Abstract
DNA vaccination using cationic polymers as carriers has the potential to be a very powerful method of immunotherapy, but typical immune responses generated have been less than robust. To better understand the details of DNA vaccine delivery in vivo, we prepared polymer/DNA complexes using three structurally distinct cationic polymers and fluorescently labeled plasmid DNA and injected them intradermally into mice. We analyzed transgene expression (luciferase) and the local tissue distribution of the labeled plasmid at the injection site at various time points (from hours to days). Comparable numbers of luciferase expressing cells were observed in the skin of mice receiving naked plasmid or polyplexes one day after transfection. At day 4, however, the polyplexes appeared to result in more transfected skin cells than naked plasmid. Live animal imaging revealed that naked plasmid dispersed quickly in the skin of mice after injection and had a wider distribution than any of the three types of polyplexes. However, naked plasmid level dropped to below detection limit after 24 h, whereas polyplexes persisted for up to 2 weeks. The PEGylated polyplexes had a significantly wider distribution in the tissue than the nonPEGylated polyplexes. PEGylated polyplexes also distributed more broadly among dermal fibroblasts and allowed greater interaction with antigen-presenting cells (APCs) (dendritic cells and macrophages) starting at around 24 h post-injection. By day 4, co-localization of polyplexes with APCs was observed at the injection site regardless of polymer structure, whereas small amounts of polyplexes were found in the draining lymph nodes. These in vivo findings demonstrate the superior stability of PEGylated polyplexes in physiological milieu and provide important insight on how cationic polymers could be optimized for DNA vaccine delivery.
Keywords: DNA vaccines, Polyplexes, PEGylation, Biodistribution
1. Introduction
The goal of vaccination is to manipulate the immune system into responding against specific antigens. Theoretically, this strategy can work both to treat ongoing infections and malignancies as well as prevent diseases by generating immunological memory. For successful immunization, antigen must be delivered to antigen-presenting cells (APCs), mainly dendritic cells (DCs) and macrophages, often along with an immunostimulatory adjuvant. These cells can then process and present the antigen and stimulate T and B cells in the lymph nodes and spleen.[1–3]
Transfection of cells with antigen-encoding plasmid DNA will result in the expression of the protein antigen by those cells. This is an attractive method of vaccination due to the high stability of plasmid DNA formulations, the potential for long-term antigen production, and the capacity of generating both humoral and cellular immune responses to multiple epitopes.[3–6] Initial attempt of DNA vaccination involved the injection of naked plasmid DNA. However, though this can result in immune responses, the efficiency of this method of delivery needs improvement.[4,5,7] Various delivery vehicles, including viral particles, liposomes, and polymeric materials, have since been used to help protect the DNA and increase transfection.[4,8] Polymeric carriers have the potential to be a very effective means of delivering antigen-encoding DNA for immunization, because polymers can be easily modified and optimized to acquire a wide range of characteristics. Delivery of DNA vaccine with various polymer-based systems has shown improvement in both humoral and cellular immune responses, but overall immune responses have not been sufficiently robust.[4,9]
Over the years a large number of polymer DNA carriers with a large variety of chemical structures have been developed, many of which have been investigated for DNA vaccine delivery. It is not clear, however, how to further improve upon current designs because little is known about the specific mechanisms of delivery in vivo, especially tissue and cellular fate of the DNA that precede immune response generation. There have been studies conducted to investigate the uptake and transfection of specific cell types using various naked DNA delivery systems in the muscle and skin.[10–13] For example, it was shown that fibroblasts, endothelial cells, and adipocytes appeared to be the primary cell types transfected after naked DNA delivery via electroporation in the skin,[11] and that myocytes were the primary recipient of DNA after intramuscular injection.[10] More recently, van den Berg and others described the transfection of primarily keratinocytes after tattooing with PEGylated polyplexes.[5] These studies also reported some, if limited, DNA uptake by, or transfection of, APCs. However, exactly which cell types are involved in the interactions with polyplex-delivered DNA, and how those interactions may affect antigen presentation as well as the timeline and magnitude of immune responses, is not well understood. Furthermore, there is a lack of systematic understanding of the structure-function relationship of the polymer carriers in the context of in vivo administration. To this end, we prepared polyplexes of plasmid DNA and three cationic polymers with distinct chemical structures: branched polyethylenimine (PEI), linear poly(2-aminoethyl methacrylate) (PAEM), and diblock copolymer PEG-b-PAEM (Fig. 1). We injected these polyplexes into mice intradermally, and analyzed the transgene expression and local biodistribution of plasmid both macroscopically on the tissue level and microscopically on the cellular level, in comparison to injections of naked plasmid. We uncovered important differences in local tissue distribution between polyplexes and naked plasmid, and between PEGylated and non-PEGylated polyplexes. This information could be highly useful for improving the design of cationic polymer carriers for DNA vaccine.
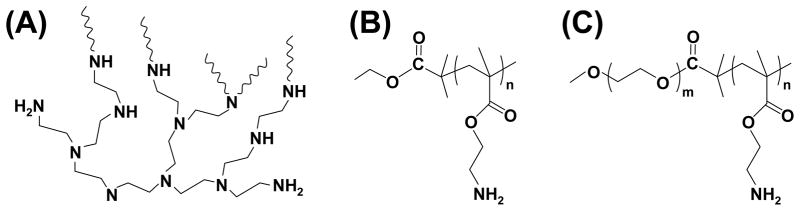
Fig. 1.
Chemical structures of branched PEI (A), PAEM (B), and PEG-b-PAEM (C).
2. Materials and Methods
2.1. Chemicals and solvents
PEI (branched, 25 kDa) was obtained from Sigma. Monomethoxy-PEG (average Mn of 5000) was from Aldrich and was used after vacuum drying at 80 °C for 2 h. Toluene (Aldrich) was dried by refluxing over sodium and distilled. The monomer N-(tert-butoxycarbonyl)aminoethyl methacrylate (tBAM) and the PEG macro-initiator for atom transfer radical polymerization (ATRP) was synthesized as described before.[14] [15] Ethyl α-bromoisobutyrate, copper (I) chloride (CuCl) and 2,2′-dipyridyl (bPy) were purchased from Sigma. Other chemicals and solvents were purchased from Sigma and used without further purification.
2.2. Polymer synthesis
The ATRP of PtBAM followed a procedure modified from Tang et al.[16] A glass two-neck flask was charged with tBAM, CuCl, bPy, and the system was degassed three times. Dried degassed toluene and ethyl α-bromoisobutyrate were added, and the mixture was heated at 80 °C for 8 h. The reaction was terminated by exposing the system to air. The reaction solution was then diluted by dichloromethane and passed through a basic aluminum oxide column to remove the copper complex. The resulting product was precipitated in hexane twice and dried in vacuum at room temperature for 2 days. To remove the Boc groups, 0.8 g of PtBAM was dissolved in 5 mL of trifluoroacetic acid and stirred for 2 h at room temperature. TFA was then removed by evaporation, and the oil residue was rinsed three times with diethyl ether. The resultant precipitate was collected by filtration, washed twice by diethyl ether, and dried overnight in vacuum. Afterwards, the polymers were washed with NaOH solution at pH 9.0, and immediately placed into dialysis tubing (MW cut-off 3500) and dialyzed against distilled water for 3 days. The final PAEM polymer was obtained by lyophilization.
PEG-b-PAEM diblock copolymer was synthesized as described in Tang et al.[16] using a 5000-Da PEG block. The final polymer was washed by NaOH solution, dialyzed and lyophilized. The average values of Mn for PtBAM and PEG-b-PtBAM were 3.37×104 and 3.96×104 with narrow distribution (PDI 1.16 and 1.20). Therefore, the average chain-length of the PAEM homopolymer (degree of polymerization, DP: 150) was the same as the PAEM segment in the PEG-b-PAEM diblock copolymer.
2.3. Polyplex preparation
Plasmid DNA encoding ovalbumin (pOVA, kindly provided by Dr. Chris Pennell) was purified from E. coli DH5α cells using an EndoFree Plasmid Maxi plasmid prep kit (Qiagen) for tissue distribution studies. The plasmid was covalently labeled with Cy3 fluorophore using a Label IT nucleic acid labeling kit (Mirus) and purified according to manufacturer’s instruction (including exhaustive dialysis to remove any free dye). Purified plasmid was verified to contain less than 0.6 EU/mg of endotoxin using the Pyrogent Gel Clot LAL assay kit (Lonza) and stored at −20°C in sterile water. Polymer stocks were first diluted in 5% glucose, filter-sterilized, and stored in aliquots at −20°C. To form polyplexes, polymer stocks were further diluted in sterile 5% glucose before DNA was added and samples were vortexed to mix. A typical batch of polyplexes was made with 12 μg of DNA and enough polymer for an N/P ratio of 8 in 36 μL total volume. Naked DNA was diluted to the same volume with 5% glucose. A luciferase plasmid (pCMV-LUC, endotoxin-free, Elim Biopharmaceuticals) was also used for in vivo gene expression experiments.
2.4 Polyplex stability in serum-containing medium
Fifteen μL of polyplex solution containing 5 μg of Cy3-labeled plasmid DNA was added to another 15 μL of either 5% glucose or cell culture medium comprised of DMEM medium (1 g/L D-glucose, L-glutamine, 110 mg/L sodium pyruvate) supplemented with 10% heat-inactivated fetal bovine serum (FBS), 100 units/mL penicillin/streptomycin, and 10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) (all cell medium components were from Gibco). Five μL of the polyplexes solution was removed immediately after dilution and after a 1-h incubation at room temperature, placed onto a glass microscope slide, covered with a glass coverslip, and was observed under an Olympus IX70 inverted microscope equipped with a standard FITC/TRITC/DAPI filter set, a 20× objective lens, an Olympus DP72 camera, and CellSens software. To see if there was any free DNA present after polyplexes were formed and after incubation in 5% glucose and serum-containing medium, polyplex solutions were analyzed on a 0.7% agarose gel stained with ethidium bromide.
2.5 Injections
Hair was plucked from a small section of skin on the hind leg of 10~16-week old male C57BL/6 mice (Jackson Labs) to mark injection site, and polyplex solutions were injected intradermally through a 29-gauge needle. For transfection experiments, 40 μg of DNA complexed with polymers at N/P ratio of 8 was prepared as described above and was injected into each mouse in a total volume of 35 μL. For tissue distribution studies, 10 μg of DNA as polyplexes in 30 μL of buffer were injected. The same volume of naked DNA and buffer only injections were also administered. Polyplexes containing all three cationic polymers (PEI, PAEM, and PEG-b-PAEM) were tested. To detect tissue distribution by Maestro live animal imaging, three mice per sample group were injected. For time course studies of transgene expression and tissue distribution by immunofluorescence, one mouse was injected with each sample and sacrificed at each time point through 4 days. All the mice were housed under specific pathogen-free conditions and cared for in accordance with the University of Minnesota and NIH guidelines.
2.6 In vivo transgene expression
Mice were sacrificed with CO2 one day or four days after injection and the skin around each injection site was removed, embedded in OCT medium and snap-frozen with liquid nitrogen. The frozen skin samples were cut into 10-μm thick sections using a cryotome and placed on SuperFrost glass slides. The cryosections were dried at room temperature for 1 h then fixed in cold acetone. After equilibrating the skin specimens to room temperature, a hydrophobic circle was drawn around the specimens with a PAP pen and the specimens were soaked in PBS for 30 min with slight agitation. The tissue sections were blocked with 5% non-fat dry milk in PBS with 0.1% Tween-20 for 30 min at room temperature in a humidity chamber, followed by staining with 1:100 diluted anti-luciferase-FITC conjugated antibody (Lifespan Biosciences) in the blocking buffer for 1 h. After washing the tissue sections 3 times with PBS, the specimens were counterstained with 1:200 diluted Hoechst 33342 (Invitrogen) and mounted with Vectashield. The tissue sections were visualized using an Olympus IX70 inverted microscope equipped with a standard FITV/TRITC/DAPI filter set and Olympus DP72 camera, and analyzed using CellSens software.
2.7 Maestro live animal imaging
To assess tissue distribution of Cy3-labeled plasmid in live animals after intradermal injection, mice were anesthetized using isofluorane at predetermined time points and imaged using a CRi Maestro live animal imaging system equipped with a Nikon AF Micro 60 mm camera. Three mice of the same experimental group were imaged together. The tails of the mice were marked to keep the order of mice (from left to right under the camera) the same for every time point. Cy3-labeled DNA fluorescence was detected using a 503–555-nm excitation filter and a 580 nm long-pass emission filter. Emission signal was collected between 550 nm and 800 nm in 10-nm steps. Three images of each group of mice were taken at each time point, rotating mice to be at different positions under the camera between images to account for any potential inconsistency due to different animal positions. All images were taken at the same stage height and light position with a constant exposure time of 5000 ms. The fluorescence signal at 560 nm was isolated and the threshold was set using Image J software. Image J was also used to quantify the total pixel area of signal for each sample at each time point.
2.8 Immunofluorescence
To determine distribution of Cy3-labeled plasmid in tissue sections, mice were euthanized using CO2 at predetermined time points after injection. The skin of each injection site and the inguinal lymph node that drains the injection site were removed, embedded in OCT medium, and frozen in a bath of 2-methylbutane using liquid nitrogen. Frozen skin samples were cut into 10-μm sections using a cryotome and placed on SuperFrost (Fischer) glass slides. Slides were allowed to dry at room temperature for 1 h and fixed by incubating in cold acetone for 10 min. For immunofluorescence staining, slides of tissues were removed from freezer and equilibrated to room temperature. A hydrophobic circle was drawn around tissue sections with a PAP pen and the slides were placed in a staining chamber and equilibrated in PBS for 30 min with slight agitation. Tissue was then blocked using Blocking Buffer (from Tyramide Signal Amplification (TSA) Biotin System, Perkin Elmer) for 30 min in a humidity chamber. Slides were also blocked with streptavidin and biotin solutions (Vector Laboratories) for 15 min each in the humidity chamber. Slides were then stained with biotin-labeled anti-CD11c (for DCs) (BioLegend), biotin-labeled anti-F4/80 (for macrophages) (BioLegend), or rabbit anti-ER-TR7 (for reticular dermal fibroblasts) (Abcam) followed by biotin-labeled anti-rabbit IgG (Invitrogen). All antibody incubation times were 1 h, except for anti-CD11c which was 30 min. Signal was developed using Tyramide Signal Amplification (TSA) Biotin System (Perkin Elmer) and a streptavidin labeled with Alexa Fluor 350 (Invitrogen). Tissue sections were then mounted using Vectashield and visualized using an Olympus IX70 inverted microscope equipped with a standard FITC/TRITC/DAPI filter set, an Olympus DP72 camera, and CellSens software. Some tissue sections were also stained with haemolysin and eosin (H&E) to observe anatomical features.
To analyze the fluorescence microscopy images, the areas of fluorescence signal from Cy3-labeled DNA and antibody staining of cell type markers were quantified in Image J. The thresholding levels on the DNA signal were varied to adjust for differences in signal intensity. Cell-DNA interaction was quantified by calculating the area of overlap between the signals of DNA and cell markers using the “co-localize” plug-in for Image J. The results on co-localization were expressed as the fraction of total DNA signal that overlapped with cell marker signal. Two to five slides were stained with each cell marker for each time point and each sample. Some of the stained tissue sections were observed at high magnification with an Olympus FV-1000 confocal microscope equipped with an Olympus 60x/1.42 NA oil-immersion lens (Center Valley, PA). Images of at least three fields of view were collected for every sample.
2.9 Statistical analysis
Statistical analysis was performed using the two-sample equal variance Student’s t-test. A probability (p) value of <0.05 was deemed statistically significant.
3. Results
3.1. Selection of cationic polymers
Three cationic polymers with distinctly different chemical structures were selected: branched 25-kDa PEI, linear PAEM, and linear PEG-b-PAEM (Fig. 1). Branched PEI is well known for efficiently transfecting cells in vitro but is quite inefficient in vivo.[8] PEI from commercial sources has heterogeneous structure and broad molecular weight distribution. On the other hand, the PAEM polymer has a different, much more defined structure than PEI. We used ATRP to synthesize a PAEM polymer with average DP of 150 and narrow molecular weight distribution. To probe the impact of PEGylation, we also synthesized PEG-b-PAEM diblock copolymer using ATRP that contained the same PAEM block (DP = 150) and a PEG block of 5000. Next, we prepared polyplexes with the three cationic polymers and plasmid DNA at an N/P ratio of 8, which yielded the highest transfection efficiency in cultured cells (data not shown), for the following in vivo transfection and tissue distribution studies.
3.2 Visual assessment of polyplex stability in simulated in vivo media
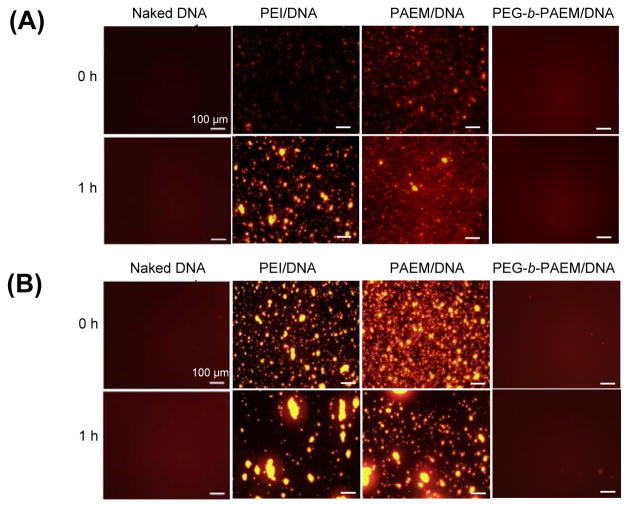
Polyplexes containing Cy3-labeled plasmid DNA were dissolved in 5% glucose injection buffer or cell medium complete with serum and buffer salts to mimic the in vivo fluid environment, and visualized by fluorescence microscopy (Fig 2). Although polyplex stability has often been analyzed using dynamic light scattering, the fluorescence microscopy method we used here enables direct visualization of the aggregation process and the aggregates without the interference of serum protein or other particles present in the media. Cy3-labeled naked DNA solutions remained clear with a slight reddish fluorescence background during the entire time of observation. Shortly after formation, aggregates of PEI and PAEM polyplexes were already visible in 5% glucose (used later for in vivo injection), especially after 1 h (Fig. 2A), presumably because of the high polyplex concentration necessary to keep the injection volume low. More severe aggregation in these polyplexes was seen in serum-containing cell culture medium with aggregates ranging from several microns to nearly a hundred microns in size after 1 h (Fig. 2B). Qualitatively, aggregates of the PAEM polyplexes appeared smaller than the PEI polyplexes. However, no visible aggregation was seen in the PEG-b-PAEM polyplexes after 1 h in either 5% glucose or complete cell culture medium (Fig. 2B). All the samples of polyplexes and naked DNA with or without incubation in complete cell medium were also analyzed by gel electrophoresis. There was no band corresponding to free plasmid DNA in any of the polyplex samples (Fig. 2C), confirming that the DNA was still bound by the polymers during incubation in serum-containing medium.
Fig. 2.
Visual assessment of polyplex stability in injection buffer (5% glucose) (A) and cell medium containing 10% serum (B) – conditions that mimic the in vivo fluid environment. Whereas PEI and PAEM polyplexes experienced much aggregation over time, PEGylated polyplexes remained stable without visible aggregation. Agarose gel electrophoresis of naked DNA and polyplexes before and after incubation in serum-containing medium (C) confirmed the absence of any free, unbound DNA. Scale bar: 100 μm.
3.3 Transgene expression in vivo
Naked luciferase plasmid and different polyplexes containing PEI, PAEM, and PEG-b-PAEM, were injected intradermally into the hind quadriceps region of mice. After day 1 and 4, the animals were sacrificed. The dermal tissue of the injection sites was harvested, cryosectioned, stained with Hoechst for cell nuclei (blue) and with a fluorescein-labeled polyclonal antibody against luciferase (green), and imaged under a fluorescence microscope. The skin sections included the epidermal and dermal tissues as shown in a representative image of H&E stained sample (Supporting Information, Fig. S1). From the representative fluorescence micrographs we can see that mice injected with the naked plasmid and the polyplexes all contained skin cells positively transfected with the luciferase gene (Fig. 3), whereas mice receiving buffer only injections did not shown any detectable luciferase signal (Supporting Information, Fig. S2). The numbers of luciferase-expressing cells in mice injected with naked plasmid and polyplexes were qualitatively comparable by day 1, but polyplexes appeared to have transfected more skin cells in mice than the naked plasmid by day 4 (Fig. 3). Possible reasons for the prolonged local gene transfection by polyplexes in vivo may include protecting the plasmid from enzymatic degradation, providing sustained presence of plasmid in the local tissue (the depot effect), and facilitating cellular uptake and intracellular trafficking of the plasmid. While it is certain that different chemical structures of cationic polymers may influence the transport processes of polyplexes both extra- and intracellularly, here we address specifically the question of local tissue distribution of the plasmid in the skin, comparing the naked plasmid with polyplexes and PEGylated with non-PEGylated polyplexes.
Fig. 3.
Transgene expression in the skin of mice after intradermal injection in the right hind quadriceps region. Each animal received 40 μg of luciferase plasmid either as naked or complexes with various cationic polymers. Shown are representative fluorescence microscopy images of mouse skin cross-sections. Luciferass-expressing cells (green) were detected by a polyclonal antibody against luciferase. Cell nuclei were stained blue (Hoechst). Images of both low (scale bar: 200 μm) and high (scale bar: 50 μm) magnification are presented. White arrows point to luciferase-expressing cells.
3.4 Tissue distribution of plasmid in live animals
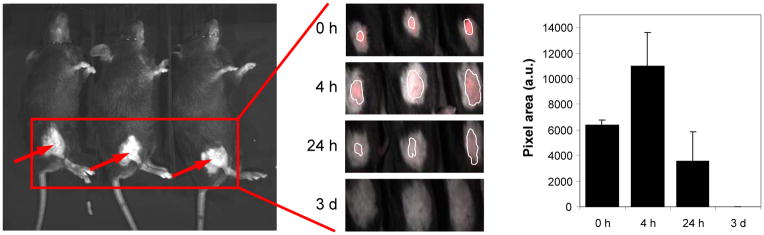
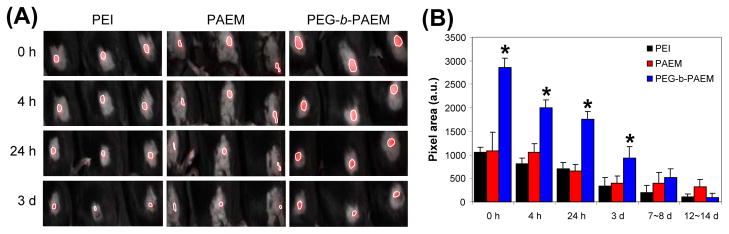
Cy3-labeled naked DNA and polyplexes were injected intradermally into the hind quadriceps region of mice and were imaged using a Maestro live animal imaging instrument. We analyzed the area of the Cy3-DNA signal at various time points after injection to determine the difference, if any, between the distribution and persistence of the naked DNA and the various polyplexes. Naked plasmid disseminated quickly in the skin in 4 h, diminished after 24 h, and completely disappeared by 3 days (Fig. 4). On the other hand, when plasmid was delivered as polyplexes, size of tissue distribution was much reduced but the signal persisted for at least 14 days (Fig. 5), and polyplexes were seen for as long as 27 days in some mice (data not shown). Furthermore, the PEGylated polyplexes appeared to have spread to significantly larger areas than the PEI and PAEM-based polyplexes during the first 3 days (Fig. 5).
Fig. 4.
Tissue distribution of naked plasmid in live animals after intradermal injection. Three mice were each injected with 10 μg of Cy3-labeled DNA in the right hind quadriceps region and were imaged together (A). Plasmid distribution in three injected mice was shown at indicated time points as marked by a white outline (B) and the area of the signal was quantified (C).
Figure 5.
Tissue distribution of polyplexes in live animals after intradermal injection. Three mice were each injected with polyplexes containing 10 μg of Cy3-labeled DNA in the right hind quadriceps region and were imaged together. Plasmid distribution in three injected mice was shown at indicated time points as marked by a white outline (A) and the area of signal was quantified (B). *PEGylated polyplexes showed statistically larger area of spreading than PEI and PAEM-based polyplexes (t test, p<0.001).
3.5. Dermal distribution of naked DNA at the injection site
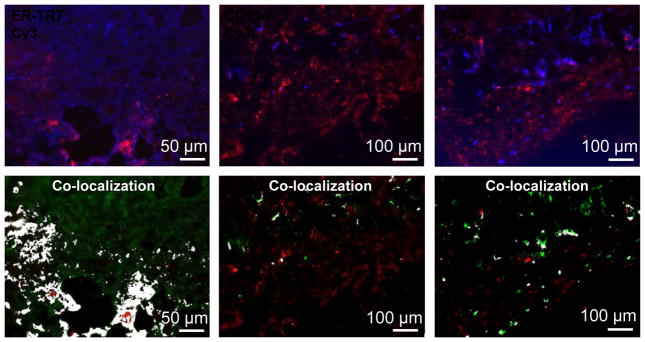
To visualize the distribution of plasmid with cellular resolution, mice were again injected in the hind quadriceps region with naked or polyplexes of Cy3-labeled plasmid DNA and sacrificed at predetermined time points. Injection sites were harvested, sectioned, and stained for ER-TR7, CD11c, and F4/80, markers for reticular dermal fibroblasts, DCs, and macrophages, respectively. The degree of interaction between the plasmid and cells 4 h after injection was estimated by determining co-localization (white) of fluorescence signals of the plasmid label (Cy3, red) with the cell markers (blue) (Fig. 6). From the representative set of images we can see that the naked plasmid spread very well throughout the injection site and had localized in close proximity to dermal fibroblasts that were abundant in the skin (Fig. 6). On the other hand, the numbers of DCs and macrophages present at the injection site 4 h post-injection were much lower than the number of fibroblasts, and the overlap between the naked plasmid and the DC and macrophage markers was also less than with that of the fibroblasts (Fig. 6). After 4 h, the signal of the naked plasmid was no longer detectable in the tissue sections.
Fig. 6.
Dermal distribution of naked plasmid at the injection site. Naked plasmid was labeled with Cy3 (red). Reticular dermal fibroblasts, DCs, and macrophages were stained for ER-TR7, CD11c, and F4/80, respectively (blue or green). Area of co-localization between the plasmid and the cell markers was painted in white. Images shown are representative fields of view. Scale bar: 50 μm (for fibroblasts), 100 μm (for DCs and macrophages).
3.6 Dermal distribution of polyplexes at the injection site
3.6.1 Dermal fibroblasts
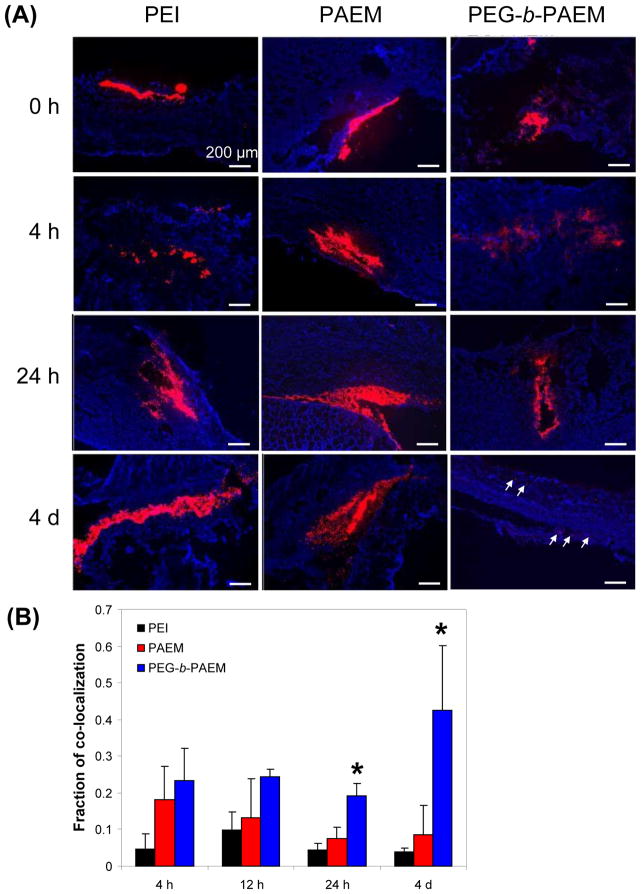
Dermal distribution of Cy3-labeled polyplexes in tissue sections of the injection site was characterized at various time points. Consistent with live animal imaging studies (Fig. 4 & 5), polyplexes persisted much longer than naked DNA in the skin. While naked DNA appeared diffusive in the tissue (Fig. 6), all the polyplexes was more aggregated, forming depots of DNA (Fig. 7A). Both PEI and PAEM polyplexes remained aggregated throughout the 4 days of observation, whereas the PEGylated polyplexes dispersed into much smaller packets with low signal intensity by day 4 (Fig. 7A, pointed by arrows). Much broader dissemination of the PEGylated polyplexes among fibroblasts was also reflected in the quantification of the fraction of co-localization, which was defined as the ratio of pixel areas between cell-marker co-localized plasmid signal and the total plasmid signal. Significantly more plasmid localized to the proximity of fibroblasts by day 1 and day 4 was seen with the PEGylated polyplexes, whereas co-localization remained low throughout 4 days with both the PEI and PAEM polyplexes (Fig. 7B). PAEM polyplexes dispersed slightly better than PEI polyplexes, although the difference was not statistically significant.
Fig. 7.
Dermal distribution of polyplexes and co-localization with dermal fibroblasts at the injection site at various time points. (A) Representative fluorescence microscopy images showing plasmid labeled with Cy3 (red), and fibroblasts stained for ER-TR7 (blue). Scale bar: 200 μm. (B) The fraction of co-localization defined as the ratio of pixel areas between co-localized plasmid signal and the total plasmid signal. *PEGylated polyplexes showed statistically more co-localization with fibroblasts than PEI and PAEM-based polyplexes (t test, p<0.05).
3.6.2 DCs
There was substantial infiltration of DCs into the injection site for all polyplexes only 24 h after injection and the cells persisted through day 4 (Fig 8A). All polyplexes again showed considerable aggregation and much co-localization with DCs was observed after DCs infiltrated. By 24 h co-localization with DCs was significantly more pronounced with the PEGylated polyplexes compared to non-PEGylated PAEM polyplexes (Fig. 8B). From day 1 to 4 there was a time-dependent increase in co-localization of both the PEI and PAEM polyplexes with DCs, and much of the co-localization between polyplexes and DCs was limited to the edges of the aggregates; apparently, DCs had little capacity of breaking apart the dense clumps of polyplex aggregates. Averaging the tissue sections through the edge of aggregates (where co-localization was high) with those through the middle of aggregates (where co-localization was rare) likely resulted in the large error range of the average values of fraction of co-localization for the PEI polyplexes at days 1 and 4 (Fig. 8B).
Fig. 8.
Dermal distribution of polyplexes and co-localization with DCs at the injection site. (A) Representative fluorescence microscopy images showing plasmid labeled with Cy3 (red), and DCs stained for CD11c (blue). Scale bar: 100 μm. (B) The fraction of co-localization defined as the ratio of pixel areas between co-localized plasmid signal and the total plasmid signal. *PEGylated polyplexes showed statistically more co-localization with DCs than PAEM-based polyplexes at 24 h (t test, p<0.05).
3.6.3 Macrophages
Similar to the observations made on DCs, infiltration of the injection site by macrophages occurred at least 1 day after injection (Supporting information, Fig. S3A). Co-localization of polyplexes with macrophages did not appear to be as substantial as with DCs, but did increase with time (Supporting information, Fig. S3B). Again, more PEGylated polyplexes co-localized with macrophages than non-PEGylated polyplexes, but the differences were not statistically significant.
3.6.4 Confocal microscopy at high magnification
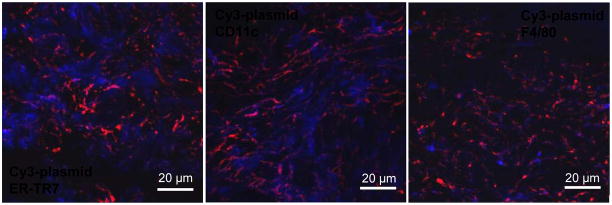
Microscopic visualization of plasmid distribution in skin tissue sections under relatively low magnification allowed large fields of view to be studied yet only revealed localization of plasmid in close proximity of the cells (Fig. 6, 7, 8). Therefore, the tissue sections were examined using confocal fluorescence microscopy at high magnification. Using PEGylated polyplexes at 24 h as an example, it is clearly that the Cy3-plasmid signal (red) was co-localizing with the signals of all three cell markers (blue), confirming without ambiguity that the polyplexes were interacting with the respective cell types (Fig. 9).
Fig. 9.
Dermal distribution of polyplexes relative to different cell types in skin tissue sections examined using confocal fluorescence microscopy at high magnification. The polyplexes shown were PEGylated and injected 24 h before imaging. Scale bar: 20 μm.
3.6 Draining lymph nodes
In addition to the injection sites, the draining lymph nodes were also harvested, sectioned, and analyzed by fluorescence microscopy to determine possible draining of Cy3-labeled naked plasmid and polyplexes. We found that naked plasmid signal was not seen at any time point. A few punctate signals were visible beginning at 12 h after polyplex injection and increased slightly 4 days after injection (Supporting information, Fig. S4). Overall, the presence of polyplexes in the draining lymph nodes was rare. The punctuate signals of DNA was not necessarily co-localized with DCs stained by CD11c (data not shown), suggesting that perhaps some polyplexes were able to migrate on their own through the lymphatic system.
4. Discussion
This study focused on examining transgene expression and local tissue distribution of naked plasmid DNA and polyplexes after intradermal injection into mice, aiming to uncover mechanisms and molecular design principles for more efficient polymer carriers for DNA vaccination in vivo. We paid special attention to the influence on dermal distribution of plasmid by using cationic polymer carriers with different molecular structures. With these studies we hope to shed light on the following three questions pertaining to polymer-mediated DNA vaccine delivery.
First, what are the benefits, if any, in using polyplexes rather than naked DNA for vaccination? Although it is well known that polymer carriers protect DNA from degradation and enhance cellular uptake in vitro, naked DNA vaccine alone (without a carrier but often with an adjuvant) has been quite effective in generating immune responses in vivo[17]. Here we have shown that at least from the standpoint of local tissue distribution of the DNA vaccine in the skin, it may be advantageous to use a cationic polymer carrier. Although naked plasmid distributed to larger areas in the skin than polyplexes (Fig. 4), it persisted for only a short period of time (a few hours) and was depleted before the arrival of substantial number of APCs (DCs and macrophages) (Fig. 6). In contrast, polyplexes formed aggregates at the injection site, serving as reservoirs of DNA that persisted for much longer periods of time (days and weeks) (Fig. 5) and enabling subsequent interactions between DNA and APCs, which infiltrated the injection site after 24 h (Fig. 7, 8, S3). Furthermore, the persistence of the plasmid delivered as polyplexes has apparently contributed to the persistence of in vivo transfection efficiency of polyplexes (Fig. 3).
Second, “to PEGylate or not to PEGylate”, that is still the question[18]. A popular drug delivery strategy, PEGylation has not yet been widely explored in DNA vaccine delivery nor has its in vivo mechanism of immune activation been clearly understood. PEGylation has long been used to enhance polyplex stability in high salt and serum environments.[8,19] Recently, Nomoto and colleagues observed directly the stabilization against aggregation by PEGylated polyplexes after intravenous injection in vivo.[20] We have carried out a similar experiment in vitro in simulated body fluid environment by incubating fluorescently labeled naked plasmid and polyplexes in complete cell culture medium containing serum. We visually observed substantial aggregation of non-PEGylated polyplexes and stabilization against aggregation by PEGylation (Fig. 2). Consistent with this in vitro finding, we found in live animals that the PEGylated polyplexes distributed to a broader area in the skin after intradermal injection than non-PEGylated polyplexes (Fig. 5). It was reported that PEGylated polyplexes and even naked DNA generated significantly more antigen expression and subsequent immune response than unprotected cationic polyplexes when delivered by tattooing the skin.[5] The proposed explanation by the authors of this report was the sequestration and deactivation of unprotected cationic polyplexes by negatively charged extracellular matrix[21]. While this is certainly a possibility, our in vitro and in vivo data here further suggest that limited diffusibility of large, highly aggregated polyplexes inside dermal tissue could be a major reason for the low efficiency of DNA vaccination reported earlier[5], and that PEGylated polyplexes would be advantageous due to their superior transport properties.
Third, what are the cells targeted in vivo by nonviral DNA vaccine? We showed that, due to its quick dispersion and transient presence at the injection site, naked plasmid has mostly co-localized with dermal fibroblasts rather than with APCs, which arrived at the site late (Fig. 6). On the other hand, aggregates of polyplexes lasted much longer and were able to interact with infiltrating APCs (Fig. 7 & 8), potentially leading to uptake and antigen expression. PEGylated polyplexes not only maintained a depot that attracted APC migration, but also appeared to spread more than non-PEGylated polyplexes, so as to encounter both incoming APCs and resident dermal fibroblasts (Fig. 9). Furthermore, in contrast to previous reports[10,22,23], we did not detect any direct draining of the naked plasmid into the lymph node. This could be due to variations of the amount of DNA injected, injection volume and pressure, tissue section thickness, and fixation method. More importantly, lymphatic draining of the polyplexes was low and was observed only after 12 h regardless which polymer was used (Fig. 9), suggesting that direct targeting of resident APCs in the lymph node was not a significant pathway for the polyplexes studied here, and perhaps for polyplexes in general either.
Taken together, several lessons can be learned from our studies that may help improve the design of cationic polymers for DNA vaccine delivery. Much attention has been given to transfecting DCs directly with antigen-encoding DNA, since endogenous expression of antigen by DCs has been shown to result in strong cellular immune response.[24–28] To target DCs directly, one could use a polymer (such as PEI or PAEM) that protects and retains DNA at the peripheral site of administration for a time period sufficiently long that allows DC infiltration and interaction. Comparing to the strategy of targeting DCs actively through DC-specific molecular ligands[4,9], creating a depot of plasmid at the periphery may be a more simple alternative approach to engage DCs. Furthermore, instead of transfecting DCs directly, cross-presentation through bystander cells has been suggested to be an important pathway in the development of a cellular immune response by DNA vaccine.[29–32] In this case, bystander cells (such as fibroblasts) can be transfected by polyplexes, express antigen, and be cross-presented by DCs through MHC-I molecules. To target the cross-presentation pathway, PEGylated polyplexes would be preferred because of their high stability, less aggregation, neutrally charged particle surface, and wider distribution among bystander cells. In fact, PEGylated carriers such as PEG-b-PAEM that combines good diffusivity in tissue with a moderate depot effect would be ideal for targeting DNA vaccine to both bystander cells and infiltrating APCs at the periphery, so as to exploit both pathways of cross-presentation and direct DC transfection. Thus, further studies should focus on exploring the relationship between different polymer carrier designs and antigen presentation as well as immune responses in vivo.
5. Conclusions
The transfection and local tissue distribution of plasmid DNA using different cationic polymers were analyzed after intradermal injection in mice. We found that naked DNA dispersed quickly in hours within the skin and showed limited co-localization with APCs. On the other hand, polyplexes formed depots at the injection site and persisted for days to engage and transfect skin cells. PEGylated polyplexes, in particular, possessed superior stability against aggregation than non-PEGylated polyplexes, and they disseminated well in the skin that promoted interaction with both the APCs and dermal fibroblasts. These findings provide in vivo evidence to support the use of PEGylated polymer carriers for DNA vaccine delivery and suggest possible approaches to further improve polymer design.
Supplementary Material
Acknowledgments
This work is partially supported by a grant from the NIH/NCI (R01CA129189) and a subcontract award from DOD (BC050156) through Washington University at St. Louis. We are grateful to the Biomedical Image Processing Lab (BIPL) at the University of Minnesota, in particular, John Oja for assistance with Maestro live animal imaging analysis, Kathy Pape and Sandy Johnson for helpful suggestions regarding tissue sectioning and immunostaining, and Prof. Bob Tranquillo for making a cryotome available for us to use.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Foged C, Sundblad A, Hovgaard L. Targeting vaccines to dendritic cells. Pharm Res. 2002;3:229–238. doi: 10.1023/a:1014474414097. [DOI] [PubMed] [Google Scholar]
- 2.Banchereau J, Briere F, Caux C, Davoust J, Lebecque S, Liu Y, et al. Immunobiology of dendritic cells. Annu Rev Immunol. 2000;18:767–811. doi: 10.1146/annurev.immunol.18.1.767. [DOI] [PubMed] [Google Scholar]
- 3.Ribas A. Genetically modified dendritic cells for cancer immunotherapy. Curr Gene Ther. 2005;5:619–628. doi: 10.2174/156652305774964758. [DOI] [PubMed] [Google Scholar]
- 4.Nguyen DN, Green JJ, Chan JM, Langer R, Anderson DG. Polymeric materials for gene delivery and DNA vaccination. Adv Mater. 2009;21:847–867. doi: 10.1002/adma.200801478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van den Berg JH, Nuijen B, Schumacher TN, Haanen JBAG, Storm G, Beijnen JH, et al. Synthetic vehicles for DNA vaccination. J Drug Targ. 2010;18:1–14. doi: 10.3109/10611860903278023. [DOI] [PubMed] [Google Scholar]
- 6.Dullaers M, Thielemans K. From pathogen to medicine: HIV-1-derived lentiviral vectors as vehicles for dendritic cell based cancer immunotherapy. J Gene Med. 2006;8:3–17. doi: 10.1002/jgm.846. [DOI] [PubMed] [Google Scholar]
- 7.O’Hagan DT, Singh M, Ulmer JB. Microparticles for the delivery of DNA vaccines. Immunol Rev. 2004;199:191–200. doi: 10.1111/j.0105-2896.2004.00153.x. [DOI] [PubMed] [Google Scholar]
- 8.Mintzer MA, Simanek EE. Nonviral vectors for gene delivery. Chem Rev. 2009;109:259–302. doi: 10.1021/cr800409e. [DOI] [PubMed] [Google Scholar]
- 9.Hubbell JA, Thomas SN, Swartz MA. Materials engineering for immunomodulation. Nature. 2009;462:449–460. doi: 10.1038/nature08604. [DOI] [PubMed] [Google Scholar]
- 10.Dupuis M, Denis-Mize K, Woo C, Goldbeck C, Selby MJ, Chen M, et al. Distribution of DNA vaccines determines their immunogenicity after intramuscular injection in mice. J Immunol. 2000;165:2850–2858. doi: 10.4049/jimmunol.165.5.2850. [DOI] [PubMed] [Google Scholar]
- 11.Drabick JJ, Glasspool-Malone J, Somiari S, King Robert AMW. Cutaneous transfection and immune responses to intradermal nucleic acid vaccination are significantly enhanced by in vivo electropermeabilization. Mol Ther. 2001;3:249–255. doi: 10.1006/mthe.2000.0257. [DOI] [PubMed] [Google Scholar]
- 12.Grønevik E, Tollefsen S, Sikkeland LIB, Haug T, Tjelle TE, Mathiesen I. DNA transfection of mononuclear cells in muscle tissue. J Gene Med. 2003;5:909–917. doi: 10.1002/jgm.416. [DOI] [PubMed] [Google Scholar]
- 13.Peachman KK, Rao M, Alving CR. Immunization with DNA through the skin. Methods. 2003;31:232–242. doi: 10.1016/s1046-2023(03)00137-3. [DOI] [PubMed] [Google Scholar]
- 14.Kuroda K, DeGrado WF. Amphiphilic polymethacrylate derivatives as antimicrobial agents. J Am Chem Soc. 2005;127:4128–4129. doi: 10.1021/ja044205+. [DOI] [PubMed] [Google Scholar]
- 15.Jankova K, Chen X, Kops J, Batsberg W. Synthesis of amphiphilic PS-b-PEG-b-PS by atom transfer radical polymerization. Macromolecules. 1998;31:538–541. [Google Scholar]
- 16.Tang R, Palumbo RN, Nagarajan L, Krogstad E, Wang C. Well-defined block copolymers for gene delivery to dendritic cells: Probing the effect of polycation chain-length. J Control Release. 2010;142:229–237. doi: 10.1016/j.jconrel.2009.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greenland JR, Letvin NL. Chemical adjuvants for plasmid DNA vaccines. Vaccine. 2007;25:3731–3741. doi: 10.1016/j.vaccine.2007.01.120. [DOI] [PubMed] [Google Scholar]
- 18.Park K. To PEGylate or not to PEGylate, that is not the question. J Controlled Release. 2010;142:147–148. doi: 10.1016/j.jconrel.2010.01.025. [DOI] [PubMed] [Google Scholar]
- 19.Lee M, Kim SW. Polyethylene glycol-conjugated copolymers for plasmid DNA delivery. Pharm Res. 2005;22:1–10. doi: 10.1007/s11095-004-9003-5. [DOI] [PubMed] [Google Scholar]
- 20.Nomoto T, Matsumoto Y, Miyata K, Oba M, Fukushima S, Nishiyama N, et al. In situ quantitative monitoring of polyplexes and polyplex micelles in the blood circulation using intravital real-time confocal laser scanning microscopy. J Controlled Release. doi: 10.1016/j.jconrel.2011.02.011. In Press, Corrected Proof. [DOI] [PubMed] [Google Scholar]
- 21.Burke RS, Pun SH. Extracellular barriers to in vivo PEI and PEGylated PEI polyplex-mediated gene delivery to the liver. Bioconjugate Chem. 2008;19:693–704. doi: 10.1021/bc700388u. [DOI] [PubMed] [Google Scholar]
- 22.Rush CM, Mitchell TJ, Garside P. A detailed characterisation of the distribution and presentation of DNA vaccine encoded antigen. Vaccine. 2010;28:1620–1634. doi: 10.1016/j.vaccine.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tuomela M, Malm M, Wallen M, Stanescu I, Krohn K, Peterson P. Biodistribution and general safety of a naked DNA plasmid, GTU®-MultiHIV, in a rat, using a quantitative PCR method. Vaccine. 2005;23:890–896. doi: 10.1016/j.vaccine.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 24.Hattori Y, Kawakami S, Suzuki S, Yamashita F, Hashida M. Enhancement of immune responses by DNA vaccination through targeted gene delivery using mannosylated cationic liposome formulations following intravenous administration. Biochem Biophys Res Comm. 2004;317:992–999. doi: 10.1016/j.bbrc.2004.03.141. [DOI] [PubMed] [Google Scholar]
- 25.Hattori Y, Kawakami S, Nakamura K, Yamashita F, Hashida M. Efficient gene transfer into macrophages and dendritic cells by in vivo gene delivery with mannosylated lipoplex via the intraperitoneal route. J Pharm Exp Ther. 2006;318:828–834. doi: 10.1124/jpet.106.105098. [DOI] [PubMed] [Google Scholar]
- 26.Un K, Kawakami S, Suzuki R, Maruyama K, Yamashita F, Hashida M. Enhanced transfection efficiency into macrophages and dendritic cells by a combination method using mannosylated lipoplexes and bubble liposomes with ultrasound exposure. Human Gene Therapy. 2010;21:65–74. doi: 10.1089/hum.2009.106. [DOI] [PubMed] [Google Scholar]
- 27.Elnekave M, Furmanov K, Nudel I, Arizon M, Clausen BE, Hovav A. Directly transfected Langerin+ dermal dendritic cells potentiate CD8+ T cell responses following intradermal plasmid DNA immunization. J Immunol. 2010;185:3463–3471. doi: 10.4049/jimmunol.1001825. [DOI] [PubMed] [Google Scholar]
- 28.Radcliffe JN, Roddick JS, Friedmann PS, Stevenson FK, Thirdborough SM. Prime-boost with alternating DNA vaccines designed to engage different antigen presentation pathways generates high frequencies of peptide-specific CD8 + T cells. J Immunol. 2006;177:6626–6633. doi: 10.4049/jimmunol.177.10.6626. [DOI] [PubMed] [Google Scholar]
- 29.Cao B, Bruder J, Kovesdi I, Huard J. Muscle stem cells can act as antigen-presenting cells: implication for gene therapy. Gene Therapy. 2004;11:1321–1330. doi: 10.1038/sj.gt.3302293. [DOI] [PubMed] [Google Scholar]
- 30.Ulmer JB, Deck RR, Dewitt CM, Donnelly JJ, Liu MA. Generation of MHC class I-restricted cytotoxic T lymphocytes by expression of a viral protein in muscle cells: antigen presentation by non-muscle cells. Immunology. 1996;89:59–67. doi: 10.1046/j.1365-2567.1996.d01-718.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cho JH, Youn JW, Sung YC. Cross-priming as a predominant mechanism for inducing CD8+ T cell responses in gene gun DNA immunization. J Immunol. 2001;167:5549–5559. doi: 10.4049/jimmunol.167.10.5549. [DOI] [PubMed] [Google Scholar]
- 32.Hon H, Oran A, Brocker T, Jacob J. B lymphocytes participate in cross-presentation of antigen following gene gun vaccination. J Immunol. 2005;174:5233–5242. doi: 10.4049/jimmunol.174.9.5233. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.