Background: Eukaryotic primase initiates replication by forming oligoribonucleotides that are elongated by DNA polymerases.
Results: Archaeal DNA primase initiates chains de novo with dNTPs, and when coupled with an archaeal polymerase and helicase, supports leading and lagging strand synthesis in vitro.
Conclusion: Archaeal primases possess DNA polymerase activity.
Significance: Factors affecting the simultaneous synthesis of leading and lagging strand can be examined in vitro.
Keywords: DNA Enzymes, DNA Polymerase, DNA Primase, DNA Replication, DNA Synthesis, Lagging Strand Synthesis, Primer Formation
Abstract
In most organisms, DNA replication is initiated by DNA primases, which synthesize primers that are elongated by DNA polymerases. In this study, we describe the isolation and biochemical characterization of the DNA primase complex and its subunits from the archaeon Thermococcus kodakaraensis. The T. kodakaraensis DNA primase complex is a heterodimer containing stoichiometric levels of the p41 and p46 subunits. The catalytic activity of the complex resides within the p41 subunit. We show that the complex supports both DNA and RNA synthesis, whereas the p41 subunit alone marginally produces RNA and synthesizes DNA chains that are longer than those formed by the complex. We report that the T. kodakaraensis primase complex preferentially interacts with dNTP rather than ribonucleoside triphosphates and initiates RNA as well as DNA chains de novo. The latter findings indicate that the archaeal primase complex, in contrast to the eukaryote homolog, can initiate DNA chain synthesis in the absence of ribonucleoside triphosphates. DNA primers formed by the archaeal complex can be elongated extensively by the T. kodakaraensis DNA polymerase (Pol) B, whereas DNA primers formed by the p41 catalytic subunit alone were not. Supplementation of reactions containing the p41 subunit with the p46 subunit leads to PolB-catalyzed DNA synthesis. We also established a rolling circle reaction using a primed 200-nucleotide circle as the substrate. In the presence of the T. kodakaraensis minichromosome maintenance (MCM) 3′ → 5′ DNA helicase, PolB, replication factor C, and proliferating cell nuclear antigen, long leading strands (>10 kb) are produced. Supplementation of such reactions with the DNA primase complex supported lagging strand formation as well.
Introduction
DNA polymerases (Pols)2 require a 3′-hydroxyl primed template to elongate DNA chains, and these primers are synthesized by DNA primases. In general, DNA primases initiate replication by synthesizing small RNA primers on both leading and lagging strands that are used by replicative Pols for subsequent elongation events. In Escherichia coli, DNA primase consists of a single subunit, the DnaG protein, that associates with both the replicative helicase (DnaB) and the replicative Pol (Pol III). In eukaryotes, DNA primase is a heterodimer containing a catalytic p48 subunit that associates tightly with a noncatalytic regulatory subunit (p58), which stabilizes and modulates the activity of the p48 subunit (1). This heterodimer is found complexed to the other proteins, the Pol α catalytic (180 kDa) and B (70 kDa) subunits, that collectively form the Pol α-primase complex (2). Typically, in the presence of single-stranded DNA templates, the primase complex synthesizes relatively short oligoribonucleotides 10–15 nucleotides (nt) in length that are elongated by the Pol α complex, generating covalently linked RNA-DNA oligonucleotide chains ∼30–40 nt long (3). These short chains are recognized by the clamp loader RFC, which loads PCNA onto a primer-template junction that tethers the replicative Pol δ and Pol ϵ, which catalyze the synthesis of lagging and leading strands, respectively (4).
Archaea have been shown to contain both the DnaG and the eukaryotic-type DNA primases. The DnaG-like primase was reported to play a role in RNA degradation rather than DNA replication (5, 6), and genetic studies suggest that the genes encoding each of the eukaryotic-like primase subunits are essential, whereas the dnaG gene is dispensable for cell growth (7). The archaeal primase contains a two-subunit structure with significant homology to the eukaryotic p58 and p48 subunits. To date, homologues corresponding to the eukaryotic Pol α (180 kDa) or B subunits have not been detected in archaeal genomes (8). Similar to the eukaryotic primase heterodimer, the archaeal p41 subunit contains DNA primase activity, and the p46 subunit appears to alter its primase activity as well as influence its properties (9). In some archaea, RNA primed intermediates, similar to those observed in eukaryotic Okazaki fragments, have been reported (10–12). However, the properties of DNA primase complexes isolated from different archaea vary widely. Studies with the heterodimeric DNA primase complex isolated from Pyrococcus furiosus revealed that the catalytic p41 subunit alone preferentially utilized dNTPs and formed DNA chains de novo that were extended up to several kilobases (9, 13). However, the P. furiosus two-subunit p41-p46 complex synthesized shorter DNA chains than the catalytic subunit alone and was also capable of synthesizing RNA chains. Furthermore, in vitro replication of M13 DNA by the P. furiosus complex was stimulated by ATP, and [γ-32P]ATP was reported to start DNA chains (9). In part, similar findings have been reported for the primase complex isolated from Pyrococcus abyssi (7). A variety of archaea primases have been shown to be multifunctional enzymes, capable of supporting deoxynucleotide incorporation in a gap-filling reaction, strand displacement synthesis, and terminal transferase activity (7, 14–16).
In this study, we describe the isolation and characterization of the primase complex and its individual subunits from Thermococcus kodakaraensis and show that it initiates oligonucleotide chains de novo from either rNTPs or dNTPs. We show that oligonucleotide chains initiated by the two-subunit primase complex on M13 DNA can be extended to full-length products by T. kodakaraensis PolB. Using a small (200 nt) primed circular DNA, we have devised a rolling circle assay by coupling T. kodakaraensis primase activity with the T. kodakaraensis proteins, PolB, MCM helicase, RFC, and PCNA and demonstrate that this system supports the synthesis of both leading and lagging strands.
EXPERIMENTAL PROCEDURES
Expression Constructs
The genes encoding PolB (TK0001) and the large subunit of primase (PriL, TK1790, p46), the large and small subunits of RFC (RFC-L, TK2219, and RFC-S, TK2218, respectively) were amplified using PCR from the T. kodakaraensis genomic DNA and cloned into pET-21a (Novagen). The gene encoding the small primase subunit (PriS, TK1791, p41) was amplified using PCR from the T. kodakaraensis genomic DNA and cloned into pET-28a (Novagen). All constructs, except for RFC-S, encode proteins containing an in-frame His6 tag at the C terminus. The two inteins found in the gene encoding PolB and the single intein found in the gene encoding RFC-S were removed using the PCR-based approach as described previously (17). A variant form of PriS in which the conserved Asp-97 and Asp-99 were replaced by Ala (D97A/D99A) was generated using QuikChange (Stratagene). This variant is referred to as mutant (m) p41, and the T. kodakaraensis primase complex (p41-p46) containing this variant is referred to as the mutant complex (m p41-p46).
A vector that expressed both subunits of RFC, RFC-L and RFC-S, was generated as follows. A pET-21a vector containing the gene encoding RFC-S was digested with BglII and BamHI, resulting in a fragment that contained the entire coding region and the upstream regulatory sequences (the T7 promoter and the ribosome binding sites). The fragment was cloned into the BglII site on the pET21 construct containing the His-tagged RFC-L. The resulting construct expressed both subunits of RFC, but only the large subunit contained a His6 tag at the C terminus. The cloning of the genes encoding T. kodakaraensis PCNA (TK0535) and MCM (TK1361) proteins was previously described (18, 19).
Expression and Purification of Recombinant Proteins
For protein expression, plasmids encoding the various proteins were transformed into E. coli BL21 DE3 Rosetta cells (Invitrogen). Expression was induced by the addition of 0.5 mm isopropyl-1-thio-β-d-galactopyranoside when the culture reach an A600 of ∼0.6 followed by incubation for 16 h at 16 °C. Cells were collected by centrifugation, resuspended in buffer containing 50 mm Tris-HCl (pH 8.0), 500 mm NaCl, 10 mm imidazole, and 10% glycerol, and incubated at 55 °C for 30 min, followed by sonication. The lysate was clarified by centrifugation and loaded onto a Ni2+ column (GE Healthcare). The column was washed with buffer containing 50 mm Tris-HCl (pH 8.0), 500 mm NaCl, 50 mm imidazole, and 10% glycerol. The His6-tagged proteins were eluted from the column with buffer containing 50 mm Tris-HCl (pH 8.8), 500 mm NaCl, 10% glycerol, 250 mm imidazole. Proteins were stored at −80 °C. The purification of PCNA and MCM proteins was previously described (18, 19).
The PriS-PriL (p41-p46) complex was made as follows. Thirty mg of purified PriS (or PriS D97A/D99A) were incubated with 36 mg of purified PriL (resulting in 1:1 molar ratio) at 25 °C for 1 h. Following incubation, the mixture was dialyzed against buffer containing 50 mm Tris-HCl (pH 8.0), 500 mm NaCl, 2 mm DTT, and 10% glycerol. An aliquot of the complex (200 μg in 200 μl) was subjected to Superdex 200 gel filtration analysis. More than 90% of the proteins were isolated as the stoichiometric p41-p46 complex.
Construction of 200-nt Primed Circle
A circular 200-nt ssDNA (containing only C, A, and G) was prepared as follows. Two reactions (30 μl) each containing 1 nmol of the 100-nt oligonucleotides A or B (sequences in supplemental Table 1) were incubated with T4 polynucleotide kinase (40 units, New England Biolabs) and [γ-32P]ATP (1 mm, 350 cpm/pmol) for 1 h at 37 °C. The phosphorylated oligonucleotides A and B were combined and mixed with 5 nmol of the oligonucleotide bridges AB and BA in 20 mm HEPES (pH 7.5) and 150 mm NaCl in a total volume of 100 μl, and the mixture was heated to 100 °C for 10 min and then cooled slowly to 22 °C. Ligation of the oligonucleotides was carried out in reactions (525 μl) containing T4 DNA ligase (8000 units), T4 DNA ligase buffer (New England Biolabs), and 1 mm ATP at 16 °C for 14 h. The ligation reaction was monitored with ExoI and ExoIII (New England Biolabs). Following complete ligation, the products were subjected to 8 m urea-10% PAGE separation in 1× Tris-borate-EDTA at 15 watts for 75 min; the region of the gel containing the circular 200-nt ssDNA was excised, and the DNA was eluted with 0.5 m ammonium acetate, 10 mm magnesium acetate, 1 mm EDTA. After ethanol precipitation, the circular 200-nt ssDNA was resuspended in 30 μl of 1× Tris-EDTA, pH 8.0, and the solution was passed through an S300 mini column filter (GE Healthcare). The procedure yielded ∼20% of the input oligonucleotide as the circular 200-nt ssDNA.
A linear 140-nt oligonucleotide (5′ T40-linked to an 100-mer containing only G, T, and C), complementary to a portion of the 200-nt circle, was prepared as follows. A reaction (20 μl) containing the linear oligonucleotide (140-nt) was phosphorylated with T4 polynucleotide kinase and [γ-32P]ATP, as described above. The labeled oligonucleotide II (1 nmol) was annealed to oligonucleotide I (0.9 nmol) and 5 nmol of the oligonucleotide bridge I-II. The ligation reaction (300 μl) was carried out by incubating T4 DNA ligase (6000 units) with 1 mm ATP in T4 DNA ligase buffer (New England Biolabs) at 16 °C for 14 h. The ligation product was isolated as described above with a yield of 40%, based on the amount of oligonucleotide I added. The primed rolling circle substrate was prepared by annealing the 200-nt circle and the linear 140-nt (as above) at a molar ratio of 1:1 followed by slow cooling to room temperature. The sequences of the oligonucleotides used in the preparation of the primed 200-mer circle are shown in supplemental Table 1.
DNA Primase Assay
Unless indicated, reaction mixtures (20 μl) containing 40 mm Tris-HCl (pH 8.0), 2 mm DTT, 10 mm magnesium acetate, 4 mm MnCl2, 100 μg/ml BSA, dNTPs and rNTPs (as indicated), single-stranded (ss) M13 DNA, and T. kodakaraensis DNA primase fractions (diluted with a solution containing 50% glycerol, 0.02 m Tris-HCl, pH 8.0, 1 mm DTT, 10 μm EDTA, and 50 μg/ml BSA) were incubated at 60 °C for 20 min. Aliquots were treated with an equal volume of a solution containing 95% formamide, 0.1× Tris-borate-EDTA, 10 mm EDTA and heated at 95 °C for 5 min and then subjected to electrophoretic separation through a 20% (w/v) polyacrylamide-8 m urea gel run in 1× Tris-borate-EDTA for 2.5 h at 300 V. Gels were visualized by autoradiography or phosphorimaging.
Replication Assays
Reaction conditions and enzymes used were as described in the figure legends.
RESULTS
Coupling of T. kodakaraensis DNA Primase Activity with Klenow Polymerase
The purified T. kodakaraensis DNA primase complex and the catalytic subunit were assayed at various temperatures in reactions containing ssM13 DNA as the template. Although isolated from a thermophile, the enzyme was substantially active at 37 °C (Fig. 1A). For this reason, the T. kodakaraensis primase activity was coupled with the Klenow Pol, an assay initially used to measure DNA primase activity (20). In this assay, which is carried out in the presence of poly(dT) as the template, short oligonucleotides synthesized de novo by the primase are elongated by the Klenow Pol, which in the absence of a primer is inactive. As shown in Table 1, the DNA primase complex supported extensive dATP incorporation that was increased by higher levels of the primase complex (Wild-type complex (p41-p46) rows). In contrast, the primase complex containing the m p41 catalytic subunit was devoid of activity (Mutant complex (m p41-p46) rows), as was the p46 subunit alone (p46 row). The p41 subunit alone was poorly active (WT p41 rows), but this activity was increased by the addition of the p46 subunit (WT p41 + p46 (50 fmol) rows). Reactions with m p41 subunit alone (Mutant p41 row), as well as those supplemented with the p46 subunit (Mutant p41 + p46 (50 fmol) row), were inactive. Thus, under the conditions used, primer formation and its ability to support extensive poly(dA) synthesis with the Klenow Pol required an active catalytic p41 subunit as well as the p46 subunit. As these reactions were carried out in the presence of [α-32P]dATP, the only added nucleotide, de novo initiation of polynucleotide chains appeared to occur solely with dATP. In contrast to the archaeal primase, the eukaryotic DNA primase complex does not initiate DNA chains with dNTPs (1). Hence, the synthesis of poly(dA) was not observed when the purified human DNA primase complex was used in place of the T. kodakaraensis primase complex in reactions containing only dATP. The addition of ATP to such reactions, however, resulted in DNA replication (Human primase complex +ATP/−ATP rows). Collectively, these findings indicate that T. kodakaraensis primase complex initiates DNA primer synthesis with dATP in the absence of ATP.
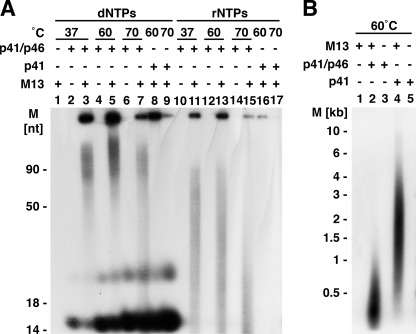
FIGURE 1.
A, influence of temperature on T. kodakaraensis primase-catalyzed synthesis of oligonucleotide. Reactions, described under “Experimental Procedures,” included 100 μm each of α-32P-labeled dATP (2200 cpm/pmol), dGTP, dCTP, and dTTP or 100 μm each of α-32P-labeled ATP (2500 cpm/pmol), GTP, CTP, and UTP, 83.4 fmol of ssM13 DNA, and 5.6 pmol of the T. kodakaraensis DNA primase complex (p41-p46) or 5.6 pmol of the T. kodakaraensis p41 subunit, as indicated. After incubation for 20 min at the temperature indicated, aliquots were used to measure the size of products by urea-PAGE analysis. M, molecular mass markers. B, size of DNA products formed by the T. kodakaraensis p41-p46 complex and T. kodakaraensis p41 subunit. Reactions were as described for panel A. Mixtures lacking primase fractions (lane 1) or ssM13 DNA (lanes 3 and 5) and those containing both M13 DNA and T. kodakaraensis primase preparations (lanes 2 and 4) were analyzed by alkaline-agarose gel electrophoresis. Approximately three times more radioactivity from reactions containing the p41 subunit than the p41-p46 complex was applied to the gel.
TABLE 1.
Coupling of T. kodakaraensis DNA Primase activity with Klenow Pol
Reaction mixtures (20 ml), containing 20 mm, Tris-HCl (pH 8.0), 2.5 mm DTT, 3 mm magnesium acetate, 100 mg/ml BSA, 50 mm [α-32P]dATP (2070 cpm/pmol), 0.4 units of Klenow, 4.54 pmol of poly(dT)221, and 50 mm NaCl were incubated for 30 min at 37 °C. Reactions with the human primase complex (p58 + p48 subunits) also included 2.5 mm ATP, where indicated. Aliquots of each reaction were plated on DEAE cellulose paper and washed three times with 0.5 m ammonium formate + 1 mm sodium pyrophosphate, dried and counted.
| Additions | Protein added | dAMP incorporated |
|---|---|---|
| fmol | pmol | |
| Wild-type complex (p41-p46) | 40 | 684 |
| 4 | 72.8 | |
| 0.4 | 10.4 | |
| Mutant complex (m p41-p46) | 40 | <1 |
| 4 | <1 | |
| WT p41 | 40 | 31 |
| 4 | 6.4 | |
| p46 | 50 | <1 |
| WT p41 + p46 (50 fmol) | 40 | 692 |
| 4 | 74.8 | |
| 0.4 | 7.6 | |
| Mutant p41 | 40 | <1 |
| Mutant p41 + p46 (50 fmol) | 40 | <1 |
| Human primase complex +ATP | 70 | 716 |
| Human primase complex −ATP | 70 | <1 |
T. kodakaraensis Primase-catalyzed Incorporation of Ribonucleotides and Deoxynucleotides
The T. kodakaraensis primase complex synthesized oligonucleotide chains with either dNTPs or rNTPs only in the presence of ssM13 DNA (Fig. 1A). In the presence of M13 and dNTPs, DNA chains were produced more extensively at 60 °C (lane 5) than 37 °C (lane 3) or 70 °C (lane 7). Under the conditions used, labeled DNA chains formed, measured by urea-PAGE analysis, varied from ∼50 nt to >90 nt in length. The amount of labeled RNA chains synthesized in the presence of rNTPs at 70 °C was lower than that formed at 37 or 60 °C, and their size varied from ∼10 to ∼75 nt. Oligonucleotide synthesis with rNTPs or dNTPs was substantially higher with the primase complex than with the catalytic subunit alone (compare lanes 8 and 9 with lanes 5 and 7; compare lane 16 with lane 13). Two products, migrating in urea-PAGE as ∼16 and ∼25 nt oligonucleotides, were observed with dNTPs (lanes 2–9) but not with rNTPs. Their synthesis did not require M13 DNA, and they were formed with both the catalytic subunit alone and the primase complex. As discussed in more detail in the accompanying article (21), these products were identified as dAMP covalently linked to glycerol and to Tris and are not oligonucleotides. The findings summarized in Fig. 1A indicate that the T. kodakaraensis primase complex utilizes dNTPs as well as rNTPs to form polynucleotide chains.
The size distribution of the labeled DNA products formed in M13 reactions was analyzed by alkaline agarose gel electrophoresis (Fig. 1B). Products formed with the primase complex were shorter in length than those produced with the T. kodakaraensis p41 subunit alone (∼0.5 and ∼2 kb, respectively). Thus, the data presented in Fig. 1 indicate that in the absence of the p46 subunit, the p41 subunit forms longer DNA products and little RNA, whereas the complex forms shorter DNA products and supports more extensive RNA synthesis. These findings were not affected by variations in the level of added DNA. These are similar to those reported previously for the P. furiosus (9) and P. abyssi primase complexes (7). Studies with the P. abyssi complex revealed that Mn2+ increased the level of RNA synthesis and decreased the length of RNA chains. RNA synthesis with the T. kodakaraensis primase complex was stimulated about 2-fold by the presence of Mn2+, whereas the size of RNA chains was marginally affected. In contrast, DNA synthesis was slightly inhibited by Mn2+ (data not presented).
Analysis of DNA and RNA Products Formed by T. kodakaraensis Primase Complex
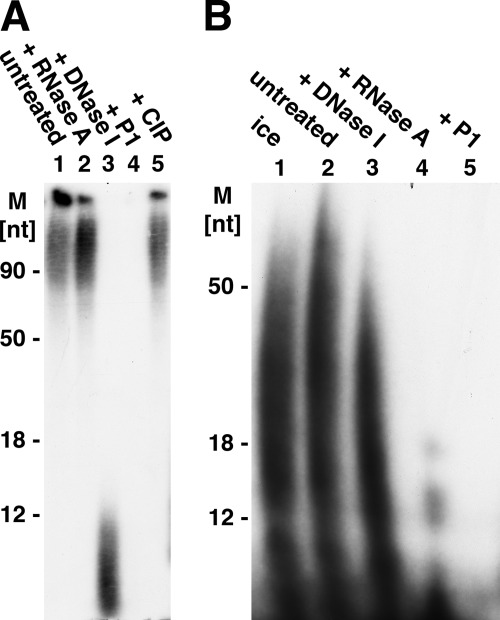
The labeled products formed by the primase complex in reactions containing M13 DNA and the four dNTPs or the four rNTPs were analyzed. Following incubation, reaction mixtures were digested with proteinase K, extracted with phenol and CHCl3, and precipitated with isopropyl alcohol. Urea-PAGE analysis of the products before and after isopropyl alcohol precipitation revealed that the prominent ∼16- and ∼25-nt bands formed with dNTPs were not recovered following isopropyl alcohol precipitation (data not presented). The isolated DNA products were subjected to digestion with pancreatic DNase I, RNase A, nuclease P1, and calf intestinal phosphatase (CIP) (Fig. 2A). Urea-PAGE analysis revealed that the DNA products were degraded by DNase I and nuclease P1 but unaffected by RNase A and CIP treatment. Digestion of the isolated RNA products revealed that they were degraded by RNase A and nuclease P1 and resistant to DNase I (Fig. 2B). These findings indicate that dNTPs and rNTPs supported the synthesis of DNA and RNA chains, respectively.
FIGURE 2.
Analysis of DNA and RNA products formed by T. kodakaraensis primase complex. Reactions (100 μl), as described in the legend for Fig. 1, containing 100 μm each of GTP, CTP, UTP, and [α-32P]ATP or 100 μm each of dGTP, dCTP, dTTP, and [α-32P]dATP, 417 fmol of ssM13, and 56 pmol of T. kodakaraensis primase complex were incubated for 20 min at 60 °C. Approximately 700 pmol of [α-32P]dATP and 500 pmol of [α-32P]ATP were incorporated. Mixtures were adjusted to 0.02 m EDTA, 1% SDS, and following the addition of 10 μg of proteinase K, they were incubated for 30 min at 37 °C and then extracted consecutively with 100 μl of phenol-CHCI3-isoamyl alcohol mixture (25:24:1, v/v) and 100 μl of CHCl3-isoamyl alcohol (24:1 v/v). Aqueous phases were adjusted to 1 m ammonium acetate and treated with 2 volumes of isopropyl alcohol. After 30 min on dry ice, the mixtures were centrifuged for 30 min at 20,000 rpm in the Eppendorf centrifuge at 4 °C, and pelleted material was washed with 50% isopropyl alcohol, dried in vacuo, and suspended in 25 μl of Tris-EDTA buffer containing 50 mm NaCl. Reactions (10 μl) containing 2 μl of the RNA or DNA products, isolated as described above, were incubated with pancreatic DNase (0.1 μg) or pancreatic RNase (1 μg) in mixtures containing 40 mm Tris-HCl (pH 7.5) and 1 mm magnesium acetate; reactions with nuclease P1 (0.3 unit) contained 40 mm sodium acetate buffer (pH 5.2); reaction with CIP (1 unit) contained 40 mm Tris-HCl (pH 8.8) and 1 mm magnesium acetate. All reactions were incubated for 30 min at 37 °C. Controls included incubation of products at 37 °C for 30 min at the indicated pH in the absence of enzymes. Aliquots of reactions were subjected to urea-PAGE separation and analysis as described in the legend for Fig. 1. A, digestion of DNA product. M, molecular mass markers. B, digestion of RNA products. The isolated RNA products were incubated on ice (lane 1) or at 37 °C in the absence of enzymes (lane 2). Incubation with the indicated enzymes were carried out as described above. Treatment of RNA products with CIP was omitted.
Template Specificity of T. kodakaraensis Primase Complex
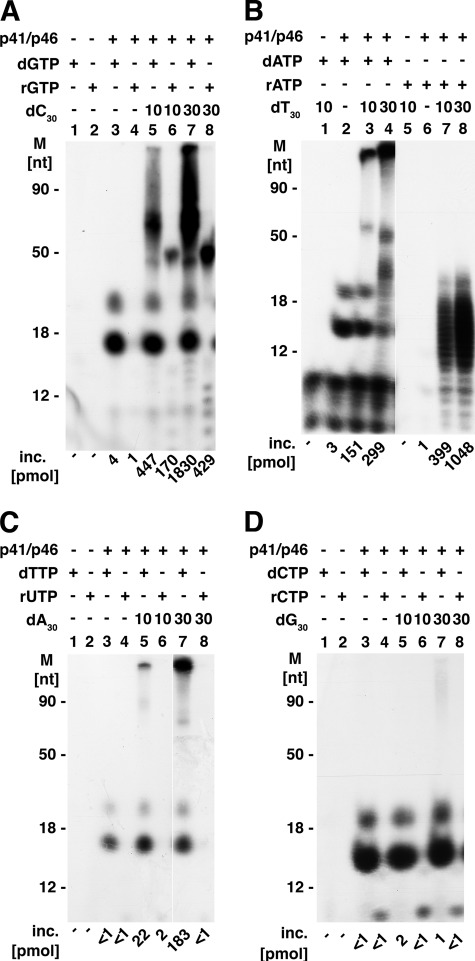
We examined the template activity of the four homo-oligodeoxynucleotides, 30 nt in length, using the T. kodakaraensis primase complex. As shown in Fig. 3, oligo(dC)30 was the most effective template (compare panel A with panels B–D), supporting extensive DNA synthesis with dGTP (lanes 5 and 7). Under the conditions used, dGTP incorporation exceeded the level of oligo(dC)30 template added, and the lengths of DNA chains, determined by urea-PAGE analyses, were >100 nt. Extensive RNA synthesis of a more homogeneous size (∼50 nt) than DNA was observed (Fig. 3A, lanes 6 and 8). (It should be noted that the autoradiogram presented in Fig. 3A was exposed for 25 min at −80 °C, whereas all other autoradiograms shown in Fig. 3 were exposed for 1 h at −80 °C.) Like oligo(dC)30, oligo(dT)30 supported extensive DNA and RNA synthesis (Fig. 3B). However, oligo(dT)30 supported the synthesis of shorter RNA chains than those formed in the presence of oligo(dC)30 (Fig. 3B, lanes 7 and 8) as well as the production of higher levels of RNA than DNA (Fig. 3B, compare lanes 7 and 8 with lanes 3 and 4). In the presence of oligo(dA)30 (Fig. 3C) or oligo(dG)30 (Fig. 3D), DNA synthesis was detected with the former but barely discernable with the latter, whereas RNA synthesis with these purine oligonucleotide templates was not observed. Thus, like eukaryotic DNA primases, the T. kodakaraensis primase complex prefers pyrimidine templates (1). DNA products formed in reactions containing oligo(dC)30 or oligo(dT)30 and the p41 subunit in lieu of the complex were similar to those shown in Fig. 3, A and B, respectively. The level of incorporation with the catalytic subunit alone was 20–30% lower than that detected with the complex.
FIGURE 3.
A–D, template specificity of T. kodakaraensis primase complex. Reaction mixtures were as described under “Experimental Procedures” and included 100 μm of indicated [α-32P]dNTP or [α-32P]rNTP, 10 or 30 pmol of oligo(dC)30 (A); oligo(dT)30 (B); oligo(A)30 (C); or oligo(dG)30 (D), and 5.6 pmol of the T. kodakaraensis primase complex. Following incubation for 20 min at 60 °C, aliquots were subjected to urea-PAGE separation. The specific activities (cpm/pmol) of the α-32P-labeled nucleotides used were: dTTP, 2580; UTP, 2020; dGTP, 2280; GTP, 1640; dCTP, 2285; CTP, 1900; dATP, 1840; ATP, 1710. M, molecular mass markers; inc., incorporation.
When M13 DNA was used in lieu of the short homopolymers (in reactions containing all four rNTPs or the four dNTPs), all labeled rNTPs and dNTPs supported RNA and DNA synthesis, respectively (data not presented). As shown in Fig. 3, DNA products formed with some of the 30-nt oligonucleotide templates were longer in length than the template added. This discrepancy could arise by mechanisms that minimally include slippage, displacement synthesis, and primer-template reannealing followed by chain extension or terminal addition of nucleotides to template ends. The latter mechanism was examined with oligo(dA)30. Incubation of the primase complex with [α-32P]dideoxy TTP and oligo(dA)30 did not result in the production of labeled DNA chains. Supplementation of reactions with terminal deoxynucleotidal transferase led to the addition of the labeled dideoxy TTP (data not presented). These findings suggest that the T. kodakaraensis primase complex did not support terminal addition under the conditions used. All products formed (with M13 or homopolymers) were hydrolyzed by nuclease P1 (>90%), a single strand-specific nuclease (data not presented), suggesting that extensive duplex DNA or duplex DNA-RNA hybrid structures were not formed under the conditions used. Because T. kodakaraensis primase reactions were carried out at 60 °C, we speculate that melting and partial reannealing reactions contributed to the synthesis of DNA chains longer than the DNA template added.
Examination of 5′ Ends of RNA and DNA Chains Formed by T. kodakaraensis Primase Complex
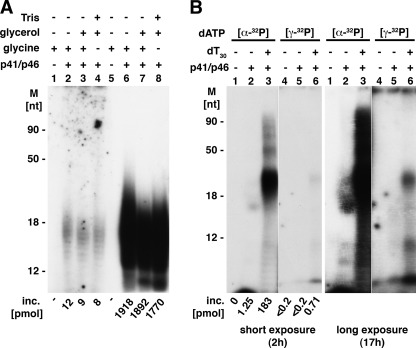
RNA chains synthesized by prokaryotic and eukaryotic primases are initiated with rNTPs with retention of the β- and γ-phosphate residues (1). We examined whether the T. kodakaraensis primase did the same. To simplify these studies, oligo(dT)30 was used as the template. Under the conditions used, the T. kodakaraensis primase complex and catalytic subunit supported extensive poly(dA) synthesis, whereas only the complex supported extensive poly(rA) synthesis (supplemental Table 2). For this reason, products formed by the T. kodakaraensis p41 catalytic subunit were not examined. As shown in Fig. 4A, reactions containing the T. kodakaraensis primase complex, oligo(dT)30, and [α-32P]ATP yielded small poly(rA) chains that averaged ∼15–20 nt in length. Based on the incorporated AMP and size of the products (lanes 6–8), we calculated that approximately ∼100 pmol of oligo(rA) chains were produced. Direct measurement of [γ-32P]ATP incorporation revealed the incorporation of ∼10 pmol, representing ∼10% of the 5′ termini calculated based on the number of oligo(rA) chains formed. These experiments were carried out in reactions containing glycine buffer (pH 8.8) and the T. kodakaraensis primase complex diluted in Tris-EDTA (no glycerol) (Fig. 4A, lanes 2 and 6), containing glycine buffer (pH 8.8) and primase diluted in glycerol (lanes 3 and 7) or Tris buffer (pH 8.0) and primase diluted in glycerol (lanes 4 and 8). These different conditions were used to determine whether the production of the glycerol and Tris-AMP products (shown in the accompanying article (21)) affect [γ-32P]ATP incorporation. As shown in Fig. 4A, they did not alter the reaction significantly. We verified that the incorporated [γ-32P]ATP was terminally located because all of the 32P (lanes 2–4) was rendered acid-soluble following treatment with CIP, whereas the 32P present in the oligo(rA) product formed from [α-32P]ATP was unaffected by CIP treatment (supplemental Table 3). Efforts to increase the level of [γ-32P]ATP incorporation relative to the amount of [α-32P]ATP incorporated were unsuccessful (including variation in the time of incubation, level of enzyme, template added, etc.). The incorporation of [γ-32P]ATP in reactions containing Mg2+ but lacking Mn2+ was reduced about 3-fold when compared with those containing both metals. The amount and size of RNA chain formed were also reduced (resulting in the formation of chains containing ∼12% of the calculated amount of [γ-32P]ATP expected to be incorporated, data not presented).
FIGURE 4.
Status of 5′ termini of RNA and DNA products formed by T. kodakaraensis DNA primase complex. A, 5′ terminus of oligo(rA) products. Reaction mixtures (20 μl) containing 40 mm glycine buffer (pH 8.8), 10 mm magnesium acetate, 4 mm MnCl2, 50 μg/ml BSA, 2 mm DTT, 100 μm [γ-32P]ATP (6150 cpm/pmol) or [α-32P]ATP (730 cpm/pmol), as indicated, 20 pmol of oligo(dT)30, and 0.56 μm T. kodakaraensis primase complex were incubated for 20 min at 60 °C. Aliquots (4 μl each) were used to measure nucleotide incorporation (inc.) and subjected to urea-SDS-PAGE analysis (4 μl for lanes 1–4 and 1 μl for lanes 5–8). Primase was omitted in lanes 1 and 5; glycerol (0.4 m final) was added to lanes 3, 4, 7, and 8; and glycine buffer (pH 8.8) was replaced by Tris buffer (pH 8.0) in lanes 4 and 8. M, molecular mass markers. B, 5′ terminus of oligo(dA) products. Reaction mixtures (10 μl), as described for panel A, contained 26 μm [γ-32P]dATP (9280 cpm/pmol) or [α-32P]dATP (5200 cpm/pmol), 10 pmol of oligo(dT)30 (lanes 3 and 6), and 0.56 μm T. kodakaraensis DNA primase complex as indicated. Lanes 2 and 4 contained enzymes but no DNA template. Following 20 min at 60 °C, aliquots were used to measure nucleotide incorporation and subjected to urea-PAGE analysis. The gel was exposed for 2 and 17 h at −80 °C.
The low level of the [γ-32P]ATP-terminated oligo(rA) chains observed could be due to the presence of a nuclease or activities that hydrolyzed the triphosphate ends following their formation in the primase reaction. We examined these notions with [γ-32P]ATP-labeled RNA products formed by the E. coli RNA polymerase (22). Incubation of these RNA preparations with the T. kodakaraensis primase complex, however, did not reduce the level of 32P present in the RNA (data not presented). We also showed that the oligo(rA) chains formed by the DNA primase complex contained 5′-phosphate termini. Alkaline hydrolysis of oligo(rA) chains formed with [α-32P]ATP in reactions as described in Fig. 4A (lanes 6–8) yielded 32P-labeled Ap (2′, 3′ mixture) and (5′, 2′(3′)-adenosine diphosphate following separation of the products by TLC on PEI plates (supplemental Table 4). Based on the level of Ap (2′, 3′ mixture) and 5′, 2′(3′)-adenosine diphosphate detected, we calculated that ∼50% of the oligo(rA) chains formed contained a 5′-phosphate terminus.
The status of the 5′ ends of oligo(dA) chains synthesized in the primase reaction was also investigated (Fig. 4B). Reaction mixtures containing 26 μm [α-32P]dATP, oligo(dT)30, and the T. kodakaraensis primase complex yielded oligo(dA) chains ∼30–40 nt long (Fig. 4B, lane 3, short exposure). Based on the level of 32P-dAMP incorporated and the size of the products, we calculated that ∼4.5–6 pmol of oligo(dA) chains were produced under these conditions. Low levels of [γ-32P]dATP were incorporated into products weakly visible after the gel was autoradiographed for 2 h (Fig. 4B, lane 6). The size of these products was similar to that of the major oligo(dA) product formed with [α-32P]dATP (compare lanes 3 and 6). The γ-32P-labeled product was more visible following longer exposure (17 h). Similar to observations made with oligo(rA), the 32P present in the [γ-32P]dATP-labeled oligo(dA) was hydrolyzed by CIP, whereas the oligo(dA) with [α-32P]dATP was unaffected by this treatment (data not presented).
The oligo(dA) products formed by the T. kodakaraensis primase complex also contained 5′-phosphate-terminated chains. This was established following degradation of [α-32P]dATP-labeled oligo(dA) chains with micrococcal nuclease and spleen phosphodiesterase. Their combined action leads to the formation of 5′, 3′-deoxyadenosine diphosphate (3′) (from the 5′-phosphate terminus) and Ap (3′) from the internucleotidic region (23). These products were separated by TLC on PEI plate and the distribution of the 32P examined (supplemental Fig. 1). Based on the level of 5′, 3′-deoxyadenosine diphosphate (3′) and dAp (3′) detected, ∼50% of the oligo(dA) chains (30–40 nt in length) contained a 5′-phosphate terminus. Incubation of the micrococcal nuclease/spleen phosphodiesterase digest with CIP converted the 5′, 3′-deoxyadenosine diphosphate and 3′-deoxyadenosine phosphate products to Pi (supplemental Fig. 1). Thus, the 5′ termini of both oligo(rA) and oligo(dA) chains formed by the T. kodakaraensis DNA primase complex contained triphosphate (∼10%) and 5′-phosphate (∼50%) termini. At present, we have no explanation for the low level of ends detected.
The P. furiosus primase complex was reported to synthesize labeled polymers following its incubation with [γ-32P]ATP and unlabeled dNTPs, indicating that chains were initiated with [γ-32P]ATP and extended by unlabeled dNTPs (9). Similar experiments with the T. kodakaraensis primase complex, however, did not lead to the incorporation of [γ-32P]ATP into products or formation of labeled polynucleotide chains (data not presented).
Collectively, these findings suggest that the de novo synthesis of RNA (and DNA) by the T. kodakaraensis primase complex occurs with retention of ∼10% of the γ-phosphate residues of ATP and dATP. We cannot rule out the possibility that chains initiated with triphosphate ends are hydrolyzed during their subsequent elongation phase. As shown in the accompanying article (21), [γ-32P]dATP and [γ-32P]rATP are hydrolyzed by the p41-p46 complex (and p41 subunit alone), generating dAMP and rAMP and a mixture of PPi and Pi. This hydrolytic activity required a catalytically active p41 subunit and was not observed with the m p41 alone or the m p41-p46 complex. A possible mechanism by which DNA and RNA chains containing 5′-phosphate termini are formed is presented in more detail under “Discussion.”
Effects of dATP and ATP on RNA and DNA Synthesis
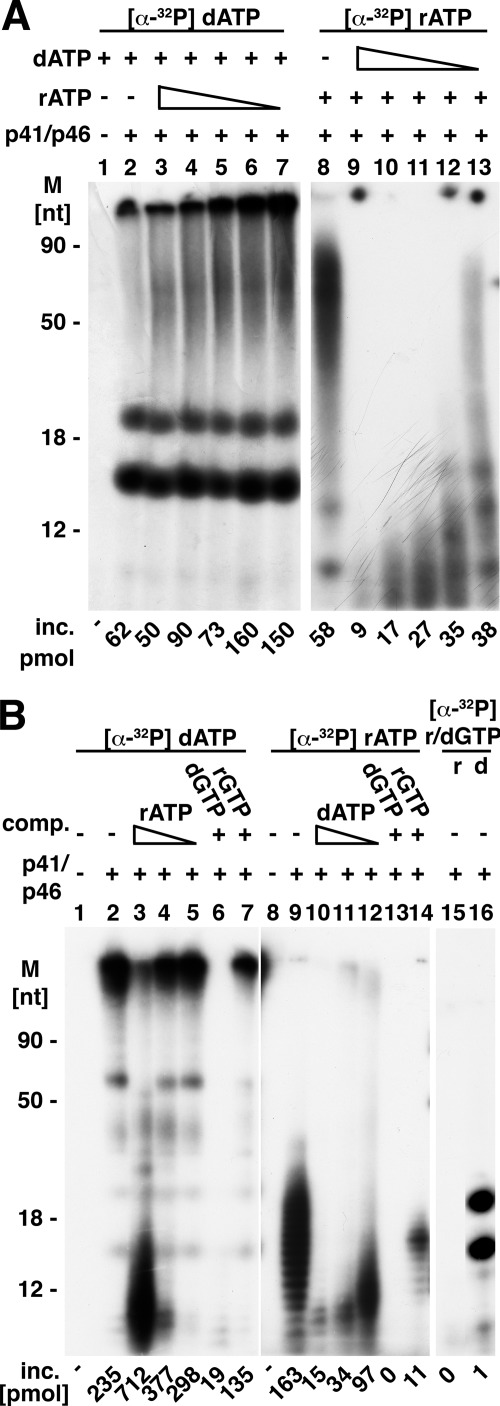
The synthesis of RNA catalyzed by the T. kodakaraensis primase complex was inhibited by dNTPs, whereas formation of DNA was hardly affected by rNTPs (Fig. 5A). Oligodeoxynucleotide synthesis was carried out in reactions containing ssM13 DNA and varying levels of unlabeled ATP in the presence of a fixed amount of α-32P-labeled dATP, dCTP, dGTP, and dTTP. At ATP:dATP molar ratios ranging from 10 to 0.1 (lanes 3–7), DNA synthesis was stimulated by ATP up to an ATP:dATP molar ratio of 3 (lane 4) and slightly inhibited at a higher molar ratio (10:1, lane 3). In contrast, incorporation of [α-32P]ATP into oligoribonucleotides was inhibited by dATP even at an ATP:dATP molar ratio of 0.1 (lane 14). These findings indicate that the T. kodakaraensis primase complex binds preferentially to dNTPs.
FIGURE 5.
A, effects of dATP and ATP on RNA and DNA synthesis. Mixtures (20 μl), as indicated under “Experimental Procedures,” contained 100 μm of each of the four dNTPs or rNTPs with [α-32P]dATP (1344 cpm/pmol) or [α-32P]ATP (1357 cpm/pmol), 83.4 fmol of M13 DNA, 5.6 pmol of the T. kodakaraensis primase complex, and varying levels of unlabeled dATP or ATP (1000, 300, 100, 30, and 10 μm), where indicated. After 20 min at 60 °C, products were subjected to urea-PAGE analysis, and the gels were exposed for 1 h at −70 °C. M, molecular mass markers; inc., incorporation. B, influence of ATP, dATP, GTP, and dGTP on polynucleotide synthesis in reactions containing oligo(dT)30 as template. Reactions were as described in the legend for Fig. 3 with 10 pmol of oligo(dT)30. Mixtures contained 100 μm [α-32P]dATP (6815 cpm/pmol) or 100 μm [α-32P]ATP (6024 cpm/pmol) and varying levels of ATP (1000, 100, and 10 μm), dATP (1000, 100, and 10 μm), GTP (1000 μm), or dGTP (1000 μm) where indicated. Following incubation, 5 μl of each reaction were subjected to urea-PAGE analysis and autoradiography. Reaction mixtures in lanes 15 and 16 contained 100 μm [α-32P]rGTP (104 cpm/pmol) or 100 μm [32P]dGTP (104 cpm/pmol), respectively. comp., complex.
We examined the relative binding affinity of the T. kodakaraensis primase complex for dNTPs and rNTPs in the presence of different DNA templates. In the presence of oligo(dT)30, the Km values for dATP and rATP were 30 and 90 μm, respectively; Km values of 50 and 250 μm for dGTP and rGTP, respectively, were observed with oligo(dC)30 as the template. Similar studies with ssM13 DNA revealed Km values for dATP and rATP of 40 and 80 μm, respectively. In the latter case, we noted that the Km values for the rNTPs and dNTPs varied depending on the level of unlabeled rNTPs or dNTPs added to reactions. The Km for dNTPs with the catalytic subunit was similar to those observed for the T. kodakaraensis complex.
The inhibitory effects of dNTPs on RNA synthesis were also investigated with oligo(dT)30 as the template (Fig. 5B). In the presence of high levels of ATP (ATP:dATP molar ratio of 10:1), dAMP incorporation was stimulated (∼3-fold), although the size of dAMP-labeled products formed was reduced (lane 3) when compared with that formed in the absence of ATP (lane 2). Alkaline hydrolysis, however, revealed that the products formed (shown in lane 3) were hybrid polynucleotide chains in which AMP and dAMP were covalently interspersed within the same chain; products formed in the absence of rATP (lane 2) were unaffected by alkali (data not presented). At lower ATP levels (lanes 4 and 5, molar ratios of ATP:dATP of 1 and 0.1, respectively), dAMP incorporation, the size of dAMP-labeled chains formed, and the stimulation of dATP incorporation by ATP were less affected. In contrast, synthesis of the short RNA chains was inhibited at all levels of dATP added, and the size of oligo(rA) chains formed and the amount of [α-32P]ATP incorporated were reduced (lanes 10–12). We also investigated whether the addition of dGTP and rGTP, nucleotides that are noncomplementary and not incorporated into DNA and RNA chains in the presence of oligo(dT)30, influenced dATP and ATP incorporation. As shown, dGTP addition (molar ratio to dATP or rATP of 10:1) inhibited both DNA and RNA synthesis (Fig. 5B, lanes 6 and 13), whereas rGTP addition reduced dATP incorporation by 43% and rATP incorporation by 92% (Fig. 5B, lanes 7 and 14). Lower molar ratios of dGTP:rATP (0.1:1) inhibited ATP incorporation by 91%, whereas dATP incorporation was reduced by only 8% (data not presented). Collectively, these findings indicate that the T. kodakaraensis primase complex binds dNTPs more effectively than rNTPs.
Primase-dependent Replication of Primed M13
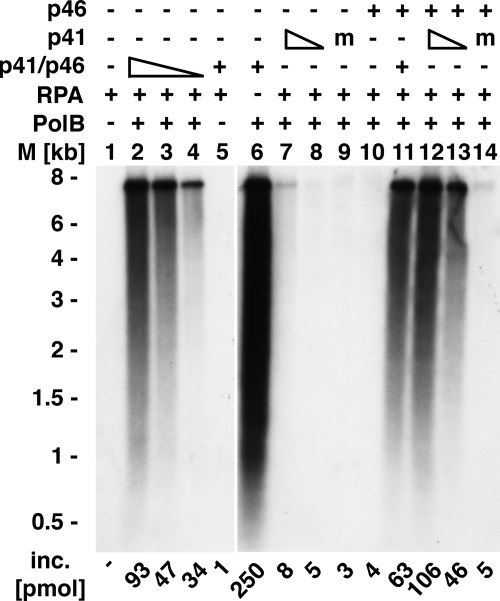
The T. kodakaraensis primase complex supported replication of unprimed ssM13 DNA in the presence of the T. kodakaraensis proteins PolB, RFC, and PCNA, which were previously shown to catalyze the replication of singly primed M13 DNA (18) (Fig. 6). Reactions carried out at 60 °C in the presence of 0.25 m NaCl that lacked either the DNA primase complex, RFC, and/or PCNA failed to support DNA synthesis (data not presented). As shown in Fig. 6, lanes 2–4, the amount of DNA synthesized depended on the level of the primase complex added, and both primase subunits were required for replication. In the absence of PolB, the primase complex did not support detectable nucleotide incorporation (lane 5). DNA synthesis was not observed in reactions containing the catalytic subunit (p41) alone (lanes 7 and 8) or the large subunit (p46) alone (lane 10). When the p41 and p46 subunits were both added, replication was observed (lanes 12 and 13). Reactions containing the m p41 catalytic subunit alone (lane 9), the m p41 subunit, plus the p46 subunit (lane 14) or the m p41-p46 complex were also inactive (data not shown). Omission of the T. kodakaraensis RPA resulted in a marked increase in DNA synthesis and formation of DNA chains that varied in length from 0.5 to 7 kb (lane 5). The level of DNA synthesized in the absence of T. kodakaraensis RPA was significantly higher (250 pmol) than the amount of DNA added (108 pmol), suggesting that in the absence of RPA, PolB catalyzed DNA displacement synthesis. The data presented in Fig. 6 indicate that primers produced by the T. kodakaraensis primase complex support DNA synthesis by PolB.
FIGURE 6.
Primase-dependent replication of M13 DNA. Reaction mixtures (20 μl) containing 40 mm Tris-HCl (pH 8.0), 10 mm magnesium acetate, 1.5 mm DTT, 50 μg/ml BSA, 20 μm [α-32P]dATP (13,000 cpm/pmol), 100 μm each of dGTP, dCTP, and dTTP, 2 mm ATP, 0.75 nm M13 DNA, 0.25 m NaCl, 360 nm T. kodakaraensis RPA, 50 nm T. kodakaraensis PCNA (TK0535), 50 nm T. kodakaraensis RFC, 20 nm T. kodakaraensis PolB, 40, 20, or 10 nm T. kodakaraensis primase complex (40 nm, lanes 2, 5, 6, and 11; 20 nm, lane 3; 10 nm, lane 4), 40 nm T. kodakaraensis p46 subunit (lanes 10–14), 20 nm m 41 subunit (lanes 9 and 14), and 20 or 10 nm wild type p41 subunit (20 nm, lanes 7 and 12; 10 nm, lanes 8 and 13) were incubated for 20 min at 60 °C. Aliquots were used to measure DNA synthesis and the size of DNA products following alkaline agarose gel (1%) electrophoresis and autoradiography. M, molecular mass markers; inc., incorporation.
Coupling of MCM Helicase, Primase, and PolB and Rolling Circle Synthesis of Leading and Lagging DNA Strands
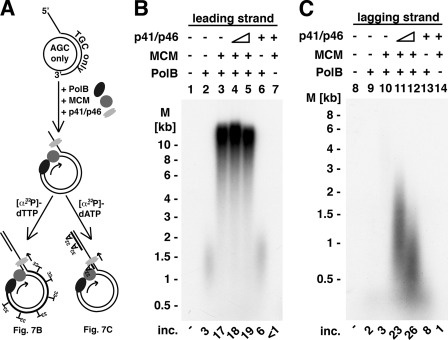
Unprimed M13 circular ssDNA was replicated in reactions containing DNA primase, PolB, RFC, and PCNA (Fig. 6). We next examined whether a rolling circle replication reaction could be established. For this purpose, a small circular 200-nt DNA was synthesized (described under “Experimental Procedures”), which contained only three deoxynucleotides (dG, dC, and dA). The circular DNA was hybridized to an oligonucleotide containing a 5′-tail of dT40 covalently linked to a 100-nt oligodeoxynucleotide complementary to a region in the 200-nt circle to generate the primed circular structure shown in Fig. 7A. As schematically shown in Fig. 7A, the nucleotide content of the circle limits [α-32P]dTTP incorporation to leading strand synthesis (Fig. 7B) and [α-32P]dATP incorporation to lagging strand formation (Fig. 7C). DNA synthesis with the primed template was observed in reactions containing the T. kodakaraensis MCM helicase (TK1361; polarity 3′ → 5′), T. kodakaraensis PolB and T. kodakaraensis RFC, and PCNA (TK0535) (Fig. 7B, lane 3). The elongation of primer ends by PolB (specifically scored by [α-32P]dTTP incorporation) generated leading single strand DNA products >10 kb long after incubation at 60 °C for 10 min. The length of the product formed represented >50 rounds of DNA synthesis. In the absence of the MCM helicase, limited DNA synthesis was observed (Fig. 7B, lane 2, ∼1.5 kb in length), suggesting that T. kodakaraensis PolB alone supported some displacement synthesis (∼7.5 turns of the 200 nt circle). The presence of the DNA primase complex in these reactions did not alter the low level of leading strand synthesis detected in the absence of the MCM helicase (Fig. 7B, lane 6). Lagging strand synthesis was examined by measuring [α-32P]dATP incorporation (carried out in the presence of all four dNTPs). As shown (Fig. 7C), lagging strand synthesis required the proteins essential for leading strand formation as well as the T. kodakaraensis primase complex (Fig. 7C, compare lane 10 with lanes 11 and 12). The size of the lagging strands formed was influenced by the level of T. kodakaraensis DNA primase added (lanes 11 and 12). The addition of increasing amounts of DNA primase yielded shorter lagging strand chains, suggesting that increased synthesis of primers reduced the level of chain extension. These findings indicated that the simultaneous synthesis of leading and lagging strands, involving a rolling circle, can be carried out with the T. kodakaraensis proteins. The data shown in Fig. 7 represent, to the best of our knowledge, the first rolling circle replication reaction catalyzed in coordination with a 3′ → 5′ DNA helicase.
FIGURE 7.
Rolling circle synthesis of leading and lagging strand. A, model of the synthesis of leading and lagging strand synthesis. The annealing of an oligonucleotide containing a 5′ oligo(T)40 tail and an 100-nt stretch (with T, G, and C) to the 200-nt circle (containing A, C, and G) yielded the DNA substrate shown. The addition of T. kodakaraensis PolB, T. kodakaraensis MCM (TK1361), T. kodakaraensis RFC, T. kodakaraensis PCNA, and dNTPs and ATP resulted in the elongation of the leading strand. Following the addition of the T. kodakaraensis DNA primase complex (p41-p46), the reaction was divided into two equal portions. One-half was supplemented with [α-32P]dTTP, and the other was supplemented with [α-32P]dATP to monitor the synthesis of leading and lagging strand synthesis as shown in panels B and C. B, leading strand synthesis. Reaction mixtures (20 μl) containing 20 mm Tris-HCl (pH 8.0), 10 mm magnesium acetate, 100 mm NaCl, 1 mm DTT, 100 μg/ml BSA, 5 nm 200-mer DNA circles hybridized to dT40-100-mer, 20 nm T. kodakaraensis RFC, and 50 nm T. kodakaraensis PCNA (TK0535) were prepared on ice. Reactions were supplemented with T. kodakaraensis PolB (44 nm), and the mixture was incubated for 5 min at 60 °C, after which T. kodakaraensis MCM (TK1361) (44 nm), T. kodakaraensis DNA primase complex (74 or 18.5 nm), ATP (2 mm), 150 μm each of dGTP, dCTP, and dATP, and 40 μm [α-32P]dTTP (18,900 cpm/pmol) was added. The level of T. kodakaraensis DNA primase complex added in lanes 6 and 7 was 74 nm. Reactions were incubated for 10 min at 60 °C, and aliquots were used to measure DNA synthesis and the size of products formed. Alkaline agarose gel (0.6%) electrophoresis was used for the analysis of leading strand synthesis. M, molecular mass markers; inc., incorporation. C, lagging strand synthesis. Reactions were as described in panel A with the exception that 150 μm each of dGTP, dCTP, and dTTP, and 40 μl of [α-32P]dATP (13,300 cpm/pmol) were added. Alkaline agarose gel (1.1%) electrophoresis was used to analyze the size of the DNA synthesized. The levels of T. kodakaraensis DNA primase added were as described in panel B.
DISCUSSION
In this study, we have described some properties of the DNA primase complex isolated from the thermophilic archaeon, T. kodakaraensis. We cloned, isolated, and characterized the T. kodakaraensis p41-p46 complex, a mutated catalytically inactive p41 subunit-p46 complex, as well as the individual subunits. In reactions containing ssM13 DNA as the template, the primase complex catalyzed the synthesis of DNA chains ∼0.5 kb in length with dNTP (in the absence of added rNTPs) and shorter RNA chains, 50–75 nt long, with rNTPs, whereas the mutant complex was inactive. The p41 catalytic subunit alone was considerably less active than the complex and supported RNA synthesis with rNTPs poorly. However, the DNA products formed by the p41 subunit were substantially longer in length (up to 6 kb) than those produced by the T. kodakaraensis primase complex. Similar to findings reported for the heterodimeric primase complex isolated from P. furiosus, oligoribonucleotide synthesis was markedly inhibited by dNTPs, whereas DNA synthesis was affected marginally by the presence of a large molar excess of rNTPs relative to the level of dNTPs added (9). These findings indicate that the T. kodakaraensis primase complex preferentially interacts with dNTPs rather than rNTPs. In accord with this notion, Km measurements indicated that the affinity of the primase for dNTPs was 2–4-fold greater than its affinity for rNTPs. These observations are in keeping with the properties of the DNA primase complex isolated from P. abyssi (7) but differ from those reported for the Sulfolobus solfataricus primase (14). In the latter case, it was reported that the affinity of rNTPs was >100-fold lower than its affinity for dNTPs. Although the intracellular concentration of nucleotides in archaea is presently unknown, the average levels reported in mammalian (24) and yeast (25) cells varied from ∼0.3 to 3 mm for rNTPs and from 10 to 50 μm for dNTPs. The much higher levels of rNTPs than dNTPs suggest that DNA primases are likely to initiate polynucleotide chains de novo with rNTPs, as reported for several archaeal species (10–12), unless other factors contribute to the initiation events. It is important to note that the eukaryotic primase complex, although capable of extending chains with dNTPs, cannot initiate chains with dNTPs (Ref. 1 and results presented in Table 1). However, as shown here and by others (7, 9), a number of the archaeal primase complexes can initiate chains with dNTPs. As shown in Table 1, the T. kodakaraensis primase complex, when coupled with the Klenow Pol, supported extensive dATP incorporation in reactions primed with poly(dT) in the absence of rATP. Although the physiological significance of extensive synthesis of DNA by DNA primase is presently unknown, the lack of a Pol α homolog in archaea suggests that a multifunctional DNA primase that initiates and extends DNA chains with dNTPs may fulfill the role played by the eukaryotic Pol α-primase complex.
The large subunit of both the archaeal and the eukaryotic primase complexes plays an important role in the formation of RNA primers (1, 7, 9). The synthesis of long DNA chains by the p41 subunit alone appears to be unique to the archaeal catalytic subunit. Although it generates DNA, albeit to a lower extent than the T. kodakaraensis p41-p46 complex, the T. kodakaraensis p41 subunit alone failed to support coupled DNA synthesis with the Klenow Pol (Table 1) or the T. kodakaraensis PolB (Fig. 6), in contrast to the T. kodakaraensis primase complex. These findings suggest that the p46 subunit contributes to this coupling event. Previous studies have shown that the initiation, translocation, and processivity of the eukaryotic primase complex are markedly affected by the large subunit (1). In keeping with these observations, disruption of the p58 subunit in yeast is lethal (26). Recent studies with the C-terminal domain of the large subunit isolated from the S. solfataricus and eukaryotes revealed the presence of an iron-sulfur cluster (Fe-S) (26, 27). A truncated version of the yeast primase complex lacking this C-terminal domain of the large subunit was incapable of RNA primer synthesis (27). We noted that the T. kodakaraensis p46 subunit, like the S. solfataricus p46 subunit, displayed a yellow-brown color and showed a broad absorption spectrum around 400 nm, in addition to the expected protein peak at 280 nm. These properties suggest that the T. kodakaraensis p46 subunit may also be a Fe-S-containing protein. It was shown that the Fe-S cluster region isolated from the eukaryotic p58 subunit selectively binds to single-stranded/double-stranded DNA junctions (27). Possibly, this interaction plays a role in a hand-off mechanism by which a Pol-DNA primase-primed DNA complex supports chain extension.
Detailed studies on the 5′-end products formed in the primase reaction were carried out with oligo(dT)30 as the template to limit the complexity of the DNA and RNA chains formed. Our analyses indicated that the oligo(rA) and oligo(dA) chains formed contained low levels of triphosphate ends (∼10%) and more substantial levels of 5′-phosphate ends (∼50%). Frick and Richardson (28) have proposed that the DNA primase initiates primer synthesis after formation of a quaternary complex with the template DNA and two nucleoside triphosphates. In this model, the NTP binding, destined to lead to the 5′-end of the oligonucleotide, is defined as the initiation site, whereas the NTP that binds to the second site is added to the 3′-end of the initiating nucleotide and is referred to as the elongation site. Subsequent steps leading to oligonucleotide synthesis (n + 1) involve the transfer of the nucleotide to the initiation site and the binding of another NTP at the elongation site. In the case of the T. kodakaraensis primase, we speculate that a substantial amount of NTPs (dNTPs) that bind to the elongation site is transferred to H2O (or Tris or glycerol) located at the initiation site. As described in the accompanying article (21), the p41-p46 complex (as well as the p41 catalytic subunit) hydrolyzes dNTPs (and NTPs) to PPi, Pi, and the corresponding nucleoside monophosphate. We suggest that the archaeal enzyme supports the stable association of the nucleoside monophosphate in the initiation site. Subsequent NTP (or dNTP) binding at the elongation site followed by the transfer reaction results in the production of an oligonucleotide chain containing a 5-phosphate terminus. Possibly, a nucleoside triphosphate located at the initiation site is used as an acceptor at a low rate, resulting in the production of an oligonucleotide containing a triphosphate end. As noted in the accompanying article (21), reactions leading to the production of the dAMP-glycerol and dAMP-Tris derivatives were only detected with dATP and not dAMP. Thus, in our proposed scheme, the transfer of the nucleoside monophosphate in the initiation site can only occur with a nucleoside triphosphate.
We examined the influence of the T. kodakaraensis DNA primase on the replication of unprimed M13 DNA in reactions containing T. kodakaraensis DNA PolB. In the presence of high salt, this highly active Pol catalyzed the elongation of primed DNA templates in the presence of RFC and PCNA (18). The generation of primers by the primase complex supported synthesis of full-length duplex M13 DNA products in reactions containing RFC, PCNA, and PolB. Both the p41 subunit and the p46 subunit were required for this activity. Surprisingly, the reaction did not require T. kodakaraensis RPA, which markedly inhibited the replication reaction at high levels. In eukaryotes, the replication of primed templates by the Pol δ or Pol ϵ is completely dependent on RPA (or a suitable DNA-binding protein). The reasons for this discrepancy remain to be further explored.
We have also devised a rolling circle assay using T. kodakaraensis proteins that leads to the synthesis of both leading and lagging strands. Leading strand synthesis required the action of T. kodakaraensis MCM 3′ → 5′ DNA helicase, PolB, and RFC and PCNA. Importantly, the long leading strands produced in this reaction supported the primase-catalyzed lagging strand synthesis. The availability of an in vitro rolling circle replication system may help to define other T. kodakaraensis proteins that contribute to the synthesis of both lagging and leading strands.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant GH034559 (to J. H.). This work was also supported by National Science Foundation Grant MCB-0815646 (to Z. K.).

This article contains supplemental Tables 1–4 and Fig. 1.
- Pol
- polymerase
- RFC
- replication factor C
- PCNA
- proliferating cell nuclear antigen
- nt
- nucleotide
- m
- mutant
- CIP
- calf intestinal phosphatase
- MCM
- minichromosome maintenance
- RPA
- replication protein A
- r
- ribo
- Ap
- 2′(3′)-adenosine monophosphate.
REFERENCES
- 1. Kuchta R. D., Stengel G. (2010) Mechanism and evolution of DNA primases. Biochim. Biophys. Acta 1804, 1180–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wang T. S. (1991) Eukaryotic DNA polymerases. Annu. Rev. Biochem. 60, 513–552 [DOI] [PubMed] [Google Scholar]
- 3. Waga S., Stillman B. (1998) The DNA replication fork in eukaryotic cells. Annu. Rev. Biochem. 67, 721–751 [DOI] [PubMed] [Google Scholar]
- 4. Nick McElhinny S. A., Gordenin D. A., Stith C. M., Burgers P. M., Kunkel T. A. (2008) Division of labor at the eukaryotic replication fork. Mol. Cell. 30, 137–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Walter P., Klein F., Lorentzen E., Ilchmann A., Klug G., Evguenieva-Hackenberg E. (2006) Characterization of native and reconstituted exosome complexes from the hyperthermophilic archaeon Sulfolobus solfataricus. Mol. Microbiol. 62, 1076–1089 [DOI] [PubMed] [Google Scholar]
- 6. Li Z., Santangelo T. J., Cuboňová L., Reeve J. N., Kelman Z. (2010) Affinity purification of an archaeal DNA replication protein network. MBio. 1, e00221–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Le Breton M., Henneke G., Norais C., Flament D., Myllykallio H., Querellou J., Raffin J. P. (2007) The heterodimeric primase from the euryarchaeon Pyrococcus abyssi: a multifunctional enzyme for initiation and repair? J. Mol. Biol. 374, 1172–1185 [DOI] [PubMed] [Google Scholar]
- 8. Lao-Sirieix S. H., Pellegrini L., Bell S. D. (2005) The promiscuous primase. Trends Genet. 21, 568–572 [DOI] [PubMed] [Google Scholar]
- 9. Liu L., Komori K., Ishino S., Bocquier A. A., Cann I. K., Kohda D., Ishino Y. (2001) The archaeal DNA primase: biochemical characterization of the p41-p46 complex from Pyrococcus furiosus. J. Biol. Chem. 276, 45484–45490 [DOI] [PubMed] [Google Scholar]
- 10. Matsunaga F., Norais C., Forterre P., Myllykallio H. (2003) Identification of short “eukaryotic” Okazaki fragments synthesized from a prokaryotic replication origin. EMBO Rep. 4, 154–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Robinson N. P., Dionne I., Lundgren M., Marsh V. L., Bernander R., Bell S. D. (2004) Identification of two origins of replication in the single chromosome of the archaeon Sulfolobus solfataricus. Cell 116, 25–38 [DOI] [PubMed] [Google Scholar]
- 12. Norais C., Hawkins M., Hartman A. L., Eisen J. A., Myllykallio H., Allers T. (2007) Genetic and physical mapping of DNA replication origins in Haloferax volcanii. PLoS Genet. 3, e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bocquier A. A., Liu L., Cann I. K., Komori K., Kohda D., Ishino Y. (2001) Archaeal primase: bridging the gap between RNA and DNA polymerases. Curr. Biol. 11, 452–456 [DOI] [PubMed] [Google Scholar]
- 14. Lao-Sirieix S. H., Bell S. D. (2004) The heterodimeric primase of the hyperthermophilic archaeon Sulfolobus solfataricus possesses DNA and RNA primase, polymerase, and 3′-terminal nucleotidyl transferase activities. J. Mol. Biol. 344, 1251–1263 [DOI] [PubMed] [Google Scholar]
- 15. Beck K., Lipps G.. (2007) Properties of an unusual DNA primase from an archaeal plasmid. Nucleic Acids Res. 35, 5635–5645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. De Falco M., Fusco A., De Felice M., Rossi M., Pisani F. M. (2004) The DNA primase of Sulfolobus solfataricus is activated by substrates containing a thymine-rich bubble and has a 3′-terminal nucleotidyl-transferase activity. Nucleic Acids Res. 32, 5223–5230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kasiviswanathan R., Shin J. H., Melamud E., Kelman Z. (2004) Biochemical characterization of the Methanothermobacter thermautotrophicus minichromosome maintenance (MCM) helicase N-terminal domains. J. Biol. Chem. 279, 28358–28366 [DOI] [PubMed] [Google Scholar]
- 18. Ladner J. E., Pan M., Hurwitz J., Kelman Z. (2011) Crystal structures of two active proliferating cell nuclear antigens (PCNAs) encoded by Thermococcus kodakaraensis. Proc. Natl. Acad. Sci. U.S.A. 108, 2711–2716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pan M., Santangelo T. J., Li Z., Reeve J. N., Kelman Z. (2011) Thermococcus kodakarensis encodes three MCM homologs, but only one is essential. Nucleic Acids Res. 39, 9671–9680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Conaway R. C., Lehman I. R. (1982) A DNA primase activity associated with DNA polymerase α from Drosophila melanogaster embryos. Proc. Natl. Acad. Sci. U.S.A. 79, 2523–2527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chemnitz Galal W., Pan M., Giulian G., Yuan W., Li S., Edwards J. L., Marino J. P., Kelman Z., Hurwitz J. (March 16, 2012) Formation of dAMP-glycerol and dAMP-Tris derivatives by the Thermococcus kodakaraensis DNA primase. J. Biol. Chem. 287, 16220–16229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Maitra U., Hurwitz H. (1965) The role of DNA in RNA synthesis. IX. Nucleoside triphosphate termini in RNA polymerase products. Proc. Natl. Acad. Sci. U.S.A. 54, 815–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Josse J., Kaiser A. D., Kornberg A. (1961) Enzymatic synthesis of deoxyribonucleic acid. VIII. Frequencies of nearest neighbor base sequences in deoxyribonucleic acid. J. Biol. Chem. 236, 864–875 [PubMed] [Google Scholar]
- 24. Traut T. W. (1994) Physiological concentrations of purines and pyrimidines. Mol. Cell Biochem. 140, 1–22 [DOI] [PubMed] [Google Scholar]
- 25. Nick McElhinny S. A., Watts B. E., Kumar D., Watt D. L., Lundström E. B., Burgers P. M., Johansson E., Chabes A., Kunkel T. A. (2010) Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases. Proc. Natl. Acad. Sci. U.S.A. 107, 4949–4954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Klinge S., Hirst J., Maman J. D., Krude T., Pellegrini L. (2007) An iron-sulfur domain of the eukaryotic primase is essential for RNA primer synthesis. Nat. Struct. Mol. Biol. 14, 875–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Weiner B. E., Huang H., Dattilo B. M., Nilges M. J., Fanning E., Chazin W. J. (2007) An iron-sulfur cluster in the C-terminal domain of the p58 subunit of human DNA primase. J. Biol. Chem. 282, 33444–33451 [DOI] [PubMed] [Google Scholar]
- 28. Frick D. N., Richardson C. C. (2001) DNA primases. Ann. Rev. Biochem. 70, 39–80 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.