Abstract
Vertebrate development requires progressive commitment of embryonic cells into specific lineages through a continuum of signals that play off differentiation versus multipotency. In mammals, Nanog is a key transcription factor that maintains cellular pluripotency by controlling competence to respond to differentiation cues. Nanog orthologs are known in most vertebrates examined to date, but absent from the Anuran amphibian Xenopus. Interestingly, in silico analyses and literature scanning reveal that basal vertebrate ventral homeobox (ventxs) and mammalian Nanog factors share extensive structural, evolutionary and functional properties. Here, we reassess the role of ventx activity in Xenopus laevis embryos and demonstrate that they play an unanticipated role as guardians of high developmental potential during early development. Joint over-expression of Xenopus ventx1.2 and ventx2.1-b (ventx1/2) counteracts lineage commitment towards both dorsal and ventral fates and prevents msx1-induced ventralization. Furthermore, ventx1/2 inactivation leads to down-regulation of the multipotency marker oct91 and to premature differentiation of blastula cells. Finally, supporting the key role of ventx1/2 in the control of developmental potential during development, mouse Nanog (mNanog) expression specifically rescues embryonic axis formation in ventx1/2 deficient embryos. We conclude that during Xenopus development ventx1/2 activity, reminiscent of that of Nanog in mammalian embryos, controls the switch of early embryonic cells from uncommitted to committed states.
Introduction
In vertebrates, early embryonic cells remain undifferentiated prior to gastrulation. As such, they are not restricted to a predetermined fate but can enter a number of differentiation pathways leading to all the cell types of the adult organism. This capacity is referred to as pluripotency. Examples of pluripotent cells include cells of the inner cell mass and epiblast of the mammalian blastocysts [1], [2], [3], [4] and those of the animal pole of Xenopus blastulae [5], [6]. During gastrulation the gene network that maintains the undifferentiated state is rewired and embryonic cells gradually lose their initial high developmental potential, which causes lineage restriction and allows the progressive building of organs [7]. In this process, cell fate is tightly controlled by signals that either promote the entry into given differentiation paths, or restrict this capacity and maintain cellular developmental potential.
Studies of pluripotency have uncovered key signals and factors that promote maintenance of the uncommitted state or lineage specification [8], [9], [10]. In mammals, these signals are thought to converge on the POU5F1/SOX2/NANOG triumvirate of transcription factors that constitutes the core network controlling pluripotency [9], [11], [12]. Nanog, which encodes a homeodomain-bearing transcription factor of the NKL class, was first identified in mammals as being essential for early embryonic development and germ-line establishment through its capacity to restrain premature differentiation of embryonic stem cells [3], [13], [14], [15]. NANOG activity indeed protects undifferentiated cells against the differentiation-inducing effects of extracellular signals and transcriptional noise [15], [16]. Though Nanog was initially thought to be a mammal-specific gene, orthologs have been characterized in most vertebrate species, including birds [17], [18], teleosts [19], [20] and non-anuran (urodele) amphibians [21], [22]. Constitutive expression of a modified axolotl Nanog ortholog was shown to sustain pluripotency in mouse ES cells cultured in the absence of LIF [18], [20], [21]; further, functional assays have shown that chick as well as zebrafish Nanog orthologs could restore the capacity to reprogram Nanog-deficient murine cells to fully pluripotent iPS cells [23], suggesting that this factor controls developmental potential across osteichtyes.In Xenopus, uncommitted embryonic cells maintain high developmental potential until the onset of gastrulation [5], [6], similar to Nanog-expressing epiblastic cells in amniote embryos [3], [9], [24]. However, much less is known about the molecular mechanism that underlies this cellular property in Xenopus. Interestingly, in all tetrapods, including Xenopus, uncommitted embryonic cells express transcription factors of the POU5F1 family [18], [21], [25], [26], [27]. In mammals, chick and amphibians, POU5F1s maintain embryonic cells in an uncommitted state, preventing their differentiation [18], [25], [28], [29]. However, if most of the major players in the mammalian pluripotency network are structurally conserved in the Xenopus genome [20], [30], so far no Nanog ortholog has been identified in anuran amphibians [31]. Thus, either Nanog remains to be characterized in anurans or other(s) factor(s) must maintain the high developmental potential of uncommitted embryonic cells in this taxon [5], [6]. Here, we present evidence suggesting that this function is carried out by ventx transcription factors in Xenopus.
Members of the VENTX family of NKL transcription factors were first identified in Xenopus and owe their name (ventral homeobox) to their ventral marginal zone expression domain in Xenopus gastrulae [32], [33]. They form a small multigenic family organized in a compact cluster in most chordate genomes, except mammals where either a single or no ventx ortholog is found. Xenopus species possess at least 6 ventx paralogs, which can be grouped in 3 subclasses: ventx1s, ventx2s and ventx3s. There is longstanding agreement that all ventx1s and ventx2s function in a similar fashion [34] and the less studied ventx3s seem to follow this pattern as well [35], [36]. All ventx factors are known to act as transcriptional repressors and to be expressed in roughly overlapping territories during early and late development, ventx2s being more broadly expressed in spatial and temporal terms [32], [33], [34], [35]. More specifically, they are all expressed in the animal hemisphere of blastulae and the ventral side of early gastrulae, where they participate in the bmp4-controlled gene network that acts in the establishment of dorso-ventral patterning [33]. In this “ventral center”, they antagonize dorsalization induced by the Spemann organizer, opposing the spread of the organizer in ventro-lateral domains by regionalizing the expression of organizer-specific genes, such as gsc [37]. When overexpressed in Xenopus embryos they give rise to ventralized phenotypes, characterized at tailbud stage by anterior truncations, short and/or bent tails and absent or defective axial structures such as notochord and floor plate [32], [33], [34], [38]. Conversely, expression of dominant-negative ventx constructs leads to double axis formation [34], whereas ventxs knock-down causes severe dorsalization, characterized by the loss of caudal territories and increased neuralization of the ectoderm [37]. Here, we propose a reinterpreted role for ventx factors, as guardians of high developmental potential during early Xenopus development. This conclusion is based on the key observation that ventx factors repress differentiation towards dorsal as well as ventral fates and that their knockdown can be rescued by ectopic expression of the mouse pluripotency regulator Nanog. We suggest that this crucial activity protects the future ventral territories from premature commitment towards dorsal fates in order to ensure proper spatio-temporal patterning of the embryo.
Results
Ventx and Nanog Factors Share Common Properties
We set out to identify a putative Nanog ortholog in Xenopus. In silico screening of sequence repositories resulted in the detection of annotated or putative Nanog orthologs in all gnathostomes, except Xenopus species. Degenerate PCR-based approaches were also unsuccessful (data not shown and see Supporting Information S1 for Extended Experimental Procedures). Moreover, the synthenic region where Nanog orthologs are found in other tetrapods, including axolotl, is conserved in Xenopus tropicalis albeit split over two scaffolds in the current state of the genome assembly (see ensembl scaffolds GL173371 and GL173015). These scaffolds contain no Nanog-related sequence, strongly arguing that the absence of Nanog from the Xenopus genus is due to secondary loss. Others have recently reached a similar conclusion [20]. Therefore, we tested the alternative hypothesis that other Xenopus transcription factors might be capable of functionally replacing Nanog.
As Nanog belongs to the NKL subclass of homeodomain-containing proteins, we focused on this group to identify putative candidates. Phylogenetic reconstruction showed NKL families to be monophyletic, except for NK4 and VENTX (Fig. S1 A). Surprisingly, the amphioxus Ventx orthologs [39] appear at the base of the NANOG group, suggesting that VENTX and NANOG families might be closely related (Fig. S1 C). Furthermore, these families share multiple features that are unique among NKLs. Notably, VENTX and NANOG are the only NKL families to have been lost in specific vertebrate lineages: Nanog is absent in the Xenopus genus whereas, inversely, rodents lack Ventx. Also, VENTX and NANOG are the only NKL to have numerous processed pseudogenes in the human genome (6 and 10 respectively) [40], which often correlates with expression in the germline or its embryonic precursors, and is a proposed signature of genes involved in the maintenance of pluripotency [41]. Finally, VENTX and NANOG have long branches when compared to other NKL families (e.g. NK1 or LBX, see Fig. S1), indicating that the homeodomains from these two families are less conserved among vertebrates than those from other NKLs (see also Table S1). These shared features make Xenopus ventxs good candidates for serving Nanog-like functions.
In line with this hypothesis, mammalian Nanog and Xenopus ventx genes encode transcriptional repressors [34], [42], [43] and share striking functional similarities (summarized in Table S2). First, the orthologs of many genes regulated by ventxs in Xenopus are regulated by NANOG in mammals; second, Nanog and ventxs are regulated by the same signalling pathways and transcription factors; third, ventxs and NANOG interact with orthologous proteins. One of the most significant parallels is that, in Xenopus and teleosts, endogenous ventx and pou5f1 transcription factors interact physically and genetically during early development [44], [45], [46], as do mammalian NANOG and POU5F1 [9].
Mouse Nanog and ventx1/2 Overexpression have Similar Effects
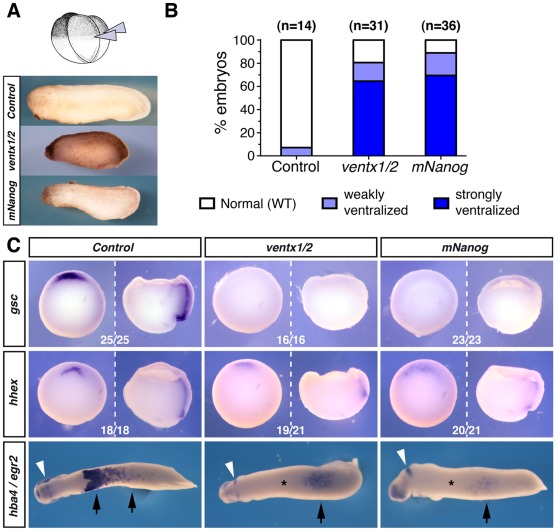
These extensive similarities prompted us to compare the effects of overexpression of mouse Nanog (mNanog) to combined Xenopus ventx1.2 [32] and ventx2.1-b [47] (referred to as ventx1/2 from now on) overexpression on Xenopus embryonic development. The relevant mRNAs were dorsally injected at the 4-cell stage (NF3 [48]), using previously described doses for ventx1/2 (0.5 ng per blastomere [34]) and half the lethal dose for mNanog (0.6 ng per blastomere, see Fig. S2). As expected [34], ventx1/2 overexpression led at tailbud stage (NF28) to severely ventralized phenotypes with truncated anterior structures ( Fig. 1 A ). Remarkably, mNanog overexpression produced similar defects with comparable penetrance ( Fig. 1 , A and B , and Fig. S3 C). In contrast, overexpression of the medaka ortholog OlNanog led to phenotypes clearly distinct from those obtained with mNanog, no ventralization being observed at any of the doses assayed, with all embryos displaying clear head features (Fig. S3A).
Figure 1. mNanog and ventx1/2 overexpression cause similar effects in Xenopus embryos.
(A) Four-cell stage embryos (NF3) were injected in both dorsal blastomeres, with a 1∶3 mix of ventx1.2 and ventx2.1-b mRNAs (ventx1/2; 0.5 ng per blastomere), with mouse Nanog mRNA (mNanog; 0.15 ng/blastomere), or with water for control. Representative phenotypes observed at tailbud stage (NF28) are shown (lateral views, anterior to the left, dorsal to the top). (B) Percentages of observed phenotypes in three independent experiments for mock (n = 14), ventx1/2 (n = 31) and mNanog (n = 36) mRNAs injections. (C) Embryos injected as in (A) were collected at early gastrulae (NF10.5; whole embryos: ventral view, dorsal side to the top; hemisected embryos: lateral view, dorsal to the left, animal side to the top) and tailbud (NF28; ventral view, anterior to the left) stages and processed for whole-mount in situ hybridization (WISH) with a gsc or hhex probe, or with hba4 (black arrowheads) and egr2 (white arrowheads), respectively. The number of embryos showing staining similar to the one photographed over the total number of embryos assayed is indicated.
We next checked if ventx1/2 and mNanog dorsal overexpression had similar impacts on gene expression. As previously reported, ventx1/2 overexpression strongly repressed the transcription of the dorsal organizer markers gsc [34] and hhex [49] at gastrula stage (NF10.5) and the blood island marker hba4 (also known as alpha-T4 globin) [50] at stage NF28 ( Fig. 1 C and Fig. S3 E). Remarkably, similar effects were observed in embryos injected with mNanog ( Fig. 1 C ), while OlNanog injection did not lead to repression of gsc at gastrula stage (Fig. S3 E), or hba4 at NF28 (data not shown). The results support the hypothesis that mammalian Nanog and Xenopus ventx1/2 share functional properties, and are coherent with the observation that OlNanog seems to share only limited functional similarities with its mammalians orthologs [19].
ventx1/2 and mNanog Overexpression Down-regulate Specification Markers for all Germ Layers and Embryonic Territories
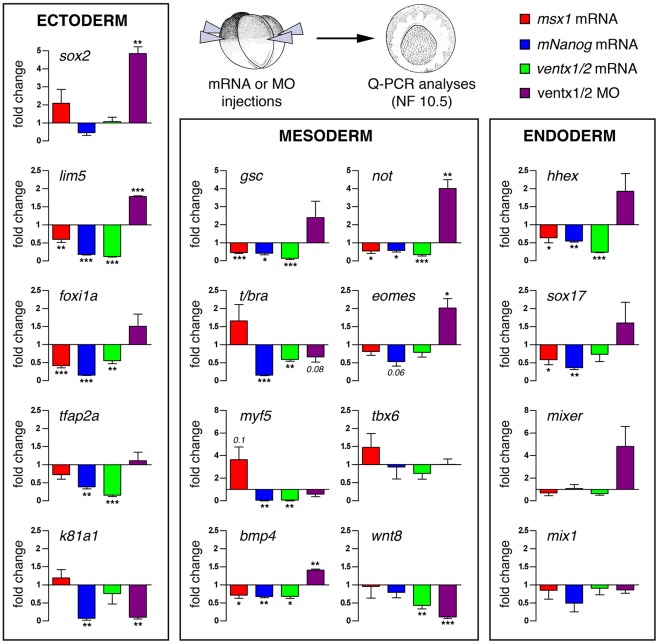
We next assessed whether the ventralizing effects of ventx1/2 and mNanog overexpression result from similar impacts on developmental gene expression. msx1 codes for another ventralizing NKL transcription factor and was used as a control at a dose (600 pg/embryo) known to efficiently ventralize embryos [51], [52]. Radial injections of ventx1/2, mNanog or msx1 mRNAs in NF3-embryos were performed and expression levels of ectodermal, mesodermal, and endodermal markers were analyzed by RT Q-PCR at stage NF10.5 ( Fig. 2 ). Strikingly, ventx1/2 and mNanog repressed most markers analysed, which was not the case for msx1. More specifically, all three factors had comparable anti-dorsalizing activities, as revealed by the robust repression of the organizer markers gsc, hhex, and not. Remarkably, they also repressed in a similar fashion the ventral mesoderm marker bmp4 and the early ectoderm markers lim5 and foxi1a. In contrast, other genes involved in epidermis (tfap2), axial (t/bra) and paraxial mesoderm (myf5) commitment were significantly down-regulated by ventx1/2 and mNanog but not msx1. Overall, msx1, ventx1/2 and mNanog overexpression repress early markers of various cell fates in all germ layers, but marked differences appear.
Figure 2. mNanog, ventx1/2, and msx1 cause distinct effects on early patterning gene expression.
For gain-of-function experiments, NF3-embryos were injected radially in all blastomeres with water, msx1 mRNA (0.3 ng/blastomere, red), mNanog mRNA (0.15 ng/blastomere, blue) or ventx1/2 mRNAs (0.5 ng/blastomere, green); For loss-of-function experiments, NF-2 embryos were injected twice radially in both blastomeres with control MO (30 ng/blastomere), or a 1∶1 mix of ventx1/2 MOs (30 ng/blastomere, purple). All embryos were collected at stage NF10.5 and processed for RT-QPCRs. Ectodermal, mesodermal and endodermal markers were assayed (each quantification was performed at least 3 times independently). For all RT-QPCR, graphs represent means of the fold-change calculated versus the appropriate control (fldx injected embryos in cases of overexpression and control MO for ventx1/2 knock-down) +/− s.e.m, and significance was assessed using paired t-test (*p≤0.05, **p≤0.005, ***p≤0.0005).
We also assessed the impact of ventx1/2 loss of function on the same markers. For this, morpholino oligonucleotides directed against ventx1 and ventx2 pseudoalleles (ventx1/2 MOs) [37] or control MO were injected radially in NF3-embryos and gene expression was analyzed at stage NF10.5. We observed that ventx1/2 knock-down led to significant overexpression of a number of genes repressed by ventx1/2 (not, lim5, bmp4). This inverse regulation did not quite reach significance for all markers, perhaps reflecting the redundant activity of ventx3s. However, we noted that the mean level of gsc RNA induction in our experiment is within the range reported elsewhere [37]. Similarly, mean levels of eomes, hhex, foxi1a, sox17 and mixer were raised, though significance was not reached. Unexpectedly, epidermal keratin (k81a1) was repressed both upon ventx1/2 knock-down, and ventx1/2 or mNanog overexpression. This might be explained by the fact that ventx1/2-deficient ectodermal cells tend to become neural, as suggested by sox2 up-regulation. A similar line of reasoning can be applied to the ventral marker wnt8.
Altogether, these data suggest that msx1, ventx1/2 and mNanog may regulate differentially early developmental networks, though causing similar ventralized phenotypes.
ventx1/2 and mNanog Repress Fate Commitment
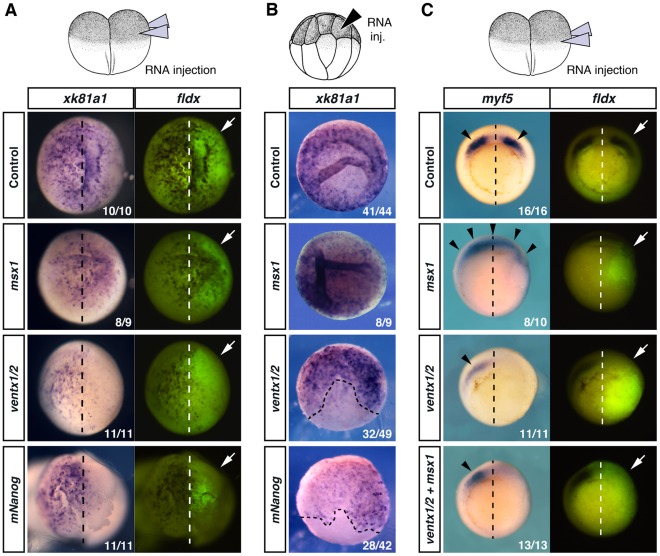
To evaluate the above hypothesis, we focused on epidermal differentiation, which is known to require Msx1 function [52]. We injected msx1, ventx1/2 or mNanog mRNAs either unilaterally, in one blastomere of 2-cell stage embryos (NF2, Fig. 3 A ) or at the 16-cell stage (NF5, Fig. 3 B ) in one AB4 blastomere fated to give rise only to epidermis. Both mNanog and ventx1/2 repressed expression of the committed ectoderm marker k81a1 in comparable fashion at NF10.5, whereas msx1 had no effect on epidermal differentiation. This result indicates that ventx1/2, unlike msx1, do not favour, but rather impede epidermal differentiation.
Figure 3. ventx1/2 overexpression prevents multiple lineage commitment.
(A) NF2-embryos were injected twice in one blastomere, either with msx1 mRNAs (0.6 ng/blastomere), ventx1/2 mRNAs (1 ng/blastomere), mNanog mRNA (0,3 ng/blastomere), or with water for control; fldx was used as a lineage tracer. WISH with a k81a1 probe were performed at stage NF10.5 (left panels, animal views, dorsal side to the top). The progeny of the injected blastomere was revealed by fluorescence; white arrows indicate the injected side (right panels). (B) Sixteen-cell stage embryos (NF5) were injected in one AB4 blastomere with msx1 mRNA (0.15 ng), ventx1/2 mRNAs (0.5 ng), mNanog mRNA (0.15 ng), or water, collected at stage NF10.5 and processed for WISH with a k81a1 probe (animal views). Black stripped lines mark the border between injected and uninjected domains. (C) NF2-embryos were injected twice in one blastomere with msx1 mRNA (5 ng/blastomere), ventx1/2 mRNAs (5 ng/blastomere), ventx1/2+msx1 mRNAs (5 ng +5 ng/blastomere), or with water. WISH with a myf5 probe were performed at stage NF10.5; black arrowheads point to myf5-expressing territories (left panels, ventral views, dorsal side to the top). The progeny of the injected blastomere was revealed by fldx fluorescence; white arrows point to the injected side (right panels).
To further assess if ventx1/2 restrain commitment in Xenopus, the “ventralizing” activity of msx1 was tested in the presence or excess of ventx1/2. Thus, msx1 and ventx1/2 mRNAs were injected separately or co-injected radially at the same concentration in one blastomere in NF2-embryos. Expression of the mesodermal marker myf5 was then assessed by whole-mount in situ hybridization (WISH) at stage NF10.5 ( Fig. 3C ), as it is known that msx1 positively regulates paraxial mesoderm differentiation [53]. In control embryos, myf5 is expressed in two dorso-lateral patches around the organizer. As expected, myf5 expression expanded into the organizer in the presence of msx1. Conversely, myf5 was repressed in the ventx1/2-injected side, in agreement with its previously described activity on paraxial mesoderm [54]. Strikingly, joint overexpression of ventx1/2 and msx1 also resulted in myf5 repression, demonstrating that ventx1/2 are able to antagonize msx1 activity during mesoderm commitment. Similar results were obtained using half as much ventx1/2 mRNAs (data not shown), suggesting that the difference of activity between ventx1/2 and msx1 is qualitative rather than quantitative.
The above data suggest that commitment into specific lineages, even of ventral origin, is not possible in the presence of high levels of ventx activity.
ventx1/2 Knockdown Represses Pluripotency Genes and Induces Premature Commitment
As pro-differentiation genes were up-regulated in ventx1/2 morphants ( Fig. 2 ), we hypothesized that these factors are involved in the temporal restriction of commitment in early embryos. In line with this hypothesis, we detected by RT-QPCR ventx1.2 and ventx2.1-b messengers in ovaries, unfertilized eggs and embryos from stages NF1 to NF10.5 (data not shown) and ventx2.1-a is known to be maternally expressed [55]. Furthermore, we found that ventx2.1-b is present in both animal and vegetal halves of 8-cell embryos (NF4, Fig. S4).
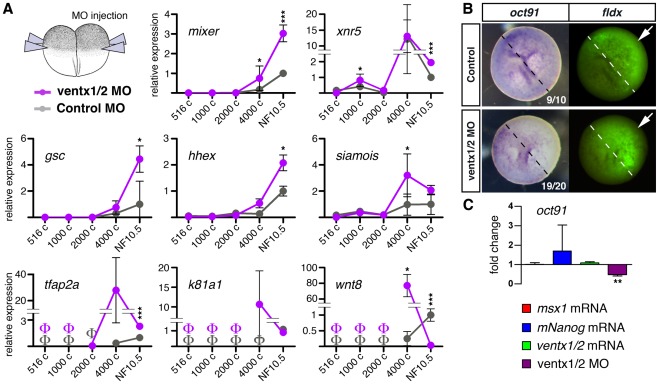
To test whether ventx1/2 are functionally required to restrict cellular commitment during early Xenopus development, we next performed RT-QPCR to monitor kinetics of expression following ventx1/2 knockdown ( Fig. 4 A ). Time-course experiments revealed that in ventx1/2 morphant embryos, expression levels of early dorsal mesendoderm (siamois, gsc, hhex), ventral mesendoderm (wnt8), pan-endodermal (mixer) and ventral ectoderm (tfap2a, k81a1) markers are higher at the 4000-cell stage when activation of zygotic transcription or “mid-blastula transition” (MBT) occurs. Remarkably, stronger expression was also observed at pre-MBT for xnr5, a nodal-related factor that participates in primary germ layer induction at these stages and pre-patterns the dorsal side of the embryo [56]. Overall, the expression profiles seem to be shifted to earlier time-points and to reach higher levels in ventx1/2 morphant embryos. These results suggest that in morphant embryos, cells are no longer protected against premature commitment. Interestingly, embryos in which the POU5F1 family member oct91 is knocked-down also fail to maintain a multipotent uncommitted cell population [25], [29]. We thus tested whether up-regulation of pro-differentiation markers in ventx1/2 LOF is accompanied by down-regulation of this marker of the uncommitted state. The expression of oct91 was significantly down-regulated after ventx1/2 knockdown, as assessed by RT-Q-PCR and WISH ( Fig. 4 , B and C). However, no significant up-regulation of oct91 was seen following ventx1/2 overexpression.
Figure 4. ventx1/2 activity is necessary to maintain an uncommitted cell population in early gastrulae.
(A) NF2-embryos were injected radially twice in both blastomeres with control MO (30 ng/blastomere), or a 1∶1 mix of ventx1/2 MOs (30 ng/blastomere). Variations of gene expression at 516-, 1000-, 2000-, 4000-cell and NF10.5 stages were assessed by RT-QPCR as in Fig. 2. Dorsal (siamois, gsc, hhex), and ventral (wnt8) mesendoderm, endoderm (xnr5, mixer) and ectoderm (tfap2a, k81a1) markers were monitored. Kinetic graphs represent means of fold-change relative to NF10.5 controls +/− s.e.m, and significance was assessed using paired t-test (*p≤0.05, **p≤0.005, ***p≤0.0005), and undetectable levels of transcript noted as Φ. (B) Animal injections were performed twice in a single blastomere NF2-embryos, using MO conditions described in (A); fldx was used as a lineage tracer. WISH with an oct91 probe (left panel) were performed at stage NF10.5 and the progeny of the injected blastomere was revealed by fluorescence (right panel). Embryos are positioned with the animal side upwards; white arrows indicate the injected side. (C) Injections were performed using mRNA and MO conditions described in Fig. 2. All embryos were collected at stage NF10.5 and processed for RT-QPCRs using the pluripotency marker oct91. Data and graphs are presented as in Fig. 2.
We conclude that ventx1/2 activity is necessary, but not sufficient, to maintain the uncommitted status of embryonic cells during Xenopus early development.
Ectopic Expression of Mouse Nanog but not msx1 Rescues ventx1/2 Morphant Embryos
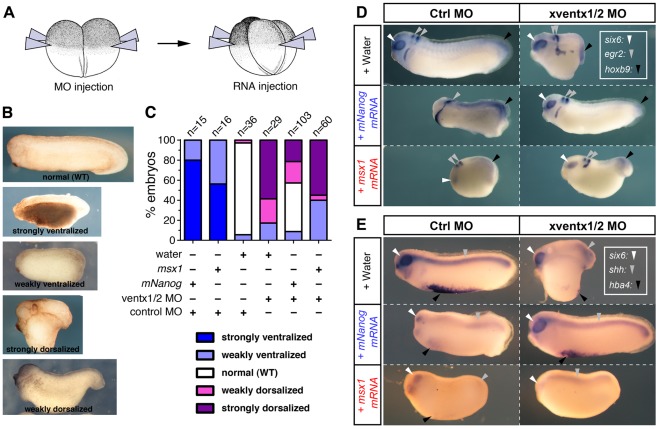
Based on the above data, we tested the ability of mNanog to rescue development of ventx1/2 knock-downed embryos. MOs directed against ventx1/2 [37] or control MO were first injected radially in each blastomere at the 2-cell stage (NF2), followed by radial injections in all blastomeres at stage NF3 of either mNanog or msx1 mRNAs ( Fig. 5 A ). Injections of control MO+mNanog and control MO+msx1 led to a high proportion of ventralized embryos ( Fig. 5 , B and C, about 80% and 60% respectively). As described [37], ventx1/2 MOs injection caused dorsalization defects in 80% of embryos, whereas control injections (control MO+water) yielded 90% of normal embryos. Quite remarkably, the addition of mNanog mRNA to ventx1/2 MOs produced 50% of embryos with normal morphology. Antero-posterior ( Fig. 5 D ) and dorso-ventral axes ( Fig. 5 E ) were correctly restored in such embryos, as revealed by whole mount in situ hybridization for markers of notochord (shh), ventral blood islands (hba4), spinal cord (hoxb9) and brain (six6 and egr2, also known as optx2 and krox-20 respectively). In contrast, msx1 overexpression failed to restore a normal morphology in ventx1/2 morphant embryos. In this condition, about 60% of embryos remained dorsalized and about 40% became ventralized, confirming that msx1 ventralizes embryos through mechanisms distinct from the ones induced by ventx1/2 and mNanog ( Fig. 5 E ). Altogether, these results demonstrate that mNanog is able to substitute for ventx1/2 in Xenopus development and that this effect is specific.
Figure 5. mNanog expression rescues specifically ventx1/2 morphant embryos.
(A) Two-cell stage embryos (NF2) were first injected radially twice with control MO (30 ng/blastomere), or a 1∶1 mix of ventx1 and ventx2 MOs (ventx1/2 MOs; 30 ng/blastomere), and subsequently injected radially at NF3 in all blastomeres with mNanog mRNA (0.15 ng/blastomere), msx1 mRNA (0.15 ng/blastomere), or with water. (B) Range of phenotypes observed in rescue of ventx1/2 knockdown experiment. (C) Percentages observed for each phenotypic category in three independent replicates of the rescue experiment. The combined injections performed are indicated at the bottom of the graph, and the number of injected embryos for each condition is indicated on the top of each bar. NF28 embryos were processed for WISH with six6, egr2 and hoxb9 (D), or six6, shh and hba4 (E) probes (anterior to the left, dorsal to the top).
Discussion
In this study, we report a number of properties of ventx1/2 in the Xenopus embryo that call for a reassessment of their biological role. Although ventx1/2 and msx1 are thought to be ventralizing regulators, their overexpression has distinct effects. msx1 suppresses dorsal but not ventral markers [52,53 and our results], whereas ventx1/2 suppress both dorsal and ventral markers in mesoderm and in epidermis [50,55, and our results]. Consequently, ventx1/2 cannot be considered bona fide ventralizing factors. Instead, we propose that ventx1/2 are guardians of developmental potential in the early embryo, a function that is necessary to achieve proportioned and progressive building of the body.
Supporting this view, we found that, in gastrulae, increased ventx1/2 activity represses the expression of transcription factors involved in early cell commitment in all germ layers, including a significant down-regulation of hhex, gsc, not, bmp4, t/bra, wnt8, lim5, foxi1a, tfap2a, and myf5 ( Fig. 1 and Fig. 2 ). Conversely, knockdown of ventx1/2 results in the significant up regulation of the differentiation markers sox2, lim5, bmp4, not, eomes ( Fig. 2 ). Further, we show that ventx1/2 are necessary to maintain normal expression levels of the well-identified pluripotency effector oct91 during gastrulation ( Fig. 4 , B and C). Thus, ventx1/2 appear to be involved in an active mechanism protecting blastula/gastrula cells from the pro-differentiation cues that establish germ layers and pattern the embryonic axis. The earliest role of ventx1/2 would therefore not be the establishment of ventral identity, but rather to prevent premature commitment, similar to Xenopus pou5f1s [25], [29], [44], [57], [58]. In line with this hypothesis, it is important to note i) that cells in early ventx1/2 expressing territories (ectoderm and ventral mesoderm) remain multipotent until late gastrulation [59], [60], ii) that active clearance of ventx proteins coincides with loss of multipotency at mid-gastrula stages [61], iii) that post-gastrula ventx2.1 expression territories coincide with stem cell-containing niches such as the dorsal ciliary margin of the eye [47] and the tailbud [62] and iv) that ventx2.1 is re-expressed together with oct91 in Xenopus somatic cells reprogrammed to an iPS-like state in vivo [63].
Our work thus highlights the role of ventx1/2 in linking cell commitment to embryonic axis patterning and agrees with experimental and theoretical works suggesting that the dorso-ventral genetic system, to which ventx1/2 belong, functions primarily as a regulator of the timing of cell commitment [64]. Indeed, according to our hypothesis, ventx1/2 would maintain early embryonic cells in an undetermined state and limit their competence to respond to differentiation-inducing signals, as NANOG does to maintain pluripotency in mammalian embryos [16], [65]. The ability of mNanog to rescue the ventx1/2 morphant phenotype in Xenopus embryos strongly supports this contention ( Fig. 5 ). In our re-interpretation of their role, ventx1/2 factors, as regulators of timing of commitment, control the progressive allocation of embryonic cells to the developing body axis. Loss of ventx1/2 allows cellular commitment and most cells precociously adopt dorsal and anterior positional identities, similar to the cells that first become negative for ventx1/2 in normal embryos. Consequently, the pool of cells available to build posterior territories is depleted, resulting in minute trunk-tail structures [37]. Conversely, ectopic ventx1/2 activity represses early commitment factors and thus causes dorso-anterior truncations [34]. As such embryos do develop posterior structures, we surmise that the uncommitted cellular state is only transient, in agreement with the reported loss of cellular competence at the end of gastrulation in normal embryos. Importantly, our data supports a role for ventx1/2 in restricting cell commitment starting at pre-MBT stages. Recent work underlines the importance of Wnt/βcat and Nodal signalling in priming cells for induction of mesendoderm and establishment of dorsal identity as early as the 1000-cell stage, well before MBT [56], [66]. Here, we show that knockdown of ventx1/2 results in premature and/or increased expression of a number of developmental genes including the nodal-related xnr5, as well as the dorsal organizer genes hhex, gsc and siamois at or before the MBT ( Fig. 4 A ). Taken together, the evidence presented here supports the concept that control of developmental potential is a strategy that ensures correct germ layer formation and body patterning, common to all gnathostomes [25].
Our proposed role of ventx1/2 in the control of cellular differentiation echoes earlier studies performed by W. Knöchel and collaborators [44]. Interestingly, these researchers recently tested the hypothesis that Xenopus ventxs could be functional homologs of Nanog [20]. However, their results contrast sharply with ours, as they did not observe rescue of morphant ventx1/2 embryos by mNanog. The reason for this discrepancy is unclear but may possibly reflect differences in experimental setup. The positive evidence of rescue of ventx1/2 knockdown by mNanog in this paper supports the view that ventx and Nanog genes are related and that Xenopus ventx1/2 and mammalian Nanog serve comparable developmental functions through the regulation of overlapping transcription programs (see Table S2). Indeed, while genetic responses induced by mNanog and ventx1/2 overexpression do not perfectly match at early stages (NF10.5, Fig. 2 ), these differences seem to be buffered by the regulation networks at work during germ layer specification and embryonic patterning, up to the point that mNanog is able to substitute for ventx1/2 to generate morphologically normal larvae ( Fig. 5 ).
Other amphibians possess Nanog orthologs [21], [22], raising the question of whether the role of ventxs in developmental potential maintenance is ancestral, or is an innovation specific to Xenopus. Functional data strongly support the ancestrality of ventxs involvement in this process, since teleost and Xenopus ventxs serve the same function during development [45], [67], [68]. Unfortunately, data concerning amniote Ventx genes is scarce, probably because they are absent from the genome of the mouse, the main experimental model in this taxon. Partial functional redundancy between Ventx and Nanog might explain the loss of the former in rodents and of the latter in Xenopus. Some indirect evidence supports this notion in mammals. The human ventx ortholog (VENTX) located next to the stem-cell marker UTF-1, shares features with its counterparts in Xenopus and fish [69]. Both human and Xenopus ventx orthologs [70] are direct targets of POU5F1 transcription factors [44], [71], [72], and human VENTX displays ventralizing activity in zebrafish embryos [69]. As mentioned earlier, VENTX retropseudogenes are unusually frequent in the human genome [40], a feature that is proposed to be a specific signature of genes involved in pluripotency maintenance such as POU5F1 and NANOG [41]. In line with this idea, VENTX is co-expressed with NANOG and POU5F1 in pluripotent-embryonal carcinomas [73], a subtype of human male germ cell tumours constituted of cells highly similar to early zygotic and ES cells [74], and these three genes are strongly down-regulated when tumour cell differentiation is forced in vitro [73]. Furthermore, a genome-wide RNA interference screen has shown that in human ES cells, VENTX or NANOG knockdown results in reduced expression of a POU5F1-GFP reporter construct in a comparable way (see Supplemental Information in [75]). Finally, VENTX expression is under the control of the POU5F1/SOX2/NANOG triumvirate, (http://biit.cs.ut.ee/escd/index.cgi?gene=ventx [72], [76]) suggesting that it may be part of the human pluripotency-regulating network [72], [76].
In conclusion, our data strongly support the concept that ventx1/2 act as guardians of high developmental potential during Xenopus early development. We propose that this role of Ventx genes is ancestral and conserved in gnathostomes, a question for future research with high biomedical relevance.
Materials and Methods
Ethics Statement
The care and treatment of animals used in this study were in accordance with institutional and national guidelines (Commission de Génie Génétique, “Direction Départementale des Services Vétérinaires”, European Union Directive 2010/63, registered as No. 4654 for the agreement decision, and as No. B 75-05-01 for the vertebrate living animals experimentation; this commission specifically approved this study).
In Silico Screening
Homeodomain sequences from all reported Nanog genes were retrieved from public repositories (http://www.ncbi.nlm.gov/; http://www.ensembl.org/) and used as queries to perform several rounds of TBLASTN screening on the Xenopus tropicalis genome assembly (http://genome.jgi-psf.org/Xentr4/Xentr4.home.html), as well as on available expressed sequence tags and cDNA sequences from Xenopus tropicalis and Xenopus laevis (http://www.ncbi.nlm.gov/). Control searches using the same queries to screen published gnathostome genomes allowed the identification of at least one likely candidate in each case.
Reverse Transcriptase - PCR Screening
Total RNAs were extracted from Xenopus laevis ovaries, unfertilized and fertilized eggs (NF1), blastulae (NF8) and early gastrulae (NF10.5). The RT-PCR protocol and primers used are detailed in supporting information.
Phylogenetic and Conservation Analyses
The methodologies used to perform molecular phylogenetic and conservation analyses of the NKL family are detailed in supporting information.
Xenopus Embryo Manipulations
Sacrifices and animal studies were conducted according to the principles and procedures described in Guidelines for Care and Use of Experimental Animals. Xenopus laevis were obtained by in vitro fertilization and staged according to Nieuwkoop and Faber [48] and cultured according to Slack et al. [77]. Embryos were injected with RNAs and/or morpholino oligonucleotides (MOs) as described in the relevant figure legends. We determined that the lethal dose for mNanog is 1 ng per embryo (data not shown). Dextran fluorescein (fldx; Molecular Probes) was used as a lineage label. In order to rule out possible interference of MOs mixed with mRNAs before injection, we performed rescue assays through injections of MOs at the 2-cell stage (NF2), followed by mRNAs injections at the 4-cell stage (NF3). All injections were performed at least three times to assess reproducibility.
In vitro Translation and Morpholino Oligonucleotides
Synthetic capped mRNAs were transcribed with the mMessage mMachine SP6 kit (Ambion) using the following templates: pCS2+-Vent1 and pCS2+-Xbr1b, both linearised with NotI (gifts of N. Papalopulu, University of Manchester, UK and collectively referred to as ventx1/2 in this work); pSP64T-xMsx1, linearised with EcoRI; pCS2+OlNanog, linearised with SacII (Gift of JL. Mullor, Centro de Investigación Príncipe Felipe, Valencia, Spain). To express mNanog, the ORF of a commercial clone (Geneservice) was PCR amplified and cloned into pCS2+, linearised with NotI and transcribed with mMESSAGE mMACHINE SP6 Kit (Ambion). Previously described morpholino oligonucleotides (MOs) directed against ventx1 and ventx2 pseudoalleles [37] were obtained from GeneTools.
Whole Mount in situ Hybridization
Injected embryos were processed for whole-mount in situ hybridization (WISH) with digoxigenin-labelled probes (Roche) using standard procedures and staining was done with BM purple (Roche). Embryos were bleached with hydrogen peroxide 4% (Carlo Erba Reagenti) and photographed with a MZ16F binocular (Leica).
Real-time Quantitative PCR and Statistical Analyses
For real-time quantitative PCR (RT-QPCR) total RNAs were extracted from 10 post-MBT ( Figs. 2 and 4 C ) or 5 pre-MBT ( Fig. 4 A ) embryos using RNeasy Plus Mini kit (Qiagen) and reverse transcribed using Superscript II reverse transcriptase (Invitrogen). Independent biological replicates were collected and RT-QPCR reactions were performed in duplicate for each sample using Power SYBR® master mix on a 7300 Real-Time PCR System (Applied Biosystems), following manufacturer recommendations. Primers (MWG Biotech) were described in previous publications or designed using Primer Express Software (Applied Biosystems), the relevant sequences and references are listed in Table S3. Primers for the housekeeping gene DNA elongation factor type 1 α (ef1a1) or ornithine decarboxylase 1 (odc1) were used as loading control for samples collected post-MBT ( Figs. 2 and 4 C ) and pre-MBT ( Fig. 4 A ), respectively. Ct data were collected using 7300 system software (Applied Biosystem) and analyzed using Excel (Microsoft). For Figs. 2 and 4 C , the Ct for each technical duplicate was averaged and normalized against ef1a1. Variations of expression were quantified using the ΔΔCts method, using the control condition as reference for each experimental replicate and fold changes were computed as 2ΔΔCt. Data from independent experiments were averaged, plotted and significance was assessed (two-way paired Student’s t-test) using Prism 5.03 (GraphPad). For Fig. 4 A , the Ct were normalized against odc1. Levels of expression were quantified using the ΔCts method averaged, plotted and significance was assessed (two-way paired Student’s t-test) using Prism 5.03 (GraphPad).
Supporting Information
Phylogenic reconstruction of the NKL group homeodomains relationships using Maximum Likelihood. (A) Global view of an unrooted maximum likelihood tree obtained with the homeodomain sequences of all known NKL members found in the genomes of the fly, amphioxus and a representative selection of vertebrates (see Supporting Information for Extended Experimental Procedures). NKL families are highlighted in different shades of grey except for NANOG (red) and VENTX (blue). Relationships between NKL families remain elusive; however all are monophyletic and well supported by bootstrap analysis with three exceptions: the NK4 (paraphyletic) VENTX (polyphyletic) and NANOG (monophyletic, but poorly supported, bootstrap: 54,4%). (B) Close-up of the region of the tree where most VENTX orthologs are found. (C) Close-up of the region of the tree containing the monophyletic NANOG group. Note that amphioxus VENTX homeodomains (VENT1 Branchiostoma floridae and VENT2 Branchiostoma floridae) are found at the root of the NANOG subtree. However, this association is not supported by bootstrap analysis (bootstrap: 18,7%) and the interpretation of amphioxus VENTXs as NANOG orthologs is at odds with the literature [39]. Both NANOG and the main VENTX group have longer branches than typical NKL-class members (e.g. NK1 and LBX groups on panels B and C, see also Table S1).
(TIF)
Determination of lethal doses of mNanog and OlNanog. NF3-embryos were injected radially in all blastomeres, with water, mNanog mRNA or OlNanog mRNA at multiples doses and embryonic lethality was assessed at late blastula (NF9) and early tadpole (NF31). The doses indicated correspond to the total amount of mRNA injected per embryo. (A-C) Representative NF9 embryos observed in the indicated conditions (top panels), an arrow points to the embryos shown at greater magnification (bottom panels). The number of embryos injected is indicated. (D-E) Percentage of lethality observed at NF9 and NF31 after mNanog and OlNanog injection, respectively. The numbers of dead and living embryos observed at both time points is given under the graphs. Note that a dose of 1,2 ng of mNanog results in 100% embryonic lethality at NF9, while 0,6 ng (half the lethal dose) had no toxic effect; hence this condition was retained for further study. Conversely, OlNanog overexpression led to increased lethality beyond the 2 ng injection condition of OlNanog RNA (dotted line). Embryo death arose from 5 ng injected embryos, and about 40% or 100% lethality was observed at NF31 for the 5 ng or the 10 ng conditions respectively. Hence the condition with 1,2 ng injected embryos was retained for further study.
(TIF)
OlNanog overexpression leads to phenotypes that strongly differ from those observed upon mNanog or ventx1/2 overexpression. (A) NF3 embryos were injected radially with OlNanog mRNA (0.6 ng, 1.2 ng, 2 ng and 5 ng./embryo), or with water for control. Representative phenotypes observed at early tadpole stage (NF31) are shown (lateral views, anterior to the left, dorsal to the top). (B) Percentages of observed phenotypes for the different OlNanog mRNAs doses assayed. Across the whole range of concentration used, the phenotypes obtained in OlNanog-injected embryos strongly differed from those resulting from mNanog overexpression (C and D). No cues of ventralization were observed as seen with mNanog-injected embryos (see black arrowheads in C), the embryos retaining distinguishable head structures. The main effect was a shortened axis, resulting from defects in blastopore closure (see white arrowheads in A).
(TIF)
ventx2.1 mRNAs are present in animal and vegetal cells of 8-cell stage Xenopus embryos. (A) 8-cell stage Xenopus embryos were separated in animal and vegetal halves, which were separately processed for RT-QPCR. (A) ventx2.1 (green) mRNA abundance in the two territories was estimated relative to the odc loading control marker, while vegt (purple) was used as a positive control. (B) As expected, we observed that vegt mRNA is almost exclusively localised in the vegetal blastomeres, while in contrast ventx2.1 mRNA is predominantly found in animal blastomeres but is also significantly present in vegetal blastomeres.
(TIF)
Nanog and Ventx homeodomains are less conserved than other NKL families. For each NKL family conserved among vertebrates (1st column) the homeodomains (HDs) of all Homo sapiens, Xenopus tropicalis, Danio rerio and Takifugu rubripes paralogs were retrieved (see Supporting Information S1 for Extended Experimental Procedures). When a given paralog was unknown in a given species but present in a closely related one, this alternate sequence was used instead. More specifically: (£) EMX1 being unknown in Takifugu rubripes, the Tetraodon nigroviridis sequence was used; (&) NANOG being unknown in Xenopus species the Ambystoma mexicanum sequence was used. For each group of orthologs, the percentage of identity along the HD of the four relevant sequences was computed. For families with multiple paralogs, only the least conserved are shown here (2nd column). The consensus sequence and percentage of identity thus obtained are indicated (3rd and 4th columns). The VENTX and NANOG families (in bold) present the lowest sequence identity in the HD, and are the only NKL families for which numerous processed pseudogenes are found in the human genome (5th column) [40,78]. This similarity extends to functional properties (see Table S2).
(TIF)
Mammalian Nanog and Xenopus ventxs share striking functional similarities. Mammalian Nanog (left) and Xenopus ventx1/2 (right) are “regulated by” (A and B), “regulate” (C) and “interact” (D) with homologous pathways, transcription factors, genes and proteins, respectively. Most of these factors are known to regulate pluripotency and/or cell commitment and differentiation in mammals (indicated by P/C), while their counterparts in frog are known to be involved in dorso/ventral patterning during embryogenesis (indicated by D/V). References 78–110 are listed as Supplemental References in Supporting Information.
(TIF)
Primer pairs used for RT-QPCR experiments in this study. For each primer pair, the forward and reverse sequences are listed, as well as the original publications (references 111–125 are listed as Supplemental References in Supporting Information).
(TIF)
The Supporting Information file contains Extended Experimental Procedures and Supplemental References.
(DOC)
Acknowledgments
We are grateful to V. Thomé for excellent technical assistance, to J.F. Riou for providing access to technical facilities, as well as to Pr P.W. Holland and T. Butts for kindly sharing sequence data that proved instrumental in the initial stages of this work.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by Ministère de l’Enseignement Supérieur et de la Recherche PhD fellowships to PS, GVM and GL; by Fondation pour la Recherche Médicale (FRM: http://www.frm.org) PhD fellowships to PS and GVM; by an Association pour la Recherche sur le Cancer (ARC: http://www.recherche-cancer.net) PhD fellowship to GL; by EU contract no. 018652 CRESCENDO; by Programme National de Recherche sur les Perturbateurs Endocriniens of Ministère de L’Ecologie et du Développement Durable Contrat 2009 n°0007065 (PNRPE: http://www.pnrpe.fr/) and by l’Agence Nationale de la Recherche (ANR: http://www.agence-nationale-recherche.fr/). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Chenoweth JG, McKay RD, Tesar PJ. Epiblast stem cells contribute new insight into pluripotency and gastrulation. Dev Growth Differ. 2010;52:293–301. doi: 10.1111/j.1440-169X.2010.01171.x. [DOI] [PubMed] [Google Scholar]
- 2.Hanna J, Cheng AW, Saha K, Kim J, Lengner CJ, et al. Human embryonic stem cells with biological and epigenetic characteristics similar to those of mouse ESCs. Proc Natl Acad Sci U S A. 2010;107:9222–9227. doi: 10.1073/pnas.1004584107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, et al. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113:631–642. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- 4.Niwa H. How is pluripotency determined and maintained? Development. 2007;134:635–646. doi: 10.1242/dev.02787. [DOI] [PubMed] [Google Scholar]
- 5.Kaneko K, Sato K, Michiue T, Okabayashi K, Ohnuma K, et al. Developmental potential for morphogenesis in vivo and in vitro. J Exp Zool B Mol Dev Evol. 2008;310:492–503. doi: 10.1002/jez.b.21222. [DOI] [PubMed] [Google Scholar]
- 6.Snape A, Wylie CC, Smith JC, Heasman J. Changes in states of commitment of single animal pole blastomeres of Xenopus laevis. Dev Biol. 1987;119:503–510. doi: 10.1016/0012-1606(87)90053-4. [DOI] [PubMed] [Google Scholar]
- 7.Thomson M, Liu SJ, Zou LN, Smith Z, Meissner A, et al. Pluripotency factors in embryonic stem cells regulate differentiation into germ layers. Cell. 2011;145:875–889. doi: 10.1016/j.cell.2011.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Watabe T, Miyazono K. Roles of TGF-beta family signaling in stem cell renewal and differentiation. Cell Res. 2009;19:103–115. doi: 10.1038/cr.2008.323. [DOI] [PubMed] [Google Scholar]
- 9.Chambers I, Tomlinson SR. The transcriptional foundation of pluripotency. Development. 2009;136:2311–2322. doi: 10.1242/dev.024398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vallier L, Touboul T, Brown S, Cho C, Bilican B, et al. Signaling pathways controlling pluripotency and early cell fate decisions of human induced pluripotent stem cells. Stem Cells. 2009;27:2655–2666. doi: 10.1002/stem.199. [DOI] [PubMed] [Google Scholar]
- 11.Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 13.Chambers I, Colby D, Robertson M, Nichols J, Lee S, et al. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- 14.Chambers I, Silva J, Colby D, Nichols J, Nijmeijer B, et al. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450:1230–1234. doi: 10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- 15.Silva J, Nichols J, Theunissen TW, Guo G, van Oosten AL, et al. Nanog is the gateway to the pluripotent ground state. Cell. 2009;138:722–737. doi: 10.1016/j.cell.2009.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kalmar T, Lim C, Hayward P, Munoz-Descalzo S, Nichols J, et al. Regulated fluctuations in nanog expression mediate cell fate decisions in embryonic stem cells. PLoS Biol. 2009;7:e1000149. doi: 10.1371/journal.pbio.1000149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Canon S, Herranz C, Manzanares M. Germ cell restricted expression of chick Nanog. Dev Dyn. 2006;235:2889–2894. doi: 10.1002/dvdy.20927. [DOI] [PubMed] [Google Scholar]
- 18.Lavial F, Acloque H, Bertocchini F, Macleod DJ, Boast S, et al. The Oct4 homologue PouV and Nanog regulate pluripotency in chicken embryonic stem cells. Development. 2007;134:3549–3563. doi: 10.1242/dev.006569. [DOI] [PubMed] [Google Scholar]
- 19.Camp E, Sanchez-Sanchez AV, Garcia-Espana A, Desalle R, Odqvist L, et al. Nanog regulates proliferation during early fish development. Stem Cells. 2009;27:2081–2091. doi: 10.1002/stem.133. [DOI] [PubMed] [Google Scholar]
- 20.Schuff M, Siegel D, Philipp M, Bundschu K, Heymann N, et al. Stem Cells Dev; 2011. Characterization of Danio rerio Nanog and Functional Comparison to Xenopus Vents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dixon JE, Allegrucci C, Redwood C, Kump K, Bian Y, et al. Axolotl Nanog activity in mouse embryonic stem cells demonstrates that ground state pluripotency is conserved from urodele amphibians to mammals. Development. 2010;137:2973–2980. doi: 10.1242/dev.049262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maki N, Suetsugu-Maki R, Tarui H, Agata K, Del Rio-Tsonis K, et al. Expression of stem cell pluripotency factors during regeneration in newts. Dev Dyn. 2009;238:1613–1616. doi: 10.1002/dvdy.21959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Theunissen TW, Costa Y, Radzisheuskaya A, van Oosten AL, Lavial F, et al. Reprogramming capacity of Nanog is functionally conserved in vertebrates and resides in a unique homeodomain. Development. 2011;138:4853–4865. doi: 10.1242/dev.068775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O’Farrell PH, Stumpff J, Su TT. Embryonic cleavage cycles: how is a mouse like a fly? Curr Biol. 2004;14:R35–45. doi: 10.1016/j.cub.2003.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morrison GM, Brickman JM. Conserved roles for Oct4 homologues in maintaining multipotency during early vertebrate development. Development. 2006;133:2011–2022. doi: 10.1242/dev.02362. [DOI] [PubMed] [Google Scholar]
- 26.Niwa H, Sekita Y, Tsend-Ayush E, Grutzner F. Platypus Pou5f1 reveals the first steps in the evolution of trophectoderm differentiation and pluripotency in mammals. Evol Dev. 2008;10:671–682. doi: 10.1111/j.1525-142X.2008.00280.x. [DOI] [PubMed] [Google Scholar]
- 27.Frankenberg S, Pask A, Renfree MB. The evolution of class V POU domain transcription factors in vertebrates and their characterisation in a marsupial. Dev Biol. 2010;337:162–170. doi: 10.1016/j.ydbio.2009.10.017. [DOI] [PubMed] [Google Scholar]
- 28.Abu-Remaileh M, Gerson A, Farago M, Nathan G, Alkalay I, et al. Oct-3/4 regulates stem cell identity and cell fate decisions by modulating Wnt/beta-catenin signalling. EMBO J. 2010;29:3236–3248. doi: 10.1038/emboj.2010.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cao Y, Siegel D, Knochel W. Xenopus POU factors of subclass V inhibit activin/nodal signaling during gastrulation. Mech Dev. 2006;123:614–625. doi: 10.1016/j.mod.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 30.Hellsten U, Harland RM, Gilchrist MJ, Hendrix D, Jurka J, et al. The genome of the Western clawed frog Xenopus tropicalis. Science. 2010;328:633–636. doi: 10.1126/science.1183670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koziol MJ, Garrett N, Gurdon JB. Tpt1 activates transcription of oct4 and nanog in transplanted somatic nuclei. Curr Biol. 2007;17:801–807. doi: 10.1016/j.cub.2007.03.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gawantka V, Delius H, Hirschfeld K, Blumenstock C, Niehrs C. Antagonizing the Spemann organizer: role of the homeobox gene Xvent-1. EMBO J. 1995;14:6268–6279. doi: 10.1002/j.1460-2075.1995.tb00317.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Onichtchouk D, Gawantka V, Dosch R, Delius H, Hirschfeld K, et al. The Xvent-2 homeobox gene is part of the BMP-4 signalling pathway controlling [correction of controling] dorsoventral patterning of Xenopus mesoderm. Development. 1996;122:3045–3053. doi: 10.1242/dev.122.10.3045. [DOI] [PubMed] [Google Scholar]
- 34.Onichtchouk D, Glinka A, Niehrs C. Requirement for Xvent-1 and Xvent-2 gene function in dorsoventral patterning of Xenopus mesoderm. Development. 1998;125:1447–1456. doi: 10.1242/dev.125.8.1447. [DOI] [PubMed] [Google Scholar]
- 35.Shapira E, Marom K, Yelin R, Levy A, Fainsod A. A role for the homeobox gene Xvex-1 as part of the BMP-4 ventral signaling pathway. Mech Dev. 1999;86:99–111. doi: 10.1016/s0925-4773(99)00120-3. [DOI] [PubMed] [Google Scholar]
- 36.Shapira E, Marom K, Levy V, Yelin R, Fainsod A. The Xvex-1 antimorph reveals the temporal competence for organizer formation and an early role for ventral homeobox genes. Mech Dev. 2000;90:77–87. doi: 10.1016/s0925-4773(99)00283-x. [DOI] [PubMed] [Google Scholar]
- 37.Sander V, Reversade B, De Robertis EM. The opposing homeobox genes Goosecoid and Vent1/2 self-regulate Xenopus patterning. EMBO J. 2007;26:2955–2965. doi: 10.1038/sj.emboj.7601705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ault KT, Dirksen ML, Jamrich M. A novel homeobox gene PV.1 mediates induction of ventral mesoderm in Xenopus embryos. Proc Natl Acad Sci U S A. 1996;93:6415–6420. doi: 10.1073/pnas.93.13.6415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kozmik Z, Holland LZ, Schubert M, Lacalli TC, Kreslova J, et al. Characterization of Amphioxus AmphiVent, an evolutionarily conserved marker for chordate ventral mesoderm. Genesis. 2001;29:172–179. doi: 10.1002/gene.1021. [DOI] [PubMed] [Google Scholar]
- 40.Holland PW, Booth HA, Bruford EA. Classification and nomenclature of all human homeobox genes. BMC Biol. 2007;5:47. doi: 10.1186/1741-7007-5-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pain D, Chirn GW, Strassel C, Kemp DM. Multiple retropseudogenes from pluripotent cell-specific gene expression indicates a potential signature for novel gene identification. J Biol Chem. 2005;280:6265–6268. doi: 10.1074/jbc.C400587200. [DOI] [PubMed] [Google Scholar]
- 42.Friedle H, Rastegar S, Paul H, Kaufmann E, Knochel W. Xvent-1 mediates BMP-4-induced suppression of the dorsal-lip-specific early response gene XFD-1' in Xenopus embryos. EMBO J. 1998;17:2298–2307. doi: 10.1093/emboj/17.8.2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liang J, Wan M, Zhang Y, Gu P, Xin H, et al. Nanog and Oct4 associate with unique transcriptional repression complexes in embryonic stem cells. Nat Cell Biol. 2008;10:731–739. doi: 10.1038/ncb1736. [DOI] [PubMed] [Google Scholar]
- 44.Cao Y, Knochel S, Donow C, Miethe J, Kaufmann E, et al. The POU factor Oct-25 regulates the Xvent-2B gene and counteracts terminal differentiation in Xenopus embryos. J Biol Chem. 2004;279:43735–43743. doi: 10.1074/jbc.M407544200. [DOI] [PubMed] [Google Scholar]
- 45.Reim G, Brand M. Maternal control of vertebrate dorsoventral axis formation and epiboly by the POU domain protein Spg/Pou2/Oct4. Development. 2006;133:2757–2770. doi: 10.1242/dev.02391. [DOI] [PubMed] [Google Scholar]
- 46.Belting HG, Wendik B, Lunde K, Leichsenring M, Mossner R, et al. Pou5f1 contributes to dorsoventral patterning by positive regulation of vox and modulation of fgf8a expression. Dev Biol. 2011;356:323–336. doi: 10.1016/j.ydbio.2011.05.660. [DOI] [PubMed] [Google Scholar]
- 47.Papalopulu N, Kintner C. A Xenopus gene, Xbr-1, defines a novel class of homeobox genes and is expressed in the dorsal ciliary margin of the eye. Dev Biol. 1996;174:104–114. doi: 10.1006/dbio.1996.0055. [DOI] [PubMed] [Google Scholar]
- 48.Nieuwkoop PD, Faber J. Normal table of Xenopus laevis (Daudin): a systematical and chronological survey of the development from the fertilized egg till the end of metamorphosis. 1994.
- 49.Rankin SA, Kormish J, Kofron M, Jegga A, Zorn AM. A gene regulatory network controlling hhex transcription in the anterior endoderm of the organizer. Dev Biol. 2011;351:297–310. doi: 10.1016/j.ydbio.2010.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kumano G, Belluzzi L, Smith WC. Spatial and temporal properties of ventral blood island induction in Xenopus laevis. Development. 1999;126:5327–5337. doi: 10.1242/dev.126.23.5327. [DOI] [PubMed] [Google Scholar]
- 51.Maeda R, Kobayashi A, Sekine R, Lin JJ, Kung H, et al. Xmsx-1 modifies mesodermal tissue pattern along dorsoventral axis in Xenopus laevis embryo. Development. 1997;124:2553–2560. doi: 10.1242/dev.124.13.2553. [DOI] [PubMed] [Google Scholar]
- 52.Suzuki A, Ueno N, Hemmati-Brivanlou A. Xenopus msx1 mediates epidermal induction and neural inhibition by BMP4. Development. 1997;124:3037–3044. doi: 10.1242/dev.124.16.3037. [DOI] [PubMed] [Google Scholar]
- 53.Takeda M, Saito Y, Sekine R, Onitsuka I, Maeda R, et al. Xenopus msx-1 regulates dorso-ventral axis formation by suppressing the expression of organizer genes. Comp Biochem Physiol B Biochem Mol Biol. 2000;126:157–168. doi: 10.1016/s0305-0491(00)00194-2. [DOI] [PubMed] [Google Scholar]
- 54.Polli M, Amaya E. A study of mesoderm patterning through the analysis of the regulation of Xmyf-5 expression. Development. 2002;129:2917–2927. doi: 10.1242/dev.129.12.2917. [DOI] [PubMed] [Google Scholar]
- 55.Schmidt JE, von Dassow G, Kimelman D. Regulation of dorsal-ventral patterning: the ventralizing effects of the novel Xenopus homeobox gene Vox. Development. 1996;122:1711–1721. doi: 10.1242/dev.122.6.1711. [DOI] [PubMed] [Google Scholar]
- 56.Skirkanich J, Luxardi G, Yang J, Kodjabachian L, Klein PS. An essential role for transcription before the MBT in Xenopus laevis. Dev Biol. 2011;357:478–491. doi: 10.1016/j.ydbio.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cao Y, Siegel D, Donow C, Knochel S, Yuan L, et al. POU-V factors antagonize maternal VegT activity and beta-Catenin signaling in Xenopus embryos. EMBO J. 2007;26:2942–2954. doi: 10.1038/sj.emboj.7601736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cao Y, Siegel D, Oswald F, Knochel W. Oct25 represses transcription of nodal/activin target genes by interaction with signal transducers during Xenopus gastrulation. J Biol Chem. 2008;283:34168–34177. doi: 10.1074/jbc.M803532200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Domingo C, Keller R. Cells remain competent to respond to mesoderm-inducing signals present during gastrulation in Xenopus laevis. Dev Biol. 2000;225:226–240. doi: 10.1006/dbio.2000.9769. [DOI] [PubMed] [Google Scholar]
- 60.Okabayashi K, Asashima M. Tissue generation from amphibian animal caps. Curr Opin Genet Dev. 2003;13:502–507. doi: 10.1016/s0959-437x(03)00111-4. [DOI] [PubMed] [Google Scholar]
- 61.Zhu Z, Kirschner M. Regulated proteolysis of Xom mediates dorsoventral pattern formation during early Xenopus development. Dev Cell. 2002;3:557–568. doi: 10.1016/s1534-5807(02)00270-8. [DOI] [PubMed] [Google Scholar]
- 62.Davis RL, Kirschner MW. The fate of cells in the tailbud of Xenopus laevis. Development. 2000;127:255–267. doi: 10.1242/dev.127.2.255. [DOI] [PubMed] [Google Scholar]
- 63.Vivien C, Scerbo P, Girardot F, Le Blay K, Demeneix BA, et al. J Biol Chem; 2012. Non-viral expression of mouse Oct4, Sox2 and Klf4 factors efficiently reprograms tadpole muscle fibers in vivo. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lane MC, Sheets MD. Heading in a new direction: implications of the revised fate map for understanding Xenopus laevis development. Dev Biol. 2006;296:12–28. doi: 10.1016/j.ydbio.2006.04.447. [DOI] [PubMed] [Google Scholar]
- 65.Vallier L, Mendjan S, Brown S, Chng Z, Teo A, et al. Activin/Nodal signalling maintains pluripotency by controlling Nanog expression. Development. 2009;136:1339–1349. doi: 10.1242/dev.033951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Blythe SA, Cha SW, Tadjuidje E, Heasman J, Klein PS. beta-Catenin primes organizer gene expression by recruiting a histone H3 arginine 8 methyltransferase, Prmt2. Dev Cell. 2010;19:220–231. doi: 10.1016/j.devcel.2010.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Flores MV, Lam EY, Crosier KE, Crosier PS. Osteogenic transcription factor Runx2 is a maternal determinant of dorsoventral patterning in zebrafish. Nat Cell Biol. 2008;10:346–352. doi: 10.1038/ncb1697. [DOI] [PubMed] [Google Scholar]
- 68.Imai Y, Gates MA, Melby AE, Kimelman D, Schier AF, et al. The homeobox genes vox and vent are redundant repressors of dorsal fates in zebrafish. Development. 2001;128:2407–2420. doi: 10.1242/dev.128.12.2407. [DOI] [PubMed] [Google Scholar]
- 69.Moretti PA, Davidson AJ, Baker E, Lilley B, Zon LI, et al. Molecular cloning of a human Vent-like homeobox gene. Genomics. 2001;76:21–29. doi: 10.1006/geno.2001.6574. [DOI] [PubMed] [Google Scholar]
- 70.Gao H, Le Y, Wu X, Silberstein LE, Giese RW, et al. VentX, a novel lymphoid-enhancing factor/T-cell factor-associated transcription repressor, is a putative tumor suppressor. Cancer Res. 2010;70:202–211. doi: 10.1158/0008-5472.CAN-09-2668. [DOI] [PubMed] [Google Scholar]
- 71.Jung M, Peterson H, Chavez L, Kahlem P, Lehrach H, et al. A data integration approach to mapping OCT4 gene regulatory networks operative in embryonic stem cells and embryonal carcinoma cells. PLoS One. 2010;5:e10709. doi: 10.1371/journal.pone.0010709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Korkola JE, Houldsworth J, Chadalavada RS, Olshen AB, Dobrzynski D, et al. Down-regulation of stem cell genes, including those in a 200-kb gene cluster at 12p13.31, is associated with in vivo differentiation of human male germ cell tumors. Cancer Res. 2006;66:820–827. doi: 10.1158/0008-5472.CAN-05-2445. [DOI] [PubMed] [Google Scholar]
- 74.Clark AT. The stem cell identity of testicular cancer. Stem Cell Rev. 2007;3:49–59. doi: 10.1007/s12015-007-0002-x. [DOI] [PubMed] [Google Scholar]
- 75.Chia NY, Chan YS, Feng B, Lu X, Orlov YL, et al. A genome-wide RNAi screen reveals determinants of human embryonic stem cell identity. Nature. 2010;468:316–320. doi: 10.1038/nature09531. [DOI] [PubMed] [Google Scholar]
- 76.Greber B, Lehrach H, Adjaye J. Control of early fate decisions in human ES cells by distinct states of TGFbeta pathway activity. Stem Cells Dev. 2008;17:1065–1077. doi: 10.1089/scd.2008.0035. [DOI] [PubMed] [Google Scholar]
- 77.Slack JM, Dale L, Smith JC. Analysis of embryonic induction by using cell lineage markers. Philos Trans R Soc Lond B Biol Sci. 1984;307:331–336. doi: 10.1098/rstb.1984.0135. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phylogenic reconstruction of the NKL group homeodomains relationships using Maximum Likelihood. (A) Global view of an unrooted maximum likelihood tree obtained with the homeodomain sequences of all known NKL members found in the genomes of the fly, amphioxus and a representative selection of vertebrates (see Supporting Information for Extended Experimental Procedures). NKL families are highlighted in different shades of grey except for NANOG (red) and VENTX (blue). Relationships between NKL families remain elusive; however all are monophyletic and well supported by bootstrap analysis with three exceptions: the NK4 (paraphyletic) VENTX (polyphyletic) and NANOG (monophyletic, but poorly supported, bootstrap: 54,4%). (B) Close-up of the region of the tree where most VENTX orthologs are found. (C) Close-up of the region of the tree containing the monophyletic NANOG group. Note that amphioxus VENTX homeodomains (VENT1 Branchiostoma floridae and VENT2 Branchiostoma floridae) are found at the root of the NANOG subtree. However, this association is not supported by bootstrap analysis (bootstrap: 18,7%) and the interpretation of amphioxus VENTXs as NANOG orthologs is at odds with the literature [39]. Both NANOG and the main VENTX group have longer branches than typical NKL-class members (e.g. NK1 and LBX groups on panels B and C, see also Table S1).
(TIF)
Determination of lethal doses of mNanog and OlNanog. NF3-embryos were injected radially in all blastomeres, with water, mNanog mRNA or OlNanog mRNA at multiples doses and embryonic lethality was assessed at late blastula (NF9) and early tadpole (NF31). The doses indicated correspond to the total amount of mRNA injected per embryo. (A-C) Representative NF9 embryos observed in the indicated conditions (top panels), an arrow points to the embryos shown at greater magnification (bottom panels). The number of embryos injected is indicated. (D-E) Percentage of lethality observed at NF9 and NF31 after mNanog and OlNanog injection, respectively. The numbers of dead and living embryos observed at both time points is given under the graphs. Note that a dose of 1,2 ng of mNanog results in 100% embryonic lethality at NF9, while 0,6 ng (half the lethal dose) had no toxic effect; hence this condition was retained for further study. Conversely, OlNanog overexpression led to increased lethality beyond the 2 ng injection condition of OlNanog RNA (dotted line). Embryo death arose from 5 ng injected embryos, and about 40% or 100% lethality was observed at NF31 for the 5 ng or the 10 ng conditions respectively. Hence the condition with 1,2 ng injected embryos was retained for further study.
(TIF)
OlNanog overexpression leads to phenotypes that strongly differ from those observed upon mNanog or ventx1/2 overexpression. (A) NF3 embryos were injected radially with OlNanog mRNA (0.6 ng, 1.2 ng, 2 ng and 5 ng./embryo), or with water for control. Representative phenotypes observed at early tadpole stage (NF31) are shown (lateral views, anterior to the left, dorsal to the top). (B) Percentages of observed phenotypes for the different OlNanog mRNAs doses assayed. Across the whole range of concentration used, the phenotypes obtained in OlNanog-injected embryos strongly differed from those resulting from mNanog overexpression (C and D). No cues of ventralization were observed as seen with mNanog-injected embryos (see black arrowheads in C), the embryos retaining distinguishable head structures. The main effect was a shortened axis, resulting from defects in blastopore closure (see white arrowheads in A).
(TIF)
ventx2.1 mRNAs are present in animal and vegetal cells of 8-cell stage Xenopus embryos. (A) 8-cell stage Xenopus embryos were separated in animal and vegetal halves, which were separately processed for RT-QPCR. (A) ventx2.1 (green) mRNA abundance in the two territories was estimated relative to the odc loading control marker, while vegt (purple) was used as a positive control. (B) As expected, we observed that vegt mRNA is almost exclusively localised in the vegetal blastomeres, while in contrast ventx2.1 mRNA is predominantly found in animal blastomeres but is also significantly present in vegetal blastomeres.
(TIF)
Nanog and Ventx homeodomains are less conserved than other NKL families. For each NKL family conserved among vertebrates (1st column) the homeodomains (HDs) of all Homo sapiens, Xenopus tropicalis, Danio rerio and Takifugu rubripes paralogs were retrieved (see Supporting Information S1 for Extended Experimental Procedures). When a given paralog was unknown in a given species but present in a closely related one, this alternate sequence was used instead. More specifically: (£) EMX1 being unknown in Takifugu rubripes, the Tetraodon nigroviridis sequence was used; (&) NANOG being unknown in Xenopus species the Ambystoma mexicanum sequence was used. For each group of orthologs, the percentage of identity along the HD of the four relevant sequences was computed. For families with multiple paralogs, only the least conserved are shown here (2nd column). The consensus sequence and percentage of identity thus obtained are indicated (3rd and 4th columns). The VENTX and NANOG families (in bold) present the lowest sequence identity in the HD, and are the only NKL families for which numerous processed pseudogenes are found in the human genome (5th column) [40,78]. This similarity extends to functional properties (see Table S2).
(TIF)
Mammalian Nanog and Xenopus ventxs share striking functional similarities. Mammalian Nanog (left) and Xenopus ventx1/2 (right) are “regulated by” (A and B), “regulate” (C) and “interact” (D) with homologous pathways, transcription factors, genes and proteins, respectively. Most of these factors are known to regulate pluripotency and/or cell commitment and differentiation in mammals (indicated by P/C), while their counterparts in frog are known to be involved in dorso/ventral patterning during embryogenesis (indicated by D/V). References 78–110 are listed as Supplemental References in Supporting Information.
(TIF)
Primer pairs used for RT-QPCR experiments in this study. For each primer pair, the forward and reverse sequences are listed, as well as the original publications (references 111–125 are listed as Supplemental References in Supporting Information).
(TIF)
The Supporting Information file contains Extended Experimental Procedures and Supplemental References.
(DOC)