Abstract
Two potent inhibitors (compounds 1 and 2) of malarial aspartyl protease, plasmepsin-II, were evaluated against wild type (NL4-3) and multidrug-resistant clinical isolate 769 (MDR) variants of human immunodeficiency virus type-1 (HIV-1) aspartyl protease. Enzyme inhibition assays showed that both 1 and 2 have better potency against NL4-3 than against MDR protease. Crystal structures of MDR protease in complex with 1 and 2 were solved and analyzed. Crystallographic analysis revealed that the MDR protease exhibits a typical wide-open conformation of the flaps (Gly48 to Gly52) causing an overall expansion in the active site cavity, which, in turn caused unstable binding of the inhibitors. Due to the expansion of the active site cavity, both compounds showed loss of direct contacts with the MDR protease compared to the docking models of NL4-3. Multiple water molecules showed a rich network of hydrogen bonds contributing to the stability of the ligand binding in the distorted binding pockets of the MDR protease in both crystal structures. Docking analysis of 1 and 2 showed a decrease in the binding affinity for both compounds against MDR supporting our structure-function studies. Thus, compounds 1 and 2 show promising inhibitory activity against HIV-1 protease variants and hence are good candidates for further development to enhance their potency against NL4-3 as well as MDR HIV-1 protease variants.
1. INTRODUCTION
AIDS (acquired immunodeficiency syndrome) is one of the major causes of global human morbidity [1]. Rapid and error prone replication of HIV-1 (human immunodeficiency virus type-1) leads to mutations that are incorporated into viral proteins resulting in drug resistance [2]. HIV-1 protease [3, 4] is a homo-dimeric aspartyl protease with two catalytic aspartic acid residues (Asp25 and Asp125) and has been validated as an important drug target [5] due to its critical role in the viral maturation and virulence [3, 4]. In spite of the availability of numerous potent HIV-1 protease inhibitors (PIs), multidrug-resistant (MDR) clinical isolates such as MDR 769 [6] show resistance against most of the PIs [7, 8]. Accumulation of mutations can cause changes in the three-dimensional structure of the viral proteins including the protease [9, 10]. Such conformational changes were later confirmed by other groups [11]. Thus, new therapeutic agents are needed to gain an efficient control over the inhibition of such MDR strains.
Previously, HIV-1 protease inhibitors have been shown to exhibit antimalarial activity [12]. In the current study, two highly potent inhibitors, compounds 1 and 2 (Figure 1), of malarial aspartyl protease, plasmepsin-II (PLM-II) [13, 14] were evaluated against wild type (NL4-3) and MDR 769 HIV-1 protease variants. 1 and 2 were the first examples of fully characterized mechanism-based inhibitors of aspartyl protease, PLM-II [15]. Both compounds were previously shown to have good therapeutic index with low nanomolar potency against PLM-II (Table 1) making them potential new treatments for malaria. Based on the similarities between the catalytic mechanism [16] and the inhibitor design [17] for PLM-II and HIV-1 protease, we hypothesized that 1 and 2 might inhibit the NL4-3 and MDR 769 HIV-1 protease variants. This is the first report of evaluating anti-malarial compounds against the MDR 769 HIV-1 protease.
Figure 1.
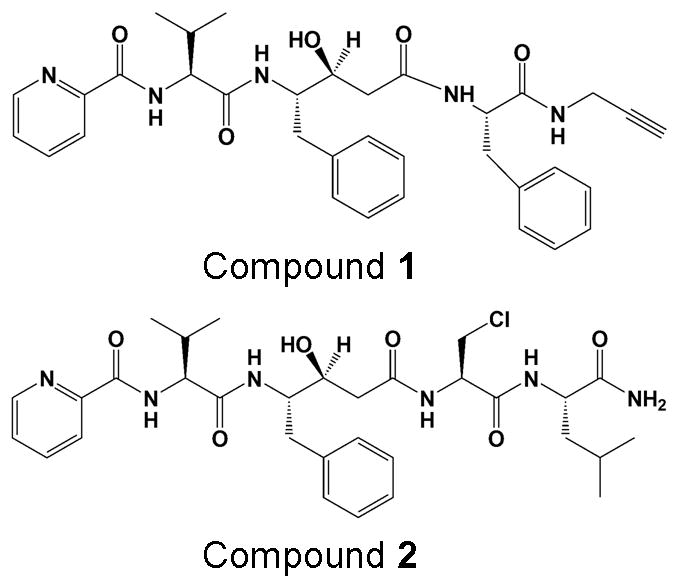
Structures of compounds 1 and 2.
Table 1. IC50 (μM) values of compounds 1 and 2.
The IC50 values of compounds 1 and 2 against the wild type (NL4-3) and MDR769 HIV-1 protease variants were obtained in the current study and the IC50 values against PLM-II were taken from Gupta et. al. [15]. IC50 value for the control, acetyl pepstatin against PLM-II was taken from Parikh et. al. [12]. Both compounds show better inhibition profiles against NL4-3 as well as MDR769 HIV-1 protease variants compared to that of the control, acetyl pepstatin.
| Compound | IC50 (μM) ± standard deviation
|
||
|---|---|---|---|
| NL4-3 | MDR769 | PLM-II | |
| 1 | 0.99 ± 0.13 | 14.53 + 1.65 | 0.02 |
| 2 | 5.7 ± 0.33 | 30.30 ± 1.97 | 0.025 |
| acetyl pepstatin | 25.3 ± 1.56 | 56.75 ± 3.17 | 7.5 |
Both 1 and 2 feature a transition-state mimic hydroxyl group in a statine based core that is known to enhance the affinity of the compound for the enzyme [18]. Potencies of both compounds were evaluated against NL4-3 and MDR 769 variants of HIV-1 protease. Crystal structures of MDR7 769 A82T mutant HIV-1 protease in complex with 1 and 2 were solved and analyzed. Both compounds were further docked into the active site cavity of NL4-3 protease using AutoDock Vina [19]. A detail crystallographic and modeling analysis was performed to understand the binding profiles of both the compounds.
2. MATERIALS AND METHODS
2.1. Compounds and HIV-1 protease
Compounds 1 and 2 were synthesized using peptide coupling method as reported previously [15]. Catalytically active NL4-3 protease was purchased from Bachem (California, USA) at a concentration of 0.3 mg/ml. Expression and purification of catalytically active (D25/D125) MDR769 and inactive (N25/N125) MDR769 A82T HIV-1 protease variants were performed as described previously [20]. The inactive (D25N/D125N) variant of the MDR769 A82T protease was chosen to avoid the auto proteolysis for crystallographic studies. Briefly, both the active and inactive variants of MDR protease were over expressed and purified from the inclusion bodies by solublizing them in 6 M urea. Following the ion-exchange chromatographic purification, the protease species were refolded and concentrated. The final yield of the active MDR protease was low (0.5 mg/ml) compared to that of the inactive A82T mutant (1.5 mg/ml) as predicted.
2.2. Enzyme inhibition assay
FRET-based enzyme inhibition assay was performed using the fluorescent HIV-1 substrate (purchased from Molecular probes – California, USA) with excitation and emission wavelengths, 340 nm and 490 nm respectively. Substrate was dissolved in DMSO at a stock concentration of 0.5 mM. The emission signal was recorded using the Model-M5 plate reader from Molecular Devices (California, USA). Reaction buffer (0.1 M sodium acetate, 1.0 M sodium chloride, 1.0 mM dithiothreitol (DTT), 1.0 mM ethylenediaminetetraacetic acid (EDTA), 10% DMSO (dimethyl sulfoxide) and 1 mg/ml bovine serum albumin (BSA)) was made fresh each time before the assay. The final pH was adjusted to 4.7. Both NL4-3 and MDR protease variants were diluted by 1600 and 3200 fold respectively, with fresh reaction buffer before assay in order to achieve proper enzyme activity. Ligand was diluted serially by two-fold with DMSO. 99 μl of diluted HIV-1 protease and 1 μl of ligand at various concentrations were incubated at 37° for 10 min. 1 μl of fluorescent substrate at stock concentration was added right before reading at 37°. The fluorescent signal was recorded for 20 min. with 1 min. intervals. The final IC50 values are average of three experiments.
2.3. X-ray crystallography
A twenty-fold molar excess of either 1 or 2 was added to the catalytically inactive A82T variant of MDR769 HIV-1 protease and incubated on ice for one hour before crystallization. Hanging drop, vapor diffusion method was used for crystallization in the pH range of 2.0 to 12.0. For each condition, 1 μl of enzyme-drug complex was mixed with 1 μl of well solution. Diffraction quality crystals were obtained in the range of pH 7.0 – 8.5 at a sodium chloride concentration of 0.3 M to 0.5 M. Diffraction datasets were collected at the LS-CAT beam line 21 at the APS (advanced photon source), IL. Diffraction data was processed using HKL2000 [21]. Structure solutions were obtained using MOLREP [22] from CCP4 [23] suite of programs with MDR769 HIV-1 protease (PDB ID: 1TW7) as a search model. The models were then refined further using REFMAC5 [24]. Electron density maps were calculated using XtalView/Xfit [25] program. The difference electron density (calculated using XtalView/Xfit program) was contoured between 1.8 to 2.0 σ while fitting the ligand. REFMAC libraries for the ligands were obtained from PRODRG server [26]. Refined coordinates for the ligand conformation with highest occupancy and reasonable thermal factor values were included in the final coordinate file. Solvent molecules were then built in using ARP/wARP [27, 28] from CCP4. Validation of the final structures was performed by PROCHECK [29, 30]. Coordinates for crystal structures of MDR769 A82T HIV-1 protease in complex with compounds 1 and 2 were deposited in RCSB (Regional Collaboratory for Structural Bioinformatics) PDB (protein data bank) with accession codes 3R0Y and 3R0W respectively. Analysis of the protease-inhibitor contacts was performed using CONTACT/ACT [31] module from CCP4 suite of programs.
2.4. Docking
Crystal structures of wild type HIV-1 protease in complex with darunavir (PDB ID: 2IEN) and PLM-II in complex with TIT-1330 (PDB ID: 1W6H) were chosen as starting structures. The coordinates for ligand, water and other small molecules were deleted from both the structures and apo-enzyme coordinates were then used as receptors to build the docking grid. All the polar hydrogen atoms were added to the receptors before generating the docking grid. The docking grid was generated in and around the active site cavity for all three receptors using AutoDock tools [32] GUI (graphic-user interface). The ligands were prepared in the same way using AutoDock tool GUI. Initially, darunavir taken from the crystal structure was re-docked back into the active site of the HIV-1 protease taken from 2IEN as a positive control using AutoDock Vina. Similar positive control was performed for TIT-1330 against PLM-II taken from 2W6H. For the MDR769 HIV-1 protease, crystal structures 3R0Y and 3R0W were used. Mutations N25D and N125D were introduced into the MDR protease (to represent catalytically active protease) model before receptor grid generation. All the graphics in this article were prepared using the open source molecular graphics program, PyMol (v0.99rc6) (www.pymol.org).
3. RESULTS AND DISCUSSION
3.1. Compounds 1 and 2 show better potency against wild type HIV-1 protease than the MDR 769 HIV-1 protease
Fluorescence resonance energy transfer (FRET)-based enzyme inhibition assays revealed that both 1 and 2 show inhibitory activity with better potency against NL4-3 than MDR 769 HIV-1 protease. As shown in Table 1, the IC50 values show that 1 is 14-fold and 2 is 5-fold more potent against NL4-3 than MDR 769 HIV-1 protease. Compound 1 is at least 25-fold (against NL4-3) and 4-fold (against MDR 769) more potent compared to the control, acetyl pepstatin (a general aspartyl protease inhibitor). Similarly, 2 is at least 4-fold (against NL4-3) and almost 2-fold (against MDR 769) more potent compared to acetyl pepstatin.
3.2. Crystal structures of MDR 769 HIV-1 protease in complex with compounds 1 and 2 show altered binding orientations of the compounds
Crystal structures of MDR 769 HIV-1 protease variant in complex with 1 and 2 were solved in the space group P41 with one protease dimer per asymmetric unit. Diffraction data and refinement statistics are given in Table 2. Crystallographic analysis suggested that both compounds bind the MDR protease in two possible orientations each. The major binding orientation for each compound with higher occupancy and lower thermal factors was determined to be the final binding orientation and were refined to obtain the final structures. The MDR protease in both structures showed the typical wide-open conformation of the flaps contributing to the expansion in the volume of the active site cavity. Due to this wide-open conformation of the flaps, the overall binding orientation of either compound looks altered when compared to other HIV-1 protease inhibitors. Analysis of protease – inhibitor contacts suggests that both 1 and 2 are involved in three direct polar contacts each with MDR protease and multiple polar contacts with active site water molecules. As shown in Figure 2, the transition-state mimic hydroxyl moiety from either compound is involved in hydrogen bonding with Asn25 and Asn125. Details of polar contacts are given in Table 3. As shown in Table 4, compound 1 is involved in a total of 13 hydrophobic contacts while compound 2 is involved in a total of 15 hydrophobic contacts.
Table 2.
X-ray diffraction data and refinement statistics.
| Parameters | Compound 1 | Compound 2 |
|---|---|---|
| PDB entry | 3R0Y | 3R0W |
| Crystal parameters: | ||
| Resolution (Å) | 18.95 – 1.65 | 18.98 – 1.7 |
| Unit cell (Å) | a = b = 45.64, c = 102.00, α = β = γ = 90 | a = b = 45.73, c = 102.15 α = β = γ = 90 |
| Space group | P41 | P41 |
| Solvent content (%) | 47.0 | 47.0 |
| Data processing: | ||
| No. of unique reflections | 25092 (2505) | 22795 (2281) |
| I/σ(I) | 30.81 (5.0) | 40.62 (6.9) |
| Rmerge‡ (%) | 5.7 (45.6) | 4.0 (26.6) |
| Data redundancy | 7.6 (7.5) | 4.0 (4.0) |
| Completeness (%) | 100 (100) | 99.3 (100) |
| Refinement statistics: | ||
| No. of reflections used | 23751 | 21607 |
| Rcryst† (%) | 19.78 | 19.78 |
| Rfree (%) | 23.06 | 24.54 |
| No. of protein atoms | 1512 | 1512 |
| No. of ligand atoms | 44 | 44 |
| No. of water molecules | 192 | 208 |
| Mean temperature factors (Å2): | ||
| Protein | 21.34 | 21.25 |
| Main chains | 20.08 | 20.20 |
| Side chains | 22.72 | 22.41 |
| Ligand | 50.52 | 59.66 |
| Waters | 37.63 | 35.44 |
| R.M.S.D. bond lengths (Å) | 0.019 | 0.013 |
| R.M.S.D. bond angles (°) | 1.67 | 1.50 |
| Ramachandran plot: | ||
| Most favored (%) | 92.9 | 93.6 |
| Additional allowed (%) | 7.1 | 6.4 |
| Generously allowed (%) | 0 | 0 |
| Disallowed (%) | 0 | 0 |
(Values in parentheses are for the highest resolution shell)
Rmerge = Σ |I − <I>|/Σ I
Rcryst = Σ ||Fobs| − |Fcalc||/Σ|Fobs|
Figure 2.
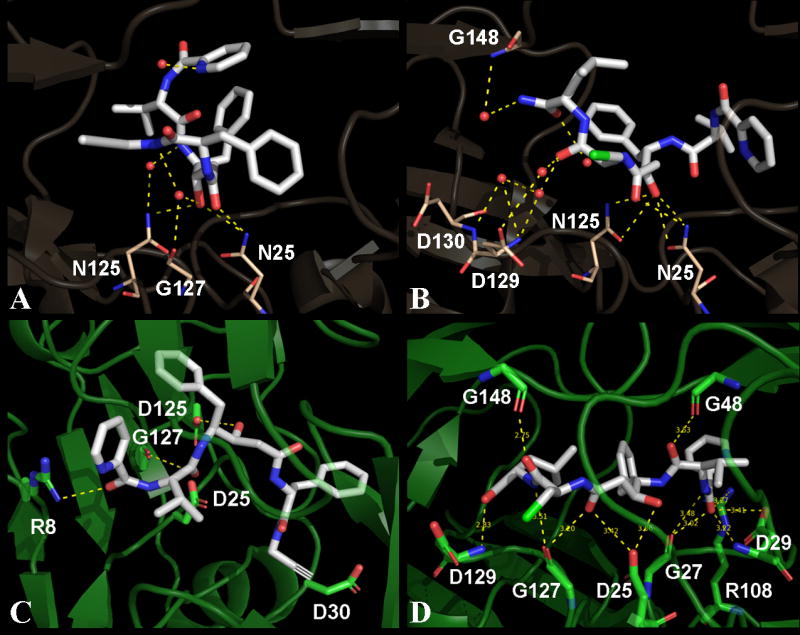
Three-dimensional view of contacts for compounds 1 and 2. Panels A and B show contacts made by compounds 1 and 2 respectively with the MDR protease in their corresponding crystal structures. Panels C and D show docked models of wild type HIV-1 protease with compounds 1 and 2 respectively. The compounds are represented as white sticks in all panels while the protease residues that are involved in polar contacts are highlighted as sticks. The protease residues are colored in baize and green for the MDR and wild type protease respectively. All the polar contacts are represented as yellow dashed lines and the active site water molecules are represented as small red spheres. Three waters that have direct contacts with compound 1 are shown in panel A. Out of the three water molecules in panel A, two bridge the compound to N125 and G127 of the MDR protease. Five waters are shown in panel B out of which, four waters are directly in contact with compound 2. Three waters bridge the compound to D129 and G148 in panel B. The binding orientation of compound 2 is more stretched-out, with more contacts compared to that of the compound 1.
Table 3. Protease inhibitor polar contacts.
Polar contacts made by compounds 1 and 2. Amino acid residues (represented as single letter amino acid codes) of MDR 769 A82T HIV-1 protease involved in polar contacts with compounds 1 and 2 are listed with their corresponding number of contacts in parenthesis. In the case of NL4-3 and MDR769, the protease residues are numbered 1–99 for monomer-A and 101–199 for monomer-B. Active site water molecules that are involved in polar contacts with either compound are listed along with the protease residues. Compound 2 shows more contacts than that of compound 1 in all three cases. Both compounds show highest number of polar contacts with PLM-II.
| Enzyme | Compound 1 | Total | compound 2 | Total |
|---|---|---|---|---|
| MDR769 | N25 (2), N125 (1), H2O (3) | 6 | N25 (3), N125 (2), H2O (4) | 9 |
| NL4-3 | R8 (1), D125 (1), G127 (1) | 3 | D25 (2), G27(2), D29(2), G48 (1), R108 (1), G127 (2), D129 (1), G148 (1) | 12 |
| PLM-II | D34 (2), G36 (1), S79 (2), Y192 (1), G216 (1), T217 (1) | 8 | D34 (3), G36 (1), V78 (1), S79 (3), Y192 (2), D214 (1), T217 (1), S218 (2) | 14 |
Table 4. Protease inhibitor hydrophobic contacts.
Hydrophobic contacts made by compounds 1 and 2. Amino acid residues (represented as single letter amino acid codes) involved in hydrophobic contacts with compounds 1 and 2 are listed for MDR769, NL4-3 and PLM-II. In the case of NL4-3 and MDR769, the protease residues are numbered 1–99 for monomer-A and 101–199 for monomer-B. Both compounds show almost equal number of hydrophobic contacts with the MDR769 HIV-1 protease. Compound 1 shows more hydrophobic contacts than 2 in the docked models of NL4-3 HIV-1 protease. Both compounds show highest number of hydrophobic contacts with PLM-II.
| Enzyme | Amino acid residues involved in hydrophobic contacts
|
|
|---|---|---|
| Compound 1 | Compound 2 | |
| MDR769 | G27, I50, P81, T82, V84, I147, P181, T182 | A28, V32, V84, G148, I150, P181, T182, V184 |
| NL4-3 | R8, A28, D30, V32, I47, I50, I84, A128, D129, D130, V132, I147, G149, I150, L176, P181, V182, I184 | L23, A28, V82, I84, A128, D129, I147, G149, I150, V182, I184 |
| PLM-II | I14, M15, Y17, I32, D34, G36, S37, M75, N76, Y77, V78, S79, F111, T114, S118, F120, I123, L131, Y192, D214, G216, T217, S218, A219, T221, F244, Q275, N288, I290, L292, F294, I300 | I14, M15, Y17, I32, D34, G36, S37, A38, M75, N76, Y77, V78, S79, F111, T114, A117, S118, F120, I123, L131, Y192, I212, D214, G216, T217, S218, A219, T221, F244, I290, L292, F294, I300 |
3.3. Crystallographic water molecules compensate for the loss of direct contacts in the expanded active site cavity of the MDR 769 HIV-1 protease
Multiple water molecules were observed in both crystal structures surrounding the ligand in the expanded active site cavity of the MDR protease. Both structures show a rich network of hydrogen bonding among the waters in and around the active site area contributing to the stability of the compounds in the expanded active site cavity. Out of the three water molecules that showed direct polar contacts with compound 1, two were observed to be bridging between the protease (one with N125 and the other with G127) and the compound (Figure 2A). Similarly, out of the four water molecules that showed direct polar contacts with compound 2, three were observed to be bridging between the protease (one with G148 and the other two with D129 backbone amide nitrogen and side chain oxygen atoms) and the compound (Figure 2B). Typically one conserved water molecule is observed in most of the HIV-1 protease crystal structures that bridges the inhibitor to the protease flaps but in the current study, due to the wide-open conformation of the flaps, the conserver water molecule was lost. Both structures showed a conserved trend of inter-flap water triad consisting of three water molecules bridging the wide-open flaps. The tips of the flaps (Gly48 to Gly52) show a curly conformation with one water molecule bound in the concavity of either flap (intra-flap water). The intra-flap water molecules showed a conserved trend of hydrogen bonding with the backbone of residues Gly49, Ile50, Gly51 and Gly52 in each monomer from both structures.
3.4. Docking solutions suggest loss of binding affinity for either compound against the MDR 769 HIV-1 protease
Both compounds 1 and 2 were individually docked into the active site of NL4-3. Docking solutions were analyzed for contacts between the protease and compounds. As shown in Figure 2, compound 2 showed more contacts with the NL4-3 receptor in the docked model compared to compound 1. Both compounds show a stretched-out binding orientation in the docked models of NL4-3 compared to their corresponding crystal structures with MDR protease. As shown in Figure 3, both compounds show better binding affinity values against NL4-3 and PLM-II compared to MDR protease. Compounds 1 and 2 show a loss of 2.8 kcal/mol and 2.5 kcal/mol respectively in their binding affinities against the MDR protease. Availability of larger chemical space in the expanded active site cavity of the MDR protease causes multiple possible binding orientations of the inhibitor until stable binding is achieved. This increase in the chemical space in part explains the loss of binding affinity for compounds 1 and 2 docked against the MDR 769 HIV-1 protease receptor.
Figure 3.
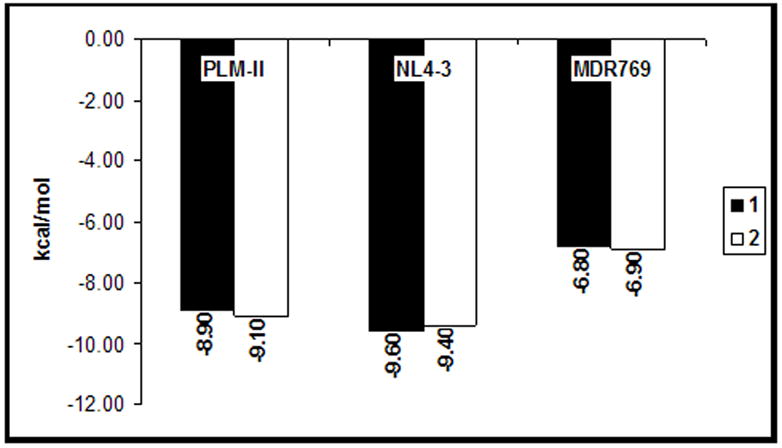
Binding affinities of compounds 1 and 2. Binding affinity values in kcal/mol (y-axis) are plotted for compounds 1 (black bars) and 2 (white bars) against Plasmepsin-II (PLM-II), wild type (NL4-3) and MDR 769 HIV-1 protease receptors. 1 and 2 show equal or better binding affinity values against the NL4-3 receptor compared to their corresponding PLM-II models. The MDR 769 HIV-1 protease models show loss (>2 kcal/mol) of binding affinity for both compounds.
3.5. Compound 2 shows less fold-resistance against MDR 769 HIV-1 protease compared to compound 1
Compounds 1 and 2 showed promising inhibition profiles against the NL4-3 protease. Previously, MDR clinical isolates such as MDR 769 HIV-1 protease were shown to be 14 to more than 100-fold resistant against FDA approved and experimental PIs [6]. Recently, it has been shown that MDR 769 HIV-1 protease exhibits 2 to almost 600-fold resistance against FDA approved PIs [8]. In the current study, the MDR 769 showed 14 and 5-fold resistance against 1 and 2 respectively suggesting that both compounds are within the range of the regular PIs with respect to the fold-change in their potency. Crystal structures and docking models showed that compound 2 is bound in a stretched-out fashion with relatively more contacts with the protease than compound 1 suggesting that compound 2 is less prone to loss of potency due to mutations in the protease compared to compound 1.
In summary, the crystals structures and docking models support the inhibitory activity of 1 and 2 against the NL4-3 as well as against MDR 769 HIV-1 protease variants suggesting that further development of these compounds would enhance their potency. We are currently in the process of evaluating new compounds designed based on the crystal structures reported here.
Highlights.
Potent anti-malarial compounds, 1 & 2, are active against HIV-1 protease variants.
Potency of both compounds was better against wild type than MDR HIV-1 protease.
Crystal structures of MDR HIV-1 protease show altered binding orientation of 1 & 2.
Active site waters compensate for the loss of direct contacts with MDR protease.
Docking shows better binding affinity of compounds against wild type protease.
Acknowledgments
We thank the National Institutes of Health for funding to LCK (Grant # AI65294). We thank the APS-LS-CAT for x-ray diffraction data collection. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357. Use of the LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor for the support of this research program (Grant 085P1000817).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.UNAIDS. The Joint United Nations Programme on HIV/AIDS. 2010 ( www.unaids.org) [PubMed]
- 2.Preston BD, Dougherty JP. Mechanisms of retroviral mutation. Trends Microbiol. 1996;4:16–21. doi: 10.1016/0966-842x(96)81500-9. [DOI] [PubMed] [Google Scholar]
- 3.Kohl NE, Emini EA, Schleif WA, et al. Active Human Immunodeficiency Virus Protease is Required for Viral Infectivity. Proc Natl Acad Sci. 1988;85:4686–4690. doi: 10.1073/pnas.85.13.4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peng C, Ho BK, Chang TW, et al. Role of human immunodeficiency virus type 1-specific protease in core protein maturation and viral infectivity. J Virol. 1989;63:2550–2556. doi: 10.1128/jvi.63.6.2550-2556.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mitsuya H, Maeda K, Das D, et al. Development of Protease Inhibitors and the Fight with Drug-Resistant HIV-1 Variants. Adv Pharmacol. 2008;56:169–197. doi: 10.1016/S1054-3589(07)56006-0. [DOI] [PubMed] [Google Scholar]
- 6.Palmer SS, Robert W, Merigan TC. Highly drug-resistant HIV-1 clinical isolates are cross-resistant to many antiretroviral compounds in current clinical development. AIDS. 1999;13:611–667. doi: 10.1097/00002030-199904160-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Logsdon BC, Vickrey JF, Martin P, et al. Crystal Structures of a Multidrug-Resistant Human Immunodeficiency Virus Type 1 Protease Reveal an Expanded Active-Site Cavity. J Virol. 2004;78:3123–3132. doi: 10.1128/JVI.78.6.3123-3132.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y, Liu Z, Brunzelle JS, et al. The higher barrier of darunavir and tipranavir resistance for HIV-1 protease. Biochem Biophys Res Commun. 2011;412:737–742. doi: 10.1016/j.bbrc.2011.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin P, Vickrey JF, Proteasa G, et al. “Wide-Open” 1.3 Å Structure of a Multidrug-Resistant HIV-1 Protease as a Drug Target. Structure. 2005;13:1887–1895. doi: 10.1016/j.str.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 10.Yedidi RS, Proteasa G, Martinez JL, et al. Contribution of the 80s loop of HIV-1 protease to the multidrug-resistance mechanism: crystallographic study of MDR769 HIV-1 protease variants. Acta Crystallogr D Biol Crystallogr. 2011;67:524–532. doi: 10.1107/S0907444911011541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heaslet H, Rosenfeld R, Giffin M, et al. Conformational flexibility in the flap domains of ligand-free HIV protease. Acta Crystallogr D Biol Crystallogr. 2007;63:866–875. doi: 10.1107/S0907444907029125. [DOI] [PubMed] [Google Scholar]
- 12.Parikh S, Liu J, Sijwali P, et al. Antimalarial effects of human immunodeficiency virus type 1 protease inhibitors differ from those of the aspartic protease inhibitor pepstatin. Antimicrob Agents Chemother. 2006;50:2207–2209. doi: 10.1128/AAC.00022-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Banerjee R, Liu J, Beatty W, et al. Four plasmepsins are active in the Plasmodium falciparum food vacuole, including a protease with an active-site histidine. Proc Natl Acad Sci. 2002;99:990–995. doi: 10.1073/pnas.022630099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klemba M, Goldberg DE. Biological roles of proteases in parasitic protozoa. Annu Rev Biochem. 2002;71:275–305. doi: 10.1146/annurev.biochem.71.090501.145453. [DOI] [PubMed] [Google Scholar]
- 15.Gupta D, Yedidi RS, Varghese S, et al. Mechanism-based inhibitors of the aspartyl protease plasmepsin II as potential antimalarial agents. J Med Chem. 2010;53:4234–4247. doi: 10.1021/jm100233b. [DOI] [PubMed] [Google Scholar]
- 16.Northrop DB. Follow the protons: a low-barrier hydrogen bond unifies the mechanisms of the aspartic proteases. Acc Chem Res. 2001;34:790–797. doi: 10.1021/ar000184m. [DOI] [PubMed] [Google Scholar]
- 17.Abdel-Rahman HM, Kimura T, Hidaka K, et al. Design of inhibitors against HIV, HTLV-I, and Plasmodium falciparum aspartic proteases. Biol Chem. 2004;385:1035–1039. doi: 10.1515/BC.2004.134. [DOI] [PubMed] [Google Scholar]
- 18.Mimoto T, Imai J, Tanaka S, et al. KNI-102, a novel tripeptide HIV protease inhibitor containing allophenylnorstatine as a transition-state mimic. Chem Pharm Bull (Tokyo) 1991;39:3088–3090. doi: 10.1248/cpb.39.3088. [DOI] [PubMed] [Google Scholar]
- 19.Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J Comput Chem. 2010;30:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vickrey JF, Logsdon BC, Proteasa G, et al. HIV-1 protease variants from 100-fold drug resistant clinical isolates: expression, purification, and crystallization. Protein Expression and purification. 2003;28:165–172. doi: 10.1016/s1046-5928(02)00650-2. [DOI] [PubMed] [Google Scholar]
- 21.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. Macromolecular Crystallography, partA. [DOI] [PubMed] [Google Scholar]
- 22.Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement. J Appl Cryst. 1997;30:1022–1025. [Google Scholar]
- 23.Collaborative Computational Project Number 4. Acta Crystallographica Section D. 1994;50:760–763. [Google Scholar]
- 24.Murshudov GN, Vagin AA, Dodson EJ. Refinement of Macromolecular Structures by the Maximum-Likelihood Method. Acta Crystallographica Section D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 25.McRee DE. XtalView/Xfit – A Versatile Program for Manipulating Atomic Coordinates and Electron Density. J Structural Biology. 1999;125:156–165. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- 26.Schuttelkopf AW, van Aalten DMF. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallographica Section D. 2004;60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 27.Lamzin VS, Wilson KS. Automated refinement of protein models. Acta Crystallographica Section D. 1993;49:129–147. doi: 10.1107/S0907444992008886. [DOI] [PubMed] [Google Scholar]
- 28.Perrakis A, Morris R, Lamzin VS. Automated protein model building combined with iterative structure refinement. Nat Struct Mol Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 29.Laskowski RA, MacArthur MW, Moss DS, et al. PROCHECK: a program to check the Stereochemical quality of protein structures. Journal of Applied Crystallography. 1993;26:283–291. [Google Scholar]
- 30.Morris AL, MacArthur MW, Hutchinson EG, et al. Stereochemical quality of protein structure coordinates. Proteins: Structure, Function, and Genetics. 1992;12:345–364. doi: 10.1002/prot.340120407. [DOI] [PubMed] [Google Scholar]
- 31.Kabsch W, Sander C. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 32.Sanner MF. Python: A Programming Language for Software Integration and Development. Journal of Molecular Graphics and Modelling. 1999;17:55–84. [PubMed] [Google Scholar]


