Abstract
Polycomb repressive complex 2 (PRC2) is a conserved multisubunit enzyme that methylates histone H3 on lysine-27. This chromatin modification is a hallmark of target genes transcriptionally silenced by the Polycomb system. At its core, PRC2 activity depends upon the SET domain active site of its catalytic subunit, EZH2, as well as critical stimulatory inputs from noncatalytic subunits, especially EED and SU(Z)12. We review recent progress on this core PRC2 machinery, including key features of the active site, control mechanisms that operate via EZH2 phosphorylation, and subunit elements and architectures that influence PRC2 function. Among these, we highlight work identifying an EED regulatory site that enables PRC2 to bind pre-existing methylated H3-K27 and stimulate enzyme output. These advances illuminate basic inner workings of PRC2 and also provide insights that could aid design of PRC2 inhibitors. The chromatin landscape that PRC2 encounters in vivo is decorated with many histone modifications that accompany active transcription, such as H3-K4 methylation. It has long been assumed that these "active" modifications oppose PRC2 at some level but, until recently, mechanisms of this antagonistic cross-talk have been elusive. We discuss new findings that illuminate how H3-K4 and H3-K36 methylation, H3-K27 acetylation, and H3-S28 phosphorylation each exert a negative impact on PRC2 function. The emerging picture presents PRC2 as a cooperative multipart machine, intricately outfitted to sense and respond to the local chromatin environment and other cues. This PRC2 design ensures flexibility and finetuning of its fundamental gene silencing roles in diverse biological contexts.
Keywords: chromatin, Polycomb, histone methyltransferase gene silencing, epigenetics
I. Introduction
Polycomb repressive complex 2 (PRC2) is an essential chromatin modifier conserved from plants to flies to humans (Pien and Grossniklaus 2007; Schuettengruber et al. 2007; Schwartz and Pirrotta 2007; Simon and Kingston 2009; Sawarkar and Paro 2010; Margueron and Reinberg 2011). Its central and best-studied function is to methylate histone H3 on lysine-27 (K27; Cao et al. 2002; Czermin et al. 2002; Kuzmichev et al. 2002; Muller et al. 2002), with the tri-methylated reaction product, H3-K27me3, constituting a common feature of repressed chromatin (Cao and Zhang 2004b; Schwartz et al. 2006; Schuettengruber et al. 2009; Filion et al. 2010). Located at over 2,000 sites in the mouse genome (Ku et al. 2008), PRC2 and H3-K27me3 have widespread roles in developmental processes of multicellular organisms and they are implicated in fundamental chromatin mechanisms that underlie stem cell regulatory circuits and cancer progression.
H3-K27 methylation is achieved through the highly cooperative action of four core PRC2 subunits (Fig. 1a). The catalytic subunit, E(Z) in Drosophila or EZH2 in humans, bears a SET domain which houses the enzyme active site (Rea et al. 2000; Dillon et al. 2005; Joshi et al. 2008). However, E(Z)/EZH2 is essentially inactive on its own and requires critical inputs from its partner subunits ESC/EED, SU(Z)12, and NURF55/RpAp48 (Cao and Zhang 2004a; Pasini et al. 2004; Ketel et al. 2005; Nekrasov et al. 2005). A major goal here is to review the inner workings and core organization of PRC2, including advances on understanding the PRC2 active site and mechanisms that control PRC2 activity through its catalytic or noncatalytic subunits. Comprehensive knowledge about PRC2 inner workings is essential to inform efforts to design small molecules that can specifically inhibit or modulate this key chromatin-modifying enzyme in cultured cells, animal models and, potentially, patients.
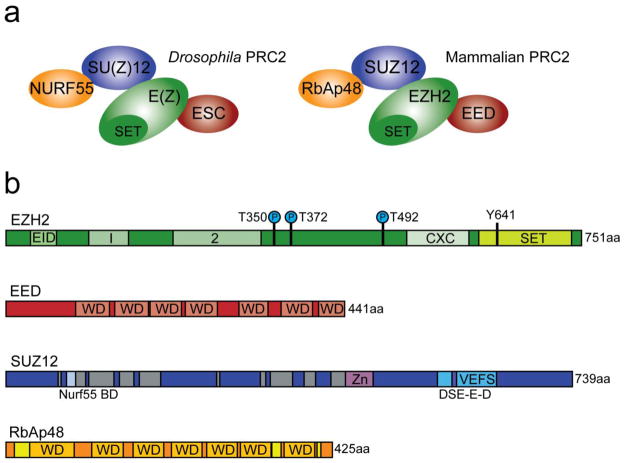
Fig. 1. PRC2 core complex.
a) Models of core fly and human PRC2 complexes. Subunit compositions and established contacts between subunits are depicted. Variants of human PRC2 resulting from alternative subunit usage are described elsewhere (Simon and Kingston 2009; Margueron and Reinberg 2011). b) Domain organizations of human PRC2 subunits. EZH2 contains a C-terminal SET domain, an adjacent cysteine-rich CXC domain, and additional conserved regions as indicated. EID denotes EED-interacting domain. EZH2 phosphorylation sites and the Y641 mutation discussed here are indicated. Both EED and RbAp48 are β-propeller proteins built from an array of seven WD repeats. RbAp48 regions in yellow contribute to the side pocket that can bind SUZ12 or histone H4. SUZ12 contains a C-terminal VEFS domain, C2H2 zinc finger, and an extended N-terminal region with conserved blocks (gray) from plants to human. An N-terminal SUZ12 region that binds NURF55 (Schmitges et al. 2011) is indicated. DSE-E-D indicates a conserved and highly charged subelement within the VEFS domain (Ketel et al. 2005).
The Polycomb repressed state, enriched for H3-K27me3, is functionally opposed by actively transcribed chromatin, which preferentially features "activating" modifications such as H3-K4me3, H3-K36me3, and acetylation of several histone tail lysines. Indeed, impairment of H3-K4 or H3-K36 histone-methylating enzymes leads to excess H3-K27 methylation in vivo (Papp and Muller 2006; Srinivasan et al. 2008). Widespread distribution of these alternative states is reinforced by recent genome-wide studies that define distinct Polycomb-silenced and active chromatin domains (Filion et al. 2010; Schwartz et al. 2010; Kharchenko et al. 2011). Correspondingly, there is molecular crosstalk whereby PRC2 senses activating modifications in the local chromatin environment via direct biochemical contact, and is consequently down-regulated. We will emphasize recent studies that illuminate how several antagonistic histone modifications can dampen or inhibit PRC2 function.
In this review, we focus on advances in understanding basic mechanisms and inputs to PRC2 function. This includes the core PRC2 machinery that deposits H3-K27me3, inner mechanisms that coordinate these working parts, and extrinsic signals and chromatin features that modulate PRC2 by impinging on core components. Besides the four central subunits, PRC2 is also impacted by additional partner proteins, which can be considered regulatory subunits that associate with the core complex. Chief among these are fly PCL and its mammalian homologs (Nekrasov et al. 2007; Cao et al. 2008; Sarma et al. 2008; Savla et al. 2008; Walker et al. 2010) and JARID2 (Peng et al. 2009; Shen et al. 2009; Landeira et al. 2010; Li et al. 2010; Pasini et al. 2010a), which help target PRC2 to chromatin sites of action as well as influence PRC2 enzyme activity. Another key class of PRC2 binding partners are non-coding RNAs (ncRNAs), such as HOTAIR and XIST, which are also implicated in PRC2 targeting (Rinn et al. 2007; Pandey et al. 2008; Zhao et al. 2008; Zhao et al. 2010). Roles of these protein and ncRNA cohorts in PRC2 chromatin recruitment have been recently reviewed (Hekimoglu and Ringrose 2009; Simon and Kingston 2009; Bracken and Helin 2009; Margueron and Reinberg 2011), so this topic will be only tangentially covered here. For recent progress on PRC2 roles in more biological and clinical contexts, readers are referred to alternative review articles that cover developmental and stem cell functions of PRC2 (Sparmann and van Lohuizen 2006; Rajasekhar and Begemann 2007; Pietersen and van Lohuizen 2008; Ito and Sun 2009) and the role of PRC2 in cancer epigenetics (Simon and Lange 2008; Bracken and Helin 2009; Chase and Cross 2011).
II: PRC2 Core Complex
Among components of the four-subunit PRC2 core complex (Fig. 1a), the SET domain of E(Z)/EZH2 is essential for enzyme function (Cao et al. 2002; Muller et al. 2002; Joshi et al. 2008) and two of the noncatalytic subunits, ESC/EED and SU(Z)12, are also vital for activity (Cao and Zhang 2004a; Pasini et al. 2004; Ketel et al. 2005; Nekrasov et al. 2005). The mechanisms by which these two critical subunits boost catalysis are not fully understood (see below). The fourth stoichiometric subunit, NURF55/RbAp48, also contributes; however, PRC2 subcomplexes lacking this subunit maintain substantial enzyme function (Cao and Zhang 2004a; Ketel et al. 2005), so it appears less central to the core methyltransferase. Another PRC2 partner, PCL/PHF1, has been implicated in specifically boosting the efficiency of the H3-K27me2 to H3-K27me3 conversion (Nekrasov et al. 2007; Sarma et al. 2008). Since recombinant PRC2 can generate H3-K27me3 without PCL/PHF1 (Nekrasov et al. 2007; Joshi et al. 2008; Sneeringer et al. 2010), this appears to be a modulatory rather than essential input to the core enzyme.
The critical inputs of multiple PRC2 subunits, together with their striking conservation from plants to humans (Whitcomb et al. 2007; Ito and Sun 2009; Sawarkar and Paro 2010), indicate that PRC2 is an ancient chromatin-modifying machine whose subunit architecture has preserved a functionally robust output. The precise nature of the subunit contacts and mechanisms that potentiate PRC2 activity have not yet been revealed. This stems partly from paucity of direct structural information on the PRC2 active site and the key functional interfaces of E(Z)/EZH2 with its partners. Nevertheless, recent progress has illuminated several aspects of the core PRC2 machine, including active site requirements, modulation by subunit phosphorylation, and the key stimulatory influence of the EED subunit. We review knowledge and advances on PRC2 inner workings by considering each core subunit.
E(Z)/EZH2
E(Z)/EZH2 contains a C-terminal SET domain plus several additional domains implicated in PRC2 function (Fig. 1b). In the context of assembled PRC2, this SET domain can perform three successive methyl transfer reactions, ultimating producing H3-K27me3. This contrasts with other types of HMTases whose capacity for methyl transfer is more limited; for example SET7/9 can only monomethylate naive histone lysine substrates (Xiao et al. 2003) and G9a, a dimethylase, lacks the ability to trimethylate the histone tail (Wu et al. 2010). Interestingly, a single amino acid change in its active site can convert G9a into a trimethyltransferase, which lacks mono- and di-methylation activity (Wu et al. 2010). This differential methylation capacity is governed by the precise configuration of the lysine substrate binding pocket, with active site tyrosine residues frequently implicated as key determinants (Collins et al. 2005; Couture et al. 2008). Correspondingly, recent progress pinpoints an active site tyrosine in determining the methylation capacity of EZH2.
Role of active site residue Y641 in differential control of mono/di/tri-methylation
Although EZH2 over-abundance has long been associated with tumor tissue samples (Varambally et al. 2002; Kleer et al. 2003; Simon and Lange 2008), somatic missense mutations that alter EZH2 in cancer cells have only recently been described. Analysis of certain lymphoma subtypes reveals recurrent mutations at a single EZH2 residue, Y641, within the SET domain (Morin et al. 2010). Based on analogy to structurally solved SET domains (Dillon et al. 2005), this active site tyrosine contacts the -amino group of the substrate lysine. Strikingly, these Y641 alterations shift the methylation capacity of EZH2; specifically, H3-K27 mono- and di-methylation is reduced but there is a concomitant surge in tri-methyltransferase activity (Sneeringer et al. 2010; Yap et al. 2011). Thus, the Y641 mutations appear to act via a gain-of-function mechanism, resulting in excess H3-K27me3 in tumor tissues. How does Y641 mutant EZH2 produce excess H3-K27me3 if it cannot efficiently generate the precursors H3-K27me1 and H3-K27me2? The answer lies in genetic coupling between wild-type and Y641 mutant EZH2, with wild-type supplying the precursor moieties and the mutant boosting the final step to H3-K27me3 (Fig. 2a). Indeed, the Y641 alleles are invariably recovered in lymphoma tissues in heterozygous form along with a wild-type allele. This fascinating disease mechanism is wholly consistent with genetic observations on Drosophila E(Z) from 20 years ago (Jones and Gelbart 1990). The mutant allele that first identified E(Z) (Wu et al. 1989) is a missense mutation that alters precisely the same active site tyrosine as Y641 (Y655; Joshi et al. 2008). Significantly, the gain-of-function behavior of this fly Y655N [E(z)1] allele requires wild-type E(z) activity in trans (Jones and Gelbart 1990), just as the oncogenic EZH2 Y641 mutations are functionally coupled with a wild-type EZH2 allele. This contrasts with another gain-of-function E(Z) active site mutation, R741K (E(z)Trm; Bajusz et al. 2001), which produces excess H3-K27me3 without coupling to wild-type E(z) (Stepanik and Harte 2012).
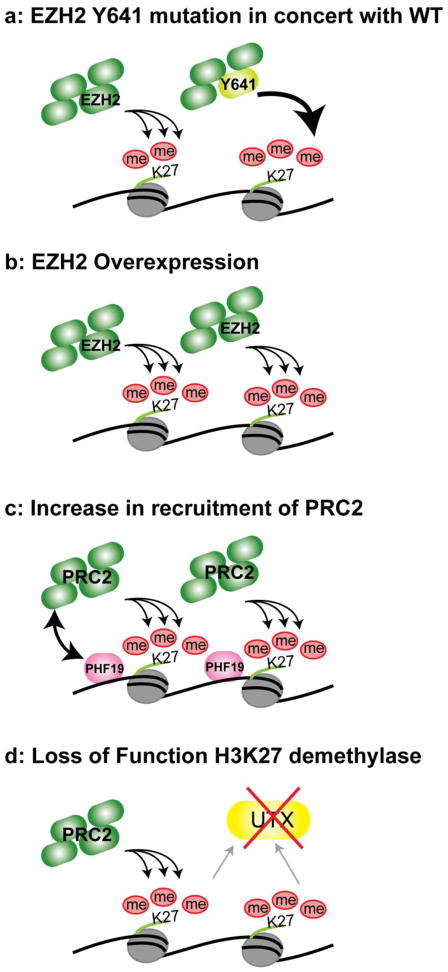
Fig. 2. Mechanisms that elevate H3K27me3 levels in cancer cells.
a) While deficient in mono- and di-methylase activity, PRC2 bearing EZH2-Y641 mutations has enhanced capacity for H3K27 tri-methylation. Acting in trans with wildtype PRC2, which can perform all three methyltransferase reactions, the Y641 gain-of-function leads to higher overall H3K27me3 levels (Sneeringer et al. 2010; Yap et al. 2011). b) Overexpression of the EZH2 catalytic subunit leads to higher overall H3K27me3 levels. As EZH2 alone is inactive, this is presumably due to a net increase in assembled PRC2 complexes. c) Overexpression of PHF19/PCL3 (Wang et al. 2004) or an interacting non-coding RNA (not shown, see Gupta et al. 2010) enhances recruitment of PRC2 to targets, leading to increased H3K27me3. d) Loss of function mutation of the demethylase UTX results in higher H3K27me3 levels (van Haaften et al. 2009). All four illustrated mechanisms can raise H3K27me3 levels and potentially contribute to hypersilencing of target genes in cancer.
These Y641 discoveries contribute to a unifying emerging view of the role of EZH2 and allied H3-K27 modifiers in cancer (Sneeringer et al. 2010; see Fig. 2). To date, cancer progression has been associated with the following related gain-of-function scenarios: 1) increases in EZH2 catalytic efficiency (Sneeringer et al. 2010; Yap et al. 2011), 2) EZH2 over-expression (Varambally et al. 2002; Kleer et al. 2003; Simon and Lange 2008), 3) overabundance of PCL family cofactors, such as PHF19, which promote H3-K27me3 accumulation (Wang et al. 2004), and 4) loss of the opposing H3-K27me3 demethylase, UTX (van Haaften et al. 2009). Thus, although diverse mechanisms are deployed, they all lead to the same outcome: abnormally elevated levels of H3-K27me3 in cancer cells.
Phosphorylation of EZH2 by cyclin-dependent kinases
An important emerging area concerns the mechanisms that modulate PRC2 activity, either positively or negatively, in response to intrinsic and extrinsic cellular cues (Sawarkar and Paro 2010). Among these cues, PRC2 activity has long been linked to cell cycle status through a role in promoting cell proliferation (Varambally et al. 2002; Bracken et al. 2003). Inputs to PRC2 activity by the cell cycle machinery have recently been described that operate via direct phosphorylation of EZH2 at multiple threonine residues (Chen et al. 2010; Kaneko et al. 2010; Wei et al. 2011; Wu and Zhang 2011). Specifically, cyclin-dependent kinases (CDKs) phosphorylate human EZH2 at T350 (T345 in mouse) and at T492 (T487 in mouse; Fig. 1b). The consequence of T350 phosphorylation is stimulation of H3-K27me3 deposition and hypersilencing at chromatin targets such as Hox loci (Chen et al. 2010). This occurs through increased PRC2 binding at target loci rather than by boosting intrinsic PRC2 enzyme activity (Chen et al. 2010; Kaneko et al. 2010). Consistent with this, T350 phosphorylation enhances interaction of EZH2 with ncRNAs, such as HOTAIR, implicated in PRC2 recruitment (Kaneko et al. 2010). Thus, CDK-mediated T350 phosphorylation positively impacts PRC2 function by augmenting its association with target chromatin. The relatively modest level of T350phos (only ~1% of EZH2 has T350phos in unphased cells) might suffice for ncRNAs to implement initial PRC2 targeting and H3-K27 methylation, which could prime subsequent binding of unmodified EZH2-PRC2 complexes that recognize and further propagate the repressive mark (Margueron et al. 2009; Kaneko et al. 2010; see below).
In contrast, EZH2 phosphorylation at T492 appears to negatively impact PRC2 function in cells (Wei et al. 2011). At the mechanistic level, T492 phosphorylation is reported to diminish PRC2 HMTase by disrupting EZH2 association with its required partner subunits, EED and SU(Z)12 (Wei et al. 2011). However, another group finds that a phospho-mimic change at the corresponding mouse residue (T487D) has no adverse effect on PRC2 assembly or HMTase (Kaneko et al. 2010). Another inconsistency is that CDK inhibition or knockdown was found to either increase (Chen et al. 2010) or diminish (Wei et al. 2011) Hox gene expression. Further work will be needed to reconcile these findings. It may be that the reported positive and negative influences due to phosphorylation on these two EZH2 residues operate in the same cell types but change dynamically, with one or the other predominating, during cell cycle progression. Alternatively, the discrepancies could reflect distinct regulatory mechanisms that control PRC2 in different cell types under study.
A new wrinkle in the EZH2 phosphorylation story is the observation that CDK phosphorylation of mouse EZH2 at T345 and T487 fosters ubiquitylation and proteosomal degradation of EZH2 (Wu and Zhang 2011). How can these seemingly disparate consequences (increased PRC2 chromatin binding versus PRC2 elimination) be reconciled? One possibility is that these outcomes reflect a multistep mechanism that limits the duration of the response to transiently elevated CDKs. Initially, T350 phosphorylation could trigger a transient increase in chromatin binding, thereby boosting local H3-K27me3, but then a rapid turndown is necessitated as CDK levels drop and the next cell cycle transition ensues. This scenario resembles "hit-and-run" control of transcription factors whereby activation is quickly followed by ubiquitylation and factor removal to finetune the response to transient cellular signals (Muratani and Tansey 2003).
Phosphorylation of EZH2 by mitogen-activated protein kinase (p38)
In addition to intrinsic cell cycle cues, examples of extrinsic signaling pathways that impact PRC2 are also emerging. The original example is the Akt pathway, which triggers EZH2 phosphorylation at serine-21, leading to reduced histone methylation by PRC2 (Cha et al. 2005). Much remains to be determined about how serine-21 phosphorylation is deployed in a physiological context since diverse cellular inputs can activate Akt kinases. More recently, another EZH2 phosphorylation site, T372, has been identified as a target of the mitogen-activated protein kinase, p38 (Palacios et al. 2010). Here the biological context is well-defined; the p38 pathway is activated in muscle stem cells in response to inflammatory cytokines that signal tissue damage. This leads to muscle regeneration by shifting gene expression programs to stimulate differentiation and dampen proliferation. At the mechanistic level, EZH2-T372 phosphorylation promotes interaction with the YY1 targeting protein, which recruits PRC2 to repressed targets, such as Pax7, in muscle cells (Caretti et al. 2004; Palacios et al. 2010). This boost in PRC2 recruitment and gene silencing resembles the outcome of T350 phosphorylation (see above), except in this case a protein rather than ncRNA recruiter is utilized.
The burst of new discoveries on EZH2 control by phosphorylation at multiple sites (summarized in Fig. 1b) has done much to connect this key chromatin modifier to upstream regulatory networks. Interestingly, mammalian EZH1, which is 65% identical to EZH2, lacks the conserved CDK phosphorylation sites found in EZH2 (Zeng et al. 2011). This differential capacity to respond to CDKs makes sense since EZH2 predominates in proliferating cells whereas EZH1 is more abundant in non-dividing cells (Margueron et al. 2008; Shen et al. 2008). It seems likely that further efforts will reveal additional mechanisms that link PRC2 epigenetic outputs to intrinsic and extrinsic cellular stimuli.
Inputs to EZH2 from core partner subunits
Among characterized SET domain proteins, there is a continuum ranging from those that can function as lysine methyltransferases on their own to those that require assembly into multiprotein complexes. At one extreme, the viral SET (vSET) protein is an active HMTase that consists essentially of an isolated SET domain (Qian et al. 2006). PRC2 lies at the other extreme, with its three noncatalytic core subunits required to stimulate activity, particularly the two (EED and SUZ12) that directly contact EZH2 (Fig. 1a). How these subunits potentiate EZH2 HMTase remains largely an open question. An instructive example is provided by another SET domain methyltransferase MLL1 which, like EZH2, depends heavily on partner subunits for optimal activity (Dou et al. 2006). Efficient MLL1 methylation of its H3-K4 target requires, at minimum, its partners RbBP5 and ASH2. A recently solved structure shows that, compared to stand-alone HMTases, the MLL1 active site is not properly configured for catalysis (Southall et al. 2009). Structural elements of the lysine substrate binding pocket are misaligned so that proper juxtaposition of methyl donor and -amino group acceptor cannot occur. Since addition of RbBP5 and ASH2 to the isolated MLL1 SET domain stimulates enzyme activity in vitro, the simplest explanation is that partner binding induces conformational changes that optimize the active site (Southall et al. 2009; Justin et al. 2010). Although comparable structural insight on the EZH2 active site is so far lacking, it seems reasonable to envision that similar mechanisms operate in PRC2. Indeed, recent work has revealed allosteric input of the EED subunit to PRC2 enzyme activity (see below).
ESC/EED
EED is a WD repeat protein (Fig. 1b) that folds into a seven-bladed β-propeller (Han et al. 2007; Margueron et al. 2009; Xu et al. 2010). β-propellers occur in functionally diverse proteins, with the donut-like structure typically providing a scaffold for interactions with partner proteins and effectors. Recent work shows that EED is equipped with a central pocket atop its β-propeller that binds specifically to trimethylated lysines (Margueron et al. 2009; Xu et al. 2010). Such a binding site is consistent with affinity for H3-K27me3 retained by PRC2 lacking a SET domain (Hansen et al. 2008). The EED top pocket, featuring a conserved aromatic cage, is configured for preferential binding to repressive chromatin marks, including H3-K27me3, H3-K9me3 and H4-K20me3, as opposed to "activating" marks such as H3-K4me3. This discrimination relies on residues flanking the methylatable lysine, thereby exploiting the ARKS motifs at repressive positions K9 and K27. PRC2 binding to tri-methylated H3 peptides boosts its HMTase activity, with the H3-K27me3 peptide delivering the most robust stimulation (Margueron et al. 2009; Xu et al. 2010). Thus, EED harbors a site for allosteric PRC2 stimulation via binding the H3-K27me3 reaction product. Importantly, this may provide a mechanism to maintain or propagate H3-K27 methylation in chromatin regions where this repressive mark pre-exists (Hansen et al. 2008; Margueron et al. 2009). Consistent with this, mutational disruption of the aromatic cage in the fly homolog, ESC, reduces H3-K27me3 levels in vivo (Margueron et al. 2009).
Several observations suggest that the EED/ESC subunit does more than just sense and convey local H3-K27me3 concentration. First, whereas PRC2 activity is boosted 3 to 7-fold by addition of H3-K27me3 peptides (Margueron et al. 2009; Xu et al. 2010), complete loss of the EED/ESC subunit causes quantitatively more severe impairment of PRC2 HMTase (Ketel et al. 2005; Nekrasov et al. 2005). A basic stimulatory role is further suggested by the requirement for ESC in PRC2 catalysis upon recombinant substrates lacking pre-methylated H3-K27 (A. Rai and J.A.S., unpubl. results). Second, (Xu et al. 2010) show that an alternative histone modification, H1-K26me3, also binds the EED aromatic cage but that, in this case, the impact is to inhibit PRC2 HMTase. Although H1-K26 methylation by PRC2 has been described (Kuzmichev et al. 2004), its potential impact on PcG silencing remains to be determined. Thus, more work is needed to fully reveal contributions of the ESC/EED subunit to PRC2 activity and regulation. To date, structure/function studies have established at least these three modules within ESC/EED: 1) the β-propeller top pocket that binds H3-K27me3 (Margueron et al. 2009; Xu et al. 2010), 2) another interface on the bottom of the β-propeller that binds an N-terminal portion of EZH2 (EID in Fig. 1b; Han et al. 2007), and 3) an N-terminal tail, outside the β-propeller, that can bind the histone-fold domain of H3 (Tie et al. 2007).
SU(Z)12
The domain organization of SU(Z)12 is depicted in Fig. 1b. The C-terminal VEFS domain mediates stable binding to EZH2/E(Z), thereby promoting PRC2 assembly. This VEFS interaction is strikingly conserved from plants to flies to mammals (Chanvivattana et al. 2004; Yamamoto et al. 2004; Ketel et al. 2005). Beyond simply providing subunit contact, the VEFS domain also likely plays an instructive or allosteric role in controlling PRC2 enzyme activity. Alteration of a conserved and highly charged subelement of the SU(Z)12 VEFS domain (Fig. 1b) disrupts PRC2 HMTase while preserving complex assembly (Ketel et al. 2005). This suggests that the VEFS domain has stimulatory influence. Furthermore, recent findings implicate SU(Z)12 in allosteric inhibition of PRC2 HMTase by H3-K4me3 (Schmitges et al. 2011; see below). This enzyme antagonism is attributed to a C-terminal portion of SU(Z)12 encompassing VEFS. Thus, the VEFS domain provides a key surface for EZH2 contact that could help optimize the PRC2 active site and also mediate regulatory inputs that control enzyme output. Structural information on VEFS domain architecture and the nature and consequences of its contact with EZH2/E(Z) should do much to illuminate underlying mechanisms.
Other SU(Z)12 domains include a single C2H2 zinc finger of unknown molecular function and an extended N-terminal region bearing scattered conserved subelements (Fig. 1b). Consistent with these conserved blocks, the N-terminal domain appears to subdivide into several functional modules; the region spanning amino acids 79–91 of fly SU(Z)12 provides a binding surface for the NURF55 subunit (Schmitges et al. 2011) and residue 274 is altered in the partial loss-of-function fly Su(z)122 mutant (Birve et al. 2001).
Besides its contributions to core PRC2, there is also evidence that SU(Z)12 mediates interactions with PRC2 cofactors such as Jarid2 (Peng et al. 2009). Moreover, SU(Z)12 is the PRC2 subunit with the strongest affinity for a set of short ncRNAs emanating from the 5' ends of repressed target genes (Kanhere et al. 2010). Further investigation of these SU(Z)12 interactions, particularly in vivo, is required to reveal how they contribute to PRC2 function and regulation.
NURF55/RpAp48
Like EED, NURF55 is a WD repeat protein that forms a seven-bladed β-propeller (Song et al. 2008). However, NURF55 is functionally distinct from the other three PRC2 subunits in two major ways. First, whereas EED, EZH2, and SUZ12 appear dedicated to PcG silencing, NURF55 is present in many chromatin-modifying complexes with diverse functions. Besides PRC2, NURF55 appears in an ISWI-class nucleosome remodeling complex (NURF), a CHD-class remodeling complex (NuRD), chromatin assembly factor 1 (CAF1), and in histone acetyltransferase and deacetylase complexes (Suganuma et al. 2008). Second, NURF55 is not essential for robust PRC2 histone methyltransferase. Loss or impairment of any of the other three PRC2 subunits causes dramatic reduction of K27 methylation in vitro (Cao and Zhang 2004a; Pasini et al. 2004; Ketel et al. 2005; Nekrasov et al. 2005), whereas removal of NURF55 yields a trimeric complex that retains robust activity, estimated at merely ~2 to 3-fold less than intact core complex on polynucleosome substrates (Cao and Zhang 2004a; Ketel et al. 2005). This dispensability for catalytic function, together with widespread deployment in other chromatin modifiers, underscores a long-standing puzzle: just what does NURF55 do in the PRC2 complex?
One long-appreciated feature of NURF55 is its ability to bind histone H4 through contact with helix 1 of the histone fold (Verreault et al. 1997). Details of this interaction were revealed by a co-crystal structure, which defines a specialized side pocket of the NURF55 β-propeller that binds this H4 helix (Song et al. 2008). Although the NURF55 side pocket contributes to activity of a HAT complex (Song et al. 2008), it was not clear how it could function in PRC2 since H4 helix 1 is not accessible when packaged in nucleosomes. This conundrum appears resolved by more recent structural studies demonstrating that this same NURF55 side pocket also binds to an N-terminal portion of SU(Z)12, in a manner that likely precludes H4 interaction (Nowak et al. 2011; Schmitges et al. 2011). Thus, in PRC2, the NURF55 side pocket is likely used for SU(Z)12 contact and complex assembly rather than histone binding.
WD repeat proteins commonly feature cavities or channels sculpted on the β-propeller top surface for protein interactions. In WD repeat chromatin proteins, these top pockets are often configured to bind histone tails (Suganuma et al. 2008), as exemplified by WDR5, which functions in H3-K4 methyltransferases (Ruthenburg et al. 2006), and EED (Margueron et al. 2009; discussed above). With satisfactory understanding of the specialized NURF55 side pocket achieved, attention has shifted to the more commonly utilized top surface. Recent structural and biochemical studies reveal a NURF55 top channel that binds to the histone H3 tail peptide, residues 1-15 (Nowak et al. 2011; Schmitges et al. 2011). Occupancy of the top and side pockets are independent such that, in PRC2, NURF55 can simultaneously bind the extreme H3 tail and SU(Z)12. What role could NURF55 binding to H3(1-15) play in a complex that methylates more proximally at H3-K27? Surprisingly, disruption of NURF55-H3 tail interaction has little or no effect on either PRC2 histone methyltransferase or overall affinity for nucleosomes (Schmitges et al. 2011). This emphasizes the intricate engineering of the PRC2 machine, which likely has several nucleosome-contacting surfaces and clearly contains multiple inputs that impact HMTase. Currently, the best guess about NURF55-H3 tail binding is that it might help PRC2 sense substrates bearing inhibitory H3-K4me3 (Schmitges et al. 2011; see below), but functional tests have yet to substantiate this.
These recent advances provide key pieces of the NURF55 puzzle. We now appreciate that NURF55 provides a multifunctional platform for histone and PRC2 subunit contacts. How these and potentially other NURF55 elements are integrated for function within PRC2 remains to be determined. In vivo tests will be needed to complement the structural and biochemical insights. In this regard, recently described fly NURF55 alleles (Anderson et al. 2011), one of which likely disrupts the SU(Z)12-binding side pocket, could be useful.
III. Control of PRC2 by Antagonistic Histone Modifications
Antagonism by H3-K4 and H3-K36 methylations
Histone methylation at positions H3-K4 and H3-K36 are hallmarks of actively transcribed genes. Their distributions along gene bodies tend to differ, with H3-K4me3 enriched at promoter/transcription start site regions, H3-K36me2 most abundant in 5'-coding regions, and H3-K36me3 peaking towards the 3' ends of coding regions (Barski et al. 2007; Bell et al. 2007; Barrand et al. 2010). H3-K36me2/3 accumulation in coding regions parallels association of H3-K36 methyltransferase with elongating RNA pol II (Li et al. 2003). There is abundant evidence that these K4 and K36 methylations oppose Polycomb silencing. Indeed, Drosophila trithorax (TRX), a K4-methyltransferase (Smith et al. 2004) and ASH1, a K36-methyltransferase (Tanaka et al. 2007; Yuan et al. 2011), were established as genetic suppressors of Polycomb mutations long before their enzyme functions were defined (Kennison and Tamkun 1988; Shearn 1989). Moreover, genetic and genome-wide studies imply that Drosophila TRX and ASH1 are dedicated antagonizers of PcG silencing, operating specifically at PcG target sites rather than as general activators of transcription (Klymenko and Muller 2004; Schwartz et al. 2010).
Two new studies shed light on molecular mechanisms underlying this antagonism (Schmitges et al. 2011; Yuan et al. 2011). The basic finding is that PRC2 methylation at H3-K27 is inhibited on substrates with pre-existing H3-K4me3 or H3-K36me2/3 (see Fig. 3a). Importantly, pre-installed H3-K4me3 does not impair PRC2 binding to substrate nucleosomes, suggesting an allosteric inhibitory mechanism that instead dampens catalytic efficiency (Schmitges et al. 2011). Another key finding is that PRC2 inhibition requires that the K4 and K36 methylations appear on the same histone tail subject to K27 methylation (Schmitges et al. 2011). Consistent with this cis requirement, mass spectrometric analysis shows that K27me3 rarely co-exists with K4me3 or K36me2/3 on the same histone tails in vivo (Young et al. 2009; Yuan et al. 2011). The cis requirement also distinguishes this inhibitory mechanism from the stimulatory effect of K27me3 on PRC2 (see above), which can operate in trans (Margueron et al. 2009). Although the PRC2 modules that mediate K4me3 and K36me2/3 inhibition have not yet been pinpointed, the evidence so far suggests that both inhibitory inputs operate through SU(Z)12 (Schmitges et al. 2011).
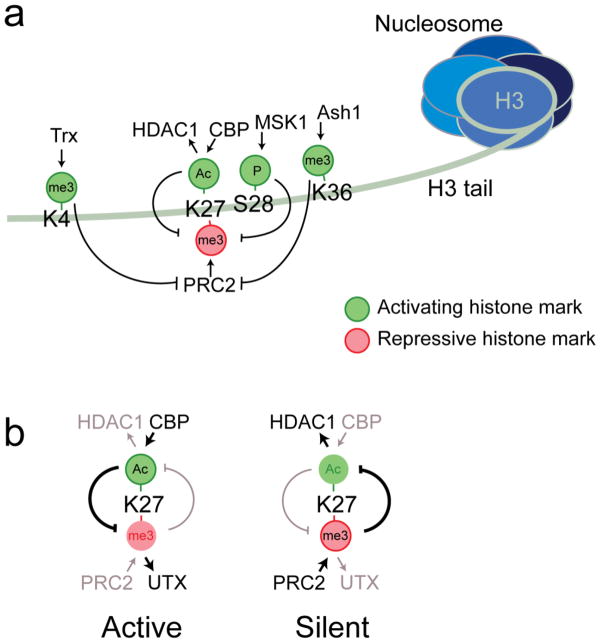
Fig. 3. Histone H3 tail modifications and crosstalk that impact PRC2.
a) PRC2 tri-methylates K27, whereas CBP acetylates K27. MSK1 phosphorylates S28, which antagonizes PRC2 and may work in concert with CBP to acetylate K27. H3K4me3 deposited by Trithorax and H3K36me2/3 deposited by Ash1 inhibit PRC2 activity. b) Methylation and acetylation of K27 is mutually exclusive. The diagram on the left depicts K27 in its active state, with CBP delivering K27Ac. UTX is required to remove K27me3 before this acetylation can occur. The diagram on the right depicts K27 in its silent state, with PRC2 delivering K27me3. HDAC1 is required to remove K27ac before PRC2 can trimethylate it.
Taken together, these recent findings reveal that PRC2 is equipped to sense its chromatin environment and adjust activity accordingly (Margueron et al. 2009; Schmitges et al. 2011; Yuan et al. 2011); if PRC2 senses actively transcribed chromatin then its methyltransferase is inhibited and, conversely, repressed local chromatin stimulates PRC2. This built-in feedback mechanism is presumably circumvented or dampened in the case of "bivalent" chromatin domains, described in embryonic stem cells, which feature concomitant accumulation of H3-K4me3 and H3-K27me3 (Bernstein et al. 2006; Mikkelsen et al. 2007; Min et al. 2011).
Antagonism by H3-K27 acetylation
Since acetylation of lysine side chains is biochemically incompatible with their methylation, a theoretically simple way to impede PRC2 is via direct H3-K27 acetylation of substrate nucleosomes. Indeed, H3-K27 acetylation is generally enriched in promoters and coding regions of active genes in metazoans (Garcia et al. 2007; Wang et al. 2008). New findings now establish that H3-K27 acetylation does functionally oppose PcG silencing in both fly and mammalian systems (Tie et al. 2009; Pasini et al. 2010b; Schwartz et al. 2010).
In Drosophila, the major HAT that targets H3-K27 is CREB-binding protein (CBP; Tie et al. 2009). Similarly, among 17 mammalian HATs tested, the two orthologs CBP and p300 were preferentially implicated in H3-K27 acetylation (Pasini et al. 2010b). This K27 HAT identification then enabled loss and over-expression studies, which generally reveal inverse accumulations of H3-K27Ac and H3-K27me3 in vivo. For example, CBP knockdown depletes K27Ac and increases K27me3 (Tie et al. 2009) whereas PRC2 loss depletes K27me3 with concomitant gain of K27Ac (Tie et al. 2009; Pasini et al. 2010b). There is also intriguing complementarity of these dueling K27 modifications during Drosophila development, with K27Ac peaking in early embryos and then declining, whereas H3-K27me3 gradually accumulates as embyogenesis proceeds (Tie et al. 2009). The opposing nature of K27Ac and K27me3 is further revealed by ChIP analysis of selected PcG targets during ES cell differentiation (Pasini et al. 2010b) and in a more extensive genome-wide analysis of alternative chromatin states (Schwartz et al. 2010).
These advances on K27 acetylation may also impact some long-standing issues in field. First, histone deacetylases (HDACs) have previously been implicated in PcG silencing (van Der Vlag and Otte 1999; Tie et al. 2001) but their mechanistic contributions have not been defined. One straightforward role for HDACs, then, would be to remove acetyl groups from K27 side chains to render them methylatable by PRC2 (Fig. 3b). Second, the first-characterized fly TRX complex, called TAC1, has both CBP HAT and H3-K4 methyltransferase activities (Petruk et al. 2001; Smith et al. 2004) and physical interaction of mammalian TRX (MLL) with CBP has long been described (Ernst et al. 2001). This raises the intriguing possibility that delivery of K4me3 and K27Ac, which both oppose PRC2, could be molecularly coordinated.
Antagonism by H3-S28 phosphorylation
H3-K27 is immediately flanked by serine-28 (S28), which is known to be phosphorylated during mitosis and also during interphase (Dunn and Davie 2005; Dyson et al. 2005). This proximity begs the question of whether S28 phosphorylation impacts PRC2 function and PcG silencing. By analogy, at the similarly arranged K9-S10 module of histone H3, S10 phosphorylation can displace heterochromatin protein 1 (HP1) recruited to the H3 tail via affinity for neighboring methylated K9 (Fischle et al. 2005; Hirota et al. 2005). Two recent studies now establish that S28 phosphorylation can profoundly disrupt PcG silencing and identify the kinase(s) responsible (Gehani et al. 2010; Lau and Cheung 2011).
The development of an antibody that recognizes doubly modified K27me3-S28phos showed that S28 phosphorylation accumulates on PcG targets in response to mitogen- and stress-activated kinase pathways (Gehani et al. 2010). This chromatin response is triggered by several pathway inputs, including retinoic acid and mitogenic stimuli, with MSK1 and MSK2 implicated as the kinases that modify S28. The molecular consequence is displacement of PRC2 and its partnering PcG complex, PRC1, with concomitant desilencing of PcG targets.
These findings are reinforced by complementary studies of an engineered MSK1 directly tethered to reporter constructs and endogenous gene targets (Lau and Cheung 2011). Here, MSK1 targeting also leads to S28 phosphorylation, displacement of PRC1 and PRC2, and gene desilencing. Since kinase-dead MSK1 cannot trigger this response, the transcriptional activator function of MSK1 is directly tied to its phosphorylation of H3-S28. Moreover, MSK1-mediated S28 phosphorylation correlates with conversion of K27me3 to K27Ac (Lau and Cheung 2011), suggesting that S28phos and K27Ac are coupled to oppose PRC2 and PcG silencing. In this scenario, K27me3-S28phos could be an intermediate which, followed by K27 demethylation and acetylation, produces an active chromatin state marked by K27Ac-S28phos. Conversely, one might also expect to find a nuclear phosphatase that removes S28phos, thereby synergizing with PRC2 in target gene silencing.
Taken together, these studies suggest a molecular switch that deploys S28 phosphorylation to convert silenced PcG targets from OFF to ON. It is tempting to speculate that such a switch could operate during ES cell differentiation since both activated MSK1 and S28 phosphorylation occur in human ES cells (Gehani et al. 2010). The mechanism by which S28phos impacts PRC2 remains to be defined. Since structural studies imply that S28 is not a key determinant of H3 peptide interactions with the EED top pocket (Margueron et al. 2009; Xu et al. 2010), it seems likely that other PRC2 sites, perhaps within the SET domain itself, are deployed to sense and respond to S28 phosphorylation.
Conclusions: Towards inner workings of PRC2 brake and accelerator mechanisms
In summary, recent progress has identified key features of the chromatin landscape that modulate PRC2 function; we now appreciate that H3-K4me3, H3-K36me2/3, H3-K27Ac, and H3-S28phos can directly antagonize PRC2 whereas pre-existing H3-K27me3 can stimulate PRC2 activity. These advances contribute to a more complete picture of how PcG target chromatin is configured for robust silencing, full activation, or more balanced states in between (Papp and Muller 2006; Schuettengruber et al. 2009; Filion et al. 2010; Schwartz et al. 2010). Furthermore, biochemical and structural studies are shedding light on the precise mechanisms by which these chromatin marks, and other extrinsic cues, apply the brakes or engage the accelerator within the PRC2 machine. It is clear that the EED and SU(Z)12 subunits are crucial cogs and that they help execute slowdowns and speedups as required by cellular conditions and local chromatin terrain. Nevertheless, important puzzles remain. To fully reveal operations under the hood, we need to know the three-dimensional architecture of the EZH2 active site as well as functional elements of key EED-EZH2 and SU(Z)12-EZH2 interfaces that presumably influence this active site. Protein dynamics studies should also address how newly identified PRC2 allosteric sites could deliver functional impact via potential conformational changes. The intricate workings of PRC2 have been finetuned by more than a billion years of evolution (Whitcomb et al. 2007; Sawarkar and Paro 2010). With recent advances, the prospects are favorable for viewing a high-resolution parts diagram and operations blueprint in the not-too-distant future.
Prospectus: Consequences of PRC2 output
Despite major progress on deciphering upstream inputs that control PRC2, the mechanistic consequences of its chromatin output, H3-K27me3, remain largely unresolved. Precisely how does acquisition of H3-K27me3 on local nucleosomes lead to gene silencing? One longstanding idea is that H3-K27me3 could help create a landing pad to recruit the partnering PcG complex, Polycomb repressive complex 1 (PRC1), to local chromatin (reviewed in Simon and Kingston 2009; Margueron and Reinberg 2011). In this scenario, the central job of PRC2 could be viewed as paving the way for PRC1 association which, in turn, implements the key events in gene silencing. However, it is far from clear that PRC1 recruitment is the sole or even main function of H3-K27me3. Alternative roles for H3-K27me3 in gene silencing could include: 1) preventing accumulation of H3-K27Ac and other "activating" histone acetylations (Tie et al. 2009; Pasini et al. 2010b), 2) blocking deposition of other activating modifications, such as H3-K4me3 or H3-K36me2/3, or 3) antagonizing the functions of remodeling complexes (Wilson et al. 2010).
Ultimately, to define precise mechanisms, discrete steps in the transcription cycle impacted by PRC2 and H3-K27me3 will need to be identified (Fuda et al. 2009). Thus, mid-gene RNA pol II elongation could be impeded by H3-K27me3-modified nucleosomes and/or polII could be impacted in 5' regions at the steps of promoter binding, initiation, promoter escape, or promoter-proximal pausing. These are not mutually exclusive possibilities and there could be inhibitory effects at several steps. Indeed, the broad domains of H3-K27me3 accumulation (Schwartz et al. 2010), together with PRC2 inhibition by both promoter-enriched (H3-K4me3) and 3'-region enriched (H3-K36me3) modifications (Schmitges et al. 2011; Yuan et al. 2011; see above), suggest that PRC2 is functionally deployed along the entire extent of gene bodies.
Recently, highly regulated genes in metazoans have been shown to accumulate promoter-occupied nucleosomes and promoter-proximal paused RNA pol II (Gilchrist et al. 2010), suggesting these two features as commonly used control points. Since most PcG target genes likely belong to this class, this chromatin architecture presents a baseline for considering mechanisms of PcG silencing complexes. Indeed, recent studies in Drosophila and mammalian cells have begun to dissect the relevant transcription cycle steps, with evidence emerging for PRC2 impact upon both polII promoter occupancy and post-initiation events (Chopra et al. 2011; Min et al. 2011). These new in vivo studies, which exploit powerful combinations of genetic, genomic, and molecular approaches, provide key steps forward in the quest to reveal PcG silencing mechanisms. With progress in this field now poised for rapid advances, further insights on both inputs and outputs of PRC2 should do much to define its ancient and fundamental roles in epigenome regulation.
References
- Anderson AE, Karandikar UC, Pepple KL, Chen Z, Bergmann A, Mardon G. The enhancer of trithorax and polycomb gene Caf1/p55 is essential for cell survival and patterning in Drosophila development. Development. 2011;138(10):1957–1966. doi: 10.1242/dev.058461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajusz I, Sipos L, Gyorgypal Z, Carrington EA, Jones RS, Gausz J, Gyurkovics H. The Trithorax-mimic allele of Enhancer of zeste renders active domains of target genes accessible to polycomb-group-dependent silencing in Drosophila melanogaster. Genetics. 2001;159(3):1135–1150. doi: 10.1093/genetics/159.3.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrand S, Andersen IS, Collas P. Promoter-exon relationship of H3 lysine 9, 27, 36 and 79 methylation on pluripotency-associated genes. Biochem Biophys Res Commun. 2010;401(4):611–617. doi: 10.1016/j.bbrc.2010.09.116. [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129(4):823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Bell O, Wirbelauer C, Hild M, Scharf AN, Schwaiger M, MacAlpine DM, Zilbermann F, van Leeuwen F, Bell SP, Imhof A, Garza D, Peters AH, Schubeler D. Localized H3K36 methylation states define histone H4K16 acetylation during transcriptional elongation in Drosophila. EMBO J. 2007;26(24):4974–4984. doi: 10.1038/sj.emboj.7601926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125(2):315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Birve A, Sengupta AK, Beuchle D, Larsson J, Kennison JA, Rasmuson-Lestander A, Muller J. Su(z)12, a novel Drosophila Polycomb group gene that is conserved in vertebrates and plants. Development. 2001;128(17):3371–3379. doi: 10.1242/dev.128.17.3371. [DOI] [PubMed] [Google Scholar]
- Bracken AP, Helin K. Polycomb group proteins: navigators of lineage pathways led astray in cancer. Nat Rev Cancer. 2009;9(11):773–784. doi: 10.1038/nrc2736. [DOI] [PubMed] [Google Scholar]
- Bracken AP, Pasini D, Capra M, Prosperini E, Colli E, Helin K. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J. 2003;22(20):5323–5335. doi: 10.1093/emboj/cdg542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Wang H, He J, Erdjument-Bromage H, Tempst P, Zhang Y. Role of hPHF1 in H3K27 methylation and Hox gene silencing. Mol Cell Biol. 2008;28(5):1862–1872. doi: 10.1128/MCB.01589-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298(5595):1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- Cao R, Zhang Y. SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED-EZH2 complex. Mol Cell. 2004a;15(1):57–67. doi: 10.1016/j.molcel.2004.06.020. [DOI] [PubMed] [Google Scholar]
- Cao R, Zhang Y. The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr Opin Genet Dev. 2004b;14(2):155–164. doi: 10.1016/j.gde.2004.02.001. [DOI] [PubMed] [Google Scholar]
- Caretti G, Di Padova M, Micales B, Lyons GE, Sartorelli V. The Polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev. 2004;18(21):2627–2638. doi: 10.1101/gad.1241904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha TL, Zhou BP, Xia W, Wu Y, Yang CC, Chen CT, Ping B, Otte AP, Hung MC. Akt-mediated phosphorylation of EZH2 suppresses methylation of lysine 27 in histone H3. Science. 2005;310(5746):306–310. doi: 10.1126/science.1118947. [DOI] [PubMed] [Google Scholar]
- Chanvivattana Y, Bishopp A, Schubert D, Stock C, Moon YH, Sung ZR, Goodrich J. Interaction of Polycomb-group proteins controlling flowering in Arabidopsis. Development. 2004;131(21):5263–5276. doi: 10.1242/dev.01400. [DOI] [PubMed] [Google Scholar]
- Chase A, Cross NC. Aberrations of EZH2 in cancer. Clin Cancer Res. 2011;17(9):2613–2618. doi: 10.1158/1078-0432.CCR-10-2156. [DOI] [PubMed] [Google Scholar]
- Chen S, Bohrer LR, Rai AN, Pan Y, Gan L, Zhou X, Bagchi A, Simon JA, Huang H. Cyclin-dependent kinases regulate epigenetic gene silencing through phosphorylation of EZH2. Nat Cell Biol. 2010;12(11):1108–1114. doi: 10.1038/ncb2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra VS, Hendrix DA, Core LJ, Tsui C, Lis JT, Levine M. The polycomb group mutant esc leads to augmented levels of paused Pol II in the Drosophila embryo. Mol Cell. 2011;42(6):837–844. doi: 10.1016/j.molcel.2011.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins RE, Tachibana M, Tamaru H, Smith KM, Jia D, Zhang X, Selker EU, Shinkai Y, Cheng X. In vitro and in vivo analyses of a Phe/Tyr switch controlling product specificity of histone lysine methyltransferases. J Biol Chem. 2005;280(7):5563–5570. doi: 10.1074/jbc.M410483200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couture JF, Dirk LM, Brunzelle JS, Houtz RL, Trievel RC. Structural origins for the product specificity of SET domain protein methyltransferases. Proc Natl Acad Sci U S A. 2008;105(52):20659–20664. doi: 10.1073/pnas.0806712105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell. 2002;111(2):185–196. doi: 10.1016/s0092-8674(02)00975-3. [DOI] [PubMed] [Google Scholar]
- Dillon SC, Zhang X, Trievel RC, Cheng X. The SET-domain protein superfamily: protein lysine methyltransferases. Genome Biol. 2005;6(8):227. doi: 10.1186/gb-2005-6-8-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou Y, Milne TA, Ruthenburg AJ, Lee S, Lee JW, Verdine GL, Allis CD, Roeder RG. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat Struct Mol Biol. 2006;13(8):713–719. doi: 10.1038/nsmb1128. [DOI] [PubMed] [Google Scholar]
- Dunn KL, Davie JR. Stimulation of the Ras-MAPK pathway leads to independent phosphorylation of histone H3 on serine 10 and 28. Oncogene. 2005;24(21):3492–3502. doi: 10.1038/sj.onc.1208521. [DOI] [PubMed] [Google Scholar]
- Dyson MH, Thomson S, Inagaki M, Goto H, Arthur SJ, Nightingale K, Iborra FJ, Mahadevan LC. MAP kinase-mediated phosphorylation of distinct pools of histone H3 at S10 or S28 via mitogen- and stress-activated kinase 1/2. J Cell Sci. 2005;118(Pt 10):2247–2259. doi: 10.1242/jcs.02373. [DOI] [PubMed] [Google Scholar]
- Ernst P, Wang J, Huang M, Goodman RH, Korsmeyer SJ. MLL and CREB bind cooperatively to the nuclear coactivator CREB-binding protein. Mol Cell Biol. 2001;21(7):2249–2258. doi: 10.1128/MCB.21.7.2249-2258.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filion GJ, van Bemmel JG, Braunschweig U, Talhout W, Kind J, Ward LD, Brugman W, de Castro IJ, Kerkhoven RM, Bussemaker HJ, van Steensel B. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell. 2010;143(2):212–224. doi: 10.1016/j.cell.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischle W, Tseng BS, Dormann HL, Ueberheide BM, Garcia BA, Shabanowitz J, Hunt DF, Funabiki H, Allis CD. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature. 2005;438(7071):1116–1122. doi: 10.1038/nature04219. [DOI] [PubMed] [Google Scholar]
- Fuda NJ, Ardehali MB, Lis JT. Defining mechanisms that regulate RNA polymerase II transcription in vivo. Nature. 2009;461:186–192. doi: 10.1038/nature08449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia BA, Hake SB, Diaz RL, Kauer M, Morris SA, Recht J, Shabanowitz J, Mishra N, Strahl BD, Allis CD, Hunt DF. Organismal differences in post-translational modifications in histones H3 and H4. J Biol Chem. 2007;282(10):7641–7655. doi: 10.1074/jbc.M607900200. [DOI] [PubMed] [Google Scholar]
- Gehani SS, Agrawal-Singh S, Dietrich N, Christophersen NS, Helin K, Hansen K. Polycomb group protein displacement and gene activation through MSK-dependent H3K27me3S28 phosphorylation. Mol Cell. 2010;39(6):886–900. doi: 10.1016/j.molcel.2010.08.020. [DOI] [PubMed] [Google Scholar]
- Gilchrist DA, Dos Santos G, Fargo DC, Xie B, Gao Y, Li L, Adelman K. Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell. 2010;143(4):540–551. doi: 10.1016/j.cell.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S, Chang HY. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464(7291):1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Z, Xing X, Hu M, Zhang Y, Liu P, Chai J. Structural basis of EZH2 recognition by EED. Structure. 2007;15(10):1306–1315. doi: 10.1016/j.str.2007.08.007. [DOI] [PubMed] [Google Scholar]
- Hansen KH, Bracken AP, Pasini D, Dietrich N, Gehani SS, Monrad A, Rappsilber J, Lerdrup M, Helin K. A model for transmission of the H3K27me3 epigenetic mark. Nat Cell Biol. 2008;10(11):1291–1300. doi: 10.1038/ncb1787. [DOI] [PubMed] [Google Scholar]
- Hekimoglu B, Ringrose L. Non-coding RNAs in polycomb/trithorax regulation. RNA Biol. 2009;6(2) doi: 10.4161/rna.6.2.8178. [DOI] [PubMed] [Google Scholar]
- Hirota T, Lipp JJ, Toh BH, Peters JM. Histone H3 serine 10 phosphorylation by Aurora B causes HP1 dissociation from heterochromatin. Nature. 2005;438(7071):1176–1180. doi: 10.1038/nature04254. [DOI] [PubMed] [Google Scholar]
- Ito T, Sun B. Epigenetic regulation of developmental timing in floral stem cells. Epigenetics. 2009;4(8):564–567. doi: 10.4161/epi.4.8.10351. [DOI] [PubMed] [Google Scholar]
- Jones RS, Gelbart WM. Genetic analysis of the enhancer of zeste locus and its role in gene regulation in Drosophila melanogaster. Genetics. 1990;126(1):185–199. doi: 10.1093/genetics/126.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi P, Carrington EA, Wang L, Ketel CS, Miller EL, Jones RS, Simon JA. Dominant alleles identify SET domain residues required for histone methyltransferase of Polycomb repressive complex 2. J Biol Chem. 2008;283(41):27757–27766. doi: 10.1074/jbc.M804442200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justin N, De Marco V, Aasland R, Gamblin SJ. Reading, writing and editing methylated lysines on histone tails: new insights from recent structural studies. Curr Opin Struct Biol. 2010;20(6):730–738. doi: 10.1016/j.sbi.2010.09.012. [DOI] [PubMed] [Google Scholar]
- Kaneko S, Li G, Son J, Xu CF, Margueron R, Neubert TA, Reinberg D. Phosphorylation of the PRC2 component Ezh2 is cell cycle-regulated and up-regulates its binding to ncRNA. Genes Dev. 2010;24(23):2615–2620. doi: 10.1101/gad.1983810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennison JA, Tamkun JW. Dosage-dependent modifiers of polycomb and antennapedia mutations in Drosophila. Proc Natl Acad Sci U S A. 1988;85(21):8136–8140. doi: 10.1073/pnas.85.21.8136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketel CS, Andersen EF, Vargas ML, Suh J, Strome S, Simon JA. Subunit contributions to histone methyltransferase activities of fly and worm polycomb group complexes. Mol Cell Biol. 2005;25(16):6857–6868. doi: 10.1128/MCB.25.16.6857-6868.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharchenko PV, Alekseyenko AA, Schwartz YB, Minoda A, Riddle NC, Ernst J, Sabo PJ, Larschan E, Gorchakov AA, Gu T, Linder-Basso D, Plachetka A, Shanower G, Tolstorukov MY, Luquette LJ, Xi R, Jung YL, Park RW, Bishop EP, Canfield TK, Sandstrom R, Thurman RE, MacAlpine DM, Stamatoyannopoulos JA, Kellis M, Elgin SC, Kuroda MI, Pirrotta V, Karpen GH, Park PJ. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature. 2011;471(7339):480–485. doi: 10.1038/nature09725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleer CG, Cao Q, Varambally S, Shen R, Ota I, Tomlins SA, Ghosh D, Sewalt RG, Otte AP, Hayes DF, Sabel MS, Livant D, Weiss SJ, Rubin MA, Chinnaiyan AM. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci U S A. 2003;100(20):11606–11611. doi: 10.1073/pnas.1933744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klymenko T, Muller J. The histone methyltransferases Trithorax and Ash1 prevent transcriptional silencing by Polycomb group proteins. EMBO Rep. 2004;5(4):373–377. doi: 10.1038/sj.embor.7400111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, Presser A, Nusbaum C, Xie X, Chi AS, Adli M, Kasif S, Ptaszek LM, Cowan CA, Lander ES, Koseki H, Bernstein BE. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 2008;4(10):e1000242. doi: 10.1371/journal.pgen.1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmichev A, Jenuwein T, Tempst P, Reinberg D. Different EZH2-containing complexes target methylation of histone H1 or nucleosomal histone H3. Mol Cell. 2004;14(2):183–193. doi: 10.1016/s1097-2765(04)00185-6. [DOI] [PubMed] [Google Scholar]
- Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002;16(22):2893–2905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landeira D, Sauer S, Poot R, Dvorkina M, Mazzarella L, Jorgensen HF, Pereira CF, Leleu M, Piccolo FM, Spivakov M, Brookes E, Pombo A, Fisher C, Skarnes WC, Snoek T, Bezstarosti K, Demmers J, Klose RJ, Casanova M, Tavares L, Brockdorff N, Merkenschlager M, Fisher AG. Jarid2 is a PRC2 component in embryonic stem cells required for multi-lineage differentiation and recruitment of PRC1 and RNA Polymerase II to developmental regulators. Nat Cell Biol. 2010;12(6):618–624. doi: 10.1038/ncb2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau PN, Cheung P. Histone code pathway involving H3 S28 phosphorylation and K27 acetylation activates transcription and antagonizes polycomb silencing. Proc Natl Acad Sci U S A. 2011;108(7):2801–2806. doi: 10.1073/pnas.1012798108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Howe L, Anderson S, Yates JR, 3rd, Workman JL. The Set2 histone methyltransferase functions through the phosphorylated carboxyl-terminal domain of RNA polymerase II. J Biol Chem. 2003;278(11):8897–8903. doi: 10.1074/jbc.M212134200. [DOI] [PubMed] [Google Scholar]
- Li G, Margueron R, Ku M, Chambon P, Bernstein BE, Reinberg D. Jarid2 and PRC2, partners in regulating gene expression. Genes Dev. 2010;24(4):368–380. doi: 10.1101/gad.1886410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ, 3rd, Voigt P, Martin SR, Taylor WR, De Marco V, Pirrotta V, Reinberg D, Gamblin SJ. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature. 2009;461(7265):762–767. doi: 10.1038/nature08398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Li G, Sarma K, Blais A, Zavadil J, Woodcock CL, Dynlacht BD, Reinberg D. Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol Cell. 2008;32(4):503–518. doi: 10.1016/j.molcel.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469(7330):343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, Lee W, Mendenhall E, O'Donovan A, Presser A, Russ C, Xie X, Meissner A, Wernig M, Jaenisch R, Nusbaum C, Lander ES, Bernstein BE. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448(7153):553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min IM, Waterfall JJ, Core LJ, Munroe RJ, Schimenti J, Lis JT. Regulating RNA polymerase pausing and transcription elongation in embryonic stem cells. Genes Dev. 2011;25(7):742–754. doi: 10.1101/gad.2005511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin RD, Johnson NA, Severson TM, Mungall AJ, An J, Goya R, Paul JE, Boyle M, Woolcock BW, Kuchenbauer F, Yap D, Humphries RK, Griffith OL, Shah S, Zhu H, Kimbara M, Shashkin P, Charlot JF, Tcherpakov M, Corbett R, Tam A, Varhol R, Smailus D, Moksa M, Zhao Y, Delaney A, Qian H, Birol I, Schein J, Moore R, Holt R, Horsman DE, Connors JM, Jones S, Aparicio S, Hirst M, Gascoyne RD, Marra MA. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet. 2010;42(2):181–185. doi: 10.1038/ng.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, Miller EL, O'Connor MB, Kingston RE, Simon JA. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell. 2002;111(2):197–208. doi: 10.1016/s0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- Muratani M, Tansey WP. How the ubiquitin-proteasome system controls transcription. Nat Rev Mol Cell Biol. 2003;4(3):192–201. doi: 10.1038/nrm1049. [DOI] [PubMed] [Google Scholar]
- Nekrasov M, Klymenko T, Fraterman S, Papp B, Oktaba K, Kocher T, Cohen A, Stunnenberg HG, Wilm M, Muller J. Pcl-PRC2 is needed to generate high levels of H3-K27 trimethylation at Polycomb target genes. EMBO J. 2007;26(18):4078–4088. doi: 10.1038/sj.emboj.7601837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrasov M, Wild B, Muller J. Nucleosome binding and histone methyltransferase activity of Drosophila PRC2. EMBO Rep. 2005;6:348–353. doi: 10.1038/sj.embor.7400376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak AJ, Alfieri C, Stirnimann CU, Rybin V, Baudin F, Ly-Hartig N, Lindner D, Muller CW. Chromatin-modifying complex component Nurf55/p55 associates with histones H3 and H4 and polycomb repressive complex 2 subunit Su(z)12 through partially overlapping binding sites. J Biol Chem. 2011;286(26):23388–23396. doi: 10.1074/jbc.M110.207407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios D, Mozzetta C, Consalvi S, Caretti G, Saccone V, Proserpio V, Marquez VE, Valente S, Mai A, Forcales SV, Sartorelli V, Puri PL. TNF/p38alpha/polycomb signaling to Pax7 locus in satellite cells links inflammation to the epigenetic control of muscle regeneration. Cell Stem Cell. 2010;7(4):455–469. doi: 10.1016/j.stem.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, Komorowski J, Nagano T, Mancini-Dinardo D, Kanduri C. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell. 2008;32(2):232–246. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- Papp B, Muller J. Histone trimethylation and the maintenance of transcriptional ON and OFF states by trxG and PcG proteins. Genes Dev. 2006;20(15):2041–2054. doi: 10.1101/gad.388706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Bracken AP, Jensen MR, Lazzerini Denchi E, Helin K. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J. 2004;23(20):4061–4071. doi: 10.1038/sj.emboj.7600402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Cloos PA, Walfridsson J, Olsson L, Bukowski JP, Johansen JV, Bak M, Tommerup N, Rappsilber J, Helin K. JARID2 regulates binding of the Polycomb repressive complex 2 to target genes in ES cells. Nature. 2010a;464(7286):306–310. doi: 10.1038/nature08788. [DOI] [PubMed] [Google Scholar]
- Pasini D, Malatesta M, Jung HR, Walfridsson J, Willer A, Olsson L, Skotte J, Wutz A, Porse B, Jensen ON, Helin K. Characterization of an antagonistic switch between histone H3 lysine 27 methylation and acetylation in the transcriptional regulation of Polycomb group target genes. Nucleic Acids Res. 2010b;38(15):4958–4969. doi: 10.1093/nar/gkq244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng JC, Valouev A, Swigut T, Zhang J, Zhao Y, Sidow A, Wysocka J. Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell. 2009;139(7):1290–1302. doi: 10.1016/j.cell.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petruk S, Sedkov Y, Smith S, Tillib S, Kraevski V, Nakamura T, Canaani E, Croce CM, Mazo A. Trithorax and dCBP Acting in a Complex to Maintain Expression of a Homeotic Gene. Science. 2001;294(5545):1331–1334. doi: 10.1126/science.1065683. [DOI] [PubMed] [Google Scholar]
- Pien S, Grossniklaus U. Polycomb group and trithorax group proteins in Arabidopsis. Biochim Biophys Acta. 2007;1769(5–6):375–382. doi: 10.1016/j.bbaexp.2007.01.010. [DOI] [PubMed] [Google Scholar]
- Pietersen AM, van Lohuizen M. Stem cell regulation by polycomb repressors: postponing commitment. Curr Opin Cell Biol. 2008;20(2):201–207. doi: 10.1016/j.ceb.2008.01.004. [DOI] [PubMed] [Google Scholar]
- Qian C, Wang X, Manzur K, Sachchidanand, Farooq A, Zeng L, Wang R, Zhou MM. Structural insights of the specificity and catalysis of a viral histone H3 lysine 27 methyltransferase. J Mol Biol. 2006;359(1):86–96. doi: 10.1016/j.jmb.2006.03.006. [DOI] [PubMed] [Google Scholar]
- Rajasekhar VK, Begemann M. Concise review: roles of polycomb group proteins in development and disease: a stem cell perspective. Stem Cells. 2007;25(10):2498–2510. doi: 10.1634/stemcells.2006-0608. [DOI] [PubMed] [Google Scholar]
- Rea S, Eisenhaber F, O'Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, Jenuwein T. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406(6796):593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129(7):1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruthenburg AJ, Wang W, Graybosch DM, Li H, Allis CD, Patel DJ, Verdine GL. Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex. Nat Struct Mol Biol. 2006;13(8):704–712. doi: 10.1038/nsmb1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarma K, Margueron R, Ivanov A, Pirrotta V, Reinberg D. Ezh2 requires PHF1 to efficiently catalyze H3 lysine 27 trimethylation in vivo. Mol Cell Biol. 2008;28(8):2718–2731. doi: 10.1128/MCB.02017-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savla U, Benes J, Zhang J, Jones RS. Recruitment of Drosophila Polycomb-group proteins by Polycomblike, a component of a novel protein complex in larvae. Development. 2008 doi: 10.1242/dev.016006. [DOI] [PubMed] [Google Scholar]
- Sawarkar R, Paro R. Interpretation of developmental signaling at chromatin: the Polycomb perspective. Dev Cell. 2010;19(5):651–661. doi: 10.1016/j.devcel.2010.10.012. [DOI] [PubMed] [Google Scholar]
- Schmitges FW, Prusty AB, Faty M, Stutzer A, Lingaraju GM, Aiwazian J, Sack R, Hess D, Li L, Zhou S, Bunker RD, Wirth U, Bouwmeester T, Bauer A, Ly-Hartig N, Zhao K, Chan H, Gu J, Gut H, Fischle W, Muller J, Thoma NH. Histone methylation by PRC2 is inhibited by active chromatin marks. Mol Cell. 2011;42(3):330–341. doi: 10.1016/j.molcel.2011.03.025. [DOI] [PubMed] [Google Scholar]
- Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128(4):735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- Schuettengruber B, Ganapathi M, Leblanc B, Portoso M, Jaschek R, Tolhuis B, van Lohuizen M, Tanay A, Cavalli G. Functional anatomy of polycomb and trithorax chromatin landscapes in Drosophila embryos. PLoS Biol. 2009;7(1):e13. doi: 10.1371/journal.pbio.1000013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz YB, Kahn TG, Nix DA, Li XY, Bourgon R, Biggin M, Pirrotta V. Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nat Genet. 2006;38(6):700–705. doi: 10.1038/ng1817. [DOI] [PubMed] [Google Scholar]
- Schwartz YB, Kahn TG, Stenberg P, Ohno K, Bourgon R, Pirrotta V. Alternative epigenetic chromatin states of polycomb target genes. PLoS Genet. 2010;6(1):e1000805. doi: 10.1371/journal.pgen.1000805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz YB, Pirrotta V. Polycomb silencing mechanisms and the management of genomic programmes. Nat Rev Genet. 2007;8(1):9–22. doi: 10.1038/nrg1981. [DOI] [PubMed] [Google Scholar]
- Shearn A. The ash-1, ash-2 and trithorax genes of Drosophila melanogaster are functionally related. Genetics. 1989;121(3):517–525. doi: 10.1093/genetics/121.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Kim W, Fujiwara Y, Simon MD, Liu Y, Mysliwiec MR, Yuan GC, Lee Y, Orkin SH. Jumonji modulates polycomb activity and self-renewal versus differentiation of stem cells. Cell. 2009;139(7):1303–1314. doi: 10.1016/j.cell.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Liu Y, Hsu YJ, Fujiwara Y, Kim J, Mao X, Yuan GC, Orkin SH. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol Cell. 2008;32(4):491–502. doi: 10.1016/j.molcel.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol. 2009;10(10):697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- Simon JA, Lange CA. Roles of the EZH2 histone methyltransferase in cancer epigenetics. Mutat Res. 2008;647(1–2):21–29. doi: 10.1016/j.mrfmmm.2008.07.010. [DOI] [PubMed] [Google Scholar]
- Smith ST, Petruk S, Sedkov Y, Cho E, Tillib S, Canaani E, Mazo A. Modulation of heat shock gene expression by the TAC1 chromatin-modifying complex. Nat Cell Biol. 2004;6(2):162–167. doi: 10.1038/ncb1088. [DOI] [PubMed] [Google Scholar]
- Sneeringer CJ, Scott MP, Kuntz KW, Knutson SK, Pollock RM, Richon VM, Copeland RA. Coordinated activities of wild-type plus mutant EZH2 drive tumor-associated hypertrimethylation of lysine 27 on histone H3 (H3K27) in human B-cell lymphomas. Proc Natl Acad Sci U S A. 2010;107(49):20980–20985. doi: 10.1073/pnas.1012525107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JJ, Garlick JD, Kingston RE. Structural basis of histone H4 recognition by p55. Genes Dev. 2008;22(10):1313–1318. doi: 10.1101/gad.1653308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southall SM, Wong PS, Odho Z, Roe SM, Wilson JR. Structural basis for the requirement of additional factors for MLL1 SET domain activity and recognition of epigenetic marks. Mol Cell. 2009;33(2):181–191. doi: 10.1016/j.molcel.2008.12.029. [DOI] [PubMed] [Google Scholar]
- Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer. 2006;6(11):846–856. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- Srinivasan S, Dorighi KM, Tamkun JW. Drosophila Kismet regulates histone H3 lysine 27 methylation and early elongation by RNA polymerase II. PLoS Genet. 2008;4(10):e1000217. doi: 10.1371/journal.pgen.1000217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanik VA, Harte PJ. A mutation in the E(Z) methyltransferase that increases trimethylation of histone H3 lysine 27 and causes inappropriate silencing of active Polycomb target genes. Dev Biol. 2012 doi: 10.1016/j.ydbio.2011.12.007. in press. [DOI] [PubMed] [Google Scholar]
- Suganuma T, Pattenden SG, Workman JL. Diverse functions of WD40 repeat proteins in histone recognition. Genes Dev. 2008;22(10):1265–1268. doi: 10.1101/gad.1676208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Katagiri Z, Kawahashi K, Kioussis D, Kitajima S. Trithorax-group protein ASH1 methylates histone H3 lysine 36. Gene. 2007;397(1–2):161–168. doi: 10.1016/j.gene.2007.04.027. [DOI] [PubMed] [Google Scholar]
- Tie F, Banerjee R, Stratton CA, Prasad-Sinha J, Stepanik V, Zlobin A, Diaz MO, Scacheri PC, Harte PJ. CBP-mediated acetylation of histone H3 lysine 27 antagonizes Drosophila Polycomb silencing. Development. 2009;136(18):3131–3141. doi: 10.1242/dev.037127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tie F, Furuyama T, Prasad-Sinha J, Jane E, Harte PJ. The Drosophila Polycomb Group proteins ESC and E(Z) are present in a complex containing the histone-binding protein p55 and the histone deacetylase RPD3. Development. 2001;128(2):275–286. doi: 10.1242/dev.128.2.275. [DOI] [PubMed] [Google Scholar]
- Tie F, Stratton CA, Kurzhals RL, Harte PJ. The N terminus of Drosophila ESC binds directly to histone H3 and is required for E(Z)-dependent trimethylation of H3 lysine 27. Mol Cell Biol. 2007;27(6):2014–2026. doi: 10.1128/MCB.01822-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329(5992):689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Der Vlag J, Otte AP. Transcriptional repression mediated by the human polycomb-group protein EED involves histone deacetylation. Nat Genet. 1999;23(4):474–478. doi: 10.1038/70602. [DOI] [PubMed] [Google Scholar]
- van Haaften G, Dalgliesh GL, Davies H, Chen L, Bignell G, Greenman C, Edkins S, Hardy C, O'Meara S, Teague J, Butler A, Hinton J, Latimer C, Andrews J, Barthorpe S, Beare D, Buck G, Campbell PJ, Cole J, Forbes S, Jia M, Jones D, Kok CY, Leroy C, Lin ML, McBride DJ, Maddison M, Maquire S, McLay K, Menzies A, Mironenko T, Mulderrig L, Mudie L, Pleasance E, Shepherd R, Smith R, Stebbings L, Stephens P, Tang G, Tarpey PS, Turner R, Turrell K, Varian J, West S, Widaa S, Wray P, Collins VP, Ichimura K, Law S, Wong J, Yuen ST, Leung SY, Tonon G, DePinho RA, Tai YT, Anderson KC, Kahnoski RJ, Massie A, Khoo SK, Teh BT, Stratton MR, Futreal PA. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nat Genet. 2009;41(5):521–523. doi: 10.1038/ng.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt RG, Otte AP, Rubin MA, Chinnaiyan AM. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419(6907):624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- Verreault A, Kaufman PD, Kobayashi R, Stillman B. Nucleosomal DNA regulates the core-histone-binding subunit of the human Hat1 acetyltransferase. Curr Biol. 1997;8(2):96–108. doi: 10.1016/s0960-9822(98)70040-5. [DOI] [PubMed] [Google Scholar]
- Walker E, Chang WY, Hunkapiller J, Cagney G, Garcha K, Torchia J, Krogan NJ, Reiter JF, Stanford WL. Polycomb-like 2 associates with PRC2 and regulates transcriptional networks during mouse embryonic stem cell self-renewal and differentiation. Cell Stem Cell. 2010;6(2):153–166. doi: 10.1016/j.stem.2009.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Robertson GP, Zhu J. A novel human homologue of Drosophila polycomblike gene is up-regulated in multiple cancers. Gene. 2004;343(1):69–78. doi: 10.1016/j.gene.2004.09.006. [DOI] [PubMed] [Google Scholar]
- Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, Zhao K. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40(7):897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Chen YH, Li LY, Lang J, Yeh SP, Shi B, Yang CC, Yang JY, Lin CY, Lai CC, Hung MC. CDK1-dependent phosphorylation of EZH2 suppresses methylation of H3K27 and promotes osteogenic differentiation of human mesenchymal stem cells. Nat Cell Biol. 2011;13(1):87–94. doi: 10.1038/ncb2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitcomb SJ, Basu A, Allis CD, Bernstein E. Polycomb Group proteins: an evolutionary perspective. Trends Genet. 2007;23(10):494–502. doi: 10.1016/j.tig.2007.08.006. [DOI] [PubMed] [Google Scholar]
- Wilson BG, Wang X, Shen X, McKenna ES, Lemieux ME, Cho YJ, Koellhoffer EC, Pomeroy SL, Orkin SH, Roberts CW. Epigenetic antagonism between polycomb and SWI/SNF complexes during oncogenic transformation. Cancer Cell. 2010;18(4):316–328. doi: 10.1016/j.ccr.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CT, Jones RS, Lasko PF, Gelbart WM. Homeosis and the interaction of zeste and white in Drosophila. Mol Gen Genet. 1989;218(3):559–564. doi: 10.1007/BF00332424. [DOI] [PubMed] [Google Scholar]
- Wu H, Min J, Lunin VV, Antoshenko T, Dombrovski L, Zeng H, Allali-Hassani A, Campagna-Slater V, Vedadi M, Arrowsmith CH, Plotnikov AN, Schapira M. Structural biology of human H3K9 methyltransferases. PLoS One. 2010;5(1):e8570. doi: 10.1371/journal.pone.0008570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SC, Zhang Y. Cyclin-dependent kinase 1 (CDK1)-mediated phosphorylation of enhancer of zeste 2 (Ezh2) regulates its stability. J Biol Chem. 2011;286(32):28511–28519. doi: 10.1074/jbc.M111.240515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao B, Jing C, Wilson JR, Walker PA, Vasisht N, Kelly G, Howell S, Taylor IA, Blackburn GM, Gamblin SJ. Structure and catalytic mechanism of the human histone methyltransferase SET7/9. Nature. 2003;421(6923):652–656. doi: 10.1038/nature01378. [DOI] [PubMed] [Google Scholar]
- Xu C, Bian C, Yang W, Galka M, Ouyang H, Chen C, Qiu W, Liu H, Jones AE, MacKenzie F, Pan P, Li SS, Wang H, Min J. Binding of different histone marks differentially regulates the activity and specificity of polycomb repressive complex 2 (PRC2) Proc Natl Acad Sci U S A. 2010;107(45):19266–19271. doi: 10.1073/pnas.1008937107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K, Sonoda M, Inokuchi J, Shirasawa S, Sasazuki T. Polycomb group suppressor of zeste 12 links heterochromatin protein 1alpha and enhancer of zeste 2. J Biol Chem. 2004;279(1):401–406. doi: 10.1074/jbc.M307344200. [DOI] [PubMed] [Google Scholar]
- Yap DB, Chu J, Berg T, Schapira M, Cheng SW, Moradian A, Morin RD, Mungall AJ, Meissner B, Boyle M, Marquez VE, Marra MA, Gascoyne RD, Humphries RK, Arrowsmith CH, Morin GB, Aparicio SA. Somatic mutations at EZH2 Y641 act dominantly through a mechanism of selectively altered PRC2 catalytic activity, to increase H3K27 trimethylation. Blood. 2011;117(8):2451–2459. doi: 10.1182/blood-2010-11-321208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young NL, DiMaggio PA, Plazas-Mayorca MD, Baliban RC, Floudas CA, Garcia BA. High throughput characterization of combinatorial histone codes. Mol Cell Proteomics. 2009;8(10):2266–2284. doi: 10.1074/mcp.M900238-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan W, Xu M, Huang C, Liu N, Chen S, Zhu B. H3K36 methylation antagonizes PRC2-mediated H3K27 methylation. J Biol Chem. 2011;286(10):7983–7989. doi: 10.1074/jbc.M110.194027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X, Chen S, Huang H. Phosphorylation of EZH2 by CDK1 and CDK2: a possible regulatory mechanism of transmission of the H3K27me3 epigenetic mark through cell divisions. Cell Cycle. 2011;10(4):579–583. doi: 10.4161/cc.10.4.14722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Ohsumi TK, Kung JT, Ogawa Y, Grau DJ, Sarma K, Song JJ, Kingston RE, Borowsky M, Lee JT. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell. 2010;40(6):939–953. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322(5902):750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]