Abstract
Drosophila is a well-established model organism for studying innate immunity because of its high resistance against microbial infections and lack of adaptive immunity. In addition, the immune signaling cascades found in Drosophila are evolutionarily conserved. Upon infection, activation of the immune signaling pathways, Toll and Imd, leads to the expression of multiple immune response genes, such as the antimicrobial peptides (AMPs). Previously, we identified an uncharacterized gene edin among the genes, which were strongly induced upon stimulation with Escherichia coli in Drosophila S2 cells. Edin has been associated with resistance against Listeria monocytogenes, but its role in Drosophila immunity remains elusive. In this study, we examined the role of Edin in the immune response of Drosophila both in vitro and in vivo. We report that edin expression is dependent on the Imd-pathway NF-κB transcription factor Relish and that it is expressed upon infection both in vitro and in vivo. Edin encodes a pro-protein, which is further processed in S2 cells. In our experiments, Edin did not bind microbes, nor did it possess antimicrobial activity to tested microbial strains in vitro or in vivo. Furthermore, edin RNAi did not significantly affect the expression of AMPs in vitro or in vivo. However, edin RNAi flies showed modestly impaired resistance to E. faecalis infection. We conclude that Edin has no potent antimicrobial properties but it appears to be important for E. faecalis infection via an uncharacterized mechanism. Further studies are still required to elucidate the exact role of Edin in the Drosophila immune response.
Introduction
Innate immunity is the first line of defense in all multicellular organisms. During the last few decades, the fruit fly Drosophila melanogaster has proven to be well suited for studying innate immune responses. In contrast to vertebrates, Drosophila only has an innate immune system, which is highly sophisticated and in part conserved among higher organisms [1]. In Drosophila, effective innate immune responses are based on the ability of several pattern-recognition receptors to recognize and bind common microbial surface structures. One main outcome of this initial microbial recognition is the activation of NF-κB immune signaling pathways, which leads to the production of several potent antimicrobial peptides (AMPs).
In Drosophila, the production of AMPs is mainly regulated by two NF-κB signaling pathways: the Imd (immune deficiency) pathway [2] reviewed in [3] and the Toll pathway [4] reviewed in [5]. Both of these pathways are highly conserved from fly to man. The Imd pathway is activated by diaminopimelic acid-type peptidoglycan (DAP) [6], present in most or all Gram-negative bacteria, but also in some Gram-positive bacteria like Listeria monocytogenes. The Toll pathway is activated mainly by the lysine-type peptidoglycan present in many other Gram-positive bacteria [7], reviewed in [5]. Both of these signaling pathways can also be induced by different fungi [8], [9]. Activation of the Imd and Toll signaling pathways upon microbial infection ultimately causes the nuclear translocation of the NF-κB transcription factors, Relish or Dif/Dorsal respectively, leading to the expression of dozens of NF-κB responsive genes [10], [11], [12], [13], [14]. The molecular function of many of these genes still remains unknown.
Earlier, we identified a gene, CG32185, to be highly induced in S2 cells in response to heat-killed Escherichia coli [14]. Later, Gordon et al. called the gene edin and found it to be associated with Listeria monocytogenes resistance [15]. In addition, it has been shown that Edin is secreted into the hemolymph in Drosophila third instar larvae upon infection [16]. Because the molecular function of Edin and the signaling pathways involved are still mainly unknown, in our current study we set out to examine the role of Edin in the Drosophila immune response both in vitro and in vivo.
Results
Edin expression is Relish-dependent in vitro and in vivo upon Gram-negative bacterial infection
When Drosophila encounters microbes, several signaling pathways are activated leading to transcriptional modifications. This response varies depending on the microbe and the site of infection. During a systemic infection, the expression of dozens of genes is induced [11], [12] leading to very effective defense responses. Upon infection, most of the highly induced genes are known to be AMP genes, DIMs (Drosophila immune-induced molecules) or genes related to signal regulation. Nevertheless, the molecular function of several of the induced genes is yet to be characterized. Previously, we studied which genes are induced in response to heat-killed Escherichia coli in Drosophila macrophage-like S2 cells [14]. Table I represents the oligonucleotide microarray data of the most strongly induced genes (data collected from [14]). The eight most strongly induced genes encode five known AMPs, one peptidoglycan recognition protein (PGRP-LB), a negative regulator of the Imd pathway (pirk) [17] and edin (CG32185). According to the microarray results, the expression of edin is strongly induced within hours after the bacterial challenge and the induction pattern of edin resembles that of known antimicrobial peptides (Table I).
Table 1. Induction of Drosophila antimicrobial peptide genes and edin in E. coli -challenged S2 cells (data collected from [14]).
| Gene | #CG | 0 h | 0.5 h | 1 h | 4 h | 24 h | Relish RNAi 4 h |
| Attacin B | CG18372 | 1±0.1 | 1.5 | 6.0 | 60.6±15.1 | 87.2±87.2 | 0.1±0.0 |
| Diptericin B | CG10794 | 1±0.0 | 2.3 | 3.6 | 52.4±4.3 | 78.1±1.8 | 0.2±0.1 |
| Attacin D | CG7629 | 1±0.0 | 1.1 | 2.6 | 47.5±6.3 | 92.5±2.1 | 0.1±0.1 |
| Metchnikowin | CG8175 | 1±0.0 | 1.7 | 6.5 | 41.3±15.7 | 52.2±2.0 | 0.4±0.1 |
| Edin | CG32185 | 1±0.1 | 0.9 | 3.5 | 29.8±6.4 | 48.5±1.1 | 0.0±0.0 |
| Pirk | CG15678 | 1±0.2 | 2.0 | 15.5 | 15.1±0.6 | 5.4±0.0 | 0.4±0.1 |
| PGRP-LB | CG14704 | 1±0.0 | 1.2 | 2.0 | 8.5±2.0 | 20.7±0.3 | 0.7±0.2 |
| Cecropin B | CG1878 | 1±0.0 | 1.2 | 3.2 | 7.4±0.9 | 4.0±0.3 | 0.6±0.0 |
In S2 cells, the response to E. coli is known to be predominantly mediated via the Imd pathway [13]. To verify whether the induction of edin is dependent on the Imd pathway, we silenced the Imd pathway by knocking down the transcription factor Relish by RNAi. The induction of edin was completely abolished in Relish dsRNA treated S2 cells at the 4 h time point (Table I) indicating that edin expression is regulated via the Imd pathway in S2 cells after induction with heat-killed E. coli.
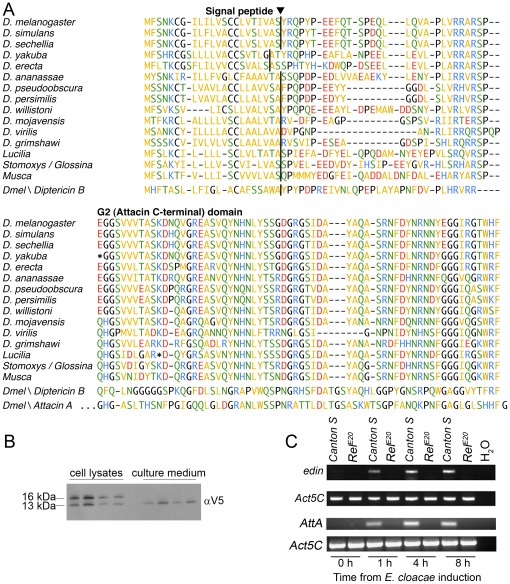
The edin gene encodes a short peptide of 115 amino acids including an N-terminal signal sequence (amino acids 1–22) (Figure 1A). The predicted signal peptidase cleavage site is supported by proteomic data from Verleyen et al. [16], who identified the predicted amino terminal of the mature protein in peptide fragments from hemolymph. Likely orthologs of the edin gene can be found in other brachyrecan flies, including all sequenced Drosophila species, but not in other insects (Figure 1A). For Musca domestica, three isoforms are represented in the EST databases (not shown). A tendency for pseudogenisation of the edin genes can be noted, as stop codons are present in the D. yakuba and D. mojavensis homologs. For the latter, an apparently functional allele is represented by an EST sequence (Figure 1A). A stop codon interrupts the open reading frame in the EST from Lucilia sericata, but this could be a sequencing error.
Figure 1. Edin is a Relish-dependently synthesized peptide, which is secreted from S2 cells.
(A–B) Edin contains a signal sequence and is secreted from S2 cells. (A) Edin sequences are aligned from 12 Drosophila species and three other dipterans. Diptericin B and Attacin A from D. melanogaster are also included in the alignment. The predicted signal peptidase cleavage sites [31] are marked. The sequences from the 12 Drosophila species are all from Clark et al. 2007 [32], except the D. mojavensis sequence which is derived from an EST sequence (EB600147). Modified gene models without introns were used for D. yakuba and D. willistoni. The Lucilia sericata sequence is derived from a single EST (FG360503). Three Stomoxys calcitrans ESTs (DN952426, DN952940, EZ048833) and one Glossina morsitans EST (AF368915) appear to contain overlapping sequence from the same gene. The Musca domestica sequence is an isoform represented by one EST (ES608713). (B) The signal sequence of Edin is cleaved before the peptide is secreted to the cell culture medium. S2 cells were transfected with a pMT-edin-V5 construct and the cell culture medium and cell lysates were analyzed with western blotting. Both full-length and cleaved forms were observed in the lysates while only the cleaved form was present in the medium. The V5 tag is located at the C-terminus of Edin. The blot represents 4 independent samples from which both cell lysates and culture medium were analyzed. (C) Edin is induced upon Enterobacter cloacae infection in Canton S flies but not in RelE20 flies. Canton S flies and RelE20-mutant flies were pricked with E. cloacae and total RNAs were extracted at the indicated time points. RT-PCR was performed and samples were electrophoresed on an agarose gel. Actin5C was used as a loading control and Attacin A as a positive control.
Iterated PSI-BLAST searches indicate that Edin is related to the Attacin/Diptericin superfamily of glycine-rich antibacterial peptides. The best hits were to Drosophila virilis Diptericin B (E = 8e-20) and Hyalophora cecropia Attacin E (E = 2e-18). Figure 1A shows an alignment to Diptericin B and the C-terminal (G2) domain of Attacin A from D. melanogaster.
Since Edin has a predicted signal sequence, we next examined if Edin is actually secreted from cells. To test this, we cloned edin cDNA into the heavy metal-inducible expression vector pMT/V5, transfected S2 cells with the construct and analyzed the presence of the protein both in the cell culture medium and cell extracts by western blotting using an anti-V5 antibody. In the S2 cells, both shorter and longer forms of Edin were detected, corresponding to V5-tagged peptides with and without the signal sequence, respectively. In the cell culture medium, only the shorter, C-terminal form, without the signal sequence could be observed (Figure 1B). This result suggests that Edin has a functional signal sequence, which is cleaved before the peptide is secreted. These results are in line with the report of Verleyen and coworkers [16], who detected amino-terminal fragments of Edin with mass spectrometry in the hemolymph of Drosophila larvae infected with a mixture of Gram-negative and Gram-positive bacteria.
Since the expression of edin is Relish-dependent in vitro, we next investigated whether edin is also induced upon microbial challenge in vivo. We infected wild-type Canton S and Relish null mutant adult flies (RelE20) with the Gram-negative bacteria Enterobacter cloacae. Total RNAs were extracted and the transcript levels of edin were determined with RT-PCR and agarose gel electrophoresis. As shown in Figure 1C, edin is induced in Canton S but not in RelE20 mutant flies. Attacin A was used as a positive control and showed a similar expression pattern to edin (Figure 1C). These results together with the previously published microarray data indicate that edin expression is strongly and rapidly induced upon a Gram-negative bacterial infection in a Relish-dependent manner both in vitro and in vivo. These results together propose that Edin has a function related to microbial resistance. Thus, we next subjected Edin to further functional characterization both in vitro and in vivo.
Edin has no significant effect on bacterial binding
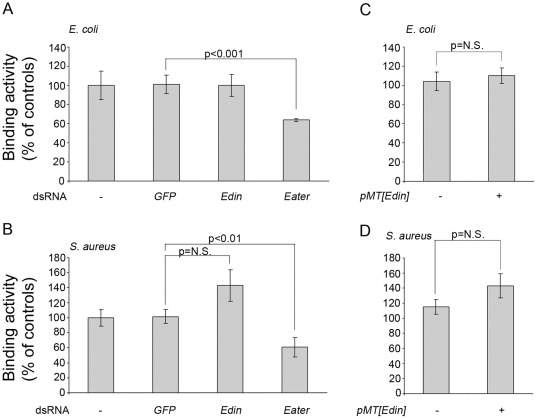
The phagocytosis of invading microbes is an essential component of Drosophila immunity [18], [19]. To this end we tested whether Edin has a role in bacterial binding or opsonization. Plasmatocyte-like S2 cells that are capable of binding and phagocytosing microbes [20] were treated with edin dsRNA and the ability of the cells to bind heat-killed, fluorescently labeled E. coli and Staphylococcus aureus was analyzed with flow cytometry. As a positive control, we used a dsRNA treatment targeting eater, which codes for an important phagocytic receptor for bacteria both in S2 cells and in Drosophila in vivo [18], [19], [21]. GFP dsRNA was used as a negative control. Edin RNAi did not affect the ability of S2 cells to bind E. coli (Figure 2A). Likewise, edin dsRNA treatments did not compromise the ability of S2 cells to bind S. aureus (Figure 2B) but rather seemed to modestly enhance the binding activity of S2 cells.
Figure 2. Edin does not affect the ability of S2 cells to bind microbes.
(A–B) The effect of edin RNAi on the binding of E. coli and S. aureus in Drosophila S2 cells. Drosophila S2 cells were soaked for three days in dsRNAs and thereafter exposed to bacteria at +4°C. GFP dsRNA was used as a negative and eater dsRNA as a positive control. (C–D) The effect of edin overexpression on the binding of E. coli and S. aureus. S2 cells were transiently transfected with a pMT construct expressing edin and endogenous edin expression was knocked down with dsRNA treatments. The ability of S2 cells to bind heat-killed E. coli (A, C) or S. aureus (B, D) was measured using flow cytometry.
To test the effect of edin overexpression on bacterial binding, S2 cells were first transiently transfected with a pMT[edin]V5 construct. An empty pMT/V5 plasmid was transfected as a control. 24 h after transfection, CuSO4 was added to the cell culture medium to induce the expression of the construct. Two days later, the medium was collected and transferred to other S2 cells which were pre-treated with edin dsRNA to block endogenous edin expression. Thereafter, FITC-labeled, heat-killed E. coli or S. aureus were added and the amount of cell-associated bacteria was monitored using flow cytometry. In line with the results of edin RNAi experiments, edin overexpression had no effect on the binding of E. coli (Fig. 2C) or S. aureus (Fig. 2D). The presence of Edin in the cell-culture medium was confirmed by western blotting using an anti-V5 antibody (data not shown).
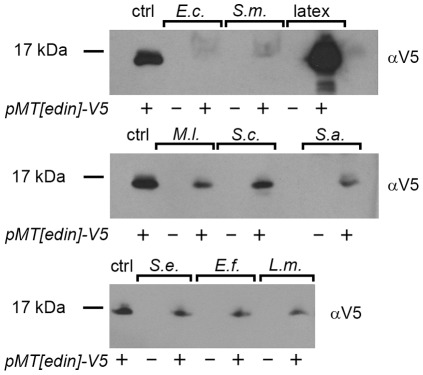
To investigate in a more direct way if Edin binds microbes, we incubated Edin-containing cell culture medium with live E. coli, Serratia marcescens, Staphylococcus epidermidis, Enterococcus faecalis, Listeria monocytogenes, Micrococcus luteus, Saccharomyces cerevisiae and S. aureus. Latex beads (carboxylated polystyrene), which are expected to bind all kinds of proteins to some extent, were used as a positive control. The microbial suspensions were incubated with 500 µl of Edin-containing medium at +4°C after which the microbes were pelleted and washed with PBS. Finally, the pellets were suspended and boiled in an SDS-PAGE sample buffer to detach bound Edin from the microbes before electrophoresis. Next, the proteins were transferred onto nitrocellulose membranes and Edin was detected using an anti-V5 antibody. As a reference, 20 µl of Edin-containing medium was loaded into the first lane. Therefore, if Edin attached efficiently to the indicated microbe, much more Edin should be detected in the samples (500 µl Edin-containing medium used) compared to the reference lane (20 µl Edin-containing medium). As shown in Figure 3 (the rightmost lanes), carboxylated latex beads, i.e. the positive control, bound Edin. In contrast, virtually no Edin was bound to the tested Gram-negative bacteria, E. coli and S. marcescens. Furthermore, only a faint signal was detected with the Gram-positive bacteria S. epidermidis, E. faecalis, L. monocytogenes, M. luteus and S. aureus, and with the baker's yeast S. cerevisiae as compared to the reference lane (ctrl in Figure 3). Based on these results, we conclude that Edin does not strongly bind any of the tested microbes.
Figure 3. The effect of Edin on microbial binding.
500 µl of Edin-V5 containing medium were incubated with 1 ml of a bacterial suspension of live E. coli (E.c.), Serratia marcescens (S.m), Staphylococcus epidermidis (S.e.), Enterococcus faecalis (E.f.), Listeria monocytogenes (L.m.), Micrococcus luteus (M.l.), Saccharomyces cerevisiae (S.c.) or S. aureus (S.a) for 1 h with mild agitation at +4°C. Latex beads treated with BSA were used as a control. The samples were then centrifuged and the pellet was washed. Edin bound to microbes was detached by adding 20 µl of SDS-PAGE loading buffer, boiled for 10 minutes, electrophoresed on SDS-PAGE and detected using a V5 antibody. The first lane of each blot is a control sample containing 20 µl of Edin-V5 medium. The following lanes contain 30 µl of the medium incubated with the indicated microbe.
The effect of Edin on immune signaling
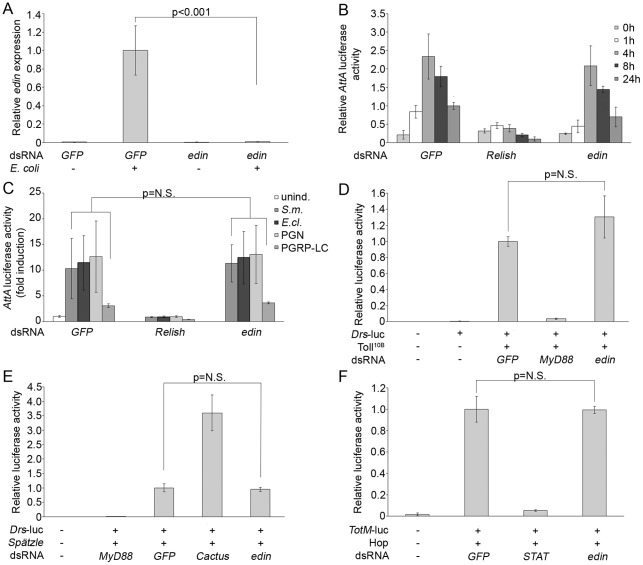
Next, we investigated whether Edin is involved in modulating the activity of Drosophila innate immune signaling cascades. S2 cells were transfected with luciferase-reporter constructs together with edin dsRNA as well as with negative and positive control dsRNAs, and the luciferase activities of the cell lysates were analyzed. Transfection efficacy and cell viability were assessed with an Actin 5C-β-galactosidase reporter. GFP dsRNA was used as a negative control in all assays. First, we tested the effectiveness of edin RNAi in vitro by treating S2 cells with GFP or edin dsRNAs, and analyzing the relative expression levels of edin. As shown in Figure 4A, edin RNAi abolishes the endogenous edin expression.
Figure 4. Effect of edin RNAi on Drosophila immune signaling in vitro.
A) Edin RNAi is effective in S2 cells. S2 cells were treated with GFP and edin dsRNA and the cells were induced by adding heat-killed E. coli. Relative expression levels of edin were analyzed from total RNAs with qRT-PCR. n = 4 for each sample. (B) Edin expression is not required for the Imd pathway signaling in vitro. S2 cells were transfected with an Attacin A-luciferase reporter together with GFP (negative control), Relish (positive control) and edin dsRNAs. The Imd pathway was activated by adding heat-killed E. coli to the cell culture medium and samples were collected at indicated time points. Edin RNAi causes a 30% decrease in the Imd pathway activity at the 24 h time point. The data for the 0 h and 24 h time points are pooled from 5 indepent experiments (n = 17 per sample). For 1 h, 4 h and 8 h time points n = 4 per sample. (C) Edin RNAi does not decrease the Imd pathway activity when the pathway is induced with S. marcescens, E. cloacae, peptidoglycan or PGRP-LC. S2 cells were transfected with an AttA-luciferase reporter and edin dsRNA and the Imd pathway was activated with S. marcescens (S.m.), E. cloacae (E.cl.), peptidoglycan (PGN) or a pMT[PGRP-LC] construct. CuSO4 was used to induce the expression of PGRP-LC. GFP and Relish dsRNAs were used as negative and positive controls, respectively. Unind. = no induction. The data for S.m., E.cl. and PGN are pooled from 3 independent experiments (n = 12 per sample). For PGRP-LC, n = 3 per sample. (D) Edin RNAi does not affect the Toll pathway activity. S2 cells were transfected with a Drosomycin-luciferase reporter together with GFP, edin and MyD88 (positive control) dsRNAs. A constitutively active form of the Toll receptor, Toll10B, was used to activate the pathway. The data are pooled from 3 independent experiments, n = 10 for each sample. (E) Edin has no effect on the Spätzle-induced Toll-pathway activity. S2 cells were transfected with a Drosomycin-luciferase reporter together with GFP, edin, MyD88 (control) and Cactus (control) dsRNAs. The Toll pathway was activated with the cleaved, active Spätzle ligand (SpzC106). n = 4 for each sample. (F) Edin RNAi has no effect on the JAK/STAT pathway. S2 cells were transfected with a Turandot M-reporter and GFP, STAT (positive control) and edin dsRNAs. The JAK/STAT pathway was activated by overexpressing HopTum-l. n = 4 for each sample.
In order to analyze the Imd pathway activity, an Attacin A-luciferase reporter and Relish dsRNA as a positive control were used and the pathway was activated by adding heat-killed E. coli to the cell culture medium. The samples were collected 0 h (no induction), 1 h, 4 h, 8 h and 24 h after E. coli induction. As expected, Relish RNAi strongly decreases the Imd-pathway activity at all time points (Figure 4B). On the contrary, edin RNAi had minor or no effect in this setting, although at the 24 h time point there was a trend for reduced Attacin A promoter driven luciferace activity (Figure 4B). Because edin RNAi appeared to have a minor effect on the Imd pathway activity when induced with heat-killed E. coli at the 24 h time point, we next investigated the effect of the edin dsRNA with other pathway elicitors. To this end, heat-killed S. marcescens, heat-killed E. cloacae, peptidoglycan and overexpression of the cytoplasmic tail of the PGRP-LC receptor were used. As shown in Figure 4C, edin RNAi had no effect on the AttA-luciferase activity in this experimental setting. These results indicate that Edin does not have an important role in the regulation of the Imd pathway activity in S2 cells.
To investigate the role of Edin in the Toll pathway signaling, we used a Drosomycin-luciferase reporter, and MyD88 dsRNA as a positive control, and activated the pathway by transfecting the cells with a constitutively active form of the Toll receptor, Toll10B (Figure 4D) or with the cleaved, active Spätzle ligand (Figure 4E). For the JAK/STAT signaling pathway, we used TurandotM-luciferase reporter and STAT dsRNA as a positive control (Figure 4F). The pathway was activated by overexpressing hopscotchTum-l, the active form of Drosophila Jak. Edin RNAi did not significantly affect the signaling via the Toll pathway (Figure 4D–E), or the JAK/STAT pathway (Figure 4F). These results indicate that Edin has no central role in regulating immune signaling in vitro.
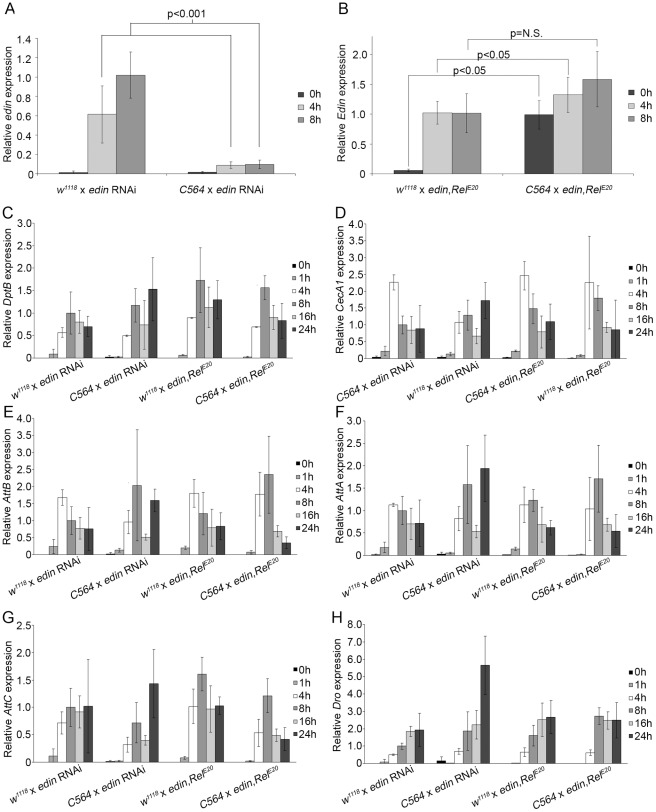
To test the role of Edin in Imd pathway regulation in vivo, we monitored the Imd pathway-mediated AMP gene expression levels with qRT-PCR in edin RNAi flies and in edin overexpression flies we created. The overexpression flies were created by microinjecting the pUAST-edin construct into RelE20 mutant embryos. To analyze Imd pathway activity, edin RNAi (VDRC #14289) and UAS-edin,RelE20 flies were crossed with the C564-GAL4 driver that targets transgene expression to the fat body in addition to some other organs [22]. The Imd pathway was then activated in week-old offspring by septic injury with E. cloacae. Flies crossed with w1118 flies were used as controls. As shown in Figure 5A, in vivo RNAi of edin using the C564-GAL4 driver strongly suppresses edin expression in whole flies, indicating that the UAS-RNAi construct is effective. UAS-edin,RelE20 flies crossed with the C564-GAL4 driver showed expression levels comparable to the E. cloacae infected control flies (Figure 5B).
Figure 5. The effect of Edin on AMP production in vivo.
Edin RNAi and overexpression flies (edin,RelE20) were crossed with C564-GAL4 flies or w1118 flies as a control, their offspring was infected with E. cloacae, total RNAs were extracted at indicated time points and qRT-PCR for the indicated genes was performed. (A) Expression of edin is knocked down in edin RNAi flies crossed to C564-GAL4 driver flies. (B) Edin overexpression flies express edin at a physiological level. Edin overexpression flies crossed with C564-GAL4 have slightly higher levels of edin compared to flies crossed with w1118. For (A–B) the data are pooled from 2 independent experiments, and n = 8 for each sample at each time point. (C–H) The effect of edin RNAi and overexpression on the production of Diptericin B (C), Cecropin A1 (D), Attacin B (E), Attacin A (F), Attacin C (G) and Drosocin (H). n = 4 for each sample at each time point. Error bars represent the standard deviation of each sample.
In agreement with our in vitro results, in vivo RNAi of edin did not show any clear effect in the expression levels of the tested AMP genes (two left-most panels, Figure 5C–H). There is a trend towards a minor decrease at the 4 h time points of the tested AMPs, excluding Drosocin (Figure 5H), but the decrease was statistically significant only with Cecropin A1 (Figure 5D) and Attacin B (Figure 5E). We next tested whether overexpression of edin affects the production of AMPs via the Imd pathway. We compared AMP expression after septic injury with E. cloacae between UAS-edin flies crossed with C564-GAL4 and UAS-edin flies crossed with w1118 flies. We observed moderate increase only in Drosocin expression at the 8 h time point (68% increase for p<0.05) (Figure 5H). Noteworthy, edin expression did not activate AMP gene expression without a microbial challenge (see the 0 h time point in the rightmost panel in Figure 5C–H). This is in line with the results in S2 cells and rules out the possibility that Edin would function as a cytokine mediating immune response from the site of induction to other tissues (for example from hemocytes to the fat body). Based on these results, we conclude that Edin has no important role in the regulation of the Imd pathway activity either in vitro or in vivo.
Edin has no potent antimicrobial properties in vitro or in vivo
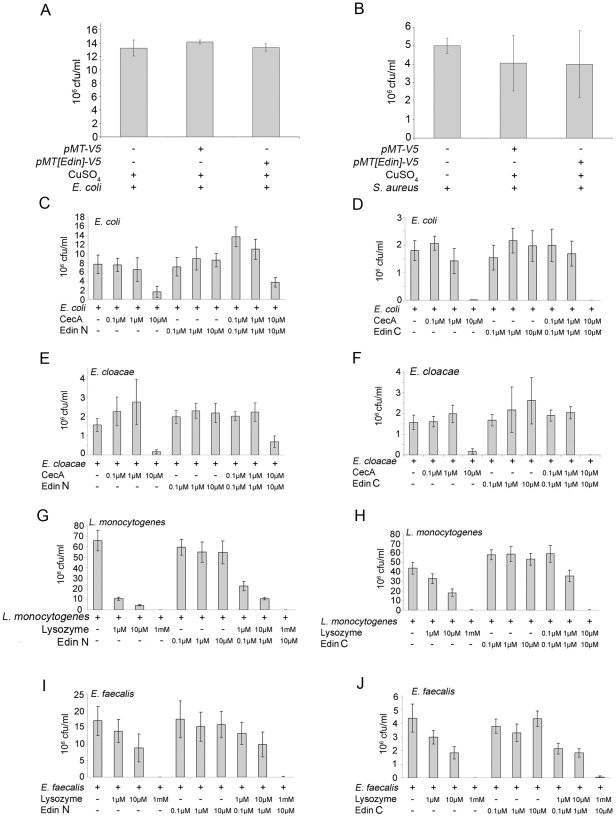
The kinetics of edin expression closely resembles those of known AMP genes, which led us to examine whether Edin has antimicrobial properties in vitro or in vivo. To study this, we first analyzed whether Edin was able to limit bacterial growth in vitro. We overexpressed edin in S2 cells, collected the cell culture medium and incubated the medium either with E. coli or S. aureus. Medium from S2 cells transfected with an empty vector was used as a control. As shown in Figure 6A and 6B, E . coli and S. aureus grew equally well in control medium and in medium containing Edin.
Figure 6. Edin has no broad antimicrobial properties against Gram positive or Gram negative bacteria in vitro.
(A–B) Edin does no limit the growth of E. coli or S. aureus in S2 cell culture medium. S2 cells were transfected with a copper-inducible pMT-edin-V5 or an empty pMT vector, and the abilities of E. coli and S. aureus to proliferate in these mediums were analyzed. (C–G) Synthetic forms of Edin do not limit the growth of E. coli (C), E. cloacae (D), L. monocytogenes (E), E. faecalis (F) or S. aureus (G). Both N-terminal and C-terminal forms of Edin were tested. Bacteria were cultured to an OD600 nm of 0.33, incubated with synthetic Edin and the ability of the bacteria to grow was analyzed. Cecropin A and Lysozyme were used as positive controls for Gram-negative and Gram-positive bacteria, respectively. Left column, N-terminal Edin; right column, C-terminal Edin.
To further investigate the antimicrobial properties of Edin in vitro, we designed synthetic peptides containing the amino acids 22–45 (Edin C-terminal form) or 50–115 (Edin N-terminal form. The peptides were tested for their ability to reduce bacterial growth in vitro. Cecropin A and Lysozyme were used as positive controls for Gram-negative and Gram-positive bacteria, respectively. The peptides were incubated with E. coli (Figure 6C–D), E. cloacae (Figure 6E–F), L. monocytogenes (Figure 6G–H) or E. faecalis (Fig. 6I–J) and colony forming units were determined. As shown in Figure 6C–J, Cecropin A and Lysozyme at their highest concentrations almost abolished the growth of the tested microbes whereas neither the synthetic C-terminal or N-terminal form of Edin was able to affect the growth of the bacteria. Moreover, no synergistic effects were observed when Edin was incubated together with either Cecropin A or Lysozyme (three rightmost columns in Figure 6 panels C–J).
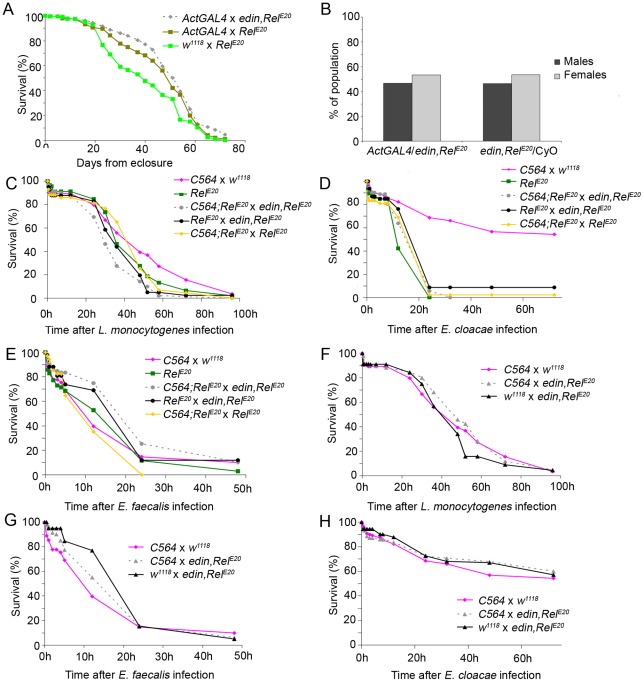
To test the antimicrobial properties of Edin in a more physiological context, the effect of edin overexpression on the survival of flies after bacterial infections was analyzed. First, to test whether overexpressing edin affects survival or lifespan, the UAS-edin,RelE20 overexpression line was crossed with the Act5C-GAL4/CyO driver line and the lifespan of the offspring was monitored. As shown in Figure 7A, overexpression of edin did not affect the lifespan of the flies and was comparable to that of the control flies. Furthermore, edin expression did not compromise the development of flies since equal amounts of UAS-edin,RelE20/ActGAL4 and UAS-edin,RelE20/CyO flies were obtained from the crosses (Figure 7B). Similar results were obtained when edin overexpression flies where crossed with either the C564-GAL4 driver line or the ubiquitous daughterless-GAL4 driver line (data not shown).
Figure 7. Overexpressing Edin has no effect on fly survival after Gram-positive or Gram-negative bacterial challenge in vivo.
(A) Overexpressing edin does not negatively affect lifespan. UAS-edin overexpression flies were crossed with Actin5C-GAL4 driver lines and the lifespan of their offspring was followed. w1118 crossed with RelE20 mutants and Act5C-GAL4 crossed with RelE20 were used as controls. The data represent one experiment, n = 100 for each cross. (B) Survival is not negatively affected in UAS-edin overexpressing flies. Equal amounts of edin,RelE20/Act5C-GAL4 and edin,RelE20/CyO genotypes were obtained from the crosses. (C–H) Flies were pricked with the indicated microbe and survival was followed. (C–E) Overexpressing edin in the RelE20 background does not protect the flies from L. monocytogenes (C), E. cloacae (D) or E. faecalis (E) infection. In C–E RelE20 crossed with edin,RelE20 and C564;RelE20 crossed with RelE20 were used as controls. (F–H) Overexpressing edin in a heterozygous w1118 background does not protect the flies from L. monocytogenes (F), E. cloacae (G) or E. faecalis (H) infection. Edin overexpression flies were pricked with E. faecalis, E. faecalis or L. monocytogenes. C564-GAL4 flies crossed with w1118 and UAS-edin,RelE20 crossed with w1118 flies were used as controls. Data are pooled from 2–3 experiments which showed similar trends, for each cross (D–J) n = 34–118.
An earlier study has shown that the expression of a single AMP can restore antimicrobial activity in Drosophila [23]. To test whether the expression of edin is sufficient to enhance resistance against septic infection in adult flies, we expressed edin in a homozygous RelE20 mutant background using a C564-GAL4;RelE20 line. In the homozygous RelE20 background, AMP production via the Imd pathway is eliminated making the flies very sensitive to infections with Gram-negative bacteria [24]. To test whether Edin had antimicrobial properties against Gram-negative or Gram-positive bacteria in vivo, we infected the UAS-edin,RelE20 flies crossed with the C564-GAL4;RelE20 driver with the Gram-positive bacterium L. monocytogenes (Figure 7C), which has a DAP-type peptidoglycan, with the Gram-negative bacterium E. cloacae (Figure 7D), and with the Gram-positive bacterium E. faecalis (Figure 7E). In this homozygous RelE20 background, overexpression of edin did not affect the survival rate upon septic injury with any of these microbes. In addition, no rescue was observed after a septic E. coli infection (data not shown). According to the results, edin overexpression was not sufficient to rescue the flies from succumbing to bacterial infection (Figure 7C–E) indicating that Edin alone does not possess sufficient antimicrobial properties against Gram-negative or Gram-positive bacteria.
To test whether Edin has antimicrobial properties in the context of a normal functioning immune response in Drosophila, we overexpressed edin in a heterozygous RelE20 mutant background. Edin overexpression flies crossed with C564-GAL4 were infected with L. monocytogenes (Figure 7F), E. cloacae (Figure 7G) and E. faecalis (Figure 7H) and monitored for survival. As shown in Figure 7F–H, overexpressing edin did not protect the flies from the bacterial infection. Together these results indicate that Edin has no antimicrobial properties against either Gram negative or Gram positive bacteria in vitro or in vivo. These results argue that Edin has another immune response modulating function.
Edin is required for normal resistance against bacteria
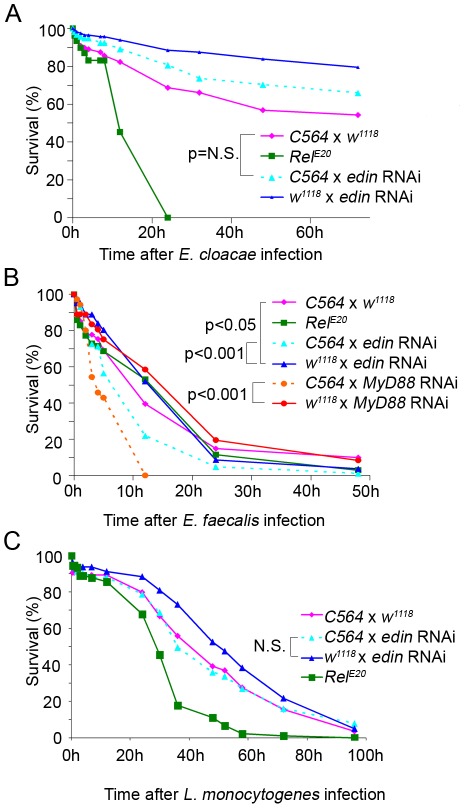
Next, we investigated whether Edin is required for normal resistance against septic infection. To this end edin RNAi flies were crossed with the C564-GAL4 driver or w1118 flies as a control, and the one-week-old offspring were infected with E. cloacae, E faecalis or L. monocytogenes. RelE20 mutant flies were used as a positive control in the E. cloacae and L. monocytogenes infection model, and UAS-MyD88 RNAi flies crossed with the C564-GAL4 driver as a positive control in the E. faecalis infection model. When infected with the Gram-negative bacterium E. cloacae, RelE20 mutant flies succumbed to the infection within 24 h. Edin RNAi flies crossed with C564-GAL4 flies showed a mild decrease in survival after E. cloacae infection compared to edin RNAi flies crossed with w1118 (Figure 8A) but this it not significant because the C564-GAL4 driver flies crossed to w1118 are more susceptible to the infection. However, a decrease in survival was observed in edin RNAi flies infected with the Gram-positive bacterium E. faecalis (Figure 8B). However, no statistically significant difference in survival was seen after an L. monocytogenes infection (Figure 8C), although a similar trend in survival could be observed, which is in line with the results by Gordon et al. [15]. These results imply that the expression of edin might be required for normal resistance against some bacterial infections.
Figure 8. Edin RNAi impairs survival in vivo after E. faecalis infection.
A–C. Healthy adult flies were pricked with a needle dipped either into a culture of E. cloacae, E. faecalis or L. monocytogenes and the survival of the flies was monitored. RelE20 mutants and/or MyD88 RNAi flies were used as positive controls. (A) Effect of edin RNAi after E. cloacae infection. Data are pooled from 3 independent experiments which showed similar trends, n = 112–117 for each cross. (B) Edin RNAi flies crossed with the C564-GAL4 driver are more susceptible to E. faecalis infection than uninduced edin RNAi flies crossed with w1118. Data are pooled from 2 independent experiments which showed similar trends n = 81–87 for each cross. For MyD88 RNAi crossed to w1118 and C564, data represents one experiment and n = 35 for both crosses. (C) Edin RNAi does not have a significant effect on fly survival against L. monocytogenes challenge. Data are pooled from 3 independent experiments which showed similar trends, n = 78–90 for each cross.
Discussion
In Drosophila, the expression of many genes is induced in response to microbial infection. In this study, we examined the role of the infection-inducible gene edin in the immune response of Drosophila melanogaster both in vitro and in vivo. We show that edin is highly induced in S2 cells by E. coli and its expression is dependent on the NF-κB transcription factor Relish both in vitro and in vivo. In line with the results of Verleyen and coworkers [16], we observe that Edin has a functional signal sequence leading to its cleavage and secretion from S2 cells. Despite the fact that edin is highly induced upon infection and that its expression pattern resembles that of known AMPs, we were not able to observe any antimicrobial properties in vitro or in vivo. Nor were we able to see any bacterial binding or opsonization when these properties of Edin were studied. Edin expression also was dispensable for AMP expression via the Imd pathway both in vitro and in vivo. However, interestingly edin RNAi flies showed decreased survival after bacterial infection with E. faecalis.
Traditionally, most studies on Drosophila AMPs have been successfully carried out in vitro. However, Drosophila is also a powerful model system for studying the activity of antimicrobial peptides in vivo, since it is easy to produce immunocompromised mutant fly lines, which are viable and fertile. Earlier studies have shown that Drosophila mutants of the Toll and Imd pathway, that have impaired production of AMPs via these signaling pathways, are highly susceptible to microbial infections [2], [4], [24] and even a single bacterial cell can be enough to kill a mutant fly [24]. The antimicrobial properties and the microbial specificity of a gene product can be studied by overexpressing the gene of interest in the mutant background of choice. It has been reported that the overexpression of a single antimicrobial peptide in Toll and Imd pathway double mutant flies can restore the resistance to a microbial infection to a level comparable to that of wild-type flies [23]. In our current study, we were not able to demonstrate a broad antimicrobial role for Edin in vitro or in vivo. In vitro, we observed no effect on the colony forming of bacterial cells when Edin was produced in S2 cells or when synthetic peptides were used.
In vivo, the effect of edin overexpression on the resistance against microbial infection was analyzed both in a homozygous RelE20 mutant background and in a heterozygous background. RelE20 mutants were selected since they are highly sensitive to Gram-negative bacterial infections. However, no increase in survival after septic injury could be observed in either one of these backgrounds. Therefore it is likely that Edin does not have an antimicrobial role in Drosophila although it is highly expressed upon bacterial infection. However, it is also possible that Edin is effective only against a specific microbe which we did not test in our current study. The in vivo analysis of antimicrobial properties of a certain peptide is further complicated by the production of a large battery of AMPs that can be partially redundant in their specificities. For instance, Edin alone might not be sufficient to fight against microbial infections, but it may require the presence of another AMP(s), or other immune effector molecules, for full activity.
Previously, Gordon and coworkers [15] have reported that high expression levels of edin are detrimental to fly survival and lifespan. We carried out lifespan experiments with our edin overexpression fly line and analyzed the proportions of the eclosed progeny. In contrast to Gordon et al., we did not observe a negative effect of edin overexpression on fly survival or lifespan. This difference in results could be due to different expression levels of edin or different genetic backgrounds of the flies used in these studies. According to our results, the edin overexpression fly line used in this study shows expression levels comparable to expression levels upon septic infection (Figure 5B). Furthermore, Gordon et al. [15] reported that Edin is required for resistance against Listeria monocytogenes infections. L. monocytogenes is a DAP-type peptidoglycan containing intracellular bacterium which can infect both mammals and Drosophila [25], [26]. Gordon et al. [15] report a significant decrease in survival after L. monocytogenes infection with two independent edin RNAi lines indicating that the normal edin expression is required for an efficient host response against the pathogen. In our current study, we did not observe a statistically significant reduction in the survival of edin RNAi flies after L. monocytogenes infection. However, the trend in the survival curve of edin RNAi flies is similar to that reported by Gordon et al. Since Listeria is an intracellular pathogen, Edin might also have an intracellular function although it is processed and secreted from the cell (Figure 1B). The processed form of Edin is also observed inside the cells (Figure 1B) which would support this hypothesis. However, further studies on the mechanisms involved in resistance against Listeria are required to elucidate the role of Edin in the infection.
We also analyzed the role of Edin as a modulator of innate immune signaling cascades. Nevertheless, our experiments indicate that Edin no strong effect on Imd pathway activity either in vitro or in vivo.
We conclude that the expression of edin is Relish-dependent both in vitro and in vivo but further studies are required to elucidate the exact role of Edin in the immune response in Drosophila. Also the mechanisms and signaling pathways involved in the Listeria monocytogenes infection remain to be studied.
Materials and Methods
Oligonucleotide microarrays
Oligonucleotide microarray expression data of S2 cells was collected from [14].
Microbial culture
Listeria monocytogenes (strain 10403S), Enterococcus faecalis, Staphylococcus aureus and Staphylococcus epidermidis were cultured in BHI. Enterobacter cloacae (strain β12) and Micrococcus luteus were cultured in LB supplemented with either 15 ng/ml of nalidixic acid (Sigma-Aldrich, St. Louis, Missouri, USA) or 100 µg/ml of streptomycin (Sigma-Aldrich), respectively. Serratia marcescens (strain Db11) and Escherichia coli were cultured in LB supplemented with 100 µg/ml of ampicillin. The baker's yeast Saccharomyces cerevisiae (AH109) was grown overnight in YPDA medium (Gibco/Life Technologies, Carlsbad, CA, USA) supplemented with 15 µg/ml of kanamycin at +30°C with shaking.
Semi-quantitative and quantitative RT-PCR
Semi-quantitative RT-PCR reactions for edin, Attacin A and Act5C were performed using Super-Script™ II One-Step RT-PCR with Platinum Taq kit (Invitrogen/Life Technologies, Carlsbad, CA, USA). The following primers were used: Edin: 5′-GTTCTCCAACAAGTGCGG-3′ (forward), and 5′- CAGAAATGCCAGGTGCCC-3′ (reverse); Attacin A: 5′-TTTGGCCTACAACAATGCTG-3′ (forward), and 5′-GCTTCTGGTTGGCAAACG-3′ (reverse); Act5C: 5′-CGAAGAAGTTGCTGCTCTGG-3′ (forward), and 5′-AGAACGATACCGGTGGTACG-3′ (reverse).
Quantitative RT-PCR was carried out using the QuantiTect SYBR Green RT-PCR kit (Qiagen) and an ABI7000 (Applied Biosystems) instrument according to the manufacturer's instructions. Results were analyzed with the ABI 7000 System SDS software version 1.2.3. The following primers were used: AttB, 5′-CAGTTCCCACAACAGGACC-3′ (forward) and 5′-CTCCTGCTGGAAGACATCC-3′ (reverse); Drosocin, 5′-TTCCTGCTGCTTGCTTGCG-3′ (forward) and 5′-TGGCAGCTTGAGTCAGGTG-3′ (reverse); AttA, 5′-GCATCCTAATCGTGGCC-3′ (forward) and 5′-GCTTCTGGTTGGCAAACG-3′ (reverse); AttC, 5′-CATCGTTGGCGTACTTGGC-3′ (forward) and 5′-TTGCTGGAAGCTATCCCGC-3′ (reverse); CecA1, 5′-CGTCGCTCTCATTCTGGC-3′ (forward) and 5′-GTTGCGGCGACATTGGC-3′ (reverse); DptB, 5′-GACTGGCTTGTGCCTTC-3′, and 5′-CCTGAAGGTATACACTCC-3′ (reverse); and Edin, 5′-CTCGTGTCCTGCTGTCTG-3′ (forward), and 5′-GCCTTCGTAGTTGTTCCG-3′(reverse).
S2 cell culture and transfections
Drosophila hemocyte-like S2 cells [27] (obtained from Invitrogen/Life Technologies) were maintained in Schneider's Insect Cell Culture Medium (Sigma-Aldrich, St. Louis, Missouri, USA) supplemented with 10% FBS, 100 U/ml Penicillin and 100 µg/ml Streptomycin (Sigma-Aldrich) at +25°C. The cells were transfected using the Fugene® transfection reagent (Roche Applied Science, Penzberg, Germany) according to the manufacturer's instructions.
Cloning and constructs
Edin was cloned into the pMT/V5-HisA (Invitrogen/Life Technologies) and pUAST [28] vectors using S2 cell cDNA as a template. The primers used were 5′-CAGAATTCATGTTCTCCAACAAGTGC-3′ and 5′-CAGGTACCTCAGAAATGCCAGGTGCC-3′ for pUAST, and 5′-CAGCGGCCGCATGTTCTCCAACAAGTGC-3′ and 5′-CACTCGAGGAAATGCCAGGTGCCCCG-3′ for pMT/V5-His.
Western blotting
S2 cells were transfected with 0.5 µg of pMT[edin]-V5. Cells were harvested, pelleted and lyzed 24 h after addition of CuSO4. 25 µg of cell lysate and supernatant were electrophoresed in NuPAGE 12% Bis-Tris gel (Invitrogen Life Technologies), blotted on a nitrocellulose membrane, and detected by Western blotting using mouse anti-V5 primary Ab (Invitrogen/Life Technologies) and goat anti-mouse Ab HRP conjugates (Molecular Probes) together with ECL Plus Western blotting detection system (GE Healthcare Life Sciences, Uppsala, Sweden).
Synthetic peptides
Two forms of synthetic Edin were ordered from Peptide 2.0. (Chantilly, VA, USA). Amino acid sequences: N-terminal form, SYRQ PYPEEF QTSPE QLLQ VAPLV; C-terminal form, SPEGG SVVVT ASKDNQ VGREAS VQYNHN LYSSG DGRGS IDAYA QASRN FDYNR NNYEG GIRGT WHF. The peptides were dissolved in H2O according to the manufacturer's instructions.
Colony forming unit assay
Edin-V5 expressed in S2 cells: S2 cells in 48-well plates in an antibiotic-free medium were transfected with 0.5 µg of pMT-edin-V5 plasmid or an empty plasmid. Expression of the plasmid was induced 48 h later by adding CuSO4 to a final concentration of 300 µM. 100 µl of overnight grown bacterial suspension (OD600 nm = 0.33, ∼1*106 bacteria/ml) was centrifuged and resuspended in 1 ml of Schneider medium supplemented with 10% FBS. 50 µl of E. coli and S. aureus suspension were added to the wells 24 h after CuSO4 and incubated for 2 h at +25°C. Serial dilutions of the bacterial suspensions were made in sterile water. 20 µl droplets of each dilution were pipetted on LB (E. coli) or BHI (S. aureus) agar plates, the plates incubated overnight at +37°C and the bacterial colonies counted.
Synthetic forms of Edin: An overnight grown bacterial suspension (∼1*106 bacteria/ml) was centrifuged and washed as above and resuspended in 5% DMSO. 5 µl of E. coli, E. cloacae, L. monocytogenes and E. faecalis suspension were added on the 96-well plates containing synthetic Edin at concentrations of 10 µM, 1 µM and 100 nM. Suspensions were incubated for 2 h at +25°C, after which serial dilutions were made in sterile water. Dilutions were plated as above and the bacterial colonies counted. Lysozyme and Cecropin A (Sigma-Aldrich, St. Louis, Missouri, USA) were used as a positive control for Gram-positive and Gram-negative bacteria, respectively.
Luciferase reporter assays and dsRNA treatments
Luciferase reporter assays to analyze the Imd, Toll and JAK/STAT pathways, and dsRNA treatments were carried out as described earlier [29], [30].
Drosophila stocks
The edin RNAi line (stock #14289) was obtained from VDRC and the C564-GAL4 flies were a kind gift from from Prof. Bruno Lemaitre (Global Health Institute, EPFL, Switzerland). CG32185 transgenic flies were generated by microinjecting the pUAST-edin construct to the RelE20 background in the Umeå Fly and Worm Transgene Facility. The genotype of the edin overexpression fly line is w;+;UAS-edin,RelE20.
Lifespan experiments
UAS-edin flies were crossed with C564-GAL4, Actin5C-GAL4/CyO and Daughterless-GAL4 driver flies. RelE20 crossed with driver flies were used as a control. The lifespan of the offspring of the crosses was monitored at +25°C. Flies were moved to vials containing 5 ml of fresh fly food twice a week and their survival was monitored. Males and females were kept in separate vials, 10 to 20 flies per vial.
Infection experiments
Infections were carried out by pricking one week-old healthy flies with a thin tungsten needle dipped in a concentrated pellet of either Escherichia coli, Enterobacter cloacae (strain β12), Enterococcus faecalis or Listeria monocytogenes (strain 10403S) which were grown overnight on culture plates.
RNA extraction from flies
Quadruplicates of five flies (2 females and 3 males) were snap frozen on dry ice 0 h, 1 h, 4 h or 8 h post-infection. Flies were homogenized in TRIsure reagent (Bioline, London, UK) and total RNAs were extracted according to the manufacturer's instructions.
Statistical analysis
Statistical analyses of results were carried out using one-way ANOVA. For survival experiments, Log Rank analysis was carried out and p<0.05 was considered to be significant.
Flow cytometry
The amount of cell-associated microbes was analyzed using flow cytometry as described earlier [20].
Binding assay
The binding assay for Edin was carried out essentially as described earlier [20] with minor modifications. In brief, S2 cells were seeded onto 24-well plates and transfected with 0.5 µg of pMT-edin-V5 plasmid or an empty pMT-V5 plasmid. CuSO4 was added 48 h later to a final concentration of 500 µM. Cells were harvested the next day and the supernatant was collected. 1 ml of overnight grown microbial culture was centrifuged and the pellet was washed 5 times with 1× PBS. 500 µl of medium containing either pMT-edin-V5 or empty pMT-V5 was added in the tubes containing the microbial pellets or latex beads treated with 0.4 M N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide (EDC) and 0.1 M N-hydroxysuccinimide (NHS) and coated with BSA (10 mg/ml in PBS, pH 7.4). The samples were incubated for 1 h in an end-to-end rotator at +4°C. Thereafter the samples were washed five times with 1 ml of 1× PBS and the pellet was suspended in 20 µl of PBS. To detach the bound Edin from the microbial cells, SDS-PAGE sample buffer was added and the suspension was boiled for 10 min. The samples were centrifuged and 30 µl of the supernatant was loaded on to a 12% NuPAGE BisTris gel, electrophoresed and the proteins were transferred to a nitrocellulose membrane as described above.
Acknowledgments
We thank other members of our laboratory, Prof. Shoichiro Kurata for the L. monocytogenes strain and VDRC for the edin RNAi line. We are also grateful to Ingrid Dacklin in Umeå Fly and Worm Transgene Facility for performing microinjections to fly embryos. The fly work was done at University of Tampere Drosophila Core Facility funded by Biocenter Finland. All authors approved the final version of the text.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This research was supported by grants from the Academy of Finland, the Sigrid Juselius Foundation (to MR), the Tampere Tuberculosis Foundation (to MR and SV), Competitive Research Funding of the Pirkanmaa Hospital District (to MR and SV), the Tampere Graduate Program in Biomedicine and Biotechnology (LMV) and the Orion-Farmos Research Foundation (LMV). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Lemaitre B, Hoffmann J. The host defense of Drosophila melanogaster. Annu Rev Immunol. 2007;25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615. [DOI] [PubMed] [Google Scholar]
- 2.Lemaitre B, Kromer-Metzger E, Michaut L, Nicolas E, Meister M, et al. A recessive mutation, immune deficiency (imd), defines two distinct control pathways in the Drosophila host defense. Proc Natl Acad Sci U S A. 1995;92:9465–9469. doi: 10.1073/pnas.92.21.9465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Valanne S, Kallio J, Kleino A, Rämet M. Large-scale RNAi screens add both clarity and complexity to Drosophila NF-κB signaling. Dev Comp Immunol. 2012;37:9–18. doi: 10.1016/j.dci.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 4.Lemaitre B, Nicolas E, Michaut L, Reichhart JM, Hoffmann JA. The dorsoventral regulatory gene cassette spätzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell. 1996;86:973–983. doi: 10.1016/s0092-8674(00)80172-5. [DOI] [PubMed] [Google Scholar]
- 5.Valanne S, Wang JH, Rämet M. The Drosophila Toll signaling pathway. J Immunol. 2011;186:649–656. doi: 10.4049/jimmunol.1002302. [DOI] [PubMed] [Google Scholar]
- 6.Leulier F, Parquet C, Pili-Floury S, Ryu JH, Caroff M, et al. The Drosophila immune system detects bacteria through specific peptidoglycan recognition. Nat Immunol. 2003;4:478–484. doi: 10.1038/ni922. [DOI] [PubMed] [Google Scholar]
- 7.Michel T, Reichhart JM, Hoffmann JA, Royet J. Drosophila Toll is activated by Gram-positive bacteria through a circulating peptidoglycan recognition protein. Nature. 2001;414:756–759. doi: 10.1038/414756a. [DOI] [PubMed] [Google Scholar]
- 8.Gottar M, Gobert V, Matskevich AA, Reichhart JM, Wang C, et al. Dual detection of fungal infections in Drosophila via recognition of glucans and sensing of virulence factors. Cell. 2006;127:1425–1437. doi: 10.1016/j.cell.2006.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hedengren-Olcott M, Olcott MC, Mooney DT, Ekengren S, Geller BL, et al. Differential activation of the NF-κB-like factors Relish and Dif in Drosophila melanogaster by fungi and Gram-positive bacteria. J Biol Chem. 2004;279:21121–21127. doi: 10.1074/jbc.M313856200. [DOI] [PubMed] [Google Scholar]
- 10.Boutros M, Agaisse H, Perrimon N. Sequential activation of signaling pathways during innate immune responses in Drosophila. Dev Cell. 2002;3:711–722. doi: 10.1016/s1534-5807(02)00325-8. [DOI] [PubMed] [Google Scholar]
- 11.De Gregorio E, Spellman PT, Rubin GM, Lemaitre B. Genome-wide analysis of the Drosophila immune response by using oligonucleotide microarrays. Proc Natl Acad Sci U S A. 2001;98:12590–12595. doi: 10.1073/pnas.221458698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Irving P, Troxler L, Heuer TS, Belvin M, Kopczynski C, et al. A genome-wide analysis of immune responses in Drosophila. Proc Natl Acad Sci U S A. 2001;98:15119–15124. doi: 10.1073/pnas.261573998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rämet M, Manfruelli P, Pearson A, Mathey-Prevot B, Ezekowitz RA. Functional genomic analysis of phagocytosis and identification of a Drosophila receptor for E. coli. Nature. 2002;416:644–648. doi: 10.1038/nature735. [DOI] [PubMed] [Google Scholar]
- 14.Valanne S, Kleino A, Myllymäki H, Vuoristo J, Rämet M. Iap2 is required for a sustained response in the Drosophila Imd pathway. Dev Comp Immunol. 2007;31:991–1001. doi: 10.1016/j.dci.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 15.Gordon MD, Ayres JS, Schneider DS, Nusse R. Pathogenesis of listeria-infected Drosophila wntD mutants is associated with elevated levels of the novel immunity gene edin. PLoS Pathog. 2008;4:e1000111. doi: 10.1371/journal.ppat.1000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Verleyen P, Baggerman G, D'Hertog W, Vierstraete E, Husson SJ, et al. Identification of new immune induced molecules in the haemolymph of Drosophila melanogaster by 2D-nanoLC MS/MS. J Insect Physiol. 2006;52:379–388. doi: 10.1016/j.jinsphys.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 17.Kleino A, Myllymäki H, Kallio J, Vanha-aho LM, Oksanen K, et al. Pirk is a negative regulator of the Drosophila Imd pathway. J Immunol. 2008;180:5413–5422. doi: 10.4049/jimmunol.180.8.5413. [DOI] [PubMed] [Google Scholar]
- 18.Kocks C, Cho JH, Nehme N, Ulvila J, Pearson AM, et al. Eater, a transmembrane protein mediating phagocytosis of bacterial pathogens in Drosophila. Cell. 2005;123:335–346. doi: 10.1016/j.cell.2005.08.034. [DOI] [PubMed] [Google Scholar]
- 19.Ulvila J, Vanha-aho LM, Rämet M Drosophila phagocytosis – still many unknowns under the surface. Apmis. 119:651–662. doi: 10.1111/j.1600-0463.2011.02792.x. [DOI] [PubMed] [Google Scholar]
- 20.Rämet M, Pearson A, Manfruelli P, Li X, Koziel H, et al. Drosophila scavenger receptor CI is a pattern recognition receptor for bacteria. Immunity. 2001;15:1027–1038. doi: 10.1016/s1074-7613(01)00249-7. [DOI] [PubMed] [Google Scholar]
- 21.Chung YS, Kocks C Recognition of pathogenic microbes by the Drosophila phagocytic pattern recognition receptor EaterJBiolChem286:26524–2. 6532 doi: 10.1074/jbc.M110.214007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harrison DA, Binari R, Nahreini TS, Gilman M, Perrimon N. Activation of a Drosophila Janus kinase (JAK) causes hematopoietic neoplasia and developmental defects. Embo J. 1995;14:2857–2865. doi: 10.1002/j.1460-2075.1995.tb07285.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tzou P, Reichhart JM, Lemaitre B. Constitutive expression of a single antimicrobial peptide can restore wild-type resistance to infection in immunodeficient Drosophila mutants. Proc Natl Acad Sci U S A. 2002;99:2152–2157. doi: 10.1073/pnas.042411999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hedengren M, Åsling B, Dushay MS, Ando I, Ekengren S, et al. Relish, a central factor in the control of humoral but not cellular immunity in Drosophila. Mol Cell. 1999;4:827–837. doi: 10.1016/s1097-2765(00)80392-5. [DOI] [PubMed] [Google Scholar]
- 25.Yano T, Mita S, Ohmori H, Oshima Y, Fujimoto Y, et al. Autophagic control of listeria through intracellular innate immune recognition in drosophila. Nat Immunol. 2008;9:908–916. doi: 10.1038/ni.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mansfield BE, Dionne MS, Schneider DS, Freitag NE. Exploration of host-pathogen interactions using Listeria monocytogenes and Drosophila melanogaster. Cell Microbiol. 2003;5:901–911. doi: 10.1046/j.1462-5822.2003.00329.x. [DOI] [PubMed] [Google Scholar]
- 27.Schneider I. Cell lines derived from late embryonic stages of Drosophila melanogaster. J Embryol Exp Morphol. 1972;27:353–365. [PubMed] [Google Scholar]
- 28.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 29.Kallio J, Leinonen A, Ulvila J, Valanne S, Ezekowitz RA, et al. Functional analysis of immune response genes in Drosophila identifies JNK pathway as a regulator of antimicrobial peptide gene expression in S2 cells. Microbes Infect. 2005;7:811–819. doi: 10.1016/j.micinf.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 30.Kallio J, Myllymäki H, Grönholm J, Armstrong M, Vanha-aho LM, et al. Eye transformer is a negative regulator of Drosophila JAK/STAT signaling. FASEB J. 2010;24:4467–4479. doi: 10.1096/fj.10-162784. [DOI] [PubMed] [Google Scholar]
- 31.Petersen TN, Brunak S, von Heijne G, Nielsen H SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 32.Clark AG, Eisen MB, Smith DR, Bergman CM, Oliver B, et al. Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]