Abstract
The genetic bases of neuropsychiatric disorders are beginning to yield to scientific inquiry. Genome-wide studies of copy number variation (CNV) have given rise to a new understanding of disease etiology, bringing rare variants to the forefront. A proportion of risk for schizophrenia, bipolar disorder and Autism can be explained by rare mutations. Such alleles arise by de novo mutation in the individual or in recent ancestry. Alleles can have specific effects on behavioral and neuroanatomical traits; however expressivity is variable, particularly for neuropsychiatric phenotypes. Knowledge from CNV studies reflects the nature of rare alleles in general and will serve as a guide as we move forward into a new era of whole genome sequencing.
Genetic Variation’s Other Half
Early surveys of genetic variation found that two human chromosomes in the population differ at a rate of 0.1% on average (Consortium, 2005). Individual base changes, called single nucleotide polymorphisms (SNPs), are by far the most numerous variants in the genome, but SNPs are only half of the story. In 2004, two landmark studies (Iafrate et al., 2004; Sebat et al., 2004) demonstrated that submicroscopic variations (<500 kb in size) in DNA copy number (CNVs) are widespread in normal human genomes. On average, there are >1000 CNVs in the genome, accounting for ~4 million base pairs of genomic difference (Conrad et al., 2010; Mills et al., 2011). Although SNPs outnumber CNVs in the genome by three orders of magnitude, their relative contributions to genomic variation (as measured in nucleotides) are similar. Thus, in addition to 0.1% of genetic difference at the nucleotide sequence level, we now recognize another 0.1% of genetic difference at the structural level.
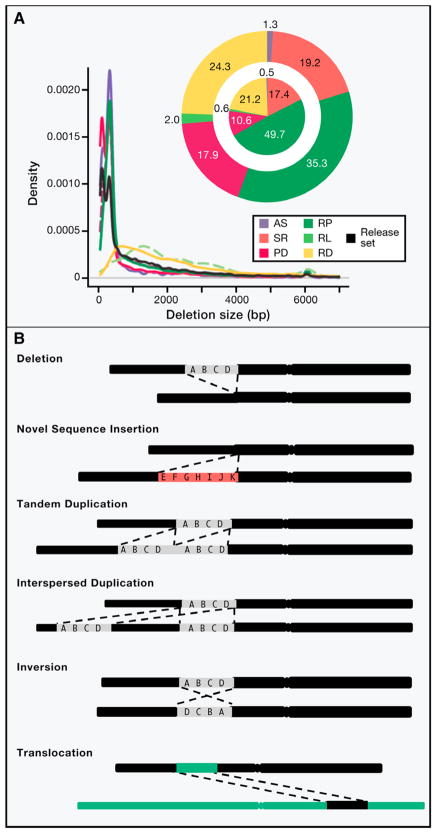
The definition of structural variation (SV) has evolved as new technologies capture an ever-widening spectrum of alleles. SVs are sometimes defined operationally as deletions, duplications, insertions and inversions that are greater than 1 kb in size (Alkan et al., 2011; Zhang et al., 2009b), but in reality SVs follow a continuous distribution of size (Figure 1A) and can include simple insertion/deletion or complex rearrangements (Figure 1B).
Figure 1. Types of structural variation and its evolving definition.
(A) Size distribution of 22,025 deletions (release set) identified from whole genome sequencing of 179 unrelated individuals and 2 trios (i.e., child-mother-father) by the 1000 genomes project. Deletions were identified by four different types of structural variation detection approaches, which all rely on mapping sequenced reads to reference human genome sequence and subsequently identifying discordant patterns that are characteristic of deletions. Read pair (RP) methods detect clusters of “discordant” paired end reads in which mapping span and/or orientation is inconsistent with the reference genome. Read depth (RD) methods detect CNVs based on the regional depth of coverage. Split read (SR) methods detect individual reads that span the breakpoint of an SV. Assembly (AS) methods detect differences between sequence contigs assembled from the sample genome and the reference genome sequence. Paired depth (PD) refers to hybrid methods that combine RP and RD. Pie charts display the contribution of different SV detection approaches to the release set. Outer pie = based on number of SV calls; inner pie = based on total number of variable nucleotides. (B) Schematic representation of deletion, novel sequence insertion (red color bar), tandem duplication, interspersed duplication, inversion and translocation in test genome (lower black bar) compared to human reference genome sequence (upper black bar). Green colored bar represent a different chromosome from reference genome. Figure adapted from Mills et al 2010.
Mutational Mechanisms
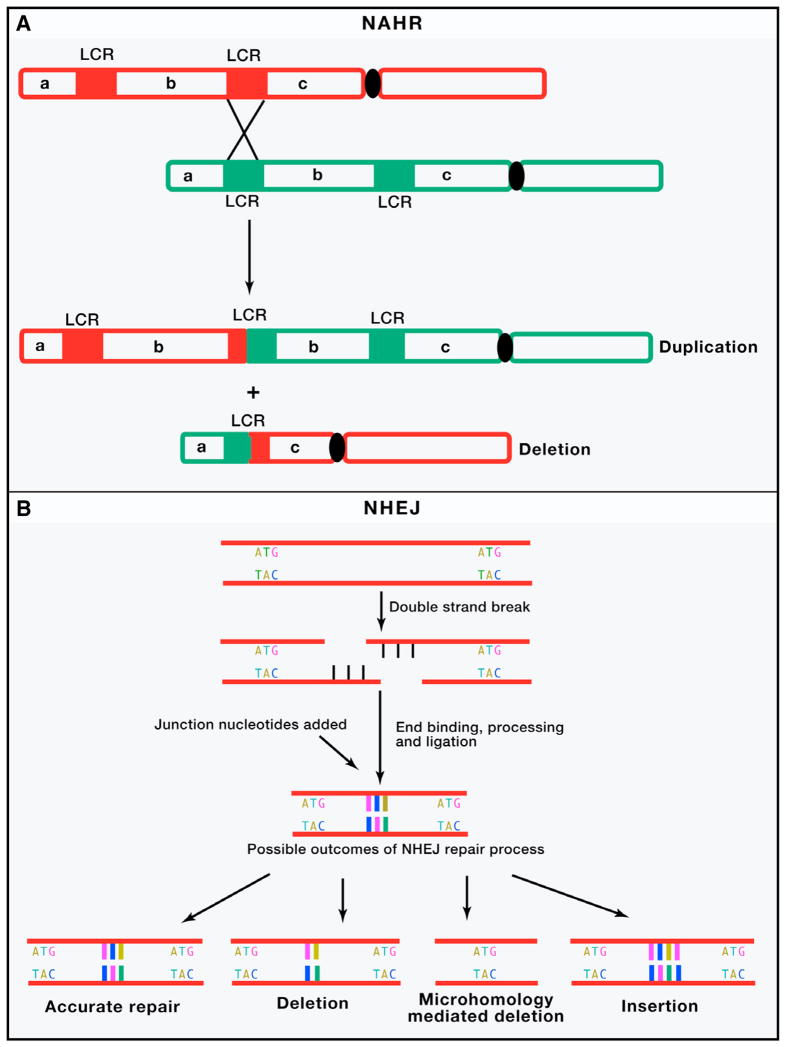
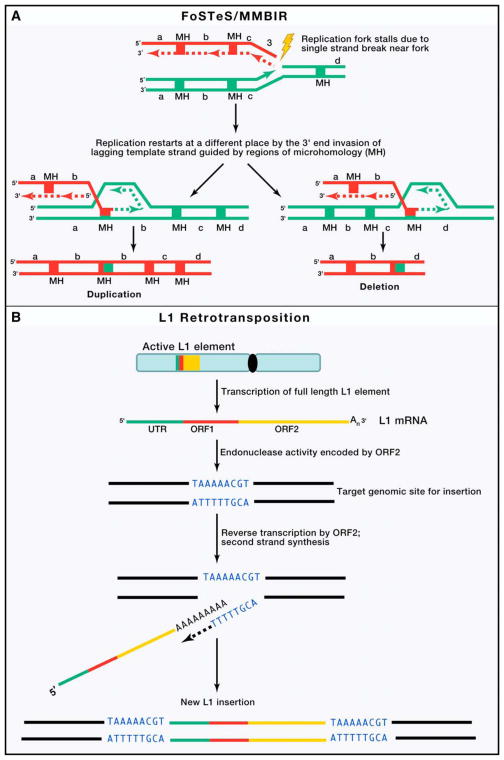
The mechanisms of structural mutation are generally inferred from sequence information at junction/breakpoint of the rearrangements. Four major mechanisms can account for the majority of SVs: non allelic homologous recombination (NAHR), non homologous end joining (NHEJ), fork stalling and template switching (FoSTeS), and L1-mediated retrotransposition (Zhang et al., 2009b)(Figure 2).
Figure 2. Four major mechanisms underlying human genomic rearrangements and CNV formation.
(A) Non allelic homologous recombination (NAHR) occurs by unequal crossing over between flanking segmental duplications (SDs represented by two red and two green bars on respective homologous chromosomes), which result in reciprocal deletion and duplication of intervening sequence (b). These homologous chromosomes segregate from each other at the next cell division, thus leading to a change in copy number in both daughter cells.(B) In classical non homologous end joining (NHEJ) double-strand break repair pathway, the ends of DNA double-strand breaks are repaired through many rounds of enzymatic activity (including tethering of DNA ends by the Ku protein, followed by recruitment of DNA-dependent protein kinase, DNA-PKcs by Ku and DNA-PKcs mediated activation of the Artemis nuclease, which trims back overhangs in preparation for ligation). The different types of DNA double strand breaks fixed by NHEJ combined with other sundry alternate repair mechanisms including microhomology mediated end joining (MMEJ) leads to diverse repaired products. Although limited base pairing can guide accurate repair, deletions of variable size, and to a lesser extent insertions, are formed. (C) A simple model of FoSTeS/MMBIR is described. When a replication fork encounters a nick (striking arrowhead) in a template strand, one arm of the fork breaks off, producing a collapsed fork. At the single double-strand end, the 5′ end of the lagging strand (dashed red colored lines) is resected, giving a 3′ overhang. The 3′ single-strand end of lagging strand template (solid red colored lines) invades the sister leading strand DNA (green colored lines) guided by regions of microhomology (MH, red and green colored boxes), forming a new low processivity replication fork. The extended end dissociates repeatedly (due to migration of holiday junction or some other helicase activity) with 5′ ends resected and reforms the fork. Whether the template switch occurs in front of or behind the position of the original collapse determines whether there is a deletion or duplication respectively. The 3′ end invasion of lagging strand template can reform replication forks on different genomic templates (>100kb apart), before returning to the original sister chromatid and forming a processive replication fork that completes replication. Thus, the final product usually contains sequence from different genomic regions (not shown). Each line represents a DNA nucleotide strand. Polarity is indicated by arrows on 3′ end. New DNA synthesis is shown by dashed lines. (D) LINE-1 retrotransposition. A full-length L1 (red, green and orange bar on gray chromosome) is transcribed and translation of ORF1 (red) and ORF2(orange) protein encoded by the L1 messenger RNA (mRNA) leads to ribonucleoprotein (RNP) formation. L1 RNP is transported to the nucleus, and retrotransposition occurs by target-site primed reverse transcription (TPRT). During TPRT, the L1 endonuclease (EN) activity of ORF2 nicks target genomic DNA (black lines), exposing a free 3′-OH that serves as a primer for reverse transcription of the L1 RNA. The mechanistic details of target site second-strand cleavage, second-strand complementary DNA (cDNA) synthesis, and completion of L1 integration require further elucidation. TPRT results in the insertion of a new, often 5′-truncated L1 copy at a new genomic location that generally is flanked by target-site duplications. Alu, SINE-R/VNTR/Alu (SVA), and cellular mRNAs can also hijack the L1-encoded protein(s) in the cytoplasm to mediate their transmobilization. ORF: open reading frame; FoSTeS: fork stalling and template switching; MMBIR: microhomology mediated break induced replication.
Non-Allelic Homologous Recombination (NAHR)
NAHR involves the alignment and subsequent crossing over between two sites in the genome that share region of sequence homology (Figure 2A). NAHR can occur both in meiosis (Turner et al., 2008) and, at lower frequency, in mitotically-dividing cells as well (Lam and Jeffreys, 2006, 2007). NAHR can involve genomic rearrangements between paralogs on homologous chromosomes (interchromosomal), sister chromatids (interchromatid), and within chromatid (intrachromatid) (Gu et al., 2008). The relative positions and extent of these homologies influence the rate of NAHR events (Liu et al., 2011b). Regions of the genome that possess tandemly-arranged segmental duplications (SDs), which are also called low copy repeats (LCRs), are more prone to frequent rearrangements between specific LCRs due to NAHR. As a result, multiple rearrangements, which are nearly identical to each other, can arise independently in different individuals (Gu et al., 2008). Such recurrent de novo CNVs can occur at rates as high as 1/4,000 newborns (Devriendt et al., 1998).
Non-Homologous End Joining (NHEJ)
NHEJ occurs as a result of the aberrant repair of DNA double strand breaks and is guided entirely by the information contained within or near the DNA lesion for repair, which makes it error prone as compared to NAHR (Lieber, 2008) (Figure 2B). The breakpoints of CNVs formed by NHEJ are frequently observed within repetitive elements, such as long terminal repeats (LTRs), short interspersed repeat elements (SINEs), long interspersed nuclear elements (LINEs), and mammalian interspersed repeats (MIRs). This suggests that NHEJ may be stimulated by certain genomic architectures, but extensive sequence homology is not required (Toffolatti et al., 2002). Breakpoints of some non-recurrent deletion CNVs have sequences with very short (2–20 basepairs) stretches of nucleotide identity. These are predicted to be formed by an alternative microhomology mediated end joining (MMEJ) mechanism (Lieber, 2010).
Fork stalling and template switching (FoSTeS)
FoSTeS is a replication-based genomic rearrangement mechanism that is induced by errors (single strand breaks) during DNA replication process (Lee et al., 2007). Hastings et al (2009a) proposed a further generalization of the FoSTeS mechanism, which is known as the MMBIR (microhomology mediated break induced replication) model (Hastings et al., 2009a) (Figure 2C). Genomic rearrangements generated by FoSTeS/MMBIR can vary greatly in size and complexity (Hastings et al., 2009b; Zhang et al., 2009c). In addition to microhomolgy mediated rearrangements (Lee et al., 2007; Liu et al., 2011a; Zhang et al., 2009c), FoSTeS mediated by large inverted repeats (>300 kb apart) and coupled with NHEJ is proposed as the predominant mechanism for complex rearrangements with duplication-triplication/inversion-duplication structures (Carvalho et al., 2011).
Retrotransposition
Long interspersed nuclear elements-1 (LINE1 or L1), the only currently active class of retrotransposons in humans, occupy nearly 20% of the genomic real estate (Goodier and Kazazian, 2008). Although ~500,000 copies are present in the genome, only 80–100 are active full length (6 kilobases) elements that can transpose to new genomic locations by a target primed reverse transcription (TPRT) mechanism (Goodier and Kazazian, 2008) (Figure 2D). Both germline and somatic L1 activity contribute significantly to structural variation in human genomes (Lupski, 2010).
The extent to which all four mutational mechanisms contribute to CNV formation is highlighted in recent findings from the 1000 genomes project (Mills et al., 2011). Approximately 70.8% of the deletions were attributed to either a non homology based mechanism (i.e., NHEJ) or MMBIR. 89.6% of small insertions were attributable to retrotransposition activity. Most tandem duplications displayed microhomology of 2–17 basepairs at breakpoints, indicating that they are likely formed by FoSTeS/MMBIR. Large deletions or duplications showed extensive stretches of sequence of >95% identity at breakpoints, suggesting that they were generated by NAHR.
De novo SVs: A Small Force that Packs a Large Punch
The rate of nucleotide substitutions genome-wide is estimated at 30–100 per generation (Conrad et al., 2011) and ~1 per exome. In contrast, the global rate of structural mutation events is lower: CNVs >10 Kb in size occur at a rate of ~0.01–0.02 per generation.(Itsara et al., 2010; Levy et al., 2011; Marshall et al., 2008; Sanders et al., 2011; Sebat et al., 2007). New retrotransposon insertions probably account for the majority of smaller events, with short (300 basepairs) Alu repeat insertions occurring at a rate of 0.05 per generation (Cordaux and Batzer, 2009) and longer (1000–9000 bp) L1 insertions occurring at a rate of 0.01–0.05. per generation (Beck et al., 2011). Thus, we estimate that the rate of the multiple classes of structural mutation combined is 0.07–0.12 per generation.
Although the absolute rate of structural mutation is low, individual mutations may affect tens or thousands of kilobases. Therefore, the overall rate of genomic change (as measured in nucleotides) is high, on the order of 1,000 bp per generation, and the functional impact per site is large.
This has important implications for the allelic architecture of disease. Based on sheer numbers, nucleotide substitutions probably account for the majority of disease risk alleles, but based on sheer size and potential to impact genes (or multiple genes), structural mutations are more pathogenic on average. Thus, we expect that CNVs as a class, and de novo CNVs in particular, will be more enriched in variants that have large effect on disease risk. Perhaps naturally, the early insights into the rare genetic causes of common disease have emerged from these classes of variants.
The Genetics of Mental Illness
The success of a particular genetic approach depends on the genetic architecture of the disease under investigation- that is, the total number of disease genes and the number and frequency of risk alleles within each gene. For diseases with a relatively simple genetic architecture, in which there is one or a few genes of major effect, linkage analysis (Botstein and Risch, 2003) and homozygosity mapping (Alkuraya, 2010) in families have proven to be highly effective approaches.
For psychiatric disorders, such as autism, schizophrenia and bipolar disorder, genetic architectures have proven to be complex, spawning a lively debate as to the nature of this complexity (Klein et al., 2010; McClellan and King, 2010). This debate has focused on the relative merits of two contrasting (but conceptually-related) hypotheses: the common variant common disease (CVCD) and rare variant common disease (RVCD) models.
The CVCD model posits that genetic risk in an individual (and in the population) is attributable to many high-frequency variants, each conferring modest level of risk (Risch and Merikangas, 1996).
By contrast, the RVCD model posits that genetic risk in an individual can be explained by rare mutations that confer significant risk. Thus, the common disease might reflect a large number (hundreds or thousands) of different causes, having low frequencies (typically less than 1/1,000 individuals), but accounting for a large proportion of attributable risk in aggregate (Bodmer and Bonilla, 2008).
Formal tests of the CVCD and RVCD hypotheses have been carried out in the form of genome-wide association studies (GWAS) (Manolio et al., 2008) and CNV studies (International schizophrenia Consortium, 2008; Sebat et al., 2007; Walsh et al., 2008) respectively. In the following sections, we discuss findings of CNV studies in autism, schizophrenia and bipolar disorder.
CNV Studies Put the Rare Variant-Common Disease Model to the Test
Within the context of psychiatric genetic studies, “CNV” has come to be virtually synonymous with “rare variant.” In truth, structural variants come in many shapes, sizes and allele frequencies, and a majority of variants present in an individual genome are common alleles (Conrad et al., 2010; McCarroll et al., 2008; Mills et al., 2011; Sudmant et al., 2010). However, it is the rare CNVs that have garnered great attention (Sebat et al., 2009).
The focus on rare CNVs is in part based on a precedent from cytogenetic studies. Cytogenetic rearrangements were reported in ~6–7% of autism spectrum disorder (ASD) cases (Folstein and Rosen-Sheidley, 2001). In addition, large cytogenetically-detectable chromosomal abnormalities, including maternally inherited duplication of chromosome 15q11-13 and microdeletions of 22q11.2, were also known to occur recurrently in a small proportion of idiopathic autism cases (Gillberg, 1998) and in schizophrenia (Murphy et al., 1999) respectively.
A CNV-based approach is also attractive for methodological reasons. Microarrays continue to be a mainstay technology platform for large scale genetic studies. Such dense oligonucleotide arrays are well suited to the detection of a predetermined panel of SNPs and for detection of large-scale copy number variants. Current genotyping platforms and CNV discovery algorithms enable the genotyping of ~1000 common copy number polymorphisms (CNPs) and the discovery of additional “novel” CNVs, including mutations that are rare or unique to an individual (Alkan et al., 2011). It is these rare CNVs that have provided the first glimpse into the many rare mutations that contribute to common psychiatric disease.
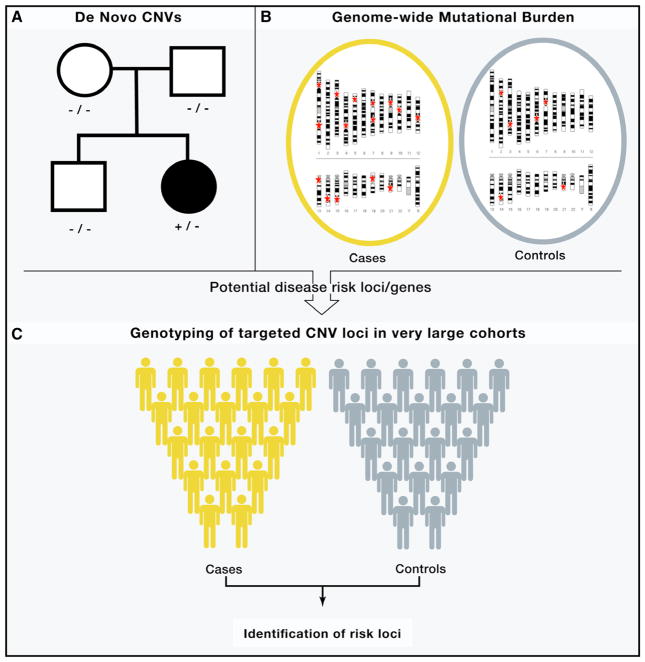
New findings have begun to emerge from genome-wide studies of CNV in three major psychiatric disorders: Autism Spectrum Disorders (ASDs), schizophrenia and bipolar disorder. Within CNV research, three study designs in particular have been used widely and to great effect (Figure 3).
Figure 3. Key Plays from the CNV Playbook.
(A) Family based studies of de novo mutation, (B) and Case-control studies of genome wide CNV burden, with CNV positions denoted by a red star (C) followed by Single marker tests for association in large cohorts.
Family-Based Studies of de novo CNVs
The central focus of these studies has been to determine the frequency of spontaneous (de novo) mutation and to determine the association of de novo CNVs with disease.
Case-control analysis of CNV burden
Similar to the family based studies, a contribution of rare CNVs to disease is evident in the overall genome-wide burden of rare variants (i.e., the number of CNVs carried by an individual). An enrichment of large (>100 kb) CNVs in patients as compared with controls has been reported in schizophrenia, autism and bipolar disorder.
Single marker or association of target regions or genes
Specific genes or genomic regions have been implicated by association in large case-control cohorts.
Although these approaches were popularized in the context of CNV studies, the same principles apply to any mutation discovery platform, including exome and whole genome sequencing, as exemplified by the first exome studies in ASD and schizophrenia (Girard et al., 2011; O’Roak et al., 2011; Xu et al., 2011). A lengthy review of genetic studies could be written about each of the following disorders. Here, we will focus on the key concepts that form our current understanding of psychiatric genetics.
Autism: A Complex Genomic Disorder
ASDs represent a heterogeneous group of disorders that share a set of common characteristics. These include core deficits in social communication and language development that are accompanied by highly restricted interests, stereotypic behaviors or both (Volkmar et al., 2009). ASDs are defined as having an age at onset younger than three. Males have a >3-fold higher risk for ASDs as compared to females (Volkmar et al., 2004).
Heritability estimates based on studies of clinically ascertained twin samples (Bailey et al., 1995; Folstein and Rutter, 1977; Hallmayer et al., 2011; Steffenburg et al., 1989) vary widely, from 38% to 90%, but it is clear that genes play a major role in ASD. Early twin studies observed 80–90% concordance for ASDs in monozygotic (MZ) twins and 5–15% concordance in dizygotic (DZ) twins and siblings. Two recent studies report somewhat higher rates of concordance in DZ twins (31%), suggesting, that the contribution of shared environmental factors could be greater than had been previously estimated (Hallmayer et al., 2011; Rosenberg et al., 2009).
Despite high heritability, the genetic basis of ASDs is complex. Early linkage studies detected numerous loci with modest levels of statistical support, and patterns of segregation in families did not appear to be consistent with classical Mendelian patterns of inheritance. Although, rare Mendelian causes of ASD had been identified (Miles, 2011), it was not known whether rare mutations of large effect contributed to idiopathic ASD.
Family-based studies
With this in mind, a series of CNV studies have been carried out to look systematically for non-Mendelian causes of ASD, focusing on de novo mutations. In 2007, Sebat et al. investigated the global frequency of de novo CNVs in trios (i.e., child-mother-father), comparing the frequencies of mutations in offspring between sporadic cases of ASD (i.e., “simplex” families with only a single affected offspring), familial cases (i.e., “multiplex” families with multiple affected offspring) and healthy control offspring (Sebat et al., 2007). In this study, a high rate of de novo CNVs in idiopathic ASD cases from simplex families (10%) was observed compared to the rate in cases from multiplex families (2%) or unaffected controls (1%). The striking 10-fold higher rate of mutations in cases suggested that a majority of mutations identified were contributing to risk.
Subsequent studies in larger samples have confirmed a high (5–10%) rate of de novo CNVs in ASD and further elucidated the extent of genetic heterogeneity in ASD (Itsara et al., 2010; Levy et al., 2011; Marshall et al., 2008; Pinto et al., 2010; Sanders et al., 2011). A detailed analysis of large ASD cohorts of simplex autism cases using very high resolution arrays was recently performed by two independent groups (Levy et al., 2011; Sanders et al., 2011). These studies reported that the burden of rare de novo CNVs is significantly greater in simplex cases (5.8–7.9%) than in unaffected siblings (1.7–1.9%) with regard to the total number of events, the size of each event, and their gene content. Affected cases on average had 16-fold excess of genes impacted by de novo CNVs compared to healthy sibs (30-fold for deletions). Based on the number of recurrent de novo CNVs and the estimated proportion of de novo CNVs ascertained, Levy et al estimated around 250–300 target loci for ASDs and Sanders et al. estimated between 130–234 loci.
Case control studies
The contribution of rare CNVs (including both de novo and inherited variants) to ASDs is also apparent from case-control studies. A large-scale CNV study was undertaken by the Autism Genome Project (AGP) (Pinto et al., 2010). When comparing 996 ASD individuals of European ancestry to 1,287 matched controls, cases were found to carry a higher global burden of rare, genic copy number variants (CNVs) (1.19 fold, P = 0.012), especially so for genomic regions previously implicated in ASD and/or intellectual disability (1.69 fold, P = 3.4 × 10−4). These findings were independently replicated by Sanders et al (Sanders et al., 2011), when CNV burden analysis included both rare transmitted and de novo CNVs; however, a significant enrichment of CNVs was not observed exclusively among variants that were inherited (Levy et al., 2011; Sanders et al., 2011).
Statistical evidence for specific risk loci
Several CNV regions have been firmly implicated in ASDs. Notably CNVs have been identified at several loci that are linked to known microdeletion syndromes including 16p11.2 (Levy et al., 2011; Sanders et al., 2011; Weiss et al., 2008), Williams Syndrome locus at 7q11.23 (Sanders et al.), Prader-Willi Angelman Syndrome at 15q11-13 (Glessner et al., 2009; Sanders et al., 2011), VCFS DiGeorge Syndrome at 22q11.2 (Sanders et al., 2011) and 1q21.1 (Sanders et al., 2011).
Pinpointing the specific genes involved in ASDs has been a challenge. The most frequent recurrent CNVs tend to be large (>500 kb) and contain multiple genes. Rare or de novo CNVs have been identified that are smaller (<100 Kb) in size, sometimes disrupting a single gene, but strong statistical evidence is lacking. There are a few genes in which mutations have been consistently detected in multiple studies, and thus these genes are recognized as bona fide risk factors for ASDs. These genes include NGLN4X (Jamain et al., 2003; Laumonnier et al., 2004), SHANK3 (Durand et al., 2007; Gauthier et al., 2009; Moessner et al., 2007), NRXN1(Bucan et al., 2009; Kim et al., 2008; Szatmari et al., 2007), SHANK2 (Berkel et al., 2010; Pinto et al., 2010), CNTN4 (Fernandez et al., 2004, 2008; Glessner et al., 2009; Roohi et al., 2009) and CNTNAP2 (Bakkaloglu et al., 2008; Strauss et al., 2006). Other novel ASD candidate genes include DPYD and DPP6 (Marshall et al., 2008), RFWD2, NLGN1, and ASTN2 (Glessner et al., 2009), SYNGAP1, DLGAP2, and the X-linked DDX53-PTCHD1 locus (Noor et al., 2010; Pinto et al., 2010).
Neurodevelopmental pathways implicated in ASD
Pathway-based analysis of CNVs is fraught with difficulty (Webber, 2011). However, some patterns have emerged and are becoming increasingly difficult to dismiss. Glessner et al. (2009) observed an enrichment of CNVs at multiple sites, and some of their top hits were genes involved in ubiquitin pathways, including UBE3A, PARK2, RFWD2 and FBXO40. Pinto et al. (2010) observed an enrichment of CNVs within gene sets involved in cellular proliferation, projection and motility, and GTPase/Ras signaling. Gilman et al. (2011) found an enrichment of CNVs in gene sets related to synapse development, axon targeting, and neuron motility. Although synaptic proteins and ubiquitin pathways were already implicated in ASDs based on small-scale studies (Bourgeron, 2009; Ehlers, 2003), these results suggest that the diversity of rare mutations in ASD affect larger sets of functionally-related genes.
Schizophrenia
Schizophrenia is formally characterized by three symptom clusters: positive, negative, and cognitive (van Os and Kapur, 2009). The positive-symptom dimension includes psychosis (i.e., paranoid delusions and auditory hallucinations); the negative-symptom dimension includes social withdrawal, lack of motivation, and difficulties in social interaction; and, the cognitive-symptom dimension refer to problems in attention, thought, perception, learning, and memory. Within the cluster of these symptoms-based diagnostic categories, which include other psychotic disorders, the term schizophrenia is used to define a syndrome characterized by prolonged periods of psychosis with bizarre delusions, negative symptoms and few affective (mania or depression) symptoms. The age at onset is typically in adolescence or early adulthood. Current medications provide relief only from positive symptoms without effective improvements in negative and cognitive symptoms (Leucht et al., 2009).
Case control studies
CNV studies have now established a significant role for rare (<1% in frequency) and large (>100kb) CNVs in risk for schizophrenia (Sebat et al., 2009). Early findings from our group observed a 3-fold enrichment of rare genic CNVs in cases as compared with controls (Walsh et al., 2008). In a larger study by the International schizophrenia Consortium, a 1.1–1.5 fold enrichment was observed in cases (International schizophrenia Consortium, 2008). These findings have been supported by several subsequent studies (Buizer-Voskamp et al., 2011; Kirov et al., 2009), confirming that rare CNVs are collectively more common in schizophrenia cases compared to controls.
Family-based studies
In the first systematic study of de novo CNVs in schizophrenia, Xu et al. observed a high rate in sporadic cases (10%) as compared with “familial” cases (defined as having an affected first or second degree relative) and a high rate compared with controls (Xu et al., 2008). Subsequent studies by Kirov et al. (Kirov et al., 2011) and by our group (Malhotra et al., 2011), also observed a high rate of de novo CNVs in schizophrenia (5% in both studies) as compared with controls; however, neither study observed a significant difference in rate between in sporadic and famial cases.
CNV regions that are implicated in schizophrenia
In schizophrenia, a large (3Mb) deletion at chromosome 22q11.21 has long been known as a significant risk factor for schizophrenia (Karayiorgou et al., 1995). Approximately 25% of 22q11.2 deletion carriers manifest symptoms of psychosis. Recent genome-wide studies have found strong evidence of association for other loci including deletions at chr1q21.1, deletions at chr3q29, duplications of chr16p11.2, deletions at chr15q13.3, exonic deletions at chr2p16.3 (NRXN1) and duplications at chr7q36.3 (VIPR2), with schizophrenia (Table 1; Supplemental Information).
Table 1. Analysis of pathogenic CNVs across multiple diagnostic categories: intellectual ability, Autism Spectrum Disorders, schizophrenia and bipolar disorder.
We analyzed 11 CNV loci where replicated evidence of association with schizophrenia and/or developmental disorders is available from multiple large-scale studies. We compared the frequency of each CNV loci in four major categories of neuropsychiatric disorders including intellectual disability (ID)/developmental delay (DD)/congenital malformations (CM), autism spectrum disorders (ASD), schizophrenia (SD) and bipolar disorder (BD)/Recurrent Depression, with its frequency in a large combined set of controls. We did not attempt a formal meta-analysis, as some studies did not include CNV data on both cases and controls. We present results with a pooled analysis of available CNV frequency data from different studies by performing a simple Fisher’s Exact Test and reporting odds ratio. We excluded overlapping individuals in different studies before doing tests for individual loci. Formal meta-analyses results are available for some of the published loci (Levinson et al., 2011) and the odds ratios (OR) and p-values reported for loci in the previous studies are similar to results reported here. Exonic deletions at the NRXN1 gene, also implicated as a risk factor for schizophrenia, were not analyzed as the CNVs affecting them are usually non-recurrent and much smaller than 500 kb and can be missed with the lower-resolution array studies included in the current analysis. See Supplemental Information for the references in the table.
| CNV Locus | Position (Mb) | Disease category | Frequency in combined cases | Frequency in combined controls | P value | Odds Ratio [95% CI] | Case References | Control References |
|---|---|---|---|---|---|---|---|---|
| 1q21.1 deletion | 145.0–146.3 | MR/DD/CM | 0.27% (102/ 38,330) | 0.02% (16/ 75,505) | 1.9 × 10−32 | 12.6 [7.4–21.3] | Mefford et al 2008; Brunettipierri et al 2008; Cooper et al 2011 | Vacic et al 2011; Grozeva et al 2011; Grozeva et al 2010; Rucker et al 2011; Sanders et al 2011; Levinson et al 2011; Cooper et al 2011 |
| ASD | 0.03% (1/ 3032) | 0.02% (16/ 75,505) | 0.49 | 1.6 [0.2–11.7] | Bucan et al 2009; Sanders et al 2011; Pinto et al 2010 | |||
| SCZ | 0.17% (21/ 12,174) | 0.02% (16/ 75,505) | 1.3 × 10−9 | 8.1 [4.3–15.6] | Vacic et al 2011; Levinson et al 2011 | |||
| BD & Recurrent Depression | 0.02% (2/ 8264) | 0.02% (16/ 75,505) | 0.69 | 1.1 [0.3–5.0] | Malhotra et al 2011; Priebe et al 2011; Grozeva et al 2010; BiGS; Rucker et al 2011 | |||
| 1q21.1 duplication | 145.0–146.3 | MR/DD/CM | 0.14% (55/ 38,330) | 0.03% (19/ 57,730) | 2.7 × 10−9 | 4.4 [2.6–7.4] | Mefford et al 2008; Brunetti-pierri et al 2008; Cooper et al 2011 | Vacic et al 2011; Grozeva et al 2010; Rucker et al 2011; Sanders et al 2011; Levinson et al 2011; Cooper et al 2011 |
| ASD | 0.26% (8/ 3032) | 0.03% (19/ 57,730) | 3.6 × 10−5 | 8.0 [3.5–18.4] | Bucan et al 2009; Sanders et al 2011; Pinto et al 2010 | |||
| SCZ | 0.14% (13/ 9365) | 0.03% (19/ 57,730) | 0.0002 | 4.2 [2.1–8.6] | Vacic et al 2011; Levinson et al 2011 | |||
| BD & Recurrent Depression | 0.10% (7/ 7382) | 0.03% (19/ 57,730) | 0.02 | 2.9 [1.2–6.9] | Malhotra et al 2011; Grozeva et al 2010; BiGS; Rucker et al 2011 | |||
| 3q29 deletion | 197.2–198.4 | MR/DD/CM | 0.07% (20/ 30465) | 0.002% (1/ 63,649) | 2.3 × 10−9 | 41.8 [5.6–311.6] | Ballif et al 2008; Cooper et al 2011 | Vacic et al 2011; Grozeva et al 2011; Grozeva et al 2010; Sanders et al 2011; Levinson et al 2011; Mulle et al 2010 |
| ASD | 0.05% (1/ 2120) | 0.002% (1/ 63,649) | 0.06 | 30.0 [1.9–480.4] | Sanders et al, 2011; Pinto et al 2010 | |||
| SCZ | 0.10% (10/ 10,123) | 0.002% (1/ 63,649) | 2.3 × 10−8 | 63.0 [8.1–491.7] | Vacic et al 2011; Levinson et al 2011; Mulle et al 2010 | |||
| BD | 0.04% (2/ 4659) | 0.002% (1/ 63,649) | 0.01 | 27.3 [2.5–301.5] | Malhotra et al 2011; Grozeva et al 2010; BiGS | |||
| 7q11.23 WBS deletion | 72.4–73.8 | MR/DD/CM | 0.27% (42/15767) | 0% (0/ 16,257) | 1.2 × 10−13 | Cooper et al 2011 | Vacic et al 2011; Grozeva et al 2010; Sanders et al 2011; Cooper et al 2011; BiGS | |
| ASD | 0% (0/2120) | 0% (0/ 16,257) | 1 | Sanders et al, 2011; Pinto et al 2010 | ||||
| SCZ | 0% (0/8701) | 0% (0/ 16,257) | 1 | Vacic et al 2011; Levinson et al 2011; | ||||
| BD | 0% (0/4659) | 0% (0/ 16,257) | 1 | Malhotra et al 2011; Grozeva et al 2010; BiGS | ||||
| 7q11.23 WBS duplication | 72.4–73.8 | MR/DD/CM | 0.10% (16/15767) | 0% (1/ 16,257) | 0.0001 | 16.5 [2.2–124.5] | Cooper et al 2011 | Vacic et al 2011; Grozeva et al 2010; Sanders et al 2011; Cooper et al 2011; BiGS |
| ASD | 0.19% (4/2120) | 0% (1/ 16,257) | 0.0008 | 30.7 [3.4–275.1] | Sanders et al, 2011; Pinto et al 2010 | |||
| SCZ | 0.05% (4/8701) | 0% (1/ 16,257) | 0.053 | 7.5 [0.8–66.9] | Vacic et al 2011; Levinson et al 2011; | |||
| BD | 0% (0/4659) | 0% (1/ 16,257) | 1 | Malhotra et al 2011; Grozeva et al 2010; BiGS | ||||
| VIPR2 (7q36.3) duplication | 158.4–158.8 | MR/DD/CM | 0.06% (11/ 18,499) | Sanders et al 2011; Levinson et al 2011; Grozeva et al 2010. | ||||
| ASD | 0.13% (3/ 2234) | 0.06% (11/ 18,499) | 0.18 | 2.3 [0.63–8.1] | Sanders et al 2011; Vacic et al 2011 | |||
| SCZ | 0.19% (14/ 7336) | 0.06% (11/ 18,499) | 0.006 | 3.2 [1.5–7.1] | Levinson et al 2011 | |||
| BD | 0.04% (2/ 4659) | 0.06% (11/ 18,499) | 1 | Malhotra et al 2011; Vacic et al 2011; Grozeva et al 2010 | ||||
| 15q11.2 deletion | 20.3–20.6 | MR/DD/CM | 0.51% (180/ 35,353) | 0.27% (201/ 75,486) | 4.9 × 10−10 | 1.9 [1.6–2.3] | Doornbos et al 2009; Mefford et al 2009; Burnside et al 2011; Cooper et al 2011 | Vacic et al 2011; Sanders et al 2011; Grozeva et al 2011; Rucker et al 2011; Cooper et al 2011; Kirov et al 2009; Levinson et al 2011 |
| ASD | 0.09% (2/ 2120) | 0.27% (201/ 75,486) | 0.19 | 0.3 [0.1–1.4] | Pinto et al 2010; Sanders et al 2011 | |||
| SCZ | 0.57% (72/ 12,665) | 0.27% (201/ 75,486) | 2.2 × 10−7 | 2.1 [1.6–2.8] | Vacic et al 2011; Kirov et al 2009; Levinson et al 2011 | |||
| BD & Recurrent Depression | 0.22% (18/8264) | 0.27% (201/ 75,486) | 0.49 | Priebe et al 2011; Malhotra et al 2011; BiGS; Grozeva et al 2010; Rucker et al 2011 | ||||
| 15q11.2-13.1 duplication including PWS critical region | 20.8–26.2 | MR/DD/CM | 0.17% (27/ 15767) | 0.009% (5/ 53,832) | 2.2 × 10−13 | 18.5 [7.1–47.9] | Cooper et al 2011 | Vacic et al 2011; Cooper et al 2011; Glessner et al 2009; Sanders et al 2011; Ingason et al 2011 |
| ASD | 0.39% (17/ 4315) | 0.009% (5/ 53,832) | 1.1 × 10−15 | 42.6 [15.7–115.5] | Glessner et al 2009; Sanders et al 2011; Pinto et al 2010 | |||
| SCZ | 0.05% (4/ 8384) | 0.009% (5/ 53,832) | 0.02 | 5.1 [1.4–19.1] | Vacic et al 2011; Ingason et al 2011 | |||
| BD | 0% (0/ 4659) | 0.009% (5/ 53,832) | 1 | Malhotra et al 2011; Grozeva et al 2010; BiGS | ||||
| 15q13.3 deletion | 28.7–30.2 | MR/DD/CM | 0.26% (68/ 25,647) | 0.02% (13/ 74,106) | 6.1 × 10−28 | 15.1 [8.4–27.4] | Van bon et al 2009; Sharp et al 2008; Miller et al 2009; Cooper et al 2011 | Vacic et al 2011; Grozeva et al 2011; Grozeva et al 2010; Rucker et al 2011; Sanders et al 2011; Cooper et al 2011; Levinson et al 2011 |
| ASD | 0.19% (4/ 2120) | 0.02% (13/ 74,106) | 0.001 | 10.8 [3.5–33.1] | Sanders et al 2011; Pinto et al 2010 | |||
| SCZ | 0.19% (22/ 11,689) | 0.02% (13/ 74,106) | 2.1 × 10−11 | 10.7 [5.4–21.3] | Vacic et al 2011; Levinson et al 2011 | |||
| BD & Recurrent Depression | 0.01% (1/ 7382) | 0.02% (13/ 74,106) | 1 | Malhotra et al 2011; Grozeva et al 2010; Rucker et al 2011 | ||||
| 16p13.11 duplication | 15.4–16.2 | MR/DD/CM | 0.30% (99/ 32,413) | 0.13% (81/ 62,973) | 8.4 × 10−9 | 2.4 [1.8–3.2] | Ramalingam et al 2011; Nagamani et al 2011; Mefford et al 2009; Cooper et al 2011 | Vacic et al 2011; Grozeva et al 2011; Rucker et al 2011; Sanders et al 2011; Cooper et al 2011; Ingason et al 2009 |
| ASD | 0.19% (4/ 2120) | 0.13% (81/ 62,973) | 0.36 | 1.5 [0.5–4.0] | Sanders et al 2011; Pinto et al 2010 | |||
| SCZ | 0.25% (13/ 5147) | 0.13% (81/ 62,973) | 0.03 | 2.0 [1.1–3.5] | Vacic et al 2011; Ingason et al 2009 | |||
| BD & Recurrent Depression | 0.07% (6/ 7382) | 0.13% (81/ 62,973) | 0.38 | 0.8 [0.4–1.9] | Malhotra et al 2011; BiGS; Grozeva et al 2010; Rucker et al 2011 | |||
| 16p11.2 deletion | 29.5–30.2 | MR/DD/CM | 0.41% (64/ 15767) | 0.04% (25/ 56,752) | 7.3 × 10−24 | 9.2 [5.8–14.7] | Cooper et al 2011 | Grozeva et al 2011; Glessner et al 2009; Rucker et al Sanders et al 2011; Cooper et al 2011; Levinson et al 2011 |
| ASD | 0.42% (18/ 4315) | 0.04% (25/ 56,752) | 2.0 × 10−10 | 9.5 [5.2–17.4] | Glessner et al 2009; Sanders et Pinto et al 2010 | |||
| SCZ | 0.04% (4/ 9890) | 0.04% (25/ 56,752) | 1 | 0.9 [0.3–2.6] | Levinson et al 2011 | |||
| BD & Recurrent Depression | 0.07% (6/ 8427) | 0.04% (25/ 56,752) | 0.28 | 1.6 [0.7–3.9] | Priebe et al 201; Mccarthy et al 2009; Rucker et al 2011 | |||
| 16p11.2 duplication | 29.5–30.2 | MR/DD/CM | 0.18% (18/ 15,767) | 0.03% (19/ 56,752) | 2.2 × 10−8 | 3.4 [1.8–6.5] | Cooper et al 2011 | Grozeva et al 2011; Glessner et al 2009; Rucker et al Sanders et al 2011; Cooper et al 2011; Levinson et al 2011 |
| ASD | 0.39% (17/ 4315) | 0.03% (19/ 56,752) | 6.2 × 10−11 | 11.8 [6.1–22.7] | Glessner et al 2009; Sanders et Pinto et al 2010 | |||
| SCZ | 0.31% (31/ 9890) | 0.03% (19/ 56,752) | 3.2 × 10−14 | 9.4 [5.3–16.6] | Levinson et al 2011 | |||
| BD & Recurrent Depression | 0.13% (11/ 8427) | 0.03% (19/ 56,752) | 0.0008 | 3.9 [1.9–8.2] | Priebe et al 2011; Mccarthy et al 2009; Rucker et al 2011 | |||
| 17p12/HNPP deletion | 14–15.4 | MR/DD/CM | 0.02% (3/ 15767) | 0.02% (14/ 59,086) | 1 | 0.8 [0.2–2.8] | Cooper et al 2011 | Vacic et al 2011; Cooper et al 2011; Grozeva et al 2011; Sanders et al 2011; Kirov et al 2009 |
| ASD | 0.09% (2/ 2120) | 0.02% (14/ 59,086) | 0.1 | 4.0 [0.9–17.5] | Sanders et al 2011; Pinto et al 2010 | |||
| SCZ | 0.14% (8/ 5891) | 0.02% (14/ 59,086) | 0.0004 | 5.7 [2.4–13.7] | Vacic et al 2011; Kirov et al 2009 | |||
| BD | 0.05% (3/ 5541) | 0.02% (14/ 59,086) | 0.17 | 2.3 [0.7–8.0] | Priebe et al 2011; Malhotra et al 2011; BiGS; Grozeva et al 2010 | |||
| 17q12 deletion | 31.9–33.2 | MR/DD/CM | 0.10% (32/ 31,516) | 0.006% (4/ 68,131) | 1.4 × 10−12 | 17.3 [6.1–49.0] | Cooper et al 2011; Moreno-deluca 2010 | Vacic et al 2011; Grozeva et al 2011; Sanders et al 2011; Moreno-deluca 2010; Cooper et al 2011. |
| ASD | 0.09% (2/ 2120) | 0.006% (4/ 68,131) | 0.01 | 16.0 [2.9–87.9] | Sanders et al 2011; Pinto et al 2011 | |||
| SCZ | 0.06% (4/ 7142) | 0.006% (4/ 68,131) | 0.004 | 9.5 [2.4–38.2] | Vacic et al 2011; Moreno-deluca 2010 | |||
| BD & Recurrent Depression | 0% (0/ 4659) | 0.006% (4/ 68,131) | 1 | Malhotra et al 2011; BiGS; Grozeva et al 2010 | ||||
| 22q11.21 deletion | 17.1–18.7 | MR/DD/CM | 0.61% (96/ 15767) | 0% (0/ 70,739) | 8.4 × 10−72 | Cooper et al 2011 | Vacic et al 2011; Grozeva et al 2011; Rucker et al 2011; Sanders et al 2011; Cooper et al 2011; Levinson et al 2011 | |
| ASD | 0.07% (2/ 3032) | 0% (0/ 70,739) | 0.002 | Bucan et al 2009; Sanders et al 2011; Pinto et al 2010 | ||||
| SCZ | 0.30% (36/ 12,202) | 0% (0/ 70,739) | 1.0 × 10−30 | Vacic et al 2011; Levinson et al 2011 | ||||
| BD & Recurrent Depression | 0.05% (4/ 7382) | 0% (0/ 70,739) | 8.0 × 10−5 | Malhotra et al 2011; Grozeva et al 2010; BiGS; Rucker et al 2011 | ||||
| 22q11.2 duplication | 17.1–18.7 | MR/DD/CM | 0.32% (50/ 15767) | 0.08% (23/ 27,133) | 5.9 × 10−8 | 3.7 [2.3–6.1] | Cooper et al 2011 | Vacic et al 2011; Grozeva et al 2010; Sanders et al 2011; Cooper et al 2011; Glessner et al 2009; ISC et al 2008; Rucker et al 2011 |
| ASD | 0.28% (12/ 4315) | 0.08% (23/ 27,133) | 0.002 | 3.3 [1.6–6.6] | Glessner et al 2009; Sanders et al 2011; Pinto et al 2010 | |||
| SCZ | 0.03% (3/ 8701) | 0.08% (23/ 27,133) | 0.17 | 0.4 [0.1–1.4] | Vacic et al 2011; MGS; ISC et al 2008 | |||
| BD & Recurrent Depression | 0.05% (4/ 7382) | 0.08% (23/ 27,133) | 0.49 | 0.6 [0.2–1.8] | Malhotra et al 2011; Grozeva et al 2010; BiGS; Rucker et al 2011 |
Neurodevelopmental pathways implicated in schizophrenia
Early on, it was apparent that rare CNVs tended to impact genes involved in neuronal function (Walsh et al., 2008). These included functional categories related to synaptic activity and neurodevelopment (Malhotra et al., 2011; Walsh et al., 2008). Kirov et al. interrogated, at a finer level, specific protein complexes and noted that de novo CNVs were significantly enriched for components of the N-methyl-d-aspartate receptor (NMDAR) and neuronal activity-regulated cytoskeleton-associated protein postsynaptic signaling complexes as well as other components of the postsynaptic density (Kirov et al., 2011).
Bipolar Disorder
Bipolar disorder, also known as manic–depressive illness, is a category of mood disorders defined by the presence of one or more episodes of abnormally elevated energy levels, cognition, and mood (mania), which often alternate with depressive episodes (Leibenluft, 2011). Unlike other major psychiatric disorders, severe cognitive or social deficits are not defining features of bipolar disorder. To the contrary, cognitive function may fluctuate in parallel with mood episodes, and periods of “hypomania” can be associated with enhanced function (Judd et al., 2005).
Results from early CNV studies suggest that rare variants play a role in bipolar disorder (Malhotra et al., 2011; Priebe et al., 2011; Zhang et al., 2009a). However, the pattern that is emerging appears to differ somewhat from the patterns now evident in schizophrenia and ASD. Current evidence suggests that CNVs have a role to play (Malhotra et al., 2011), but some, particularly large deletions appear to play a very limited role (Grozeva et al., 2010; Malhotra et al., 2011).
Case control studies
The results of case-control studies have been inconsistent. Two studies have reported an enrichment of rare CNVs in bipolar disorder (Priebe et al., 2011; Zhang et al., 2009a). In both studies, the observed effect was greatest in subjects with an early age-at-onset. However, the observed effects were still quite small (OR~1.5), and results from two other studies (Grozeva et al., 2010; McQuillin et al., 2011) did not support these findings. Notably, very few of the CNVs that contribute to risk for schizophrenia are also associated with bipolar disorder, the possible exceptions being microduplications of 16p11.2 (McCarthy et al., 2009) and microdeletions of 3q29 (Clayton-Smith et al., 2010; Malhotra et al., 2011), which have been reported in multiple cases (Table 1).
Family-based studies
Given the strong and reproducible associations that have been observed for de novo CNVs in ASD and Schizophrenia, it would be logical to investigate this class of mutation in mood disorders as well. In the first of such studies (Malhotra et al., 2011), we examined the rate of de novo CNVs in bipolar disorder. Frequencies of de novo CNVs were significantly higher (4.3%) in bipolar disorder as compared with healthy individuals(0.09%). The rate of de novo CNVs among cases with an age at onset younger than 18 was higher still (5.6%), and comparable to the rate we observed in schizophrenia (4.5%) using the same methods.
There is evidence to suggest that bipolar disorder consists of multiple distinct subtypes. One measure that appears to stratify some of these subtypes is age at onset (Faraone et al., 2003; Potash et al., 2007). The enrichment of inherited or de novo CNVs in subjects with an early onset of mania (Malhotra et al., 2011; Priebe et al., 2011; Zhang et al., 2009a) is consistent with the notion of distinct subtypes and suggest that individuals with an early onset of mania might constitute a subclass of bipolar disorder in which there is a greater contribution from rare alleles of large effect. Also consistent with this notion, a previous study found that segregation of early-onset bipolar disorder in families was consistent with major gene effects, whereas familial segregation of late-onset bipolar disorder was consistent with a multifactorial etiology (Grigoroiu-Serbanescu et al., 2001).
As yet, there is limited CNV evidence implicating specific genes or genomic regions in bipolar disorder. Likewise, pathway enrichment analyses have not shown clear patterns. Pathway enrichment analyses of CNV in Zhang et al reported enrichment of genes associated with psychological disorders and genes involved in learning (Zhang et al., 2009a). We examined pathways enriched among de novo CNVs in bipolar disorder and observed an enrichment of genes involved in regulation of cell shape, but we did not observe a significant enrichment of genes involved in neuronal function or development (Malhotra et al., 2011).
The Emerging Genetic Architecture of Neuropsychiatric Disease
A rare variant/heterogeneity model of common disease and its negative implications for GWAS had been acknowledged as a possibility early on (Reich and Lander, 2001). However, family data did not appear to be consistent with major gene effects. When we take into consideration some key observations of CNV studies, a rare variant model is now plausible and consistent with the genetic data. Two key aspects to consider are de novo mutation and variable expressivity.
De Novo (or Recent) Mutation
Genome-wide screens for de novo mutation have become an essential approach for gene discovery in psychiatric disease. (Kirov et al., 2011; Levy et al., 2011; Malhotra et al., 2011; Marshall et al., 2008; Sanders et al., 2011; Sebat et al., 2007; Xu et al., 2008). Some fraction of disease alleles occurs as de novo mutations, and overall this class of mutations has a low frequency (30–100 nucleotide substitutions per generation and 0.07–0.12 SVs per generation). Hence, the numbers of neutral variants in the genome are small, and de novo mutations have consistently shown the strongest genetic effect (Kirov et al., 2011; Levy et al., 2011; Malhotra et al., 2011; Marshall et al., 2008; Sanders et al., 2011; Sebat et al., 2007; Xu et al., 2008).
De novo mutation offers a possible explanation for the lack of mendelian consistency observed in family studies and the discrepancy between high monozygotic and low dizygotic twin concordance rates (Zhao et al., 2007). De novo mutation also offers a plausible explanation for the elevated incidence of psychiatric disorders observed in the offspring of men of advanced paternal age (Hultman et al., 2011).
De novo CNV is a contributing factor in 5–10% of patients. The contribution of de novo point mutation has not been fully explored but preliminary studies suggest the contribution of exomic de novo mutations to ASD to be similar (Ben Neale, Evan Eichler, Matthew State, http://www.broadinstitute.org/scientific-community/science/programs/psychiatric-disease/symposium/session-videos). All told, de novo mutation in coding regions appears to contribute in a significant but minor fraction (<20%) of ASD cases.
Of course, this is accounting only for mutations that occur spontaneously in the affected individual. Despite strong selection, rare risk alleles may persist over multiple generations. Very rarely does this persistence manifest as a near-mendelian trait (Millar et al., 2000). More typically, the phenotypic expression of the recent mutation is variable.
Variable Expressivity of CNV Genotype: Genes Don’t Code for Behavior
One of the most interesting as well as challenging observations has been the degree of phenotypic variability associated with individual CNVs, i.e. the “expressivity” of the genotype. Virtually every CNV allele that is associated with a psychiatric disorder is present at a low frequency in populations of healthy controls, and virtually every CNV is also associated with a wide variety of other neuropsychiatric or neurodevelopmental conditions including bipolar disorder, seizure disorder, intellectual disability, attention deficit hyperactivity disorder (ADHD) etc (Cooper et al., 2011; Elia et al., 2011; Girirajan and Eichler, 2010; Sahoo et al., 2011; Williams et al., 2011). Several examples of variable expressivity of CNV genotypes are described in Table 1
Some well characterized examples of variable expressivity are the clinical phenotypes associated with rearrangements at two loci, 1q21.1 (Class I/1 Mb) (Brunetti-Pierri et al., 2008; Mefford et al., 2008) and 16p11.2 (Class I/600 kb) (Bijlsma et al., 2009; Fernandez et al., 2010; Jacquemont et al., 2011; Shinawi et al., 2010). The clinical phenotypes associated with a single allele are diverse and include pediatric neurodevelopmental disorders and adult psychiatric conditions. Psychiatric diagnoses of individuals carrying identical microduplications of 1q21.1 include autism or schizophrenia (Table 1). Likewise microduplications of 16p11.2 are associated with autism, schizophrenia or bipolar disorder (McCarthy et al., 2009; Weiss et al., 2008). Both can also be carried by apparently asymptomatic individuals. Thus, even the rare subtype of a disorder (as defined by a CNV genotype) is complex.
Phenotypic variability can be attributed to other aspects of nature and nurture. Undoubtedly, the phenotypic expression of rare high-penetrance alleles is modulated by other genetic factors, including rare variants, as well as common (polygenic) variation (Purcell et al., 2009) or epigenetic regulation (Hirasawa and Feil, 2010). Indeed, evidence from CNV studies supports an oligogenic model where multiple rare variants contribute to genetic risk (Girirajan et al., 2010). Another model has been proposed that attributes phenotypic variability to a combination of locus heterogeneity and pleiotropic effects of the individual alleles (State and Levitt, 2011).
How exactly does CNV genotype relate to psychiatric phenotype? One possibility worth considering is that CNVs may not be at all specific in their effects. It has been postulated that CNVs linked to ASD are primarily associated with intellectual disability rather than with aspects of social cognition (Skuse, 2007). According to this theory, the CNV confers risk simply because clinically recognizable psychiatric conditions are more likely to arise among individuals with low intelligence. Indeed a number of large deletions are strongly associated with intellectual disability or developmental delay (Table 1). However, not all genetic findings are consistent with this model. Intellectual disability is itself a highly variable trait, and does not appear to be a primary characteristic for a number of disease-associated CNVs. Some CNV alleles have no association with intellectual disability (e.g. 17p12/HNPP) or a relatively weak one compared with the association with psychiatric phenotypes (e.g., microduplications of 1q21.1 and 16p11.2), see Table 1. In addition, a recent study of de novo CNVs in ASD has found that de novo CNVs are not a strong predictor of low intelligence quotient (Sanders et al., 2011). These observations suggest that the degree of risk conferred for a psychiatric disorder is related to specific genes within the CNV region and how changes in gene dosage influence neurodevelopment.
For some of the more well-characterized genomic disorders, a relationship between CNV genotype and clinical phenotype is beginning to emerge (Brunetti-Pierri et al., 2008; McCarthy et al., 2009). For instance, reciprocal rearrangements of 1q21.1 and 16p11.2 influence neuropsychiatric traits, susceptibility to epilepsy and head size in humans. Furthermore, deletions and duplications of each region have contrasting effects on head size and psychiatric features (McCarthy et al., 2009) (Table 1). While the underlying molecular, cellular, neuroanatomical mechanisms are still unclear, these results suggest that the psychiatric features associated with a mutation might relate to specific effects of the mutation on brain growth.
Behavioral abnormalities associated with CNVs have been confirmed in animal models (Horev et al., 2011; Nakatani et al., 2009; Peca et al., 2011; Tabuchi et al., 2007; Tamada et al., 2010). Mice with a paternal duplication of 15q11-13 display poor social interaction, behavioral inflexibility, abnormal ultrasonic vocalizations, and correlates of anxiety (Nakatani et al., 2009). Mice with reciprocal deletions and duplication of 16p11.2 have contrasting effects on mobility, grooming and repetitive behaviors (Horev et al., 2011). Mice lacking neurexin-1α display a decrease in pre-pulse inhibition, an increase in grooming behaviors, impairment in nest-building activity, and an improvement in motor learning (Etherton et al., 2009). Mice lacking Contactin-associated protein 2 (Cntnap2) display deficits in social interaction and communication, hyperactivity, and seizures (Penagarikano et al., 2011). These observations confirm some effects of CNV genotype on behavior; however determining the genes responsible for specific behavioral phenotypes in mouse and relating this to human phenotypes will be a challenge.
Compared to behavior, neuroanatomical features are more analogous between model organisms and human, and the neuroanatomical effects of CNVs might be as well. For example reciprocal deletion and duplication of 16p11.2 result in similar brain structural alterations in human and mouse, the deletion associated with brain overgrowth and the duplication associated with reduced brain volume (Horev et al., 2011; McCarthy et al., 2009; Shinawi et al., 2010), and structural alterations appear to be widely distributed across multiple brain regions. A recent study has shown that over expression of human genes from the 16p11.2 CNV region in zebrafish influences brain size (Nicholas Katsanis, http://www.schizophreniaforum.org/new/detail.asp?id=1694), consistent with the observations in human and mouse.
Relating CNV genotype to neurobiology
Specific abnormalities at the cellular level have also been linked to CNVs. Mice lacking neurexin-1α have defects in synaptic calcium channel function and neurotransmitter release (Missler et al., 2003). Mice lacking Shank3 have defects in striatal synapses and cortico-striatal circuits (Peca et al., 2011). Mice lacking Cntnap2 exhibit neuronal migration abnormalities, reduced number of interneurons, and abnormal neuronal network activity (Penagarikano et al., 2011). Furthermore, temporal lobe sections from human subjects lacking Cntnap2 display abnormal patterns of neuronal migration (Strauss et al., 2006).
Characterization of cellular phenotypes in humans is becoming tractable with the use of induced pluripotent stem cell (iPSC) technology (Dolmetsch and Geschwind, 2011). Human-derived iPSCs, which can be differentiated into a variety of neuronal cell types, offer great promise in understanding of innate cellular and molecular defects that contribute to the initiation and progression of neuropsychiatric disorders. Unlike genetically-engineered model systems, neuronal cell cultures derived from patients captures the complete set of risk alleles present in the patient germline and the genetic diversity of the patient population.
As a proof-of-principle, several recent studies have now shown that hiPSC-derived neurons from patients with psychiatric disorders exhibit significant aberrations in neuronal connectivity, synapse maturation, and synaptic function compared with those of healthy controls (Brennand et al., 2011; Cheung et al., 2011; Marchetto et al., 2010; Pasca et al., 2011). Brennand et al (2011) studied hiPSC-derived neurons from four schizophrenia patients with unknown disease etiologies. Schizophrenia-hiPSC-derived neurons had significantly reduced neuronal connectivity, reduced neurite outgrowth, reduced dendritic levels of PSD95, and altered gene expression profiles. Defects in neuronal connectivity and gene expression were ameliorated following treatment with the dopamine receptor antagonist loxapine. These early studies provide clues into the neurobiological processes that underlie schizophrenia, but without information on the genetic contributors in these patients, a clear mechanistic understanding is lacking.
hiPSC-models of monogenic disorders have begun to facilitate a mechanistic understanding of how genes contribute to disease. Pasca et al (2011) showed that human mutations in the Timothy Syndrome gene Cav1.2 influence calcium signaling and the differentiation of cortical neurons, and the observed defects on calcium (Ca2+) signaling were reversible with the L-type calcium channel blocker roscovitine. Marchetto et al (Marchetto et al., 2010) showed that cultured neurons derived from humans with mutations in the Rett Syndrome gene MeCP2 had fewer synapses, reduced spine density, smaller soma size, and exhibited a reduction in the intracellular calcium response and decrease in the frequency and amplitude of spontaneous excitatory and inhibitory postsynaptic currents.
Implications for Clinical Care
Genetic testing has value in establishing a biologically-based diagnosis. A CNV genotype may be associated with a variety of clinical features, including some that are not commonly evaluated in the psychiatric clinic. Therefore, genotype information has clear potential to influence clinical practice. The International Standard Cytogenomic Array (ISCA) consortium and American College of Medical Genetics has now established clinical guidelines for the use of chromosomal microarray analysis as a first tier diagnostic test for individuals with developmental disabilities or congenital anomalies (Miller et al., 2010). However, genetic testing has not yet been established as the nationwide standard of care for ASD, schizophrenia or bipolar disorder. Given the rapid pace of discovery in psychiatric genetics, it is likely that these new discoveries will have significant impact on clinical diagnosis and care in the coming decade.
CNV studies have directly implicated specific genes in psychiatric disease. This presents new challenges and new opportunities for the development of novel drugs. In particular, there could soon be numerous new therapeutic targets to examine. Rare subtypes of autism have spawned investigations into therapeutic mechanisms, such as the use of mGluR5 antagonists in fragile-X syndrome (Krueger and Bear, 2011). One new drug target recently identified in schizophrenia is the Vasoactive Intestinal Peptide Receptor-2 (VIPR2). Rare microduplications of VIPR2 are significantly associated with schizophrenia (Vacic et al., 2011). The VIPR2 gene encodes a class-II G-protein coupled receptor VPAC2. Disease associated variants result in overexpression of VIPR2 and increased cyclic-AMP accumulation (Vacic et al., 2011). VIPR2 has several important roles in regulating neurodevelopment and behavior (Chaudhury et al., 2008; Harmar et al., 2002; Waschek, 1995). The over expression of this receptor could have a direct relationship to the pathogenic mechanisms underlying schizophrenia. These results also suggest that a selective antagonist of VPAC2 could have therapeutic value in the treatment of schizophrenia.
New challenges also exist for drug development. A single target might contribute genetic risk to only a small fraction (<1%) of patients, and a compound active against a single target might benefit only patients with that mutation. Hence, drug discovery for neuropsychiatric diseases could involve developing a catalogue of drugs targeting a variety of orphan diseases (Braun et al., 2010). More optimistically, one target gene might represent one component of a pathway that is dysregulated in a larger proportion of cases. Thus, the “orphan” drug designed to treat a rare disorder might turn out to have efficacy in a broader class of patients.
Future Directions
The majority of CNV contribution to disease remains unknown. The genetic associations listed in Table 1 consist almost entirely of genomic hotspots (Mefford and Eichler, 2009). These represent the largest and most pathogenic risk alleles. However, in studies of de novo CNV these hotspots represent 25% of mutations and thus, probably represent a minority of the risk variants. The majority are non-recurrent mutations, which have lower mutation rates and lower frequencies and will require larger studies to unequivocally demonstrate an association with disease. Such large scale studies are underway through International efforts including by the Psychiatric Genomics Consortium (PGC) (Ripke et al., 2011) and Wellcome Trust Case Control Consortium (WTCCC). Large Scale meta-analysis of GWAS has obtained statistically convincing evidence for common variants in schizophrenia (Ripke et al., 2011) and bipolar disorder (Sklar et al., 2011). These efforts are accompanied by ongoing CNV studies of the same cohorts, and will be well powered to capture additional risk genes.
A growing body of research on CNV provides a compelling rationale for undertaking a complementary sequencing approach to psychiatric disease. The era of high-throughput sequencing is now in full swing, with efforts currently under way to sequence exomes and whole genomes in all major psychiatric disorders (Girard et al., 2011; Najmabadi et al., 2011; O’Roak et al., 2011; Vissers et al., 2010; Xu et al., 2011). These efforts promise to capture a larger fraction of rare genetic variation and increase the proportion of genetic risk that can be explained.
The nature of rare CNV alleles in psychiatric disease -risk alleles arising by recent de novo mutation, conferring significant disease risks, and having highly variable phenotypic expression - is likely to be the nature of rare alleles in general. This knowledge will serve as a guide as we move forward into the era of complete genome sequencing. Genetic approaches that have worked well for CNVs (Figure 3) should adapt well to sequencing platforms. Indeed the pioneering exome studies of ASD and schizophrenia have begun with a strong focus on de novo mutations in trios, with compelling preliminary results (Girard et al., 2011; O’Roak et al., 2011; Vissers et al., 2010; Xu et al., 2011). As always, success will depend on statistical power and sample size. However, as we move ahead, success will increasingly depend on our ability to integrate the signal from de novo, inherited, common, and rare forms of variation in the genome.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alkan C, Coe BP, Eichler EE. Genome structural variation discovery and genotyping. Nat Rev Genet. 2011;12:363–376. doi: 10.1038/nrg2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkuraya FS. Homozygosity mapping: one more tool in the clinical geneticist’s toolbox. Genet Med. 2010;12:236–239. doi: 10.1097/GIM.0b013e3181ceb95d. [DOI] [PubMed] [Google Scholar]
- Bailey A, Le Couteur A, Gottesman I, Bolton P, Simonoff E, Yuzda E, Rutter M. Autism as a strongly genetic disorder: evidence from a British twin study. Psychol Med. 1995;25:63–77. doi: 10.1017/s0033291700028099. [DOI] [PubMed] [Google Scholar]
- Bakkaloglu B, O’Roak BJ, Louvi A, Gupta AR, Abelson JF, Morgan TM, Chawarska K, Klin A, Ercan-Sencicek AG, Stillman AA, et al. Molecular cytogenetic analysis and resequencing of contactin associated protein-like 2 in autism spectrum disorders. Am J Hum Genet. 2008;82:165–173. doi: 10.1016/j.ajhg.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck CR, Garcia-Perez JL, Badge RM, Moran JV. LINE-1 elements in structural variation and disease. Annu Rev Genomics Hum Genet. 2011;12:187–215. doi: 10.1146/annurev-genom-082509-141802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkel S, Marshall CR, Weiss B, Howe J, Roeth R, Moog U, Endris V, Roberts W, Szatmari P, Pinto D, et al. Mutations in the SHANK2 synaptic scaffolding gene in autism spectrum disorder and mental retardation. Nat Genet. 2010;42:489–491. doi: 10.1038/ng.589. [DOI] [PubMed] [Google Scholar]
- Bijlsma EK, Gijsbers AC, Schuurs-Hoeijmakers JH, van Haeringen A, Fransen van de Putte DE, Anderlid BM, Lundin J, Lapunzina P, Jurado LA, Delle Chiaie B, et al. Extending the phenotype of recurrent rearrangements of 16p11.2: deletions in mentally retarded patients without autism and in normal individuals. Eur J Med Genet. 2009 doi: 10.1016/j.ejmg.2009.03.006. [DOI] [PubMed] [Google Scholar]
- Bodmer W, Bonilla C. Common and rare variants in multifactorial susceptibility to common diseases. Nat Genet. 2008;40:695–701. doi: 10.1038/ng.f.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botstein D, Risch N. Discovering genotypes underlying human phenotypes: past successes for mendelian disease, future approaches for complex disease. Nat Genet. 2003;33(Suppl):228–237. doi: 10.1038/ng1090. [DOI] [PubMed] [Google Scholar]
- Bourgeron T. A synaptic trek to autism. Curr Opin Neurobiol. 2009;19:231–234. doi: 10.1016/j.conb.2009.06.003. [DOI] [PubMed] [Google Scholar]
- Braun MM, Farag-El-Massah S, Xu K, Cote TR. Emergence of orphan drugs in the United States: a quantitative assessment of the first 25 years. Nat Rev Drug Discov. 2010;9:519–522. doi: 10.1038/nrd3160. [DOI] [PubMed] [Google Scholar]
- Brennand KJ, Simone A, Jou J, Gelboin-Burkhart C, Tran N, Sangar S, Li Y, Mu Y, Chen G, Yu D, et al. Modelling schizophrenia using human induced pluripotent stem cells. Nature. 2011;473:221–225. doi: 10.1038/nature09915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunetti-Pierri N, Berg JS, Scaglia F, Belmont J, Bacino CA, Sahoo T, Lalani SR, Graham B, Lee B, Shinawi M, et al. Recurrent reciprocal 1q21.1 deletions and duplications associated with microcephaly or macrocephaly and developmental and behavioral abnormalities. Nat Genet. 2008;40:1466–1471. doi: 10.1038/ng.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucan M, Abrahams BS, Wang K, Glessner JT, Herman EI, Sonnenblick LI, Alvarez Retuerto AI, Imielinski M, Hadley D, Bradfield JP, et al. Genome-wide analyses of exonic copy number variants in a family-based study point to novel autism susceptibility genes. PLoS Genet. 2009;5:e1000536. doi: 10.1371/journal.pgen.1000536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buizer-Voskamp JE, Muntjewerff JW, Strengman E, Sabatti C, Stefansson H, Vorstman JA, Ophoff RA. Genome-wide analysis shows increased frequency of copy number variation deletions in dutch schizophrenia patients. Biol Psychiatry. 2011;70:655–662. doi: 10.1016/j.biopsych.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho CM, Ramocki MB, Pehlivan D, Franco LM, Gonzaga-Jauregui C, Fang P, McCall A, Pivnick EK, Hines-Dowell S, Seaver LH, et al. Inverted genomic segments and complex triplication rearrangements are mediated by inverted repeats in the human genome. Nat Genet. 2011;43:1074–1081. doi: 10.1038/ng.944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhury D, Loh DH, Dragich JM, Hagopian A, Colwell CS. Select cognitive deficits in vasoactive intestinal peptide deficient mice. BMC Neurosci. 2008;9:63. doi: 10.1186/1471-2202-9-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung AY, Horvath LM, Grafodatskaya D, Pasceri P, Weksberg R, Hotta A, Carrel L, Ellis J. Isolation of MECP2-null Rett Syndrome patient hiPS cells and isogenic controls through X-chromosome inactivation. Hum Mol Genet. 2011;20:2103–2115. doi: 10.1093/hmg/ddr093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton-Smith J, Giblin C, Smith RA, Dunn C, Willatt L. Familial 3q29 microdeletion syndrome providing further evidence of involvement of the 3q29 region in bipolar disorder. Clin Dysmorphol. 2010;19:128–132. doi: 10.1097/MCD.0b013e32833a1e3c. [DOI] [PubMed] [Google Scholar]
- Conrad DF, Keebler JE, DePristo MA, Lindsay SJ, Zhang Y, Casals F, Idaghdour Y, Hartl CL, Torroja C, Garimella KV, et al. Variation in genome-wide mutation rates within and between human families. Nat Genet. 2011;43:712–714. doi: 10.1038/ng.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad DF, Pinto D, Redon R, Feuk L, Gokcumen O, Zhang Y, Aerts J, Andrews TD, Barnes C, Campbell P, et al. Origins and functional impact of copy number variation in the human genome. Nature. 2010;464:704–712. doi: 10.1038/nature08516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium IH. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper GM, Coe BP, Girirajan S, Rosenfeld JA, Vu TH, Baker C, Williams C, Stalker H, Hamid R, Hannig V, et al. A copy number variation morbidity map of developmental delay. Nat Genet. 2011;43:838–846. doi: 10.1038/ng.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nat Rev Genet. 2009;10:691–703. doi: 10.1038/nrg2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devriendt K, Fryns JP, Mortier G, van Thienen MN, Keymolen K. The annual incidence of DiGeorge/velocardiofacial syndrome. J Med Genet. 1998;35:789–790. doi: 10.1136/jmg.35.9.789-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolmetsch R, Geschwind DH. The human brain in a dish: the promise of iPSC-derived neurons. Cell. 2011;145:831–834. doi: 10.1016/j.cell.2011.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand CM, Betancur C, Boeckers TM, Bockmann J, Chaste P, Fauchereau F, Nygren G, Rastam M, Gillberg IC, Anckarsater H, et al. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat Genet. 2007;39:25–27. doi: 10.1038/ng1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers MD. Activity level controls postsynaptic composition and signaling via the ubiquitin-proteasome system. Nat Neurosci. 2003;6:231–242. doi: 10.1038/nn1013. [DOI] [PubMed] [Google Scholar]
- Elia J, Glessner JT, Wang K, Takahashi N, Shtir CJ, Hadley D, Sleiman PM, Zhang H, Kim CE, Robison R, et al. Genome-wide copy number variation study associates metabotropic glutamate receptor gene networks with attention deficit hyperactivity disorder. Nat Genet. 2011;44:78–84. doi: 10.1038/ng.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etherton MR, Blaiss CA, Powell CM, Sudhof TC. Mouse neurexin-1alpha deletion causes correlated electrophysiological and behavioral changes consistent with cognitive impairments. Proc Natl Acad Sci U S A. 2009;106:17998–18003. doi: 10.1073/pnas.0910297106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faraone SV, Glatt SJ, Tsuang MT. The genetics of pediatric-onset bipolar disorder. Biol Psychiatry. 2003;53:970–977. doi: 10.1016/s0006-3223(02)01893-0. [DOI] [PubMed] [Google Scholar]
- Fernandez BA, Roberts W, Chung B, Weksberg R, Meyn S, Szatmari P, Joseph-George AM, Mackay S, Whitten K, Noble B, et al. Phenotypic spectrum associated with de novo and inherited deletions and duplications at 16p11.2 in individuals ascertained for diagnosis of autism spectrum disorder. J Med Genet. 2010;47:195–203. doi: 10.1136/jmg.2009.069369. [DOI] [PubMed] [Google Scholar]
- Fernandez T, Morgan T, Davis N, Klin A, Morris A, Farhi A, Lifton RP, State MW. Disruption of contactin 4 (CNTN4) results in developmental delay and other features of 3p deletion syndrome. Am J Hum Genet. 2004;74:1286–1293. doi: 10.1086/421474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez T, Morgan T, Davis N, Klin A, Morris A, Farhi A, Lifton RP, State MW. Disruption of Contactin 4 (CNTN4) results in developmental delay and other features of 3p deletion syndrome. Am J Hum Genet. 2008;82:1385. doi: 10.1016/j.ajhg.2008.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folstein S, Rutter M. Genetic influences and infantile autism. Nature. 1977;265:726–728. doi: 10.1038/265726a0. [DOI] [PubMed] [Google Scholar]
- Folstein SE, Rosen-Sheidley B. Genetics of autism: complex aetiology for a heterogeneous disorder. Nat Rev Genet. 2001;2:943–955. doi: 10.1038/35103559. [DOI] [PubMed] [Google Scholar]
- Freitag CM, Staal W, Klauck SM, Duketis E, Waltes R. Genetics of autistic disorders: review and clinical implications. Eur Child Adolesc Psychiatry. 2010;19:169–178. doi: 10.1007/s00787-009-0076-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier J, Spiegelman D, Piton A, Lafreniere RG, Laurent S, St-Onge J, Lapointe L, Hamdan FF, Cossette P, Mottron L, et al. Novel de novo SHANK3 mutation in autistic patients. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:421–424. doi: 10.1002/ajmg.b.30822. [DOI] [PubMed] [Google Scholar]
- Gillberg C. Chromosomal disorders and autism. J Autism Dev Disord. 1998;28:415–425. doi: 10.1023/a:1026004505764. [DOI] [PubMed] [Google Scholar]
- Gilman SR, Iossifov I, Levy D, Ronemus M, Wigler M, Vitkup D. Rare de novo variants associated with autism implicate a large functional network of genes involved in formation and function of synapses. Neuron. 2011;70:898–907. doi: 10.1016/j.neuron.2011.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard SL, Gauthier J, Noreau A, Xiong L, Zhou S, Jouan L, Dionne-Laporte A, Spiegelman D, Henrion E, Diallo O, et al. Increased exonic de novo mutation rate in individuals with schizophrenia. Nat Genet. 2011;43:860–863. doi: 10.1038/ng.886. [DOI] [PubMed] [Google Scholar]
- Girirajan S, Eichler EE. Phenotypic variability and genetic susceptibility to genomic disorders. Hum Mol Genet. 2010;19:R176–187. doi: 10.1093/hmg/ddq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girirajan S, Rosenfeld JA, Cooper GM, Antonacci F, Siswara P, Itsara A, Vives L, Walsh T, McCarthy SE, Baker C, et al. A recurrent 16p12.1 microdeletion supports a two-hit model for severe developmental delay. Nat Genet. 2010;42:203–209. doi: 10.1038/ng.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glessner JT, Wang K, Cai G, Korvatska O, Kim CE, Wood S, Zhang H, Estes A, Brune CW, Bradfield JP, et al. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature. 2009 doi: 10.1038/nature07953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodier JL, Kazazian HH., Jr Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell. 2008;135:23–35. doi: 10.1016/j.cell.2008.09.022. [DOI] [PubMed] [Google Scholar]
- Grigoroiu-Serbanescu M, Martinez M, Nothen MM, Grinberg M, Sima D, Propping P, Marinescu E, Hrestic M. Different familial transmission patterns in bipolar I disorder with onset before and after age 25. Am J Med Genet. 2001;105:765–773. doi: 10.1002/ajmg.10047. [DOI] [PubMed] [Google Scholar]
- Grozeva D, Kirov G, Ivanov D, Jones IR, Jones L, Green EK, St Clair DM, Young AH, Ferrier N, Farmer AE, et al. Rare copy number variants: a point of rarity in genetic risk for bipolar disorder and schizophrenia. Arch Gen Psychiatry. 2010;67:318–327. doi: 10.1001/archgenpsychiatry.2010.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Zhang F, Lupski JR. Mechanisms for human genomic rearrangements. Pathogenetics. 2008;1:4. doi: 10.1186/1755-8417-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallmayer J, Cleveland S, Torres A, Phillips J, Cohen B, Torigoe T, Miller J, Fedele A, Collins J, Smith K, et al. Genetic heritability and shared environmental factors among twin pairs with autism. Arch Gen Psychiatry. 2011;68:1095–1102. doi: 10.1001/archgenpsychiatry.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmar AJ, Marston HM, Shen S, Spratt C, West KM, Sheward WJ, Morrison CF, Dorin JR, Piggins HD, Reubi JC, et al. The VPAC(2) receptor is essential for circadian function in the mouse suprachiasmatic nuclei. Cell. 2002;109:497–508. doi: 10.1016/s0092-8674(02)00736-5. [DOI] [PubMed] [Google Scholar]
- Hastings PJ, Ira G, Lupski JR. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet. 2009a;5:e1000327. doi: 10.1371/journal.pgen.1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings PJ, Lupski JR, Rosenberg SM, Ira G. Mechanisms of change in gene copy number. Nat Rev Genet. 2009b;10:551–564. doi: 10.1038/nrg2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirasawa R, Feil R. Genomic imprinting and human disease. Essays Biochem. 2010;48:187–200. doi: 10.1042/bse0480187. [DOI] [PubMed] [Google Scholar]
- Horev G, Ellegood J, Lerch JP, Son YE, Muthuswamy L, Vogel H, Krieger AM, Buja A, Henkelman RM, Wigler M, et al. Dosage-dependent phenotypes in models of 16p11.2 lesions found in autism. Proc Natl Acad Sci U S A. 2011;108:17076–17081. doi: 10.1073/pnas.1114042108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hultman CM, Sandin S, Levine SZ, Lichtenstein P, Reichenberg A. Advancing paternal age and risk of autism: new evidence from a population-based study and a meta-analysis of epidemiological studies. Mol Psychiatry. 2011;16:1203–1212. doi: 10.1038/mp.2010.121. [DOI] [PubMed] [Google Scholar]
- Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, Scherer SW, Lee C. Detection of large-scale variation in the human genome. Nat Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- International schizophrenia Consortium. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itsara A, Wu H, Smith JD, Nickerson DA, Romieu I, London SJ, Eichler EE. De novo rates and selection of large copy number variation. Genome Res. 2010;20:1469–1481. doi: 10.1101/gr.107680.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquemont S, Reymond A, Zufferey F, Harewood L, Walters RG, Kutalik Z, Martinet D, Shen Y, Valsesia A, Beckmann ND, et al. Mirror extreme BMI phenotypes associated with gene dosage at the chromosome 16p11.2 locus. Nature. 2011;478:97–102. doi: 10.1038/nature10406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamain S, Quach H, Betancur C, Rastam M, Colineaux C, Gillberg IC, Soderstrom H, Giros B, Leboyer M, Gillberg C, et al. Mutations of the X-linked genes encoding neuroligins NLGN3 and NLGN4 are associated with autism. Nat Genet. 2003;34:27–29. doi: 10.1038/ng1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judd LL, Akiskal HS, Schettler PJ, Endicott J, Leon AC, Solomon DA, Coryell W, Maser JD, Keller MB. Psychosocial disability in the course of bipolar I and II disorders: a prospective, comparative, longitudinal study. Arch Gen Psychiatry. 2005;62:1322–1330. doi: 10.1001/archpsyc.62.12.1322. [DOI] [PubMed] [Google Scholar]
- Karayiorgou M, Morris MA, Morrow B, Shprintzen RJ, Goldberg R, Borrow J, Gos A, Nestadt G, Wolyniec PS, Lasseter VK, et al. schizophrenia susceptibility associated with interstitial deletions of chromosome 22q11. Proc Natl Acad Sci U S A. 1995;92:7612–7616. doi: 10.1073/pnas.92.17.7612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HG, Kishikawa S, Higgins AW, Seong IS, Donovan DJ, Shen Y, Lally E, Weiss LA, Najm J, Kutsche K, et al. Disruption of neurexin 1 associated with autism spectrum disorder. Am J Hum Genet. 2008;82:199–207. doi: 10.1016/j.ajhg.2007.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirov G, Grozeva D, Norton N, Ivanov D, Mantripragada KK, Holmans P, Craddock N, Owen MJ, O’Donovan MC. Support for the involvement of large cnvs in the pathogenesis of schizophrenia. Hum Mol Genet. 2009 doi: 10.1093/hmg/ddp043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirov G, Pocklington AJ, Holmans P, Ivanov D, Ikeda M, Ruderfer D, Moran J, Chambert K, Toncheva D, Georgieva L, et al. De novo CNV analysis implicates specific abnormalities of postsynaptic signalling complexes in the pathogenesis of schizophrenia. Mol Psychiatry. 2011 doi: 10.1038/mp.2011.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein RJ, Xu X, Mukherjee S, Willis J, Hayes J. Successes of genome-wide association studies. Cell. 2010;142:350–351. doi: 10.1016/j.cell.2010.07.026. author reply 353–355. [DOI] [PubMed] [Google Scholar]
- Krueger DD, Bear MF. Toward fulfilling the promise of molecular medicine in fragile X syndrome. Annu Rev Med. 2011;62:411–429. doi: 10.1146/annurev-med-061109-134644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam KW, Jeffreys AJ. Processes of copy-number change in human DNA: the dynamics of {alpha}-globin gene deletion. Proc Natl Acad Sci U S A. 2006;103:8921–8927. doi: 10.1073/pnas.0602690103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam KW, Jeffreys AJ. Processes of de novo duplication of human alpha-globin genes. Proc Natl Acad Sci U S A. 2007;104:10950–10955. doi: 10.1073/pnas.0703856104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laumonnier F, Bonnet-Brilhault F, Gomot M, Blanc R, David A, Moizard MP, Raynaud M, Ronce N, Lemonnier E, Calvas P, et al. X-linked mental retardation and autism are associated with a mutation in the NLGN4 gene, a member of the neuroligin family. Am J Hum Genet. 2004;74:552–557. doi: 10.1086/382137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JA, Carvalho CM, Lupski JR. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell. 2007;131:1235–1247. doi: 10.1016/j.cell.2007.11.037. [DOI] [PubMed] [Google Scholar]
- Leibenluft E. Severe mood dysregulation, irritability, and the diagnostic boundaries of bipolar disorder in youths. Am J Psychiatry. 2011;168:129–142. doi: 10.1176/appi.ajp.2010.10050766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leucht S, Corves C, Arbter D, Engel RR, Li C, Davis JM. Second-generation versus first-generation antipsychotic drugs for schizophrenia: a meta-analysis. Lancet. 2009;373:31–41. doi: 10.1016/S0140-6736(08)61764-X. [DOI] [PubMed] [Google Scholar]
- Levy D, Ronemus M, Yamrom B, Lee YH, Leotta A, Kendall J, Marks S, Lakshmi B, Pai D, Ye K, et al. Rare de novo and transmitted copy-number variation in autistic spectrum disorders. Neuron. 2011;70:886–897. doi: 10.1016/j.neuron.2011.05.015. [DOI] [PubMed] [Google Scholar]
- Lieber MR. The mechanism of human nonhomologous DNA end joining. J Biol Chem. 2008;283:1–5. doi: 10.1074/jbc.R700039200. [DOI] [PubMed] [Google Scholar]
- Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Erez A, Nagamani SC, Dhar SU, Kolodziejska KE, Dharmadhikari AV, Cooper ML, Wiszniewska J, Zhang F, Withers MA, et al. Chromosome catastrophes involve replication mechanisms generating complex genomic rearrangements. Cell. 2011a;146:889–903. doi: 10.1016/j.cell.2011.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Lacaria M, Zhang F, Withers M, Hastings PJ, Lupski JR. Frequency of nonallelic homologous recombination is correlated with length of homology: evidence that ectopic synapsis precedes ectopic crossing-over. Am J Hum Genet. 2011b;89:580–588. doi: 10.1016/j.ajhg.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupski JR. Retrotransposition and structural variation in the human genome. Cell. 2010;141:1110–1112. doi: 10.1016/j.cell.2010.06.014. [DOI] [PubMed] [Google Scholar]
- Malhotra D, McCarthy S, Michaelson JJ, Vacic V, Burdick KE, Yoon S, Cichon S, Corvin A, Gary S, Gershon ES, et al. High Frequencies of De Novo CNVs in bipolar disorder and Schizophrenia. Neuron. 2011;72:951–963. doi: 10.1016/j.neuron.2011.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio TA, Brooks LD, Collins FS. A HapMap harvest of insights into the genetics of common disease. J Clin Invest. 2008;118:1590–1605. doi: 10.1172/JCI34772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetto MC, Carromeu C, Acab A, Yu D, Yeo GW, Mu Y, Chen G, Gage FH, Muotri AR. A model for neural development and treatment of Rett syndrome using human induced pluripotent stem cells. Cell. 2010;143:527–539. doi: 10.1016/j.cell.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall CR, Noor A, Vincent JB, Lionel AC, Feuk L, Skaug J, Shago M, Moessner R, Pinto D, Ren Y, et al. Structural Variation of Chromosomes in Autism Spectrum Disorder. The American Journal of Human Genetics. 2008;82:477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarroll SA, Kuruvilla FG, Korn JM, Cawley S, Nemesh J, Wysoker A, Shapero MH, de Bakker PI, Maller JB, Kirby A, et al. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat Genet. 2008;40:1166–1174. doi: 10.1038/ng.238. [DOI] [PubMed] [Google Scholar]
- McCarthy SE, Makarov V, Kirov G, Addington AM, McClellan J, Yoon S, Perkins DO, Dickel DE, Kusenda M, Krastoshevsky O, et al. Microduplications of 16p11.2 are associated with schizophrenia. Nat Genet. 2009;41:1223–1227. doi: 10.1038/ng.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClellan J, King MC. Genetic heterogeneity in human disease. Cell. 2010;141:210–217. doi: 10.1016/j.cell.2010.03.032. [DOI] [PubMed] [Google Scholar]
- McQuillin A, Bass N, Anjorin A, Lawrence J, Kandaswamy R, Lydall G, Moran J, Sklar P, Purcell S, Gurling H. Analysis of genetic deletions and duplications in the University College London bipolar disorder case control sample. Eur J Hum Genet. 2011;19:588–592. doi: 10.1038/ejhg.2010.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mefford HC, Eichler EE. Duplication hotspots, rare genomic disorders, and common disease. Curr Opin Genet Dev. 2009;19:196–204. doi: 10.1016/j.gde.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mefford HC, Sharp AJ, Baker C, Itsara A, Jiang Z, Buysse K, Huang S, Maloney VK, Crolla JA, Baralle D, et al. Recurrent rearrangements of chromosome 1q21.1 and variable pediatric phenotypes. N Engl J Med. 2008;359:1685–1699. doi: 10.1056/NEJMoa0805384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles JH. Autism spectrum disorders--a genetics review. Genet Med. 2011;13:278–294. doi: 10.1097/GIM.0b013e3181ff67ba. [DOI] [PubMed] [Google Scholar]
- Millar JK, Wilson-Annan JC, Anderson S, Christie S, Taylor MS, Semple CA, Devon RS, Clair DM, Muir WJ, Blackwood DH, et al. Disruption of two novel genes by a translocation co-segregating with schizophrenia. Hum Mol Genet. 2000;9:1415–1423. doi: 10.1093/hmg/9.9.1415. [DOI] [PubMed] [Google Scholar]
- Miller DT, Adam MP, Aradhya S, Biesecker LG, Brothman AR, Carter NP, Church DM, Crolla JA, Eichler EE, Epstein CJ, et al. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet. 2010;86:749–764. doi: 10.1016/j.ajhg.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills RE, Walter K, Stewart C, Handsaker RE, Chen K, Alkan C, Abyzov A, Yoon SC, Ye K, Cheetham RK, et al. Mapping copy number variation by population-scale genome sequencing. Nature. 2011;470:59–65. doi: 10.1038/nature09708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missler M, Zhang W, Rohlmann A, Kattenstroth G, Hammer RE, Gottmann K, Sudhof TC. Alpha-neurexins couple Ca2+ channels to synaptic vesicle exocytosis. Nature. 2003;423:939–948. doi: 10.1038/nature01755. [DOI] [PubMed] [Google Scholar]
- Moessner R, Marshall CR, Sutcliffe JS, Skaug J, Pinto D, Vincent J, Zwaigenbaum L, Fernandez B, Roberts W, Szatmari P, et al. Contribution of SHANK3 Mutations to Autism Spectrum Disorder. The American Journal of Human Genetics. 2007;81:1289–1297. doi: 10.1086/522590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy KC, Jones LA, Owen MJ. High rates of schizophrenia in adults with velo-cardio-facial syndrome. Arch Gen Psychiatry. 1999;56:940–945. doi: 10.1001/archpsyc.56.10.940. [DOI] [PubMed] [Google Scholar]
- Najmabadi H, Hu H, Garshasbi M, Zemojtel T, Abedini SS, Chen W, Hosseini M, Behjati F, Haas S, Jamali P, et al. Deep sequencing reveals 50 novel genes for recessive cognitive disorders. Nature. 2011;478:57–63. doi: 10.1038/nature10423. [DOI] [PubMed] [Google Scholar]
- Nakatani J, Tamada K, Hatanaka F, Ise S, Ohta H, Inoue K, Tomonaga S, Watanabe Y, Chung YJ, Banerjee R, et al. Abnormal behavior in a chromosome-engineered mouse model for human 15q11-13 duplication seen in autism. Cell. 2009;137:1235–1246. doi: 10.1016/j.cell.2009.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor A, Whibley A, Marshall CR, Gianakopoulos PJ, Piton A, Carson AR, Orlic-Milacic M, Lionel AC, Sato D, Pinto D, et al. Disruption at the PTCHD1 Locus on Xp22.11 in Autism spectrum disorder and intellectual disability. Sci Transl Med. 2010;2:49ra68. doi: 10.1126/scitranslmed.3001267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Roak BJ, Deriziotis P, Lee C, Vives L, Schwartz JJ, Girirajan S, Karakoc E, Mackenzie AP, Ng SB, Baker C, et al. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nat Genet. 2011;43:585–589. doi: 10.1038/ng.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasca SP, Portmann T, Voineagu I, Yazawa M, Shcheglovitov A, Pasca AM, Cord B, Palmer TD, Chikahisa S, Nishino S, et al. Using iPSC-derived neurons to uncover cellular phenotypes associated with Timothy syndrome. Nat Med. 2011;17:1657–1662. doi: 10.1038/nm.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peca J, Feliciano C, Ting JT, Wang W, Wells MF, Venkatraman TN, Lascola CD, Fu Z, Feng G. Shank3 mutant mice display autistic-like behaviours and striatal dysfunction. Nature. 2011;472:437–442. doi: 10.1038/nature09965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penagarikano O, Abrahams BS, Herman EI, Winden KD, Gdalyahu A, Dong H, Sonnenblick LI, Gruver R, Almajano J, Bragin A, et al. Absence of CNTNAP2 leads to epilepsy, neuronal migration abnormalities, and core autism-related deficits. Cell. 2011;147:235–246. doi: 10.1016/j.cell.2011.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D, Pagnamenta AT, Klei L, Anney R, Merico D, Regan R, Conroy J, Magalhaes TR, Correia C, Abrahams BS, et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466:368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potash JB, Toolan J, Steele J, Miller EB, Pearl J, Zandi PP, Schulze TG, Kassem L, Simpson SG, Lopez V, et al. The bipolar disorder phenome database: a resource for genetic studies. Am J Psychiatry. 2007;164:1229–1237. doi: 10.1176/appi.ajp.2007.06122045. [DOI] [PubMed] [Google Scholar]
- Priebe L, Degenhardt FA, Herms S, Haenisch B, Mattheisen M, Nieratschker V, Weingarten M, Witt S, Breuer R, Paul T, et al. Genome-wide survey implicates the influence of copy number variants (CNVs) in the development of early-onset bipolar disorder. Mol Psychiatry. 2011 doi: 10.1038/mp.2011.8. [DOI] [PubMed] [Google Scholar]
- Purcell SM, Wray NR, Stone JL, Visscher PM, O’Donovan MC, Sullivan PF, Sklar P. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich DE, Lander ES. On the allelic spectrum of human disease. Trends Genet. 2001;17:502–510. doi: 10.1016/s0168-9525(01)02410-6. [DOI] [PubMed] [Google Scholar]
- Ripke S, Sanders AR, Kendler KS, Levinson DF, Sklar P, Holmans PA, Lin DY, Duan J, Ophoff RA, Andreassen OA, et al. Genome-wide association study identifies five new schizophrenia loci. Nat Genet. 2011 doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1516–1517. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- Roohi J, Montagna C, Tegay DH, Palmer LE, DeVincent C, Pomeroy JC, Christian SL, Nowak N, Hatchwell E. Disruption of contactin 4 in three subjects with autism spectrum disorder. J Med Genet. 2009;46:176–182. doi: 10.1136/jmg.2008.057505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg RE, Law JK, Yenokyan G, McGready J, Kaufmann WE, Law PA. Characteristics and concordance of autism spectrum disorders among 277 twin pairs. Arch Pediatr Adolesc Med. 2009;163:907–914. doi: 10.1001/archpediatrics.2009.98. [DOI] [PubMed] [Google Scholar]
- Sahoo T, Theisen A, Rosenfeld JA, Lamb AN, Ravnan JB, Schultz RA, Torchia BS, Neill N, Casci I, Bejjani BA, et al. Copy number variants of schizophrenia susceptibility loci are associated with a spectrum of speech and developmental delays and behavior problems. Genet Med. 2011;13:868–880. doi: 10.1097/GIM.0b013e3182217a06. [DOI] [PubMed] [Google Scholar]
- Sanders SJ, Ercan-Sencicek AG, Hus V, Luo R, Murtha MT, Moreno-De-Luca D, Chu SH, Moreau MP, Gupta AR, Thomson SA, et al. Multiple Recurrent De Novo CNVs, Including Duplications of the 7q11.23 Williams Syndrome Region, Are Strongly Associated with Autism. Neuron. 2011;70:863–885. doi: 10.1016/j.neuron.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, Yamrom B, Yoon S, Krasnitz A, Kendall J, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Troge J, Alexander J, Young J, Lundin P, Maner S, Massa H, Walker M, Chi M, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- Sebat J, Levy DL, McCarthy SE. Rare structural variants in schizophrenia: one disorder, multiple mutations; one mutation, multiple disorders. Trends Genet. 2009;25:528–535. doi: 10.1016/j.tig.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinawi M, Liu P, Kang SH, Shen J, Belmont JW, Scott DA, Probst FJ, Craigen WJ, Graham BH, Pursley A, et al. Recurrent reciprocal 16p11.2 rearrangements associated with global developmental delay, behavioural problems, dysmorphism, epilepsy, and abnormal head size. J Med Genet. 2010;47:332–341. doi: 10.1136/jmg.2009.073015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sklar P, Ripke S, Scott LJ, Andreassen OA, Cichon S, Craddock N, Edenberg HJ, Nurnberger JI, Jr, Rietschel M, Blackwood D, et al. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat Genet. 2011 doi: 10.1038/ng.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skuse DH. Rethinking the nature of genetic vulnerability to autistic spectrum disorders. Trends Genet. 2007;23:387–395. doi: 10.1016/j.tig.2007.06.003. [DOI] [PubMed] [Google Scholar]
- State MW, Levitt P. The conundrums of understanding genetic risks for autism spectrum disorders. Nat Neurosci. 2011 doi: 10.1038/nn.2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffenburg S, Gillberg C, Hellgren L, Andersson L, Gillberg IC, Jakobsson G, Bohman M. A twin study of autism in Denmark, Finland, Iceland, Norway and Sweden. J Child Psychol Psychiatry. 1989;30:405–416. doi: 10.1111/j.1469-7610.1989.tb00254.x. [DOI] [PubMed] [Google Scholar]
- Strauss KA, Puffenberger EG, Huentelman MJ, Gottlieb S, Dobrin SE, Parod JM, Stephan DA, Morton DH. Recessive symptomatic focal epilepsy and mutant contactin-associated protein-like 2. N Engl J Med. 2006;354:1370–1377. doi: 10.1056/NEJMoa052773. [DOI] [PubMed] [Google Scholar]
- Sudmant PH, Kitzman JO, Antonacci F, Alkan C, Malig M, Tsalenko A, Sampas N, Bruhn L, Shendure J, Eichler EE. Diversity of human copy number variation and multicopy genes. Science. 2010;330:641–646. doi: 10.1126/science.1197005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szatmari P, Paterson AD, Zwaigenbaum L, Roberts W, Brian J, Liu XQ, Vincent JB, Skaug JL, Thompson AP, Senman L, et al. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat Genet. 2007;39:319–328. doi: 10.1038/ng1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabuchi K, Blundell J, Etherton MR, Hammer RE, Liu X, Powell CM, Sudhof TC. A neuroligin-3 mutation implicated in autism increases inhibitory synaptic transmission in mice. Science. 2007;318:71–76. doi: 10.1126/science.1146221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamada K, Tomonaga S, Hatanaka F, Nakai N, Takao K, Miyakawa T, Nakatani J, Takumi T. Decreased exploratory activity in a mouse model of 15q duplication syndrome; implications for disturbance of serotonin signaling. PLoS One. 2010;5:e15126. doi: 10.1371/journal.pone.0015126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toffolatti L, Cardazzo B, Nobile C, Danieli GA, Gualandi F, Muntoni F, Abbs S, Zanetti P, Angelini C, Ferlini A, et al. Investigating the mechanism of chromosomal deletion: characterization of 39 deletion breakpoints in introns 47 and 48 of the human dystrophin gene. Genomics. 2002;80:523–530. [PubMed] [Google Scholar]
- Turner DJ, Miretti M, Rajan D, Fiegler H, Carter NP, Blayney ML, Beck S, Hurles ME. Germline rates of de novo meiotic deletions and duplications causing several genomic disorders. Nat Genet. 2008;40:90–95. doi: 10.1038/ng.2007.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacic V, McCarthy S, Malhotra D, Murray F, Chou HH, Peoples A, Makarov V, Yoon S, Bhandari A, Corominas R, et al. Duplications of the neuropeptide receptor gene VIPR2 confer significant risk for schizophrenia. Nature. 2011;471:499–503. doi: 10.1038/nature09884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Os J, Kapur S. Schizophrenia. Lancet. 2009;374:635–645. doi: 10.1016/S0140-6736(09)60995-8. [DOI] [PubMed] [Google Scholar]
- Vissers LE, de Ligt J, Gilissen C, Janssen I, Steehouwer M, de Vries P, van Lier B, Arts P, Wieskamp N, del Rosario M, et al. A de novo paradigm for mental retardation. Nat Genet. 2010;42:1109–1112. doi: 10.1038/ng.712. [DOI] [PubMed] [Google Scholar]
- Volkmar FR, Lord C, Bailey A, Schultz RT, Klin A. Autism and pervasive developmental disorders. J Child Psychol Psychiatry. 2004;45:135–170. doi: 10.1046/j.0021-9630.2003.00317.x. [DOI] [PubMed] [Google Scholar]
- Volkmar FR, State M, Klin A. Autism and autism spectrum disorders: diagnostic issues for the coming decade. J Child Psychol Psychiatry. 2009;50:108–115. doi: 10.1111/j.1469-7610.2008.02010.x. [DOI] [PubMed] [Google Scholar]
- Walsh T, McClellan JM, McCarthy SE, Addington AM, Pierce SB, Cooper GM, Nord AS, Kusenda M, Malhotra D, Bhandari A, et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–543. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- Waschek JA. Vasoactive intestinal peptide: an important trophic factor and developmental regulator? Dev Neurosci. 1995;17:1–7. doi: 10.1159/000111268. [DOI] [PubMed] [Google Scholar]
- Webber C. Functional enrichment analysis with structural variants: pitfalls and strategies. Cytogenet Genome Res. 2011;135:277–285. doi: 10.1159/000331670. [DOI] [PubMed] [Google Scholar]
- Weiss LA, Shen Y, Korn JM, Arking DE, Miller DT, Fossdal R, Saemundsen E, Stefansson H, Ferreira MA, Green T, et al. Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med. 2008;358:667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- Williams NM, Franke B, Mick E, Anney RJ, Freitag CM, Gill M, Thapar A, O’Donovan MC, Owen MJ, Holmans P, et al. Genome-Wide Analysis of Copy Number Variants in Attention Deficit Hyperactivity Disorder: The Role of Rare Variants and Duplications at 15q13.3. Am J Psychiatry. 2011 doi: 10.1176/appi.ajp.2011.11060822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Roos JL, Dexheimer P, Boone B, Plummer B, Levy S, Gogos JA, Karayiorgou M. Exome sequencing supports a de novo mutational paradigm for schizophrenia. Nat Genet. 2011;43:864–868. doi: 10.1038/ng.902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Roos JL, Levy S, van Rensburg EJ, Gogos JA, Karayiorgou M. Strong association of de novo copy number mutations with sporadic schizophrenia. Nat Genet. 2008;40:880–885. doi: 10.1038/ng.162. [DOI] [PubMed] [Google Scholar]
- Zhang D, Cheng L, Qian Y, liey-Rodriguez N, Kelsoe JR, Greenwood T, Nievergelt C, Barrett TB, McKinney R, Schork N, et al. Singleton deletions throughout the genome increase risk of bipolar disorder. MolPsychiatry. 2009a;14:376–380. doi: 10.1038/mp.2008.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Gu W, Hurles ME, Lupski JR. Copy number variation in human health, disease, and evolution. Annu Rev Genomics Hum Genet. 2009b;10:451–481. doi: 10.1146/annurev.genom.9.081307.164217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Khajavi M, Connolly AM, Towne CF, Batish SD, Lupski JR. The DNA replication FoSTeS/MMBIR mechanism can generate genomic, genic and exonic complex rearrangements in humans. Nat Genet. 2009c;41:849–853. doi: 10.1038/ng.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Leotta A, Kustanovich V, Lajonchere C, Geschwind DH, Law K, Law P, Qiu S, Lord C, Sebat J, et al. A unified genetic theory for sporadic and inherited autism. Proc Natl Acad Sci U S A. 2007;104:12831–12836. doi: 10.1073/pnas.0705803104. [DOI] [PMC free article] [PubMed] [Google Scholar]