Abstract
Schizophrenia is one of the most common psychiatric disorders, but despite progress in identifying the genetic factors implicated in its development, the mechanisms underlying its etiology and pathogenesis remain poorly understood. Development of mouse models is critical for expanding our understanding of the causes of schizophrenia. However, translation of disease pathology into mouse models has proven to be challenging, primarily due to the complex genetic architecture of schizophrenia and the difficulties in the recreation of susceptibility alleles in the mouse genome. In this review we highlight current research on models of major susceptibility loci and the information accrued from their analysis. We describe and compare the different approaches that are necessitated by diverse susceptibility alleles, and discuss their advantage and drawbacks. Finally, we discuss emerging mouse models, such as second-generation pathophysiology models based on innovative approaches that are facilitated by the information gathered from the current genetic mouse models.
Keywords: schizophrenia, mouse models, candidate genes, rare mutations, common variants, disease models
I. Schizophrenia
Schizophrenia (SCZ) is a debilitating mental disorder that affects nearly 1% of the world’s population. Onset of behavioral symptoms occurs in the late teens and early twenties for most patients. These are defined as positive symptoms (hallucinations, delusions, disordered thoughts and behaviors), negative symptoms (flattened affect, asociality, avolition) and cognitive deficits such as impaired working memory (WM) and executive function. There are also structural brain abnormalities, which have been identified in patients who suffer from SCZ, such as enlargement of the ventricles and a reduction in cortical gray matter (Tandon et al., 2008). However, SCZ is a heterogeneous disease with no single defining symptom or any consistent diagnostic biological marker. Pharmacological interventions have limited effectiveness in the treatment of negative symptoms and cognitive deficits, and this poor response is further complicated by the wide array of side effects associated with antipsychotics and the need for a chronic course of treatment (Webber and Marder, 2008). Results from twin and family studies show that SCZ has a high degree of heritability at around 80% (Sullivan et al., 2003). However, identical twins are only about 50% concordant for the disease suggesting that epigenetic, environmental and very likely stochastic factors also likely play a substantial role.
II. Mouse Models of SCZ
To date, a number of animal models of psychiatric diseases have been developed to address the different aspects of these disorders (Arguello and Gogos, 2006; Nestler and Hyman, 2010). The most proximal brain dysfunction representing the clinically observed psychopathology has been recapitulated through pathophysiology models. These include primarily models that test the hypothesis that because psychoactive drugs produce a psychopathology similar to that seen in individuals with a certain psychiatric disease, the neurotransmitter system affected by the drug is dysfunctional in the disorder (Svenningsson et al., 2003). Examples of these include models of dopaminergic and glutamatergic dysregulation that can be modeled through pharmacological (Dawe et al., 2009) and genetic interventions (Mohn et al., 1999; Kellendonk et al., 2006). Models that attempt to recapitulate the processes that lead to the pathophysiology of a disorder are models of pathogenesis (Weinberger, 1995). In this paradigm, prenatal or postnatal perturbations that interfere with normal brain maturation are assessed for their capacity to produce the pathophysiological deficits later in development. Examples are hippocampal lesion models (Lipska and Weinberger, 2000) or exposure to a methylating agent that introduces various neuroanatomical and behavioral abnormalities (Moore et al., 2006). There are a number of challenges for interpreting such pathophysiology and pathogenesis-oriented approaches in the absence of accompanying human genetic evidence, and the question whether they represent legitimate disease models remains a matter of debate (Nestler and Hyman, 2010). Finally, models designed on experimentally proven risk factors or casuative agents of human disease are referred to as etiological models. This review will exclusively focus on this last approach, exemplified by specific genetic mouse models. In the last section of this paper we will also discuss the potential of generating novel pathophysiology models based on knowledge accrued by analysis of genetic models.
III. Genetic Architecture of SCZ
Two principle theories have been proposed to explain the genetic etiology of the disorder. The common disease/common allele (CDCA) hypothesis (Pritchard and Cox, 2002) proposes that the presence of common mutations with low penetrance in many genes acting in concert leads to the disease. Conversely, the common disease/rare allele (CDRA) hypothesis assumes an association with rare, but highly penetrant mutations that increase vulnerability for the disease (Cirulli and Goldstein, 2010). Evidence suggests that with SCZ, as with many other diseases, these scenarios are not mutually exclusive and that both common and rare mutations are likely involved in the etiology of the disease.
Genetic association studies provide a powerful approach for identifying risk genes in feasible sample sizes. These studies are based primarily on the CDCA hypothesis and typically examine if common genetic variants are associated with a certain trait or disorder. The simplest design compares the frequencies of genetic variants between groups of non-related cases and controls. Family-based studies that compare the frequencies of the transmitted alleles to non-transmitted alleles from parents to affected offspring are also used to examine the relationship of genetic variants to the disease. Candidate genes in this type of studies are typically identified based on a priori evidence, by focusing on candidates derived from neurobiological hypotheses (functional candidate genes) or by attempting to identify positional candidate genes either through systematic follow-up of linkage signals or based on possible biological functions (Gogos and Gerber, 2006). More recently, genome-wide association studies (GWAS) have allowed for an unbiased investigation of polymorphisms throughout the entire genome (Owen et al., 2010). GWAS have opened a window into the biology of common complex diseases and yielded several genes of small effect showing strong association with a number of complex diseases or traits (McCarthy et al., 2008). Results from GWAS in SCZ have been promising but remain controversial (McClellan and King, 2010). Moreover, support for all previously identified top candidate genes has not been found in such agnostic GWAS (Sanders et al., 2008). Overall, although common variants of small effect almost certainly contribute to the genetic risk of psychiatric disorders, genetic association studies have had only limited success so far in identifying them in an unequivocal manner. This is likely due to the complexity of the affected organ (the brain) as well as a number of technical confounds that limit the power of such assays. In that respect, it is worth noting that the Schizophrenia Research Forum (szgene.org) (Allen et al., 2008) lists 1008 susceptibility genes and 8788 polymorphisms as genetic risk loci identified primarily by candidate genetic association studies. Many of these are considered strong susceptibility genes, but none have unequivocal support.
One interesting irony of recent psychiatric genetics is that when these large GWAS data sets, collected initially to test the CDCA hypothesis, were used to determine the prevalence of large structural variations (chromosomal microdeletions and microduplications, also named copy number variations or CNVs) in the genome, new evidence emerged demonstrating the importance of rare large-effect variants in the genetic etiology of the disease in both familial and sporadic cases (Sebat et al., 2009). Specifically, several studies found an enrichment of CNVs in patients with SCZ (Xu et al., 2008; International Schizophrenia Consortium, 2008; Walsh et al., 2008; Stefansson et al., 2008), providing strong empirical evidence supporting the notion that multiple rare genetic variants contribute to the genetic risk of the disease. Notably, several rare variants with large effects on the development of SCZ have been known for a long time. In particular, the association between recurrent 22q11.2 microdeletions and SCZ, described over fifteen years ago (Karayiorgou et al., 1995), represented a shift in our understanding of the genetic architecture of SCZ, highlighting the role that rare and highly penetrant mutations play in the disease risk. This view is further strengthened by the recent identification of a widespread role of CNVs in determining susceptibility to SCZ as well as other psychiatric and neurodevelopmental disorder.
A number of bioinformatics approaches have been employed to test whether the various genes identified via GWAS or disrupted by CNVs found in patients converge to a relatively small number of affected genetic pathways (Xu et al., 2008; Glessner et al., 2010; O’Dushlaine et al., 2011; Walsh et al., 2008). Suggestive evidence has been provided for processes ranging from neurodevelopment, neurotransmission, and cell adhesion, to RNA processing. Additional approaches tested for affected functional modules, focusing on identifying among SCZ candidate genes ones displaying primate-accelerated evolution and their functional interactions (Loe-Mie et al., 2010; Bayes et al., 2011). Such approaches, as well as additional sophisticated network-based analyses (Gilman et al., 2011), can in principle provide insights into affected molecular pathways and inspire pathway-based modeling strategies. However, it remains unclear, at this point, whether functional convergence should be expected at the level of genes and molecular pathways or, rather, at the level of synapses and neural circuits affected by the SCZ susceptibility genes. This is an important issue that will influence interpretation of findings emerging from investigation of existing mouse lines and which will facilitate the design of second generation mouse models, which will be discussed in more detail below.
IV. Genetic Mouse Models of SCZ
Modeling susceptibility alleles in mice holds tremendous promise for uncovering the function of a gene and its contribution to the pathophysiology of the associated disease or disease-related endophenotypes (i.e., heritable disease-related traits found not only in patients but also in unaffected relatives) (Arguello and Gogos, 2006). Genetic mouse models are indispensable for analysis of the effects of disease-associated mutations on neural circuits and behaviors as well as the underlying molecular and cellular pathways. In addition, they offer a unique opportunity to examine early mutational effects and their developmental progression as well as allowing a thorough investigation of interactions among susceptibility genes and between genes and environmental factors.
To fully exploit these advantages, the design and analysis of mouse models needs to be carefully considered. Modeling psychiatric disorders presents a unique set of challenges, primarily stemming from the need to translate specific phenotypic traits relevant for the disease and often associated with distinctly “human” processes, into etiologically valid paradigms in the mouse where the molecular-, synaptic, cellular, and circuit-level consequences of a given mutation can be analyzed with a high degree of confidence. The difficulty of this task is further complicated by the complex nature of susceptibility alleles that may be difficult to model and often have unknown or unexpected functional outcomes. Within this context, the most important consideration in developing genetic mouse models has to do with the nature of the disease-associated genetic variants. The feasibility to translate genetic findings varies greatly between highly penetrant rare alleles and for common alleles of small effect. Common alleles are generally considered more difficult to model. One reason is that, with possibly few exceptions, studies have not uncovered common genetic variants with a clear effect on gene expression or its function. Common alleles usually have no obvious effect on protein structure (Rebbeck et al., 2004), and in some cases may only serve as proxies for physically linked, true risk variants residing within the identified gene or a nearby gene (Newton-Cheh and Hirschhorn, 2005). An additional consideration when modeling common alleles is the degree of gene sequence conservation between species which dictates the ability to model small genetic changes in mouse orthologues of human genes. Consequently, modeling approaches for these types of genetic lesions typically aim at approximating the predicted effects of the mutation, rather than attempting to faithfully reproduce the risk allele. Therefore, several interesting candidate genes have been modeled, primarily through constitutive or conditional knockout approaches or generation of dominant-negative transgenic mice. These models have proved indispensable in deciphering the functions of candidate genes and the molecular pathways they participate in, as well as in generating initial hypotheses on how their disruption contributes to disease risk. However, it is important to keep in mind that such approaches are typically not able to convey the complexities of the original mutations. In particular, the effects of the subtle variations in patient’s risk alleles cannot be easily compared to the consequences of the typically robust lesions introduced by, for example, gene knockout technologies. Examples that demonstrate the complexities pertaining to models of polymorphisms come from recent studies of autism, cognition and epilepsy associated alleles. Point mutations in neuroligin-3 resulting in amino acid substitutions were associated with autism-spectrum disorder in humans, and modeled by a knock-in approach in the endogenous mouse orthologue of the gene (Tabuchi et al., 2007). Introduction of such a mutation in mice resulted in several behavioral changes considered as models of autistic behavior in mice, accompanied by an increase in inhibitory synaptic transmission. In contrast, a complete knockout of neuroligin-3 did not cause such changes, indicating a gain-of-function mutation that cannot be modeled by traditional knockout approaches. A similar approach was used to explore the Met/Val polymorphism in the BDNF gene sequence that has been associated with various cognitive deficits in humans. In contrast to the neuroligin-3 study, the functional effects of the polymorphism (which affects activity-dependent BDNF secretion) on certain behavioral and cellular phenotypes was also observed in heterozygous Bdnf knockout mice (Cao et al., 2007; Chen et al., 2006). However whether this will be true for a wider spectrum of behavioral and cellular phenotypes remains to be determined. Finally even for alleles with well-established loss-of-function properties heterozygous and homozygous mice may have qualitatively different phenotypes. Since many risk alleles are heterozygous, phenotypes of homozygous mice may be poor predictors of the disease pathophysiology. An epilepsy-related sodium channel gene, SCN1A, which is expressed in both hippocampal pyramidal cells and interneurons, provides an instructive example given that heterozygous loss-of-function reduces sodium currents only in interneurons (Yu et al., 2006).
Studies based on the CDRA hypothesis have been more fruitful in identifying well-defined genetic lesions with clear effects on gene function or expression (Stefansson et al., 2008; Vacic et al., 2011; International Schizophrenia Consortium, 2008; Xu et al., 2008). This significantly facilitated modeling efforts, especially with highly penetrant loss or gain-of-function alleles that can be faithfully modeled by appropriate genetic manipulations. Indeed, this potential for an accurate reproduction of the disease allele is a particular advantage of this approach because it is more likely to recapitulate the physiological effects relevant to disease pathogenesis. Thus, despite their rarity and apparent heterogeneity, such mutations may be helpful in identifying cellular pathways and neural circuits affected in SCZ. Moreover, it is expected that the rapidly increasing number of models based on such lesions will facilitate uncovering convergent signaling pathways and neural circuits affected in mental disorders and thus provide downstream targets for drug discovery (Kvajo et al., 2010).
When modeling psychiatric disorders, in addition to generating a valid construct that mimics the genetic mutation in humans, it is equally important to carefully choose an appropriate phenotyping strategy. SCZ, in particular, is a complex disorder with some uniquely human traits, thus only certain aspects of its pathology can be faithfully modeled in mice (Arguello and Gogos, 2006; Arguello and Gogos, 2010). For instance, auditory hallucinations and delusions accompanying psychosis may be impossible to measure in mouse models. However, the basic foundations of perception, attention and memory are present and these can be readily assessed through objective behavioral tests. In this context, it is important to emphasize here that the etiological validity of a mouse model should be dissociated from its behavioral phenotype, simply because, with the exception of cognitive symptoms, most of the cardinal symptoms of SCZ cannot be reliably modeled in mice. Given that there are no cognitive symptoms that are specific to SCZ, behavioral assays should rather be viewed as tools that can be used to pinpoint brain areas and neural circuits affected by the engineered bona fide mutation. This realization highlights the importance of choosing phenotyping approaches based on well-established behavioral paradigms whose neural underpinnings are well characterized.
IVa. Mouse Models Based on the Rare Variant Hypothesis
The finding that a significant fraction of individuals with SCZ carry rare mutations represent a genuine paradigm shift and a decisive step towards understanding the pathogenesis and pathophysiology of the disease via the development of mouse models. In this section we summarize findings from animal models of two rare mutations that have been the most extensively characterized to date. These structural mutations arguably represent the tip of the iceberg and results from additional genetic models of psychosis based on newly identified CNVs, such as deletions in the CNTNAP2 locus (Friedman et al., 2008) or 16p.11.2 duplications (McCarthy et al., 2009) are expected to appear shortly in the literature.
1. Mouse Models of the 22q11.2 SCZ Susceptibility Locus
Occurring predominantly de novo, chromosomal microdeletions of the 22q11.2 locus are among the most common chromosomal abnormalities, with a frequency of 1 in every 4000 live births, and the cause of the 22q11.2 deletion syndrome (22q11.2DS). The phenotype of this syndrome is highly variable and can affect multiple organs and tissues. Common physical manifestations of the disorder include craniofacial and cardiovascular anomalies, immunodeficiency, short stature and hypocalcanemia (Karayiorgou et al., 2010). Individuals with 22q11.2DS also have cognitive and behavioral impairments, with a high risk for developing SCZ; long-term medical care and prenatal screening are increasingly being directed towards the early recognition and treatment of these symptoms. More specifically, individuals with the 22q11.2 microdeletion exhibit a spectrum of deficits in cognitive abilities linked to activity in the hippocampus and prefrontal cortex, such as measures of attention, WM, executive function and short-term verbal memory (Sobin et al., 2005a; Woodin et al., 2001; Bearden et al., 2001). Moreover, 22q11.2 microdeletion carriers display a range of subtle neuroanatomical deficits, including reductions in the volume of cortical and subcortical structures, enlarged lateral ventricles, and decreased cortical complexity and disorganization of the white matter (Barnea-Goraly et al., 2003; Bearden et al., 2009; Eliez et al., 2001). Up to one third of children with the microdeletion develop SCZ or schizoaffective disorder in adolescence or early adulthood (Murphy et al., 1999; Pulver et al., 1994; Chow et al., 2006). Indeed, these microdeletions account for up to 1–2% of sporadic SCZ cases in Caucasian populations (Xu et al., 2008, Karayiorgou et al., 2010). Although atypical microdeletions have been described, most of the microdeletions found in patients with SCZ are either 3 megabases (Mb) in size (including approximately 60 known genes) or 1.5 Mb in size (including approximately 35 known genes). Most of the genes affected by the 1.5 Mb deletion are expressed in the brain and it has been suggested that this region contains all key genes responsible for the increased risk of psychiatric illness (Karayiorgou et al., 1995).
There are a number of issues pertaining to the behavioral and psychiatric phenotypes associated with the 22q11.2 microdeletions, which clearly demonstrate the validity of using the 22q11.2DS as a model for SCZ and impinge upon interpretation of results obtained by relevant mouse models (for a more detailed account of these issues see a review by Karayiorgou et al., 2010). First, the core symptoms of the disease in 22q11.2 carriers are indistinguishable from other patients with non-22q11.2-associated SCZ or schizoaffective disorder. Second, earlier claims that bipolar disorder is also prevalent among 22q11.2 carriers have been safely dismissed by subsequent studies. Third, more recent claims, coinciding with emerging interest in autism genetics, that 22q11.2 microdeletions are associated with high rates of autism or autism spectrum disorders (ASDs), are highly controversial. It has been argued that diagnoses of autism or autism spectrum disorders, which are normally made at an age well before the first manifestations of SCZ, may simply reflect misdiagnosis of social impairments associated with premorbid phases of SCZ. Importantly, no significant enrichment of 22q11.2 deletions have been identified among individuals with either ASD or bipolar disorder from the general population in large genome-wide studies, further arguing against a genuine association between these disorders and 22q11.2 microdeletions. Fourth, claims that 22q11.2DS-associated psychiatric phenotypes can be confounded by the presence of mental retardation also seem unjustified. While most school-aged children with 22q11.2DS have lower than typical full scale IQ (as is also the case with many patients with SCZ), a very small percentage of children fall into the low average intelligence range. Furthermore, the pattern of cognitive dysfunction is relatively specific with most children with 22q11.2DS achieving higher scores in verbal tasks than in nonverbal tasks. Indeed, due to high phenotypic variability, many individuals with 22q11.2DS who develop SCZ have no serious intellectual disability or any of the congenital malformation abnormalities that are associated with the syndrome. Even if congenital malformations (such as facial dysmorphisms) are present, they are typically subtle and easily missed on evaluation, making these individuals clinically indistinguishable from other SCZ patients. As a result, individuals with 22q11.2 microdeletions and SCZ have been consistently recruited into research samples (including the large-scale patient cohorts used in recent GWAS), despite intensive prescreening. Overall, while carriers of the 22q11.2 microdeletion have high rates of certain behavioral disorders during childhood similar to children with other developmental disabilities, in late adolescence and early adulthood the picture changes and settles into a very specific pattern where up to one-third of all patients carrying this deletion ultimately develop psychotic symptoms that consistently meet the diagnostic criteria for SCZ or schizoaffective disorder (Gothelf et al., 2007; Murphy et al., 1999; Pulver et al., 1994). This inordinately high risk of developing SCZ is not associated with any other neurogenetic syndrome and indicates a strong etiological validity of relevant mouse models.
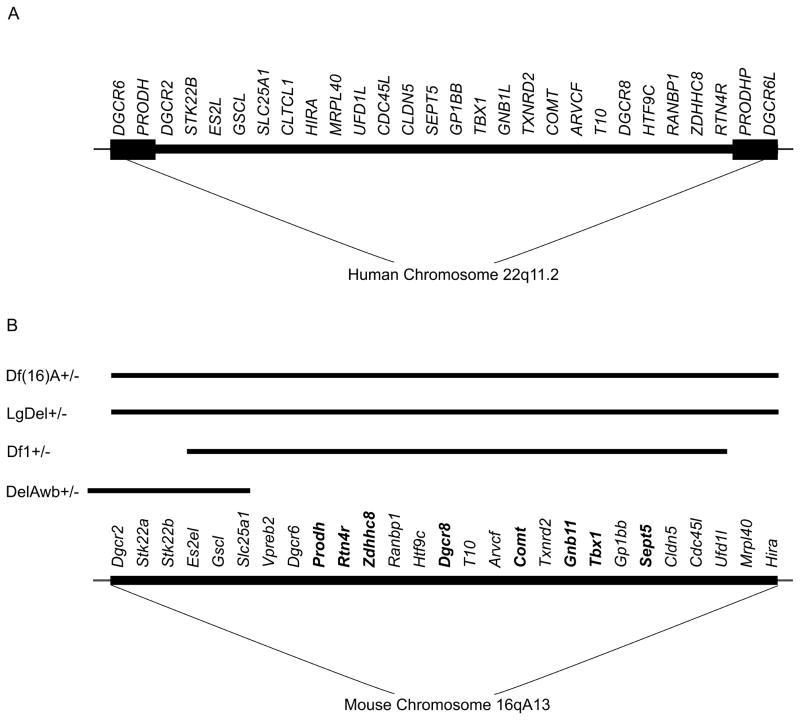
The 22q11.2 region is syntenic with a region of mouse chromosome 16. The mouse equivalent of the human 1.5 Mb 22q11.2 locus harbours orthologues of all the human functional genes except clathrin heavy polypeptide-like 1 (CLTCL1). Another difference is that humans (but not mice) carry two functional genes (DiGeorge syndrome critical region 6 (DGCR6) and DGCR6-like (DGCR6L)) in the deleted locus as a result of intralocus duplication. Overall, there is a high degree of conservation within the 22q11.2 region, and this provides a unique opportunity to generate mouse models with strong etiological validity (Drew et al., 2010; Karayiorgou et al., 2010). Accordingly, the 22q11.2 microdeletion has been modeled through long-range deletions encompassing the whole locus, as well through knockouts of individual genes located within the 22q11.2 region (Table 1, Table 2, Table 3 and Figure 1). These two approaches are complementary: long-range deletions recapitulate the complexity of the human lesion and capture interactions between genes in the region, while analysis of the individual gene knockouts provides information about their individual contributions to the molecular, synaptic, and cellular pathways involved in the syndrome.
Table 1.
Behavioral changes in 22q11 mutant mice.
| Mouse/Behavior | Df(16)A+/− Stark et al. 2008, Mukai et al. 2008, Sigurdsson et al. 2010, Drew et al. 2011 |
LgDel+/− Long et al. 2006 |
Df1 +/− Paylor et al. 2001, 2006, Earls et al. 2010 |
DelAwb +/− Kimber et al. 2009 |
|
|---|---|---|---|---|---|
| Locomotor Activity | ↑ | n.c. | n.c. | n.c. | |
| Anxiety | Open Field | ↑margin ♂ | n.c. | n.c. | n.c. |
| Light/Dark Transition | ↑dark ♂ | N/A | n.c. | n.c. | |
| Hotplate Test/Pain Sensitivity | N/A | ↑sensitivity | n.c. | n.c. | |
| Sensory Motor Gating (PPI) | ↓ | ↓ | ↓ | ↑ | |
| Startle Response | n.c. | ↑ ♂ | n.c.** | n.c. | |
| Acoustic Startle Habituation | N/A | N/A | n.c. | n.c. | |
| Cognition | Contextual/Cued Fear Conditioning | ↓/↓ | n.c./n.c. | n.c. (1 hr), ↓ (24hr)/n.c. | n.c./n.c. |
| T-maze, Spatial Working Memory | ↓acquisition | N/A | N/A | N/A | |
| MWM, Spatial Memory | n.c. | N/A | ↓ | N/A | |
| Locomotor Response to Drug | N/A | N/A | N/A | N/A | |
Abbreviations: N/A (not available), ↓(decrease), ↑(increase), n.c. (not changed), PPI (prepulse inhibition), ♀(females only), ♂ (males only)
Note: In Paylor et al. 2001 there is an increase in startle response in ♂ that is not seen in Paylor et al. 2006.
Table 2.
Morphological changes in brains of 22q11 mutant mice and 22q11 region single-gene mutant mice.
| Mouse/Morphology | Df(16)A+/− Stark et al. 2008, Mukai et al. 2008, Sigurdsson et al. 2010 Drew et al. 2011 |
LgDel+/− Long et al. 2006 |
Prodh−/− Gogos et al. 1999, Paterlini et al. 2005 |
DGCR8+/− Stark et al. 2008, Fenelon et al. 2011 |
Zdhhc8 +/−, −/− Mukai et al. 2004, 2008 |
Df1 +/− Earls et al. 2010 |
|
|---|---|---|---|---|---|---|---|
| Cortex | Dendritic Complexity | N/A | N/A | N/A | n.c. mPFC | N/A | N/A |
| Spines | N/A | N/A | N/A | mPFC: ↓width | N/A | N/A | |
| Layers | N/A | ↓cells in layer II-IV | N/A | ↓layer II/IV markers | N/A | N/A | |
| Interneurons | N/A | Altered distribution PV/CB cells in medial CTX | N/A | N/A | N/A | N/A | |
| Proliferation | Progenitor cells | N/A | ↓basal, n.c. apical | N/A | ↓basal, n.c. apical | N/A | N/A |
| BrdU Labeling | N/A | ↓cells labeled at e13.5 | N/A | ↓cells labeled at e16.5 | N/A | N/A | |
| Cell Cycle Regulatory Genes | N/A | ↓ CyclinD1, E2f2, Sestrin 2 | N/A | N/A | N/A | N/A | |
| Hippocampus | Dendritic Complexity | ↓ CA1 | N/A | N/A | ↓ CA1 | ↓ CA1 | n.c. |
| Spines | ↓density & width, n.c. length (mushroom/CA1) | N/A | N/A | CA1 mushroom: ↓width, n.c. length & density | CA1 mushroom: ↓density, n.c. length & width | n.c. | |
| Signaling | Glutamatergic Synapses | ↓PSD95 & VGlut1 in CA1 | N/A | N/A | N/A | ↓PSD95 (−/−) & VGlut1 in CA1 | n.c. AMPA-R to NMDA-R ratio in CA1, ↑ size vesicles, n.c. number of synapses or vesicles |
| COMT | N/A | N/A | ↑ levels | N/A | N/A | N/A | |
| Dopamine & Assoc. Proteins | N/A | N/A | n.c. Basal DA ↑DA response to amphetamine, ↓DRD1, ↓DARPP32, ↑PPP3C, n.c. DRD2 |
N/A | N/A | N/A | |
| Other Neurotransmitters | N/A | N/A | ↓Glut, GABA, aspartate in Hypothalamus ↓GABA, aspartate in CTX |
N/A | N/A | N/A | |
| Electrophysiology | Hippocampus | ↓CA1 inhibition ↓LTP ↓CA3 activation |
N/A | CA1: ↑synaptic transmission, ↑glutamate release, ↓PPR, ↓LTP | CA1: n.c. synaptic transmission, STD | N/A | CA1: ↑LTP, PTP, STP, PPF, synaptic facilitation, Altered Ca2+ signaling, n.c. synaptic transmission or excitability |
| Cortex | N/A | N/A | N/A | mPFC: n.c. synaptic transmission, paired pulse facilitation. ↑STD, ↓EPSP summation, potentiation | N/A | N/A | |
| Synchrony | ↓HPC/mPFC | N/A | N/A | N/A | N/A | N/A | |
| miRNA Processing | ↑pri forms, ↓mature forms of some | N/A | N/A | ↑pri forms, ↓mature forms of some | N/A | N/A | |
Abbreviations: N/A (not available), ↓(decrease), ↑(increase), not (not changed), HPC (Hippocampus), CTX (cortex), mPFC (medial prefrontal cortex), PV (parvalbumin), CB (calbindin), DA (Dopamine), PPR (pair pulse ratio), LTP (long term potentiation), STD (short term depression), PTP (posttetanic potentiation), STP (short term potentiation), PPF (paired pulse facilitation).
Table 3.
Behavioral changes in 22q11 region single-gene mutant mice.
| Mouse/Behavior | Prodh−/− Gogos et al. 1999, Paterlini et al 2005 |
DGCR8+/− Stark et al. 2008 |
Zdhhc8 +/− and −/− Mukai et al. 2004 |
RTN4R +/− and −/− Hsu et al. 2007 |
COMT−/−, +/− # Gogos et al. 1998, * Papaleo et al. 2008 |
Tbx1 +/− ^Paylor et al. 2006† Long et al. 2006 |
Gnb1l +/− Paylor et al. 2006 |
Sept5 +/−, −/− (aka Cdcrel1) Paylor et al. 2006, Suzuki et al. 2009 |
|
|---|---|---|---|---|---|---|---|---|---|
| Locomotor Activity | ↓ | n.c. | ↓activity, rears | ↓distance, rears | n.c.#,* | n.c. † | N/A | n.c. | |
| Anxiety/Depression | Open Field | n.c. | n.c. | ↑margin ♀ | n.c. | n.c. #,* | n.c. † | N/A | n.c. |
| Light/Dark Transition | n.c. | N/A | N/A | n.c. | ↓ ♀ (−/−)# | N/A^, † | N/A | N/A | |
| TST | N/A | N/A | N/A | n.c. | N/A#,* | N/A^, † | N/A | n.c. | |
| EPM | N/A | N/A | N/A | N/A | ↓ open arm* | N/A^, † | N/A | ↑ | |
| Pain Sensitivity | N/A | N/A | N/A | n.c. on hot plate | n.c. on hot plate, ↓ tail flick * | n.c. hot plate † | N/A | N/A | |
| Sensory Motor Gating (PPI) | ↓ | ↓ | ↓ ♀ (−/−) | n.c. | n.c. #,* | ↓^, n.c. † | ↓ | ↑ (−/−) | |
| Startle Response | n.c. | n.c. | N/A | n.c. | n.c. # ↑ (−/−) * |
↑^, n.c. † | n.c. | ↑ (+/−) | |
| Acoustic Startle Habituation | n.c. | N/A | N/A | N/A | N/A #,* | N/A^, † | N/A | N/A | |
| Cognition | Contextual/Cued Fear Conditioning | ↓/↓ | n.c. | N/A | n.c. | N/A #,* | n.c. † | N/A | N/A |
| T-maze, Spatial Working Memory | n.c. | ↓ acquisition | N/A | n.c. | ↑ (−/−)* | n.c. † | N/A | n.c. | |
| Aggression | N/A | N/A | N/A | N/A | ↑♂ (+/−) # | N/A^, † | N/A | ↓ (−/−) | |
| Behavioral Response to Drug | amphetamine, MK801: ↓activity | N/A | MK801: ↓activity | MK801: ↓activity, n.c.Y maze, n.c. PPI, & n.c. startle | MK801: ↓♀# | N/A^, † | N/A | N/A | |
| Effect of COMT Inhibition on Behavior | Response to amphetamine ↓locomotion, T-maze, PPI | N/A | N/A | N/A | N/A#,* | N/A^, † | N/A | N/A | |
Abbreviations: N/A(not available), ↓(decrease), ↑(increase), n.c. (not changed), PPI (prepulse inhibition), TST (tail suspension test), EPM (elevated plus maze), ♀(females only), ♂ (males only),
(Gogos et al. 1998),
(Papaleo et al. 2008),
(Long et al 2006).
Figure 1.
The chromosomal location and genetic organization of the 22q11.2 locus in humans (A) and the syntenic mouse region (B). Almost all of the functional human genes in this segment are represented in the mouse, organized in a different order. The minimal 1.5 Mb deletion is mediated by the low copy repeat sequences, illustrated as black boxes in the human chromosome. PRODH-P and DGCR6L indicate pseudogenes. Black lines above the mouse region denote the extent of the deletions in mouse models discussed in the text and summarized in Table 1. Single-gene deletion models that are discussed in the text are indicated in bold.
Two of the long-range deletion models (the Df(16)A+/− model (Stark et al., 2008) and the LgDel/+ model (Merscher et al., 2001), lack the entire 1.5 Mb region on one chromosome and thus most closely mimic human 22q11.2 microdeletion. These and other models have been subjected to extensive assessments of behavioral performance in an attempt to discern and compare the psychiatric endophenotypes associated with the loss of the genes in this region (Table 1). Due to the nature of the neuroanatomical and behavioral deficits in human carriers, these studies focused primarily on hippocampal- and prefrontal cortex- dependent behavioral functions. Assessment of these domains showed a number of behavioral abnormalities (Stark et al., 2008; Earls et al., 2010; Paylor et al., 2006). Some of the behavioral alterations, such as impairments in WM and prepulse inhibition (PPI), correspond with the neuropsychiatric phenotype seen in individuals with the 22q11.2DS.
WM refers to the short-term retention of information aiming at planning and organizing a forthcoming action that can be delayed. Impairments in WM have become increasingly recognized as a key deficit in SCZ (Green et al., 2000), and deficits in spatial WM tasks have been found in children and adolescents with the 22q11.2 microdeletion (Kates et al., 2007). Such WM abnormalities may reflect a more general disruption of the dynamics of neural networks that play a role sensory perception and cognition. Df(16)A+/− mice show a deficit in acquiring a WM -dependent task (the T-maze delayed non-match to place task) the successful execution of which depends on the frontal regions of the mouse neocortex and their interaction with the hippocampus. Interestingly, the Df(16)A+/− mice show normal spatial reference memory as assayed by the Morris Water Maze (MWM) test (Drew et al., 2011) As discussed below, this specific profile of cognitive deficits is also observed in other mouse models of rare mutations and may point to common affected neural circuits and synaptic processes. It should be noted that Earls et al. (2010) reported that another 22q11.2 deletion mouse model (Paylor et al., 2001) that carries a smaller size deletion, shows impaired performance in a MWM assay (Earls et al., 2010). The reasons for this discrepancy between these two strains (which appears to also extend to other physiological alterations) are unclear. They may be related to procedural differences or, more likely, they may reflect an important role of one or more of the differentially targeted genes. Specifically, the model analyzed by Earls et al., (2010) has 5 fewer genes (Dgcr2, Stk22a, Stk22b, Mrpl40 and Hira) deleted than the Df(16)A+/− mice and the typical human 1.5Mb 22q11.2 deletion (Karayiorgou et al., 1995; Karayiorgou et al., 2010) and one of them (Dgcr2) has recently emerged as a potential key factor underlying the high risk for SCZ associated with this locus (Xu et al., 2011).
PPI refers to a reduction in the magnitude of the startle reflex that occurs when an organism is presented with a non-startling stimulus (a prepulse) before being presented with the startling stimulus (Braff et al., 2001). It is considered a measure of sensorimotor gating and reflects the ability to inhibit the processing of irrelevant sensory information. Deficits in PPI have been observed in patients with SCZ as well as in patients with other psychiatric and neurological disorders (Braff et al., 2001). PPI deficits have also been reported in children with the 22q11.2 microdeletion (Sobin et al., 2005b). The fact that core behavioral features of the phenotype described in humans with 22q11.2DS-associated SCZ (such as deficits in PPI or WM) can be recapitulated in mutant mice supports the view that 22q11.2 mouse models can be used to dissect the underlying abnormalities on a molecular-, synaptic-, cellular- and neural circuitry-level.
Interestingly, in addition to deficits in WM and PPI, mice with a hemizygous deletion spanning all of the orthologous genes in the 1.5 Mb region exhibit robust deficits in both cued and contextual fear memory (Stark et al., 2008). The observation of deficits in fear memory indicates a previously largely unappreciated amygdala circuit dysfunction in 22q11.2DS, which may have a crucial role in the pathophysiology of 22q11.2-associated anxiety symptoms. However, the relation of this finding to increased SCZ risk remains unknown.
The notion that behavioral deficits can help guide us to identify and dissect relevant neural circuits affected by the pathogenic mutations is further supported by a recent analysis of the contribution of aberrant neural synchrony to spatial WM deficits in the Df(16)A+/− model. This work, capitalizing on the observation of impaired WM performance in 22q11.2 animal models, has helped to provide important new insights into the nature of altered brain connectivity that emerges as a result of the 22q11.2 microdeletion. Sigurdsson et al. (2010) assessed connectivity by measuring synchrony in neuronal firing between the dorsal hippocampus and medial prefrontal cortex (mPFC) during learning and execution of a WM task, and found a reduction in phase-locking of PFC neuron spiking to the theta rhythm in the dorsal hippocampus in Df(16)A+/− mice (Sigurdsson et al., 2010). Moreover, there was a correlation between the degree of synchrony and the time it took mutant mice to learn the WM task, suggesting that the impaired connectivity in the mutant mice hampers the flow of spatial information from HPC to mPFC during spatial WM-dependent tasks. Interestingly, parallel analysis of the CA1 region of the hippocampus of the Df(16)A+/− mice revealed only modest deficits in local physiology, more prominent among them a reduction in the level of inhibition of CA1 pyramidal neurons, implying a decrease in interneuron activity. Notably, induction of c-Fos expression by exploration of a novel environment confirmed a relative sparing of CA1 but showed a robust decrease in the number of activated CA3 pyramidal neurons (Drew et al., 2011).
There has been a great deal of interest for the development of structural and imaging techniques that would allow a direct comparison of the neuroanatomical changes reported in human carriers with the mouse models and possibly provide insights on the neural substrates underlying the observed behavioral and functional connectivity alterations. While no results from such studies are yet available, a considerable effort to examine the developmental processes thought to underlie such changes is under way (Table 2). Indeed, subtle but significant deficits in the process of corticogenesis resulting in decreased layer II-III pyramidal neuron density and misplaced interneurons were found in in LgDel/+ mice (Meechan et al., 2009). Moreover, in vivo and in vitro analysis in the Df(16)A+/− model showed a decrease in dendritic complexity and the number of spines and excitatory synapses in the hippocampal pyramidal cells of mutant mice (Mukai et al., 2008). Thus, it is tempting to speculate that such changes may at least in part account for the reduction in cortical thickness or regional decreases in grey matter volumes observed in some individuals with 22q11.2, and that this may be also relevant to similar phenotypes in patients with non-22q11.2DS-associated SCZ (Ide and Lewis, 2010).
The contributions of individual genes to the psychiatric aspects of the 22q11.2 syndrome have been studied in a number of mouse models (Table 3). Among them, PRODH is a leading candidate gene (Li et al., 2008; Jacquet et al., 2005; Bender et al., 2005), which encodes a mitochondrial enzyme that metabolizes L-proline, a putative neuromodulator of glutamateric and GABAergic transmission. A Prodh knock-down mouse strain (Gogos et al., 1999; Paterlini et al., 2005) showed deficits in PPI and fear memory, as well as hypersensitivity to the locomotor effects of amphetamine. The increased sensitivity to amphetamine was also reflected in increased cortical dopamine efflux in the mutant mice following acute, systemic amphetamine administration. Transcriptional profiling in the prefrontal cortex of Prodh-deficient mice revealed an upregulation in COMT, an enzyme involved in degradation of dopamine and encoded by a SCZ candidate gene (Gothelf et al., 2005) also located within the 22q11.2 microdeletion locus. There is strong evidence that COMT modulates clearance of extracellular dopamine in the prefrontal cortex (Yavich et al., 2007), suggesting that the increase in Comt expression in Prodh-deficient mice may be a homeostatic response to buffer excessive dopamine signaling in the frontal cortex. This was confirmed by experiments showing that pharmacological inhibition of Comt activity potentiated the effect of amphetamine on locomotor activity, exaggerated deficits in PPI and induced deficits in a WM dependent task in Prodh-deficient mice (Paterlini et al., 2005). Thus, this animal model revealed a genetic feedback loop that involves two interacting 22q11.2 genes, which in turn affect both dopaminergic transmission and modulate the risk of SCZ associated with this locus. Notably, these animal-model based predictions found support in recent studies of individuals with 22q11.2 microdeletion (Raux et al., 2007).
The structural changes observed in the long-range deletion models appear to be contributed by several distinct candidate genes. In mice lacking the gene for the palmitoyltransferase Zdhhc8, behavioral deficits in PPI and exploratory activity were coupled with decreased dendritic growth and formation of excitatory spines (Mukai et al., 2004; Mukai et al., 2008). Zdhhc8 is required for the process of palmitoylation, a reversible post-translational protein modification critical for protein trafficking and the functional modulation of diverse membrane and cytosolic proteins (Resh, 1999). Changes in the palmitoylation of PSD95, a key postsynaptic protein, were observed in both Zdhhc8 deficient mice and deletion mutants (Ho et al., 2011; Mukai et al., 2008), suggesting that such deficits may underlie at least some of the morphological changes, although additional substrates remain to be identified (El-Husseini and Bredt, 2002). By contrast, analysis of heterozygous knockout mice of Nogo Receptor 1 (Hsu et al., 2007), revealed no major behavioral or structural deficits suggesting that this gene is not a major contributor to the syndrome, but may modulate the risk conferred by other genes in the region.
Finally, TBX1, GNB1L and Septin 5 are additional genes located at the minimal critical regions that have been implicated in some of the behavioral deficits found in the 22q11.2DS through studies of animal models (Paylor et al., 2006; Suzuki et al., 2009). Tbx1 is a transcription factor and its mutation is sufficient to cause most of the physical features of the syndrome (Lindsay et al., 2001; Jerome and Papaioannou, 2001; Merscher et al., 2001). Heterozygous deletions of Gnb1l and Tbx1 result in PPI deficits in mice, while homozygous deficiency in Septin 5 has been shown to enhance PPI as well as modulate affective behaviors and cognitive performance.
Possibly the most striking and unexpected finding from the analysis of mouse models of individual genes is the demonstration that the 22q11.2 microdeletion results in dysregulation in the level of miRNAs (Stark et al., 2008), a class of small, non-coding RNAs that regulate the stability and translation of mRNA. This is due to the fact that the deleted region of human chromosome 22 and mouse chromosome 16 contains Dgcr8, a key component of the “microprocessor” complex essential for miRNA production. The Dgcr8 gene is haploinsufficient and an approximate halving of its expression results in the down-regulation (by ~20–80%) of a specific subset of mature brain miRNAs that may account for a portion of the transcript mis-regulation observed in the brain of Df(16)A+/− mice. As mentioned above, Df(16)A+/− mice show a deficit in acquiring a WM-dependent task. Interestingly, it was determined that this deficit arises, in part, due to deficiency of Dcgr8 and miRNA biogenesis, since heterozygous deficiency of Dgcr8 alone affects acquisition of the task, without affecting associative memory. Morphological analysis of the prefrontal cortex in Dgcr8+/− mutant mice revealed subtle changes in neuronal density stemming from impaired corticogenesis, as well as a modest decrease in the size of dendritic spines of pyramidal neurons (Fenelon et al., 2011). Electrophysiological analysis of these neurons revealed normal intrinsic membrane properties and basal synaptic transmission, but increased short-term synaptic depression and less potentiation following sustained high frequency stimulation within a physiologically-relevant range. The basis for these neurophysiological phenotypes are not known, but they are predicted to be presynaptic in nature (Fenelon et al., 2011). It is widely hypothesized that sustained activity of PFC neurons, during a delay period between the presentation of a brief, informative cue and the generation of a behavioral response, underlies the transient retention of information in WM tasks. In this respect, the finding that Dgcr8 mutants display increased synaptic depression upon sustained activation of afferent fibers in the PFC suggests that similar changes would occur during persistent bursts of synaptic inputs in vivo during WM tasks and may contribute to the deficits in WM performance observed in Dgcr8+/− and Df(16)A+/− mutant mice. Interestingly, the CA1/CA3 synapse in the hippocampus displayed normal basic synaptic transmission and synaptic plasticity. Thus, it appears that miRNA dysregulation contributes to the cognitive impairments observed in mouse models of 22q11.2DS and in patients with the 22q11.2 microdeletion. Alterations in miRNA biogenesis are expected to have considerable impact on transcript stability, but it remains unknown at this point how many genes are affected. Finding the targets of the dysregulated miRNAs will uncover additional mechanisms contributing to the SCZ endophenotypes and explain how impaired miRNA formation alters the structure and function of neural circuits thought to underlie susceptibility to SCZ and other psychiatric disorders.
Taken together, studies in the 22q11.2 microdeletion models and individual candidate genes from this region suggest that deficits associated with the mutation result from the combined effects of genes acting both individually as well as interactively. Future research aims at uncovering additional gene contributions and establishing how changes in circuit formation, excitatory and inhibitory transmission, and neuromodulation by dopamine and L-proline (Paterlini et al., 2005; Mukai et al., 2008; Stark et al., 2008) all translate into behavioral changes in mice and SCZ in humans.
2. Mouse Models of Disrupted in Schizophrenia-1 (DISC1)
A mutation in DISC1 was first discovered in a large Scottish family with a history of psychiatric disorders including SCZ, bipolar disease, major depression, and anxiety disorders (St Clair D. et al., 1990). A balanced chromosomal translocation t(1;11) (q42;q14) segregating with the disorders was found to disrupt two genes named Disrupted in Schizophrenia 1 (DISC1) and Disrupted in Schizophrenia 2 (DISC2) (Millar et al., 2000). While DISC1 has become one of the most promising and certainly most studied SCZ susceptibility genes, DISC2 transcript is a non-coding RNA antisense to DISC1 and its function is currently unknown.
Unlike the 22q11.2 deletion, which is recurrent in the general population, the truncating lesion in DISC1 has been so far identified only in the Scottish family. Moreover, no additional rare mutations in DISC1 have been found by either GWAS of common variants or CNV scans. Linkage and association studies linked common variants in the DISC1 locus with psychiatric disorders in karyotypically normal patient populations (Allen et al., 2008), but the possibility that these results are merely due to chance cannot be excluded. Several studies also provide suggestive evidence that healthy subjects and patients with SCZ carrying DISC1 variants display changes in gray matter volume in the hippocampus, prefrontal cortex and other brain regions (Callicott et al., 2005; Cannon et al., 2005; Di Giorgio A. et al., 2008; Szeszko et al., 2008), as well as deficits in working, short-term, and long-term memory (Burdick et al., 2005; Cannon et al., 2005; Hennah et al., 2005), attention (Hennah et al., 2005; Liu et al., 2006) and hippocampal and prefrontal cortical activation (Callicott et al., 2005; Prata et al., 2008). It should be noted that like other associations studies, these studies of DISC1 have not produced any statistically unequivocal results, and there have been multiple studies which have found no association between DISC1 and SCZ or other mental diseases (Devon et al., 2001; Chen et al., 2007; Kim et al., 2008; Sanders et al., 2008; Hayesmoore et al., 2008; Lim et al., 2009; Zhang et al., 2005).
An impediment to the study of DISC1-associated pathogenesis is that, aside from the associated psychiatric phenotypes, the effects of the Scottish mutation on brain function or structure in patients are completely unknown. Moreover, little is known about the impact of this mutation on DISC1 expression and protein production. The splicing of DISC1 results in numerous isoforms (Nakata et al., 2009), thus the mutation is predicted to have complex functional outcomes. The chromosomal translocation affects the C-terminal of the three major isoforms, but leaves intact many of the shorter ones (Millar et al., 2000). It is unknown if the C-terminally truncated proteins are expressed in the Scottish family; however, it has been postulated that they could act as a dominant negative, and thus have an affect beyond loss of function. While the evidence for such a scenario is currently lacking, many of the early in vitro studies investigated the effects of C-terminally truncated Disc1 and indeed found that their expression likely acts in a dominant negative fashion and affects the outgrowth of neurites as well as the localization of the endogenous Disc1 protein and its putative binding partners (Chubb et al., 2008). These findings strongly influenced the development of several mouse models expressing various truncated versions of Disc1, which are described in more detail below.
The DISC1 gene has no sequence homology to other genes, thus exhaustive in vitro and in vivo efforts to uncover its functions are currently taking place. So far, suggestive but still uncertain evidence has been offered implicating the Disc1 gene in a number of roles, including centrosomal and mitochondrial functions, neuronal outgrowth and migration, and regulation of several intracellular signaling cascades including cAMP and GSK3β signaling (Kvajo et al., 2010; Chubb et al., 2008). The Disc1 expression pattern is developmentally regulated with the highest levels occurring in the hippocampus, thalamus and cortex during embryonic and early postnatal periods (Austin et al., 2004). However, Disc1 expression quickly decreases in most areas of the brain, and during adulthood is highly expressed primarily in the hippocampus, specifically the dentate gyrus, with lower expression levels in the cortex. In vivo approaches have primarily focused on elucidating the roles of Disc1 in these structures. Several high profile studies utilized RNA interference to transiently knock down Disc1 in individual neurons. Knockdown of Disc1 in the embryonic cortex leads to extensive disruption of migration and mispositioning of neurons and affects the localization of proteins such as NudEL and Lis1 at the centrosome (Kamiya et al., 2005). Using the same technique, Mao et al. (2009) found that Disc1 knockdown decreases progenitor cell proliferation and also affects neuronal distribution in the developing cortex and adult hippocampus through the Wnt/β catenin/GSK3β pathways. Interestingly, alterations in cell positioning in this model are thought to arise not from a migration deficit, but from premature exit from the mitotic cycle, which in turn results in more neurons in layers 2/3 at expense of the progenitor cell population (Mao et al., 2009). This illustrates how differences in experimental designs, neither one of which models the actual susceptibility allele, can lead to conflicting results. A set of studies also looked at the effect of Disc1 knockdown in adult born cells in the mature dentate gyrus, and found that decreased Disc1 led to enhanced migration, exuberant overgrowth of dendrites and axons, and a generally faster maturation of adult born cells (Duan et al., 2007; Faulkner et al., 2008). Surprisingly, the pattern of enhanced migration was found only in adult born neurons; knockdown in developing granule cells during late embryonic stages resulted in fewer cells migrating into the granule cell layer (Meyer and Morris, 2009). While the reasons for these differences are not known, the effect of Disc1 knockdown in adult born neurons was tentatively attributed to its interaction with Girdin/KIAA1212 and subsequent alteration of Akt signaling (Kim et al., 2009; Enomoto et al., 2009).
Interestingly, and as described in more detail below, the prominent effects of shRNA-mediated knockdown on neuronal migration and maturation were not found in mice modeling the Disc1 allele found in the Scottish family (Kvajo et al., 2008) nor in any transgenic models (Clapcote et al., 2007; Ayhan et al., 2011; Pletnikov et al., 2008a; Hikida et al., 2007; Pletnikov et al., 2008b; Shen et al., 2008), raising an important general issue pertaining to the information obtained by models based on shRNA-mediated approaches as opposed to models based on germ-line genetic lesions. Discrepant findings from such models are not surprising and are well established in the literature (see for example the work of Khelfaoui et al., 2007; Govek et al., 2004) where transient suppression of oligophrenin-1 in neurons using RNAi and antisense RNA produces phenotypes that are distinct from those induced by a germ-line mutation of this gene). While shRNA-mediated approaches may provide valuable information about cell-autonomous functions, they have significant limitations when utilized to model in vivo effects of a gene disruption. Targeting of a small portion of cells within the brain cannot reproduce the timing and magnitude of a germ-line genetic disruption, and may result in unpredictable interactions with the “wild-type” context surrounding them. However, in the case of Disc1, results from studies utilizing RNA interference techniques may need to be reconsidered in light of a recent report showing that mouse strains carrying an intragenic 25 bp deletion of Disc1, leading to a premature stop codon and abolishing multiple Disc1 isoforms (Koike et al., 2006), show neuronal migration deficits when treated with a shRNA targeting these abolished isoforms (Kubo et al., 2010). The authors appear to rationalize these finding by referring to putative splicing isoforms postulated in a previous study by the same group (Ishizuka et al., 2007). Inherent in this later study was the assumption that Disc1 isoforms exist that have identical sizes but a sufficiently divergent sequence, so that they are not recognized by antibodies sharing the same epitopes. However, this scenario is not only biologically implausible, it is also not supported by existing data from the various Disc1 isoforms identified the last five years (Nakata et al., 2009). The most logical and parsimonious view is that the study from Kubo et al. (2010) indicates the existence of off-target effects for at least some of the shRNA sequences that have been previously utilized, thus raising concerns about this approach in general. This, together with the fact that many of the commercially available anti-Disc1 antibodies appear to be non-specific (Kvajo et al., 2008), argues for carefully designed mouse model-based analysis and emphasizes the necessity for etiologically valid approaches to uncover which of the many postulated cellular roles attributed to Disc1 play a role in the disease pathogenesis.
Mouse Models Expressing Truncated Disc1
As previously mentioned, a number of transgenic mouse models have been created with the aim of investigating the effects of the truncated Disc1 protein. These include mouse lines are overexpressing a truncated N-terminal sequence of the human DISC1 under the control of the αCAMKII promoter (named DN-DISC1) (Hikida et al., 2007) or the αCAMKII promoter and a TET-off design that allows for inducible expression of the protein (named inducible hDISC1) (Ayhan et al., 2011; Pletnikov et al., 2008b; Pletnikov et al., 2008a). A third mouse model carries a bacterial artificial chromosome with the first eight exons of the mouse Disc1 sequence (named Disc1tr) (Shen et al., 2008). All three models displayed a range of behavioral abnormalities. However, a comparison of phenotypes is somewhat limited, not only because of differences in their design, but also because of the diverse and non-overlapping battery of behaviors that each has been tested for (Table 4). DN-DISC1 mice are hyperactive and have deficits in PPI and olfaction (Hikida et al., 2007). They also have a depression-like phenotype in the forced swim test but normal social interaction. However, cognitive function was intact with normal spatial and WM. Conversely, the inducible model overexpressing truncated hDISC1 throughout development and adulthood (Pletnikov et al., 2008a) displayed normal PPI and olfaction, with sexually dimorphic deficits in spatial memory and locomotion. Moreover, these mice have increased aggressive behavior in social interactions. In a follow up study using the same mouse line, Ayhan et al. examined the effect of truncated DISC1 overexpression during distinct developmental stages (Ayhan et al., 2011). Overexpression during the pre- and postnatal period resulted in depression-like phenotype in the tail suspension test, increased locomotor response to MK801 or D-amphetamine and alterations in social interaction. If hDISC1 was expressed only during the prenatal period, there were no alterations in behavior. Expression restricted to the postnatal period increased immobility in the forced swim test and decreased social interactions. Disc1tr mice also showed changes in latent inhibition as well as depression-associated phenotypes in the forced swim and tail suspension tests, but their social behaviors have not been examined (Shen et al., 2008). Morphological analysis revealed some overlapping structural changes in the three mouse lines, primarily enlarged ventricles (Hikida et al., 2007; Shen et al., 2008; Pletnikov et al., 2008b), as well as decreased numbers of interneurons in the cortex (Hikida et al., 2007; Shen et al., 2008) and hippocampus (Shen et al., 2008). Additionally, thinning of the cortex and the corpus callosum, along with an accompanying decrease in neurogenesis were reported in the Disc1tr mice (Shen et al., 2008) (Table 5).
Table 4.
Behavioral changes in Disc1 mutant mice.
| Mouse/Behavior | Disc1Tm1Kara Koike et al. 2006, Kvajo et al. 2008 |
DN- DISC1 Hikidia et al. 2007 |
Inducible hDISC1 Pletnikov et al. 2008, Ayhan et al. 2010 |
Disc1tr Shen et al. 2008 |
DISC1-cc Li et al. 2007 |
L100P Clapcote et al. 2007, Lee et al. 2011 |
Q31L Clapcote et al. 2007, Lee et al. 2011 |
|
|---|---|---|---|---|---|---|---|---|
| Locomotor Activity | n.c. | ↑ | ↑♂ | n.c. | N/A | ↑ | n.c. | |
| Sensory Motor Gating (PPI) | n.c. | ↓ | n.c. | N/A | N/A | ↓ | ↓ | |
| Latent Inhibition | N/A | N/A | N/A | ↓ | N/A | ↓ | ↓ | |
| Cognition | DNMP | ↓(1 & 2 choice) | N/A | N/A | N/A | ↓ | ↓ | ↓ |
| MWM | n.c. | n.c. | ↑♀ | N/A | N/A | n.c. | n.c. | |
| Win-Shift | n.c. | N/A | N/A | N/A | N/A | N/A | N/A | |
| Novel Object | n.c. | N/A | N/A | N/A | N/A | N/A | N/A | |
| Fear Conditioning | n.c. | N/A | N/A | N/A | N/A | N/A | N/A | |
| Depression | FST | N/A | N/A | ↑ Immobility ♀(Post) |
↑ immobility | ↑ immobility | n.c. | ↑ immobility |
| TST | N/A | N/A | ↑ Immobility ♀(Pre+Post) |
↑ immobility | N/A | N/A | N/A | |
| Sucrose Reward | N/A | N/A | N/A | N/A | N/A | n.c. | ↓ | |
| Anxiety | Elevated Plus Maze | N/A | n.c. | n.c. | N/A | N/A | n.c. | n.c. |
| Stress Calls | N/A | N/A | N/A | ↑ | N/A | N/A | N/A | |
| Olfaction | N/A | ↓ | n.c. | N/A | N/A | n.c. | n.c. | |
| Social Interaction | N/A | n.c. | ↓ non- aggressive (Pre+Post), (Post) ↑aggressive (Pre+Post) |
N/A | ↓ | n.c. | ↓ | |
| Locomotor Response to Drug | N/A | N/A | Response to amphetamine, MK801: ↑♂ (Pre+Post) |
N/A | N/A | N/A | N/A | |
Abbreviations: N/A (not available), ↓(decrease), ↑(increase), n.c. (not changed), PPI (prepulse inhibition), DNMP (delayed non match to place test), MWM (morris water maze), FST (forced swim test), TST (tail suspension test), ♀(females only), ♂ (males only), Pre+Post (expression of inducible hDISC1 throughout prenatal and postnatal period), Post (expression of inducible hDISC1 only during postnatal period).
Table 5.
Morphological changes in brains of Disc1 mutant mice.
| Mouse/Morphology | Disc1Tm1Kara Koike et al. 2006, Kvajo et al. 2008 |
DN-DISC1 Hikidia et al. 2007 |
Inducible hDISC1 Pletnikov et al. 2008, Ayhan et al. 2010 |
Disc1tr Shen et al. 2008 |
DISC1-cc Li et al. 2007 |
L100P Clapcote et al. 2007, Lee et al. 2011 |
Q31L Clapcote et al. 2007, Lee et al. 2011 |
|
|---|---|---|---|---|---|---|---|---|
| Volume | Brain | N/A | N/A | ↓ (Pre), (Post) | ↓in ♂ | N/A | ↓13% | ↓6% |
| Cortex | ↓14% | N/A | ↓ (Pre+Post), (Post) | ↓ layers II/III | N/A | ↓ | ↓ | |
| Ventricle | N/A | ↑ | ↑ (Pre+Post), (Post) | ↑ | N/A | N/A | N/A | |
| Corpus Callosum | N/A | N/A | N/A | ↓ | N/A | N/A | N/A | |
| Cellular Changes | Dentate Gyrus/Granule Cells | ↓dendritic complexity, ↑angle of 1° dendrite, ↑migration | N/A | ↑spine density (Pre+Post) | N/A | ↓dendritic complexity | N/A | N/A |
| CA1/Pyramidal Cells | n.c. dendritic complexity, n.c. synaptic transmission, LTP in CA1 | N/A | N/A | N/A | ↓ basal synaptic transmission n.c. LTP | n.c. dendritic complexity ↓spine density |
n.c. dendritic complexity ↓spine density |
|
| Cortex | ↓length of apical dendrite mPFC layer V | N/A | ↑spine density (Pre) | N/A | N/A | ↓ cortical density & migration, altered cortical layers, ↓dendritic length & spine density | ↓ cortical density & migration, altered cortical layers, ↓dendritic length & spine density | |
| Inter- neurons | n.c. Parvalbumin, Calbindin in mPFC | ↓ Parvalbumin, n.c. Calbindin, Calretinin in mPFC | ↓ Parvalbumin(Pre) (Pre+Post), (Post) in CTX n.c. Calretinin |
↓Parvalbumin in mPFC, HPC, altered localization in DLFC | N/A | N/A | N/A | |
| Neurogenesis | ↓adult in HPC | N/A | N/A | ↓in CTX | N/A | ↓in CTX | ↓in CTX | |
| Protein Expression | N/A | N/A | ↓mDisc1, Lis1, Snap25, n.c. NdeL1, PSD95 | N/A | N/A | n.c. mDisc1, PDE4 | n.c. mDisc1, PDE4 | |
| PDE4 Activity | N/A | N/A | ND | N/A | N/A | n.c. | ↓by 50% | |
Abbreviations: N/A (not available), ↓(decrease), ↑(increase), n.c. (not changed), HPC (Hippocampus), CTX (cortex), mPFC (medial prefrontal cortex), DLFC (dorsal lateral frontal cortex), ♀(females only), ♂ (males only). Pre (expression of inducible hDISC1 only during prenatal period), Pre+Post (expression of inducible hDISC1 throughout prenatal and postnatal period), Post (expression of inducible hDISC1 only during postnatal period).
Another mouse model used the αCAMKII promoter to express a C-terminal fragment of the human DISC1 gene in an inducible and reversible fashion (Li et al., 2007). This modeling strategy aims at disrupting the putative protein-protein interactions mediated by the C-terminal part of the DISC1 protein. While the relevance of this genetic model to human pathology is limited, the mice displayed some psychiatric disease-related phenotypes, including deficits in WM and social interaction, as well as depression-like phenotypes. Interestingly, these phenotypes were only seen when C terminal DISC1 expression was induced during development, and not during adulthood.
ENU-induced Disc1 Mutant Mice
Two mutant mouse strains produced by N-ethyl-N-nitrosourea-mediated mutagenesis were recently described, carrying point mutations in the Disc1 sequence resulting in Q31L and L100P amino acid substitutions (Clapcote et al., 2007). No point mutations in DISC1 have been found in human populations, thus their relevance for modeling SCZ pathology is not clear. Interestingly, these mouse lines show distinct clusters of behavioral phenotypes representing depression- and SCZ-like deficits. The Q31L mouse has a depression-like phenotype with increased immobility in the forced swim test, decreased motivation in the sucrose reward task, and decreased social interaction, which could be reversed by bupropion, an antidepressant. The L100P mice are hyperactive and both the Q31L and the L100P mice showed decreases in paired pulse inhibition and latent inhibition as well as deficits in spatial WM during the delayed non-match to place task. Moreover, the reductions in PPI and LI were partially alleviated in L100P but not Q31L mice when they were treated with an antipsychotic drug. At least some of these phenotypes have been attributed to impaired interaction of Disc1 with phosphodiasterase 4b (PDE4b) (Millar et al., 2005), the enzyme critical for the degradation of cAMP in the brain. A recent follow up study investigating the cortical development of these mice (Lee et al., 2011) found an alteration in the formation of cortical layers resulting from decreased progenitor proliferation and impaired neuronal migration. Moreover, the dendritic complexity of cortical, but not hippocampal neurons, was also decreased.
Mouse Model of the Scottish Pedigree Mutation
A disease-oriented approach was used to generate mutant mice carrying a truncating lesion in the endogenous Disc1 mouse gene, modeling the mutation found in the Scottish family (Koike et al., 2006). This approach preserves the endogenous spatial and temporal expression pattern of the allele, thus recapitulating the effects of the human lesion without introduction of potential transgene artifacts. This mutant mouse strain carries a Disc1 allele (Disc1Tm1Kara) with two termination codons (in exons 7 and 8) and a premature polyadenylation site in intron 8. Western blot analysis of brain extracts revealed the elimination of the two major Disc1 isoforms, and the retention of the short N-terminal isoforms, thus recapitulating the predicted effects of the Scottish mutation (Kvajo et al., 2008; Koike et al., 2006).
Behavioral characterization of these mice revealed no change in PPI and normal cognitive performance in a battery of tests, except for a robust and highly specific deficit in WM observed in two independent assays (Table 4) (Koike et al., 2006; Kvajo et al., 2008). Importantly, the behavioral deficits were present in the same magnitude in both homozygous and heterozygous mice. The Scottish allele in humans is a heterozygous mutation, and although gene dosage correlations may not be evolutionary conserved, alterations found in heterozygous mutant mice may be particularly useful in determining the phenotypes more relevant for the pathology. These behavioral deficits point to a hippocampal and/or prefrontal cortex impairment, and thus a comprehensive investigation of these structures was performed (Table 5) (Kvajo et al., 2008). While the gross brain morphology appeared normal, analysis at a cellular level has shown variations in a number of hippocampal and a few prefrontal cortex parameters. Analysis of adult neurogenesis in the dentate gyrus revealed a decrease in the proliferation of neuronal precursors and a slightly enhanced migration of adult born neurons, as well as an alteration in the alignment of the cells. Mature granule cells also displayed this misalignment, as well as a decrease in dendritic complexity and the number of dendritic spines. Interestingly, these morphological deficits appear to be largely confined to the dentate gyrus, as analysis of the CA1 region of the hippocampus revealed no changes in the morphology of pyramidal cells. However, electrophysiological recordings of the CA3/CA1 synapse showed alterations in short-term plasticity, while long-term potentiation and release probability were unaffected. Pyramidal neurons in the prefrontal cortex displayed normal dendritic complexity; however, a 10% decrease in the length of the apical dendrites was found, coupled with a small decrease in the overall volume of the prefrontal cortex. Quantification of the number of interneurons revealed no changes in this structure. Overall, analysis in these mice suggests that modest yet widespread alterations in structure and short-term plasticity could underlie the development of SCZ in the Scottish family.
IVb. Mouse Models Based on the Common Allele Hypothesis
In light of the inordinate number of candidate SCZ susceptibility genes identified by genetic association studies using common variants and the serious shortcomings of such approaches, we will focus here on a limited number of relevant models included within the “Top 30” candidates genes of the SchizophreniaGene database (http://www.szgene.org) (Table 6). This is an unbiased database of candidate risk genes compiled through meta-analyses of genetic association studies. The list is by no means definitive and is constantly evolving with the ranking of individual genes fluctuating over time. As such, this list may not represent an accurate indicator of relevance for each candidate susceptibility gene and we use it only to avoid a biased representation of genes. Table 6 summarizes the behavioral and structural brain-related phenotypes found in available mouse models of these genes. As previously discussed, these mouse models (primarily knockouts and overexpressors) do not recapitulate the polymorphisms associated with the risk of SCZ, and their phenotypes primarily act as a general guidance in identifying the processes regulated by these genes. In addition, as an example of a strong positional candidate gene within the “Top 30” list, we will discuss in some detail the results of mouse models addressing the contributions of Neuregulin 1 (NRG1), highlighting a number of shortcomings in the interpretation of the results from such models. It should be noted that while NRG1 received a lot of attention in the past, recent agnostic GWA studies do not demonstrate this gene in their top results (Purcell et al., 2009).
Table 6.
Behavioral and structural alterations in available mouse models from the “Top 30” common alleles (Based on the SRF Schizophrenia susceptibility list from March 15th 2011). Representative references for genetic studies and key genetic models and are annotated.
| Locus | Representative Genetic Studies | Gene Function | Mutant Models | Behavioral Phenotypes | Physiological Phenotypes | Representative Models |
|---|---|---|---|---|---|---|
| PRSS16 | (Stefansson et al., 2009) | Serine protease expressed in the thymus, playing a role in the antigen presenting pathway | N/A | N/A | N/A | N/A |
| PGBD1 | (Stefansson et al., 2009) | Transposase specifically expressed in the brain. No known function | N/A | N/A | N/A | N/A |
| NRGN | (Stefansson et al., 2009) | Postsynaptic protein kinase substrate that binds calmodulin in the absence of calcium | KO | ↓Spatial learning (MWM), anxiety-like phenotype (light-dark test, open field) | ↑LTD ↓LTP, STP (paired pulse facilitation) |
(Pak et al., 2000) |
| NOTCH4 | (Stefansson et al., 2009) | Member of the Notch family, playing a role in a variety of developmental processes | N/A | N/A | N/A | N/A |
| HIST1H2 BJ |
(Stefansson et al., 2009) | Member of the histone 2B family, responsible for the nucleosome structure of the chromosomes | N/A | N/A | N/A | N/A |
| PDE4B | (Pickard et al., 2007) | Member of the type IV, cyclic AMP (cAMP)- specific, cyclic nucleotide phosphodiesterase (PDE) family. Specifically hydrolyzes cAMP in the brain | KO | ↓PPI ↓locomotor activity ↓AMPH response anxiety-like phenotypes (holeboard test, light-dark test, open field) anti-depressant-like behavior (tail suspension test, forced swim test) |
↓DA, 5-HT ↑ adult neurogenesis |
(Zhang et al., 2008) |
| TCF4 | (Stefansson et al., 2009) | A basic helix-turn-helix transcription factor | KO | N/A | Perinatally lethal due to deficits in a subpopulation of progenitor cells | (Brinkmeier et al., 2007) |
| OE | ↓PPI ↓contextual and cued fear memory |
N/A | (Brzozka et al., 2010) | |||
| GWA_16 p13.12 |
(O’Donovan et al., 2008) | Unknown | N/A | N/A | N/A | N/A |
| ZNF804A | (O’Donovan et al., 2008) | Zinc finger protein 804A | N/A | N/A | N/A | |
| DRD4 | (Weiss et al., 1996) | Encodes the D4 dopamine receptor subtype | KO | ↑sensitivity to stimulants ↓exploratory behaviors (emergence, novel object) anxiety-like phenotypes (elevated plus maze, light-dark test) |
↑spontaneous synaptic activity ↓glutamate density associated with asymmetrical synaptic contacts |
(Rubinstein et al., 1997) |
| DRD2 | (Shaikh et al., 1994) | Encodes the D2 dopamine receptor subtype | KO | ↓locomotor activity ↑sensitivity to stimulants |
N/A | (Palmer et al., 2003) |
| BAC OE | ↑locomotor activity ↓sensitivity to stimulants |
N/A | (Kramer et al., 2011) | |||
| DAOA | (Chumakov et al., 2002) | Primate-specific protein with a mitochondrial function | BAC | ↓PPI ↑compulsive behaviors ↑PCP sensitivity ↓aggression ↓olfactory discrimination ↓PPI ↓motor learning ↓spatial learning |
Mitochondrial dysfunction ↓ STP (paired pulse facilitation) ↓high-frequency transmission |
(Otte et al., 2009) |
| AHI1 | (Amann- Zalcenstein et al., 2006) | Mutated in the Joubert syndrome. Required for both cerebellar and cortical development in humans |
KO | N/A | Retinal degeneration, vesicle trafficking deficits | (Westfall et al., 2010) |
| Neuron- specific KO | Depression-like phenotypes(forced swim test, tail suspension test) | ↓TrkB signaling | (Xu et al., 2010) | |||
| GWA_11 p14.1 |
(O’Donovan et al., 2008) | Unknown | N/A | N/A | N/A | N/A |
| TPH1 | (Hong et al., 2001) | Catalyzes the biosynthesis of serotonin outside the brain | N/A | N/A | N/A | N/A |
| HTR2A | (Williams et al., 1996) | Serotonin receptors | KO | Sleep-wakefulness disturbances | N/A | (Popa et al., 2005) |
| RPP21 | (Purcell et al., 2009) | Subunit of nuclear ribonuclease P | N/A | N/A | N/A | N/A |
| CCKAR | (Tachikawa et al., 2001) | Cholecystokinin receptor that regulates the release of beta-endorphin and dopamine | Natural KO in rat strain | Anxiety-like phenotypes (elevated plus maze, black and white test box) | N/A | (Yamamoto et al., 2000) |
| GABRB2 | (Lo et al., 2004) | GABA receptor, mediating the fast inhibitory synaptic transmission | KO | ↓sensitivity to sedative drugs ↑locomotor activity |
N/A | (Blednov et al., 2003) |
| DTNBP1 | (Straub et al., 2002) | Organelle biogenesis | KO | ↓working memory (DNMP, T- maze) ↓spatial memory (MWM) ↓novel object recognition ↓habituation ↓social interactions |
↓number of synaptic vesicles and probability of release ↑vesicle size ↑NR2a in the hpc ↑LTP in the hpc ↑D2 in the PFC ↓inhibitory input, PPF, EPSC in the PFC |
(Jentsch et al., 2009) |
| C6orf217 | (Amann- Zalcenstein et al., 2006) | Primate-specific gene with unknown function | N/A | N/A | N/A | N/A |
| RELN | (Shifman et al., 2008) | Extracellular matrix protein critical for cell positioning and neuronal migration | KO | Ataxia | Deficits in cell migration and other systems | (D’Arcangelo et al., 1995) |
| HET | ↓task acquisition ↑PPI |
↓spine density ↑ neuronal packing density ↓GABAergic signaling |
(Liu et al., 2001) | |||
| MDGA1 | (Kahler et al., 2008) | GPI-anchored immunoglobulin superfamily | KO | N/A | Migration deficits | (Ishikawa et al., 2011) |
| CMYA5 | (Chen et al., 2010) | Z-disc-related protein | N/A | N/A | N/A | N/A |
| DISC1 | (Hennah et al., 2009) | Scaffold protein, involved in cellular proliferation and migration as well as intracellular signaling | Mouse models of Disc1 are discussed in detail in the text and Table 2 | |||
| NRG1 | (Stefansson et al., 2002) | Signaling protein that mediates cell-cell interactions in multiple cell systems | Mouse models of Nrg1 are discussed in detail in the text | |||
| MTHFR | (Arinami et al., 1997) | Folate metabolism | HET | hyperactivity ↓motor ability ↓ recognition memory (object recognition) |
Cerebellar abnormalities | (Levav-Rabkin et al., 2011) |
| AKT1 | (Emamian et al., 2004) | Serine-threonine protein kinase involved in cellular survival, signaling and neuromodulation | KO | ↓working memory retention ↓contextual fear conditioning ↓spatial learning (MWM) PPI* ↑anxiety (elevated zero maze) ↑sensitivity to stimulants |
↓dendritic development ↓adult neurogenesis ↓LTP and STP in the hpc altered gene expression |
(Emamian et al., 2004) |
| RGS4 | (Chowdari et al., 2002) | GTPase activating protein | KO | ↓social interaction | N/A | (Pacey et al., 2011) |
| PPP3CC | (Gerber et al., 2003) | Calmodulin-dependent protein phosphatase involved in a wide range of biologic activities | Forebrain specific KO | ↓working memory (DMTP, radial maze) Hyperlocomotion, ↓social interactions, ↓PPI ↓latent inhibition ↓nesting behavior ↑sensitivity to stimulants |
↓LTD in the hpc | (Zeng et al., 2001) |
| Inducible KO | ↑short-term and long- term memory (object recognition) | ↑LTP in the hpc ↑STP (paired pulse facilitation) in the hpc |
(Malleret et al., 2001) | |||
| OE | ↓long term memory (Barnes maze) | (Mansuy et al., 1998) | ||||
No changes in basal PPI were observed in Emamian et al. 2008, while Balu et. al. 2008 reported decreased PPI in a different mutant strain.
Abbreviations: KO: knockout; OE: overexpressor; Het/+: heterozygote, BAC: bacterial artificial chromosome, N/A: not available
Mouse Models of Neuregulin 1
Neuregulin 1 (NRG1), and to a lesser extend its receptors ErbB2 and ErbB4, are among the leading candidate susceptibility genes for SCZ (Stefansson et al., 2002; Law et al., 2007). NRG1 is a pleitropic gene which regulates diverse biological functions including neuronal migration, neurite outgrowth, synaptic transmission, glial cell proliferation, myelination, as well as both glutamatergic and GABAergic signaling (Corfas et al., 2004). A number of polymorphisms in NRG1, ErbB2 and ErbB4 have been associated with SCZ as well as with variations in cognitive functions and sensorimotor gating in normal populations (Silberberg et al., 2006; Hong et al., 2004; Roussos et al., 2011) even though the possibility that these results are merely due to chance cannot be excluded. The majority of these polymorphisms are located in the intronic parts of the gene, thereby not directly affecting the properties of the protein product. It has been hypothesized that they may alter NRG1 expression, which is supported by some studies suggesting both increased and decreased NRG1 expression in the presence of certain polymorphisms (Law et al., 2006; Nicodemus et al., 2009). Analysis of NRG1 expression in patient populations has presented a similarly mixed picture, with both reductions and increases in protein and mRNA having been reported (Hashimoto et al., 2004; Chong et al., 2008; Bertram et al., 2007; Law et al., 2006; Silberberg et al., 2006; Law et al., 2007). Most recently, a single rare deletion in ErbB4 has been found in a whole genome scan for CNVs (Walsh et al., 2008), but this association did not reach genome-wide significance and its effects on neuregulin signaling as well as its relevance for the disease are presently unknown.
The lack of a consistent effect of polymorphisms on NRG1 expression or function makes their modeling a challenging task. So far, the majority of approaches focused on examining the effects of altered Nrg1/ErbB signaling by either knockdown or overexpression of the target genes. These models typically generate a significant amount of information on the cellular, structural and behavioral deficits induced by impaired Nrg1 function. However, the value of these models for studying the underlying pathology of SCZ is limited due to the disproportionate magnitude of these genetic lesions when compared to the NRG1 and ErbB-associated polymorphisms. Mice with targeted deletions of Nrg1 and ErbB4 die during embryogenesis (Gassmann and Lemke, 1997; Meyer and Birchmeier, 1995). Thus, Nrg1 and ErbB4 heterozygous mice as well as Nrg1 hypomorphic mice lacking various domains of the gene have been used to study the effects of decreased Nrg1 levels. Nrg1 deficient mice show largely normal brain morphology, but display deficits in several neurotransmitter systems, including glutamate signaling through NMDA receptors, as well as serotonin signaling (Stefansson et al., 2002). Behavioral analyses revealed deficits in PPI and latent inhibition, alterations in locomotor motor activity and anxiety. Nrg1 mutants also showed disrupted social interactions, altered behavioral responses to NMDA receptor antagonists, and deficits in contextual fear conditioning but normal spatial learning and WM (Ehrlichman et al., 2009; Karl et al., 2007; O’Tuathaigh et al., 2006; O’Tuathaigh et al., 2007; O’Tuathaigh et al., 2008; O’Tuathaigh et al., 2010; Dean et al., 2008; Rimer et al., 2005). Interestingly, ErbB4 heterozygous mice displayed only some of the neuregulin-associated behavioral phenotypes. For instance, they were found to have increased locomotor activity, but no alterations in PPI (Stefansson et al., 2002).
The roles of Nrg1 signaling in specific cell populations have been examined in mouse strains with conditionally inactivated ErbB receptor signaling. While these models uncovered cell-specific functions of Nrg1, they do not represent valid disease models, because cell-specific contributions of neuregulin to disease phenotypes have not been described. Barros et al. (2009) deleted ErbB2/ErbB4 specifically in neuronal progenitors, which was predicted to abolish all neuregulin-dependent signaling in developing neurons (Barros et al., 2009). Unexpectedly, these mice showed normal development of cortical and hippocampal layers, challenging the proposed role of Nrg1 in neuronal migration. However, they had prominent deficits in spine maturation, which is in line with in vitro data suggesting a role for Nrg1 in synaptic stabilization (Huang et al., 2000). Similar to some Nrg1 hypomorphs, these mutant mice displayed reduced PPI and increased aggression. Both morphological and behavioral defects could be rescued by treatment with the antipsychotic clozapine. It should be noted, however, that the outcome of pharmacological treatments cannot be used to either infer underlying disease mechanisms or ascribe etiological validity in a mouse model of SCZ.
Nrg1 is prominently expressed by oligodendrocytes and its signaling is thought to be critical for their development (Nave and Salzer, 2006). This led researchers to examine the role of Nrg1 by oligodendrocyte-specific expression of a dominant–negative form of ErbB4. These mice displayed changes in oligodendrocyte number and morphology, reduced myelin thickness, and slower conduction velocity in CNS axons, as well as increased levels of dopamine receptors and transporters (Roy et al., 2007). The mutation also led to behavioral deficits, including increased anxiety and impaired social behaviors. Behavioral phenotypes such as hyperactivity, deficits in prepulse inhibition, and impaired contextual and WM were also found in mice modeling a disruption of neuregulin signaling in inhibitory interneurons (Wen et al., 2010). Some of these phenotypes could be ameliorated by the GABA enhancer diazepam, suggesting that neuregulin-mediated disruption of inhibitory circuits directly contributed to their development. The relationship of these phenotypes to SCZ is uncertain: while changes in inhibitory neurons have been reported in schizophrenic patients (Daskalakis et al., 2007), they have been also associated with neurodevelopmental disorders such as autism (Tabuchi et al., 2007) and Rett syndrome (Chao et al., 2010). A few studies examined the effects of increased Nrg1 expression using transgenic approaches. These models have the most obvious drawbacks, stemming primarily from the non-physiological spatial and temporal expression of Nrg1. Kato et al. (2010) reported that mice overexpressing Nrg1 in all tissues displayed increased locomotor activity, decreased context-dependent fear conditioning, and impaired social interaction (Kato et al., 2010). Moreover, they also showed increases in parvalbumin interneuron and myelination markers in the prefrontal cortex as well as marked decreases in tyrosine hydroxylase levels and dopamine content in the hippocampus. Mice overexpressing Nrg1 in postmitotic neurons displayed a myelination phenotype affecting their motor performance and possible changes in PPI (Deakin et al., 2009).
Recently, a missense mutation in the transmembrane part of Nrg1 has been associated with SCZ (Walss-Bass et al., 2006). This mutation was subsequently implicated in deficient proteolytic processing of neuregulin by the BACE1 protease as well as with impaired dendritic growth in cultured neurons (Chen et al., 2010). Possible changes in processing of Nrg1 precursors have also been found in schizophrenic patients (Barakat et al., 2010). BACE1 deficient mice displayed altered Nrg1 proteolysis and downstream ErbB signaling (Savonenko et al., 2008) as well defects in prepulse inhibition, cognitive impairments and social recognition, and reduced spine density in the hippocampus. However, the contributions of neuregulin to these phenotypes are difficult to dissect out because BACE1 has numerous other physiological targets and has been implicated in non-psychiatric brain pathologies such as Alzheimer’s disease (Vassar et al., 2009).
The studies summarized above provide initial insight into the basic function of NRG1 and have uncovered cell-specific contributions to its function. However, whether such models can recapitulate the relevant clinical aspects of common alleles is a matter of debate. While dysregulation of neuregulin signaling appears to result in a relatively coherent set of behavioral abnormalities, their relevance for the development of SCZ is uncertain, and, provided that the association between Nrg1 and SCZ is indeed a true finding, a more disease-focused approach is necessary to identify those changes that are most relevant for the expression of the pathology in humans.
V. Common Themes Emerging From Current Genetic Mouse Models
The rapidly expanding number of genetic mouse models of SCZ has led to an increase in expectations of finding shared phenotypes and points of conversion that will ultimately help uncover the cellular pathways and aberrant neurocircuitry underlying the disorder. This view is pertinent for efforts to integrate findings from various models of the same gene, as with the various Disc1 or neuregulin models summarized above. However, it became even more urgent with the advent of human genetic studies, which exposed the large functional diversity of susceptibility genes carrying rare mutations (Xu et al., 2008; Xu et al., 2009; Xu et al., 2011). Despite these expectations, the knowledge so far accrued does not appear to support the idea of phenotypic convergence and only few common themes emerge from comparison of existing mouse models.
There are several possible reasons for such an outcome. First, as previously discussed, robust genetic findings are critical for the creation of reliable mouse models. So far there have been few alleles, especially among the common variants, that were consistently associated with SCZ in multiple studies. Larger GWAS for common and rare variants as well as deep sequencing studies that are under way are expected to provide more reliable information and stronger candidates for modeling, at least for rare mutations. Next, the design of animal models critically impacts the results obtained from these studies. “Generic” models built by conventional transgenic approaches which do not reproduce the bona fide risk allele are typically associated with a spectrum of phenotypes, many of which are only tangentially relevant for the disease. Indeed, a deletion of a given gene will typically affect multiple processes, making it necessary to dissect out the specific processes pertinent for the pathology. Similarly, overexpression of a dominant negative form of a protein may result in non-specific cellular toxicity which will generate deficits that may have nothing to do with how that gene contributes to the disease. Finally, introduction of mutations that have not been well characterized functionally and have not been associated with the disease will result in yet more inconclusive phenotypes. It is becoming increasingly apparent from the diverse sets of phenotypes elicited by various modeling approaches that carefully designed, etiologically valid models are best positioned to uncover the unique and specific contributions of susceptibility alleles. Attempts to integrate findings from several types of models (even those not relevant for the disorder) in order to reach a consensus, are likely to be counterproductive. Overall, emphasis should be placed on a few well-designed models of robust and well-understood genetic lesions that will provide information pertinent to the disease pathology.
However, it should be noted that even the most carefully designed mouse models have limitations when recreating a complex disorder such as SCZ. Due to the genetic architecture of SCZ, a mutation in a single candidate gene may convey only a particular aspect of the disorder. Similarly, it has been proposed that certain aspects of the disease may stem from interplay with environmental factors (Meyer and Feldon, 2010), thus necessitating additional efforts to model such interactions. Finally, emergence of efficient compensatory responses such as changes in activity and expression of other genes (Anholt et al., 2003) may suppress some phenotypes, requiring additional genetic, pharmacological, or environmental manipulations for them to become fully penetrant at the behavioral level.
Despite these problems, some themes are beginning to emerge, primarily through comparison of etiologically designed models. While analysis of molecular, cellular and synaptic alterations is only now beginning, comprehensive behavioral surveys have amassed enough information to allow a dissection of common behavioral traits. In this regard, the highly specific profile of cognitive deficits in the Disc1 mouse model of the Scottish mutation, with a robust impairment in spatial WM in the absence of equally robust changes in short-term-, associative-, reference- and recognition memory is particularly valuable, pointing to a single affected cognitive domain with well defined anatomical substrates. A similar pattern of deficits (impaired WM but normal spatial reference memory) were found in the Df(16)A+/− mouse model of the 22q11.2 microdeletion, despite the fact that it displays a broader pattern of behavioral alterations (Table 1). This observation suggested that common deficits in Disc1 and 22q11.2 mouse models may reside in the prefrontal cortex-hippocampal circuit, which is thought to modulate spatial WM. Follow-up work showed that spatial WM deficits in the 22q11 mice may be due to alterations in intrinsic prefrontal cortex circuits (Fenelon et al., 2011) as well as in the communication between the prefrontal cortex and the hippocampus (Sigurdsson et al., 2010). Within the prefrontal cortex, aberrant short-term plasticity was identified as a candidate synaptic mechanism (Fenelon et al., 2011) and it was suggested that similar alterations in short-term dynamics may underlie some of the SCZ-associated deficits, including dysconnectivity, cognitive dysfunction, and inability to accurately interpret and integrate sensory information. Moreover, the same processes underlying these quantifiable deficits may be also contribute to symptoms such as hallucinations and delusions, which are currently out of reach for modeling efforts. Interestingly, deficits in short term plasticity and/or WM are also shared by several mouse models of prominent candidate genes including neurogranin, dysbindin, Akt1 and calcineurin (Table 6), supporting the notion that impaired modulation of these processes may be at the core of SCZ susceptibility.
In addition to possible convergence at the level of neural circuitry and behavior, it is worth noting that a number of common themes emerge at the level of intracellular signaling pathways (Kvajo et al., 2010). Within the “Top 30” genes three common pathways stand out: cAMP-dependent signaling, which has been associated with PDE4b (Siuciak et al., 2008), Disc1 (Clapcote et al., 2007), DRD4 (Blasi and Bertolino, 2006), neurogranin (Wu et al., 2002), and GABA receptor function (Chalifoux and Carter, 2010); Akt1- dependent signaling which is downstream of Akt1, Disc1 (Enomoto et al., 2009), DRD2 (Beaulieu et al., 2007), Nrg1 (Flores et al., 2000) and GABA receptor signaling (Wang et al., 2003), and calcineurin signaling which is downstream of calcineurin, GABAR (Greengard et al., 1998), Nrg1 (Kao et al., 2009) and neurogranin (Diez-Guerra, 2010).
Although it is still early for definitive conclusions, analysis of existing mouse models suggests that despite of the underlying genetic complexity, SCZ pathology may stem from alterations in a relatively small number of neural processes and signaling pathways. It also further emphasizes the importance of avoiding a heavy handed approach in the design of animal models, and the need to faithfully replicate the allele-specific phenotypes in order to be able to integrate in a more efficient and definitive way results from multiple mouse models of susceptibility alleles.
VI. Emerging Models
During the past few years the field of psychiatric genetics has witnessed both a renewed interest in rare SCZ risk alleles (especially rare pathogenic CNVs) as well as some unexpected difficulties in establishing genetic causality in psychiatric disorders via genetic association studies based on common variants. The finding that a significant fraction of individuals with SCZ carry pathogenic CNVs could represent a decisive step towards understanding the pathogenesis of the disease. Arguably, the identified structural mutations represent the tip of the iceberg, and more mutations are expected to be found by carrying out deep sequencing studies (Xu et al., 2011). These efforts promise to reveal deleterious rare point mutation and genetic truncations of current candidate genes, as well as previously unsuspected novel genes, providing a unique opportunity for engineering genetically faithful and thus etiologically valid animal and cellular models (Arguello & Gogos, 2006).
Considering the great functional variety among the alleles uncovered by recent screens, choosing the best candidates will be a demanding task. The primary criteria for the identification of these alleles need to involve strong statistical support, preferably from multiple, well-designed studies. In addition, finding candidates that are good pharmacological targets will be particularly helpful for moving towards development of new treatments for psychosis. An example of such a candidate allele are the recently uncovered SCZ-associated duplications in the VIPR2 locus (Vacic et al., 2011). VIPR2 encodes the vasoactive intestinal peptide (VIP) receptor VPAC2, a G-protein-coupled receptor that is expressed in a variety of tissues including the suprachiasmatic nucleus, hippocampus, amygdala and hypothalamus (Dickson and Finlayson, 2009). VPAC2 binds VIP and activates cyclic AMP signaling and downstream PKA-dependent processes. While very little is known about the specific roles of this receptor in the brain, genetic studies in mice showed its importance in sustaining normal circadian oscillations in the suprachiasmatic nucleus (Cutler et al., 2003; Harmar et al., 2002). VIPR2 is a novel candidate gene and the robustness and consistency of the association between the described CNVs and SCZ make it a forerunner for future modeling attempts. Moreover, the well-described downstream effects of the mutation, resulting in increased cAMP levels, make it a particularly attractive candidate for pharmacological interventions. In fact, VPAC2-mediated signaling has been already targeted in non-neurobiological pathologies, including COPD (Groneberg et al., 2006) and diabetes (Tsutsumi et al., 2002), making the transition to psychiatric pharmacology less arduous.
Another avenue to be explored by emerging genetic models will deal with the compound effects of genetic mutations. It has been generally accepted that susceptibility to SCZ is likely conveyed by a combination of multiple genetic lesions, and as discussed above, this represents one of the limitations of the “single allele” mouse models. Thus, models addressing the interactions of structural variants with each other or with environmental factors will bridge these limitations and provide a more complete picture of the complex interplay among the susceptibility factors.
Yet another important category of emerging SCZ models will be novel models of pathophysiology. It has been argued that the most proximal cause of SCZ symptoms are abnormalities in neuronal circuits rather than genetic alterations. Therefore, modeling disruptions in circuits, instead of focusing efforts in susceptibility alleles, may ultimately be a more productive approach towards understanding the structural alterations leading to pathology. Although this is a valid argument, until recently this strategy has been hampered by our rudimentary understanding of the anatomical framework of SCZ. Focusing on neuronal circuits based on largely unfounded hypotheses and without evidence for robust and consistent SCZ-associated structural and functional abnormalities are likely to be counterproductive. On the other hand it is becoming increasingly clear that the complexity of the neural substrates affected in SCZ offers a large mutational target comprised of possibly hundreds genes (which along with the relatively high rate of protein-altering new mutations, provides a plausible explanation for both the high global incidence and the persistence of SCZ) (Xu et al., 2011). The size of the mutational target implies that it may be equally counterproductive to generate and analyze genetic models for each one these culprit genes. Recent advances using genetic mouse models highlighted several affected circuits and synaptic processes, suggesting that focusing into a small number of valid and robust genetic models may be sufficient, at least initially, to generate the type of convergent data necessary to pave the way towards efficient second-generation pathophysiology models. Production of such models is facilitated by the recent development of powerful tools, which allow modeling of the structural and functional alterations implicated in circuit dysfunction. The emerging field of optogenetics, for example, which combines optical and genetic techniques to probe and modulate neural circuits within intact animals, has been successfully used to dissect circuits involved in neurological disorders such as Parkinson’s disease (Gradinaru et al., 2009) and behaviors that may be related to psychiatric disorders (Tye et al., 2011). Such approaches can be used, for intance, to model the prefrontal-cortical and hippocampal contributions (Sigurdsson et al., 2010; Fenelon et al., 2011) to the spatial WM deficits in the 22q11.2 mouse model by modulating the activity of hippocampal/prefrontal cortex circuits. They may also be used to model specific deficits in hippocampal circuits revealed by analysis of the etiologically valid mouse model of the DISC1 mutation. As mentioned above, recent studies showed that Disc1 deficiency as well as the 22q11.2 microdeletion impact on the function of dentate gyrus and/or the CA3 region. These are structures of interest for the pathogenesis of mental disorders (Tamminga et al., 2010) that modulate cognitive processes such as WM and pattern separation, both of which have been proposed as potential mechanisms for the development of psychotic thought content. These hypotheses can be tested by using optogenetic approaches to mimic the alterations in activity of individual segments of the DG/CA3/CA1 circuit that are observed in animal models and thus provide a more complete picture of the contributions of multiple circuits to SCZ-associated deficits.
In summary, the recent advancements in the genetics of SCZ have opened new avenues for the development of etiologically valid mouse models. A complementation of the knowledge generated from genetic mouse models with the exploration of the underlying circuit alterations will allow a more stringent parsing of the current ideas about the neurobiology of SCZ, and help the development of new innovative diagnostic tools and therapies.
Highlights.
We review the progress in the development of mouse models of schizophrenia
Current models of susceptibility genes are described and their advantages and disadvantages are discussed
We describe emerging mouse models which are based on novel findings and technologies
Acknowledgments
We apologize to all whose original work could not be cited due to space constraints. We would like to thank Drs. Kimberly Stark and Sander Markx for critical readings of the manuscript and helpful feedback. The authors wish to acknowledge current grant support for their work from the US National Institutes of Health and from the Simons Foundation. M.K is supported by the Essel Foundation NARSAD Young Investigator Award.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- Allen NC, Bagade S, McQueen MB, Ioannidis JP, Kavvoura FK, Khoury MJ, Tanzi RE, Bertram L. Systematic meta-analyses and field synopsis of genetic association studies in schizophrenia: the SzGene database. Nat Genet. 2008;40:827–834. doi: 10.1038/ng.171. [DOI] [PubMed] [Google Scholar]
- Amann-Zalcenstein D, Avidan N, Kanyas K, Ebstein RP, Kohn Y, Hamdan A, Ben-Asher E, Karni O, Mujaheed M, Segman RH, Maier W, Macciardi F, Beckmann JS, Lancet D, Lerer B. AHI1, a pivotal neurodevelopmental gene, and C6orf217 are associated with susceptibility to schizophrenia. Eur J Hum Genet. 2006;14:1111–1119. doi: 10.1038/sj.ejhg.5201675. [DOI] [PubMed] [Google Scholar]
- Anholt RR, Dilda CL, Chang S, Fanara JJ, Kulkarni NH, Ganguly I, Rollmann SM, Kamdar KP, Mackay TF. The genetic architecture of odor-guided behavior in Drosophila: epistasis and the transcriptome. Nat Genet. 2003;35:180–184. doi: 10.1038/ng1240. [DOI] [PubMed] [Google Scholar]
- Arguello PA, Gogos JA. Modeling madness in mice: one piece at a time. Neuron. 2006;52:179–196. doi: 10.1016/j.neuron.2006.09.023. [DOI] [PubMed] [Google Scholar]
- Arguello PA, Gogos JA. Cognition in mouse models of schizophrenia susceptibility genes. Schizophr Bull. 2010;36:289–300. doi: 10.1093/schbul/sbp153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arinami T, Yamada N, Yamakawa-Kobayashi K, Hamaguchi H, Toru M. Methylenetetrahydrofolate reductase variant and schizophrenia/depression. Am J Med Genet. 1997;74:526–528. doi: 10.1002/(sici)1096-8628(19970919)74:5<526::aid-ajmg14>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- Austin CP, Ky B, Ma L, Morris JA, Shughrue PJ. Expression of Disrupted-In-Schizophrenia-1, a schizophrenia-associated gene, is prominent in the mouse hippocampus throughout brain development. Neuroscience. 2004;124:3–10. doi: 10.1016/j.neuroscience.2003.11.010. [DOI] [PubMed] [Google Scholar]
- Ayhan Y, Abazyan B, Nomura J, Kim R, Ladenheim B, Krasnova IN, Sawa A, Margolis RL, Cadet JL, Mori S, Vogel MW, Ross CA, Pletnikov MV. Differential effects of prenatal and postnatal expressions of mutant human DISC1 on neurobehavioral phenotypes in transgenic mice: evidence for neurodevelopmental origin of major psychiatric disorders. Mol Psychiatry. 2011;16:293–306. doi: 10.1038/mp.2009.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barakat A, Dean B, Scarr E, Evin G. Decreased Neuregulin 1 C-terminal fragment in Brodmann’s area 6 of patients with schizophrenia. Schizophr Res. 2010;124:200–207. doi: 10.1016/j.schres.2010.09.001. [DOI] [PubMed] [Google Scholar]
- Barnea-Goraly N, Menon V, Krasnow B, Ko A, Reiss A, Eliez S. Investigation of white matter structure in velocardiofacial syndrome: a diffusion tensor imaging study. Am J Psychiatry. 2003;160:1863–1869. doi: 10.1176/appi.ajp.160.10.1863. [DOI] [PubMed] [Google Scholar]
- Barros CS, Calabrese B, Chamero P, Roberts AJ, Korzus E, Lloyd K, Stowers L, Mayford M, Halpain S, Muller U. Impaired maturation of dendritic spines without disorganization of cortical cell layers in mice lacking NRG1/ErbB signaling in the central nervous system. Proc Natl Acad Sci U S A. 2009;106:4507–4512. doi: 10.1073/pnas.0900355106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayes A, van de Lagemaat LN, Collins MO, Croning MD, Whittle IR, Choudhary JS, Grant SG. Characterization of the proteome, diseases and evolution of the human postsynaptic density. Nat Neurosci. 2011;14:19–21. doi: 10.1038/nn.2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bearden CE, van Erp TG, Dutton RA, Lee AD, Simon TJ, Cannon TD, Emanuel BS, Donald-McGinn D, Zackai EH, Thompson PM. Alterations in midline cortical thickness and gyrification patterns mapped in children with 22q11.2 deletions. Cereb Cortex. 2009;19:115–126. doi: 10.1093/cercor/bhn064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bearden CE, Woodin MF, Wang PP, Moss E, Donald-McGinn D, Zackai E, Emannuel B, Cannon TD. The neurocognitive phenotype of the 22q11.2 deletion syndrome: selective deficit in visual-spatial memory. J Clin Exp Neuropsychol. 2001;23:447–464. doi: 10.1076/jcen.23.4.447.1228. [DOI] [PubMed] [Google Scholar]
- Beaulieu JM, Tirotta E, Sotnikova TD, Masri B, Salahpour A, Gainetdinov RR, Borrelli E, Caron MG. Regulation of Akt signaling by D2 and D3 dopamine receptors in vivo. J Neurosci. 2007;27:881–885. doi: 10.1523/JNEUROSCI.5074-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender HU, Almashanu S, Steel G, Hu CA, Lin WW, Willis A, Pulver A, Valle D. Functional consequences of PRODH missense mutations. Am J Hum Genet. 2005;76:409–420. doi: 10.1086/428142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram I, Bernstein HG, Lendeckel U, Bukowska A, Dobrowolny H, Keilhoff G, Kanakis D, Mawrin C, Bielau H, Falkai P, Bogerts B. Immunohistochemical evidence for impaired neuregulin-1 signaling in the prefrontal cortex in schizophrenia and in unipolar depression. Ann N Y Acad Sci. 2007;1096:147–156. doi: 10.1196/annals.1397.080. [DOI] [PubMed] [Google Scholar]
- Blasi G, Bertolino A. Imaging genomics and response to treatment with antipsychotics in schizophrenia. NeuroRx. 2006;3:117–130. doi: 10.1016/j.nurx.2005.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blednov YA, Jung S, Alva H, Wallace D, Rosahl T, Whiting PJ, Harris RA. Deletion of the alpha1 or beta2 subunit of GABAA receptors reduces actions of alcohol and other drugs. J Pharmacol Exp Ther. 2003;304:30–36. doi: 10.1124/jpet.102.042960. [DOI] [PubMed] [Google Scholar]
- Braff DL, Geyer MA, Swerdlow NR. Human studies of prepulse inhibition of startle: normal subjects, patient groups, and pharmacological studies. Psychopharmacology (Berl) 2001;156:234–258. doi: 10.1007/s002130100810. [DOI] [PubMed] [Google Scholar]
- Brinkmeier ML, Potok MA, Davis SW, Camper SA. TCF4 deficiency expands ventral diencephalon signaling and increases induction of pituitary progenitors. Dev Biol. 2007;311:396–407. doi: 10.1016/j.ydbio.2007.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzozka MM, Radyushkin K, Wichert SP, Ehrenreich H, Rossner MJ. Cognitive and sensorimotor gating impairments in transgenic mice overexpressing the schizophrenia susceptibility gene Tcf4 in the brain. Biol Psychiatry. 2010;68:33–40. doi: 10.1016/j.biopsych.2010.03.015. [DOI] [PubMed] [Google Scholar]
- Burdick KE, Hodgkinson CA, Szeszko PR, Lencz T, Ekholm JM, Kane JM, Goldman D, Malhotra AK. DISC1 and neurocognitive function in schizophrenia. Neuroreport. 2005;16:1399–1402. doi: 10.1097/01.wnr.0000175248.25535.f6. [DOI] [PubMed] [Google Scholar]
- Callicott JH, Straub RE, Pezawas L, Egan MF, Mattay VS, Hariri AR, Verchinski BA, Meyer-Lindenberg A, Balkissoon R, Kolachana B, Goldberg TE, Weinberger DR. Variation in DISC1 affects hippocampal structure and function and increases risk for schizophrenia. Proc Natl Acad Sci U S A. 2005;102:8627–8632. doi: 10.1073/pnas.0500515102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon TD, Hennah W, van Erp TG, Thompson PM, Lonnqvist J, Huttunen M, Gasperoni T, Tuulio-Henriksson A, Pirkola T, Toga AW, Kaprio J, Mazziotta J, Peltonen L. Association of DISC1/TRAX haplotypes with schizophrenia, reduced prefrontal gray matter, and impaired short- and long-term memory. Arch Gen Psychiatry. 2005;62:1205–1213. doi: 10.1001/archpsyc.62.11.1205. [DOI] [PubMed] [Google Scholar]
- Cao L, Dhilla A, Mukai J, Blazeski R, Lodovichi C, Mason CA, Gogos JA. Genetic modulation of BDNF signaling affects the outcome of axonal competition in vivo. Curr Biol. 2007;17:911–921. doi: 10.1016/j.cub.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalifoux JR, Carter AG. GABAB receptors modulate NMDA receptor calcium signals in dendritic spines. Neuron. 2010;66:101–113. doi: 10.1016/j.neuron.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao HT, Chen H, Samaco RC, Xue M, Chahrour M, Yoo J, Neul JL, Gong S, Lu HC, Heintz N, Ekker M, Rubenstein JL, Noebels JL, Rosenmund C, Zoghbi HY. Dysfunction in GABA signalling mediates autism-like stereotypies and Rett syndrome phenotypes. Nature. 2010;468:263–269. doi: 10.1038/nature09582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen QY, Chen Q, Feng GY, Lindpaintner K, Wang LJ, Chen ZX, Gao ZS, Tang JS, Huang G, He L. Case-control association study of Disrupted-in-Schizophrenia-1 (DISC1) gene and schizophrenia in the Chinese population. J Psychiatr Res. 2007;41:428–434. doi: 10.1016/j.jpsychires.2006.01.001. [DOI] [PubMed] [Google Scholar]
- Chen Y, Hancock ML, Role LW, Talmage DA. Intramembranous valine linked to schizophrenia is required for neuregulin 1 regulation of the morphological development of cortical neurons. J Neurosci. 2010;30:9199–9208. doi: 10.1523/JNEUROSCI.0605-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZY, Jing D, Bath KG, Ieraci A, Khan T, Siao CJ, Herrera DG, Toth M, Yang C, McEwen BS, Hempstead BL, Lee FS. Genetic variant BDNF (Val66Met) polymorphism alters anxiety-related behavior. Science. 2006;314:140–143. doi: 10.1126/science.1129663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Lee G, Maher BS, Fanous AH, Chen J, Zhao Z, Guo A, van den OE, Sullivan PF, Shi J, Levinson DF, Gejman PV, Sanders A, Duan J, Owen MJ, Craddock NJ, O’Donovan MC, Blackman J, Lewis D, Kirov GK, Qin W, Schwab S, Wildenauer D, Chowdari K, Nimgaonkar V, Straub RE, Weinberger DR, O’Neill FA, Walsh D, Bronstein M, Darvasi A, Lencz T, Malhotra AK, Rujescu D, Giegling I, Werge T, Hansen T, Ingason A, Noethen MM, Rietschel M, Cichon S, Djurovic S, Andreassen OA, Cantor RM, Ophoff R, Corvin A, Morris DW, Gill M, Pato CN, Pato MT, Macedo A, Gurling HM, McQuillin A, Pimm J, Hultman C, Lichtenstein P, Sklar P, Purcell SM, Scolnick E, St CD, Blackwood DH, Kendler KS. GWA study data mining and independent replication identify cardiomyopathy-associated 5 (CMYA5) as a risk gene for schizophrenia. Mol Psychiatry. 2010 doi: 10.1038/mp.2010.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong VZ, Thompson M, Beltaifa S, Webster MJ, Law AJ, Weickert CS. Elevated neuregulin-1 and ErbB4 protein in the prefrontal cortex of schizophrenic patients. Schizophr Res. 2008;100:270–280. doi: 10.1016/j.schres.2007.12.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow EW, Watson M, Young DA, Bassett AS. Neurocognitive profile in 22q11 deletion syndrome and schizophrenia. Schizophr Res. 2006;87:270–278. doi: 10.1016/j.schres.2006.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdari KV, Mirnics K, Semwal P, Wood J, Lawrence E, Bhatia T, Deshpande SN, BKT, Ferrell RE, Middleton FA, Devlin B, Levitt P, Lewis DA, Nimgaonkar VL. Association and linkage analyses of RGS4 polymorphisms in schizophrenia. Hum Mol Genet. 2002;11:1373–1380. doi: 10.1093/hmg/11.12.1373. [DOI] [PubMed] [Google Scholar]
- Chubb JE, Bradshaw NJ, Soares DC, Porteous DJ, Millar JK. The DISC locus in psychiatric illness. Mol Psychiatry. 2008;13:36–64. doi: 10.1038/sj.mp.4002106. [DOI] [PubMed] [Google Scholar]
- Chumakov I, Blumenfeld M, Guerassimenko O, Cavarec L, Palicio M, Abderrahim H, Bougueleret L, Barry C, Tanaka H, La RP, Puech A, Tahri N, Cohen-Akenine A, Delabrosse S, Lissarrague S, Picard FP, Maurice K, Essioux L, Millasseau P, Grel P, Debailleul V, Simon AM, Caterina D, Dufaure I, Malekzadeh K, Belova M, Luan JJ, Bouillot M, Sambucy JL, Primas G, Saumier M, Boubkiri N, Martin-Saumier S, Nasroune M, Peixoto H, Delaye A, Pinchot V, Bastucci M, Guillou S, Chevillon M, Sainz-Fuertes R, Meguenni S, urich-Costa J, Cherif D, Gimalac A, Van DC, Gauvreau D, Ouellette G, Fortier I, Raelson J, Sherbatich T, Riazanskaia N, Rogaev E, Raeymaekers P, Aerssens J, Konings F, Luyten W, Macciardi F, Sham PC, Straub RE, Weinberger DR, Cohen N, Cohen D. Genetic and physiological data implicating the new human gene G72 and the gene for D-amino acid oxidase in schizophrenia. Proc Natl Acad Sci U S A. 2002;99:13675–13680. doi: 10.1073/pnas.182412499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirulli ET, Goldstein DB. Uncovering the roles of rare variants in common disease through whole-genome sequencing. Nat Rev Genet. 2010;11:415–425. doi: 10.1038/nrg2779. [DOI] [PubMed] [Google Scholar]
- Clapcote SJ, Lipina TV, Millar JK, Mackie S, Christie S, Ogawa F, Lerch JP, Trimble K, Uchiyama M, Sakuraba Y, Kaneda H, Shiroishi T, Houslay MD, Henkelman RM, Sled JG, Gondo Y, Porteous DJ, Roder JC. Behavioral phenotypes of Disc1 missense mutations in mice. Neuron. 2007;54:387–402. doi: 10.1016/j.neuron.2007.04.015. [DOI] [PubMed] [Google Scholar]
- Corfas G, Roy K, Buxbaum JD. Neuregulin 1-erbB signaling and the molecular/cellular basis of schizophrenia. Nat Neurosci. 2004;7:575–580. doi: 10.1038/nn1258. [DOI] [PubMed] [Google Scholar]
- Cutler DJ, Haraura M, Reed HE, Shen S, Sheward WJ, Morrison CF, Marston HM, Harmar AJ, Piggins HD. The mouse VPAC2 receptor confers suprachiasmatic nuclei cellular rhythmicity and responsiveness to vasoactive intestinal polypeptide in vitro. Eur J Neurosci. 2003;17:197–204. doi: 10.1046/j.1460-9568.2003.02425.x. [DOI] [PubMed] [Google Scholar]
- D’Arcangelo G, Miao GG, Chen SC, Soares HD, Morgan JI, Curran T. A protein related to extracellular matrix proteins deleted in the mouse mutant reeler. Nature. 1995;374:719–723. doi: 10.1038/374719a0. [DOI] [PubMed] [Google Scholar]
- Daskalakis ZJ, Fitzgerald PB, Christensen BK. The role of cortical inhibition in the pathophysiology and treatment of schizophrenia. Brain Res Rev. 2007;56:427–442. doi: 10.1016/j.brainresrev.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Dawe GS, Hwang EH, Tan CH. Pathophysiology and animal models of schizophrenia. Ann Acad Med Singapore. 2009;38:425–426. [PubMed] [Google Scholar]
- Deakin IH, Law AJ, Oliver PL, Schwab MH, Nave KA, Harrison PJ, Bannerman DM. Behavioural characterization of neuregulin 1 type I overexpressing transgenic mice. Neuroreport. 2009;20:1523–1528. doi: 10.1097/WNR.0b013e328330f6e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean B, Karl T, Pavey G, Boer S, Duffy L, Scarr E. Increased levels of serotonin 2A receptors and serotonin transporter in the CNS of neuregulin 1 hypomorphic/mutant mice. Schizophr Res. 2008;99:341–349. doi: 10.1016/j.schres.2007.10.013. [DOI] [PubMed] [Google Scholar]
- Devon RS, Anderson S, Teague PW, Burgess P, Kipari TM, Semple CA, Millar JK, Muir WJ, Murray V, Pelosi AJ, Blackwood DH, Porteous DJ. Identification of polymorphisms within Disrupted in Schizophrenia 1 and Disrupted in Schizophrenia 2, and an investigation of their association with schizophrenia and bipolar affective disorder. Psychiatr Genet. 2001;11:71–78. doi: 10.1097/00041444-200106000-00003. [DOI] [PubMed] [Google Scholar]
- Di Giorgio A, Blasi G, Sambataro F, Rampino A, Papazacharias A, Gambi F, Romano R, Caforio G, Rizzo M, Latorre V, Popolizio T, Kolachana B, Callicott JH, Nardini M, Weinberger DR, Bertolino A. Association of the SerCys DISC1 polymorphism with human hippocampal formation gray matter and function during memory encoding. Eur J Neurosci. 2008;28:2129–2136. doi: 10.1111/j.1460-9568.2008.06482.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson L, Finlayson K. VPAC and PAC receptors: From ligands to function. Pharmacol Ther. 2009;121:294–316. doi: 10.1016/j.pharmthera.2008.11.006. [DOI] [PubMed] [Google Scholar]
- Diez-Guerra FJ. Neurogranin, a link between calcium/calmodulin and protein kinase C signaling in synaptic plasticity. IUBMB Life. 2010;62:597–606. doi: 10.1002/iub.357. [DOI] [PubMed] [Google Scholar]
- Drew LJ, Crabtree GW, Markx S, Stark KL, Chaverneff F, Xu B, Mukai J, Fenelon K, Hsu PK, Gogos JA, Karayiorgou M. The 22q11.2 microdeletion: Fifteen years of insights into the genetic and neural complexity of psychiatric disorders. Int J Dev Neurosci. 2010 doi: 10.1016/j.ijdevneu.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew LJ, Stark KL, Fenelon K, Karayiorgou M, Macdermott AB, Gogos JA. Evidence for altered hippocampal function in a mouse model of the human 22q11.2 microdeletion. Mol Cell Neurosci. 2011 doi: 10.1016/j.mcn.2011.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan X, Chang JH, Ge S, Faulkner RL, Kim JY, Kitabatake Y, Liu XB, Yang CH, Jordan JD, Ma DK, Liu CY, Ganesan S, Cheng HJ, Ming GL, Lu B, Song H. Disrupted-In-Schizophrenia 1 Regulates Integration of Newly Generated Neurons in the Adult Brain. Cell. 2007 doi: 10.1016/j.cell.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earls LR, Bayazitov IT, Fricke RG, Berry RB, Illingworth E, Mittleman G, Zakharenko SS. Dysregulation of presynaptic calcium and synaptic plasticity in a mouse model of 22q11 deletion syndrome. J Neurosci. 2010;30:15843–15855. doi: 10.1523/JNEUROSCI.1425-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlichman RS, Luminais SN, White SL, Rudnick ND, Ma N, Dow HC, Kreibich AS, Abel T, Brodkin ES, Hahn CG, Siegel SJ. Neuregulin 1 transgenic mice display reduced mismatch negativity, contextual fear conditioning and social interactions. Brain Res. 2009;1294:116–127. doi: 10.1016/j.brainres.2009.07.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Husseini A, Bredt DS. Protein palmitoylation: a regulator of neuronal development and function. Nat Rev Neurosci. 2002;3:791–802. doi: 10.1038/nrn940. [DOI] [PubMed] [Google Scholar]
- Eliez S, Blasey CM, Schmitt EJ, White CD, Hu D, Reiss AL. Velocardiofacial syndrome: are structural changes in the temporal and mesial temporal regions related to schizophrenia? Am J Psychiatry. 2001;158:447–453. doi: 10.1176/appi.ajp.158.3.447. [DOI] [PubMed] [Google Scholar]
- Emamian ES, Hall D, Birnbaum MJ, Karayiorgou M, Gogos JA. Convergent evidence for impaired AKT1-GSK3beta signaling in schizophrenia. Nat Genet. 2004;36:131–137. doi: 10.1038/ng1296. [DOI] [PubMed] [Google Scholar]
- Enomoto A, Asai N, Namba T, Wang Y, Kato T, Tanaka M, Tatsumi H, Taya S, Tsuboi D, Kuroda K, Kaneko N, Sawamoto K, Miyamoto R, Jijiwa M, Murakumo Y, Sokabe M, Seki T, Kaibuchi K, Takahashi M. Roles of disrupted-in-schizophrenia 1-interacting protein girdin in postnatal development of the dentate gyrus. Neuron. 2009;63:774–787. doi: 10.1016/j.neuron.2009.08.015. [DOI] [PubMed] [Google Scholar]
- Faulkner RL, Jang MH, Liu XB, Duan X, Sailor KA, Kim JY, Ge S, Jones EG, Ming GL, Song H, Cheng HJ. Development of hippocampal mossy fiber synaptic outputs by new neurons in the adult brain. Proc Natl Acad Sci U S A. 2008;105:14157–14162. doi: 10.1073/pnas.0806658105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenelon K, Mukai J, Xu B, Hsu PK, Drew LJ, Karayiorgou M, Fischbach GD, Macdermott AB, Gogos JA. Deficiency of Dgcr8, a gene disrupted by the 22q11.2 microdeletion, results in altered short-term plasticity in the prefrontal cortex. Proc Natl Acad Sci U S A. 2011;108:4447–4452. doi: 10.1073/pnas.1101219108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores AI, Mallon BS, Matsui T, Ogawa W, Rosenzweig A, Okamoto T, Macklin WB. Akt-mediated survival of oligodendrocytes induced by neuregulins. J Neurosci. 2000;20:7622–7630. doi: 10.1523/JNEUROSCI.20-20-07622.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman JI, Vrijenhoek T, Markx S, Janssen IM, van d V, Faas BH, Knoers NV, Cahn W, Kahn RS, Edelmann L, Davis KL, Silverman JM, Brunner HG, van Kessel AG, Wijmenga C, Ophoff RA, Veltman JA. CNTNAP2 gene dosage variation is associated with schizophrenia and epilepsy. Mol Psychiatry. 2008;13:261–266. doi: 10.1038/sj.mp.4002049. [DOI] [PubMed] [Google Scholar]
- Gassmann M, Lemke G. Neuregulins and neuregulin receptors in neural development. Curr Opin Neurobiol. 1997;7:87–92. doi: 10.1016/s0959-4388(97)80125-0. [DOI] [PubMed] [Google Scholar]
- Gerber DJ, Hall D, Miyakawa T, Demars S, Gogos JA, Karayiorgou M, Tonegawa S. Evidence for association of schizophrenia with genetic variation in the 8p21.3 gene, PPP3CC, encoding the calcineurin gamma subunit. Proc Natl Acad Sci U S A. 2003;100:8993–8998. doi: 10.1073/pnas.1432927100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilman SR, Iossifov I, Levy D, Ronemus M, Wigler M, Vitkup D. Rare de novo variants associated with autism implicate a large functional network of genes involved in formation and function of synapses. Neuron. 2011;70:898–907. doi: 10.1016/j.neuron.2011.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glessner JT, Reilly MP, Kim CE, Takahashi N, Albano A, Hou C, Bradfield JP, Zhang H, Sleiman PM, Flory JH, Imielinski M, Frackelton EC, Chiavacci R, Thomas KA, Garris M, Otieno FG, Davidson M, Weiser M, Reichenberg A, Davis KL, Friedman JI, Cappola TP, Margulies KB, Rader DJ, Grant SF, Buxbaum JD, Gur RE, Hakonarson H. Strong synaptic transmission impact by copy number variations in schizophrenia. Proc Natl Acad Sci U S A. 2010;107:10584–10589. doi: 10.1073/pnas.1000274107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogos JA, Gerber DJ. Schizophrenia susceptibility genes: emergence of positional candidates and future directions. Trends Pharmacol Sci. 2006;27:226–233. doi: 10.1016/j.tips.2006.02.005. [DOI] [PubMed] [Google Scholar]
- Gogos JA, Santha M, Takacs Z, Beck KD, Luine V, Lucas LR, Nadler JV, Karayiorgou M. The gene encoding proline dehydrogenase modulates sensorimotor gating in mice. Nat Genet. 1999;21:434–439. doi: 10.1038/7777. [DOI] [PubMed] [Google Scholar]
- Gothelf D, Eliez S, Thompson T, Hinard C, Penniman L, Feinstein C, Kwon H, Jin S, Jo B, Antonarakis SE, Morris MA, Reiss AL. COMT genotype predicts longitudinal cognitive decline and psychosis in 22q11.2 deletion syndrome. Nat Neurosci. 2005;8:1500–1502. doi: 10.1038/nn1572. [DOI] [PubMed] [Google Scholar]
- Gothelf D, Feinstein C, Thompson T, Gu E, Penniman L, Van SE, Kwon H, Eliez S, Reiss AL. Risk factors for the emergence of psychotic disorders in adolescents with 22q11.2 deletion syndrome. Am J Psychiatry. 2007;164:663–669. doi: 10.1176/ajp.2007.164.4.663. [DOI] [PubMed] [Google Scholar]
- Govek EE, Newey SE, Akerman CJ, Cross JR, van d V, van AL. The X-linked mental retardation protein oligophrenin-1 is required for dendritic spine morphogenesis. Nat Neurosci. 2004;7:364–372. doi: 10.1038/nn1210. [DOI] [PubMed] [Google Scholar]
- Gradinaru V, Mogri M, Thompson KR, Henderson JM, Deisseroth K. Optical deconstruction of parkinsonian neural circuitry. Science. 2009;324:354–359. doi: 10.1126/science.1167093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green MF, Kern RS, Braff DL, Mintz J. Neurocognitive deficits and functional outcome in schizophrenia: are we measuring the “right stuff”? Schizophr Bull. 2000;26:119–136. doi: 10.1093/oxfordjournals.schbul.a033430. [DOI] [PubMed] [Google Scholar]
- Greengard P, Nairn AC, Girault JA, Ouimet CC, Snyder GL, Fisone G, Allen PB, Fienberg A, Nishi A. The DARPP-32/protein phosphatase-1 cascade: a model for signal integration. Brain Res Brain Res Rev. 1998;26:274–284. doi: 10.1016/s0165-0173(97)00057-x. [DOI] [PubMed] [Google Scholar]
- Groneberg DA, Rabe KF, Fischer A. Novel concepts of neuropeptide-based drug therapy: vasoactive intestinal polypeptide and its receptors. Eur J Pharmacol. 2006;533:182–194. doi: 10.1016/j.ejphar.2005.12.055. [DOI] [PubMed] [Google Scholar]
- Harmar AJ, Marston HM, Shen S, Spratt C, West KM, Sheward WJ, Morrison CF, Dorin JR, Piggins HD, Reubi JC, Kelly JS, Maywood ES, Hastings MH. The VPAC(2) receptor is essential for circadian function in the mouse suprachiasmatic nuclei. Cell. 2002;109:497–508. doi: 10.1016/s0092-8674(02)00736-5. [DOI] [PubMed] [Google Scholar]
- Hashimoto R, Straub RE, Weickert CS, Hyde TM, Kleinman JE, Weinberger DR. Expression analysis of neuregulin-1 in the dorsolateral prefrontal cortex in schizophrenia. Mol Psychiatry. 2004;9:299–307. doi: 10.1038/sj.mp.4001434. [DOI] [PubMed] [Google Scholar]
- Hayesmoore JB, Bray NJ, Owen MJ, O’Donovan MC. DISC1 mRNA expression is not influenced by common Cis-acting regulatory polymorphisms or imprinting. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:1065–1069. doi: 10.1002/ajmg.b.30715. [DOI] [PubMed] [Google Scholar]
- Hennah W, Tuulio-Henriksson A, Paunio T, Ekelund J, Varilo T, Partonen T, Cannon TD, Lonnqvist J, Peltonen L. A haplotype within the DISC1 gene is associated with visual memory functions in families with a high density of schizophrenia. Mol Psychiatry. 2005;10:1097–1103. doi: 10.1038/sj.mp.4001731. [DOI] [PubMed] [Google Scholar]
- Hennah W, Thomson P, McQuillin A, Bass N, Loukola A, Anjorin A, Blackwood D, Curtis D, Deary IJ, Harris SE, Isometsa ET, Lawrence J, Lonnqvist J, Muir W, Palotie A, Partonen T, Paunio T, Pylkko E, Robinson M, Soronen P, Suominen K, Suvisaari J, Thirumalai S, St CD, Gurling H, Peltonen L, Porteous D. DISC1 association, heterogeneity and interplay in schizophrenia and bipolar disorder. Mol Psychiatry. 2009;14:865–873. doi: 10.1038/mp.2008.22. [DOI] [PubMed] [Google Scholar]
- Hikida T, Jaaro-Peled H, Seshadri S, Oishi K, Hookway C, Kong S, Wu D, Xue R, Andrade M, Tankou S, Mori S, Gallagher M, Ishizuka K, Pletnikov M, Kida S, Sawa A. From the Cover: Dominant-negative DISC1 transgenic mice display schizophrenia-associated phenotypes detected by measures translatable to humans. Proc Natl Acad Sci U S A. 2007;104:14501–14506. doi: 10.1073/pnas.0704774104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho GP, Selvakumar B, Mukai J, Hester LD, Wang Y, Gogos JA, Snyder SH. S-Nitrosylation and S-Palmitoylation Reciprocally Regulate Synaptic Targeting of PSD-95. Neuron. 2011;71:131–141. doi: 10.1016/j.neuron.2011.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong CJ, Tsai SJ, Wang YC. Association between tryptophan hydroxylase gene polymorphism (A218C) and schizophrenic disorders. Schizophr Res. 2001;49:59–63. doi: 10.1016/s0920-9964(00)00039-6. [DOI] [PubMed] [Google Scholar]
- Hong CJ, Huo SJ, Liao DL, Lee K, Wu JY, Tsai SJ. Case-control and family-based association studies between the neuregulin 1 (Arg38Gln) polymorphism and schizophrenia. Neurosci Lett. 2004;366:158–161. doi: 10.1016/j.neulet.2004.05.027. [DOI] [PubMed] [Google Scholar]
- Hsu R, Woodroffe A, Lai WS, Cook MN, Mukai J, Dunning JP, Swanson DJ, Roos JL, Abecasis GR, Karayiorgou M, Gogos JA. Nogo Receptor 1 (RTN4R) as a candidate gene for schizophrenia: analysis using human and mouse genetic approaches. PLoS ONE. 2007;2:e1234. doi: 10.1371/journal.pone.0001234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YZ, Won S, Ali DW, Wang Q, Tanowitz M, Du QS, Pelkey KA, Yang DJ, Xiong WC, Salter MW, Mei L. Regulation of neuregulin signaling by PSD-95 interacting with ErbB4 at CNS synapses. Neuron. 2000;26:443–455. doi: 10.1016/s0896-6273(00)81176-9. [DOI] [PubMed] [Google Scholar]
- Ide M, Lewis DA. Altered cortical CDC42 signaling pathways in schizophrenia: implications for dendritic spine deficits. Biol Psychiatry. 2010;68:25–32. doi: 10.1016/j.biopsych.2010.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Schizophrenia Consortium. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa T, Gotoh N, Murayama C, Abe T, Iwashita M, Matsuzaki F, Suzuki T, Yamamoto T. IgSF molecule MDGA1 is involved in radial migration and positioning of a subset of cortical upper-layer neurons. Dev Dyn. 2011;240:96–107. doi: 10.1002/dvdy.22496. [DOI] [PubMed] [Google Scholar]
- Ishizuka K, Chen J, Taya S, Li W, Millar JK, Xu Y, Clapcote SJ, Hookway C, Morita M, Kamiya A, Tomoda T, Lipska BK, Roder JC, Pletnikov M, Porteous D, Silva AJ, Cannon TD, Kaibuchi K, Brandon NJ, Weinberger DR, Sawa A. Evidence that many of the DISC1 isoforms in C57BL/6J mice are also expressed in 129S6/SvEv mice. Mol Psychiatry. 2007;12:897–899. doi: 10.1038/sj.mp.4002024. [DOI] [PubMed] [Google Scholar]
- Jacquet H, Demily C, Houy E, Hecketsweiler B, Bou J, Raux G, Lerond J, Allio G, Haouzir S, Tillaux A, Bellegou C, Fouldrin G, Delamillieure P, Menard JF, Dollfus S, D’Amato T, Petit M, Thibaut F, Frebourg T, Campion D. Hyperprolinemia is a risk factor for schizoaffective disorder. Mol Psychiatry. 2005;10:479–485. doi: 10.1038/sj.mp.4001597. [DOI] [PubMed] [Google Scholar]
- Jentsch JD, Trantham-Davidson H, Jairl C, Tinsley M, Cannon TD, Lavin A. Dysbindin modulates prefrontal cortical glutamatergic circuits and working memory function in mice. Neuropsychopharmacology. 2009;34:2601–2608. doi: 10.1038/npp.2009.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerome LA, Papaioannou VE. DiGeorge syndrome phenotype in mice mutant for the T-box gene, Tbx1. Nat Genet. 2001;27:286–291. doi: 10.1038/85845. [DOI] [PubMed] [Google Scholar]
- Kamiya A, Kubo K, Tomoda T, Takaki M, Youn R, Ozeki Y, Sawamura N, Park U, Kudo C, Okawa M, Ross CA, Hatten ME, Nakajima K, Sawa A. A schizophrenia-associated mutation of DISC1 perturbs cerebral cortex development. Nat Cell Biol. 2005;7:1167–1178. doi: 10.1038/ncb1328. [DOI] [PubMed] [Google Scholar]
- Kao SC, Wu H, Xie J, Chang CP, Ranish JA, Graef IA, Crabtree GR. Calcineurin/NFAT signaling is required for neuregulin-regulated Schwann cell differentiation. Science. 2009;323:651–654. doi: 10.1126/science.1166562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karayiorgou M, Morris MA, Morrow B, Shprintzen RJ, Goldberg R, Borrow J, Gos A, Nestadt G, Wolyniec PS, Lasseter VK. Schizophrenia susceptibility associated with interstitial deletions of chromosome 22q11. Proc Natl Acad Sci U S A. 1995;92:7612–7616. doi: 10.1073/pnas.92.17.7612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karayiorgou M, Simon TJ, Gogos JA. 22q11.2 microdeletions: linking DNA structural variation to brain dysfunction and schizophrenia. Nat Rev Neurosci. 2010;11:402–416. doi: 10.1038/nrn2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karl T, Duffy L, Scimone A, Harvey RP, Schofield PR. Altered motor activity, exploration and anxiety in heterozygous neuregulin 1 mutant mice: implications for understanding schizophrenia. Genes Brain Behav. 2007;6:677–687. doi: 10.1111/j.1601-183X.2006.00298.x. [DOI] [PubMed] [Google Scholar]
- Kates WR, Krauss BR, Abdulsabur N, Colgan D, Antshel KM, Higgins AM, Shprintzen RJ. The neural correlates of non-spatial working memory in velocardiofacial syndrome (22q11.2 deletion syndrome) Neuropsychologia. 2007;45:2863–2873. doi: 10.1016/j.neuropsychologia.2007.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato T, Kasai A, Mizuno M, Fengyi L, Shintani N, Maeda S, Yokoyama M, Ozaki M, Nawa H. Phenotypic characterization of transgenic mice overexpressing neuregulin-1. PLoS ONE. 2010;5:e14185. doi: 10.1371/journal.pone.0014185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellendonk C, Simpson EH, Polan HJ, Malleret G, Vronskaya S, Winiger V, Moore H, Kandel ER. Transient and selective overexpression of dopamine D2 receptors in the striatum causes persistent abnormalities in prefrontal cortex functioning. Neuron. 2006;49:603–615. doi: 10.1016/j.neuron.2006.01.023. [DOI] [PubMed] [Google Scholar]
- Khelfaoui M, Denis C, van GE, de BF, Schmitt A, Houbron C, Morice E, Giros B, Ramakers G, Fagni L, Chelly J, Nosten-Bertrand M, Billuart P. Loss of X-linked mental retardation gene oligophrenin1 in mice impairs spatial memory and leads to ventricular enlargement and dendritic spine immaturity. J Neurosci. 2007;27:9439–9450. doi: 10.1523/JNEUROSCI.2029-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Park HJ, Jung KH, Ban JY, Ra J, Kim JW, Park JK, Choe BK, Yim SV, Kwon YK, Chung JH. Association study of polymorphisms between DISC1 and schizophrenia in a Korean population. Neurosci Lett. 2008;430:60–63. doi: 10.1016/j.neulet.2007.10.010. [DOI] [PubMed] [Google Scholar]
- Kim JY, Duan X, Liu CY, Jang MH, Guo JU, Pow-anpongkul N, Kang E, Song H, Ming GL. DISC1 regulates new neuron development in the adult brain via modulation of AKT-mTOR signaling through KIAA1212. Neuron. 2009;63:761–773. doi: 10.1016/j.neuron.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike H, Arguello PA, Kvajo M, Karayiorgou M, Gogos JA. Disc1 is mutated in the 129S6/SvEv strain and modulates working memory in mice. Proc Natl Acad Sci U S A. 2006;103:3693–3697. doi: 10.1073/pnas.0511189103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer PF, Christensen CH, Hazelwood LA, Dobi A, Bock R, Sibley DR, Mateo Y, Alvarez VA. Dopamine D2 receptor overexpression alters behavior and physiology in Drd2-EGFP mice. J Neurosci. 2011;31:126–132. doi: 10.1523/JNEUROSCI.4287-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo K, Tomita K, Uto A, Kuroda K, Seshadri S, Cohen J, Kaibuchi K, Kamiya A, Nakajima K. Migration defects by DISC1 knockdown in C57BL/6, 129X1/SvJ, and ICR strains via in utero gene transfer and virus-mediated RNAi. Biochem Biophys Res Commun. 2010;400:631–637. doi: 10.1016/j.bbrc.2010.08.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvajo M, McKellar H, Arguello PA, Drew LJ, Moore H, Macdermott AB, Karayiorgou M, Gogos JA. A mutation in mouse Disc1 that models a schizophrenia risk allele leads to specific alterations in neuronal architecture and cognition. Proc Natl Acad Sci U S A. 2008;105:7076–7081. doi: 10.1073/pnas.0802615105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvajo M, McKellar H, Gogos JA. Molecules, signaling, and schizophrenia. Curr Top Behav Neurosci. 2010;4:629–656. doi: 10.1007/7854_2010_41. [DOI] [PubMed] [Google Scholar]
- Law AJ, Kleinman JE, Weinberger DR, Weickert CS. Disease-associated intronic variants in the ErbB4 gene are related to altered ErbB4 splice-variant expression in the brain in schizophrenia. Hum Mol Genet. 2007;16:129–141. doi: 10.1093/hmg/ddl449. [DOI] [PubMed] [Google Scholar]
- Law AJ, Lipska BK, Weickert CS, Hyde TM, Straub RE, Hashimoto R, Harrison PJ, Kleinman JE, Weinberger DR. Neuregulin 1 transcripts are differentially expressed in schizophrenia and regulated by 5′ SNPs associated with the disease. Proc Natl Acad Sci U S A. 2006;103:6747–6752. doi: 10.1073/pnas.0602002103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee FH, Fadel MP, Preston-Maher K, Cordes SP, Clapcote SJ, Price DJ, Roder JC, Wong AH. Disc1 Point Mutations in Mice Affect Development of the Cerebral Cortex. J Neurosci. 2011;31:3197–3206. doi: 10.1523/JNEUROSCI.4219-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levav-Rabkin T, Blumkin E, Galron D, Golan HM. Sex-dependent behavioral effects of Mthfr deficiency and neonatal GABA potentiation in mice. Behav Brain Res. 2011;216:505–513. doi: 10.1016/j.bbr.2010.08.031. [DOI] [PubMed] [Google Scholar]
- Li T, Ma X, Hu X, Wang Y, Yan C, Meng H, Liu X, Toulopoulou T, Murray RM, Collier DA. PRODH gene is associated with executive function in schizophrenic families. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:654–657. doi: 10.1002/ajmg.b.30648. [DOI] [PubMed] [Google Scholar]
- Li W, Zhou Y, Jentsch JD, Brown RA, Tian X, Ehninger D, Hennah W, Peltonen L, Lonnqvist J, Huttunen MO, Kaprio J, Trachtenberg JT, Silva AJ, Cannon TD. Specific developmental disruption of disrupted-in-schizophrenia-1 function results in schizophrenia-related phenotypes in mice. Proc Natl Acad Sci U S A. 2007;104:18280–18285. doi: 10.1073/pnas.0706900104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim SM, Kim HJ, Nam M, Chung JH, Park YH. Association study of DISC1 in Korean population with autism spectrum disorders. Psychiatr Genet. 2009;19:160. doi: 10.1097/YPG.0b013e32832a9bd1. [DOI] [PubMed] [Google Scholar]
- Lindsay EA, Vitelli F, Su H, Morishima M, Huynh T, Pramparo T, Jurecic V, Ogunrinu G, Sutherland HF, Scambler PJ, Bradley A, Baldini A. Tbx1 haploinsufficieny in the DiGeorge syndrome region causes aortic arch defects in mice. Nature. 2001;410:97–101. doi: 10.1038/35065105. [DOI] [PubMed] [Google Scholar]
- Lipska BK, Weinberger DR. To model a psychiatric disorder in animals: schizophrenia as a reality test. Neuropsychopharmacology. 2000;23:223–239. doi: 10.1016/S0893-133X(00)00137-8. [DOI] [PubMed] [Google Scholar]
- Liu WS, Pesold C, Rodriguez MA, Carboni G, Auta J, Lacor P, Larson J, Condie BG, Guidotti A, Costa E. Down-regulation of dendritic spine and glutamic acid decarboxylase 67 expressions in the reelin haploinsufficient heterozygous reeler mouse. Proc Natl Acad Sci U S A. 2001;98:3477–3482. doi: 10.1073/pnas.051614698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YL, Fann CS, Liu CM, Chen WJ, Wu JY, Hung SI, Chen CH, Jou YS, Liu SK, Hwang TJ, Hsieh MH, Ouyang WC, Chan HY, Chen JJ, Yang WC, Lin CY, Lee SF, Hwu HG. A single nucleotide polymorphism fine mapping study of chromosome 1q42.1 reveals the vulnerability genes for schizophrenia, GNPAT and DISC1: Association with impairment of sustained attention. Biol Psychiatry. 2006;60:554–562. doi: 10.1016/j.biopsych.2006.04.024. [DOI] [PubMed] [Google Scholar]
- Lo WS, Lau CF, Xuan Z, Chan CF, Feng GY, He L, Cao ZC, Liu H, Luan QM, Xue H. Association of SNPs and haplotypes in GABAA receptor beta2 gene with schizophrenia. Mol Psychiatry. 2004;9:603–608. doi: 10.1038/sj.mp.4001461. [DOI] [PubMed] [Google Scholar]
- Loe-Mie Y, Lepagnol-Bestel AM, Maussion G, Doron-Faigenboim A, Imbeaud S, Delacroix H, Aggerbeck L, Pupko T, Gorwood P, Simonneau M, Moalic JM. SMARCA2 and other genome-wide supported schizophrenia-associated genes: regulation by REST/NRSF, network organization and primate-specific evolution. Hum Mol Genet. 2010;19:2841–2857. doi: 10.1093/hmg/ddq184. [DOI] [PubMed] [Google Scholar]
- Malleret G, Haditsch U, Genoux D, Jones MW, Bliss TV, Vanhoose AM, Weitlauf C, Kandel ER, Winder DG, Mansuy IM. Inducible and reversible enhancement of learning, memory, and long-term potentiation by genetic inhibition of calcineurin. Cell. 2001;104:675–686. doi: 10.1016/s0092-8674(01)00264-1. [DOI] [PubMed] [Google Scholar]
- Mansuy IM, Mayford M, Jacob B, Kandel ER, Bach ME. Restricted and regulated overexpression reveals calcineurin as a key component in the transition from short-term to long-term memory. Cell. 1998;92:39–49. doi: 10.1016/s0092-8674(00)80897-1. [DOI] [PubMed] [Google Scholar]
- Mao Y, Ge X, Frank CL, Madison JM, Koehler AN, Doud MK, Tassa C, Berry EM, Soda T, Singh KK, Biechele T, Petryshen TL, Moon RT, Haggarty SJ, Tsai LH. Disrupted in schizophrenia 1 regulates neuronal progenitor proliferation via modulation of GSK3beta/beta-catenin signaling. Cell. 2009;136:1017–1031. doi: 10.1016/j.cell.2008.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, Ioannidis JP, Hirschhorn JN. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- McCarthy SE, Makarov V, Kirov G, Addington AM, McClellan J, Yoon S, Perkins DO, Dickel DE, Kusenda M, Krastoshevsky O, Krause V, Kumar RA, Grozeva D, Malhotra D, Walsh T, Zackai EH, Kaplan P, Ganesh J, Krantz ID, Spinner NB, Roccanova P, Bhandari A, Pavon K, Lakshmi B, Leotta A, Kendall J, Lee YH, Vacic V, Gary S, Iakoucheva LM, Crow TJ, Christian SL, Lieberman JA, Stroup TS, Lehtimaki T, Puura K, Haldeman-Englert C, Pearl J, Goodell M, Willour VL, DeRosse P, Steele J, Kassem L, Wolff J, Chitkara N, McMahon FJ, Malhotra AK, Potash JB, Schulze TG, Nothen MM, Cichon S, Rietschel M, Leibenluft E, Kustanovich V, Lajonchere CM, Sutcliffe JS, Skuse D, Gill M, Gallagher L, Mendell NR, Craddock N, Owen MJ, O’Donovan MC, Shaikh TH, Susser E, DeLisi LE, Sullivan PF, Deutsch CK, Rapoport J, Levy DL, King MC, Sebat J. Microduplications of 16p11.2 are associated with schizophrenia. Nat Genet. 2009;41:1223–1227. doi: 10.1038/ng.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClellan J, King MC. Genetic heterogeneity in human disease. Cell. 2010;141:210–217. doi: 10.1016/j.cell.2010.03.032. [DOI] [PubMed] [Google Scholar]
- Meechan DW, Tucker ES, Maynard TM, Lamantia AS. Diminished dosage of 22q11 genes disrupts neurogenesis and cortical development in a mouse model of 22q11 deletion/DiGeorge syndrome. Proc Natl Acad Sci U S A. 2009;106:16434–16445. doi: 10.1073/pnas.0905696106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merscher S, Funke B, Epstein JA, Heyer J, Puech A, Lu MM, Xavier RJ, Demay MB, Russell RG, Factor S, Tokooya K, Jore BS, Lopez M, Pandita RK, Lia M, Carrion D, Xu H, Schorle H, Kobler JB, Scambler P, Wynshaw-Boris A, Skoultchi AI, Morrow BE, Kucherlapati R. TBX1 is responsible for cardiovascular defects in velo-cardio-facial/DiGeorge syndrome. Cell. 2001;104:619–629. doi: 10.1016/s0092-8674(01)00247-1. [DOI] [PubMed] [Google Scholar]
- Meyer D, Birchmeier C. Multiple essential functions of neuregulin in development. Nature. 1995;378:386–390. doi: 10.1038/378386a0. [DOI] [PubMed] [Google Scholar]
- Meyer KD, Morris JA. Disc1 regulates granule cell migration in the developing hippocampus. Hum Mol Genet. 2009 doi: 10.1093/hmg/ddp266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer U, Feldon J. Epidemiology-driven neurodevelopmental animal models of schizophrenia. Prog Neurobiol. 2010;90:285–326. doi: 10.1016/j.pneurobio.2009.10.018. [DOI] [PubMed] [Google Scholar]
- Millar JK, Pickard BS, Mackie S, James R, Christie S, Buchanan SR, Malloy MP, Chubb JE, Huston E, Baillie GS, Thomson PA, Hill EV, Brandon NJ, Rain JC, Camargo LM, Whiting PJ, Houslay MD, Blackwood DH, Muir WJ, Porteous DJ. DISC1 and PDE4B are interacting genetic factors in schizophrenia that regulate cAMP signaling. Science. 2005;310:1187–1191. doi: 10.1126/science.1112915. [DOI] [PubMed] [Google Scholar]
- Millar JK, Wilson-Annan JC, Anderson S, Christie S, Taylor MS, Semple CA, Devon RS, Clair DM, Muir WJ, Blackwood DH, Porteous DJ. Disruption of two novel genes by a translocation co-segregating with schizophrenia. Hum Mol Genet. 2000;9:1415–1423. doi: 10.1093/hmg/9.9.1415. [DOI] [PubMed] [Google Scholar]
- Mohn AR, Gainetdinov RR, Caron MG, Koller BH. Mice with reduced NMDA receptor expression display behaviors related to schizophrenia. Cell. 1999;98:427–436. doi: 10.1016/s0092-8674(00)81972-8. [DOI] [PubMed] [Google Scholar]
- Moore H, Jentsch JD, Ghajarnia M, Geyer MA, Grace AA. A neurobehavioral systems analysis of adult rats exposed to methylazoxymethanol acetate on E17: implications for the neuropathology of schizophrenia. Biol Psychiatry. 2006;60:253–264. doi: 10.1016/j.biopsych.2006.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai J, Dhilla A, Drew LJ, Stark KL, Cao L, Macdermott AB, Karayiorgou M, Gogos JA. Palmitoylation-dependent neurodevelopmental deficits in a mouse model of 22q11 microdeletion. Nat Neurosci. 2008;11:1302–1310. doi: 10.1038/nn.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai J, Liu H, Burt RA, Swor DE, Lai WS, Karayiorgou M, Gogos JA. Evidence that the gene encoding ZDHHC8 contributes to the risk of schizophrenia. Nat Genet. 2004;36:725–731. doi: 10.1038/ng1375. [DOI] [PubMed] [Google Scholar]
- Murphy KC, Jones LA, Owen MJ. High rates of schizophrenia in adults with velo-cardio-facial syndrome. Arch Gen Psychiatry. 1999;56:940–945. doi: 10.1001/archpsyc.56.10.940. [DOI] [PubMed] [Google Scholar]
- Nakata K, Lipska BK, Hyde TM, Ye T, Newburn EN, Morita Y, Vakkalanka R, Barenboim M, Sei Y, Weinberger DR, Kleinman JE. DISC1 splice variants are upregulated in schizophrenia and associated with risk polymorphisms. Proc Natl Acad Sci U S A. 2009;106:15873–15878. doi: 10.1073/pnas.0903413106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nave KA, Salzer JL. Axonal regulation of myelination by neuregulin 1. Curr Opin Neurobiol. 2006;16:492–500. doi: 10.1016/j.conb.2006.08.008. [DOI] [PubMed] [Google Scholar]
- Nestler EJ, Hyman SE. Animal models of neuropsychiatric disorders. Nat Neurosci. 2010;13:1161–1169. doi: 10.1038/nn.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton-Cheh C, Hirschhorn JN. Genetic association studies of complex traits: design and analysis issues. Mutat Res. 2005;573:54–69. doi: 10.1016/j.mrfmmm.2005.01.006. [DOI] [PubMed] [Google Scholar]
- Nicodemus KK, Law AJ, Luna A, Vakkalanka R, Straub RE, Kleinman JE, Weinberger DR. A 5′ promoter region SNP in NRG1 is associated with schizophrenia risk and type III isoform expression. Mol Psychiatry. 2009;14:741–743. doi: 10.1038/mp.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donovan MC, Craddock N, Norton N, Williams H, Peirce T, Moskvina V, Nikolov I, Hamshere M, Carroll L, Georgieva L, Dwyer S, Holmans P, Marchini JL, Spencer CC, Howie B, Leung HT, Hartmann AM, Moller HJ, Morris DW, Shi Y, Feng G, Hoffmann P, Propping P, Vasilescu C, Maier W, Rietschel M, Zammit S, Schumacher J, Quinn EM, Schulze TG, Williams NM, Giegling I, Iwata N, Ikeda M, Darvasi A, Shifman S, He L, Duan J, Sanders AR, Levinson DF, Gejman PV, Cichon S, Nothen MM, Gill M, Corvin A, Rujescu D, Kirov G, Owen MJ, Buccola NG, Mowry BJ, Freedman R, Amin F, Black DW, Silverman JM, Byerley WF, Cloninger CR. Identification of loci associated with schizophrenia by genome-wide association and follow-up. Nat Genet. 2008;40:1053–1055. doi: 10.1038/ng.201. [DOI] [PubMed] [Google Scholar]
- O’Dushlaine C, Kenny E, Heron E, Donohoe G, Gill M, Morris D, Corvin A. Molecular pathways involved in neuronal cell adhesion and membrane scaffolding contribute to schizophrenia and bipolar disorder susceptibility. Mol Psychiatry. 2011;16:286–292. doi: 10.1038/mp.2010.7. [DOI] [PubMed] [Google Scholar]
- Otte DM, Bilkei-Gorzo A, Filiou MD, Turck CW, Yilmaz O, Holst MI, Schilling K, bou-Jamra R, Schumacher J, Benzel I, Kunz WS, Beck H, Zimmer A. Behavioral changes in G72/G30 transgenic mice. Eur Neuropsychopharmacol. 2009;19:339–348. doi: 10.1016/j.euroneuro.2008.12.009. [DOI] [PubMed] [Google Scholar]
- O’Tuathaigh CM, Babovic D, O’Sullivan GJ, Clifford JJ, Tighe O, Croke DT, Harvey R, Waddington JL. Phenotypic characterization of spatial cognition and social behavior in mice with ‘knockout’ of the schizophrenia risk gene neuregulin 1. Neuroscience. 2007;147:18–27. doi: 10.1016/j.neuroscience.2007.03.051. [DOI] [PubMed] [Google Scholar]
- O’Tuathaigh CM, Harte M, O’Leary C, O’Sullivan GJ, Blau C, Lai D, Harvey RP, Tighe O, Fagan AJ, Kerskens C, Reynolds GP, Waddington JL. Schizophrenia-related endophenotypes in heterozygous neuregulin-1 ‘knockout’ mice. Eur J Neurosci. 2010;31:349–358. doi: 10.1111/j.1460-9568.2009.07069.x. [DOI] [PubMed] [Google Scholar]
- O’Tuathaigh CM, O’Connor AM, O’Sullivan GJ, Lai D, Harvey R, Croke DT, Waddington JL. Disruption to social dyadic interactions but not emotional/anxiety-related behaviour in mice with heterozygous ‘knockout’ of the schizophrenia risk gene neuregulin-1. Prog Neuropsychopharmacol Biol Psychiatry. 2008;32:462–466. doi: 10.1016/j.pnpbp.2007.09.018. [DOI] [PubMed] [Google Scholar]
- O’Tuathaigh CM, O’Sullivan GJ, Kinsella A, Harvey RP, Tighe O, Croke DT, Waddington JL. Sexually dimorphic changes in the exploratory and habituation profiles of heterozygous neuregulin-1 knockout mice. Neuroreport. 2006;17:79–83. doi: 10.1097/01.wnr.0000192738.31029.0a. [DOI] [PubMed] [Google Scholar]
- Owen MJ, Craddock N, O’Donovan MC. Suggestion of roles for both common and rare risk variants in genome-wide studies of schizophrenia. Arch Gen Psychiatry. 2010;67:667–673. doi: 10.1001/archgenpsychiatry.2010.69. [DOI] [PubMed] [Google Scholar]
- Pacey LK, Doss L, Cifelli C, van der KD, Heximer SP, Hampson DR. Genetic deletion of regulator of G-protein signaling 4 (RGS4) rescues a subset of fragile X related phenotypes in the FMR1 knockout mouse. Mol Cell Neurosci. 2011;46:563–572. doi: 10.1016/j.mcn.2010.12.005. [DOI] [PubMed] [Google Scholar]
- Pak JH, Huang FL, Li J, Balschun D, Reymann KG, Chiang C, Westphal H, Huang KP. Involvement of neurogranin in the modulation of calcium/calmodulin-dependent protein kinase II, synaptic plasticity, and spatial learning: a study with knockout mice. Proc Natl Acad Sci U S A. 2000;97:11232–11237. doi: 10.1073/pnas.210184697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer AA, Low MJ, Grandy DK, Phillips TJ. Effects of a Drd2 deletion mutation on ethanol-induced locomotor stimulation and sensitization suggest a role for epistasis. Behav Genet. 2003;33:311–324. doi: 10.1023/a:1023450625826. [DOI] [PubMed] [Google Scholar]
- Paterlini M, Zakharenko SS, Lai WS, Qin J, Zhang H, Mukai J, Westphal KG, Olivier B, Sulzer D, Pavlidis P, Siegelbaum SA, Karayiorgou M, Gogos JA. Transcriptional and behavioral interaction between 22q11.2 orthologs modulates schizophrenia-related phenotypes in mice. Nat Neurosci. 2005;8:1586–1594. doi: 10.1038/nn1562. [DOI] [PubMed] [Google Scholar]
- Paylor R, Glaser B, Mupo A, Ataliotis P, Spencer C, Sobotka A, Sparks C, Choi CH, Oghalai J, Curran S, Murphy KC, Monks S, Williams N, O’Donovan MC, Owen MJ, Scambler PJ, Lindsay E. Tbx1 haploinsufficiency is linked to behavioral disorders in mice and humans: implications for 22q11 deletion syndrome. Proc Natl Acad Sci U S A. 2006;103:7729–7734. doi: 10.1073/pnas.0600206103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickard BS, Thomson PA, Christoforou A, Evans KL, Morris SW, Porteous DJ, Blackwood DH, Muir WJ. The PDE4B gene confers sex-specific protection against schizophrenia. Psychiatr Genet. 2007;17:129–133. doi: 10.1097/YPG.0b013e328014492b. [DOI] [PubMed] [Google Scholar]
- Pletnikov MV, Ayhan Y, Nikolskaia O, Xu Y, Ovanesov MV, Huang H, Mori S, Moran TH, Ross CA. Inducible expression of mutant human DISC1 in mice is associated with brain and behavioral abnormalities reminiscent of schizophrenia. Mol Psychiatry. 2008a;13:173–186. doi: 10.1038/sj.mp.4002079. [DOI] [PubMed] [Google Scholar]
- Pletnikov MV, Ayhan Y, Xu Y, Nikolskaia O, Ovanesov M, Huang H, Mori S, Moran TH, Ross CA. Enlargement of the lateral ventricles in mutant DISC1 transgenic mice. Mol Psychiatry. 2008b;13:115. doi: 10.1038/sj.mp.4002144. [DOI] [PubMed] [Google Scholar]
- Popa D, Lena C, Fabre V, Prenat C, Gingrich J, Escourrou P, Hamon M, Adrien J. Contribution of 5-HT2 receptor subtypes to sleep-wakefulness and respiratory control, and functional adaptations in knock-out mice lacking 5-HT2A receptors. J Neurosci. 2005;25:11231–11238. doi: 10.1523/JNEUROSCI.1724-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prata DP, Mechelli A, Fu CH, Picchioni M, Kane F, Kalidindi S, McDonald C, Kravariti E, Toulopoulou T, Miorelli A, Murray R, Collier DA, McGuire PK. Effect of disrupted-in-schizophrenia-1 on pre-frontal cortical function. Mol Psychiatry. 2008;13:915–7. 909. doi: 10.1038/mp.2008.76. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Cox NJ. The allelic architecture of human disease genes: common disease-common variant…or not? Hum Mol Genet. 2002;11:2417–2423. doi: 10.1093/hmg/11.20.2417. [DOI] [PubMed] [Google Scholar]
- Pulver AE, Nestadt G, Goldberg R, Shprintzen RJ, Lamacz M, Wolyniec PS, Morrow B, Karayiorgou M, Antonarakis SE, Housman D. Psychotic illness in patients diagnosed with velo-cardio-facial syndrome and their relatives. J Nerv Ment Dis. 1994;182:476–478. doi: 10.1097/00005053-199408000-00010. [DOI] [PubMed] [Google Scholar]
- Purcell SM, Wray NR, Stone JL, Visscher PM, O’Donovan MC, Sullivan PF, Sklar P. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raux G, Bumsel E, Hecketsweiler B, van AT, Zinkstok J, Manouvrier-Hanu S, Fantini C, Breviere GM, Di RG, Pustorino G, Vogels A, Swillen A, Legallic S, Bou J, Opolczynski G, Drouin-Garraud V, Lemarchand M, Philip N, Gerard-Desplanches A, Carlier M, Philippe A, Nolen MC, Heron D, Sarda P, Lacombe D, Coizet C, Alembik Y, Layet V, Afenjar A, Hannequin D, Demily C, Petit M, Thibaut F, Frebourg T, Campion D. Involvement of hyperprolinemia in cognitive and psychiatric features of the 22q11 deletion syndrome. Hum Mol Genet. 2007;16:83–91. doi: 10.1093/hmg/ddl443. [DOI] [PubMed] [Google Scholar]
- Rebbeck TR, Spitz M, Wu X. Assessing the function of genetic variants in candidate gene association studies. Nat Rev Genet. 2004;5:589–597. doi: 10.1038/nrg1403. [DOI] [PubMed] [Google Scholar]
- Resh MD. Fatty acylation of proteins: new insights into membrane targeting of myristoylated and palmitoylated proteins. Biochim Biophys Acta. 1999;1451:1–16. doi: 10.1016/s0167-4889(99)00075-0. [DOI] [PubMed] [Google Scholar]
- Rimer M, Barrett DW, Maldonado MA, Vock VM, Gonzalez-Lima F. Neuregulin-1 immunoglobulin-like domain mutant mice: clozapine sensitivity and impaired latent inhibition. Neuroreport. 2005;16:271–275. doi: 10.1097/00001756-200502280-00014. [DOI] [PubMed] [Google Scholar]
- Roussos P, Giakoumaki SG, Adamaki E, Bitsios P. The influence of schizophrenia-related neuregulin-1 polymorphisms on sensorimotor gating in healthy males. Biol Psychiatry. 2011;69:479–486. doi: 10.1016/j.biopsych.2010.09.009. [DOI] [PubMed] [Google Scholar]
- Roy K, Murtie JC, El-Khodor BF, Edgar N, Sardi SP, Hooks BM, oit-Marand M, Chen C, Moore H, O’Donnell P, Brunner D, Corfas G. Loss of erbB signaling in oligodendrocytes alters myelin and dopaminergic function, a potential mechanism for neuropsychiatric disorders. Proc Natl Acad Sci U S A. 2007;104:8131–8136. doi: 10.1073/pnas.0702157104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein M, Phillips TJ, Bunzow JR, Falzone TL, Dziewczapolski G, Zhang G, Fang Y, Larson JL, McDougall JA, Chester JA, Saez C, Pugsley TA, Gershanik O, Low MJ, Grandy DK. Mice lacking dopamine D4 receptors are supersensitive to ethanol, cocaine, and methamphetamine. Cell. 1997;90:991–1001. doi: 10.1016/s0092-8674(00)80365-7. [DOI] [PubMed] [Google Scholar]
- Sanders AR, Duan J, Levinson DF, Shi J, He D, Hou C, Burrell GJ, Rice JP, Nertney DA, Olincy A, Rozic P, Vinogradov S, Buccola NG, Mowry BJ, Freedman R, Amin F, Black DW, Silverman JM, Byerley WF, Crowe RR, Cloninger CR, Martinez M, Gejman PV. No significant association of 14 candidate genes with schizophrenia in a large European ancestry sample: implications for psychiatric genetics. Am J Psychiatry. 2008;165:497–506. doi: 10.1176/appi.ajp.2007.07101573. [DOI] [PubMed] [Google Scholar]
- Savonenko AV, Melnikova T, Laird FM, Stewart KA, Price DL, Wong PC. Alteration of BACE1-dependent NRG1/ErbB4 signaling and schizophrenia-like phenotypes in BACE1-null mice. Proc Natl Acad Sci U S A. 2008;105:5585–5590. doi: 10.1073/pnas.0710373105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaikh S, Collier D, Arranz M, Ball D, Gill M, Kerwin R. DRD2 Ser311/Cys311 polymorphism in schizophrenia. Lancet. 1994;343:1045–1046. [PubMed] [Google Scholar]
- Sebat J, Levy DL, McCarthy SE. Rare structural variants in schizophrenia: one disorder, multiple mutations; one mutation, multiple disorders. Trends Genet. 2009;25:528–535. doi: 10.1016/j.tig.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S, Lang B, Nakamoto C, Zhang F, Pu J, Kuan SL, Chatzi C, He S, Mackie I, Brandon NJ, Marquis KL, Day M, Hurko O, McCaig CD, Riedel G, St CD. Schizophrenia-related neural and behavioral phenotypes in transgenic mice expressing truncated Disc1. J Neurosci. 2008;28:10893–10904. doi: 10.1523/JNEUROSCI.3299-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shifman S, Johannesson M, Bronstein M, Chen SX, Collier DA, Craddock NJ, Kendler KS, Li T, O’Donovan M, O’Neill FA, Owen MJ, Walsh D, Weinberger DR, Sun C, Flint J, Darvasi A. Genome-wide association identifies a common variant in the reelin gene that increases the risk of schizophrenia only in women. PLoS Genet. 2008;4:e28. doi: 10.1371/journal.pgen.0040028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigurdsson T, Stark KL, Karayiorgou M, Gogos JA, Gordon JA. Impaired hippocampal-prefrontal synchrony in a genetic mouse model of schizophrenia. Nature. 2010;464:763–767. doi: 10.1038/nature08855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silberberg G, Darvasi A, Pinkas-Kramarski R, Navon R. The involvement of ErbB4 with schizophrenia: association and expression studies. Am J Med Genet B Neuropsychiatr Genet. 2006;141B:142–148. doi: 10.1002/ajmg.b.30275. [DOI] [PubMed] [Google Scholar]
- Siuciak JA, McCarthy SA, Chapin DS, Martin AN. Behavioral and neurochemical characterization of mice deficient in the phosphodiesterase-4B (PDE4B) enzyme. Psychopharmacology (Berl) 2008;197:115–126. doi: 10.1007/s00213-007-1014-6. [DOI] [PubMed] [Google Scholar]
- Sobin C, Kiley-Brabeck K, Daniels S, Khuri J, Taylor L, Blundell M, nyane-Yeboa K, Karayiorgou M. Neuropsychological characteristics of children with the 22q11 Deletion Syndrome: a descriptive analysis. Child Neuropsychol. 2005a;11:39–53. doi: 10.1080/09297040590911167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobin C, Kiley-Brabeck K, Karayiorgou M. Lower prepulse inhibition in children with the 22q11 deletion syndrome. Am J Psychiatry. 2005b;162:1090–1099. doi: 10.1176/appi.ajp.162.6.1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Clair D, Blackwood D, Muir W, Carothers A, Walker M, Spowart G, Gosden C, Evans HJ. Association within a family of a balanced autosomal translocation with major mental illness. Lancet. 1990;336:13–16. doi: 10.1016/0140-6736(90)91520-k. [DOI] [PubMed] [Google Scholar]
- Stark KL, Xu B, Bagchi A, Lai WS, Liu H, Hsu R, Wan X, Pavlidis P, Mills AA, Karayiorgou M, Gogos JA. Altered brain microRNA biogenesis contributes to phenotypic deficits in a 22q11-deletion mouse model. Nat Genet. 2008;40:751–760. doi: 10.1038/ng.138. [DOI] [PubMed] [Google Scholar]
- Stefansson H, Ophoff RA, Steinberg S, Andreassen OA, Cichon S, Rujescu D, Werge T, Pietilainen OP, Mors O, Mortensen PB, Sigurdsson E, Gustafsson O, Nyegaard M, Tuulio-Henriksson A, Ingason A, Hansen T, Suvisaari J, Lonnqvist J, Paunio T, Borglum AD, Hartmann A, Fink-Jensen A, Nordentoft M, Hougaard D, Norgaard-Pedersen B, Bottcher Y, Olesen J, Breuer R, Moller HJ, Giegling I, Rasmussen HB, Timm S, Mattheisen M, Bitter I, Rethelyi JM, Magnusdottir BB, Sigmundsson T, Olason P, Masson G, Gulcher JR, Haraldsson M, Fossdal R, Thorgeirsson TE, Thorsteinsdottir U, Ruggeri M, Tosato S, Franke B, Strengman E, Kiemeney LA, Melle I, Djurovic S, Abramova L, Kaleda V, Sanjuan J, de FR, Bramon E, Vassos E, Fraser G, Ettinger U, Picchioni M, Walker N, Toulopoulou T, Need AC, Ge D, Yoon JL, Shianna KV, Freimer NB, Cantor RM, Murray R, Kong A, Golimbet V, Carracedo A, Arango C, Costas J, Jonsson EG, Terenius L, Agartz I, Petursson H, Nothen MM, Rietschel M, Matthews PM, Muglia P, Peltonen L, St CD, Goldstein DB, Stefansson K, Collier DA. Common variants conferring risk of schizophrenia. Nature. 2009;460:744–747. doi: 10.1038/nature08186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefansson H, Rujescu D, Cichon S, Pietilainen OP, Ingason A, Steinberg S, Fossdal R, Sigurdsson E, Sigmundsson T, Buizer-Voskamp JE, Hansen T, Jakobsen KD, Muglia P, Francks C, Matthews PM, Gylfason A, Halldorsson BV, Gudbjartsson D, Thorgeirsson TE, Sigurdsson A, Jonasdottir A, Jonasdottir A, Bjornsson A, Mattiasdottir S, Blondal T, Haraldsson M, Magnusdottir BB, Giegling I, Moller HJ, Hartmann A, Shianna KV, Ge D, Need AC, Crombie C, Fraser G, Walker N, Lonnqvist J, Suvisaari J, Tuulio-Henriksson A, Paunio T, Toulopoulou T, Bramon E, Di FM, Murray R, Ruggeri M, Vassos E, Tosato S, Walshe M, Li T, Vasilescu C, Muhleisen TW, Wang AG, Ullum H, Djurovic S, Melle I, Olesen J, Kiemeney LA, Franke B, Sabatti C, Freimer NB, Gulcher JR, Thorsteinsdottir U, Kong A, Andreassen OA, Ophoff RA, Georgi A, Rietschel M, Werge T, Petursson H, Goldstein DB, Nothen MM, Peltonen L, Collier DA, St CD, Stefansson K. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefansson H, Sigurdsson E, Steinthorsdottir V, Bjornsdottir S, Sigmundsson T, Ghosh S, Brynjolfsson J, Gunnarsdottir S, Ivarsson O, Chou TT, Hjaltason O, Birgisdottir B, Jonsson H, Gudnadottir VG, Gudmundsdottir E, Bjornsson A, Ingvarsson B, Ingason A, Sigfusson S, Hardardottir H, Harvey RP, Lai D, Zhou M, Brunner D, Mutel V, Gonzalo A, Lemke G, Sainz J, Johannesson G, Andresson T, Gudbjartsson D, Manolescu A, Frigge ML, Gurney ME, Kong A, Gulcher JR, Petursson H, Stefansson K. Neuregulin 1 and susceptibility to schizophrenia. Am J Hum Genet. 2002;71:877–892. doi: 10.1086/342734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub RE, Jiang Y, MacLean CJ, Ma Y, Webb BT, Myakishev MV, Harris-Kerr C, Wormley B, Sadek H, Kadambi B, Cesare AJ, Gibberman A, Wang X, O’Neill FA, Walsh D, Kendler KS. Genetic variation in the 6p22.3 gene DTNBP1, the human ortholog of the mouse dysbindin gene, is associated with schizophrenia. Am J Hum Genet. 2002;71:337–348. doi: 10.1086/341750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan PF, Kendler KS, Neale MC. Schizophrenia as a complex trait: evidence from a meta-analysis of twin studies. Arch Gen Psychiatry. 2003;60:1187–1192. doi: 10.1001/archpsyc.60.12.1187. [DOI] [PubMed] [Google Scholar]
- Suzuki G, Harper KM, Hiramoto T, Sawamura T, Lee M, Kang G, Tanigaki K, Buell M, Geyer MA, Trimble WS, Agatsuma S, Hiroi N. Sept5 deficiency exerts pleiotropic influence on affective behaviors and cognitive functions in mice. Hum Mol Genet. 2009;18:1652–1660. doi: 10.1093/hmg/ddp086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svenningsson P, Tzavara ET, Carruthers R, Rachleff I, Wattler S, Nehls M, McKinzie DL, Fienberg AA, Nomikos GG, Greengard P. Diverse psychotomimetics act through a common signaling pathway. Science. 2003;302:1412–1415. doi: 10.1126/science.1089681. [DOI] [PubMed] [Google Scholar]
- Szeszko PR, Hodgkinson CA, Robinson DG, DeRosse P, Bilder RM, Lencz T, Burdick KE, Napolitano B, Betensky JD, Kane JM, Goldman D, Malhotra AK. DISC1 is associated with prefrontal cortical gray matter and positive symptoms in schizophrenia. Biol Psychol. 2008;79:103–110. doi: 10.1016/j.biopsycho.2007.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabuchi K, Blundell J, Etherton MR, Hammer RE, Liu X, Powell CM, Sudhof TC. A neuroligin-3 mutation implicated in autism increases inhibitory synaptic transmission in mice. Science. 2007;318:71–76. doi: 10.1126/science.1146221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachikawa H, Harada S, Kawanishi Y, Okubo T, Suzuki T. Linked polymorphisms (-333G>T and -286A>G) in the promoter region of the CCK-A receptor gene may be associated with schizophrenia. Psychiatry Res. 2001;103:147–155. doi: 10.1016/s0165-1781(01)00276-1. [DOI] [PubMed] [Google Scholar]
- Tamminga CA, Stan AD, Wagner AD. The hippocampal formation in schizophrenia. Am J Psychiatry. 2010;167:1178–1193. doi: 10.1176/appi.ajp.2010.09081187. [DOI] [PubMed] [Google Scholar]
- Tandon R, Keshavan MS, Nasrallah HA. Schizophrenia, “Just the Facts”: what we know in 2008 part 1: overview. Schizophr Res. 2008;100:4–19. doi: 10.1016/j.schres.2008.01.022. [DOI] [PubMed] [Google Scholar]
- Tsutsumi M, Claus TH, Liang Y, Li Y, Yang L, Zhu J, Dela CF, Peng X, Chen H, Yung SL, Hamren S, Livingston JN, Pan CQ. A potent and highly selective VPAC2 agonist enhances glucose-induced insulin release and glucose disposal: a potential therapy for type 2 diabetes. Diabetes. 2002;51:1453–1460. doi: 10.2337/diabetes.51.5.1453. [DOI] [PubMed] [Google Scholar]
- Tye KM, Prakash R, Kim SY, Fenno LE, Grosenick L, Zarabi H, Thompson KR, Gradinaru V, Ramakrishnan C, Deisseroth K. Amygdala circuitry mediating reversible and bidirectional control of anxiety. Nature. 2011;471:358–362. doi: 10.1038/nature09820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacic V, McCarthy S, Malhotra D, Murray F, Chou HH, Peoples A, Makarov V, Yoon S, Bhandari A, Corominas R, Iakoucheva LM, Krastoshevsky O, Krause V, Larach-Walters V, Welsh DK, Craig D, Kelsoe JR, Gershon ES, Leal SM, Dell AM, Morris DW, Gill M, Corvin A, Insel PA, McClellan J, King MC, Karayiorgou M, Levy DL, DeLisi LE, Sebat J. Duplications of the neuropeptide receptor gene VIPR2 confer significant risk for schizophrenia. Nature. 2011;471:499–503. doi: 10.1038/nature09884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassar R, Kovacs DM, Yan R, Wong PC. The beta-secretase enzyme BACE in health and Alzheimer’s disease: regulation, cell biology, function, and therapeutic potential. J Neurosci. 2009;29:12787–12794. doi: 10.1523/JNEUROSCI.3657-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh T, McClellan JM, McCarthy SE, Addington AM, Pierce SB, Cooper GM, Nord AS, Kusenda M, Malhotra D, Bhandari A, Stray SM, Rippey CF, Roccanova P, Makarov V, Lakshmi B, Findling RL, Sikich L, Stromberg T, Merriman B, Gogtay N, Butler P, Eckstrand K, Noory L, Gochman P, Long R, Chen Z, Davis S, Baker C, Eichler EE, Meltzer PS, Nelson SF, Singleton AB, Lee MK, Rapoport JL, King MC, Sebat J. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–543. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- Walss-Bass C, Liu W, Lew DF, Villegas R, Montero P, Dassori A, Leach RJ, Almasy L, Escamilla M, Raventos H. A novel missense mutation in the transmembrane domain of neuregulin 1 is associated with schizophrenia. Biol Psychiatry. 2006;60:548–553. doi: 10.1016/j.biopsych.2006.03.017. [DOI] [PubMed] [Google Scholar]
- Wang Q, Liu L, Pei L, Ju W, Ahmadian G, Lu J, Wang Y, Liu F, Wang YT. Control of synaptic strength, a novel function of Akt. Neuron. 2003;38:915–928. doi: 10.1016/s0896-6273(03)00356-8. [DOI] [PubMed] [Google Scholar]
- Webber MA, Marder SR. Better pharmacotherapy for schizophrenia: what does the future hold? Curr Psychiatry Rep. 2008;10:352–358. doi: 10.1007/s11920-008-0056-8. [DOI] [PubMed] [Google Scholar]
- Weinberger DR. From neuropathology to neurodevelopment. Lancet. 1995;346:552–557. doi: 10.1016/s0140-6736(95)91386-6. [DOI] [PubMed] [Google Scholar]
- Weiss J, Magert HJ, Cieslak A, Forssmann WG. Association between different psychotic disorders and the DRD4 polymorphism, but no differences in the main ligand binding region of the DRD4 receptor protein compared to controls. Eur J Med Res. 1996;1:439–445. [PubMed] [Google Scholar]
- Wen L, Lu YS, Zhu XH, Li XM, Woo RS, Chen YJ, Yin DM, Lai C, Terry AV, Jr, Vazdarjanova A, Xiong WC, Mei L. Neuregulin 1 regulates pyramidal neuron activity via ErbB4 in parvalbumin-positive interneurons. Proc Natl Acad Sci U S A. 2010;107:1211–1216. doi: 10.1073/pnas.0910302107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westfall JE, Hoyt C, Liu Q, Hsiao YC, Pierce EA, Page-McCaw PS, Ferland RJ. Retinal degeneration and failure of photoreceptor outer segment formation in mice with targeted deletion of the Joubert syndrome gene, Ahi1. J Neurosci. 2010;30:8759–8768. doi: 10.1523/JNEUROSCI.5229-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams J, Spurlock G, McGuffin P, Mallet J, Nothen MM, Gill M, Aschauer H, Nylander PO, Macciardi F, Owen MJ. Association between schizophrenia and T102C polymorphism of the 5-hydroxytryptamine type 2a-receptor gene. European Multicentre Association Study of Schizophrenia (EMASS) Group. Lancet. 1996;347:1294–1296. doi: 10.1016/s0140-6736(96)90939-3. [DOI] [PubMed] [Google Scholar]
- Woodin M, Wang PP, Aleman D, Donald-McGinn D, Zackai E, Moss E. Neuropsychological profile of children and adolescents with the 22q11.2 microdeletion. Genet Med. 2001;3:34–39. doi: 10.1097/00125817-200101000-00008. [DOI] [PubMed] [Google Scholar]
- Wu J, Li J, Huang KP, Huang FL. Attenuation of protein kinase C and cAMP-dependent protein kinase signal transduction in the neurogranin knockout mouse. J Biol Chem. 2002;277:19498–19505. doi: 10.1074/jbc.M109082200. [DOI] [PubMed] [Google Scholar]
- Xu B, Roos JL, Dexheimer P, Boone B, Plummer B, Levy S, Gogos JA, Karayiorgou M. Exome sequencing supports a de novo mutational paradigm for schizophrenia. Nat Genet. 2011 doi: 10.1038/ng.902. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Roos JL, Levy S, van Rensburg EJ, Gogos JA, Karayiorgou M. Strong association of de novo copy number mutations with sporadic schizophrenia. Nat Genet. 2008;40:880–885. doi: 10.1038/ng.162. [DOI] [PubMed] [Google Scholar]
- Xu B, Woodroffe A, Rodriguez-Murillo L, Roos JL, van Rensburg EJ, Abecasis GR, Gogos JA, Karayiorgou M. Elucidating the genetic architecture of familial schizophrenia using rare copy number variant and linkage scans. Proc Natl Acad Sci U S A. 2009;106:16746–16751. doi: 10.1073/pnas.0908584106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Yang H, Lin YF, Li X, Cape A, Ressler KJ, Li S, Li XJ. Neuronal Abelson helper integration site-1 (Ahi1) deficiency in mice alters TrkB signaling with a depressive phenotype. Proc Natl Acad Sci U S A. 2010;107:19126–19131. doi: 10.1073/pnas.1013032107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y, Akiyoshi J, Kiyota A, Katsuragi S, Tsutsumi T, Isogawa K, Nagayama H. Increased anxiety behavior in OLETF rats without cholecystokinin-A receptor. Brain Res Bull. 2000;53:789–792. doi: 10.1016/s0361-9230(00)00407-x. [DOI] [PubMed] [Google Scholar]
- Yavich L, Forsberg MM, Karayiorgou M, Gogos JA, Mannisto PT. Site-specific role of catechol-O-methyltransferase in dopamine overflow within prefrontal cortex and dorsal striatum. J Neurosci. 2007;27:10196–10209. doi: 10.1523/JNEUROSCI.0665-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu FH, Mantegazza M, Westenbroek RE, Robbins CA, Kalume F, Burton KA, Spain WJ, McKnight GS, Scheuer T, Catterall WA. Reduced sodium current in GABAergic interneurons in a mouse model of severe myoclonic epilepsy in infancy. Nat Neurosci. 2006;9:1142–1149. doi: 10.1038/nn1754. [DOI] [PubMed] [Google Scholar]
- Zeng H, Chattarji S, Barbarosie M, Rondi-Reig L, Philpot BD, Miyakawa T, Bear MF, Tonegawa S. Forebrain-specific calcineurin knockout selectively impairs bidirectional synaptic plasticity and working/episodic-like memory. Cell. 2001;107:617–629. doi: 10.1016/s0092-8674(01)00585-2. [DOI] [PubMed] [Google Scholar]
- Zhang HT, Huang Y, Masood A, Stolinski LR, Li Y, Zhang L, Dlaboga D, Jin SL, Conti M, O’donnell JM. Anxiogenic-like behavioral phenotype of mice deficient in phosphodiesterase 4B (PDE4B) Neuropsychopharmacology. 2008;33:1611–1623. doi: 10.1038/sj.npp.1301537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Tochigi M, Ohashi J, Maeda K, Kato T, Okazaki Y, Kato N, Tokunaga K, Sawa A, Sasaki T. Association study of the DISC1/TRAX locus with schizophrenia in a Japanese population. Schizophr Res. 2005;79:175–180. doi: 10.1016/j.schres.2005.05.023. [DOI] [PubMed] [Google Scholar]