Background: Cdx transcription factors are known to convey the posteriorizing signals from the canonical Wnt pathway.
Results: Cdx proteins integrate canonical Wnt signals on a Pax3 neural crest enhancer.
Conclusion: Cdx proteins are involved in Wnt-mediated induction of Pax3 at the neural plate border.
Significance: Our data suggest that Cdx proteins are important novel players within the neural crest gene regulatory network.
Keywords: beta-Catenin, Gene Regulation, Neurodevelopment, Transcription/Developmental Factors, Wnt Signaling, Cdx, Pax3, Neural Crest Cells, Neural Tube
Abstract
One of the earliest events in neural crest development takes place at the neural plate border and consists in the induction of Pax3 expression by posteriorizing Wnt·β-catenin signaling. The molecular mechanism of this regulation is not well understood, but several observations suggest a role for posteriorizing Cdx transcription factors (Cdx1/2/4) in this process. Cdx genes are known as integrators of posteriorizing signals from Wnt, retinoic acid, and FGF pathways. In this work, we report that Wnt-mediated regulation of murine Pax3 expression is indirect and involves Cdx proteins as intermediates. We show that Pax3 transcripts co-localize with Cdx proteins in the posterior neurectoderm and that neural Pax3 expression is reduced in Cdx1-null embryos. Using Wnt3a-treated P19 cells and neural crest-derived Neuro2a cells, we demonstrate that Pax3 expression is induced by the Wnt-Cdx pathway. Co-transfection analyses, electrophoretic mobility shift assays, chromatin immunoprecipitation, and transgenic studies further indicate that Cdx proteins operate via direct binding to an evolutionarily conserved neural crest enhancer of the Pax3 proximal promoter. Taken together, these results suggest a novel neural function for Cdx proteins within the gene regulatory network controlling neural crest development.
Introduction
Pax3 is a paired-box homeodomain transcription factor essential for normal neural crest (NC),5 neural tube (NT) and skeletal muscle development. In mice, homozygous Pax3 loss of function, as seen in Splotch (Sp) mutants, leads to early embryonic lethality due to its role in NC cells (NCC). Although Pax3+/Sp mice only display pigmentation anomalies (white belly spot), most Pax3Sp/Sp embryos die around embryonic day 14.0 (e14.0) due to a severe NC defect leading to lack of outflow tract septation and heart failure (1). Pax3Sp/Sp mice also display other severe anomalies, including spinal ganglia malformations, intestinal aganglionosis, and posterior NT defects (spina bifida) (2). At the molecular level, Pax3 plays a critical role in the gene regulatory network controlling NCC development downstream of canonical Wnt signals. Together with members of the Msx, Dlx, and Zic families, Pax3 specifies the neural plate border and promotes induction of NCC (3–5). At later stages, Pax3 also controls survival of dorsal NT progenitors through stimulation of p53 degradation (6, 7). In humans, heterozygous PAX3 mutations have been associated with Waardenburg syndrome, which is characterized by NC defects such as cranio-facial and pigmentary anomalies (8–10).
Neural Pax3 expression begins at the early somite stage (around e8.25) prior to initiation of NT closure. At this stage, Pax3 transcripts are detected on the lateral borders of both the anterior neural plate and posterior neural plate (PNP) (11). Following neural plate bending in a closed NT (from e8.5 onward), Pax3 transcripts are then detected in the dorsal neurectoderm, including pre-migratory NCC, in an almost continuous manner along the anterior-posterior (AP) axis. Indeed, strong Pax3 expression is detected in two large domains extending 1) from the forebrain down to rhombomere 4 and 2) from rhombomere 6 down to the rostral half of the PNP. Although detectable, Pax3 expression is clearly much weaker in rhombomere 5. At later stages, caudal Pax3 expression follows progression of posterior elongation and is maintained in the dorsal half of the closed neural tube until e14.5. Pax3 expression is also detected in a subset of migratory NCC contributing to the cardiac outflow tract, peripheral, and enteric nervous systems as well as in melanocytes but is generally down-regulated as NCC differentiate.
Regulatory sequences sufficient to mediate induction and dorsal restriction of Pax3 expression in the hindbrain and trunk are contained in the proximal 1.6-kb promoter (12). Deletion analysis of this promoter has revealed that a block of 674 bp containing two evolutionarily conserved regions of ∼250 bp, called neural crest enhancer 1 and 2 (NCE1 and -2), is sufficient to drive expression in the dorsal NT and NCC (13). NCE1 bears Pbx binding sites (activated by Pbx1-containing transcriptional complexes), which appear to be specifically required to control Pax3 expression in the hindbrain (14, 15). NCE2 contains a Tead binding site that was shown to be required for the activity of the whole 674-bp NCE in e10.5 transgenic embryos (13). In addition, both NCE1 and NCE2 contain a binding site for Pou class III members, and mutation of these sites leads to reduced NT activity of the 1.6-kb promoter in e9.5 transgenic embryos (15). On the other hand, Pax3 expression is induced by posteriorizing Wnt signals and dorsally restricted in response to dorso-ventral patterning signals, such as Sonic Hedgehog (Shh) (5, 16–20). However, how the canonical Wnt and Shh signals are integrated at the transcriptional level is still unclear.
The vertebrate Cdx genes (Cdx1, Cdx2, and Cdx4) are related to Drosophila caudal (cad) (21), and their gene products have conserved the ancestral ability to specify the posterior embryo and pattern the AP axis. Murine Cdx genes are sequentially activated in ectodermal and mesodermal cells of the primitive streak around e7.25, with Cdx1 activated first and Cdx4 activated last (22–24). At e8.5, all Cdx genes are expressed in the caudal embryo and form a nested set along the AP axis. Whereas Cdx1 and Cdx2 exhibit an almost perfect overlap in expression around the hindbrain/spinal cord boundary, Cdx4 is expressed slightly more posterior at this stage. The most anterior domain of Cdx expression appears to be restricted to the dorsal NT, with Cdx1 protein expressed in NCC emigrating from this domain (23, 25). This anterior limit of expression regresses concomitantly with axial elongation, persisting in the caudal embryo until e10.5 for Cdx1 and Cdx4 and until e12.5 for Cdx2. All three Cdx genes are also expressed in the developing hindgut epithelium with Cdx1 and Cdx2 expression maintained postnatally (26).
Cdx gene expression is regulated by posteriorizing signals from Wnt, retinoic acid (RA), and fibroblast growth factor (FGF) pathways in multiple species (27–38). Among these posteriorizing signals, the evolutionarily conserved role of the canonical Wnt pathway in Cdx regulation is the best characterized. Indeed, both Cdx1 and Cdx4 have been clearly identified as direct targets of the Wnt·β-catenin pathway (27, 29, 30). Moreover, other data suggest that Cdx2 is also responsive to canonical Wnt signals, although evidence for a direct regulation is sparse (39–45). In addition, Cdx proteins can interact with the Lef1-β-catenin transcriptional effector of the canonical Wnt pathway (46).
Our understanding of Cdx function has long been hampered by the functional redundancy between Cdx members and the vital role of Cdx2 during implantation (47, 48). Recent development of a conditional Cdx2 allele (49, 50) and analysis of Cdx double mutants (51–54) has demonstrated important regulatory roles for Cdx proteins in several processes during mouse embryogenesis. In the mesoderm, Cdx proteins regulate axial patterning, axial elongation, and somitogenesis in addition to placentogenesis and hematopoiesis. An important part of the Cdx function is executed through direct regulation of diverse Hox genes (28, 51–53, 55, 56). However, recent work has shown that a significant proportion of the Cdx function is also fulfilled via direct regulation of several non-Hox targets (49, 57). In the endoderm, Cdx proteins are involved in intestinal patterning and cell differentiation via Hox-dependent and -independent mechanisms (50, 58–61).
The function of Cdx members in the neurectoderm is less well understood. As in the mesoderm and endoderm, Cdx proteins control neurectoderm AP patterning via Hox-dependent and -independent mechanisms (37, 43, 62–64). Multiple gain- or loss-of-function mouse models have also demonstrated that Cdx proteins are important for proper formation of the NT and NC-derived spinal ganglia, but whether they do so in a tissue-autonomous or -non-autonomous (via the mesoderm) manner remains an open question (53, 54, 65–67). Nevertheless, analysis of neurectoderm-specific Cdx loss of function in ascidian embryos demonstrates that Cdx proteins may impact NT and NC formation in a tissue-autonomous manner. Indeed, such mutant embryos display NT defects as well as an absence of pigment cells, which are derived from NC-like cells (68, 69).
In this study, we investigated the possibility that Cdx proteins might convey the posteriorizing Wnt signals to Pax3. We found that murine Pax3 is in fact an indirect Wnt target and that Cdx proteins can directly activate neural Pax3 expression at least via the previously described NCE2 (13). Altogether, our data strengthen the idea that Cdx members are involved in both NT and NCC development in a tissue-autonomous manner downstream of Wnt signals.
EXPERIMENTAL PROCEDURES
Ethics Statement
Experiments involving mice were performed following Canadian Council of Animal Care guidelines for the care and manipulation of animals used in medical research. Protocols involving the manipulation of animals were approved by the institutional ethics committee of the University of Quebec at Montreal (Comité Institutionnel de Protection des Animaux; reference number 0511-R2-648-0512).
Generation and Analysis of Mice
Cdx1−/− mutant mice (56) were kindly provided by D. Lohnes (University of Ottawa, Canada). Pax3+/Sp mice were obtained from Jackson Laboratories (Bar Harbor, ME). Cdx1+/−Pax3+/Sp mutants were generated by Pax3+/Sp and Cdx1−/− intercrosses. Cdx1−/−Pax3+/Sp mutants were then generated by crossing Cdx1+/−Pax3+/Sp and Cdx1−/− mice.
Transgenes carrying the lacZ gene under control of the Hsp68 minimal promoter (70) and either a wild type or Cdx binding site (CdxBS)-mutated NCE2 were prepared, and transgenic embryos were generated from injected B6C3 oocytes according to standard techniques (71). In order to facilitate identification of transgenic embryos and provide a positive control for transgene expression, a previously described Gata4p-GFP transgene (72) was co-injected with each lacZ construct. Nine days after microinjection, foster mothers were sacrificed, and embryos were collected and individually analyzed for GFP and β-galactosidase activity. For X-gal staining, embryos were fixed in 2% paraformaldehyde for 15 min, washed twice with PBS, and incubated overnight in staining solution (5 mm K3Fe(CN)6, 5 mm K4Fe(CN)6, 2 mm MgCl2, 0.01% Nonidet P-40, 0.01% sodium deoxycholate in PBS) containing 1 mg/ml X-gal. Staining reactions were carried out overnight at 37 °C.
Whole-mount in situ hybridization and immunohistochemistry were performed as previously described (29, 48). Mice were mated overnight, and noon of the day of vaginal plug detection was considered as e0.5. Embryos to be compared were stage-matched according to established criteria (73) and processed in parallel. The probe for in situ hybridization of Pax3 mRNA was generated from a previously described plasmid (11), kindly provided by J. Epstein. Immunohistochemistry was performed using polyclonal antisera to Cdx1 and Cdx2 (48). All embryo images were taken with a Leica DFC 495 camera mounted on a Leica M205 FA stereomicroscope (Leica Microsystems Canada).
Chromatin Immunoprecipitation (ChIP) Analysis
ChIP assays in Neuro2a (N2a) cells were performed as described previously (29) using anti-FLAG M2 antibody (Sigma). PCR amplifications were performed with GoTaq DNA polymerase (Promega) and consisted of 30 cycles of 30 s at 96 °C, 30 s at 60 °C, and 30 s at 72 °C. The primers used for this study were Pax3 NCE2 Forward (5′-GGCACAATGGTACCTTCTCTAAGG) and Reverse (5′- AAGCTTCCCTTCTGAGAAGCGGGGACTTTAAA) and Pax3 exon 7 Forward (5′- CCGTGTCAGATCCCAGTAGCAC) and Reverse (5′-CTGAGGTGAAAGGCCATTGCCG). PCR products were resolved on a 1.5% agarose gel.
Electrophoretic Mobility Shift Assays (EMSA)
Pax3 NCE2 was scanned by using eight double-stranded end-labeled oligonucleotides and tested for Cdx binding by EMSA. Five micrograms of nuclear extracts from mock- or FLAG-Cdx1-transfected COS7 cells were used in each reaction as described previously (29). Supershifts were performed using 1 μg of anti-FLAG M2 antibody (Sigma). Specificity of binding was assessed by competition with a 100-fold excess of unlabeled WT or mutated cold probes. The oligonucleotides comprising mutated CdxBS1 and CdxBS3 sequences were as described for site-directed mutagenesis. The upper strands of each wild type or mutated double-stranded probe as well as of a probe harboring a consensus Cdx binding site (as positive control) are summarized in supplemental Table S1.
Plasmid Constructs
For generation of the Engrailed-Cdx1 (EnRCdx1) fusion construct, the coding sequence of the Drosophila Engrailed repressor domain (74) was subcloned upstream of a PCR product corresponding to the DNA binding domain (homeodomain) of Cdx1 ended by a Stop codon (see supplemental Fig. S1). FLAG-tagged Cdx1 expression vector has been described previously (46). FLAG-tagged EnRCdx1 and EnR constructs were generated by subcloning the relevant sequences into a modified pCEP4 plasmid (Invitrogen) (46). Cdx1, Cdx2, Cdx4, EnR, and EnRCdx1 bicistronic expression vectors were generated by subcloning the respective cDNA into the pIRES2-EGFP vector (Clontech).
Pax3 proximal promoter sequences were obtained by PCR amplification in accordance with previous work (1, 13). Oligonucleotide sequences are available upon request. These PCR products were cloned in pGEM-T vector and validated by sequencing. Luciferase reporter constructs bearing the Pax3 minimal promoter with or without various lengths of the 1.6-kb 5′ upstream sequences were generated by subcloning these sequences into pXP2 (75).
Point mutations were introduced into each of the three CdxBS of NCE2 by using the QuikChange multisite-directed mutagenesis kit (Stratagene), according to the manufacturer's instructions. Sequences of the oligonucleotides used for site-directed mutagenesis were as follows: CdxBS1, 5′-CAGCAGTTTAGTCTGAATGCCATAATAccTTCCTGAGAAC; CdxBS2, 5′-CTAGCCAAGACGTTGCTTCTTcgATTTTTCCAGCAGTTTA; CdxBS3, 5′-AAGGACAGACAGTCTcgACAACACTCCTGGCGTCATATCC (point mutations are denoted in lowercase letters).
Cell Culture and Transfection Analysis
P19 cells were propagated in AMEM supplemented with 7.5% FBS and 2.5% NCS, N2a cells were propagated in EMEM supplemented with 10% FBS, and COS7 cells were propagated in DMEM supplemented with 10% FBS. All transfections were performed using GeneJuice reagents (Novagen) in accordance with the manufacturer's instructions.
For time course analysis, P19 cells were seeded in 6-well plates (2 × 105 cells/well), and the following day, standard culture medium was replaced by a Wnt3a- or a control-conditioned medium (29). Cells were harvested before treatment (t = 0 h) and 2, 4, 8, 12, or 18 h post-treatment; snap frozen; and stored at −80 °C prior to RT-PCR analysis. To assess the requirement for de novo protein synthesis, P19 cells were pretreated for 30 min with 30 μg/ml cycloheximide (CHX) or with the vehicle (DMSO) alone and then treated for 24 h with Wnt3a- or control-conditioned medium in the presence of 1 μg/ml CHX. Afterward, cells were harvested, snap frozen, and stored as described above prior to RT-PCR analysis. To inhibit Cdx function, P19 cells were seeded in 100-mm plates (2 × 106 cells/plate) and transiently transfected with 6 μg of EnRCdx1-IRES-GFP or the negative control EnR-IRES-GFP expression vector. Thirty hours after transfection, cells were treated with Wnt3a- or control-conditioned medium and cultured for another 20 h. GFP-positive cells were then recovered by FACS and analyzed by RT-PCR.
To modulate Cdx activity in N2a cells, transient transfections with 6 μg of EnRCdx1-IRES-GFP, Cdx1-IRES-GFP, Cdx2-IRES-GFP, or Cdx4-IRES-GFP expression vectors were performed in 100-mm tissue culture plates (2 × 106 cells/plate). Forty-eight hours after transfection, GFP-positive cells were recovered by FACS and analyzed by RT-PCR.
For luciferase reporter assays, N2a (8 × 104 cells/well) and/or P19 (3 × 104 cells/well) cells were transfected in 24-well plates with 100 ng of Pax3p-luciferase reporter construct alone or with increasing amounts (1.25–5 ng) of Cdx expression vectors or with the maximum amount of Cdx expression vector and increasing amounts (2.5–20 ng) of EnRCdx1 expression vector. When required, an empty expression vector was also included to ensure a total of 125 ng of DNA/well. For analysis of deleted or mutated Pax3p-luciferase reporter constructs, a fixed concentration (5 ng) of Cdx2 expression vector was used. All transfections were performed at least three times in triplicate. Forty-eight hours after transfection, cells were disrupted in 100 μl of lysis buffer (0.1 m Tris (pH 8.0), 1% Igepal, 1 mm dithiothreitol) and assessed for luciferase activity with a Berthold LB9507 luminometer (Berthold Technologies).
RNA Extraction and RT-PCR Analysis
Total RNA was isolated from frozen cell pellets by using the RNeasy kit (Qiagen), and cDNA was synthesized using a poly(dT) oligonucleotide and Superscript II reverse transcriptase (Invitrogen) in accordance with manufacturer's protocols. PCR amplifications were then performed with Platinum Taq DNA polymerase (Invitrogen) and consisted of 25–35 cycles of 30 s at 96 °C, 30 s at 60 °C, and 45 s at 68 °C. Amplified bands were size-fractioned on a 2% agarose gel. The primers used in this study were Cdx1 (forward, 5′-GCAAGTCCGAGCTGGCTGCTA; reverse, 5′-GGGTAGAAACTCCTCCTTGACG), Cdx2 (forward, 5′-CCACACTTGGGCTCTCCGAGA; reverse, 5′-GGGTCACTGGGTGACAGTGGA); Cdx4 (forward, 5′-AGTTAACCTGGGCCTTTCTGA; reverse, 5′-ATTCAGAAACTATGACCTGCTGTATC), Pax3 (forward, 5′-CCTGCCAACATACCAGCTGTCG; reverse, 5′-CTGAGGTGAAAGGCCATTGCCG); Gapdh (forward, 5′-TCCTGCACCACCAACTGCTTAGC; reverse, 5′-AGGTCCACCACCCTGTTGCTGTA). All oligonucleotides were designed to encompass an intron allowing the detection of contaminating genomic DNA by the presence of a larger band. As an additional control, PCR was also performed with RNA that had not been reverse-transcribed. Statistical analysis was carried out using GraphPad Prism software version 5.0. For the comparison of groups, a paired Student's t test was used (two-tailed p value, α = 0.05). Differences between means were classed as not significant (p > 0.05) or significant (*, p < 0.05).
RESULTS
Cdx Members and Pax3 Are Co-expressed in Caudal Neurectoderm
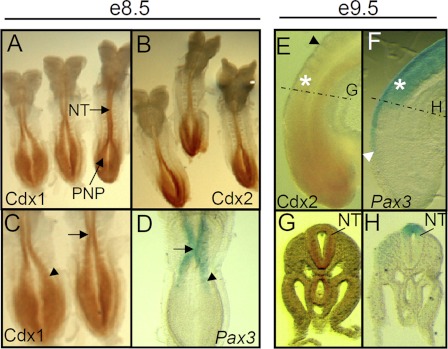
To validate our hypothesis that Cdx proteins are good candidates to induce Pax3 expression in the caudal neurectoderm, we compared the expression pattern of Cdx proteins and Pax3 mRNA. This analysis revealed extensive Cdx-Pax3 overlap in the caudal neurectoderm of e8.5 and e9.5 embryos (Fig. 1), in accordance with the well documented Cdx and Pax3 expression patterns (11, 22, 23). In e8.5 embryos, Pax3 transcripts are detected at both the PNP border and the site of initiation of NT closure (around the level of the fifth somite), where Cdx proteins are strongly expressed (Fig. 1, C and D). In e9.5 embryos, Pax3 and Cdx overlapping expression domains are displaced caudally in parallel with progression of axial elongation, NCC induction, and NT closure (Fig. 1, E–H). In brief, Cdx spatio-temporal expression patterns are consistent with a role in the induction of Pax3 expression in the caudal neurectoderm.
FIGURE 1.
Cdx members and Pax3 are co-expressed in the caudal neurectoderm during the early steps of NT and NCC formation. A and B, whole-mount immunohistochemistry showing Cdx1 (A) and Cdx2 (B) protein distribution in e8.5 embryos. Note the overlap in the PNP and NT. C and D, higher magnification view of Cdx1 protein distribution and comparison with Pax3 gene expression domain as detected by whole-mount in situ hybridization. Note the overlap at the site of initiation of NT closure (arrow) and at the lateral PNP (arrowhead). E–H, comparison of Cdx2 (E and G) protein distribution with Pax3 (F and H) gene expression domain in the caudal end of e9.5 embryos (lateral views). For comparative purposes, the location of the last formed somite is indicated by an asterisk. The black arrowhead in E indicates the anterior limit of Cdx2 expression; note the overlap with the posterior domain of Pax3 gene expression (white arrowhead in F). The dotted lines in E and F indicate the level at which the transverse sections shown in G and H were cut. The Cdx2 protein is widely detected in the tailbud region (E), but this pattern becomes more restricted anteriorly, with notable strong detection in the whole NT (G). Note the overlapping detection of Cdx2 protein (G) and Pax3 mRNA (H) in the posterior NT.
Pax3 Neural Expression Is Regulated by Cdx Proteins
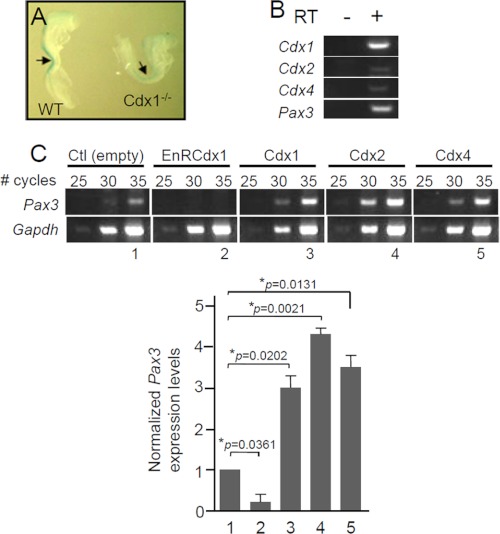
To determine whether Pax3 is a Cdx target gene, we evaluated its expression in e8.5 Cdx1-null embryos. This analysis revealed that Pax3 expression is slightly reduced in the dorsal NT of 4–6-somite stage Cdx1−/− embryos (Fig. 2A). Our data also suggest that this effect is transient because no difference in Pax3 expression is noted at later stages (>8 somites) (data not shown). To evaluate if this slight and transient reduction in Pax3 expression is functionally significant, we generated and analyzed an allelic series of Cdx1-Pax3 compound mutants. Cdx1+/−Pax3+/Sp and Cdx1−/−Pax3+/Sp animals were obtained at expected mendelian ratios and were similar to Pax3+/Sp animals, exhibiting a white belly spot (data not shown). Given the known functional redundancy between Cdx members (36, 48, 52, 53), this raises the possibility that the presence of Cdx2 and Cdx4 ensures that Pax3 expression levels do not significantly fall below 50% in these compound mutants. Therefore, the transient reduction in Pax3 mRNA levels observed in the dorsal NT of Cdx1−/− embryos most likely reflects the fact that Cdx1 is expressed slightly earlier and more anteriorly than other Cdx members. In this regard, it is noteworthy that a similar transient effect has been recently reported for expression of the Cdx neural target Mafb in Cdx1-null embryos (62). Taken together, this suggests that all Cdx proteins might be involved in the control of Pax3 expression.
FIGURE 2.
Regulation of Pax3 expression by Cdx proteins. A, whole-mount in situ hybridization analysis of Pax3 expression in 6-somite (e8.5) wild type (left) and Cdx1-null (right) mouse embryos. Embryos were processed and stained in parallel. Slightly fewer Pax3 transcripts are detected in the dorsal NT (arrow) of the Cdx1-null embryo. B, RT-PCR analysis showing co-expression of Pax3 with the Cdx genes in N2a cells. C, semiquantitative RT-PCR analysis showing alteration of endogenous Pax3 expression levels upon modulation of Cdx activity in N2a cells. Prior to RT-PCR analysis, cells were transiently transfected with the indicated expression vector (GFP is co-expressed as a bicistronic transcript, allowing FACS-mediated recovery of transfected cells). Pax3 expression levels are normalized against Gapdh expression. Note that Pax3 expression is drastically reduced in cells expressing EnRCdx1, whereas it is significantly increased in cells overexpressing a Cdx member. Numbers 1–5 represent the samples for which signal intensity was assessed by densitometry, and results were used to generate the graph at the bottom. Note that similar results were obtained from three independent experiments. Error bars, S.E.
To circumvent the functional redundancy of Cdx members, we generated a FLAG-tagged dominant negative Cdx protein consisting of the repressor domain of Drosophila Engrailed fused to the DNA binding domain (homeodomain) of Cdx1 (EnRCdx1; see supplemental Fig. S1). Given that the homeodomain is highly conserved (more than 90%) among Cdx members, EnRCdx1 is expected to recognize all Cdx target genes and inhibit their expression. We used the EnRCdx1 dominant negative tool to assess whether endogenous Pax3 expression is affected by modulation of Cdx activity in N2a cells. NC-derived N2a cells are a good model to study the regulation of the endogenous Pax3 promoter by Cdx because they co-express all three Cdx members as well as Pax3 (Fig. 2B). N2a cells were transiently transfected with expression constructs encoding Cdx1, Cdx2, Cdx4, or the dominant negative EnRCdx1, and endogenous Pax3 expression was evaluated by semiquantitative RT-PCR. As shown in Fig. 2C, Pax3 expression levels are significantly altered by modulation of Cdx activity in N2a cells. Overexpression of any of the three Cdx members results in a robust increase, whereas overexpression of EnRCdx1 leads to a complete knockdown of Pax3 expression. Therefore, these data indicate that Pax3 is a Cdx target gene in N2a cells.
Pax3 Is Induced by Wnt-Cdx Pathway in Undifferentiated P19 Cells
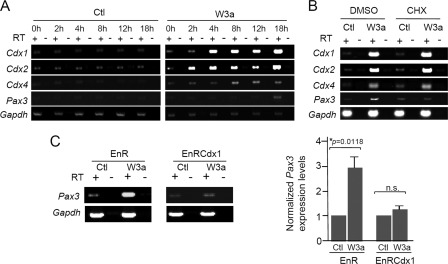
Previous studies have reported that neural Pax3 expression is induced by canonical Wnt signals (5, 16, 19, 20). To evaluate the potential contribution of Cdx proteins in this process, we performed a series of RT-PCR analyses in P19 embryocarcinoma cells cultured in the presence or absence of Wnt3a-conditioned medium, which is known for activating the canonical pathway (29, 76). The N2a cell line could not be used for this assay because the canonical Wnt pathway is constitutively activated in these cells due to Wnt7a autocrine regulation (77, 78). On the other hand, P19 cells have been used previously to study the regulation of Pax3 expression in the context of neural differentiation (12, 13, 15), and activation of the Wnt·β-catenin pathway (79). As shown in Fig. 3A, a time course analysis first revealed that expression of each Cdx member is rapidly induced (2–4 h) following treatment with Wnt3a-conditioned medium. Under the same conditions, Pax3 expression is also induced but considerably delayed (18 h). This delay being suggestive of an indirect regulation, we then directly assessed the necessity for an intermediary factor by inhibiting de novo protein synthesis with the protein synthesis inhibitor CHX. Cdx1 and Cdx4 are known direct targets of Wnt·β-catenin signaling (27–30). Accordingly, Cdx1 and Cdx4 induction by Wnt3a-conditioned medium is not affected by inhibition of protein synthesis (Fig. 3B). Interestingly, Cdx2 induction was also found to be independent of de novo protein synthesis, suggesting that Cdx2 is a direct target of Wnt·β-catenin signaling in this model. In marked contrast, Pax3 induction is blocked by treatment with CHX, demonstrating that a protein intermediary is needed to convey canonical Wnt signals to the Pax3 promoter (Fig. 3B). Of note, such an outcome is also supported by transient co-transfection assays indicating that Lef1·β-catenin complexes are very weak activators of the proximal 1.6-kb promoter of Pax3 in N2a or P19 cells (1.6-fold; data not shown).
FIGURE 3.
Regulation of Pax3 expression by the Wnt-Cdx pathway. A–C, analyses of P19 cells cultured in Wnt3a-conditioned medium (W3a) or control-conditioned medium (Ctl). After treatment, expression of Cdx1, Cdx2, Cdx4, and Pax3 was assessed by RT-PCR. Gapdh was used as a loading control. Note that expression of Cdx1, Cdx2, Cdx4, and Pax3 is specifically induced by treatment with W3a, whereas no induction is seen with Ctl. Shown are representative results obtained from three independent experiments. A, time course analysis in cells incubated for the indicated time with Ctl (left panels) or W3a (right panels); note that expression of all Cdx members is induced by W3a much earlier than Pax3. B, dependence of de novo protein synthesis for Pax3 induction. Cells were pretreated for 30 min with vehicle (DMSO) or 30 μg/ml CHX and then cultured for 24 h in Ctl or W3a in the presence of 1 μg/ml CHX. Note that Pax3 induction is affected by CHX treatment, whereas expression of Cdx1, Cdx2, and Cdx4 is not. C, semiquantitative RT-PCR analysis showing alteration of endogenous Pax3 expression level in the presence of the Cdx dominant negative fusion protein EnRCdx1. Prior to RT-PCR analysis, cells were transiently transfected with EnR (control) or EnRCdx1 expression vector, which co-expresses GFP as a bicistronic transcript. Approximately 30 h after transfection, cells were cultured for another 24 h in the presence or absence of W3a. GFP-positive cells were then recovered by FACS, total RNA was extracted, and endogenous Pax3 expression was assessed by RT-PCR. Note that Pax3 induction is reduced in cells expressing EnRCdx1 but not in cells expressing EnR. Pax3 expression levels were normalized against Gapdh expression. n.s., not significant. Error bars, S.E.
To determine whether Cdx members act as intermediaries between canonical Wnt signals and Pax3, we knocked down the Cdx function in P19 cells using the EnRCdx1 dominant negative protein. This analysis indicated that overexpression of EnRCdx1, but not that of EnR alone, strongly impairs induction of Pax3 by Wnt3a (Fig. 3C), suggesting that Cdx function is required for this regulation. Taken together, these results indicate that the neural plate border specifier Pax3 is an indirect target of Wnt·β-catenin signaling and implicate Cdx members as mediators of Wnt inductive signals.
Identification of Cdx Binding Sites in Proximal Pax3 Promoter
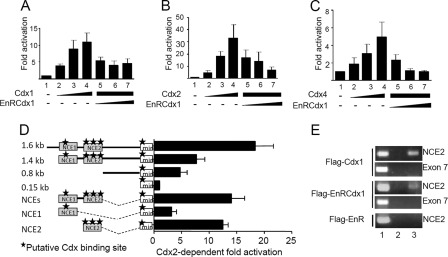
Previous studies have demonstrated that the proximal 1.6-kb promoter of Pax3 recapitulates endogenous posterior expression of Pax3 in the dorsal NT and NCC (12, 13). In order to understand the mechanism of Cdx-mediated regulation of Pax3 expression, we performed a series of co-transfection assays in N2a cells. To assess whether Cdx proteins transactivate the proximal 1.6-kb Pax3 promoter, we generated a luciferase reporter construct driven by these regulatory sequences (Pax3p1.6kb-Luc) and evaluated the effect of Cdx1, Cdx2, Cdx4, or EnRCdx1 expression on its activity. As shown in Fig. 4, A–C, all three Cdx proteins robustly induce this reporter in a dose-dependent manner, and each induction is strongly repressed by EnRCdx1. It is also interesting to note that, although each Cdx is equally expressed (data not shown), Cdx2 elicits the strongest response (32-fold), followed by Cdx1 (11-fold) and Cdx4 (5-fold). Such an observation is in accordance with previous work and suggests that Cdx members do not exhibit the same strength of transactivation (80).
FIGURE 4.
Identification of Cdx-responsive regions in the Pax3 proximal promoter. A–D, co-transfection assays in N2a cells using Luciferase reporter constructs driven by 5′-flanking sequences of Pax3. The results (mean ± S.E. (error bars) of four or five independent experiments performed in triplicate) are expressed as -fold induction compared with the relevant reporter vector alone. A–C, Cdx-dependent transactivation of a Pax3p1.6kb-luciferase construct. Note that the addition of Cdx1 (A), Cdx2 (B), or Cdx4 (C) expression construct results in strong dose-dependent transactivation (lanes 2–4), which is repressed by increasing amounts of EnRCdx1 (lanes 5–7). D, detection of Cdx-responsive regions in the Pax3 promoter via co-transfection assays in N2a cells. Luciferase reporter constructs consisting of the Pax3 150-bp minimal promoter (min) with or without various lengths or regions of the Pax3 promoter were assayed for Cdx2 transactivation. Potential Cdx binding sites are indicated by black stars. Note that Cdx2 responsiveness correlates with the presence of potential Cdx binding sites, which are located in previously identified NCEs, and especially with the three putative binding sites located in NCE2. E, chromatin immunoprecipitation assays in N2a cells showing the presence of Cdx1 and EnRCdx1 proteins on the endogenous Pax3 promoter. Primers flanking Pax3 NCE2 or exon 7 (as a negative control) were used to amplify anti-FLAG-immunoprecipitated DNA from FLAG-Cdx1-transfected (top panels), FLAG-EnRCdx1-transfected (middle panels), or FLAG-EnR-transfected (bottom panel) cells. Lane 1, input; lane 2, preimmune serum IP; lane 3, anti-FLAG IP. Note in lane 3 that a PCR product for NCE2 is obtained from FLAG-Cdx1- and FLAG-EnRCdx1-transfected cells but not from the negative control FLAG-EnR-transfected cells.
The Cdx-dependent transcriptional response of the proximal 1.6-kb promoter correlates with the presence of five putative Cdx binding sites identified via bioinformatic (MatInspector, Genomatix) and manual analyses. Interestingly, four of these potential Cdx binding sites are located in the previously described NCE1 and NCE2. To better define the sequence elements that mediate Cdx transactivation, a promoter deletion analysis was carried out in N2a cells, and Cdx2 was used to assay Cdx responsiveness. This assay revealed that the NCE2 region, which contains three putative Cdx binding sites, mediates most of Cdx2 transactivation (Fig. 4D). Accordingly, ChIP-PCR assays performed in N2a cells indicate that both FLAG-tagged Cdx1 and EnRCdx1 proteins are present on the endogenous Pax3 NCE2 (Fig. 4E).
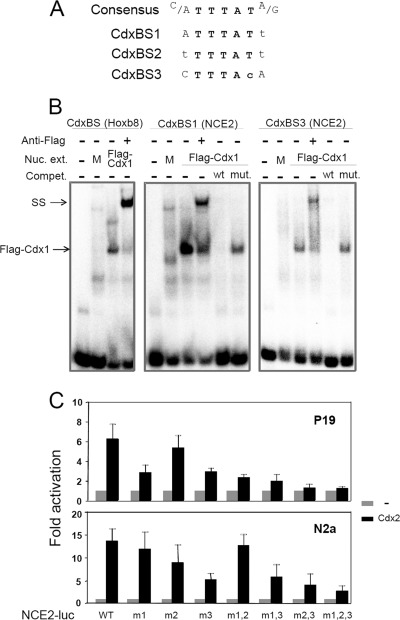
EMSA was then used to verify whether putative Cdx binding sites (named CdxBS1, CdxBS2, and CdxBS3) contained in NCE2 can be directly bound by a Cdx protein. In order to rule out the possibility that other unpredicted Cdx binding sites might also exist, we first scanned the whole NCE2 sequences with eight overlapping double-stranded oligonucleotide probes (supplemental Fig. S2 and supplemental Table S1). Incubation of these probes with nuclear extracts from COS7 cells overexpressing FLAG-Cdx1 revealed that Cdx1 preferentially binds to probes bearing either CdxBS1 or CdxBS3. Cdx1 binding to the probe bearing CdxBS2 was very weak, whereas no binding was observed for the remaining probes. The presence of FLAG-Cdx1 in the complex formed with probes containing CdxBS1 or CdxBS3 was demonstrated by supershift with an anti-FLAG antibody (Fig. 5B). Moreover, specificity of Cdx binding to CdxBS1 and CdxBS3 was confirmed by the absence of competition with cold probes bearing point mutations in the predicted elements (Fig. 5B). Taken altogether, our data demonstrate that Cdx proteins directly regulate Pax3 expression at least via functional binding sites contained in Pax3 NCE2.
FIGURE 5.
Identification and characterization of Cdx binding sites in Pax3 NCE2. A, sequence comparisons of all three putative Cdx-binding sites (CdxBS1, CdxBS2, and CdxBS3) relative to a consensus CdxBS sequence. Mismatches are denoted by lowercase letters. B, analysis of Cdx1 binding to CdxBS1 and CdxBS3 via electrophoretic mobility shift assay. All in vitro binding reactions were performed in parallel under identical conditions. M, mock. The presence of Cdx1 in the shifted bands was confirmed by the addition of 1 μg of anti-FLAG antibody to the binding reaction mix. Cdx1 binding and anti-FLAG supershifts (SS) are indicated by arrows. A probe containing a consensus CdxBS present in the Hoxb8 promoter was used as a positive control. Specificity of Cdx1 binding was assessed by preincubation of FLAG-Cdx1-containing nuclear extracts with a 100-fold excess of wild type (wt) or mutated (mut) cold probes. Note that preincubation with wild type unlabeled probes leads to inhibition of Cdx1 binding to the radiolabeled probes, whereas preincubation with mutated probes did not. C, impact of CdxBS mutation on Cdx2-mediated activation of the NCE2 reporter in cell culture. Wild type or CdxBS mutant versions of a Pax3NCE2-luciferase reporter were generated and assessed for Cdx2-mediated transactivation in P19 and N2a cells. Cells were transiently transfected with the NCE2-Luc reporter alone or with a Cdx2 expression vector. The results (mean ± S.E. (error bars) of seven or eight independent experiments performed in triplicate) are expressed as -fold induction compared with the relevant reporter vector alone. m1, m2, and m3, Pax3NCE2-luciferase reporter constructs containing point mutations in the CdxBS1, CdxBS2, and CdxBS3, respectively; m1,2, m1,3, and m2,3, double mutations of CdxBS; m1,2,3, triple mutation. Note that concomitant mutation of all three CdxBS is required to almost completely abolish the transactivation of Pax3 NCE2 by Cdx2.
Cdx Binding Sites Are Essential for Pax3 NCE2 Activity
To determine whether Cdx binding sites identified by EMSA are functionally important for Pax3 NCE2 activity, we generated a series of NCE2-luciferase reporter constructs containing the mutated elements (alone or in combination) and evaluated their responsiveness to Cdx2 in co-transfection assays. In P19 cells, single and double mutations of CdxBS reduce Cdx2-mediated transactivation of the NCE2 reporter, whereas simultaneous mutation of all three CdxBS results in complete loss of Cdx2-mediated transactivation (Fig. 5C). In accordance with our EMSA data, this analysis also revealed that the contribution of CdxBS1 and CdxBS3 to NCE2 activity is more important than that of CdxBS2. Similar results were obtained in N2a cells, although it can be noted that mutation of CdxBS1 does not considerably inhibit the Cdx2 responsiveness of Pax3 NCE2 in these cells. Interestingly, it can also be noted that the order of transactivation of the wild type NCE2 reporter is higher in N2a (∼14-fold) than in P19 cells (∼6-fold) and that the triple mutant construct remains slightly more active in N2a than in P19 cells. These observations suggest that Cdx proteins, in addition to directly binding the Pax3 NCE2, might also be recruited to and/or stabilized on this enhancer via an interaction with a neurectoderm-specific factor also required for strong activation.
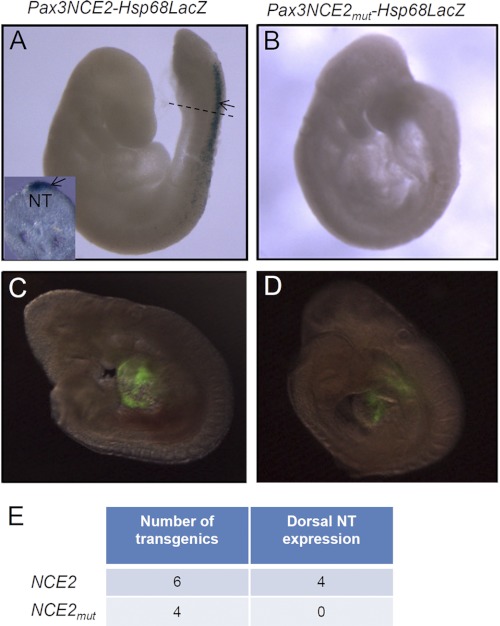
To verify the importance of all three Cdx binding sites of Pax3 NCE2 in vivo, we generated lacZ reporter constructs driven by either wild type or mutated Pax3 NCE2 upstream of the Hsp68 minimal promoter and evaluated β-galactosidase activity in transient e9.5 transgenic embryos (Fig. 6). This analysis first revealed that this short enhancer of 245 bp is sufficient to recapitulate Pax3 expression in the dorsal NT and premigratory NCC of the caudal embryo (Fig. 6A; compare with Fig. 1H). Importantly, we also found that point mutations in all three Cdx binding sites abolish NT and NCC expression of the NCE2 reporter (Fig. 6B). Of note, this lack of expression of the mutated NCE2 reporter is not due to transgene integration in a repressive chromatin region because expression of a co-injected Gata4p-GFP transgene is not affected (Fig. 6, C and D). Therefore, these results demonstrate that intact Cdx binding sites in Pax3 NCE2 are required to recapitulate caudal and dorsal neurectoderm-specific expression of Pax3 in transgenic mice.
FIGURE 6.
Cdx binding sites are required for the activity of a Pax3NCE2-lacZ reporter in transgenic embryos. A and B, whole mount β-galactosidase staining performed in e9.5 transgenic embryos generated from the wild type (Pax3NCE2-lacZ) (A) or mutated (Pax3NCE2mut-lacZ) transgene (B), the latter carrying specific point mutations in all three CdxBS of NCE2. The dotted line in A indicates where the transverse section shown in the inset was cut. C and D, direct GFP fluorescence detection of a Gata4p-GFP transgene co-injected with the wild type (C) or mutated transgene (D). Note that staining is detected in the dorsal NT (arrow) of transgenic embryos generated from the wild type construct (A). Mutation of all CdxBS abrogates β-galactosidase activity in the dorsal NT (B), whereas detection of Gata4p-GFP is not affected (D). E, table indicating the total number of transgenic embryos obtained for each construct as well as the number of embryos exhibiting β-galactosidase activity in the dorsal NT.
DISCUSSION
We presented data indicating that the dorsal NT/NCC marker Pax3 is a direct target of the Cdx proteins downstream of canonical Wnt signals. Cdx proteins convey canonical Wnt signals to the proximal Pax3 promoter through direct binding to Cdx binding sites located in the evolutionarily conserved NCE2. These Cdx binding sites are essential both for Cdx-mediated transactivation of NCE2 in cell culture experiments and for expression of a NCE2 reporter in the dorsal NT and NCC of e9.5 transgenic embryos, supporting the existence of the Wnt-Cdx-Pax3 pathway in vivo.
Wnt-mediated Induction of Pax3 Expression at Neural Plate Border
Although several studies have reported that Pax3 is a posterior Wnt-induced gene, very little is known regarding the possible mechanism of this regulation (5, 16, 19, 20). Our data now indicate that induction of murine Pax3 expression by canonical Wnt signals is indirect, involving Cdx proteins as intermediaries. In this regard, it is interesting to note that all three Cdx genes are direct targets of Wnt·β-catenin signaling in P19 cells. Although this was expected for Cdx1 and Cdx4 (27–30), such an outcome was somehow surprising for Cdx2, given a previous report indicating that exogenous Wnt3a can specifically induce Cdx1 but not Cdx2 in embryo culture (27). However, this result is in agreement with more recent work suggesting that Cdx2 is also a direct target of canonical Wnt signals (39–42, 44).
Our data challenge a recent report showing that Pax3 expression in the dorsal NT can also be regulated by another evolutionarily conserved enhancer located in intron 4 (named ECR2) and described as containing multiple putative Lef/Tcf binding sites (81). Indeed, it was reported that mutation of these putative binding sites abrogates ECR2 activity in transgenic zebrafishes. However, this mutated transgene was not assayed in mice, and these putative binding sites were not shown to be bound by Lef/Tcf proteins. Because the consensus Lef/Tcf binding site exhibits rather low binding specificity for HMG-box proteins, this raises the possibility that putative Lef/Tcf binding sites identified in Pax3 ECR2 are not bound by Lef/Tcf proteins but rather by other HMG-box proteins, such as Sox members (82–85). Alternatively, it is also possible that Pax3 is a direct Wnt target in zebrafish and not in mice. Regardless of the mechanism operating in other organisms, our CHX experiments indicate that murine Pax3 is an indirect Wnt target. Moreover, we have found that a luciferase reporter construct driven by ECR2 is, like NCE2, very poorly activated by Lef1·β-catenin complexes in transient transfection assays using P19 and N2a cells (data not shown).
Recent work in Xenopus embryos also suggests that Wnt-mediated induction of Pax3 expression at the neural plate border might be controlled by species-specific mechanisms. Indeed, de Crozé et al. (20) have reported that Pax3 expression at the neural plate border is regulated by canonical Wnt signals via both direct and indirect means. This work showed that, although Pax3 can be directly induced by canonical Wnt signals, the transcription factor AP2a is required as an intermediate for full activation (20). The existence of such a mechanism in mice is very unlikely because knock-out of all AP2 isoforms (via deletion of exon 5) has been shown to result in cranial neural crest defects that do not involve reduced Pax3 expression (86).
On the other hand, other studies in Xenopus embryos have revealed that the homeobox gene Gbx2 is a direct downstream target of Wnt·β-catenin signaling acting upstream of Pax3 at the neural plate border (87). Similarly to Cdx genes, Gbx2 is a known posteriorizing gene. However, in marked contrast to Cdx proteins, Gbx2 cannot directly activate Pax3 expression because it acts as a repressor (87, 88). Therefore, in this case, it appears that Gbx2 is required to repress an unknown repressor of Pax3 at the neural plate border. More work will be required to determine whether this mechanism is conserved in mice.
Regulation of Pax3 Expression via NCE2
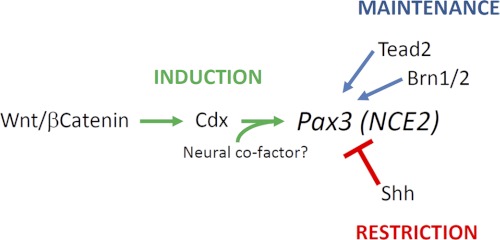
We have shown for the first time that the Pax3 NCE2 alone is sufficient to recapitulate induction as well as dorsal restriction of Pax3 expression in the caudal NT. Taken together with previous work, this suggests that NCE2 is involved in the posterior whereas NCE1 is rather involved in the anterior expression of Pax3 (13–15). As summarized in Fig. 7, our in vitro and in vivo data further indicate that the activity of NCE2 is regulated by the posteriorizing Wnt-Cdx pathway. Given the broad distribution of Cdx proteins in the posterior neurectoderm, it is currently unclear how NCE2 exhibits dorsally restricted activity. As described previously for Pax3 expression, this could first be ensured by a repressive mechanism involving Shh signals emerging from the node, notochord, and floor plate (17), implying that Shh-responsive regulatory sequences are contained within NCE2. Such restricted activity of NCE2 might also be due to an interaction between Cdx proteins and a neural plate border co-factor. In this regard, our transfection assays in N2a cells have suggested that Cdx-mediated transactivation of NCE2 might rely in part on the presence of a neural factor. The identity of such co-factor is currently unknown, and it is most likely not a transcription factor previously reported to act on NCE2 (Tead2 and Brn1/2) (13, 15). Indeed, although both Tead2 and Brn1/2 have been implicated in the regulation of Pax3 expression, their expression pattern is not consistent with a role in the induction of Pax3 expression in vivo. On one hand, Tead2 and its co-factor YAP65 are almost ubiquitously expressed at e8.5–e9.5, becoming restricted to neural tissues only after e10.0 (89, 90). On the other hand, the proneural factors Brn1 and Brn2, as well as other Pou class III members Brn4 and Tst1/Oct6, are not expressed in the PNP (91–96). Thus, these observations strongly suggest that Tead2 and Brn1/2 are involved in the maintenance rather than induction of Pax3 expression. More work will obviously be required to better understand the regulatory mechanisms involved in the dorsal restriction of Pax3 expression, and our data indicate that at least some of them are operating via NCE2.
FIGURE 7.
Control of Pax3 expression in the caudal neurectoderm via NCE2. Induction of Pax3 expression in the posterior neural plate is controlled by the Wnt-Cdx pathway. Expression in the closed neural tube is later maintained by the activity of Tead2 as well as Brn1/2 transcription factors. Restriction of Pax3 expression at the lateral neural plate and dorsal neural tube is ensured by repressive Shh signals emerging from the node, notochord, and floor plate. An unknown neuron-specific Cdx co-factor might also be involved in the spatial restriction of NCE2 activity.
Novel Function for Cdx Proteins in Caudal Neurectoderm Development
Strong Cdx expression in the caudal neurectoderm is highly conserved across chordates. However, Cdx function in this lineage is poorly understood because of functional redundancy. Until recently, Cdx proteins were mostly known for their evolutionarily conserved role in the control of neural AP patterning via Hox-dependent mechanisms (37, 64, 67, 97). Cdx proteins are now also known to regulate neural AP patterning via Hox-independent mechanisms in different species (62, 63). More recently, Cdx1-Cdx2 double knock-out mice were generated and revealed a novel redundant role for Cdx members in the control of NT closure (54). This work showed that Cdx proteins regulate the planar cell polarity gene Ptk7 and further suggested that Cdx members are involved in the regulation of convergent extension movements in the caudal embryo. This analysis involved a conditional mutagenesis approach to circumvent the peri-implantation lethality associated with the Cdx2 null mutation, via a CMV-β-actin-Cre-ERT2 transgene and a floxed allele of Cdx2. Thus, the Cdx function was lost in all three germ layers, and it is uncertain whether this novel Cdx role in neurulation is tissue-autonomous, -non-autonomous, or a combination of both. Ptk7 loss of function in Xenopus appears to affect convergent extension of the neurectoderm (98), but this has not been reported in mice. Indeed, although Ptk7−/− mouse mutants have been shown to have defective convergent extension movements in the mesoderm, an analysis of the neurectoderm has not been reported (99). Therefore, more detailed analysis of Cdx1-Cdx2 double knock-out animals and conditional approaches involving tissue-specific loss of function will be required to clarify the Cdx-dependent processes involved in NT formation.
On the other hand, our work now suggests that Cdx proteins may impact caudal neurectoderm development in a tissue-autonomous manner, at least via the regulation of Pax3 expression. As evidenced by the severe NC and NT defects observed in Pax3Sp/Sp mutants, Pax3 plays a crucial role in the neurectoderm (1, 2). Pax3 is important for NCC induction, and analysis of Pax3-deficient embryos has indicated that NC defects are due to a marked reduction in the number of NCC that emigrate from the NT at cranial levels and a progressive complete loss of NCC at more caudal levels (100–102). This progressive increase in the severity of NC defects along the AP axis reflects the cranial co-expression of the functionally redundant Pax7 (103). Pax3 is also important for NT closure, being required for the survival of dorsal progenitors via down-regulation of p53 activity (6, 7). Thus, by acting upstream of Pax3, the canonical Wnt-Cdx pathway might control cell specification and maintenance of progenitor populations required for proper NC and NT development.
A role for Cdx proteins in NC development has not been formally reported in any species. This is most likely because such a role is masked by functional redundancy and/or the presence of very severe posterior truncation phenotypes in Cdx compound mutants. However, several observations are in agreement with a role for Cdx proteins in NC development as well as the conservation of this role through evolution. In mice, an analysis of NCC in Cdx1-Cdx2 double knockouts has not been reported, but Cdx1−/−Cdx2+/− mutants are known to display abnormal and fused dorsal root ganglia (53). In zebrafish, Cdx loss of function leads to a reduced number of Rohon-Beard cells (which share a common precursor with NCC) and the absence of NC-derived spinal nerve roots (63, 104). In ascidian, Cdx loss of function results in the absence of pigment cells, which are derived from NC-like cells (68, 69). Future work focusing on a more detailed characterization of compound mutants or tissue-specific loss-of-function studies should help validate these observations and confirm a role for Cdx proteins in NC development.
In conclusion, our work suggests that Cdx proteins occupy a strategic position between canonical Wnt signals and Pax3 at the beginning of the gene regulatory cascade controlling NCC development. Because Cdx genes are not expressed in the anterior neurectoderm, the Wnt-Cdx pathway cannot impact cranial NC induction. Therefore, our data are in accordance with the general idea that NCC are intrinsically different along the AP axis (105–108) and strongly suggest that these differences are already in place during induction of NCC.
Supplementary Material
Acknowledgments
We thank Denis Flipo (University of Quebec at Montreal) for the FACS analyses as well as Qinzhang Zhu and Li Lian (Institut de Recherches Cliniques de Montréal) for microinjections. We thank Jonathan Epstein for the Pax3 probe, James B. Jaynes for the Engrailed cDNA, and David Lohnes for the Cdx antibodies as well as Cdx1-null mice.
This work was supported by Canadian Institutes of Health Research Grants DCO190GP and IHD-94366 as well as a new investigator grant from the Banting Research Foundation.

This article contains supplemental Table S1 and Figs. S1 and S2.
- NC
- neural crest
- NT
- neural tube
- NCC
- NC cell(s)
- PNP
- posterior neural plate
- NCE
- neural crest enhancer
- RA
- retinoic acid
- CdxBS
- Cdx binding site
- X-gal
- 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside
- CHX
- cycloheximide
- W3a
- Wnt3a-conditioned medium
- Ctl
- control-conditioned medium
- en
- embryonic day n.
REFERENCES
- 1. Li J., Liu K. C., Jin F., Lu M. M., Epstein J. A. (1999) Transgenic rescue of congenital heart disease and spina bifida in Splotch mice. Development 126, 2495–2503 [DOI] [PubMed] [Google Scholar]
- 2. Auerbach R. (1954) Analysis of the developmental effects of a lethal mutation in the house mouse. J. Exp. Zool. 127, 305–329 [Google Scholar]
- 3. Meulemans D., Bronner-Fraser M. (2004) Gene-regulatory interactions in neural crest evolution and development. Dev. Cell 7, 291–299 [DOI] [PubMed] [Google Scholar]
- 4. Betancur P., Bronner-Fraser M., Sauka-Spengler T. (2010) Assembling neural crest regulatory circuits into a gene regulatory network. Annu. Rev. Cell Dev. Biol. 26, 581–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Monsoro-Burq A. H., Wang E., Harland R. (2005) Msx1 and Pax3 cooperate to mediate FGF8 and WNT signals during Xenopus neural crest induction. Dev. Cell 8, 167–178 [DOI] [PubMed] [Google Scholar]
- 6. Wang X. D., Morgan S. C., Loeken M. R. (2011) Pax3 stimulates p53 ubiquitination and degradation independent of transcription. PloS One 6, e29379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pani L., Horal M., Loeken M. R. (2002) Rescue of neural tube defects in Pax-3-deficient embryos by p53 loss of function. Implications for Pax-3-dependent development and tumorigenesis. Genes Dev. 16, 676–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Baldwin C. T., Hoth C. F., Amos J. A., da-Silva E. O., Milunsky A. (1992) An exonic mutation in the HuP2 paired domain gene causes Waardenburg's syndrome. Nature 355, 637–638 [DOI] [PubMed] [Google Scholar]
- 9. Baldwin C. T., Hoth C. F., Macina R. A., Milunsky A. (1995) Mutations in PAX3 that cause Waardenburg syndrome type I. Ten new mutations and review of the literature. Am. J. Med. Genet. 58, 115–122 [DOI] [PubMed] [Google Scholar]
- 10. Tassabehji M., Read A. P., Newton V. E., Harris R., Balling R., Gruss P., Strachan T. (1992) Waardenburg's syndrome patients have mutations in the human homologue of the Pax-3 paired box gene. Nature 355, 635–636 [DOI] [PubMed] [Google Scholar]
- 11. Goulding M. D., Chalepakis G., Deutsch U., Erselius J. R., Gruss P. (1991) Pax-3, a novel murine DNA binding protein expressed during early neurogenesis. EMBO J. 10, 1135–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Natoli T. A., Ellsworth M. K., Wu C., Gross K. W., Pruitt S. C. (1997) Positive and negative DNA sequence elements are required to establish the pattern of Pax3 expression. Development 124, 617–626 [DOI] [PubMed] [Google Scholar]
- 13. Milewski R. C., Chi N. C., Li J., Brown C., Lu M. M., Epstein J. A. (2004) Identification of minimal enhancer elements sufficient for Pax3 expression in neural crest and implication of Tead2 as a regulator of Pax3. Development 131, 829–837 [DOI] [PubMed] [Google Scholar]
- 14. Chang C. P., Stankunas K., Shang C., Kao S. C., Twu K. Y., Cleary M. L. (2008) Pbx1 functions in distinct regulatory networks to pattern the great arteries and cardiac outflow tract. Development 135, 3577–3586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pruitt S. C., Bussman A., Maslov A. Y., Natoli T. A., Heinaman R. (2004) Hox/Pbx and Brn binding sites mediate Pax3 expression in vitro and in vivo. Gene Expr. Patterns 4, 671–685 [DOI] [PubMed] [Google Scholar]
- 16. Taneyhill L. A., Bronner-Fraser M. (2005) Dynamic alterations in gene expression after Wnt-mediated induction of avian neural crest. Mol. Biol. Cell 16, 5283–5293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Goulding M. D., Lumsden A., Gruss P. (1993) Signals from the notochord and floor plate regulate the region-specific expression of two Pax genes in the developing spinal cord. Development 117, 1001–1016 [DOI] [PubMed] [Google Scholar]
- 18. Bang A. G., Papalopulu N., Kintner C., Goulding M. D. (1997) Expression of Pax-3 is initiated in the early neural plate by posteriorizing signals produced by the organizer and by posterior non-axial mesoderm. Development 124, 2075–2085 [DOI] [PubMed] [Google Scholar]
- 19. Bang A. G., Papalopulu N., Goulding M. D., Kintner C. (1999) Expression of Pax-3 in the lateral neural plate is dependent on a Wnt-mediated signal from posterior nonaxial mesoderm. Dev. Biol. 212, 366–380 [DOI] [PubMed] [Google Scholar]
- 20. de Crozé N., Maczkowiak F., Monsoro-Burq A. H. (2011) Reiterative AP2a activity controls sequential steps in the neural crest gene regulatory network. Proc. Natl. Acad. Sci. U.S.A. 108, 155–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Brooke N. M., Garcia-Fernàndez J., Holland P. W. (1998) The ParaHox gene cluster is an evolutionary sister of the Hox gene cluster. Nature 392, 920–922 [DOI] [PubMed] [Google Scholar]
- 22. Beck F., Erler T., Russell A., James R. (1995) Expression of Cdx-2 in the mouse embryo and placenta. Possible role in patterning of the extra-embryonic membranes. Dev. Dyn. 204, 219–227 [DOI] [PubMed] [Google Scholar]
- 23. Meyer B. I., Gruss P. (1993) Mouse Cdx-1 expression during gastrulation. Development 117, 191–203 [DOI] [PubMed] [Google Scholar]
- 24. Gamer L. W., Wright C. V. (1993) Murine Cdx-4 bears striking similarities to the Drosophila caudal gene in its homeodomain sequence and early expression pattern. Mech. Dev. 43, 71–81 [DOI] [PubMed] [Google Scholar]
- 25. Gaunt S. J., Drage D., Trubshaw R. C. (2005) Cdx4/LacZ and Cdx2/LacZ protein gradients formed by decay during gastrulation in the mouse. Int. J. Dev. Biol. 49, 901–908 [DOI] [PubMed] [Google Scholar]
- 26. Beck F. (2002) Homeobox genes in gut development. Gut 51, 450–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Prinos P., Joseph S., Oh K., Meyer B. I., Gruss P., Lohnes D. (2001) Multiple pathways governing Cdx1 expression during murine development. Dev. Biol. 239, 257–269 [DOI] [PubMed] [Google Scholar]
- 28. Pilon N., Oh K., Sylvestre J. R., Savory J. G., Lohnes D. (2007) Wnt signaling is a key mediator of Cdx1 expression in vivo. Development 134, 2315–2323 [DOI] [PubMed] [Google Scholar]
- 29. Pilon N., Oh K., Sylvestre J. R., Bouchard N., Savory J., Lohnes D. (2006) Cdx4 is a direct target of the canonical Wnt pathway. Dev. Biol. 289, 55–63 [DOI] [PubMed] [Google Scholar]
- 30. Lickert H., Domon C., Huls G., Wehrle C., Duluc I., Clevers H., Meyer B. I., Freund J. N., Kemler R. (2000) Wnt/β-catenin signaling regulates the expression of the homeobox gene Cdx1 in embryonic intestine. Development 127, 3805–3813 [DOI] [PubMed] [Google Scholar]
- 31. Lengerke C., Schmitt S., Bowman T. V., Jang I. H., Maouche-Chretien L., McKinney-Freeman S., Davidson A. J., Hammerschmidt M., Rentzsch F., Green J. B., Zon L. I., Daley G. Q. (2008) BMP and Wnt specify hematopoietic fate by activation of the Cdx-Hox pathway. Cell Stem Cell 2, 72–82 [DOI] [PubMed] [Google Scholar]
- 32. Keenan I. D., Sharrard R. M., Isaacs H. V. (2006) FGF signal transduction and the regulation of Cdx gene expression. Dev. Biol. 299, 478–488 [DOI] [PubMed] [Google Scholar]
- 33. Ikeya M., Takada S. (2001) Wnt-3a is required for somite specification along the anteroposterior axis of the mouse embryo and for regulation of Cdx-1 expression. Mech. Dev. 103, 27–33 [DOI] [PubMed] [Google Scholar]
- 34. Houle M., Sylvestre J. R., Lohnes D. (2003) Retinoic acid regulates a subset of Cdx1 function in vivo. Development 130, 6555–6567 [DOI] [PubMed] [Google Scholar]
- 35. Houle M., Prinos P., Iulianella A., Bouchard N., Lohnes D. (2000) Retinoic acid regulation of Cdx1. An indirect mechanism for retinoids and vertebral specification. Mol. Cell. Biol. 20, 6579–6586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Faas L., Isaacs H. V. (2009) Overlapping functions of Cdx1, Cdx2, and Cdx4 in the development of the amphibian Xenopus tropicalis. Dev. Dyn. 238, 835–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bel-Vialar S., Itasaki N., Krumlauf R. (2002) Initiating Hox gene expression. In the early chick neural tube differential sensitivity to FGF and RA signaling subdivides the HoxB genes in two distinct groups. Development 129, 5103–5115 [DOI] [PubMed] [Google Scholar]
- 38. Shimizu T., Bae Y. K., Muraoka O., Hibi M. (2005) Interaction of Wnt and caudal-related genes in zebrafish posterior body formation. Dev. Biol. 279, 125–141 [DOI] [PubMed] [Google Scholar]
- 39. Benahmed F., Gross I., Gaunt S. J., Beck F., Jehan F., Domon-Dell C., Martin E., Kedinger M., Freund J. N., Duluc I. (2008) Multiple regulatory regions control the complex expression pattern of the mouse Cdx2 homeobox gene. Gastroenterology 135, 1238–1247, 1247.e1–3 [DOI] [PubMed] [Google Scholar]
- 40. He S., Pant D., Schiffmacher A., Meece A., Keefer C. L. (2008) Lymphoid enhancer factor 1-mediated Wnt signaling promotes the initiation of trophoblast lineage differentiation in mouse embryonic stem cells. Stem Cells 26, 842–849 [DOI] [PubMed] [Google Scholar]
- 41. Joo J. H., Taxter T. J., Munguba G. C., Kim Y. H., Dhaduvai K., Dunn N. W., Degan W. J., Oh S. P., Sugrue S. P. (2010) Pinin modulates expression of an intestinal homeobox gene, Cdx2, and plays an essential role for small intestinal morphogenesis. Dev. Biol. 345, 191–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Marikawa Y., Tamashiro D. A., Fujita T. C., Alarcón V. B. (2009) Aggregated P19 mouse embryonal carcinoma cells as a simple in vitro model to study the molecular regulations of mesoderm formation and axial elongation morphogenesis. Genesis 47, 93–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nordström U., Maier E., Jessell T. M., Edlund T. (2006) An early role for WNT signaling in specifying neural patterns of Cdx and Hox gene expression and motor neuron subtype identity. PLoS Biol. 4, e252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Saegusa M., Hashimura M., Kuwata T., Hamano M., Wani Y., Okayasu I. (2007) A functional role of Cdx2 in β-catenin signaling during transdifferentiation in endometrial carcinomas. Carcinogenesis 28, 1885–1892 [DOI] [PubMed] [Google Scholar]
- 45. Zhao X., Duester G. (2009) Effect of retinoic acid signaling on Wnt/β-catenin and FGF signaling during body axis extension. Gene Expr. Patterns 9, 430–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Béland M., Pilon N., Houle M., Oh K., Sylvestre J. R., Prinos P., Lohnes D. (2004) Cdx1 autoregulation is governed by a novel Cdx1-LEF1 transcription complex. Mol. Cell. Biol. 24, 5028–5038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Strumpf D., Mao C. A., Yamanaka Y., Ralston A., Chawengsaksophak K., Beck F., Rossant J. (2005) Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development 132, 2093–2102 [DOI] [PubMed] [Google Scholar]
- 48. Savory J. G., Pilon N., Grainger S., Sylvestre J. R., Béland M., Houle M., Oh K., Lohnes D. (2009) Cdx1 and Cdx2 are functionally equivalent in vertebral patterning. Dev. Biol. 330, 114–122 [DOI] [PubMed] [Google Scholar]
- 49. Savory J. G., Bouchard N., Pierre V., Rijli F. M., De Repentigny Y., Kothary R., Lohnes D. (2009) Cdx2 regulation of posterior development through non-Hox targets. Development 136, 4099–4110 [DOI] [PubMed] [Google Scholar]
- 50. Gao N., White P., Kaestner K. H. (2009) Establishment of intestinal identity and epithelial-mesenchymal signaling by Cdx2. Dev. Cell 16, 588–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Young T., Rowland J. E., van de Ven C., Bialecka M., Novoa A., Carapuco M., van Nes J., de Graaff W., Duluc I., Freund J. N., Beck F., Mallo M., Deschamps J. (2009) Cdx and Hox genes differentially regulate posterior axial growth in mammalian embryos. Dev. Cell 17, 516–526 [DOI] [PubMed] [Google Scholar]
- 52. van Nes J., de Graaff W., Lebrin F., Gerhard M., Beck F., Deschamps J. (2006) The Cdx4 mutation affects axial development and reveals an essential role of Cdx genes in the ontogenesis of the placental labyrinth in mice. Development 133, 419–428 [DOI] [PubMed] [Google Scholar]
- 53. van den Akker E., Forlani S., Chawengsaksophak K., de Graaff W., Beck F., Meyer B. I., Deschamps J. (2002) Cdx1 and Cdx2 have overlapping functions in anteroposterior patterning and posterior axis elongation. Development 129, 2181–2193 [DOI] [PubMed] [Google Scholar]
- 54. Savory J. G., Mansfield M., Rijli F. M., Lohnes D. (2011) Cdx mediates neural tube closure through transcriptional regulation of the planar cell polarity gene Ptk7. Development 138, 1361–1370 [DOI] [PubMed] [Google Scholar]
- 55. Wang Y., Yabuuchi A., McKinney-Freeman S., Ducharme D. M., Ray M. K., Chawengsaksophak K., Archer T. K., Daley G. Q. (2008) Cdx gene deficiency compromises embryonic hematopoiesis in the mouse. Proc. Natl. Acad. Sci. U.S.A. 105, 7756–7761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Subramanian V., Meyer B. I., Gruss P. (1995) Disruption of the murine homeobox gene Cdx1 affects axial skeletal identities by altering the mesodermal expression domains of Hox genes. Cell 83, 641–653 [DOI] [PubMed] [Google Scholar]
- 57. Grainger S., Lam J., Savory J. G., Mears A. J., Rijli F. M., Lohnes D. (2012) Cdx regulates Dll1 in multiple lineages. Dev. Biol. 361, 1–11 [DOI] [PubMed] [Google Scholar]
- 58. Verzi M. P., Shin H., Ho L. L., Liu X. S., Shivdasani R. A. (2011) Essential and redundant functions of caudal family proteins in activating adult intestinal genes. Mol. Cell. Biol. 31, 2026–2039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Suh E., Chen L., Taylor J., Traber P. G. (1994) A homeodomain protein related to caudal regulates intestine-specific gene transcription. Mol. Cell. Biol. 14, 7340–7351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Grainger S., Savory J. G., Lohnes D. (2010) Cdx2 regulates patterning of the intestinal epithelium. Dev. Biol. 339, 155–165 [DOI] [PubMed] [Google Scholar]
- 61. Crissey M. A., Guo R. J., Funakoshi S., Kong J., Liu J., Lynch J. P. (2011) Cdx2 levels modulate intestinal epithelium maturity and Paneth cell development. Gastroenterology 140, 517–528.e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sturgeon K., Kaneko T., Biemann M., Gauthier A., Chawengsaksophak K., Cordes S. P. (2011) Cdx1 refines positional identity of the vertebrate hindbrain by directly repressing Mafb expression. Development 138, 65–74 [DOI] [PubMed] [Google Scholar]
- 63. Skromne I., Thorsen D., Hale M., Prince V. E., Ho R. K. (2007) Repression of the hindbrain developmental program by Cdx factors is required for the specification of the vertebrate spinal cord. Development 134, 2147–2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Isaacs H. V., Pownall M. E., Slack J. M. (1998) Regulation of Hox gene expression and posterior development by the Xenopus caudal homologue Xcad3. EMBO J. 17, 3413–3427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. van de Ven C., Bialecka M., Neijts R., Young T., Rowland J. E., Stringer E. J., Van Rooijen C., Meijlink F., Nóvoa A., Freund J. N., Mallo M., Beck F., Deschamps J. (2011) Concerted involvement of Cdx/Hox genes and Wnt signaling in morphogenesis of the caudal neural tube and cloacal derivatives from the posterior growth zone. Development 138, 3451–3462 [DOI] [PubMed] [Google Scholar]
- 66. Gaunt S. J., Drage D., Trubshaw R. C. (2008) Increased Cdx protein dose effects upon axial patterning in transgenic lines of mice. Development 135, 2511–2520 [DOI] [PubMed] [Google Scholar]
- 67. Charité J., de Graaff W., Consten D., Reijnen M. J., Korving J., Deschamps J. (1998) Transducing positional information to the Hox genes. Critical interaction of cdx gene products with position-sensitive regulatory elements. Development 125, 4349–4358 [DOI] [PubMed] [Google Scholar]
- 68. Mita K., Fujiwara S. (2007) Nodal regulates neural tube formation in the Ciona intestinalis embryo. Dev. Genes Evol. 217, 593–601 [DOI] [PubMed] [Google Scholar]
- 69. Jeffery W. R., Strickler A. G., Yamamoto Y. (2004) Migratory neural crest-like cells form body pigmentation in a urochordate embryo. Nature 431, 696–699 [DOI] [PubMed] [Google Scholar]
- 70. Kothary R., Clapoff S., Darling S., Perry M. D., Moran L. A., Rossant J. (1989) Inducible expression of an hsp68-lacZ hybrid gene in transgenic mice. Development 105, 707–714 [DOI] [PubMed] [Google Scholar]
- 71. Nagy A., Gertsenstein M., Vintersten K., Behringer R. (2003) Manipulating the Mouse Embryo: A Laboratory Manual, pp. 341–358, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 72. Pilon N., Raiwet D., Viger R. S., Silversides D. W. (2008) Novel pre- and post-gastrulation expression of Gata4 within cells of the inner cell mass and migratory neural crest cells. Dev. Dyn. 237, 1133–1143 [DOI] [PubMed] [Google Scholar]
- 73. Kaufman M. H. (1992) The Atlas of Mouse Development, pp. 38–81, Academic Press, London [Google Scholar]
- 74. Jaynes J. B., O'Farrell P. H. (1991) Active repression of transcription by the engrailed homeodomain protein. EMBO J. 10, 1427–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Nordeen S. K. (1988) Luciferase reporter gene vectors for analysis of promoters and enhancers. BioTechniques 6, 454–458 [PubMed] [Google Scholar]
- 76. Shibamoto S., Higano K., Takada R., Ito F., Takeichi M., Takada S. (1998) Cytoskeletal reorganization by soluble Wnt-3a protein signaling. Genes Cells 3, 659–670 [DOI] [PubMed] [Google Scholar]
- 77. Colombres M., Henríquez J. P., Reig G. F., Scheu J., Calderón R., Alvarez A., Brandan E., Inestrosa N. C. (2008) Heparin activates Wnt signaling for neuronal morphogenesis. J. Cell. Physiol. 216, 805–815 [DOI] [PubMed] [Google Scholar]
- 78. Shi F., Cheng Y. F., Wang X. L., Edge A. S. (2010) β-Catenin up-regulates Atoh1 expression in neural progenitor cells by interaction with an Atoh1 3′ enhancer. J. Biol. Chem. 285, 392–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Petropoulos H., Skerjanc I. S. (2002) β-Catenin is essential and sufficient for skeletal myogenesis in P19 cells. J. Biol. Chem. 277, 15393–15399 [DOI] [PubMed] [Google Scholar]
- 80. Savory J. G., Mansfield M., St Louis C., Lohnes D. (2011) Cdx4 is a Cdx2 target gene. Mech. Dev. 128, 41–48 [DOI] [PubMed] [Google Scholar]
- 81. Degenhardt K. R., Milewski R. C., Padmanabhan A., Miller M., Singh M. K., Lang D., Engleka K. A., Wu M., Li J., Zhou D., Antonucci N., Li L., Epstein J. A. (2010) Distinct enhancers at the Pax3 locus can function redundantly to regulate neural tube and neural crest expressions. Dev. Biol. 339, 519–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Huang B. L., Brugger S. M., Lyons K. M. (2010) Stage-specific control of connective tissue growth factor (CTGF/CCN2) expression in chondrocytes by Sox9 and β-catenin. J. Biol. Chem. 285, 27702–27712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Kormish J. D., Sinner D., Zorn A. M. (2010) Interactions between SOX factors and Wnt/β-catenin signaling in development and disease. Dev. Dyn. 239, 56–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Kuwabara T., Hsieh J., Muotri A., Yeo G., Warashina M., Lie D. C., Moore L., Nakashima K., Asashima M., Gage F. H. (2009) Wnt-mediated activation of NeuroD1 and retro-elements during adult neurogenesis. Nat. Neurosci. 12, 1097–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Liu X., Luo M., Xie W., Wells J. M., Goodheart M. J., Engelhardt J. F. (2010) Sox17 modulates Wnt3A/β-catenin-mediated transcriptional activation of the Lef-1 promoter. Am. J. Physiol. Lung Cell. Mol. Physiol. 299, L694–L710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Schorle H., Meier P., Buchert M., Jaenisch R., Mitchell P. J. (1996) Transcription factor AP-2 essential for cranial closure and craniofacial development. Nature 381, 235–238 [DOI] [PubMed] [Google Scholar]
- 87. Li B., Kuriyama S., Moreno M., Mayor R. (2009) The posteriorizing gene Gbx2 is a direct target of Wnt signaling and the earliest factor in neural crest induction. Development 136, 3267–3278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Heimbucher T., Murko C., Bajoghli B., Aghaallaei N., Huber A., Stebegg R., Eberhard D., Fink M., Simeone A., Czerny T. (2007) Gbx2 and Otx2 interact with the WD40 domain of Groucho/Tle corepressors. Mol. Cell. Biol. 27, 340–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Yasunami M., Suzuki K., Houtani T., Sugimoto T., Ohkubo H. (1995) Molecular characterization of cDNA encoding a novel protein related to transcriptional enhancer factor-1 from neural precursor cells. J. Biol. Chem. 270, 18649–18654 [DOI] [PubMed] [Google Scholar]
- 90. Sawada A., Nishizaki Y., Sato H., Yada Y., Nakayama R., Yamamoto S., Nishioka N., Kondoh H., Sasaki H. (2005) Tead proteins activate the Foxa2 enhancer in the node in cooperation with a second factor. Development 132, 4719–4729 [DOI] [PubMed] [Google Scholar]
- 91. Bouchard M., Grote D., Craven S. E., Sun Q., Steinlein P., Busslinger M. (2005) Identification of Pax2-regulated genes by expression profiling of the mid-hindbrain organizer region. Development 132, 2633–2643 [DOI] [PubMed] [Google Scholar]
- 92. He X., Treacy M. N., Simmons D. M., Ingraham H. A., Swanson L. W., Rosenfeld M. G. (1989) Expression of a large family of POU-domain regulatory genes in mammalian brain development. Nature 340, 35–41 [DOI] [PubMed] [Google Scholar]
- 93. Heydemann A., Nguyen L. C., Crenshaw E. B., 3rd (2001) Regulatory regions from the Brn4 promoter direct LACZ expression to the developing forebrain and neural tube. Brain Res. Dev. Brain Res. 128, 83–90 [DOI] [PubMed] [Google Scholar]
- 94. Sugitani Y., Nakai S., Minowa O., Nishi M., Jishage K., Kawano H., Mori K., Ogawa M., Noda T. (2002) Brn-1 and Brn-2 share crucial roles in the production and positioning of mouse neocortical neurons. Genes Dev. 16, 1760–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Mathis J. M., Simmons D. M., He X., Swanson L. W., Rosenfeld M. G. (1992) Brain 4. A novel mammalian POU domain transcription factor exhibiting restricted brain-specific expression. EMBO J. 11, 2551–2561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Monuki E. S., Kuhn R., Weinmaster G., Trapp B. D., Lemke G. (1990) Expression and activity of the POU transcription factor SCIP. Science 249, 1300–1303 [DOI] [PubMed] [Google Scholar]
- 97. Shimizu T., Bae Y. K., Hibi M. (2006) Cdx-Hox code controls competence for responding to Fgfs and retinoic acid in zebrafish neural tissue. Development 133, 4709–4719 [DOI] [PubMed] [Google Scholar]
- 98. Wehner P., Shnitsar I., Urlaub H., Borchers A. (2011) RACK1 is a novel interaction partner of PTK7 that is required for neural tube closure. Development 138, 1321–1327 [DOI] [PubMed] [Google Scholar]
- 99. Yen W. W., Williams M., Periasamy A., Conaway M., Burdsal C., Keller R., Lu X., Sutherland A. (2009) PTK7 is essential for polarized cell motility and convergent extension during mouse gastrulation. Development 136, 2039–2048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Conway S. J., Bundy J., Chen J., Dickman E., Rogers R., Will B. M. (2000) Decreased neural crest stem cell expansion is responsible for the conotruncal heart defects within the splotch (Sp(2H))/Pax3 mouse mutant. Cardiovasc. Res. 47, 314–328 [DOI] [PubMed] [Google Scholar]
- 101. Epstein J. A., Li J., Lang D., Chen F., Brown C. B., Jin F., Lu M. M., Thomas M., Liu E., Wessels A., Lo C. W. (2000) Migration of cardiac neural crest cells in Splotch embryos. Development 127, 1869–1878 [DOI] [PubMed] [Google Scholar]
- 102. Olaopa M., Zhou H. M., Snider P., Wang J., Schwartz R. J., Moon A. M., Conway S. J. (2011) Pax3 is essential for normal cardiac neural crest morphogenesis but is not required during migration nor outflow tract septation. Dev. Biol. 356, 308–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Mansouri A., Stoykova A., Torres M., Gruss P. (1996) Dysgenesis of cephalic neural crest derivatives in Pax7−/− mutant mice. Development 122, 831–838 [DOI] [PubMed] [Google Scholar]
- 104. Epperlein H. H., Selleck M. A., Meulemans D., McHedlishvili L., Cerny R., Sobkow L., Bronner-Fraser M. (2007) Migratory patterns and developmental potential of trunk neural crest cells in the axolotl embryo. Dev. Dyn. 236, 389–403 [DOI] [PubMed] [Google Scholar]
- 105. Abzhanov A., Tzahor E., Lassar A. B., Tabin C. J. (2003) Dissimilar regulation of cell differentiation in mesencephalic (cranial) and sacral (trunk) neural crest cells in vitro. Development 130, 4567–4579 [DOI] [PubMed] [Google Scholar]
- 106. Le Douarin N. M., Creuzet S., Couly G., Dupin E. (2004) Neural crest cell plasticity and its limits. Development 131, 4637–4650 [DOI] [PubMed] [Google Scholar]
- 107. Lwigale P. Y., Conrad G. W., Bronner-Fraser M. (2004) Graded potential of neural crest to form cornea, sensory neurons, and cartilage along the rostrocaudal axis. Development 131, 1979–1991 [DOI] [PubMed] [Google Scholar]
- 108. Thibaudeau G., Holder S., Gerard P. (1998) Anterior/posterior influences on neural crest-derived pigment cell differentiation. Pigment Cell Res. 11, 189–197 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.