Abstract
Fear conditioning (FC) may provide a useful model for some components of post-traumatic stress disorder (PTSD). We used a C57BL/6J × DBA/2J F2 intercross (n = 620) and a C57BL/6J × DBA/2J F8 advanced inter-cross line (n = 567) to fine-map quantitative trait loci (QTL) associated with FC. We conducted an integrated genome-wide association analysis in QTLRel and identified five highly significant QTL affecting freezing to context as well as four highly significant QTL associated with freezing to cue. The average percent decrease in QTL width between the F2 and the integrated analysis was 59.2%. Next, we exploited bioinformatic sequence and expression data to identify candidate genes based on the existence of non-synonymous coding polymorphisms and/or expression QTLs. We identified numerous candidate genes that have been previously implicated in either fear learning in animal models (Bcl2, Btg2, Dbi, Gabr1b, Lypd1, Pam and Rgs14) or PTSD in humans (Gabra2, Oprm1 and Trkb); other identified genes may represent novel findings. The integration of F2 and AIL data maintains the advantages of studying FC in model organisms while significantly improving resolution over previous approaches.
Keywords: Advanced intercross lines, Fear conditioning, Fear learning, Genome-wide association, Post-traumatic stress disorder, Quantitative trait loci
Introduction
Translational mouse models have provided a useful strategy for understanding the genetic and biological underpinnings of the acquisition of fear and phobias (Carey 1990), as well as the etiologic processes related to fear and anxiety. Despite some limitations (Layton and Krikorian 2002), the classical conditioning paradigm known as fear conditioning (FC) has been commonly used to model various components of post-traumatic stress disorder (PTSD; Fanselow and LeDoux 1999; Amstadter et al. 2009; Jovanovic and Ressler 2010; Johnson et al. 2011). FC is a form of Pavlovian learning in which an aversive unconditioned stimulus (US) is paired with a previously neutral conditioned cue (CS). Following training, recall of the fearful memory is measured by observation of freezing, a species-specific response to fear. Freezing is used to measure fear of the CS or fear of the context in which the fearful memory was acquired. In contrast to most common tests of anxiety-like behaviors in mice, FC is highly conserved across species, is exhibited in both laboratory and natural environments, and can easily be measured in humans; and may thus be a useful intermediate phenotype for aspects of PTSD (LaBar et al. 1995; Amstadter et al. 2009). For example, the sight, sound, and smell of traumatic events become potent memories through which Pavlovian fear is acquired in both PTSD and FC (Johnson and LeDoux 2004). Additionally, PTSD patients have been shown to be more “conditionable” than individuals without PTSD and take longer to extinguish fear (Peri et al. 2000; Blechert et al. 2005; Orr et al. 2000). Lastly, Pavlovian fear is heritable in both mice and humans (Wehner et al. 1997; Grillon et al. 1998; Hettema et al. 2003) and its neurological underpinnings have been well established (LeDoux 2000; Richardson et al. 2004).
Genetic mapping studies in mice have traditionally used recombinant inbred lines (RI), backcrosses (BC), F2 intercrosses, short-term selected lines (STSL), consomic and congenic mice to identify quantitative trait loci (QTLs) for FC (Caldarone et al. 1997; Owen et al. 1997a, b; Wehner et al. 1997; Radcliffe et al. 2000; Ponder et al. 2007a; Brigman et al. 2009; Wilson et al. 2011; Sokoloff et al. 2011). Due to limited recombination, these techniques are only able to identify large genomic regions and are therefore sub-optimal for identifying the genes that underlie QTLs (Peters et al. 2007; Flint 2011; Parker and Palmer 2011). We have recently begun to address this limitation by using populations with greater numbers of accumulated recombinations such as advanced intercross lines (AILs; Cheng et al. 2010; Samocha et al. 2010; Lionikas et al. 2010; Parker et al. 2011a, b; Parker and Palmer 2011). AILs are created by successive generations of pseudo-random mating after the F2 generation. Each additional generation leads to the accumulation of new recombinations, which allows for more precise mapping due to a breakdown in linkage disequilibrium. Because AILs are derived from two inbred founders, they maintain the simplicity of more traditional crosses, possess no rare alleles, and every marker perfectly discriminates between the two founder strains.
In the present experiment, we created an F2 intercross and an F8 AIL derived from C57BL/6J (B6) × DBA/2J (D2) mice. We chose B6 and D2 inbred mice as our progenitor strains in order to take advantage of the vast amount of bioinformatics resources associated with these strains and because these strains display robust differences in fear conditioning (Owen et al. 1997b; Ponder et al. 2007a). By combining GWAS with complementary bioinformatics resources available for B6 and D2 mice, we utilized sequence data to identify coding single nucleotide polymorphisms (SNPs), and gene expression data to identify putatively causal expression polymorphisms.
Materials and methods
Animals and housing
All procedures were approved by the University of Chicago Institutional Animal Care and Use Committee (IACUC) in accordance with NIH guidelines for the care and use of laboratory animals. Inbred female C57BL/6J (B6) and male DBA/2J (D2) mice were obtained from Jackson Labs (Bar Harbor, ME). These mice were used to produce the B6 × D2 F1 mice, which were then bred to create the subsequent F2 (308 male, 317 female) and F8 (284 male, 283 female) generations. We have previously reported a genetic analysis of anxiety-like behavior and FC in the F2 but not the F8 mice (Sokoloff et al. 2011). Colony rooms were maintained on a 12:12 h light–dark cycle (lights on at 0630) in same-sex groups of two to five mice with standard lab chow and water available ad libitum. Mice were approximately 2–3 months of age at the start of testing (F2 mean age = 86.6 days, SD = 7.8, range = 70–102; F8 mean age = 74.0 days, SD = 6.4, range = 60–85).
Fear conditioning (FC)
FC procedures are identical to those described in Ponder et al. (2007a). Briefly, FC chambers were obtained from Med Associates (St. Albans, VT, USA). Chambers had inside dimensions of 29 cm × 19 cm × 25 cm with metal walls on each side, clear plastic front and back walls, clear plastic ceilings and stainless steel bars on the floor. A fluorescent light provided dim illumination (~3 lux) and a fan provided a low level of masking background noise. Chambers were cleaned with 10% isopropanol between animals. Behavior was recorded with digital video and analyzed with FreezeFrame software from Actimetrics (Evanston, IL, USA).
Testing for FC consisted of a 5 min test that occurred three times over consecutive days during the light phase, between 0800 and 1,700 h (Fig. 1). Mice were transported from the adjacent vivarium and allowed to habituate to the procedure room for 30 min in their home cages. Mice were then transferred to the FC chambers in individual holding cages with clean bedding. On test day 1, mice were placed into the chamber. 180 s later, mice were exposed twice to the conditioned stimulus (CS), which consisted of an 85 dB, 3 kHz tone that persisted for 30 s and co-terminated with the unconditioned stimulus (US), which was a 2 s, 0.5 mA foot shock delivered through the stainless steel floor. After each CS-US pairing, there was a 30 s inter-trial interval (ITI). On day 1, two measures were calculated for QTL mapping: (1) baseline freezing, defined as average percent time freezing beginning 30 s after the mice were placed into the test chambers, and ending 150 s later (30–180 s; pre-training freezing), and (2) time spent freezing to each CS presentation, calculated by averaging the percent time spent freezing to the tone presentations (180–210 s, 240–270 s; freezing to tone day 1).
Fig. 1.
Fear conditioning in F2 and F8 mice. A three-day procedure was used to phenotype each subject. Each test lasted 5 min. On day 1, pre-training freezing was measured in F2 (a) and F8 (b) mice from 30 to 180 s, after which mice were exposed to two 30-s tones (indicated by hatched bars, labeled T1 and T2) that co-terminated with a 2-s, 0.5-mA foot shock (indicated by arrows, labeled S1 and S2). On day 2, freezing to context was measured in F2 (c) and F8 (d) mice from 30 to 180 s. On day 3, freezing to the altered context was measured in F2 (e) and F8 (f) mice from 30 to 180 s after which freezing to cue was measured (180–210 + 240–270 s); the time spent freezing to each tone was averaged to obtain the freezing to tone/cue variable. Each data point represents the average % freezing calculated across the 30 s time bin. Error bars represent the standard error of the mean
Test day 2 began exactly 24 h after the start of test day 1. On test day 2, the testing environment was identical to day 1; however, neither tones (CS) nor shocks (US) were presented. On day 2, freezing to context was used for QTL mapping; this was defined as average percentage of time freezing in response to the test chamber during the same period of time as pre-training freezing (30–180 s; freezing to context). We chose this time period for QTL mapping to allow for direct comparisons to the freezing scores to tone day 1 and altered context day 3, and to avoid measuring freezing behavior during the latter part of the trial in which the mice might have predicted shocks based on the previous days test.
Test day 3 began exactly 24 h after the start of test day 2. On test day 3, the context was altered in several ways: (1) a different experimenter conducted the testing and wore a different style of gloves, (2) the transfer cages had no bedding, (3) the metal shock grid, chamber door and one wall were covered with hard white plastic, (4) yellow film was placed over the chamber lights, (5) chambers and plastic surfaces were cleaned with 0.1% acetic acid solution, and (6) the vent fan was partially obstructed to alter the background noise. On day 3, the tone (CS) was presented at the same times as on day one, but no foot shocks (US) were paired with it. On day 3, freezing to cue was defined as the average percent time spent freezing during the two 30 s CS presentations (180–210 s, 240–270 s; freezing to cue) and used for QTL mapping.
Data analysis
Initial analyses began with an independent samples t test to assess percent freezing differences between the F2 and F8 AIL populations. All analyses were conducted in PASW Statistics 18 (SPSS Inc., Chicago, IL, USA).
Genotyping
DNA from the F2 generation was extracted and genotyped by KBiosciences (Hoddesdon, Hertfordshire, UK) using KASPar, a fluorescence-based PCR assay. Markers consisted of 164 polymorphic SNPs selected from Petkov et al. (2004). The average distance between informative markers inthis panel was 15.78 Mb, range = 2.68–44.34 Mb. DNA from the F8 AIL was extracted using a salting out protocol and was genotyped using the Illumina Mouse Medium Density Linkage Panel (Illumina, San Diego, CA, USA) at the Genomics Core Facility at Northwestern University (http://web.cgm.northwestern.edu/cgm/Core-Facilities/Genomics-Core). The SNP panel consists of 1,449 SNPs, 1,060 of which were polymorphic between B6 and D2 mice. In our population, the average distance between markers was 2.77 Mb, range = 0.13 Mb to 23.27 Mb.
QTL mapping
Freezing data was converted to z-scores prior to genome-wide analysis. Genome-wide association analysis was performed in the combined population of the F2 and F8 AIL using the R package QTLRel (http://cran.r-project.org/web/packages/QTLRel/index.html). This software accounted for the complex relationships (e.g. sibling, half-sibling, cousins) among the F8 mice by using a mixed model as described previously (Cheng et al. 2010, 2011). For each analysis, P < 0.05 genome-wide significance thresholds were estimated using 1,000 permutations. Sex was included as an interactive covariate.
Bioinformatic analyses
The GeneNetwork mapping module (www.genenetwork.org; Wang et al. 2003; Chesler et al. 2004) was used to identify expression QTLs (eQTLs) in whole brain, amygdala, and hippocampal mRNA from B6 × D2 F2 and B6 × D2 RI mice that co-mapped to our behavioral QTLs (whole brain accessed on April 27, 2011, database: OHSU/VA B6D2F2 Brain mRNA Affymetrix M430 (Aug 05) RMA; Hitzemann et al. 2004, Hofstetter et al. 2008; amygdala accessed on June 28, 2011, database: INIA Amygdala Cohort Affymetrix Mouse Gene 1.0ST (Mar11) RMA; Mozhui and Williams unpublished data, hippocampus accessed on June 28, 2011, database: Hipp Consortium Affymetrix M430 (June 06) PDNN; Overall et al. 2009). Next, in order to narrow the list of candidate genes within the QTL intervals, we used high density sequence data provided by the Welcome Trust Sanger Institute (accessed on October 25th, 2011; http://www.sanger.ac.uk/cgi-bin/modelorgs/mousegenomes/snps.pl; Keane et al. 2011; Yalcin et al. 2011) to compare genomic regions between B6 and D2 mice. These strains were sequenced to an average of 25× coverage on the Illumina GAII platform (Illumina, San Diego, CA, USA) with a mixture of 54, 76, and 108 bp paired reads. We used this data to search for genes within the QTL intervals that possessed “consequential” polymorphisms between B6 and D2 mice (such as nonsynonymous coding SNPs, stop-gain SNPs, stop-loss SNPs, SNPs resulting in frameshifts and SNPs located in essential splice sites).
Results
Phenotypic analysis
The F2 and F8 AIL differed from each other for pre-training freezing (Fig. 1a, F2 mean = 2.6%, SD = 2.6%; Fig. 1b, F8 mean = 6.4%, SD = 7.2%; F1, 1,172 = 146.22; P < 0.0001). The two populations did not differ significantly in their freezing to context (Fig. 1c, F2 mean = 29.88%, SD = 18.6%; Fig. 1d, F8 mean = 23.68%, SD = 19.39%) or in freezing to cue (Fig. 1e, F2 mean = 52.16%, SD = 20.84%; Fig. 1f, F8 mean = 52.53%, SD = 21.36%). The slight disparity in pre-training freezing between populations may be due to the segregation of alleles associated with freezing behavior during the creation of the F8 AIL mice, or the result of handling effects of different testers across the ~2 year period between the F2 and F8 generations. As a result of these differences, both the F2 and the F8 AIL data were converted to z scores prior to genome-wide analysis in order to control for pre-training freezing differences that might otherwise be interpreted as differences in FC. Figure 1 displays the percent freezing across all 3 days in both the F2 and F8 AIL mice.
QTL mapping
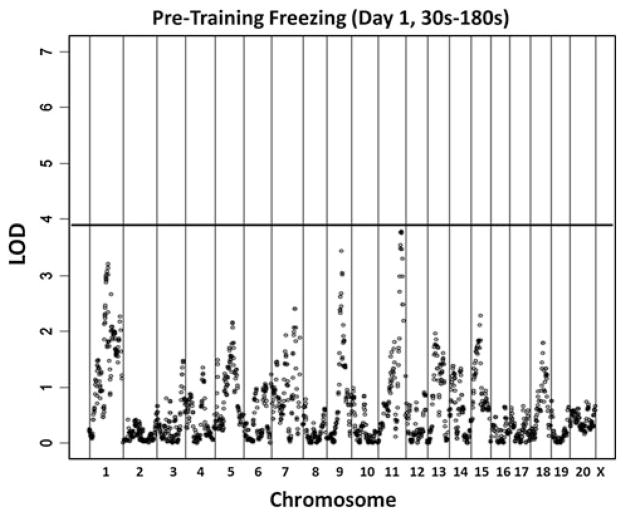
We performed genome-wide analysis on the integrated B6 × D2 F2 and B6 × D2 F8 populations for pre-training freezing, freezing to tone/shock day 1, freezing to context, and freezing to cue. Using 1,000 permutations, significance thresholds for these traits were determined to range from 3.92 to 4.03 LOD. No QTLs reached genome-wide significance for pre-training freezing. We identified two QTLs associated with freezing to tone day 1 (on chromosomes 1 and 13), five QTLs associated with freezing to context (on chromosomes 1, 2, 5, 10, and 13) and four QTLs associated with freezing to cue (on chromosomes 1, 2, 5 and 13). Figures 2, 3, 4 and 5 display the results of the integrated analysis for pre-training freezing, freezing to tone day 1, freezing to context and freezing to cue, respectively. The 1.5-LOD support intervals for these QTL ranged from 6.2 to 39.4 Mb, with an average 1.5-LOD support interval of 26.3 Mb (Table 1).
Fig. 2.
Integrated genome-wide results for percent freezing during pre-training (Day 1, 30–180 s; P < 0.05 significance threshold LOD = 3.92)
Fig. 3.
Integrated genome-wide results for percent freezing to tone day 1 (Day 1, 180–210 s, 240–270 s; P < 0.05 significance threshold LOD = 4.01)
Fig. 4.
Integrated genome-wide results for percent freezing to context (Day 2, 30–180 s; P < 0.05 significance threshold LOD = 4.01)
Fig. 5.
Integrated genome-wide results for percent freezing to cue (Day 3, 180–210 s + 240–270 s; P < 0.05 significance threshold LOD = 4.03)
Table 1.
QTLs for freezing to tone day 1, freezing to context and freezing to cue
| Behavior | Chr. | LOD | Peak LOD location (Mb) | 1.5-LOD start (Mb) | 1.5-LOD end (Mb) | Width (Mb) | SNP genotype with increased freezing |
|---|---|---|---|---|---|---|---|
| Freezing to tone day 1 | 1 | 14.0 | 109.515 | 104.264 | 127.206 | 22.94 | D2 |
| 13 | 6.4 | 55.240 | 46.459 | 73.083 | 26.62 | D2 | |
| Freezing to context | 1 | 6.9 | 132.415 | 129.953 | 136.107 | 6.15 | D2 |
| 2 | 4.6 | 91.483 | 73.315 | 107.506 | 34.19 | B6 | |
| 5 | 5.3 | 76.761 | 73.058 | 112.437 | 39.38 | D2 | |
| 10 | 4.8 | 12.931 | 3.470 | 27.855 | 24.39 | B6 | |
| 13 | 5.2 | 67.311 | 40.134 | 73.083 | 32.95 | D2 | |
| Freezing to cue | 1 | 5.5 | 111.832 | 93.792 | 129.953 | 36.16 | D2 |
| 2 | 5.4 | 84.709 | 80.456 | 108.521 | 28.07 | B6 | |
| 5 | 4.12 | 76.761 | 64.285 | 91.490 | 27.21 | D2 | |
| 13 | 6.9 | 55.240 | 47.380 | 58.826 | 11.45 | D2 |
Table includes chromosome, peak LOD score, peak SNP, Mb location and width of the QTLs as well as the SNP genotype with increased freezing
Bioinformatic analyses
Numerous genes were identified whose mRNA expression co-mapped to the behavioral QTLs for freezing to tone day 1 (Supplementary Table 1), freezing to context (Supplementary Table 2) or freezing to cue (Supplementary Table 3). Many of these differentially expressed genes have been previously reported in mice selectively bred for differences in contextual fear conditioning (Ponder et al. 2007a, b, 2008) and are listed in italics in the supplementary tables. Others have been implicated in fear learning in other animal models and/or PTSD in human subjects: B-cell leukemia/lymphoma2, Bcl2, (Ding et al. 2010; Li et al. 2010; Liu et al. 2011); B-cell translocation gene 2, anti-proliferative, Btg2, (Farioli-Vecchioli et al. 2009; Kurumaji et al. 2008); diazepam bindinding inhibitor, Dbi, (Katsura et al. 2002; Sherrin et al. 2009); gamma-aminobutyric acid (GABA) A receptor, subunit α-2, Gabra2, (Nelson et al. 2009); Ly6/Plaur domain containing 1, Lypd1, (Tekinay et al. 2009); μ-opioid receptor 1, Oprm1, (Pitman et al. 1990; Glover 1993; Good and Westbrook 1995; Liberzon et al. 2007); peptidylglycine alpha-amidating monooxygenase, Pam, (Gaier et al. 2010); neurotrophic tyrosine kinase receptor type 2, Trkb, (Takei et al. 2011). We then examined our 1.5-LOD support interval for the presence of “consequential” SNPs that had the potential to directly alter proteins (i.e. nonsynonymous coding, stop-gain, stop-lost, frameshift, splice sites). Numerous SNPs were identified in genes with known relevance to fear learning and/or PTSD: Cdh7, (Ponder et al. 2007a); Trkb, (Givalois et al. 2001; Kozlovsky et al. 2007); gamma-aminobutyric acid (GABA) A receptor, subunit β-1, Gabrb1, (Ciocchi et al. 2010); regulator of G-protein signaling 14, Rgs14, (Lee et al. 2010). Supplemental Tables 4, 5, and 6 list the location, gene names, gene symbols, and number of coding SNPs per gene for freezing to tone day 1 (Supplemental Table 4), freezing to context (Supplemental Table 5) and freezing to cue (Supplemental Table 6).
Discussion
We performed genome-wide mapping of QTL underlying fear conditioning in an F2 and F8 AIL mouse population. We identified two QTLs associated with freezing to tone on day 1 (on chromosomes 1 and 13), five QTLs associated with freezing to context (on chromosomes 1, 2, 5, 10, and 13) and four QTLs associated with freezing to cue (on chromosomes 1, 2, 5, and 13). No QTLs reached significance for baseline freezing measures. Some of the QTLs we identified were concordant with QTLs implicated in previous studies, although the direction of the allelic effect was not always replicated. For example, both we and Wehner et al. (1997) identified a QTL on chromosome 10 for freezing to context, and the B6 allele was consistently associated with increased freezing. This effect was not observed in the chromosome 10 QTL reported by Ponder et al. (2007a) in which the A/J, not the B6 strain, was responsible for increased freezing. The discrepancies may be due to extremely high freezing observed in A/Js (such that A/J > B6 > D2 freezing) or there maybe be two different alleles in this region of chromosome 10, such that B6 ≠ D2 at locus 1 and B6 ≠ AJ at locus 2. Two other QTLs we observed for freezing to context (on chromosomes 2 and 13) were also described in STSL mice derived from B6 × A/J (Ponder et al. 2008), but the direction of the effect was not reported. Importantly, the AIL provided significantly greater resolution and narrower support intervals as compared to the F2, CSS, and STSL mice.
Both contextual and cued fear mapped to similar chromosomal regions, despite known differences in their neuroanatomical substrates (Fanselow and LeDoux 1999; Jovanovic and Ressler 2010). This may indicate the presence of alleles that influence both traits; alternatively it could be due to different alleles that are located closely to each other in the genome. In support of the former explanation, we recently performed QTL mapping and factor analysis on numerous anxiety and FC traits in the same F2 B6 × D2 mice used in this study (Sokoloff et al. 2011). While factor analysis suggested that contextual and cued fear learning loaded onto separate factors, QTL mapping tended to identify the same QTLs. Talbot et al. (2003) reported that contextual and cued fear were highly correlated (r = 0.63) in HS mice, and others (Radcliffe et al. 2000; Ponder et al. 2007b) have observed that selection for freezing to context caused coincident changes in freezing to cue. Thus, it is likely that contextual and cued fear are modulated by some of the same alleles, but only gene identification can definitively show this to be the case. The chromosome 1 QTL for freezing to the tone/shock on day 1 also overlapped with the cued fear QTLs and the chromosome 13 QTL for freezing to tone/shock on day 1 overlapped with both contextual and cued QTLs. These QTLs may reflect an acute fear response that is distinct from the learned, conditioned response. In order to more closely examine this, we compared QTL mapping results from freezing to the first tone/shock pairing versus freezing to the second tone/shock pairing on day 1. The chromosome 1 QTL for freezing behavior to the tone/shock pairing on day 1 was observed in both the first and second tone/shock pairing (Tone 1 LOD score = 6.09, Tone 2 LOD score = 11.36), but the chromosome 13 QTL for freezing behavior was only seen for the second tone/shock pairing (Tone 1 LOD score = 2.65, Tone 2 LOD score = 4.98). In addition, we observed a QTL on chromosome 11 that was specific to freezing to Tone 1 and not Tone 2. Because freezing to tone 1 occurs before any shocks have been presented, the QTLs on chromosome 1 and 11 represent a response to the tone itself, whereas the chromosome 13 QTL detected during tone 2 reflects a conditioned response. Earlier work in our lab (Sokoloff et al. 2011) has shown a dissociation of the day 1 freezing response from the day 2 and day 3 freezing responses through factor analysis, but not through QTL mapping in the B6 × D2 F2 mice. Bush et al. (2007) examined the dissociation of fear reactivity (initial fear response) from fear recovery (conditioned fear response) in outbred rats. They reported that rats displaying the highest levels of freezing behavior during fear conditioning continued to have significantly increased levels of CS-elicited fear in subsequent tests, but that it did not predict fear recovery as measured by freezing during extinction. Studies of fear conditioning in humans have also begun to investigate the genetics of individual differences in the acquisition of fear memories (Hettema et al. 2003).
To further narrow our list of candidate genes we used a series of bioinformatic approaches. First, we identified eQTLs that co-mapped with our QTLs. eQTLs are genomic loci that regulate gene transcription and expression on a genome-wide scale, and are believed to underlie many QTLs for complex traits (Nicolae et al. 2010; Li and Deng 2010). While co-mapping of a QTL and an eQTL does not constitute proof that the latter causes the former, it does suggests a clear and testable hypothesis—the candidate gene can be directly manipulated using a variety of molecular or pharmacological approaches. We used an existing database (http://www.GeneNetwork.org) of mRNA expression from amygdala, hippocampus and whole brain of untreated B6 × D2 F2 or RI mice. This identified a number of genes whose expression co-mapped within the 1.5-LOD intervals of our QTLs (Supplemental Tables 1, 2, and 3). Many of these genes (C1ql2, Cd59a, Cdh7, Fryl, Hdac4, Kit, Lypd1, Mcm6, Rab3gap1, Stk25, Slc35f5, Ubxd, Zfp71rs1) have previously been shown to have differential expression in the hippocampus and/or the amygdala of B6 × D2 or B6 × A/J derived mice selectively bred for differences in contextual freezing (Ponder et al. 2007a, b, 2008).
We also identified differential expression of numerous genes that have been implicated in fear learning in other rodent models (Table 2). For example, expression of Trkb mapped to the QTL on chromosome 13 for freezing to context. Takei et al. 2011 recently demonstrated that Trkb signaling in the hippocampus is enhanced in response to fear conditioning and Musumeci et al. (2009) report that Trkb receptors modulate specific phases of fear learning and amygdalar synaptic plasticity. Another gene we identified in our eQTL region was Bcl2. Expression of Bcl2 mapped to the QTL on chromosome 1 for freezing to cue, and hippocampal Bcl2 expression is significantly upregulated in a rat model of PTSD (Li et al. 2010). We also found expression differences in Btg2 within the QTL on chromosome 1 for freezing to cue. Farioli-Vecchioli et al. (2009) have reported impaired contextual fear conditioning in a Btg2 null mouse model. Additionally, expression of Dbi mapped to the QTL on chromosome 1 for freezing to cue. Dbi is an anxiogenic neuropeptide whose expression increases following conditioned psychological stress and is functionally involved in hippocampal-dependent enhancement of contextual fear (Sherrin et al. 2009; Katsura et al. 2002). We also observed that expression of Lypd1 mapped to the QTL on chromosome 1 for freezing to cue, and Lypd1 knockout mice display increased cued fear conditioning (Tekinay et al. 2009). Lastly, expression of Pam mapped to the QTL on chromosome 1 for freezing to cue and mice heterozygous for the Pam gene are deficient in short- and long-term contextual and cued fear conditioning (Gaier et al. 2010).
Table 2.
Candidate genes
| Gene | eQTL | Coding SNP | eQTL in selected lines | KO with abnormal FC phenotype | Implicated in rodent FC | Implicated in human PTSD |
|---|---|---|---|---|---|---|
| Bcl2 | X | Li et al. (2010) | ||||
| Btg2 | X | Farioli-Vecchioli et al. (2009) | ||||
| Cdh7 | X | Ponder et al. (2007b) | Irvine et al. (2005) | |||
| Dbi | X |
Katsura et al. (2002) Sherrin et al. (2009) |
||||
| Gabra2 | X | Nelson et al. (2009) | ||||
| Gabrb1 | X |
Stork et al. (2002) Chhatwal et al. (2005) Ciocchi et al. (2010) |
||||
| Lypd1 | X | Ponder et al. (2007b) | Tekinay et al. (2009) | |||
| Oprm1 | X | Good and Westbrook (1995) |
Pitman et al. (1990) Glover (1993) Liberzon et al. (2007) |
|||
| Pam | X | Gaier et al. (2010) | ||||
| Rgs14 | X | Lee et al. (2010) | ||||
| Trkb | X | X |
Musumeci et al. (2009) Takei et al. (2011) |
Frielingsdorf et al. (2010) Soliman et al. (2010) |
The gene symbol is listed, along with an “X” to indicate if it was an eQTL or a non-synonymous coding SNP. Citations are provided for genes that have been previously reported in selectively bred mice, if a knockout mouse exists with an abnormal FC phenotype, or if additional evidence from rodent or human studies provides support
Next, we detected expression differences in genes that have known relevance to PTSD in humans (Table 2). Expression of Gabra2 co-mapped to the QTL for freezing to cue on chromosome 5. Nelson et al. (2009) reported three SNPs in Gabra2 interacted with childhood trauma exposure to predict PTSD in adulthood. The expression of Oprm1 co-mapped to the QTL on chromosome 10 for freezing to context. The opioid system has been implicated in the modulation of fear (Good and Westbrook 1995), and endogenous opioid abnormalities have been reported in patients with PTSD (Glover 1993; Pitman et al. 1990). Furthermore, treatment with opioid antagonists reduces PTSD symptoms and central μ-opioid receptor binding is altered after psychological trauma (Liberzon et al. 2007). Lastly, Trkb (known to be important for fear learning in rodents) may also be functionally relevant to PTSD in humans. Induction of brain derived neurotrophic factor (BDNF) and activation of its intracellular receptor Trkb increases neural survival, synaptic transmission, long term potentiation and long term depression (Lipsky and Marini 2007). This has significant implications for memory formation in individuals with PTSD yet few studies have been conducted examining BDNF/Trkb in human populations. Dell’osso et al. (2009) reported significantly lower levels of plasma BDNF in patients with PTSD as compared with healthy individuals and human carriers of the BDNFMet allele displayed slower suppression of a learned fear response (Frielingsdorf et al. 2010), impaired fear extinction (Soliman et al. 2010), and abnormal fronto-amygdala activity (Soliman et al. 2010). However, Zhang et al. (2006) reported no association between three BDNF gene variants and PTSD in a sample of 96 cases and 250 control subjects.
Finally, we identified coding SNPs within each of our 1.5-LOD QTL intervals (Supplemental Tables 4, 5, and 6). Some of the genes within these regions had coding SNPs known to be involved in fear learning and/or PTSD (Table 2). Cdh7 has two non-synonymous coding SNPs within the freezing to cue QTL on chromosome 1, and is thought to play a role in acquisition of fear memories, fear learning and regulation of the Pavlovian fear neural network through calcium mediated cell adhesion (Irvine et al. 2005; Ponder et al. 2007a, b; Johnson et al. 2011). Trkb has one non-synonymous coding SNP within the freezing to context QTL on chromosome 13. Trkb antagonists have anxiolytic properties in mice (Cazorla et al. 2011a, b), and Trkb has been implicated in the neuro-biological mechanisms underlying behaviorally induced stress associated with PTSD (Givalois et al. 2001; Kozlovsky et al. 2007). The gamma-aminobutyric acid (GABA) A receptor, subunit β-1 (Gabrb1) within the chromosome 5 QTL for freezing to cue contains a non-synonymous coding SNP. GABAergic neurotransmission in the amygdala have been shown vital in encoding of conditioned fear (Ciocchi et al. 2010) and alterations in GABA receptor levels are evident following fear conditioning (Chhatwal et al. 2005; Stork et al. 2002). We also identified non-synonymous coding SNPs in the regulator of G-protein signaling 14 (Rgs14) for both freezing to cue and freezing to context QTLs on chromosome 13. Rgs14 is a natural suppressor of synaptic plasticity in hippocampal neurons as well as a suppressor of hippocampal-based learning and memory (Lee et al. 2010).
Our study has several important limitations. First of all, because we have used a cross between two inbred strains, we are studying the alleles that segregate between them and not the total number of alleles that segregate among all laboratory strains or wild mice. However, we did observe significant overlap in the QTLs we identified in our population with QTLs identified in other populations of mice, which is consistent with the idea that laboratory mice are segregating a relatively limited number of alleles (Yang et al. 2007, 2011). Additionally, our gene expression data was from brains of untreated mice. Thus, examining gene expression differences in mice that underwent the FC paradigm may provide useful information regarding gene expression levels following exposure to traumatic events. However, we have not taken that approach, in part because it is difficult to determine at what time(s) after treatment gene expression should be considered. Furthermore, we did not control for the effect of SNPs located within probes. Many groups have noted the highly significant overrepresentation of transcripts for which the additive allele effect is greater for the B6 than for the D2 strain (Chesler et al. 2005; Ciobanu et al. 2010; Radcliffe et al. 2006). This is most likely due to the fact that both the Affymetrix M430 and 1.0 ST probes were designed using sequence from B6 mice, and some of the cis-QTLs are artifacts of D2 polymorphisms occurring in probe-complimentary sequence (Radcliffe et al. 2006). Thus, a systematic imbalance exists in which some fraction of our cis-QTLs is likely caused by SNPs that overlap probes rather than by genuine quantitative differences in mRNA levels. This is a particular concern for eQTLs in which the B6 allele shows higher expression. Unfortunately, even if one removes these probes from analysis, the bias in favor of B6 alleles remains; this may be due to the presence of unknown SNPs, isoform variation, and differences in alternative splicing, initiation, and termination of transcription (Ciobanu et al. 2010). Bias associated with array-based measures of expression may ultimately be resolved by a transition to next-generation sequencing of RNA samples, which allows for a more direct way to measure mRNA abundance (Ciobanu et al. 2010). In addition, a key component of PTSD is the failure to extinguish fearful associations (Amstadter et al. 2009; Johnson et al. 2011). Our FC paradigm focused on acquisition of fearful associations rather than extinction. Furthermore, the observed FC phenotype is the combined result of many other genetically influenced factors including anxiety, sensory modalities (nociception, auditory, visual, olfaction, tactile), learning and memory. Any number of these elements may be driving the association we have reported between the identified QTLs and their phenotypes. Finally, while the use of an AIL produced smaller QTL intervals, we did not obtain single gene resolution, which would clearly provide the most specific and actionable information.
In conclusion, we have mapped numerous QTLs associated with fear conditioning in an AIL. Some of the QTLs we identified correspond to QTLs identified by other researchers, and in the majority of cases we have narrowed the confidence intervals quite significantly as compared to those previous studies. The combination of high resolution mapping with sequence and expression data offers a powerful approach and permits identification of several candidate genes that may underlie differences in these phenotypes. This has allowed us to integrate multiple lines of complementary evidence from both the mouse and human literature to provide further support for particular candidates. In summary, by using an AIL we performed a GWAS in a situation where all alleles were common, and where uniform environmental conditions were maintained, which limited the interactions between genes and environment. These advantages allowed us to map QTL with a modest sample size and identify small regions that warrant further molecular evaluation.
Acknowledgments
The authors would like to thank Jackie Lim, Ryan Walters, and Garrett Birkhoff for their assistance in behavioral testing. Access to amygdala microarray data for the BXD strains was provided by Drs. Khyobeni Mozhui and Rob Williams. The authors would like to acknowledge GeneNetwork (http://www.genenetwork.org) for providing bioinformatic tools and public data that have contributed to this manuscript. This work was supported by NIH grants R01MH079103, R21DA024845, R01DA021336 (AAP), and T32DA07255 (CCP).
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s10519-011-9524-8) contains supplementary material, which is available to authorized users.
Contributor Information
Clarissa C. Parker, Department of Human Genetics, University of Chicago, 920 E 58th St., CLSC-507D, Chicago, IL 60637, USA
Greta Sokoloff, Department of Human Genetics, University of Chicago, 920 E 58th St., CLSC-507D, Chicago, IL 60637, USA.
Riyan Cheng, Department of Human Genetics, University of Chicago, 920 E 58th St., CLSC-507D, Chicago, IL 60637, USA.
Abraham A. Palmer, Email: aap@uchicago.edu, Department of Human Genetics, University of Chicago, 920 E 58th St., CLSC-507D, Chicago, IL 60637, USA. Department of Psychiatry and Behavioral Neuroscience, The University of Chicago, Chicago, IL 60637, USA
References
- Amstadter AB, Nugent NR, Koenen KC. Genetics of PTSD: fear conditioning as a model for future research. Psychiatr Am. 2009;39:358–367. doi: 10.3928/00485713-20090526-01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blechert J, Michael T, Vriends N, Margraf J, Wilhelm FH. Fear conditioning in posttraumatic stress disorder: evidence for delayed extinction of autonomic, experiential, and behavioural responses. Psychol Med. 2005;35:791–806. doi: 10.1016/j.brat.2007.02.012. [DOI] [PubMed] [Google Scholar]
- Brigman JL, Mathur P, Lu L, Williams RW, Holmes A. Genetic relationship between anxiety-related and fear-related behaviors in BXD recombinant inbred mice. Behav Pharmacol. 2009;20:204–209. doi: 10.1097/FBP.0b013e32830c368c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush DEA, Sotres-Bayon F, LeDoux JE. Individual differences in fear: isolating fear reactivity and fear recovery phenotypes. J Trauma Stress. 2007;20:413–422. doi: 10.1002/jts.20261. [DOI] [PubMed] [Google Scholar]
- Caldarone B, Saavedra C, Tartaglia K, Wehner JM, Dudek BC, Flaherty L. Quantitative trait loci analysis affecting contextual conditioning in mice. Nat Genet. 1997;17:335–337. doi: 10.1038/ng1197-335. [DOI] [PubMed] [Google Scholar]
- Carey G. Genes, fears, phobias and phobic disorders. J Couns Dev. 1990;68:628–632. [Google Scholar]
- Cazorla M, Premont J, Mann A, Girard N, Kellendonk C, Rognan D. Identification of a low-molecular weight TrkB antagonist with anxiolytic and antidepressant activity in mice. J Clin Invest. 2011a;121:1846–1857. doi: 10.1172/JCI43992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazorla M, Arrang JM, Premont J. Pharmacological characterization of six trkB antibodies reveals a novel class of functional agents for the study of the BDNF receptor. Br J Pharmacol. 2011b;162:947–960. doi: 10.1111/j.1476-5381.2010.01094.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng R, Lim JE, Samocha KE, Sokoloff G, Abney M, Skol AD, Palmer AA. Genome-wide association studies and the problem of relatedness among advanced intercross lines and other highly recombinant populations. Genetics. 2010;185:1033–1044. doi: 10.1534/genetics.110.116863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng R, Abney M, Palmer AA, Skol AD. QTLRel: an R package for genome-wide association studies in which relatedness is a concern. BMC Genet. 2011;12:66. doi: 10.1186/1471-2156-12-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesler EJ, Lu L, Wan J, Williams RW, Manly KF. WebQTL: rapid exploratory analysis of gene expression and genetic networks for brain and behavior. Nat Neurosci. 2004;7:485–486. doi: 10.1038/nn0504-485. [DOI] [PubMed] [Google Scholar]
- Chesler EJ, Lu L, Shou S, Qu Y, Gu J, Wang J, Hsu HC, Mountz JD, Baldwin NE, Langston MA, Threadgill DW, Manly KF, Williams RW. Complex trait analysis of gene expression uncovers polygenic and pleiotropic networks that modulate nervous system function. Nat Genet. 2005;37:233–242. doi: 10.1038/ng1518. [DOI] [PubMed] [Google Scholar]
- Chhatwal JP, Myers KM, Ressler KJ, Davis M. Regulation of gephyrin and GABAA receptor binding within the amygdala after fear acquisition and extinction. J Neurosci. 2005;25:502–506. doi: 10.1523/JNEUROSCI.3301-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciobanu DC, Lu L, Mozhui K, Wang X, Jagalur M, Morris JA, Taylor WL, Dietz K, Simon P, Williams RW. Detection, validation, and downstream analysis of allelic variation in gene expression. Genetics. 2010;184:119–128. doi: 10.1534/genetics.109.107474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciocchi S, Herry C, Grenier F, Wolff SB, Letzkus JJ, Vlachos I, Ehrlich I, Sprengel R, Deisseroth K, Stadler MB, Muller C, Luthi A. Encoding of conditioned fear in central amygdala inhibitory circuits. Nature. 2010;468:277–282. doi: 10.1038/nature09559. [DOI] [PubMed] [Google Scholar]
- Dell’osso L, Carmassi C, Del Debbio A, Dell’osso MC, Bianchi C, da Pozzo E, Origlia N, Domenici L, Massimetti G, Marazziti D, Piccinni A. Brain-derived neurotrophic factor plasma levels in patients suffering from post-traumatic stress disorder. Prog Neuropsychopharmacol Biol Psychiatry. 2009;33:899–902. doi: 10.1016/j.pnpbp.2009.04.018. [DOI] [PubMed] [Google Scholar]
- Ding J, Han F, Shi Y. Single-prolonged stress induces apoptosis in the amygdala in a rat model of post-traumatic stress disorder. J Psychiatr Res. 2010;44:48–55. doi: 10.1016/j.jpsychires.2009.06.001. [DOI] [PubMed] [Google Scholar]
- Fanselow MS, LeDoux JE. Why we think plasticity underlying Pavlovian fear conditioning occurs in the basolateral amygdala. Neuron. 1999;23:155–184. doi: 10.1016/s0896-6273(00)80775-8. [DOI] [PubMed] [Google Scholar]
- Farioli-Vecchioli S, Araulli D, Costanzi M, Leonardi L, Cina I, Micheli L, Nutini M, Longone P, Oh SP, Cestari V, Tirone F. Impaired terminal differentiation of hippocampal granule neurons and defective contextual memory n PC3/Tis21 knockout mice. PLoS One. 2009;4:e8339. doi: 10.1371/journal.pone.0008339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint J. Mapping quantitative traits and strategies to find quantitative trait genes. Methods. 2011;53:163–174. doi: 10.1016/j.ymeth.2010.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frielingsdorf H, Bath KG, Soliman F, Difed J, Casey BJ, Lee FS. Variant brain-derived neurotrophic factor Val66Met endophenotypes: implications for posttraumatic stress disorder. Ann NY Acad Sci. 2010;1208:150–157. doi: 10.1111/j.1749-6632.2010.05722.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaier ED, Rodriquiz RM, Ma XM, Sivaramakrishnan S, Bousquet-Moore D, Wetsel WC, Eipper BA, Mains RE. Haploin-sufficiency in peptidylglycine alpha-amidating monooxygenase leads to altered synaptic transmission in the amygdala and impaired emotional responses. J Neurosci. 2010;30:13656–13669. doi: 10.1523/JNEUROSCI.2200-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Givalois L, Marmigere F, Rage F, Ixart G, Arancibia S, Tapia-Arancibia L. Immobilization stress rapidly and differentially modulates BDNF and TrkB mRNA expression in the pituitary gland of adult male rats. Neuroendocrinology. 2001;74:148–159. doi: 10.1159/000054681. [DOI] [PubMed] [Google Scholar]
- Glover H. A preliminary trial of nalmefene for the treatment of emotional numbing in combat veterans with post-traumatic stress disorder. Isr J Psychiatry Relat Sci. 1993;30:255–263. [PubMed] [Google Scholar]
- Good AJ, Westbrook RF. Effects of a microinjection of morphine into the amygdala on the acquisition and expression of conditioned fear and hypoalgesia in rats. Behav Neurosci. 1995;109:631–641. doi: 10.1037//0735-7044.109.4.631. [DOI] [PubMed] [Google Scholar]
- Grillon C, Dierker L, Merikangas KR. Fear-potentiated startle in adolescent offspring of parents with anxiety disorders. Biol Psychiatry. 1998;44:990–997. doi: 10.1016/s0006-3223(98)00188-7. [DOI] [PubMed] [Google Scholar]
- Hettema JM, Annas P, Neale MC, Kendler KD, Fredrikson M. A twin study of the genetics of fear conditioning. Arch Gen Psychiatry. 2003;60:702–708. doi: 10.1001/archpsyc.60.7.702. [DOI] [PubMed] [Google Scholar]
- Hitzemann R, Reed C, Malmanger B, Lawler M, Hitzemann B, Cunningham B, McWeeney S, Belknap J, Harrington C, Buck K, Phillips T, Crabbe J. On the integration of alcohol-related quantitative trait loci and gene expression analyses. Alcohol Clin Exp Res. 2004;28:1437–1448. doi: 10.1097/01.alc.0000139827.86749.da. [DOI] [PubMed] [Google Scholar]
- Hofstetter JR, Hitzemann RJ, Belknap JK, Walter NAR, McWeeney SK, Mayeda AR. Characterization of the quantitative trait locus for haloperidol-induced catalepsy on distal mouse chromosome 1. Genes Brain Behav. 2008;7:214–223. doi: 10.1111/j.1601-183X.2007.00340.x. [DOI] [PubMed] [Google Scholar]
- Irvine EE, Vernon J, Giese KP. AlphaCaMKII autophosphorylation contributes to rapid learning but is not necessary for memory. Nat Neurosci. 2005;8:411–412. doi: 10.1038/nn1431. [DOI] [PubMed] [Google Scholar]
- Johnson LR, LeDoux JE. The anatomy of fear: microcircuits of the lateral amygdala. In: Gorman JM, editor. Fear and anxiety: the benefits of translational research. APPA Press; Washington: 2004. pp. 227–250. [Google Scholar]
- Johnson LR, McGuire J, Lazarus R, Palmer AA. Pavlovian fear memory circuits and phenotype models of PTSD. Neuropsychopharmacology. 2011 Jul 19; doi: 10.1016/j.neuropharm.2011.07.004. [Epub ahead of Print] [DOI] [PubMed] [Google Scholar]
- Jovanovic T, Ressler KJ. How the neurocircuitry and genetics of fear inhibition may inform our understanding of PTSD. Am J Psychiatry. 2010;167:648–662. doi: 10.1176/appi.ajp.2009.09071074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsura M, Mohri Y, Shuto K, Tsuijimura A, Ukai M, Ohkma S. Psychological stress, but not physical stress, causes increase in diazepam binding inhibitor (DBI) mRNA expression in mouse brains. Brain Res Mol Brain Res. 2002;104:103–109. doi: 10.1016/s0169-328x(02)00219-x. [DOI] [PubMed] [Google Scholar]
- Keane TM, Goodstadt L, Danecek P, White MA, Wong K, Yalcin B, Heger A, Agam A, Slater G, Goodson M, Furlotte NA, Eskin E, Nellaker C, Whitley H, Cleak J, Janowitz D, Hernandez-Pliego P, Edwards A, Belgard TG, Oliver PL, McIntyre RE, Bhomra A, Nicod J, Gan X, Yuan W, van der Weyden L, Steward CA, Bala S, Stalker J, Mott R, Durbin R, Jackson IJ, Czechanski A, Guerra-Assuncao JA, Donahue LR, Reinholdt LG, Payseur BA, Ponting CP, Birney E, Flint J, Adams DJ. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature. 2011;477:289–294. doi: 10.1038/nature10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlovsky N, Matar MA, Kaplan Z, Kotler M, Zohar J, Cohen H. Long-term down-regulation of BDNF mRNA in rat hippocampal CA1 subregion correlates with PTSD-like behavioural stress response. Int J Neuropsychopharmacol. 2007;10:741–758. doi: 10.1017/S1461145707007560. [DOI] [PubMed] [Google Scholar]
- Kurumaji A, Ito T, Ishii S, Nishikawa T. Effects of FG7142 and immobilization stress on the gene expression in the neocortex of mice. Neurosci Res. 2008;62:155–159. doi: 10.1016/j.neures.2008.08.001. [DOI] [PubMed] [Google Scholar]
- LaBar KS, LeDoux JE, Spencer D, Phelps EA. Impaired fear conditioning following unilateral temporal lobectomy in humans. J Neurosci. 1995;15:5879–5891. doi: 10.1523/JNEUROSCI.15-10-06846.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layton B, Krikorian R. Memory mechanisms in posttraumatic stress disorder. J Neuropsychiatry Clin Neurosci. 2002;14:254–261. doi: 10.1176/jnp.14.3.254. [DOI] [PubMed] [Google Scholar]
- LeDoux JE. Emotion circuits in the brain. Annu Rev Neurosci. 2000;23:155–184. doi: 10.1146/annurev.neuro.23.1.155. [DOI] [PubMed] [Google Scholar]
- Lee SE, Simons SB, Heldt SA, Zhao M, Schroeder JP, Vellano CP, Cowan DP, Ramineni S, Yates CK, Feng Y, Smith Y, Sweatt JD, Weinshenker D, Ressler KJ, Dudek SM, Hepler JR. RGS14 is a natural suppressor of both synaptic plasticity in CA2 neurons and hippocampal-based learning and memory. Proc Natl Acad Sci USA. 2010;107:16994–16998. doi: 10.1073/pnas.1005362107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Deng H. Systems genetics, bioinformatics and eQTL mapping. Genetica. 2010;138:915–924. doi: 10.1007/s10709-010-9480-x. [DOI] [PubMed] [Google Scholar]
- Li X, Han F, Liu D, Shi Y. Changes of Bax, Bcl-2 and apoptosis in hippocampus in the rat model of post-traumatic stress disorder. Neurol Res. 2010;32:579–586. doi: 10.1179/016164110X12556180206194. [DOI] [PubMed] [Google Scholar]
- Liberzon I, Taylor SF, Phan KL, Britton JC, Fig LM, Bueller JA, Koeppe RA, Zubieta JK. Altered central micro-opioid receptor binding after psychological trauma. Biol Psychiatry. 2007;61:1030–1038. doi: 10.1016/j.biopsych.2006.06.021. [DOI] [PubMed] [Google Scholar]
- Lionikas A, Cheng R, Lim JE, Palmer AA, Blizard DA. Fine-mapping of muscle weight QTL in LG/J and SM/J intercrosses. Physiol Genomics. 2010;42A:33–38. doi: 10.1152/physiolgenomics.00100.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipsky RH, Marini AM. Brain-derived neurotrophic factor in neuronal survival and behavior-related plasticity. Ann NY Acad Sci. 2007;1122:130–143. doi: 10.1196/annals.1403.009. [DOI] [PubMed] [Google Scholar]
- Liu H, Wang HT, Han F, Shi YX. Activity of 5-HT1A receptor is involved in neuronal apoptosis of the amygdala in a rat model of post-traumatic stress disorder. Mol Med Report. 2011;4:291–295. doi: 10.3892/mmr.2011.415. [DOI] [PubMed] [Google Scholar]
- Musumeci G, Sciarretta C, Rodriguez-Moreno A, Al Banchabouchi M, Negrete-Diaz V, Costanzi M, Berno V, Egorov AV, von Bohlen Und Halbach O, Cestari V, Delgado-Garcia JM, Minichiello L. TrkB modulates fear learning and amygdalar synaptic plasticity by specific docking sites. J Neurosci. 2009;29:10131–10143. doi: 10.1523/JNEUROSCI.1707-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson EC, Agrawal A, Pergadia ML, Lynskey MT, Todorov AA, Wang JC, Todd RD, Martin NG, Heath AC, Goate AM, Montgomery GW, Madden PA. Association of childhood trauma exposure and GABRA2 polymorphisms with risk of posttraumatic stress disorder in adults. Mol Psychiatry. 2009;14:234–235. doi: 10.1038/mp.2008.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolae DL, Gamazon E, Zhang W, Duan S, Dolan ME, Cox NJ. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 2010;6:e1000888. doi: 10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr SP, Metzger LJ, Lasko NB, Macklin ML, Peri T, Pitman RK. De novo conditioning in trauma-exposed individuals with and without posttraumatic stress disorder. J Abnorm Psychol. 2000;109:290–298. [PubMed] [Google Scholar]
- Overall RW, Kempermann G, Peirce J, Lu L, Goldowitz D, Gage FH, Goodwin S, Smit AB, Airey DC, Rosen GD, Schalkwyk LC, Sutter TR, Nowakowski RS, Whatley S, Williams RW. Genetics of the hippocampal transcriptome in mouse: a systematic survey and online neurogenomics resource. Front Neurosci. 2009;3:55. doi: 10.3389/neuro.15.003.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen EH, Logue SF, Rasmussen DL, Wehner JM. Assessment of learning by the Morris water task and fear conditioning in inbred mouse strains and F1 hybrids: implications of genetic background for single gene mutations and quantitative trait loci analysis. Neuroscience. 1997a;80:1087–1099. doi: 10.1016/s0306-4522(97)00165-6. [DOI] [PubMed] [Google Scholar]
- Owen EH, Christensen SC, Paylor R, Wehner JM. Identification of quantitative trait loci involved in contextual and auditory-cued fear conditioning in BXD recombinant inbred strains. Behav Neurosci. 1997b;111:292–300. doi: 10.1037//0735-7044.111.2.292. [DOI] [PubMed] [Google Scholar]
- Parker CC, Palmer AA. Dark matter: are mice the solution to missing heritability? Front Gene. 2011;2:32. doi: 10.3389/fgene.2011. 00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker CC, Cheng R, Sokoloff G, Lim JE, Skol AD, Abney M, Palmer AA. Fine-mapping alleles for body weight in LG/J × SM/J F2 and F34 advanced intercross lines. Mamm Genome. 2011a;22:563–571. doi: 10.1007/s00335-011-9349-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker CC, Cheng R, Sokoloff G, Palmer AA. Genome-wide association for methamphetamine sensitivity in an advanced intercross mouse line. Genes Brain Behav. 2011b Oct 27; doi: 10.1111/j.1601-183X.2011.00747.x. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peri T, Ben-Shakhar G, Orr SP, Shalev AY. Psychophysiologic assessment of aversive conditioning in posttraumatic stress disorder. Biol Psychiatry. 2000;47:512–519. doi: 10.1016/s0006-3223(99)00144-4. [DOI] [PubMed] [Google Scholar]
- Peters LL, Robledo RF, Bult CJ, Churchill GA, Paigen BJ, Svenson KL. The mouse as a model for human biology: a resource guide for complex trait analysis. Nat Rev Genet. 2007;8:58–69. doi: 10.1038/nrg2025. [DOI] [PubMed] [Google Scholar]
- Petkov PM, Ding Y, Cassell MA, Zhang W, Wagner G, Sargent EE, Asquith S, Crew V, Johnson KA, Robinson P, Scott VE, Wiles MV. An efficient SNP system for mouse genome scanning and elucidating strain relationships. Genome Res. 2004;9:1806–1811. doi: 10.1101/gr.2825804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitman RK, van der Kolk BA, Orr SP, Greenberg MS. Naloxone-reversible analgesic response to combat-related stimuli in posttraumatic stress disorder: a pilot study. Arch Gen Psychiatry. 1990;47:541–544. doi: 10.1001/archpsyc.1990.01810180041007. [DOI] [PubMed] [Google Scholar]
- Ponder CA, Munoz M, Gilliam TC, Palmer AA. Genetic architecture of fear conditioning in chromosome substitution strains: relationship to measures of innate (unlearned) anxiety-like behavior. Mamm Genome. 2007a;18:221–228. doi: 10.1007/s00335-007-9013-9. [DOI] [PubMed] [Google Scholar]
- Ponder CA, Kliethermes CL, Drew MR, Muller J, Das K, Risbrough VB, Crabbe JC, Gilliam TC, Palmer AA. Selection for contextual fear conditioning affects anxiety-like behaviors and gene expression. Genes Brain Behav. 2007b;6:736–749. doi: 10.1111/j.1601-183X.2007.00306.x. [DOI] [PubMed] [Google Scholar]
- Ponder CA, Huded CP, Munoz MB, Gulden FO, Gilliam TC, Palmer AA. Rapid selection response for contextual fear conditioning in a cross between C57BL/6J and A/J: behavioral, QTL and gene expression analysis. Behav Genet. 2008;38:277–291. doi: 10.1007/s10519-008-9203-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radcliffe RA, Lowe MV, Wehner JM. Confirmation of contextual fear conditioning QTLs by short-term selection. Behav Genet. 2000;30:183–191. doi: 10.1023/a:1001910107167. [DOI] [PubMed] [Google Scholar]
- Radcliffe RA, Lee MJ, Williams RW. Prediction of cis-QTLs in a pair of inbred mouse strains with the use of expression and haplotype data from public databases. Mamm Genome. 2006;17:629–642. doi: 10.1007/s00335-005-0178-9. [DOI] [PubMed] [Google Scholar]
- Richardson MP, Strange BA, Dolan RJ. Encoding of emotional memories depends on amygdala and hippocampus and their interactions. Nat Neurosci. 2004;7:278–285. doi: 10.1038/nn1190. [DOI] [PubMed] [Google Scholar]
- Samocha KE, Lim JE, Cheng R, Sokoloff G, Palmer AA. Fine mapping of QTL for prepulse inhibition in LG/J and SM/J mice using F(2) and advanced intercross lines. Genes Brain Behav. 2010;9:759–767. doi: 10.1111/j.1601-183X.2010.00613.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherrin T, Blank T, Saravana R, Rayner M, Spiess J, Todorovic C. Region specific gene expression profile in mouse brain after chronic corticotropin releasing factor receptor 1 activation: the novel role for diazepam binding inhibitor in contextual fear conditioning. Neuroscience. 2009;162:14–22. doi: 10.1016/j.neuroscience.2009.04.012. [DOI] [PubMed] [Google Scholar]
- Sokoloff G, Parker CC, Lim JE, Palmer AA. Anxiety and fear in a cross of C57BL/6J and DBA/2J mice: mapping overlapping and independent QTL for related traits. Genes Brain Behav. 2011;10:604–614. doi: 10.1111/j.1601-183X.2011.00699.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soliman F, Glatt CE, Bath KG, Levita L, Jones RM, Pattwell SS, Jing D, Tottenham N, Amso D, Somerville LH, Voss HU, Glover G, Ballon DJ, Liston C, Teslovich T, Van Kempen T, Lee FS, Casey BJ. A genetic variant BDNF polymorphism alters extinction learning in both mouse and human. Science. 2010;327:863–866. doi: 10.1126/science.1181886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stork O, Ji FY, Obata K. Reduction of extracellular GABA in the mouse amygdala during and following confrontation with a conditioned fear stimulus. Neurosci Lett. 2002;327:138–142. doi: 10.1016/s0304-3940(02)00387-7. [DOI] [PubMed] [Google Scholar]
- Takei S, Morinobu S, Yamamoto S, Fuchikami M, Matsumoto T, Yamawaki S. Enhanced hippocampal BDNF/TrkB signaling in response to fear conditioning in an animal model of posttraumatic stress disorder. J Psychiatr Res. 2011;45:460–468. doi: 10.1016/j.jpsychires.2010.08.009. [DOI] [PubMed] [Google Scholar]
- Talbot CJ, Radcliffe RA, Fullerton J, Hitzemann R, Wehner JM, Flint J. Fine scale mapping of a genetic locus for conditioned fear. Mamm Genome. 2003;14:223–230. doi: 10.1007/s00335-002-3059-5. [DOI] [PubMed] [Google Scholar]
- Tekinay AB, Nong Y, Miwa JM, Lieberam I, Ibanez-Tallon I, Greengard P, Heintz N. A role for LYNX2 in anxiety-related behavior. Proc Natl Acad Sci USA. 2009;106:4477–4482. doi: 10.1073/pnas.0813109106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Williams RW, Manly KF. WebQTL: web-based complex trait analysis. Neuroinformatics. 2003;1:299–308. doi: 10.1385/NI:1:4:299. [DOI] [PubMed] [Google Scholar]
- Wehner JM, Radcliffe RA, Rosmann ST, Christensen SC, Rasmussen DL, Fulker DW, Wiles M. Quantitative trait locus analysis of contextual fear conditioning in mice. Nat Genet. 1997;17:331–334. doi: 10.1038/ng1197-331. [DOI] [PubMed] [Google Scholar]
- Wilson YM, Brodnicki TC, Lawrence AJ, Murphy M. Congenic mouse strains enable discrimination of genetic determinants contributing to fear and fear memory. Behav Genet. 2011;41:278–287. doi: 10.1007/s10519-010-9387-4. [DOI] [PubMed] [Google Scholar]
- Yalcin B, Wong K, Agam A, Goodson M, Keane TM, Gan X, Nellaker C, Goodstadt L, Nicod J, Bhomra A, Hernandez-Pliego P, Whitley H, Cleak J, Dutton R, Janowitz D, Mott R, Adams DJ, Flint J. Sequence-based characterization of structural variation in the mouse genome. Nature. 2011;477:326–329. doi: 10.1038/nature10432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Bell TA, Churchill GA, Pardo-Manuel de Villena F. On the subspecific origin of the laboratory mouse. Nat Genet. 2007;39:1100–1107. doi: 10.1038/ng2087. [DOI] [PubMed] [Google Scholar]
- Yang H, Wang JR, Didion JP, Buus RJ, Bell TA, Welsh CE, Bonhomme F, Yu AH, Nachman MW, Pialek J, Tucker P, Boursot P, McMillan L, Churchill GA, de Villena FP. Subspecific origin and haplotype diversity in the laboratory mouse. Nat Genet. 2011;43:648–655. doi: 10.1038/ng.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Ozbay F, Lappalainen J, Kranzler HR, van Dyck CH, Charney DS, Price LH, Southwick S, Yang BZ, Rasmussen A, Gelernter J. Brain derived neurotrophic factor (BDNF) gene variants and Alzheimer’s disease, affective disorders, posttraumatic stress disorder, schizophrenia, and substance dependence. Am J Med Genet B Neuropsychiatr Genet. 2006;141B:387–393. doi: 10.1002/ajmg.b.30332. [DOI] [PMC free article] [PubMed] [Google Scholar]