Abstract
Recent studies have established an important role for rare genomic deletions and duplications in the etiology of schizophrenia. This research suggests that the genetic architecture of neuropsychiatric disorders includes a constellation of rare mutations in many different genes. Mutations that confer substantial risk for schizophrenia have been identified at several loci, most of which have also been implicated in other neurodevelopmental disorders, including autism. Genetic heterogeneity is a characteristic of schizophrenia; conversely, phenotypic heterogeneity is a characteristic of all schizophrenia-associated mutations. Both kinds of heterogeneity probably reflect the complexity of neurodevelopment. Research strategies must account for both genetic and clinical heterogeneity to identify the genes and pathways crucial for the development of neuropsychiatric disorders.
A resurgence of the field of schizophrenia genetics
Genes play an important role in the etiology of schizophrenia, with a heritability estimated at ~ 80% 1. Despite intensive effort to discover genetic risk factors for schizophrenia, causal variants have eluded definitive identification 2–5. In the past, linkage studies were confounded by an under-appreciated degree of locus heterogeneity, yielding weak signals at many locations throughout the genome, the bulk of which did not replicate consistently across studies 6. Candidate gene-based analyses of common variants were also largely unsuccessful 2–5, 7. The first wave of genome-wide association studies produced variable results,8–10 confounded by a lack of statistical power and the extremely small effect sizes of common risk alleles.
In the past two years this trend has reversed. Studies by several groups have begun to shed new light on the genetics of schizophrenia. Recent findings have established that both rare mutations of large effect 11–15 and common variants of modest effect 16–19 contribute to genetic risk for schizophrenia. Collectively, these studies show that schizophrenia is characterized by much more genetic heterogeneity than was previously thought. The risk alleles that have been implicated include rare copy number variants (CNVs) and common haplotypes based on single nucleotide polymorphisms (SNPs). Mutations that confer risk are located throughout the genome and involve many different genes.
What may represent the greatest change in our scientific understanding of schizophrenia is the recognition that individually rare genetic variants at many loci play a role in the etiology of schizophrenia. This discovery is primarily based on findings that emerged from early cytogenetic studies and recent studies of copy number variation (CNV). Here we explore the role of rare structural variants in schizophrenia and discuss the wider implications for psychiatric research.
The CNV-based approach
Copy number variants (CNVs) are large (typically >1000 bp) deletions and duplications of the genome that vary in copy number among individuals in the population 20–22. Other classes of structural variation include balanced rearrangements, such as inversions and balanced translocations. Once considered to be anomalies that were rare among healthy individuals, CNVs are now recognized as a source of inter-individual genetic variation 21–26.
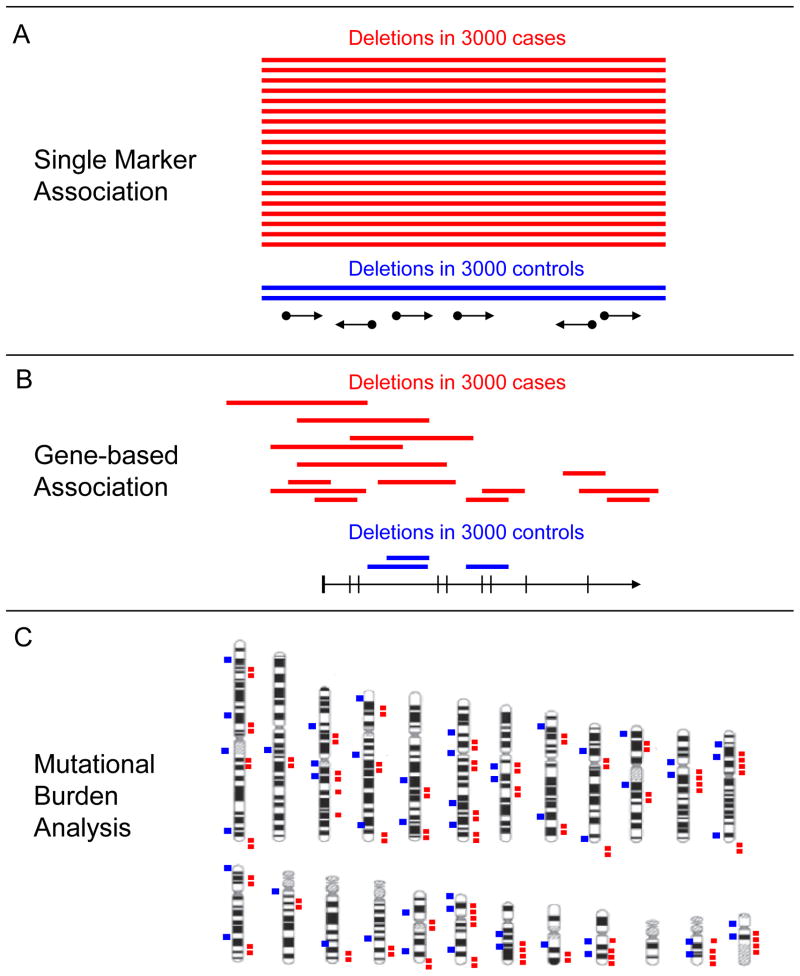
CNV analysis is a mutation discovery approach, similar in nature to other rare variant approaches, such as candidate gene re-sequencing 27–35, Because CNV analysis is typically applied genome-wide, this approach permits researchers to look for disease association in multiple ways. This includes single-marker association (examining the frequencies of individual variants); gene-based association (examining the frequency of multiple individually rare mutations in the same genes); and genome-wide mutational burden analysis (examining the aggregate frequency of all rare CNVs in patients and controls) (Figure 1). Using these technologies, recent studies have shown that individually rare structural variants are associated with schizophrenia 11–13, 36 and other neurocognitive disorders 20–22.
Figure 1. Detecting the association of rare structural variants (SVs) with disease.
Three different approaches to detecting the association of rare SVs with disease are shown. The association of rare structural variants with disease can be determined by comparing cases and controls with respect to the frequency of an individual mutation (single marker association, Panel a), the aggregate frequency of multiple mutations within a single gene or region (gene-wise mutational burden, Panel b), or the aggregate frequency of all rare structural variants (genome-wide mutational burden, Panel c). Panels A and B depict a typical genome browser view displaying tracks for SVs and annotated genes, c depicts an ideogram displaying the distribution of SVs genome-wide.
Rare structural variants play an important role in the etiology schizophrenia
Early evidence implicating structural abnormalities in schizophrenia came from cytogenetic studies 37–40. Seminal examples include a translocation of the gene disrupted in schizophrenia (DISC1) identified in a large Scottish pedigree 41 and a microdeletion of Chromosome 22q11.2 which is the underlying cause of Velocardiofacial syndrome and confers a substantial risk of schizophrenia42. The evidence for these individual variants was convincing. However, because such mutations accounted for a small proportion of cases, it was not widely anticipated that rare structural variants as a class would contribute significantly to the genetic risk for schizophrenia.
Further evidence that rare variants contributed to disease risk required the development of more sensitive microarray-based methods for analysis of CNVs 43–46. Multiple independent studies strongly suggest a role for individually rare structural variants in schizophrenia patients. This line of research has uncovered several findings. First, the genome-wide burden of rare structural variants is significantly greater in patients than in healthy controls. Second, recurrent deletions or duplications at different loci are significantly over-represented in schizophrenia. Finally, many of the structural variants that are associated with schizophrenia are not located in ’hotspots’ in the genome, but are dispersed throughout the genome and involve many different genes.
Mutational burden analysis
In 2008, a series of studies by multiple groups applied the first systematic tests of the hypothesis that rare copy number variants contribute to risk of schizophrenia. This was done by comparing the total genome-wide mutational burden of rare CNVs in patients with that of matched controls (Figure 1c). Assuming that patterns of rare non-causal variation are similar in patients and controls, an enrichment of causal rare variants in cases would be predicted to increase the total genomic burden of rare CNVs.
Several studies have shown that the mutational burden of rare structural variants is significantly greater in patients with schizophrenia than in healthy controls. One study, by our group, examined rare CNVs of >100 Kb from constitutional genomic DNA of 150 cases and 258 controls. Rare deletions and duplications were detected in 5% of healthy controls compared with 15% of adult-onset schizophrenia patients and 20% of patients with disease onset by 18 years of age, a 3.4-fold and 4.8-fold enrichment, respectively 11. This result was replicated in a sample of 83 trios (consisting of a patient and both parents), in which a 2.6-fold enrichment of rare structural variants was observed in patients compared with the untransmitted parental chromosomes. An increased mutational burden of rare CNVs in schizophrenia was also observed by two independent groups 12, 13. The International Schizophrenia Consortium (ISC)(http://pngu.mgh.harvard.edu/isc/), studying several independent cohorts, found that affected individuals overall had a 1.15-fold increased risk of harboring a rare CNV compared with ancestry-matched controls. The strongest effects were observed for variants that contained genes and for events found in only one individual 12. These findings have been replicated in more recent studies of schizophrenia 15 and bipolar disorder 47. One study did not replicate the association of rare CNVs with schizophrenia 48 and in another study, the increased frequency of rare CNVs in patients was observed for the largest (>2 Mb) CNVs, but not for all rare variants >100 Kb 49.
In a family-based study, Xu and colleagues observed an 8-fold increase in frequency of de novo CNVs in patients from families with sporadic schizophrenia compared with healthy controls 36. By contrast, rare de novo CNVs were not observed in any patients from families with multiple cases of schizophrenia. These results are consistent with previous family-based studies of autism spectrum disorders (ASDs) 50, 51, and suggest that rare de novo mutations account for some proportion of sporadic cases. In a more recent study by Xu et al, the contribution of inherited structural variants in this cohort was found to be greatest in patients with a positive family history of mental illness 52. These results are consistent with rare CNVs playing an etiological role in sporadic and familial schizophrenia.
Thus far, case-control and family-based studies 11, 12, 36, 50 indicate that mutations contributing to schizophrenia are not limited to a small number of “hotspots,” but instead involve many different genes throughout the genome. Some of these rare mutations are recurrent, but most are observed in only a single patient, and it is this latter class of mutations that shows the strongest association with disease 11, 12. These findings are consistent with the idea that individually rare mutations in many different genes, some of which are unique to a single patient, are genetic risk factors for schizophrenia.
Large deletions and duplications confer significant risk for schizophrenia
In large cohorts of patients with schizophrenia, some genomic ’hotspots‘ harbor multiple structural variants strongly associated with the disease. For example, deletions at four different loci, 1q21.112, 13, 15q11.213, 15q13.312, 13 and 22q11.2 12, 37 are significantly over-represented in schizophrenia patients compared with controls. More recent studies have identified significant associations with microduplications of 16p11.2 53, and 16p13.1 54 and microdeletions of the gene neurexin-1 (NRXN1) 14, 55 All of these structural variants were found to confer substantial risk. Reported odds ratios for deletions of 1q21.1 and 15q.13.3 were 6.6–14.8 and 11.5–17.9, respectively (see table 1 for more details). Odds ratios for the microduplications of 16p11.2 and 16p13.1 were 8.3–25.8 and 3.0, respectively. The odds ratio for deletions of neurexin-1 (NRXN1) in schizophrenia was 9.0 14. Several other candidate regions for schizophrenia have been identified based on suggestive evidence from CNV studies. These include deletions of the genes Contactin Associated Protein-like 2 (CNTNAP2) 56, a duplication spanning the Amyloid beta (A4) Precursor Protein-Binding A2 (APBA2) 55, and balanced translocations of the genes Phosphodiesterase 4B (PDE4B) 39 and Neuronal PAS-domain protein 3 (NPAS3) 40. These and other findings provide promising avenues for further research.
Table 1.
Genomic regions implicated by rare structural variants in schizophreniaa.
| Locusb | Gene(s)c,d | copy number change | Frequency in SCZ (%) | OR | Reference | Other Associated Disordersd |
|---|---|---|---|---|---|---|
| Replicated significant associations from case-control studies | ||||||
| 1q21.1 | ~10 genes | deletion | 0.23–0.32 | 6.6–14.8 | [12,13] | DD, CM60, 70 |
| 15q13.3 | ~10 genes | deletion | 0.17–0.3 | 11.5–17.9 | [12,13] | GE71, 72, MR73 |
| 16p11.2 | >25 genes | duplication | 0.3 | 8.3–25.4 | [11,53] | ASD, BD, MD, P-NOS53, 62, VCFS42, Anxiety, Depression, ADHD, OCD69 |
| 22q11.2 | >25 genes | deletion | 0.5–2.0 | 30 | [12,13,37] | |
| 2p16.3 | NRXN1 | deletion | 0.47 | 9.0 | [14,55] | ASD 65, 66, 109 |
| Significant association reported in individual studies | ||||||
| 15q11.2 | ~10 genes | deletion | 0.55 | 2.73 | [13] | ASD, DD 110 |
| 17p12 | ~10 genes | deletion | 0.13 | 7.8 | [15] | HNPP 111 |
| 16p13.1 | ~14 genes | duplication | 0.30 | 3.3 | [54] | DD64, 112 |
| 1q42.2 | DISC1 | balanced translocation | NA | NA | [38] | BD, MDD41 |
| Nonsignificant associations reported in individual studies | ||||||
| 1p31.3 | PDE4B | balanced translocation | NA | NA | [39] | |
| 14q13.1 | NPAS3 | balanced translocation | NA | NA | [40] | |
| 7q35 | CNTNAP2 | deletion | NA | NA | [56] | ADHD113, ASD 114–116 |
| 15p13.1 | APBA2 | duplication | NA | NA | [55] | |
Abbreviations: Autism Spectrum Disorder (ASD), Developmental Delay (DD), Congenital Malformations (CM), Generalized Epilepsy (GE), Mental Retardation (MR), Bipolar Disorder (BD), Psychosis Not Otherwise Specified (P-NOS), Velo-Cardio-Facial Syndrome (VCFS), Attention-Deficit Hyperactivity Disorder (ADHD), Obsessive Compulsive Disorder (OCD), Hereditary Neuropathy with Liability to Pressure Palsies (HNPP), Major Depressive Disorder (MD)
Approximate genomic coordinates (hg18) for the six large CNV regions are chr1:144,900,000–146,000,000 (1q21.1), chr15:20300000–20700000 (15q11.2), chr15:28,800,000–30,300,000 (15q13.3), chr16:29,500,000–30,100,000 (16p11.2), chr16:14,890,000–16,390,000 (16p13.1), chr17:14,000,000–15,500,000 (17p12), and chr22:17,200,000–18,700,000 (22q11.2).
Number of genes is approximate because breakpoints of the rearrangements fall within complex clusters of duplicated genes, and the exact boundaries of the rearrangements vary between patients.
The full names of genes in Table 1 are Neurexin-1 (NRXN1), Disrupted in Schizophrenia-1 (DISC1), Phosphodiesterase 4B (PDE4B), Neuronal PAS Domain Protein 3 (NPAS3), Contactin Associated Protein-2 (CNTNAP2) and Amyloid A4 Precursor Protein-Binding, Family A, Member 2 (APBA2).
All CNVs that have shown a significant and replicated association (see Table 1) are located within regions of genome that are unstable. Disease-associated mutations are observed frequently at these loci because they are subject to high rates of structural mutation (as high as one mutation in 10 000 individuals) 23. Regions of instability at 1q21.1, 15q11,1, 15q13.3, 16p11.2, 16p13.1 and 22q11.2 are flanked by tandem segmental duplications, which mediate rearrangements by non-allelic homologous recombination (NAHR) 23, 37, 57–59. Recurrent de novo rearrangements have been detected in families at all 5 of these loci 60, 61, 50, 62–64 and the same is true for the gene NRXN1 at 2p16.3 14, 65, 66. These results suggest that the high frequency of CNVs that are observed at these loci in patients is due to a high rate of spontaneous mutation.
Collectively, these seven hotspots account for a small but measurable fraction of schizophrenia cases (~2–4%, Table 1). However, these few loci do not account for most of the increased CNV burden in schizophrenia. For example, when the most significant individual loci in the ISC study were removed from the mutational burden analysis, the effect was not greatly diminished 12. This suggests that CNVs at other loci account for most of the effect. In other words ’warm spots‘, with low or intermediate rates of mutation (10−6–10−5) could outnumber the ‘hotspots’ with high mutation rates (10−5–10−4). The prevalence of causal CNVs at any one such region is likely to be less than one in 500 patients. Therefore, very large family-based and case-control studies are needed to provide statistical evidence for the involvement of any single event.
Implications for the genetic architecture of schizophrenia
One disorder involves a multiplicity of genes and risk alleles
Rare mutations at many loci throughout the genome contribute to the etiology of schizophrenia. These findings are consistent with a multiple rare variant (MRV) model of schizophrenia 2, 30, 67, and have contributed to a shift in the prevailing view. An exclusively polygenic model that explains the genetic basis of this disease by the combined action of common genetic variants with modest effects 68 is no longer realistic.
Although, the identification of common risk alleles in schizophrenia has been a challenge, this endeavor has yielded some intriguing new findings. Statistical evidence for the contribution of common genes of small effect has come from recent large GWAS studies. The major histocompatibility (MHC) locus on 6p22.1 was implicated in three different studies 16–18. In addition, one study has reported a genome-wide significant association of the transcription factor ZNF804A with schizophrenia 19. Furthermore, the study by Purcell et al 16 addressed the aggregate role of common polygenic variation. By defining a large number of ’score‘ alleles (i.e. putative risk alleles) based on modest statistical evidence from a primary sample, Purcell et al. 16 estimated that the same SNPs explained ~30% of the variance in risk in multiple independent cohorts of schizophrenia (based on Nagelkerke’s pseudo R-squared from logistic regression). These results, if correct, suggest that the population of individuals with schizophrenia is enriched for a large number (hundreds or possibly thousands) of different common alleles, each contributing a very modest effect.
The relative importance of common variants and rare variants in the etiology of schizophrenia is not known, and the scientific debate over this issue is polarized. Thus, the results of GWAS studies have been met with excitement by some and with skepticism by others in the community (http://www.schizophreniaforum.org/new/detail.asp?id=1532). The truth will ultimately be determined based on empirical evidence. Having demonstrated a genetic contribution from both classes of variation is only the first step. We must keep in mind that the value of these two genetic approaches will also be judged based on how much the risk variants they uncover teach us about disease process. It is now essential to determine the underlying biological mechanisms by identifying more of the individual loci involved and determining how variation (whether rare or common) at these loci influence the function genes and neurobiological pathways.
The rare variant and polygenic models should therefore be regarded as complementary rather than as mutually exclusive. Rare variants play a role in disease etiology, and include mutations that confer high risk, but these are not classical mendelian causes of disease. Schizophrenia is a multifactorial disorder. One or more mutations of large effect can exert a major influence on the disease phenotype in an individual patient. The disease phenotype of the same individual may[s1] [E2]also be influenced by common genetic variants and other factors, including epigenetic variation and environmental exposures. In our view, such modifiers, whether environmental, epigenetic, or genetic, will be easier to characterize after the genes of major effect are identified and their biological actions are better understood.
One mutation can lead to multiple disorders
Some structural variants confer risk for schizophrenia and for other disease phenotypes, suggesting that there is high phenotypic variability even for mutations with high odds ratios. For instance, phenotypic variability has been observed among mutation carriers within the same family. In a large Scottish pedigree harboring a translocation of DISC1, clinical presentation was variable among the 29 relatives with the translocation and included people with schizophrenia, bipolar disorder, major depressive disorder and those without mental illness 41. Within the families of five schizophrenic patients with microduplications of 16p11.2, other mutation carriers had diagnoses of schizophrenia, bipolar disorder, psychosis-NOS and major depression 53.
The clinical variability of schizophrenia-associated CNVs is also evident from the fact that some CNVs are associated with disease in studies of different neuropsychiatric or developmental disorders. All of the CNVs that are statistically over-represented in schizophrenia are also significantly associated with other disease phenotypes, as listed in table 1. For example, the 22q11 microdeletion is associated with schizophrenia and Velocardiofacial syndrome 42, as well as anxiety, depression, attention-deficit hyperactivity disorder, obsessive-compulsive disorder and autism spectrum disorders 69. This is also the case for other CNVs that have been recently identified. Deletions and duplications at 1q21 are associated with schizophrenia 12, 13 and multiple pediatric phenotypes, including developmental delay and congenital malformations 60, 70. Deletions at 15q13.3 are associated with schizophrenia, generalized epilepsy 71, 72 and mental retardation 73. Similarly, microduplications of 16p11.2 were recently found to be significantly associated with schizophrenia, autism, and bipolar disorder 53, 62. One study has examined a set of 28 candidate loci across multiple disorders and found CNVs at these loci to be significantly enriched in autism, schizophrenia and mental retardation compared with controls 74. These results are consistent with the idea that schizophrenia and other disorders share overlapping biological pathways.
It is intriguing that some of the same mutations are associated with pediatric developmental disorders and adult neuropsychiatric disorders, suggesting that the phenotype of the individual might in part be dependent on the age at ascertainment. Prior to the onset of schizophrenia, ’prodromal‘ symptoms of cognitive impairment could meet criteria for another clinical diagnosis, such as autism. This is clearly the case for patients with microdeletion of 22q11.2 and Velocardiofacial syndrome. The same might be true for other schizophrenia-associated microdeletions and microduplications discussed in this review.
Given the complexities of neurodevelopment, it is not surprising that the impact of structural variants is variable. In principle, expressivity could be modified by additional genetic events, either gene-specific or global epigenetic events, environmental exposures, or stochastic events during development. It will be important to determine how severe mutations and their modifiers combine to influence variable expressivity.
Implications for psychiatric research
Strategies for gene discovery
The growing evidence for the role of rare variants in psychiatric disorders comes predominantly from studies of relatively large structural variants (CNVs >100 Kb). A large CNV might represent only one of several types of risk alleles that can affect the same locus. The others are likely to be sequence variants, indels or smaller CNVs. Therefore, to further explore the rare genetic causes of schizophrenia, it will be necessary to capture a greater fraction of all genetic variation.
A wide spectrum of genetic variants, common and rare, can be ascertained using whole-genome sequence data from an individual 75. The availability of next generation sequencing platforms and whole genome sequence data from multiple individuals 76–78 (http:www.1000genomes.org), has spurred a rapid development of new methods for the detection of point mutations 16, 17, 76–78, indels 18, 76–78, and structural variants large and small 18, 25, 79–82. These advances have brought the goal of complete genome sequencing in large patient samples within reach. However, given the cost of a single 20X genome, which is ~ $10 000–$20 000 (as of August 2009), the cost for an ambitious ’schizophrenia genome project‘ involving thousands of samples would require an investment of hundreds of millions of dollars. Cheaper alternatives could also be considered. A more focused and more affordable approach would be to sequence a set of candidate genes. The development of new methods for capturing and sequencing targeted subsets of the genome 83, 84 has made it feasible to re-sequence either specific candidate regions or the complete human exome. The candidate gene resequencing approach has proven effective in identifying rare variants that influence colorectal cancer risk 33, HDL cholesterol levels 31 and hypertension 27. Based on the success of the CNV-based mutation discovery approach, there is now a strong rationale for applying the complementary sequencing-based approach to schizophrenia and other psychiatric disorders.
Strategies for studying the neurobiology of disease
Mutations that confer high risk for brain disorders are well suited for the development of genetic model systems. A severe mutation in a gene, such as a deletion, frameshift, or stop mutation can be readily introduced into an animal model. Furthermore, a mutation associated with a severe human phenotype, such as significant cognitive impairment, might be more likely to produce a recognizable phenotype in an animal. For these reasons, disease-associated CNVs have inspired the development of new model systems for the study of the disease process. For example, a chromosome-engineered transgenic mouse has been developed for the 22q11.2 microdeletion, and this mouse shows significant impairments in fear conditioning and spatial learning 85. In addition, analysis of gene expression in the prefrontal cortex and hippocampus of the 22q11.2 mouse has showed evidence for alterations in microRNA biogenesis that might be due to haploinsufficiency of the gene DGCR8 85. Chromosome-engineered mouse models have been made for other genomic disorders, including the deletion of 17q11.2 in Smith-Magenis Syndrome (SMS)86, duplication of 17p12 in Charcot Marie Tooth disease (CMT1A) 87 and duplication of 15q11-13 in autism 88. These models recapitulate some aspects of the human clinical phenotype associated with each mutation.
Studies of individual gene knockouts have also proven helpful in characterizing the biological effects of structural variants. For example, characterization of individual gene knockouts in 17q11.2 has identified a dosage sensitive gene, RAI, that is likely responsible for developmental abnormalities in SMS 86, 89. A mouse knockout of NRXN1 displays abnormalities in neurotransmitter release 90, and these effects could have relevance to psychiatric phenotypes associated with this gene. Mouse knockouts of Methyl-CpG-binding protein 2 (MECP2) 91 and Fragile X Mental Retardation Protein-1 (FMR1) 92 recapitulate in mouse aspects of the human phenotype of Rett’s Syndrome and Fragile X syndrome, respectively.
Animal models of structural variants have also been used to test new therapeutic strategies. For instance, it has been shown that ascorbic acid normalizes impaired locomotor activity in CMT1A mice 93 by inhibiting cAMP dependent overexpression of Peripheral Myelin Binding Protein-22 (PMP22) 94. Potential therapeutics have also been identified using single gene knockouts. Using a FMR1 knockout model of fragile-X syndrome, multiple groups have shown that inhibition of the Metabotropic Glutamate Receptor-5 (mGluR5) rescues aspects of the fragile-X phenotype 95–97. This finding has led to the identification of the mGluR5 inhibitor, MPEP, as a potential treatment for Fragile X syndrome 95. Thus, compounds that have therapeutic effects in animal models could make promising leads for drug development.
Another potential approach to studying the functional effects of disease mutations is through the use of human cells derived from patients that carry specific mutations. For instance, studies of gene expression have been carried out using lymphocytes derived from patients with Fragile-X syndrome and microduplications of 15q11-13 98. However because lymphocytes may not be the ideal tissue for the study of brain disorders, other approaches are being considered. For example, new methods for deriving pluripotent cells from human tissue have made it possible to obtain induced plutipotent stem cells (iPSCs) from humans 99, 100. Human iPS cells can in turn be differentiated into several different neuronal cell types, which would make it possible to study cultured neurons from specific patients. To this end some groups have used this approach to derive cells from patients with known disorders, such as Rett Syndrome 101. The same approach could be applied to specific groups of patients with neuropsychiatric disorders who share a risk variant identified from CNV studies.
Implications for clinical research
Phenotypic heterogeneity among patients has been a major factor confounding clinical research on brain disorders. Inconsistent results from human studies of cognition 102, 103 and brain imaging 104, 105 could in part be explained by the underlying genetic heterogeneity and corresponding phenotypic heterogeneity. Therefore, stratifying patients by specific CNV genotypes could enhance power to identify clinical correlates of individual mutations. Intensive clinical studies of patients who share a specific structural variant have helped to define novel microdeletion syndromes 73, 106, 107 that were not recognizable based on symptomatology. The same principle could be applied within any heterogenous patient group, including a schizophrenia cohort.
Two groups have recently analyzed clinical data from individuals with rearrangements of, 1q21.1 60 and 16p11.2 53. Remarkably, both studies observed that the reciprocal rearrangements at these loci were associated with changes in head circumference. Microdeletions and microduplications of 1q21.1 were associated with microcephaly and macrocephaly respectively 60, and microdeletions of 16p11.2 were associated with a significantly increased head size 53. These results suggest that changes in brain volume might be related to the psychiatric phenotype in patients that carry these mutations.
More generally, these studies demonstrate that some schizophrenia-associated CNVs can be associated with specific clinical endophenotypes. However, in this context, the analysis strategy differs from the classical endophenotype approach 108. For rare variants, researchers tend to follow a traditional cytogenetics approach, where analysis of clinical data is performed retrospectively on a small genetically-defined patient group. This approach is effective because genetic heterogeneity is reduced considerably within a small group of patients who share a disease-associated rare CNV.
Concluding remarks
The recent identification of mutations that confer substantial risk for schizophrenia has generated much excitement within the field. However, there is much more to be learned. Very few of the individual rare variants have been definitively identified. In addition, the relative contributions of rare variants of large effect and common alleles with modest effects are not known. It is clear, however, that the role of rare variants is not limited to a small percentage of patients. Further studies on a much larger scale are needed to uncover the rare genetic variants that underlie schizophrenia.
Statistical power to detect the association of rare variants in a gene depends not only on sample size, but also on the sensitivity of a method to detect mutations. To understand the contribution of rare variants to schizophrenia, it is first necessary to ascertain the full spectrum of structural variants as well as other classes of variation including point mutations and indels. This could best be achieved through whole-genome sequencing. As the power and cost of genome-wide screening technologies improves, we anticipate that many more critical genes will be identified. These discoveries may include loci that are hotspots, with mutation rates of one in 10,000, as well as many other genes and genomic regions with lower mutation rates (e.g. one in 100,000) and lower overall frequencies. Although the cost of whole-genome sequencing in a large patient sample is substantial, a significant investment could be justified given the power of the data derived from these methods to transform the field of psychiatric genetics.
The discovery of rare schizophrenia-associated CNVs and other mutations that confer substantial risk of disease would, for the first time, identify single mutations that have reasonable predictive value, with potential to improve the clinical diagnosis of patients. First, the clinical relevance of the individual mutations must be definitively established and the penetrance for a variety of clinical phenotypes must be understood. This can be achieved by intensive clinical studies of patients and their families who share a disease mutation.
The ultimate goal of psychiatric genetic research is to identify and characterize neurobiological pathways and processes that are disrupted in individuals with mental disorders. Although schizophrenia might arise from a multitude of different causes, it is likely that these will influence a more limited number of key brain systems or pathways. A thorough characterization of these critical pathways will make a substantial contribution to our understanding of pathophysiology and provide important targets for treatment.
Acknowledgments
Funding for JS was provided by grants from Ted and Vada Stanley, the Simons Foundation, J.M. and C.D. Stone and the National Institutes of Health (NIH) (MH076431, HF004222). Funding for DLL was provided by grants from NARSAD to DLL, the Essel Foundation and the Sidney R. Baer, Jr. Foundation and the NIH (MH071523; MH31340)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cardno AG, Gottesman II. Twin studies of schizophrenia: from bow-and-arrow concordances to star wars Mx and functional genomics. Am J Med Genet. 2000;97:12–17. [PubMed] [Google Scholar]
- 2.McClellan JM, et al. Schizophrenia: a common disease caused by multiple rare alleles. Br J Psychiatry. 2007;190:194–199. doi: 10.1192/bjp.bp.106.025585. [DOI] [PubMed] [Google Scholar]
- 3.Craddock N, et al. Phenotypic and genetic complexity of psychosis. Invited commentary on … Schizophrenia: a common disease caused by multiple rare alleles. Br J Psychiatry. 2007;190:200–203. doi: 10.1192/bjp.bp.106.033761. [DOI] [PubMed] [Google Scholar]
- 4.Kendler KS. “A gene for…”: the nature of gene action in psychiatric disorders. Am J Psychiatry. 2005;162:1243–1252. doi: 10.1176/appi.ajp.162.7.1243. [DOI] [PubMed] [Google Scholar]
- 5.Crow TJ. The emperors of the schizophrenia polygene have no clothes. Psychol Med. 2008;38:1681–1685. doi: 10.1017/S0033291708003395. [DOI] [PubMed] [Google Scholar]
- 6.Burmeister M, et al. Psychiatric genetics: progress amid controversy. Nat Rev Genet. 2008;9:527–540. doi: 10.1038/nrg2381. [DOI] [PubMed] [Google Scholar]
- 7.Sanders AR, et al. No significant association of 14 candidate genes with schizophrenia in a large European ancestry sample: implications for psychiatric genetics. Am J Psychiatry. 2008;165:497–506. doi: 10.1176/appi.ajp.2007.07101573. [DOI] [PubMed] [Google Scholar]
- 8.Sullivan PF, et al. Genomewide association for schizophrenia in the CATIE study: results of stage 1. Mol Psychiatry. 2008;13:570–584. doi: 10.1038/mp.2008.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kirov G, et al. A genome-wide association study in 574 schizophrenia trios using DNA pooling. Mol Psychiatry. 2009;14:796–803. doi: 10.1038/mp.2008.33. [DOI] [PubMed] [Google Scholar]
- 10.Shifman S, et al. Genome-wide association identifies a common variant in the reelin gene that increases the risk of schizophrenia only in women. PLoS Genet. 2008;4:e28. doi: 10.1371/journal.pgen.0040028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walsh T, et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–543. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- 12.Consortium TIS. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stefansson H, et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rujescu D, et al. Disruption of the neurexin 1 gene is associated with schizophrenia. Hum Mol Genet. 2009;18:988–996. doi: 10.1093/hmg/ddn351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kirov G, et al. Support for the involvement of large copy number variants in the pathogenesis of schizophrenia. Hum Mol Genet. 2009;18:1497–1503. doi: 10.1093/hmg/ddp043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Purcell SM, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stefansson H, et al. Common variants conferring risk of schizophrenia. Nature. 2009;460:744–747. doi: 10.1038/nature08186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi J, et al. Common variants on chromosome 6p22.1 are associated with schizophrenia. Nature. 2009;460:753–757. doi: 10.1038/nature08192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O’Donovan MC, et al. Identification of loci associated with schizophrenia by genome-wide association and follow-up. Nat Genet. 2008;40:1053–1055. doi: 10.1038/ng.201. [DOI] [PubMed] [Google Scholar]
- 20.Feuk L, et al. Structural variation in the human genome. Nat Rev Genet. 2006;7:85–97. doi: 10.1038/nrg1767. [DOI] [PubMed] [Google Scholar]
- 21.Iafrate AJ, et al. Detection of large-scale variation in the human genome. Nat Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 22.Sebat J, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 23.Lupski JR. Genomic rearrangements and sporadic disease. Nat Genet. 2007;39:S43–47. doi: 10.1038/ng2084. [DOI] [PubMed] [Google Scholar]
- 24.Redon R, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tuzun E, et al. Fine-scale structural variation of the human genome. Nat Genet. 2005;37:727–732. doi: 10.1038/ng1562. [DOI] [PubMed] [Google Scholar]
- 26.Eichler EE, et al. Completing the map of human genetic variation. Nature. 2007;447:161–165. doi: 10.1038/447161a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ji W, et al. Rare independent mutations in renal salt handling genes contribute to blood pressure variation. Nat Genet. 2008;40:592–599. doi: 10.1038/ng.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ahituv N, et al. Medical sequencing at the extremes of human body mass. Am J Hum Genet. 2007;80:779–791. doi: 10.1086/513471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Azzopardi D, et al. Multiple rare nonsynonymous variants in the adenomatous polyposis coli gene predispose to colorectal adenomas. Cancer Res. 2008;68:358–363. doi: 10.1158/0008-5472.CAN-07-5733. [DOI] [PubMed] [Google Scholar]
- 30.Bodmer W, Bonilla C. Common and rare variants in multifactorial susceptibility to common diseases. Nat Genet. 2008;40:695–701. doi: 10.1038/ng.f.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cohen JC, et al. Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science. 2004;305:869–872. doi: 10.1126/science.1099870. [DOI] [PubMed] [Google Scholar]
- 32.Cohen JC, et al. Multiple rare variants in NPC1L1 associated with reduced sterol absorption and plasma low-density lipoprotein levels. Proc Natl Acad Sci U S A. 2006;103:1810–1815. doi: 10.1073/pnas.0508483103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fearnhead NS, et al. Multiple rare variants in different genes account for multifactorial inherited susceptibility to colorectal adenomas. Proc Natl Acad Sci U S A. 2004;101:15992–15997. doi: 10.1073/pnas.0407187101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fearnhead NS, et al. Rare variant hypothesis for multifactorial inheritance: susceptibility to colorectal adenomas as a model. Cell Cycle. 2005;4:521–525. doi: 10.4161/cc.4.4.1591. [DOI] [PubMed] [Google Scholar]
- 35.Li B, Leal SM. Discovery of Rare Variants via Sequencing: Implications for Association Studies. 2008 doi: 10.1371/journal.pgen.1000481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu B, et al. Strong association of de novo copy number mutations with sporadic schizophrenia. Nat Genet. 2008;40:880–885. doi: 10.1038/ng.162. [DOI] [PubMed] [Google Scholar]
- 37.Karayiorgou M, et al. Schizophrenia susceptibility associated with interstitial deletions of chromosome 22q11. Proc Natl Acad Sci U S A. 1995;92:7612–7616. doi: 10.1073/pnas.92.17.7612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Millar JK, et al. Disruption of two novel genes by a translocation co-segregating with schizophrenia. Hum Mol Genet. 2000;9:1415–1423. doi: 10.1093/hmg/9.9.1415. [DOI] [PubMed] [Google Scholar]
- 39.Millar JK, et al. DISC1 and PDE4B are interacting genetic factors in schizophrenia that regulate cAMP signaling. Science. 2005;310:1187–1191. doi: 10.1126/science.1112915. [DOI] [PubMed] [Google Scholar]
- 40.Pickard BS, et al. Disruption of a brain transcription factor, NPAS3, is associated with schizophrenia and learning disability. Am J Med Genet B Neuropsychiatr Genet. 2005;136:26–32. doi: 10.1002/ajmg.b.30204. [DOI] [PubMed] [Google Scholar]
- 41.Chubb JE, et al. The DISC locus in psychiatric illness. Mol Psychiatry. 2008;13:36–64. doi: 10.1038/sj.mp.4002106. [DOI] [PubMed] [Google Scholar]
- 42.Ousley O, et al. A review of neurocognitive and behavioral profiles associated with 22q11 deletion syndrome: implications for clinical evaluation and treatment. Curr Psychiatry Rep. 2007;9:148–158. doi: 10.1007/s11920-007-0085-8. [DOI] [PubMed] [Google Scholar]
- 43.Albertson DG, Pinkel D. Genomic microarrays in human genetic disease and cancer. Hum Mol Genet. 2003;12(Spec No 2):R145–152. doi: 10.1093/hmg/ddg261. [DOI] [PubMed] [Google Scholar]
- 44.Lucito R, et al. Representational oligonucleotide microarray analysis: a high-resolution method to detect genome copy number variation. Genome Res. 2003;13:2291–2305. doi: 10.1101/gr.1349003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pollack JR, et al. Microarray analysis reveals a major direct role of DNA copy number alteration in the transcriptional program of human breast tumors. Proc Natl Acad Sci U S A. 2002;99:12963–12968. doi: 10.1073/pnas.162471999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shaw-Smith C, et al. Microarray based comparative genomic hybridisation (array-CGH) detects submicroscopic chromosomal deletions and duplications in patients with learning disability/mental retardation and dysmorphic features. J Med Genet. 2004;41:241–248. doi: 10.1136/jmg.2003.017731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang D, et al. Singleton deletions throughout the genome increase risk of bipolar disorder. Mol Psychiatry. 2009;14:376–80. doi: 10.1038/mp.2008.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shi YY, et al. A study of rare structural variants in schizophrenia patients and normal controls from Chinese Han population. Mol Psychiatry. 2008;13:911–913. doi: 10.1038/mp.2008.69. [DOI] [PubMed] [Google Scholar]
- 49.Need AC, et al. A genome-wide investigation of SNPs and CNVs in schizophrenia. PLoS Genet. 2009;5:e1000373. doi: 10.1371/journal.pgen.1000373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sebat J, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marshall CR, et al. Structural Variation of Chromosomes in Autism Spectrum Disorder. The American Journal of Human Genetics. 2008;82:477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu B, et al. Elucidating the genetic architecture of familial schizophrenia using rare copy number variant and linkage scans. Proc Natl Acad Sci U S A. 2009;106:16746–16751. doi: 10.1073/pnas.0908584106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McCarthy S, et al. Microduplications of 16p11.2 are strongly associated with schizophrenia. 2009 doi: 10.1038/ng.474. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ingason A, et al. Copy number variations of chromosome 16p13.1 region associated with schizophrenia. Mol Psychiatry. 2009 doi: 10.1038/mp.2009.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kirov G, et al. Comparative genome hybridization suggests a role for NRXN1 and APBA2 in schizophrenia. Hum Mol Genet. 2008;17:458–465. doi: 10.1093/hmg/ddm323. [DOI] [PubMed] [Google Scholar]
- 56.Friedman JI, et al. CNTNAP2 gene dosage variation is associated with schizophrenia and epilepsy. Mol Psychiatry. 2008;13:261–266. doi: 10.1038/sj.mp.4002049. [DOI] [PubMed] [Google Scholar]
- 57.Murphy KC, et al. High rates of schizophrenia in adults with velo-cardio-facial syndrome. Arch Gen Psychiatry. 1999;56:940–945. doi: 10.1001/archpsyc.56.10.940. [DOI] [PubMed] [Google Scholar]
- 58.Bassett AS, et al. 22q11 deletion syndrome in adults with schizophrenia. Am J Med Genet. 1998;81:328–337. [PMC free article] [PubMed] [Google Scholar]
- 59.Gothelf D, et al. Velocardiofacial manifestations and microdeletions in schizophrenic inpatients. Am J Med Genet. 1997;72:455–461. [PubMed] [Google Scholar]
- 60.Brunetti-Pierri N, et al. Recurrent reciprocal 1q21.1 deletions and duplications associated with microcephaly or macrocephaly and developmental and behavioral abnormalities. Nat Genet. 2008;40:1466–1471. doi: 10.1038/ng.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sharp AJ, et al. A recurrent 15q13.3 microdeletion syndrome associated with mental retardation and seizures. Nat Genet. 2008;40:322–328. doi: 10.1038/ng.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Weiss LA, et al. Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med. 2008;358:667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 63.Kumar RA, et al. Recurrent 16p11.2 microdeletions in autism. Hum Mol Genet. 2008;17:628–638. doi: 10.1093/hmg/ddm376. [DOI] [PubMed] [Google Scholar]
- 64.Hannes FD, et al. Recurrent reciprocal deletions and duplications of 16p13.11: the deletion is a risk factor for MR/MCA while the duplication may be a rare benign variant. J Med Genet. 2009;46:223–232. doi: 10.1136/jmg.2007.055202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Szatmari P, et al. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat Genet. 2007;39:319–328. doi: 10.1038/ng1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kim HG, et al. Disruption of neurexin 1 associated with autism spectrum disorder. Am J Hum Genet. 2008;82:199–207. doi: 10.1016/j.ajhg.2007.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sebat J. Major changes in our DNA lead to major changes in our thinking. Nat Genet. 2007;39:S3–5. doi: 10.1038/ng2095. [DOI] [PubMed] [Google Scholar]
- 68.Fisher RA. The correlation between relatives on the supposition of Mendelian inheritance. Philos Trans R Soc Edinb. 1918;52:399–433. [Google Scholar]
- 69.Gothelf D, et al. Genetic, developmental, and physical factors associated with attention deficit hyperactivity disorder in patients with velocardiofacial syndrome. Am J Med Genet B Neuropsychiatr Genet. 2004;126B:116–121. doi: 10.1002/ajmg.b.20144. [DOI] [PubMed] [Google Scholar]
- 70.Mefford HC, et al. Recurrent rearrangements of chromosome 1q21.1 and variable pediatric phenotypes. N Engl J Med. 2008;359:1685–1699. doi: 10.1056/NEJMoa0805384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Helbig I, et al. 15q13.3 microdeletions increase risk of idiopathic generalized epilepsy. Nat Genet. 2009;41:160–162. doi: 10.1038/ng.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dibbens LM, et al. Familial and sporadic 15q13.3 microdeletions in idiopathic generalized epilepsy: precedent for disorders with complex inheritance. Hum Mol Genet. 2009;18:3626–3631. doi: 10.1093/hmg/ddp311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sharp AJ, et al. Discovery of previously unidentified genomic disorders from the duplication architecture of the human genome. Nat Genet. 2006;38:1038–1042. doi: 10.1038/ng1862. [DOI] [PubMed] [Google Scholar]
- 74.Guilmatre A, et al. Recurrent rearrangements in synaptic and neurodevelopmental genes and shared biologic pathways in schizophrenia, autism, and mental retardation. Arch Gen Psychiatry. 2009;66:947–956. doi: 10.1001/archgenpsychiatry.2009.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mardis ER. The impact of next-generation sequencing technology on genetics. Trends Genet. 2008;24:133–141. doi: 10.1016/j.tig.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 76.Bentley DR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang J, et al. The diploid genome sequence of an Asian individual. Nature. 2008;456:60–65. doi: 10.1038/nature07484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wheeler DA, et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–876. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- 79.Korbel JO, et al. Paired-end mapping reveals extensive structural variation in the human genome. Science. 2007;318:420–426. doi: 10.1126/science.1149504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yoon S, et al. Sensitive and accurate detection of copy number variants using read depth of coverage. Genome Res. 2009;19:1586–1592. doi: 10.1101/gr.092981.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Alkan C, et al. Personalized copy number and segmental duplication maps using next-generation sequencing. Nat Genet. 2009;41:1061–1067. doi: 10.1038/ng.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chiang DY, et al. High-resolution mapping of copy-number alterations with massively parallel sequencing. Nat Methods. 2009;6:99–103. doi: 10.1038/nmeth.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hodges E, et al. Genome-wide in situ exon capture for selective resequencing. Nat Genet. 2007;39:1522–1527. doi: 10.1038/ng.2007.42. [DOI] [PubMed] [Google Scholar]
- 84.Albert TJ, et al. Direct selection of human genomic loci by microarray hybridization. Nat Methods. 2007;4:903–905. doi: 10.1038/nmeth1111. [DOI] [PubMed] [Google Scholar]
- 85.Stark A, et al. Animal MicroRNAs confer robustness to gene expression and have a significant impact on 3′UTR evolution. Cell. 2005;123:1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- 86.Walz K, et al. Behavioral characterization of mouse models for Smith-Magenis syndrome and dup(17)(p11.2p11.2) Hum Mol Genet. 2004;13:367–378. doi: 10.1093/hmg/ddh044. [DOI] [PubMed] [Google Scholar]
- 87.Lupski JR, et al. DNA duplication associated with Charcot-Marie-Tooth disease type 1A. Cell. 1991;66:219–232. doi: 10.1016/0092-8674(91)90613-4. [DOI] [PubMed] [Google Scholar]
- 88.Nakatani J, et al. Abnormal behavior in a chromosome-engineered mouse model for human 15q11-13 duplication seen in autism. Cell. 2009;137:1235–1246. doi: 10.1016/j.cell.2009.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bi W, et al. Inactivation of Rai1 in mice recapitulates phenotypes observed in chromosome engineered mouse models for Smith-Magenis syndrome. Hum Mol Genet. 2005;14:983–995. doi: 10.1093/hmg/ddi085. [DOI] [PubMed] [Google Scholar]
- 90.Missler M, et al. Alpha-neurexins couple Ca2+ channels to synaptic vesicle exocytosis. Nature. 2003;423:939–948. doi: 10.1038/nature01755. [DOI] [PubMed] [Google Scholar]
- 91.Shahbazian M, et al. Mice with truncated MeCP2 recapitulate many Rett syndrome features and display hyperacetylation of histone H3. Neuron. 2002;35:243–254. doi: 10.1016/s0896-6273(02)00768-7. [DOI] [PubMed] [Google Scholar]
- 92.Fmr1 knockout mice: a model to study fragile X mental retardation. The Dutch-Belgian Fragile X Consortium. Cell. 1994;78:23–33. [PubMed] [Google Scholar]
- 93.Passage E, et al. Ascorbic acid treatment corrects the phenotype of a mouse model of Charcot-Marie-Tooth disease. Nat Med. 2004;10:396–401. doi: 10.1038/nm1023. [DOI] [PubMed] [Google Scholar]
- 94.Kaya F, et al. Ascorbic acid inhibits PMP22 expression by reducing cAMP levels. Neuromuscul Disord. 2007;17:248–253. doi: 10.1016/j.nmd.2006.12.008. [DOI] [PubMed] [Google Scholar]
- 95.Yan QJ, et al. Suppression of two major Fragile X Syndrome mouse model phenotypes by the mGluR5 antagonist MPEP. Neuropharmacology. 2005;49:1053–1066. doi: 10.1016/j.neuropharm.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 96.Dolen G, et al. Correction of fragile X syndrome in mice. Neuron. 2007;56:955–962. doi: 10.1016/j.neuron.2007.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.de Vrij FM, et al. Rescue of behavioral phenotype and neuronal protrusion morphology in Fmr1 KO mice. Neurobiol Dis. 2008;31:127–132. doi: 10.1016/j.nbd.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nishimura Y, et al. Genome-wide expression profiling of lymphoblastoid cell lines distinguishes different forms of autism and reveals shared pathways. Hum Mol Genet. 2007;16:1682–1698. doi: 10.1093/hmg/ddm116. [DOI] [PubMed] [Google Scholar]
- 99.Yu J, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 100.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 101.Hotta A, et al. Isolation of human iPS cells using EOS lentiviral vectors to select for pluripotency. Nat Methods. 2009;6:370–376. doi: 10.1038/nmeth.1325. [DOI] [PubMed] [Google Scholar]
- 102.Kremen WS, et al. Is the Wisconsin Card Sorting Test a useful neurocognitive endophenotype? Am J Med Genet B Neuropsychiatr Genet. 2007;144B:403–406. doi: 10.1002/ajmg.b.30527. [DOI] [PubMed] [Google Scholar]
- 103.Jablensky A. Subtyping schizophrenia: implications for genetic research. Mol Psychiatry. 2006;11:815–836. doi: 10.1038/sj.mp.4001857. [DOI] [PubMed] [Google Scholar]
- 104.Waddington JL, et al. Magnetic resonance imaging and spectroscopy in schizophrenia. Br J Psychiatry Suppl. 1990:56–65. [PubMed] [Google Scholar]
- 105.Nikolaus S, et al. In vivo imaging of synaptic function in the central nervous system: II. Mental and affective disorders. Behav Brain Res. 2009;204:32–66. doi: 10.1016/j.bbr.2009.06.009. [DOI] [PubMed] [Google Scholar]
- 106.Koolen DA, et al. A new chromosome 17q21.31 microdeletion syndrome associated with a common inversion polymorphism. Nat Genet. 2006;38:999–1001. doi: 10.1038/ng1853. [DOI] [PubMed] [Google Scholar]
- 107.Shaw-Smith C, et al. Microdeletion encompassing MAPT at chromosome 17q21.3 is associated with developmental delay and learning disability. Nat Genet. 2006;38:1032–1037. doi: 10.1038/ng1858. [DOI] [PubMed] [Google Scholar]
- 108.Gottesman II, Gould TD. The endophenotype concept in psychiatry: etymology and strategic intentions. Am J Psychiatry. 2003;160:636–645. doi: 10.1176/appi.ajp.160.4.636. [DOI] [PubMed] [Google Scholar]
- 109.Yan J, et al. Neurexin 1alpha structural variants associated with autism. Neurosci Lett. 2008;438:368–370. doi: 10.1016/j.neulet.2008.04.074. [DOI] [PubMed] [Google Scholar]
- 110.Doornbos M, et al. Nine patients with a microdeletion 15q11.2 between breakpoints 1 and 2 of the Prader-Willi critical region, possibly associated with behavioural disturbances. Eur J Med Genet. 2009;52:108–115. doi: 10.1016/j.ejmg.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 111.Chance PF, et al. DNA deletion associated with hereditary neuropathy with liability to pressure palsies. Cell. 1993;72:143–151. doi: 10.1016/0092-8674(93)90058-x. [DOI] [PubMed] [Google Scholar]
- 112.Mefford HC, et al. A method for rapid, targeted CNV genotyping identifies rare variants associated with neurocognitive disease. Genome Res. 2009;19:1579–1585. doi: 10.1101/gr.094987.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Elia J, et al. Rare structural variants found in attention-deficit hyperactivity disorder are preferentially associated with neurodevelopmental genes. Mol Psychiatry. 2009 doi: 10.1038/mp.2009.57. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Bakkaloglu B, et al. Molecular cytogenetic analysis and resequencing of contactin associated protein-like 2 in autism spectrum disorders. Am J Hum Genet. 2008;82:165–173. doi: 10.1016/j.ajhg.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Arking DE, et al. A common genetic variant in the neurexin superfamily member CNTNAP2 increases familial risk of autism. Am J Hum Genet. 2008;82:160–164. doi: 10.1016/j.ajhg.2007.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Alarcon M, et al. Linkage, association, and gene-expression analyses identify CNTNAP2 as an autism-susceptibility gene. Am J Hum Genet. 2008;82:150–159. doi: 10.1016/j.ajhg.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]