Summary
Argonaute proteins are required for the biogenesis of some small RNAs (sRNAs), including the PIWI-interacting RNAs and some microRNAs. How Argonautes mediate maturation of sRNAs independent of their slicer activity is not clear. The maturation of the Neurospora miRNA-like sRNA, milR-1, requires the Argonaute protein QDE-2, Dicer, and QIP. Here, we reconstitute this Argonaute-dependent sRNA biogenesis pathway in vitro and discover that the RNA exosome is also required for milR-1 production. Our results demonstrate that QDE-2 mediates milR-1 maturation by recruiting exosome and QIP and by determining the size of milR-1. The exonuclease QIP first separates the QDE-2-bound pre-milR-1 duplex and then mediates 3’ to 5’ trimming and maturation of pre-milRNA together with exosome using a hand-over mechanism. In addition, exosome is also important for the decay of sRNAs. Together, our results establish a biochemical mechanism of an Argonaute-dependent sRNA biogenesis pathway and critical roles of exosome in sRNA processing.
Introduction
RNA interference (RNAi) is a conserved regulatory mechanism from fungi to mammals in which small non-coding RNAs (sRNAs) mediate post-transcriptional or transcriptional gene silencing (Ambros, 2004; Buhler and Moazed, 2007; Ghildiyal and Zamore, 2009; Hannon, 2002). A common theme among all known RNAi pathways is that the single-stranded sRNAs guide Argonaute family proteins to RNA targets to regulate gene expression. Argonaute proteins, the core component of all known RNAi pathways, function either by directly slicing their RNA targets or by mediating RNA degradation and translational repression. Since the discovery of the first small RNA in C. elegans, various types of sRNAs (with sizes ranging from 20-30 nt), including microRNAs (miRNAs), various small interfering RNAs (siRNAs), and PIWI-interacting RNAs (piRNAs), have been discovered in animals, plants, and fungi (Ghildiyal and Zamore, 2009; Kim et al., 2009; Lee et al., 1993).
The animal piRNAs associate with the PIWI clade of Argonaute proteins and are enriched in germline cells to suppress transposon activity (Aravin et al., 2006; Aravin et al., 2001; Batista et al., 2008; Das et al., 2008; Ghildiyal and Zamore, 2009; Girard et al., 2006; Grimson et al., 2008; Grivna et al., 2006; Lau et al., 2006; Lin, 2007; Vagin et al., 2006). Although most of known types of sRNAs are products of RNAse III ribonuclease Dicer cleavage of double-stranded RNA precursors, piRNAs with sizes ranging from 23-30 nt are produced independently of Dicer. Biogenesis of piRNAs is dependent on the PIWI proteins, and these sRNAs arise from long single-stranded RNA precursors. Although secondary piRNAs are known to be produced by a ping-pong amplification mechanism via the slicer activity of PIWI-related proteins (Brennecke et al., 2007; Gunawardane et al., 2007; Li et al., 2009; Malone et al., 2009), how primary piRNAs are produced from their RNA precursors and how PIWI proteins mediate such a process are not clear. Recently, a study using a cell-free system led to the proposal of a 3’ end trimming model for piRNA maturation (Kawaoka et al., 2011). In this model, PIWI protein binds the single-stranded piRNA precursors which are then 3’ to 5’ trimmed into maturation by an exonuclease. Whether this proposed mechanism occurs in vivo and the identity of the exonuclease are not known. Like piRNAs, the Dicer-independent primal RNAs in fission yeast are also dependent on an Argonaute protein for accumulation (Halic and Moazed, 2010).
The filamentous fungus Neurospora crassa is one of the first organisms in which RNAi-related phenomena was studied (Catalanotto et al., 2000; Cogoni and Macino, 1999a, b). As a filamentous fungus, Neurospora has a remarkable reservoir of sRNAs that are made by diverse biogenesis pathways (Li et al., 2010). We previously discovered the existence of miRNA-like sRNAs (milRNAs) in this organism and found that milRNAs are produced through at least four different pathways (Lee et al., 2010). milR-1 is the most abundant milRNA-producing loci in the Neurospora genome; the maturation of milR-1 milRNAs requires the Argonaute protein QDE-2 (Quelling Defective 2) but not its slicer activity. In addition, the production of milR-1 milRNAs also requires Dicer and a putative nuclease QIP. Dicer is required for the generation of milR-1 pre-milRNAs from primary-milRNA (pri-milRNA). In a dicer mutant, the pre-milRNAs and mature single-stranded milR-1 are not observed, and pri-milR-1 accumulates to high levels, suggesting that pri-milR-1 is cleaved by Dicer to generate double-stranded pre-milRNAs. QIP was previously identified as a QDE-2-interacting protein and is important for siRNA RISC activation by removing the nicked siRNA passenger strand from the QDE-2-bound siRNA complex (Maiti et al., 2007). In the qip mutant, the mature milR-1 milRNA is abolished, but pre-milRNAs are maintained, indicating that QIP is required for the maturation of milR-1 milRNAs (Lee et al., 2010). These results led us to propose a model in which the QDE-2 Argonaute binds to pre-milRNAs and recruits one or more trimmer enzymes to process the pre-milRNAs into mature milRNAs by 3’-5’ trimming. This model is very similar to that of the recently proposed piRNA maturation model (Kawaoka et al., 2011).
In this study, we reconstituted in vitro the Argonaute-dependent milR-1 biogenesis pathway from pri-milRNA to mature milRNAs by using recombinant proteins. In addition to QIP, we identified the RNA exosome, a 3’ to 5’ exonuclease complex, as a trimmer that is essential for milR-1 maturation. Together, our results establish the biochemical framework of an Argonaute-dependent sRNA biogenesis pathway and demonstrate the important roles of exosome in small RNA processing and degradation.
Results
The Neurospora Dicer-2 generates pre-milR-1 from pri-milRNA
The QDE-2-dependent milR-1 milRNAs are the most abundant milRNAs in Neurospora under normal growth conditions (Lee et al., 2010). The production of mature single-stranded milR-1 milRNAs requires Dicer, QDE-2, and the putative exonuclease QIP (Figure 1A). The 170-nt pri-milRNA of milR-1 is predicted to form a long stem-loop structure with milR-1 and milR-1* sequences located near the bottom of the stem structure. In the dicer (dcl-1/dcl-2) double mutant, the milR-1 pri-milRNA accumulates to high levels and the production of pre-milRNAs and the milRNAs is completely abolished (Lee et al., 2010), suggesting that Dicer is responsible for the generation of pre-milRNAs from pri-miRNAs. To demonstrate that Dicer can directly produce pre-milRNA of milR-1 from pri-milRNA, we expressed and purified the full-length recombinant Neurospora DCL-2 (His-DCL-2), which is responsible for more than 90% of the Neurospora Dicer activity (Catalanotto et al., 2004), from Sf9 cells (Figure 1B). As expected, the recombinant DCL-2 generated siRNAs around 24-25nt from a long double-stranded RNA substrate (Figure 1C), consistent with the previous result using Neurospora extracts (Catalanotto et al., 2004). To examine the activity of the recombinant DCL-2 on milR-1 pri-milRNA, we synthesized the full-length pri-milRNA in vitro. As shown in Figure 1D, the treatment of pri-milRNA with DCL-2 resulted in the production of small RNAs of the same size as the synthesized 33-nt pre-milR-1, the precursor of mature milRNAs (see below). The DCL-2 generated pre-milRNA is double stranded as indicated by its mobility in a native gel (Figure 1D, the bottom panel). This result demonstrates that DCL-2 can directly generate the precursor of milRNA by cleaving the loop structure from the milR-1 pri-milRNA.
Figure 1. Recombinant DCL-2 processes pri-milR-1 into pre-milR-1 in vitro.
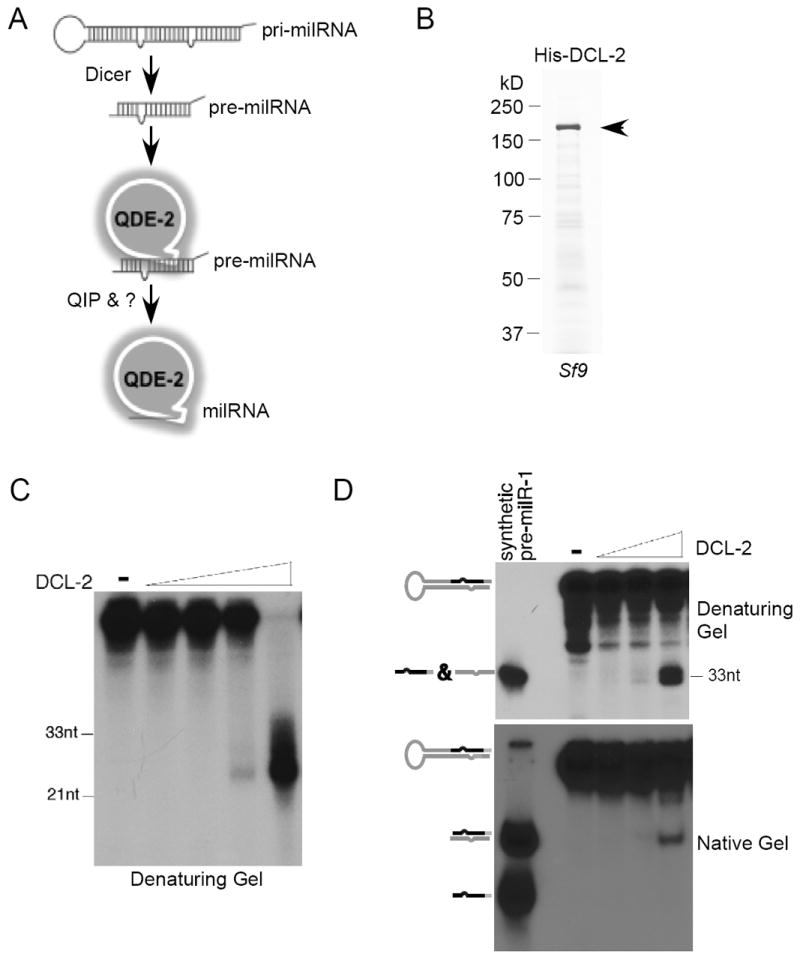
(A) A model of the milR-1 biogenesis pathway showing that the production of milR-1 requires Dicer, Argonaute protein QDE-2, and the putative exonuclease QIP.
(B) The Coomassie blue stained SDS-PAGE gel showing the purified His-DCL-2 from Sf9 cells.
(C) Dicer assay using radiolabeled dsRNA as the substrate.
(D) Dicer assay using radiolabeled pri-milR-1 as the substrate.
QIP is a 3’-5’ exonuclease in vitro
Northern blot analysis using a probe specific for milR-1 milRNAs previously showed that there are two double-stranded pre-milR-1 species (pre-milR-1 and pre-milR-1’) (Lee et al., 2010). In agreement with our previous results, the mature single-stranded milRNA species, but not the pre-milR-1, are abolished in the qde-2RIP and qipKO mutants (Figure 2A), indicating that both QDE-2 and QIP function downstream of the pre-miRNA generation and are required for milRNA maturation. In addition, the levels of pre-milR-1, but not pre-milR-1’, were increased in both qde-2 and qip mutants, suggesting that pre-milR-1 is processed by QDE-2 and QIP to generate mature milRNAs. Consistent with this notion, pre-milR-1, but not pre-milR-1’, specifically associates with QDE-2 (Lee et al., 2010).
Figure 2. QIP is a 3’-5’ exonuclease and prefers short pre-milR-1 as substrates in vitro.
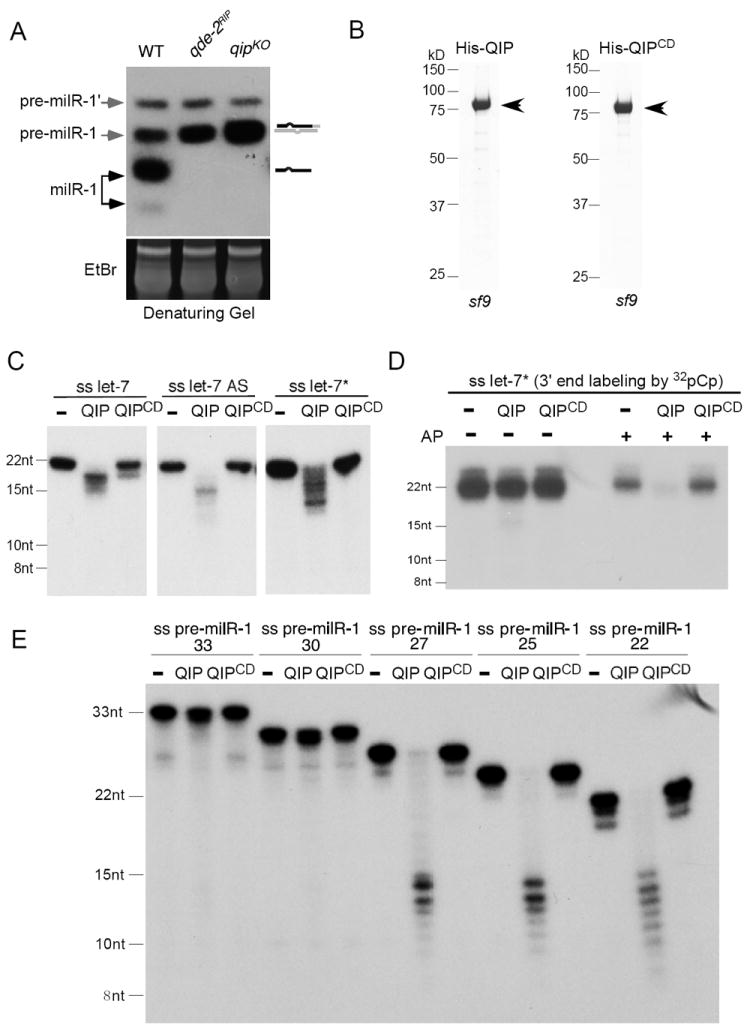
(A) Northern blot analysis showing the level of pre-milR-1 and milRNAs in the indicated strains in denaturing gels. The ethidium bromide-stained gel in the bottom panel shows equal loading of RNA samples.
(B) The Coomassie blue stained SDS-PAGE gel showing purified His-QIP and His-QIPCD from Sf9 cells.
(C) Indicated 5’ [32P] radiolabeled synthetic small RNAs incubated with buffer and His-QIP. The products were separated by a denaturing gel.
(D) Indicated 3’ [32P] radiolabeled synthetic small RNAs were treated with/without alkaline phosphatase (AP) before they were incubated with buffer and His-QIP. The products were separated by a denaturing gel.
(E) Indicated 5’ [32P] radiolabeled synthetic pre-milR-1 RNAs were incubated with buffer and His-QIP. The products were separated by a denaturing gel.
Our previous bioinformatics analysis showed that QIP contains a putative 3’-5’exonuclease domain that belongs to the DEDDh superfamily of 3’-5’ exonucleases (Maiti et al., 2007). The role of QIP in milR-1 maturation and its interaction with QDE-2 are consistent with its role as an exonuclease that trims the pre-milR-1 from the 3’ends. To directly test this hypothesis, we expressed and purified the recombinant, His-tagged wild-type QIP and QIP with its predicted catalytic sites mutated (QIPCD) from Sf9 cells (Figure 2B). Using various 21-nt, 5’ end labeled, single-stranded human let-7 miRNAs as substrates, we showed that the recombinant QIP could generate a ladder of smaller RNA products (Figure 2C). This activity was not observed with the catalytically dead QIP. These results demonstrate that QIP is indeed a nuclease that cleaves short single-stranded sRNAs.
To demonstrate that QIP is a 3’ to 5’ exonuclease and to understand its substrate requirement for 3’end nucleotide modification, we labeled the single-stranded let-7* RNA at the 3’ end by 32pCp to remove its 3’end hydroxyl group. As shown in Figure 2D, QIP could efficiently cleave the alkaline phosphatase treated RNA (to create 3’ hydroxyl group) but not the untreated RNA. This result indicates that QIP is a 3’ to 5’ exonuclease that requires 3’ hydroxyl group on its RNA substrate.
We then examined the effect of recombinant QIP on the synthetic 33-nt single-stranded pre-milR-1 and pre-milR-1*. Surprisingly, we found that QIP could not efficiently process the pre-milR-1 RNAs in vitro (Figure S1). This result raises the possibility that QIP may prefer short RNA substrates. To test this, we synthesized a series of single-stranded pre-milR-1 of different lengths with 3’ end deletions. Although QIP could not efficiently cleave the pre-milR-1 that is larger than 30 nt, it cleaved 22-27 nt pre-milR-1 RNAs efficiently into RNA fragments between 9-15 nt (Figure 2E). In addition, the cleavage by QIP was more complete for the 22 and 25 nt pre-milR-1 RNAs than that for the 27 nt RNA. This result suggests QIP prefers shorter RNA substrates. Therefore, if QIP is a nuclease that results in the maturation of milR-1, a hand-over mechanism exists and another nuclease is required for generating shorter pre-milR-1 RNAs as substrates for QIP. In addition, because QIP cleaves RNA substrates into fragments that are much smaller than the mature milR-1, a mechanism must also exist to determine the milR-1sizes.
Another nuclease can progressively process pre-milR-1 via 3’-5’ trimming
To examine whether there is another nuclease that processes pre-milR-1, we compared the milR-1 profile of the wild-type strain and the qip mutants. As shown in Figure 3A, a faint ladder of sRNAs was observed between the mature milR-1 and the pre-milR-1 in the qipKO and qipCD mutants but not in the wild-type sRNA sample. In addition, the levels of the sRNA ladder and pre-milR-1 were further increased in the qipCD mutant (Figure 3A and 3B). This ladder of sRNAs was singled-stranded because its gel mobilities did not change after the RNA samples were heat denatured. These results indicate that another Neurospora nuclease can progressively processes pre-milR-1 into mature milR-1 independent of QIP, albeit very inefficiently. The accumulation of this small RNA ladder in the qip mutants suggests that these sRNAs are substrates of QIP, which prefers smaller RNA substrates. The fact that the ladder of sRNAs accumulated to a higher level in the qipCD mutant than the qipCD mutant suggest that the catalytically dead QIP can protect these small RNAs from being degraded.
Figure 3. A QDE-2-dependent ladder of processed pre-milR-1 is produced independent of QIP.
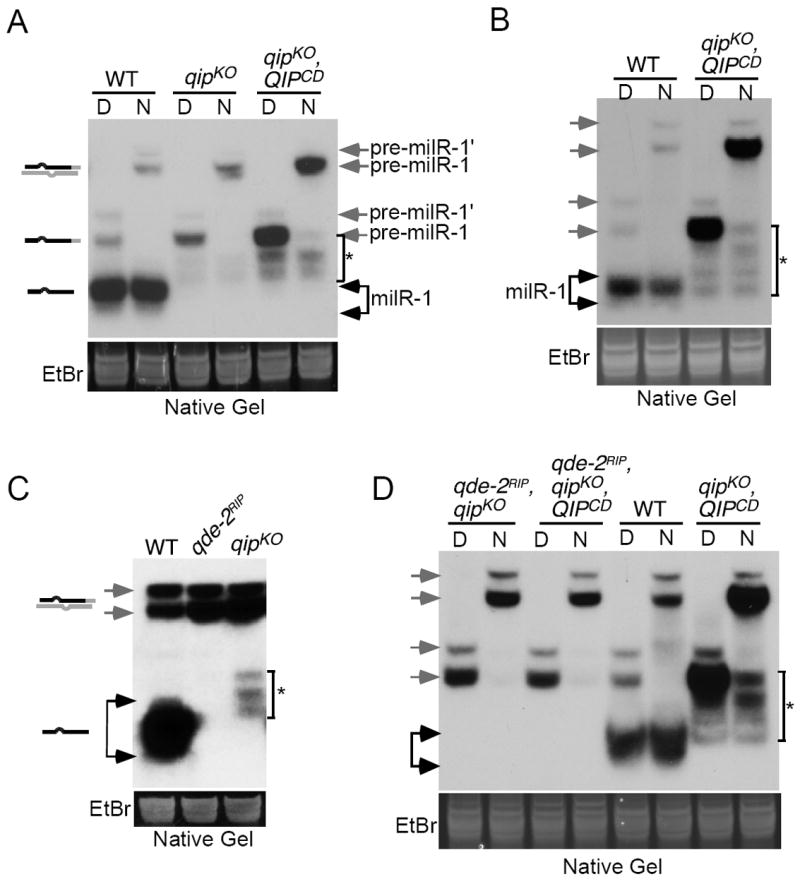
(A - D) Northern blot analysis of small RNA samples separated on native gels showing the levels of milR-1 pre-milRNAs and milRNAs in the indicated strains. “D” indicates that RNA samples were denatured by boiling. “N” represents native RNA samples.
The generation of the sRNA ladder requires the QDE-2 Argonaute since it was completely absent in the qde-2RIP mutant (Figure 3C). Furthermore, the sRNA ladder was also abolished in the qde-2RIP, qipKO double mutants with or without the expression of the catalytic dead form of QIP (Figure 3D). These results suggest that in addition to QIP, QDE-2 may recruit another nuclease to progressively process pre-milR-1 to generate the substrates for QIP.
Exosome is required for the maturation of milR-1 milRNAs
To identify the nuclease, we screened various nuclease knock-out mutants available in the Neurospora knock-out library for genes involved in milR-1 production (Colot et al., 2006). Among these mutants, the rrp6KO mutant showed a reduced level of mature milR-1 (Figure 4A). rrp6 (NCU02256) encodes the Neurospora sequence homolog of the yeast RRP6, which is a component of the exosome complex. The RNA exosome, a highly conserved large complex consisting of several 3’ to 5’ exonucleases, is a major regulator of RNA (rRNA, mRNA, and noncoding RNA) metabolism as it mediates 3’ to 5’ RNA processing and degradation (Houseley et al., 2006; LaCava et al., 2005; Vanacova et al., 2005). In the budding yeast, the exosome functions require Mtr4p, which is an essential co-factor of the exosome and part of the TRAMP (Trf4/Air2/Mtr4p Polyadenylation) complex that polyadenylates RNA substrates (Houseley and Tollervey, 2006; LaCava et al., 2005; Vanacova et al., 2005). As in yeast, the knock-out of rrp6 is not lethal in Neurospora, indicating that RRP6 is not essential for exosome functions.
Figure 4. The exosome complex is required for milR-1 maturation and is responsible for the generation of the single-stranded pre-milR-1 ladder in the qip mutants.
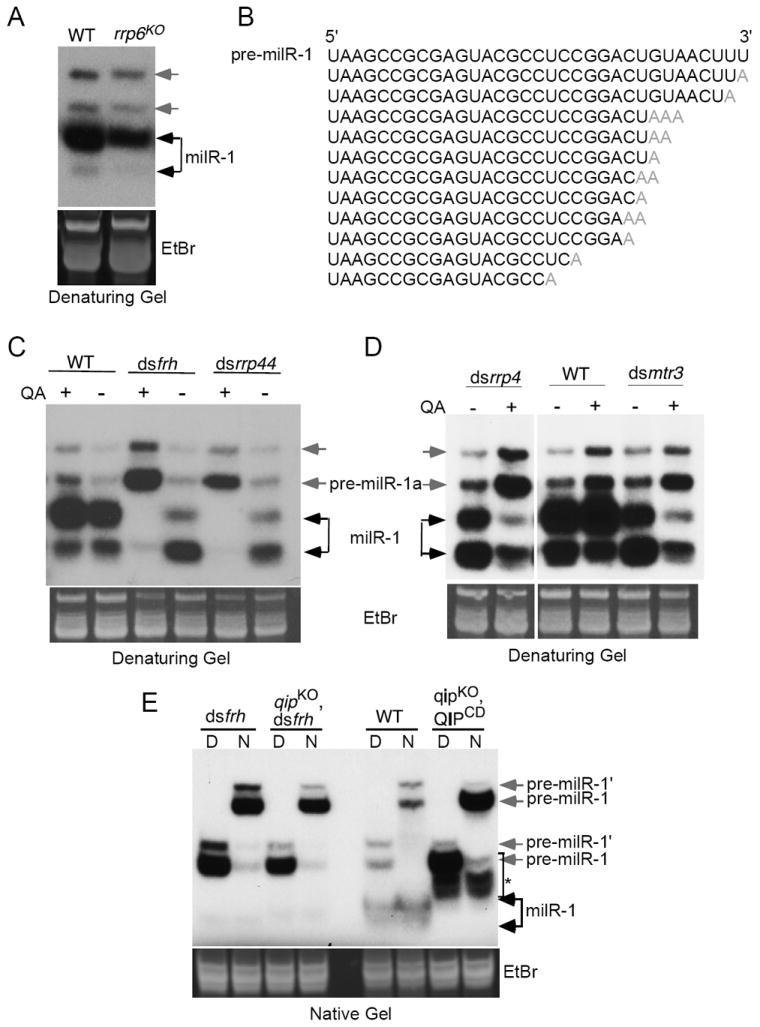
(A) Northern blot analysis of small RNA samples separated in denaturing gels showing the levels of milR-1 pre-milRNAs and milRNAs in the indicated strains.
(B) Representative small RNA species with untemplated adenosines identified by Solexa deep sequencing analysis.
(C) & (D) Northern blot analysis of small RNA samples separated in denaturing gels showing the levels of milR-1 pre-milRNAs and milRNAs in the indicated strains. dsfrh and dsrrp44 are the dsRNA knockdown strains in which frh or rrp44 dsRNA expression is induced by QA.
(E) Northern blot analysis of small RNA samples separated in native gels showing the levels of milR-1 pre-milRNAs and milRNAs in the indicated strains. “D” indicates that RNA samples were denatured by boiling. “N” represents native RNA samples.
We previously performed deep sequencing and 5’ RACE experiments on the QDE-2-associated sRNAs (Lee et al., 2010). In our analysis of the milR-1-related sRNA reads, we found that almost all pre-milR-1 and mature milRNA reads shared the same 5’ U position, but that their 3’ends were heterogeneous with sizes ranging from ~17 to 33 nt. This observation suggests that pre-milR-1 is subjected to 3’-5’ enzymatic trimming. In addition, some of pre-milR-1 and mature milRNA reads had one to three 3’ untemplated adenosines (Figure 4B). The presence of these untemplated adenosines in the milR-1 pre-milRNA and mature milR-1 reads also suggests that they are exosome substrates.
To demonstrate the role of exosome in milR-1 production, we examined the strains in which the essential Neurospora exosome components were inducibly knocked-down by expression of dsRNA specific for the gene of interest. RRP44 (also called Dis3), a member of the RNase II family, is the core catalytic subunit of the yeast and Neurospora exosome complex (Dziembowski et al., 2007; Guo et al., 2009; Schneider et al., 2007). FRH (FRQ-interacting RNA helicase) is the Neurospora sequence and functional homolog of the yeast Mtr4p (Cheng et al., 2005; Guo et al., 2009). Long inverted repeats specific for these genes were under the control of quinic acid (QA)-inducible promoter, so that in the presence of QA, the gene of interest was specifically silenced by the expression of dsRNA (Cheng et al., 2005). As shown in Figure S2A, the additional QA resulted in dramatic reduction of cell growth of both dsrrp44 and dsfrh strains, indicating the essential roles rrp44 and frh in cell growth. Northern blot analysis showed that the addition of QA resulted in a near complete disappearance of mature milR-1 and a dramatic increase of pre-milR-1 levels in both dsrrp44 and dsfrh strains (Figure 4C). To further confirm this result, we created two additional strains, dsrrp4 and dsmtr3, in which two other exosome components, rrp4 and mtr3, respectively, can be inducibly knocked-down. As expected, the silencing of rrp4 and mtr3 also resulted in the decrease of mature milR-1 and increase of pre-milR-1 levels (Figure 4D). In addition, deletion of one of the two of the Neurospora homolog of air2 (NCU04617) also resulted in a significant reduction of mature milR-1 (Figure S2B). Together, these results indicate that exosome is essential for the maturation of milR-1 milRNAs.
To examine whether the RNA exosome is responsible for the generation of the single-stranded small RNA ladder seen in the qipKO strains, we compared the milR-1 profiles in the wild-type, dsfrh, dsrrp44, qipKO, and qipCD strains. In addition, we also created a qipKO strain (qipKO, dsfrh) in which frh can be inducibly silenced. As shown in Figure 4E and Figure S2C, the silencing of frh or rrp44 abolished the production of the sRNA ladder in the wild-type and qipKO strains. These results indicate that exosome is required for processing pre-milR-1 into the single-stranded small RNA ladder seen in the qip mutants.
QDE-2 mediates the processing pre-milR-1 by the exosome
Because of the essential role of the exosome in mediating the QDE-2-dependent milR-1 maturation, we tested whether QDE-2 interacts with exosome in vivo. An immunoprecipitation assay using an anti-FRH antibody showed that Myc-QDE-2 co-precipitated with FRH in an RNA-independent manner (Figure 5A). In addition, we have previously shown that FRH associates with the core exosome components (Guo et al., 2009). There results suggest that QDE-2 recruits the exosome to process pre-milR-1.
Figure 5. QDE-2 interacts with an exosome component and promotes the trimming of single-stranded pre-milR-1 by the exosome.
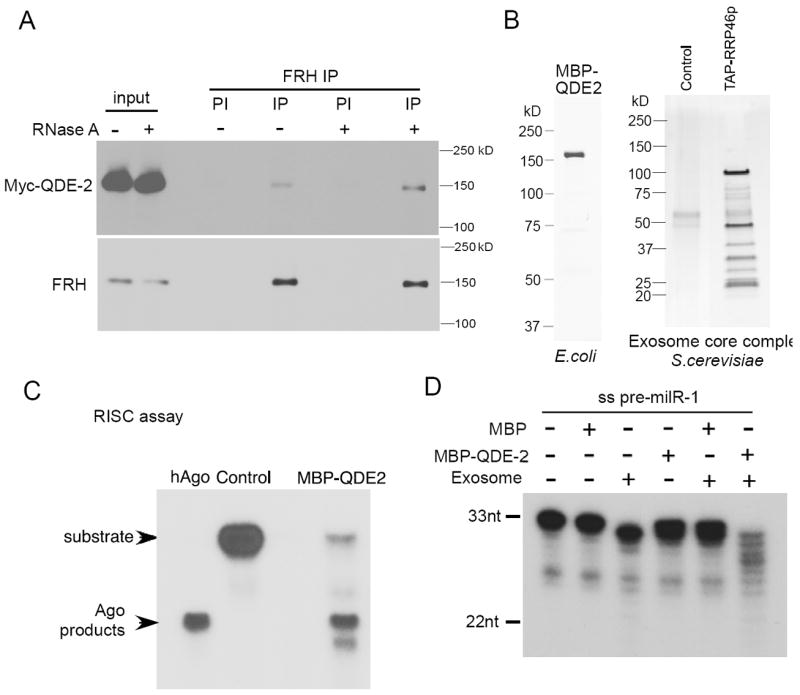
(A) Western blot analysis showing the result of immunoprecipitation with the anti-FRH antibody in the Myc-QDE-2 strain. Cell extract was incubated with/without RNase A before immunoprecipitation. Immunoprecipitation with the FRH pre-immune (PI) serum was used as the negative control.
(B) The Coomassie blue stained SDS-PAGE gel showing purified MBP-QDE-2 from E. coli, and the silver stained SDS-PAGE gel showing purified core exosome complex from S. cerevisiae.
(C) siRNA-initiated RISC assays performed with human Ago2 (hAgo) or MBP-QDE-2.
(D) 5’ [32P] radiolabeled single-stranded pre-milR-1 incubated with the indicated purified proteins. The products were separated by a denaturing gel.
To demonstrate the role of exosome in mediating the QDE-2-dependent milR-1 maturation, we expressed and purified the full-length MBP-tagged QDE-2 in E. coli (Figure 5B). The budding yeast exosome homoenzyme is currently the only active eukaryotic exosome complex that can be reconstituted in vitro (Liu et al., 2006). Therefore, to obtain a functional exosome complex, we purified the budding yeast core exosome complex by tandem-affinity purification using a TAP-tagged RRP46 strain (Callahan and Butler, 2010). To examine whether the recombinant QDE-2 is functional, we performed a RISC assay to examine whether MBP-QDE-2 was able to mediate single-stranded siRNA-dependent RNA cleavage. As shown in Figure 5C, like the human Ago2 protein, MBP-QDE-2 sliced the RNA target into the expected size, indicating that MBP-QDE-2 is functional. We then examined whether MBP-QDE-2 facilitated the processing of the single-stranded pre-milR-1 by the exosome. As shown In Figure 5D, although exosome alone only showed weak processing activity towards the synthetic single-stranded pre-milR-1, the addition of MBP-QDE-2 but not MBP to the exosome dramatically increased the 3’-5’ trimming of the pre-milR-1, resulting in a small RNA ladder similar to what we observed in the qip mutants. Although the optimal activity of exosome requires is known to require the TRAMP complex (LaCava et al., 2005), this result indicates that QDE-2 can promote the activity of exosome to pre-milR-1. In addition, exosome could not process double-stranded pre-milR-1 with or without QDE-2 (Figure S3 & Figure 6D), consistent with its known activity only on single-stranded RNAs. Together, these results suggest that QDE-2 recruits the exosome and promotes its trimming of the single-stranded pre-milRNAs. However, the double-stranded pre-milR-1 needs to be unwound into single-stranded to allow exosome-mediated processing.
Figure 6. QIP unwinds the QDE-2-bound pre-milR-1 duplex and then process it to maturation together with exosome.
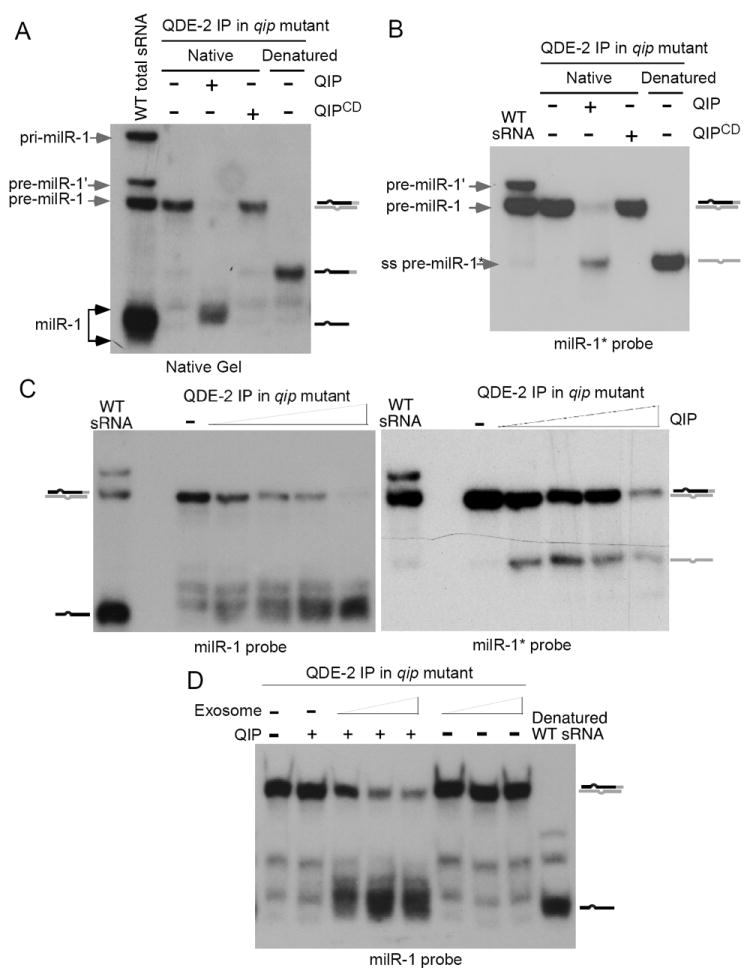
(A) Myc-QDE-2 immunoprecipitated RNAs from the qipKO strain was incubated with the QIP recombinant proteins. The products were separated by a native gel, and northern blot was probed with a milR-1 specific star-fire probe.
(B & C) Pre-milRNA processing assays using QDE-2 immunoprecipitates from the qipKO strain and the indicated recombinant proteins. The reaction products were separated by native gels, and the northern blot was probed with a milR-1* or milR-1 specific probe. For the experiment in (A & B), 0.5μg His-QIP or His-QIPCD was used. For the experiment in (C), 0.25μg, 0.5μg, 1μg, 2μg His-QIP were used. Independent QDE-2 immunoprecipitation products from the qipKO strain were used in these experiments.
(D) Exosome promotes the QIP-dependent maturation of milR-1 in vitro. Pre-milRNA processing assays were performed by using QDE-2 immunoprecipitates from the qipKO strain and different concentrations of exosome (87ng, 175ng, or 0.7μg). For reactions containing QIP, 60ng His-QIP was used. The reaction products were separated by a native gel. Northern blot was probed with a milR-1 specific probe.
QIP is an exonuclease required for the QDE-2-dependent milR-1 maturation in vitro
Because pre-milR-1 is associated with QDE-2 and QDE-2 associates with exosome factors in Neurospora, we then examined whether QIP can process the QDE-2-bound pre-milR-1 purified from Neurospora into mature milR-1. In a wild-type strain, immunoprecipitation of QDE-2 pulled down both the mature single-stranded milR-1 and the double-stranded pre-milR-1 (Figure S4A). The QDE-2-bound pre-milR-1 is double-stranded because the denatured (boiled) RNA sample exhibited significantly increased the mobility relative to a sample that was not denatured.
To specifically obtain the QDE-2-pre-milR-1 complex, we immunoprecipitated QDE-2 from the qipKO mutant extracts and used the immunoprecitates as substrates for the QIP nuclease assays (Figure S4B). The use of QDE-2-bound sRNA from the qipKO mutant ensured that only pre-milR-1 but not the mature milR-1 was immunoprecipitated. The resulting sRNAs were extracted, separated on a native gel, and analyzed by northern blot analysis with a probe specific for milR-1. As shown in Figure 6A, immunoprecipitation of QDE-2 in the qipKO mutant indeed only pulled down pre-milR-1, supporting the notion that pre-milR-1 is the precursor of mature milR-1. The addition of recombinant QIP, but not QIPCD, to the reaction resulted in the complete disappearance of the double-stranded pre-milR-1 and the appearance of single-stranded sRNA species similar to the mature milR-1 milRNAs in size. This in vitro complementation result indicates that QIP is an exonuclease that directly processes the QDE-2-bound pre-milR-1 into mature milRNAs.
QIP separates the QDE-2-associated double-stranded pre-milR-1 and collaborates with exosome to process pre-milRNA into milRNAs
Because exosome cannot process double-stranded pre-milRNAs, a factor is required to separate the pre-milR-1 duplex or remove the pre-milR-1* strand to generate single-stranded substrates for exosome. It has been long hypothesized that factors may exist that unwind the siRNA and miRNA duplexes loaded onto Argonaute proteins, but no such factor with dsRNA strand separation activity has yet been identified (Haley and Zamore, 2004; Matranga et al., 2005). We initially identified QIP as a QDE-2-interacting protein that is required for the removal of passenger strands of siRNA from the QDE-2 associated siRNA duplex (Maiti et al., 2007). Although a small amount of exosome-processed single-stranded RNAs was observed in the qip mutants (Figure 3), which is likely due to spontaneous strand separation of the QDE-2-bound pre-milR-1, the vast majority of the pre-milR-1 is in duplex form. In addition, the levels of the pre-milR-1 duplex are dramatically increased in the qip mutants (Figures 2A and 3). These observations raised the possibility that QIP has the ability to separate the QDE-2-bound pre-milR-1 duplex into single-strands. Interestingly, the budding yeast sequence homolog of QIP, Gfd2p, was previously identified as a high-copy suppressor of a DEAD-box RNA helicase mutant (Estruch and Cole, 2003).
To test this hypothesis, we examined whether the recombinant QIP separate the QDE-2-bound pre-milR-1 into single strands. The immunoprecipitates of QDE-2 from the qipKO strain was used in the assay, so that most of pre-milR-1 was in duplex form. Because the milRNA strand of the pre-milR-1 is rapidly converted into smaller RNA species (Figure 6A), we performed northern blot analysis specific for the milRNA* strand. As shown in Figure 6B, the addition of QIP in the reaction resulted in the disappearance of the pre-milR-1 duplex and the appearance of a small RNA band that was identical in size as the single-stranded full-length pre-milR-1*, indicating the separation of the QDE-2-bound double-stranded pre-milR-1. In addition, the recombinant QIP is also able to convert the synthesized double-stranded pre-milR-1 in vitro into single-stranded (Figure S4C). Together, these results indicate that QIP is a factor that can separate sRNA duplex and can also degrade single-stranded sRNA. It should be noted that the duplex separation ability of QIP also require QDE-2 in vivo, since pre-milR-1 and siRNA are maintained in duplex forms in the qde-2 mutant (Figure 2) (Maiti et al., 2007). It is likely that QIP requires QDE-2 to recruit double-stranded RNA substrates.
Titration of the recombinant QIP in the assay using QDE-2-bound pre-milR-1showed that as the concentration of QIP increased, the levels of the single-stranded pre-milR-1* first increased and then decreased at high QIP concentrations (Figure 6C, right panel). This result indicates that the duplex pre-milRNAs were first separated into single strands before being processed. We failed to detect a significant amount of smaller processed pre-milR-1* products, suggesting that the pre-milRNA* strand was processed into very small sizes that could not be detected by our northern blot analysis. In contrast, when we analyzed the milR-1 strand products, an increasing concentration of QIP resulted in increases in levels of single-stranded sRNAs similar in sizes as the mature milRNAs, and the full-length single-stranded pre-milR-1 was not easily detected (Figure 6B, left panel). These experiments indicate that after strand separation, the milR-1 strand of pre-milRNA is immediately and progressively processed by QIP and exosome to maturation. As many miRNAs in animal and plants, milR-1 is the predominant species and that little milR-1* milRNA is produced in vivo (Lee et al., 2010). Thus, our in vitro reconstitution results recapitulate this highly selective processing and maturation step.
Why is pre-milR-1 processed to maturation and pre-milR-1* degraded away? What determines the sizes of mature milR-1 milRNAs? The differences in QIP processing efficiency and end products between the milR-1 and milR-1* strands of pre-milRNA indicate that the same nucleases act on them differently. The much slower degradation rate of the full-length pre-milR-1* strand in the in vitro reactions suggests that it dissociates from the QDE-2-pre-milRNA complex after duplex separation, making it less accessible to QIP and exosome. Supporting this notion, we found that in Neurospora, while QDE-2 is associated with single-stranded processed pre-milR-1 species, no QDE-2-bound single-stranded pre-milR-1* species could be detected (Figure S4D). Thus, the different outcomes of pre-milR-1 and pre-milR-1* should be due to the binding of pre-milR-1 by QDE-2. Together, these results suggest that upon strand separation, pre-milR-1* dissociates from QDE-2 and is degraded away, while pre-milR-1 remains associated with QDE-2 and is progressively processed by QIP and exosome. Thus, the sizes of the mature milR-1 are determined by the region of pre-milRNA that is protected by QDE-2 from nuclease processing.
Because of the association of between QDE-2 and FRH, which is known to associate with RRP44 in Neurospora (Guo et al., 2009), a small amount of exosome should exist in the QDE-2 immunoprecipitates used in the QIP nuclease assays. To determine the role of exosome in milR-1 maturation, we performed the assay using a limiting amount of QIP and different concentrations of purified core exosome complex. As shown in Figure 6C, although this concentration of QIP could only modestly increase the level of mature milR-1, the addition of exosome led to a dramatic increase of mature milR-1 levels. In contrast, exosome alone had little effects on pre-milR-1 processing in the absence of QIP. This in vitro result is consistent with our genetic results and indicates that the maturation of milR-1 is a collaborative process requires QDE-2, QIP and exosome. QDE-2 recruits both QIP and exosome to pre-milRNA-1. The processing of pre-milR-1 by the exosome requires the strand-separation of pre-milRNA duplex by QIP. On the other hand, exosome can generate substrates for QIP. Afterwards, both exosome and QIP contribute to the processing and maturation of milR-1. Together, our results present above establish the biochemical basis for an Argonaute-dependent sRNA biogenesis pathway.
The role of exosome in degrading other milRNAs and siRNAs
The essential role of exosome in milR-1 production also prompted us to examine whether it regulates other Neurospora milRNAs and siRNA. Like milR-1, the production of milR-2 milRNAs also requires QDE-2, but milR-2 biogenesis requires the catalytic activity of QDE-2 and is produced independent of Dicer (Lee et al., 2010). On the other hand, milR-3, like most plant and animal miRNAs, requires only Dicer for its biogenesis. Except for milR-1, no other known milRNAs requires QIP for biogenesis. As shown in Figure 7A, the silencing of either rrp44 or frh in Neurospora resulted in significant increases in levels of milR-2 and milR-3 milRNAs. Similarly, silencing of rrp44 in an albino-2 (al-2) quelled strain also resulted in an increase of the al-2-specific siRNA (Figure 7B). These results suggest that exosome is important for the decay of siRNAs and these two milRNAs and is not required for their maturation. Together, our results indicate that the exosome has at least two different roles in sRNA regulation in Neurospora: It mediates sRNA degradation and, in the case of milR-1 and perhaps other small RNAs, is directly involved in the maturation by 3’-5’ processing.
Figure 7. Exosome is important for the decay of other milRNAs and siRNAs.
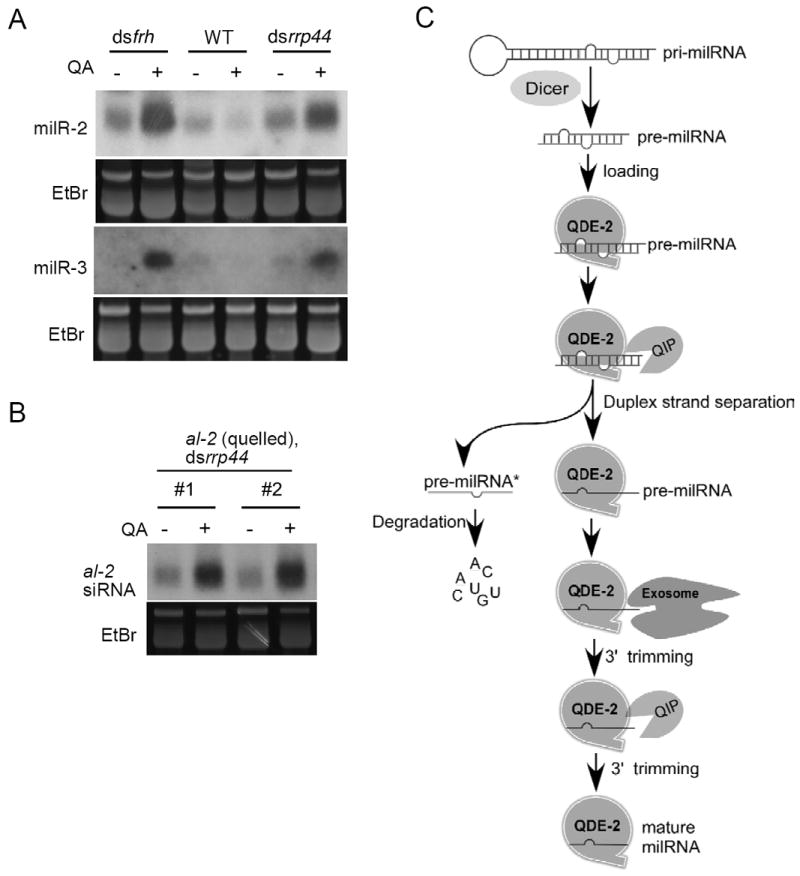
(A) Northern blot analysis of small RNA samples separated in a denaturing gel showing the levels of milR-2 and milR-3 in the indicated strains.
(B) Northern blot analysis of small RNA samples separated in a denaturing gel showing the level of al-2 siRNA in the indicated strains.
(C) A model of milR-1 biogenesis pathway from pri-milRNA. See discussion for details.
Discussion
Argonaute-dependent sRNA biogenesis are found in many organisms. Here, by using the Neurospora milR-1 milRNA as an example, we established a biochemical mechanism for an Argonaute-dependent sRNA production process and uncovered the collaborative and distinct roles of the Argonaute QDE-2, exosome and QIP in the milR-1 maturation process. Our biochemical and genetic results suggest that the maturation of milR-1 is a five-step process (Figure 7C). First, the 170-nt pri-milR-1 is directly cleaved by Dicer to generate the ~33-nt double-stranded pre-milRNA. Although we showed that the recombinant DCL-2 is sufficient to convert pri-milRNA into the functional pre-milR-1, pre-milR-1’ was not produced by Dicer cleavage in vitro, and it is likely that additional factors are involved in this step in vivo. We previously showed that the down-regulation of MRPL3, a putative RNAse III domain-containing protein, resulted in a decrease in the levels of pre-milR-1 (Lee et al., 2010), suggesting that MRPL-3 may also participate in this process.
The second step is Argonaute QDE-2 binding of the duplex pre-milRNA. Third, QIP, which is recruited by QDE-2, separates the pre-milRNA duplex into single-stranded RNAs in collaboration with QDE-2; the pre-milRNA strand remains QDE-2-associated, and the pre-milRNA* strand is released from the complex and degraded way. We demonstrated that QIP is indeed a 3’ to 5’exoribonuclease (Figure 2C) and importantly, it also posses an activity that can trigger the stand-separation of QDE-2 associated duplex RNA in vitro (Figure 6 and Figure S4C). The role of QIP in strand-separation of the pre-milR-1 duplex in vivo was further supported by the increase of pre-milR-1 duplex levels in qip mutants (Figures 2 and 3) and by its role in converting the QDE-2-associated siRNA duplex into single strands (Maiti et al., 2007). Because both pre-milR-1 and siRNA are maintained in duplex forms in qde-2 mutants (Figure 2) (Maiti et al., 2007), QDE-2 should also contributes to the duplex seapation process in vivo. This notion is consistent with the existence of low levels of single-stranded pre-milRNA in the qip mutants (Figure 3) and with previous results that the Drosophila Ago1 can passively separate miRNA duplexes in vitro (Kawamata et al., 2009). The ability of QIP to separate QDE-2-bound duplex pre-milRNA and its role in siRNA RISC activation suggest that strand-separation of Argonaute-bound duplex siRNA and miRNA is an important step in the activation of siRISC and miRISC. Our results and the fact that the budding yeast homolog of QIP, Gfd2p, was identified as a high copy suppressor of an RNA helicase mutant (Estruch and Cole, 2003) suggest that similar exoribonucleases may also posses dsRNA strand-separation activity.
The selective dissociation of the milRNA* strand of the pre-milR-1 from QDE-2 is likely pre-determined by the asymmetric loading of the duplex pre-milRNA onto QDE-2, so that the strand separation process preferentially dissociates milRNA* strand from the complex. The asymmetric loading of siRNAs, duplex-miRNAs, and pre-piRNAs into Argonaute proteins was recently suggested to be due to the selective binding of Argonaute proteins to small RNAs with a 5’ U (Kawaoka et al., 2011; Mi et al., 2008; Seitz et al., 2011). The pre-milR-1 milRNA strand also has a strong preference for 5’ U, thus, the asymmetric loading of the pre-milRNA duplex may be due to the selective binding of QDE-2 to the pre-milRNA strand.
In the fourth step, the exosome trims the QDE-2 bound pre-milRNAs from 3’ to 5’ end into sRNAs of intermediate sizes. Our conclusion that the exosome carries out this trimming is supported by several lines of evidence. Recombinant QIP cannot efficiently process the single-stranded full-length pre-milR-1and prefers shorter pre-milR-1 substrates (Figure 2). In addition, there is the accumulation of a ladder of single-stranded pre-milRNAs with sizes between the mature milR-1 and pre-milR-1 in the qip mutants (Figure 3), indicating that QIP can only efficiently process pre-processed pre-milR-1. Importantly, the silencing of the essential exosome components in Neurospora abolishes the QDE-2-dependent maturation of milR-1 and the production of the processed pre-milRNA ladder, indicating that the exosome is required for milR-1 maturation and is responsible for the generation of the prem-milR-1 substrates for QIP. Furthermore, even in the absence of QIP, a low level of the processed pre-miRNA with sizes similar to the mature milR-1 can be observed (Figures 3B and C), suggesting that exosome alone is able to convert pre-milRNA into mature milRNA, albeit very inefficiently.
In the fifth and final step, the exosome-processed pre-milRNAs are further processed into mature milRNAs in a process involving both QIP and exosome. The accumulation of the intermediate-sized partially processed pre-milRNAs in the qip mutants indicates that they are substrates of QIP. Supporting this notion, QIP can efficiently convert this ladder of processed pre-milRNAs into mature milRNA in vitro (Figure 6). In addition, we found that the addition of exosome strongly promoted the QIP-mediated maturation of QDE-2-bound pre-milRNA (Figure 6C), indicating that QIP and exosome collaborate in the 3’ to 5’ progressive trimming of pre-milR-1 into the mature milRNAs.
Our results also shed important insights in the role of Argonauate protein in small RNA maturation. Based on our results, the Argonaute QDE-2 has three essential roles in the milR-1 maturation process: 1) it binds to pre-milR-1 and determines which milR-1 strand will be matured, 2) it recruits exoribonucleases to process pre-milR-1, and 3) it determines the sizes of milR-1 by protecting the mature milR-1 from further processing. We showed that the duplex pre-milR-1 was first become single-stranded before being proceed, however, pre-milR-1 and pre-milR-1* have complete different outcome after processing: while pre-milR-1 was matured into milR-1, the pre-milR-1* was degraded away. In addition, we showed that although the single-stranded pre-milR-1 is associated with QDE-2, the single-stranded pre-milR-1* is not (Figure S4D). This result is also consistent with the much slower degradation kinetics of pre-milR-1* than that of pre-milR-1 (Figure 6C) and the fact that QIP can degrade the synthetic unprotected pre-milR-1 into very small RNA fragments (Figure 2E). Thus, the different processing outcomes of pre-milR-1 and pre-milR-1* is due to the binding of pre-milR-1 by QDE-2.
Crystal structure of an archaeal Argonaute suggested both the 5’ and 3’ ends of the guide RNA are anchored on the protein and the 5’ end of the guide RNA is anchored within a highly conserved basic pocket, suggesting that when in a complex, ~21nt from the 5’end of the small RNA are in complex with and protected by Argonaute (Ma et al., 2005; Parker et al., 2004; Wang et al., 2008). Although the association between QDE-2 and pre-milR-1 protect the region containing the mature milR-1, the 3’ ends of the large pre-milR-1 should be accessible by the nucleases. Thus, the sizes of mature milR-1 are determined by the region of pre-milR-1 that is protected by QDE-2 from further processing.
Recently, a cell-free system derived from a silkworm ovary-derived cell line was established and was shown to recapitulate key steps of animal piRNA biogenesis (Kawaoka et al., 2011). The proposed 3’ end trimming model for piRNA maturation is very similar to that we proposed for biogenesis of the Neurospora milR-1. However, the enzyme or enzymes that trim the piRNA-precursor in a PIWI-dependent manner have not been identified. Our study suggests that the highly conserved RNA exosome and DEDDh superfamily of exonucleases may function as the trimmer enzymes in piRNA and other small RNA processing. Since the submission of our paper, the putative exoribonuclease Nibbler has been shown to modify the 3’ ends of mature miR-34 miRNAs in Drosophila (Han et al., 2011; Liu et al., 2011). Interestingly, even though QIP and Nibbler are not sequence homologs, they both belong to the DEDDh superfamily of the ribonuclease. In addition, miR-451 in vertebrates is also generated by an Argonaute-dependent mechanism that is similar to that of the milR-2 in Neurospora (Cheloufi et al., 2010; Cifuentes et al., 2010; Lee et al., 2010; Yang et al., 2010). For these two miRNAs, however, the slicer activity of the Argonaute proteins is required to generate miRNA precursors before they are trimmed into maturation by un-identified nuclease.
Exosome components were previously suggested to be involved in small RNA decay (Bail et al., 2010; Halic and Moazed, 2010; Ibrahim et al., 2010), and exosome has also been shown to process the 3’ ends of some Drosophila mirtron-derived miRNAs (Flynt et al., 2010). In the fission yeast, the Argonaute-dependent priRNA levels and size distributions were found to be modestly effected in a dis3 mutant (Halic and Moazed, 2010), but a clear role for exosome in priRNA production is still unclear. In Neurospora, exosome is clearly essential for Neurospora milR-1 maturation. On the other hand, we found that the silencing of the exosome components resulted in the significant accumulation of siRNA and other milRNAs examined (Figure 7), indicating that exosome also functions in sRNA decay pathways in Neurospora. Therefore, exosome has at least two opposing roles in regulating sRNA levels.
Experimental Procedures
Strains, growth conditions, and protein purification
Please see Supplemental Experimental Procedures.
Dicer assay
Uniformly radiolabeled dsRNA or pri-milR-1 substrates were prepared and the Dicer reactions carried out essentially as previously described (Bernstein et al., 2001). The al-2 gene was used as the template for T7 in vitro transcription. Briefly, in a 10 μl reaction, 105 cpm [32P]-labeled RNA substrate and 1 mM ATP were incubated with different amounts of His-DCL-2 in buffer (100 mM potassium acetate, 10 mM HEPES (pH 7.4), 1 mM magnesium acetate) for 30 minutes at 30°C. For dsRNA substrates, 14 ng, 54 ng, 136 ng, and 1.36 μg His-DCL-2 was used. For pri-milR-1a substrates, 0.68 μg, 1.36 μg, and 4.98 μg His-DCL-2 was used. The products were resolved on 16% denaturing or native polyacrylamide gels, and gels were exposed to X-ray film.
Northern blot analysis
Small RNAs were separated on 16% denaturing or native polyacrylamide gels and transferred onto a Hybond-NX membrane (GE Healthcare). Crosslinking of RNA to Hybond-NX was performed as described (Pall et al., 2007). Northern blots were carried out as previously described (Lee et al., 2010).
Exonuclease processing assay
Single-stranded let-7 (UGAGGUAGUAGGUUGUAUAGUU), let-7 antisense (CUAUACAACCUACUACCUCAUU), let-7 star (CUAUACAAUCUACUGUCUUUC), pre-milR-1 (UAAGCCGCGAGUACGCCUCCGGACUGUAACUUU), pre-milR-1 30 (UAAGCCGCGAGUACGCCUCCGGACUGUAAC), pre-milR-1 27 (UAAGCCGCGAGUACGCCUCCGGACUGU), pre-milR-1 25 UAAGCCGCGAGUACGCCUCCGGACU), pre-milR-1 22 (UAAGCCGCGAGUACGCCUCCGGACUGUAACUUU, and pre-milR-1 star (GGUUACAGCCCCCGGGACGCACUACGGUUUAUA) were synthesized by Sigma and radiolabeled at their 5’ ends by T4 polynucleotide kinase (New England Biolabs). For 3’ end labeling, RNA oligos were radiolabled at their 3’ends by T4 RNA ligase with 32pCp (New England Biolabs).
In a 10 μl reaction, 105 cpm [32P]-labeled RNA substrate and 1 mM ATP were incubated with 0.5 μg His-QIP or His-QIPCD in buffer X1 (10 mM Tris-HCl (pH 7.4), 12.5 mM potassium acetate, 5 mM magnesium acetate, 1 mM DTT) for 30 minutes at 30°C. The reaction was stopped as described (Bernstein et al., 2001; Elbashir et al., 2001). The products were resolved on 20% denaturing polyacrylamide gels.
Small RNA immunoprecipitation
Immunopurification of the Myc-QDE-2 ribonucleoprotein complex was performed as previously described (Maiti et al., 2007) except that beads were washed with buffer X1 instead of the extraction buffer.
Co-Immunoprecipitation
Co-immunoprecipitation was performed as previously described (Cheng et al., 2005) with the following modifications: Before adding FRH antibody into the protein solution, 100 μl 20 mM DTSSP solution (Thermo #21578) was added to 1 ml protein solution (at 1 mg/ml). The reaction was quenched with 20 μl 1 M Tris solution after a 4-hour incubation at 4 °C.
siRNA-initiated RISC assay
The single-stranded let-7 was used as the guide strand to perform the siRNA-initiated RISC assay previously described (Liu et al., 2003). The mRNA substrate containing one let-7 target site was prepared as described (Liu et al., 2003).
Exosome processing assay
The exosome processing assay was performed using the procedure described for the exonuclease processing assay except that MBP-QDE-2 was incubated with [32P]-labeled RNA substrate and 1 mM ATP in buffer X1 for 30 minutes at 30 °C, and then exosome was added to the reaction for 30 minutes at 30 °C.
Supplementary Material
Acknowledgments
We thank Qiaohong Ye, Yi Yang, Liande Li, Xuechen Ye, and Jun Liao for technical assistance and Dr. Qinghua Liu for critical comments on the paper. Dr. Oded Yarden for providing the al-2-containing plasmid and Dr. J. Scott Butler for the yeast RRP46-TAP strain. This work was supported by grants from the National Institutes of Health to Yi Liu and from the Welch Foundation to Yi Liu (I-1560).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Naumova NM, Tulin AV, Vagin VV, Rozovsky YM, Gvozdev VA. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr Biol. 2001;11:1017–1027. doi: 10.1016/s0960-9822(01)00299-8. [DOI] [PubMed] [Google Scholar]
- Bail S, Swerdel M, Liu H, Jiao X, Goff LA, Hart RP, Kiledjian M. Differential regulation of microRNA stability. RNA. 2010;16:1032–1039. doi: 10.1261/rna.1851510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista PJ, Ruby JG, Claycomb JM, Chiang R, Fahlgren N, Kasschau KD, Chaves DA, Gu W, Vasale JJ, Duan S, et al. PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans. Mol Cell. 2008;31:67–78. doi: 10.1016/j.molcel.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Buhler M, Moazed D. Transcription and RNAi in heterochromatic gene silencing. Nat Struct Mol Biol. 2007;14:1041–1048. doi: 10.1038/nsmb1315. [DOI] [PubMed] [Google Scholar]
- Callahan KP, Butler JS. TRAMP complex enhances RNA degradation by the nuclear exosome component Rrp6. J Biol Chem. 2010;285:3540–3547. doi: 10.1074/jbc.M109.058396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalanotto C, Azzalin G, Macino G, Cogoni C. Gene silencing in worms and fungi. Nature. 2000;404:245. doi: 10.1038/35005169. [DOI] [PubMed] [Google Scholar]
- Catalanotto C, Pallotta M, ReFalo P, Sachs MS, Vayssie L, Macino G, Cogoni C. Redundancy of the two dicer genes in transgene-induced posttranscriptional gene silencing in Neurospora crassa. Mol Cell Biol. 2004;24:2536–2545. doi: 10.1128/MCB.24.6.2536-2545.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheloufi S, Dos Santos CO, Chong MMW, Hannon GJ. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010;465:584–589. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng P, He Q, He Q, Wang L, Liu Y. Regulation of the Neurospora circadian clock by an RNA helicase. Genes Dev. 2005;19:234–241. doi: 10.1101/gad.1266805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cifuentes D, Xue H, Taylor DW, Patnode H, Mishima Y, Cheloufi S, Ma E, Mane S, Hannon GJ, Lawson ND, et al. A Novel miRNA Processing Pathway Independent of Dicer Requires Argonaute2 Catalytic Activity. Science. 2010;328:1694–1698. doi: 10.1126/science.1190809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogoni C, Macino G. Gene silencing in Neurospora crassa requires a protein homologous to RNA-dependent RNA polymerase. Nature. 1999a;399:166–169. doi: 10.1038/20215. [DOI] [PubMed] [Google Scholar]
- Cogoni C, Macino G. Posttranscriptional gene silencing in Neurospora by a RecQ DNA helicase. Science. 1999b;286:2342–2344. doi: 10.1126/science.286.5448.2342. [DOI] [PubMed] [Google Scholar]
- Colot HV, Park G, Turner GE, Ringelberg C, Crew CM, Litvinkova L, Weiss RL, Borkovich KA, Dunlap JC. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc Natl Acad Sci U S A. 2006;103:10352–10357. doi: 10.1073/pnas.0601456103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das PP, Bagijn MP, Goldstein LD, Woolford JR, Lehrbach NJ, Sapetschnig A, Buhecha HR, Gilchrist MJ, Howe KL, Stark R, et al. Piwi and piRNAs act upstream of an endogenous siRNA pathway to suppress Tc3 transposon mobility in the Caenorhabditis elegans germline. Mol Cell. 2008;31:79–90. doi: 10.1016/j.molcel.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RL, deSerres D. Genetic and microbial research techniques for Neurospora crassa. Methods i Enzymology. 1970;27A:79–143. [Google Scholar]
- Dziembowski A, Lorentzen E, Conti E, Seraphin B. A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Nat Struct Mol Biol. 2007;14:15–22. doi: 10.1038/nsmb1184. [DOI] [PubMed] [Google Scholar]
- Estruch F, Cole CN. An Early Function during Transcription for the Yeast mRNA Export Factor Dbp5p/Rat8p Suggested by Its Genetic and Physical Interactions with Transcription Factor IIH Components. Molecular Biology of the Cell. 2003;14:1664–1676. doi: 10.1091/mbc.E02-09-0602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynt AS, Greimann JC, Chung WJ, Lima CD, Lai EC. MicroRNA biogenesis via splicing and exosome-mediated trimming in Drosophila. Mol Cell. 2010;38:900–907. doi: 10.1016/j.molcel.2010.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- Grimson A, Srivastava M, Fahey B, Woodcroft BJ, Chiang HR, King N, Degnan BM, Rokhsar DS, Bartel DP. Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals. Nature. 2008;455:1193–1197. doi: 10.1038/nature07415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivna ST, Beyret E, Wang Z, Lin H. A novel class of small RNAs in mouse spermatogenic cells. Genes Dev. 2006;20:1709–1714. doi: 10.1101/gad.1434406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC. A slicer-mediated mechanism for repeat-associated siRNA 5’ end formation in Drosophila. Science. 2007;315:1587–1590. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- Guo J, Cheng P, Yuan H, Liu Y. The exosome regulates circadian gene expression in a posttranscriptional negative feedback loop. Cell. 2009;138:1236–1246. doi: 10.1016/j.cell.2009.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley B, Zamore PD. Kinetic analysis of the RNAi enzyme complex. Nat Struct Mol Biol. 2004;11:599–606. doi: 10.1038/nsmb780. [DOI] [PubMed] [Google Scholar]
- Halic M, Moazed D. Dicer-independent primal RNAs trigger RNAi and heterochromatin formation. Cell. 2010;140:504–516. doi: 10.1016/j.cell.2010.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Bo W, Hung J-H, Weng Z, Zamore Phillip D, Ameres Stefan L. The 32-to-52 Exoribonuclease Nibbler Shapes the 32 Ends of MicroRNAs Bound to Drosophila Argonaute1. Current biology : CB. 2011;21:1878–1887. doi: 10.1016/j.cub.2011.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannon GJ. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- Houseley J, LaCava J, Tollervey D. RNA-quality control by the exosome. Nat Rev Mol Cell Biol. 2006;7:529–539. doi: 10.1038/nrm1964. [DOI] [PubMed] [Google Scholar]
- Houseley J, Tollervey D. Yeast Trf5p is a nuclear poly(A) polymerase. EMBO Rep. 2006;7:205–211. doi: 10.1038/sj.embor.7400612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim F, Rymarquis LA, Kim EJ, Becker J, Balassa E, Green PJ, Cerutti H. Uridylation of mature miRNAs and siRNAs by the MUT68 nucleotidyltransferase promotes their degradation in Chlamydomonas. Proc Natl Acad Sci U S A. 2010;107:3906–3911. doi: 10.1073/pnas.0912632107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamata T, Seitz H, Tomari Y. Structural determinants of miRNAs for RISC loading and slicer-independent unwinding. Nat Struct Mol Biol. 2009;16:953–960. doi: 10.1038/nsmb.1630. [DOI] [PubMed] [Google Scholar]
- Kawaoka S, Izumi N, Katsuma S, Tomari Y. 3’ end formation of PIWI-interacting RNAs in vitro. Mol Cell. 2011;43:1015–1022. doi: 10.1016/j.molcel.2011.07.029. [DOI] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell. 2005;121:713–724. doi: 10.1016/j.cell.2005.04.029. [DOI] [PubMed] [Google Scholar]
- Lau NC, Seto AG, Kim J, Kuramochi-Miyagawa S, Nakano T, Bartel DP, Kingston RE. Characterization of the piRNA complex from rat testes. Science. 2006;313:363–367. doi: 10.1126/science.1130164. [DOI] [PubMed] [Google Scholar]
- Lee HC, Li L, Gu W, Xue Z, Crosthwaite SK, Pertsemlidis A, Lewis ZA, Freitag M, Selker EU, Mello CC, et al. Diverse pathways generate microRNA-like RNAs and Dicer-independent small interfering RNAs in fungi. Mol Cell. 2010;38:803–814. doi: 10.1016/j.molcel.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Li C, Vagin VV, Lee S, Xu J, Ma S, Xi H, Seitz H, Horwich MD, Syrzycka M, Honda BM, et al. Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell. 2009;137:509–521. doi: 10.1016/j.cell.2009.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Chang SS, Liu Y. RNA interference pathways in filamentous fungi. Cell Mol Life Sci. 2010;67:3849–3863. doi: 10.1007/s00018-010-0471-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H. piRNAs in the germ line. Science. 2007;316:397. doi: 10.1126/science.1137543. [DOI] [PubMed] [Google Scholar]
- Liu N, Abe M, Sabin Leah R, Hendriks G-J, Naqvi Ammar S, Yu Z, Cherry S, Bonini Nancy M. The Exoribonuclease Nibbler Controls 32 End Processing of MicroRNAs in Drosophila. Current biology : CB. 2011;21:1888–1893. doi: 10.1016/j.cub.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Greimann JC, Lima CD. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell. 2006;127:1223–1237. doi: 10.1016/j.cell.2006.10.037. [DOI] [PubMed] [Google Scholar]
- Liu Q, Rand TA, Kalidas S, Du F, Kim HE, Smith DP, Wang X. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science. 2003;301:1921–1925. doi: 10.1126/science.1088710. [DOI] [PubMed] [Google Scholar]
- Ma JB, Yuan YR, Meister G, Pei Y, Tuschl T, Patel DJ. Structural basis for 5’-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature. 2005;434:666–670. doi: 10.1038/nature03514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiti M, Lee HC, Liu Y. QIP, a putative exonuclease, interacts with the Neurospora Argonaute protein and facilitates conversion of duplex siRNA into single strands. Genes Dev. 2007;21:590–600. doi: 10.1101/gad.1497607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CD, Brennecke J, Dus M, Stark A, McCombie WR, Sachidanandam R, Hannon GJ. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell. 2009;137:522–535. doi: 10.1016/j.cell.2009.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- Mi S, Cai T, Hu Y, Chen Y, Hodges E, Ni F, Wu L, Li S, Zhou H, Long C, et al. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5’ terminal nucleotide. Cell. 2008;133:116–127. doi: 10.1016/j.cell.2008.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pall GS, Codony-Servat C, Byrne J, Ritchie L, Hamilton A. Carbodiimide-mediated cross-linking of RNA to nylon membranes improves the detection of siRNA, miRNA and piRNA by northern blot. Nucleic Acids Research. 2007;35:e60. doi: 10.1093/nar/gkm112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker JS, Roe SM, Barford D. Crystal structure of a PIWI protein suggests mechanisms for siRNA recognition and slicer activity. Embo J. 2004;23:4727–4737. doi: 10.1038/sj.emboj.7600488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C, Anderson JT, Tollervey D. The exosome subunit Rrp44 plays a direct role in RNA substrate recognition. Mol Cell. 2007;27:324–331. doi: 10.1016/j.molcel.2007.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitz H, Tushir JS, Zamore PD. A 5’-uridine amplifies miRNA/miRNA* asymmetry in Drosophila by promoting RNA-induced silencing complex formation. Silence. 2011;2:4. doi: 10.1186/1758-907X-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- Vanacova S, Wolf J, Martin G, Blank D, Dettwiler S, Friedlein A, Langen H, Keith G, Keller W. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol. 2005;3:e189. doi: 10.1371/journal.pbio.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Sheng G, Juranek S, Tuschl T, Patel DJ. Structure of the guide-strand-containing argonaute silencing complex. Nature. 2008;456:209–213. doi: 10.1038/nature07315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J-S, Maurin T, Robine N, Rasmussen KD, Jeffrey KL, Chandwani R, Papapetrou EP, Sadelain M, O’Carroll D, Lai EC. Conserved vertebrate mir-451 provides a platform for Dicer-independent, Ago2-mediated microRNA biogenesis. Proceedings of the National Academy of Sciences. 2010;107:15163–15168. doi: 10.1073/pnas.1006432107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


