J. Oncol. 36, 405–414 [PubMed] [Google Scholar]
- 12. Yu G., Tseng G. C., Yu Y. P., Gavel T., Nelson J., Wells A., Michalopoulos G., Kokkinakis D., Luo J. H. (2006) CSR1 suppresses tumor growth and metastasis of prostate cancer. Am. J. Pathol. 168, 597–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chambers I., Frampton J., Goldfarb P., Affara N., McBain W., Harrison P. R. (1986) The structure of the mouse glutathione peroxidase gene: the selenocysteine in the active site is encoded by the “termination” codon, TGA. EMBO J. 5, 1221–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vogt T. M., Ziegler R. G., Graubard B. I., Swanson C. A., Greenberg R. S., Schoenberg J. B., Swanson G. M., Hayes R. B., Mayne S. T. (2003) Serum selenium and risk of prostate cancer in U.S. blacks and whites. Int. J. Cancer 103, 664–670 [DOI] [PubMed] [Google Scholar]
- 15. Polyak K., Xia Y., Zweier J. L., Kinzler K. W., Vogelstein B. (1997) A model for p53-induced apoptosis. Nature 389, 300–305 [DOI] [PubMed] [Google Scholar]
- 16. Lee J. H., Kang Y., Khare V., Jin Z. Y., Kang M. Y., Yoon Y., Hyun J. W., Chung M. H., Cho S. I., Jun J. Y., Chang I. Y., You H. J. (2010) The p53-inducible gene 3 (PIG3) contributes to early cellular response to DNA damage. Oncogene 29, 1431–1450 [DOI] [PubMed] [Google Scholar]
- 17. Venot C., Maratrat M., Dureuil C., Conseiller E., Bracco L., Debussche L. (1998) The requirement for the p53 proline-rich functional domain for mediation of apoptosis is correlated with specific PIG3 gene transactivation and with transcriptional repression. EMBO J. 17, 4668–4679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Flatt P. M., Polyak K., Tang L. J., Scatena C. D., Westfall M. D., Rubinstein L. A., Yu J., Kinzler K. W., Vogelstein B., Hill D. E., Pietenpol J. A. (2000) p53-dependent expression of PIG3 during proliferation, genotoxic stress, and reversible growth arrest. Cancer Lett. 156, 63–72 [DOI] [PubMed] [Google Scholar]
- 19. Nomdedéu J. F., Perea G., Estivill C., Badell I., Lasa A., Aventín A. (2008) Microsatellite instability may involve the pentanucleotide repeat of the PIG3 promoter in bcr/abl acute lymphoblastic leukemia. Leuk. Res. 32, 186–188 [DOI] [PubMed] [Google Scholar]
- 20. Yu Y. P., Luo J. H. (2006) Myopodin-mediated suppression of prostate cancer cell migration involves interaction with zyxin. Cancer Res. 66, 7414–7419 [DOI] [PubMed] [Google Scholar]
- 21. Alnemri E. S. (1997) Mammalian cell death proteases: a family of highly conserved aspartate specific cysteine proteases. J. Cell. Biochem. 64, 33–42 [DOI] [PubMed] [Google Scholar]
- 22. He Y., Wang Y., Li P., Zhu S., Wang J., Zhang S. (2011) Identification of GPX3 epigenetically silenced by CpG methylation in human esophageal squamous cell carcinoma. Dig. Dis. Sci. 56, 681–688 [DOI] [PubMed] [Google Scholar]
- 23. Sreekanthreddy P., Srinivasan H., Kumar D. M., Nijaguna M. B., Sridevi S., Vrinda M., Arivazhagan A., Balasubramaniam A., Hegde A. S., Chandramouli B. A., Santosh V., Rao M. R., Kondaiah P., Somasundaram K. (2010) Identification of potential serum biomarkers of glioblastoma: serum osteopontin levels correlate with poor prognosis. Cancer Epidemiol. Biomarkers Prev. 19, 1409–1422 [DOI] [PubMed] [Google Scholar]
- 24. Blomquist T., Crawford E. L., Mullins D., Yoon Y., Hernandez D. A., Khuder S., Ruppel P. L., Peters E., Oldfield D. J., Austermiller B., Anders J. C., Willey J. C. (2009) Pattern of antioxidant and DNA repair gene expression in normal airway epithelium associated with lung cancer diagnosis. Cancer Res. 69, 8629–8635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Karlsson S., Olsson B., Klinga-Levan K. (2009) Gene expression profiling predicts a three-gene expression signature of endometrial adenocarcinoma in a rat model. Cancer Cell Int. 9, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lin J. C., Kuo W. R., Chiang F. Y., Hsiao P. J., Lee K. W., Wu C. W., Juo S. H. (2009) Glutathione peroxidase 3 gene polymorphisms and risk of differentiated thyroid cancer. Surgery 145, 508–513 [DOI] [PubMed] [Google Scholar]
- 27. Jee C. D., Kim M. A., Jung E. J., Kim J., Kim W. H. (2009) Identification of genes epigenetically silenced by CpG methylation in human gastric carcinoma. Eur. J. Cancer 45, 1282–1293 [DOI] [PubMed] [Google Scholar]
- 28. Saga Y., Ohwada M., Suzuki M., Konno R., Kigawa J., Ueno S., Mano H. (2008) Glutathione peroxidase 3 is a candidate mechanism of anticancer drug resistance of ovarian clear cell adenocarcinoma. Oncol. Rep. 20, 1299–1303 [PubMed] [Google Scholar]
- 29. Peng D. F., Razvi M., Chen H., Washington K., Roessner A., Schneider-Stock R., El-Rifai W. (2009) DNA hypermethylation regulates the expression of members of the Mu-class glutathione S-transferases and glutathione peroxidases in Barrett's adenocarcinoma. Gut 58, 5–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. el-Deiry W. S. (1998) Regulation of p53 downstream genes. Semin. Cancer Biol. 8, 345–357 [DOI] [PubMed] [Google Scholar]
- 31. Brown K. M., Arthur J. R. (2001) Selenium, selenoproteins and human health: a review. Public Health Nutrition 4, 593–599 [DOI] [PubMed] [Google Scholar]
- 32. Giannattasio A., De Rosa M., Smeraglia R., Zarrilli S., Cimmino A., Di Rosario B., Ruggiero R., Colao A., Lombardi G. (2002) Glutathione peroxidase (GPX) activity in seminal plasma of healthy and infertile males. J. Endocrinol. Invest. 25, 983–986 [DOI] [PubMed] [Google Scholar]
- 33. Kho C. W., Lee P. Y., Bae K. H., Cho S., Lee Z. W., Park B. C., Kang S., Lee do H., Park S. G. (2006) Glutathione peroxidase 3 of Saccharomyces cerevisiae regulates the activity of methionine sulfoxide reductase in a redox state-dependent way. Biochem. Biophys. Res. Commun. 348, 25–35 [DOI] [PubMed] [Google Scholar]