Abstract
Objective
Smoking is a prominent risk factor for lung cancer. However, it is not an established prognostic factor for lung cancer in clinics. To date, no gene test is available for diagnostic screening of lung cancer risk or prognostication of clinical outcome in smokers. This study sought to identify a smoking associated gene signature in order to provide a more precise diagnosis and prognosis of lung cancer in smokers.
Methods and materials
An implication network based methodology was used to identify biomarkers by modeling crosstalk with major lung cancer signaling pathways. Specifically, the methodology contains the following steps: 1) identifying genes significantly associated with lung cancer survival; 2) selecting candidate genes which are differentially expressed in smokers versus non-smokers from the survival genes identified in Step 1; 3) from these candidate genes, constructing gene coexpression networks based on prediction logic for the smoker group and the non-smoker group, respectively; 4) identifying smoking-mediated differential components, i.e., the unique gene coexpression patterns specific to each group; and 5) from the differential components, identifying genes directly co-expressed with major lung cancer signaling hallmarks.
Results
A smoking-associated 6-gene signature was identified for prognosis of lung cancer from a training cohort (n=256). The 6-gene signature could separate lung cancer patients into two risk groups with distinct post-operative survival (log-rank P < 0.04, Kaplan-Meier analyses) in three independent cohorts (n=427). The expression-defined prognostic prediction is strongly related to smoking association and smoking cessation (P < 0.02; Pearson’s Chi-squared tests). The 6-gene signature is an accurate prognostic factor (hazard ratio = 1.89, 95% CI: [1.04, 3.43]) compared to common clinical covariates in multivariate Cox analysis. The 6-gene signature also provides an accurate diagnosis of lung cancer with an overall accuracy of 73% in a cohort of smokers (n=164). The coexpression patterns derived from the implication networks were validated with interactions reported in the literature retrieved with STRING8, Ingenuity Pathway Analysis, and Pathway Studio.
Conclusions
The pathway-based approach identified a smoking-associated 6-gene signature that predicts lung cancer risk and survival. This gene signature has potential clinical implications in the diagnosis and prognosis of lung cancer in smokers.
Keywords: implication networks based on prediction logic, gene coexpression networks based on formal logic, smoking, gene signature, lung cancer diagnosis and prognosis, signaling pathways
1. Introduction
Lung cancer remains the leading cause of cancer deaths in the United States [1]. Non-small cell lung cancer (NSCLC) accounts for about 80% of lung cancer cases. Two major subtypes of NSCLC are lung adenocarcinoma and squamous cell lung cancer. Smoking is a strong risk factor in lung cancer development and is responsible for about 90% of lung cancer cases [2–4]. Our previous study showed that smoking intensity at the time of diagnosis is a significant and independent prognostic factor of lung cancer[5]. Nevertheless, smoking is not an established lung cancer prognostic determinant in clinical practice, and its mechanistic effect on lung cancer progression remains unclear. In this study, we sought to identify a smoking-associated gene signature with implications in lung cancer diagnosis and prognosis by analyzing genome-wide transcriptional profiles of lung cancer patient samples.
Traditional approaches to biomarker discovery rank genes based on their association with the clinical outcome and select the top-ranked genes as signature genes [6–8]. However, these approaches do not account for the interactions among genes. It is known that genes function through a series of interactions with one another, and disease is one possible result of these interactions. Recent studies indicate that molecular network analyses could be used to improve disease classification [9–11], identify disease genes [12], discover novel therapeutic targets [13,14], and reveal disease related sub-networks [15].
Boolean networks have been used to gain insights into gene regulation functions [16–19]. The Boolean implication networks presented by Sahoo et al. [20,21]used scatter plots of expression between two genes to derive the implication relations. Their study did not use Boolean implication networks as a gene selection system. We developed an induction algorithm based on prediction logic [22] to derive implication relations. In our previous studies, implication networks were employed to model disease-mediated genome-wide coexpression networks for the identification of prognostic gene signatures [23,24]. In this study, implication networks were used to infer the relevance of signaling pathways in a set of selected genes associated with smoking and lung cancer survival.
Genes implicated in cancer initiation and progression show dysregulated interactions with their molecular partners [25], and cancer genes are more likely to actively interact with signaling proteins [26]. We hypothesized that an analysis of genes associated with smoking and major lung cancer signaling pathways could lead to the identification of a gene signature that provides a more accurate diagnosis and prognosis of lung cancer. The following steps were carried out to test the hypothesis: 1) Genes that were significantly associated with lung cancer survival were identified from genome-wide expression profiles using the training set (n=256). 2) Genes with differential expression in smokers versus non-smokers were then selected for further analysis. 3) The implication network algorithm was employed to construct smoking mediated gene coexpression networks. 4) By comparing the coexpression patterns from smoking mediated gene coexpression networks, the unique coexpression patterns that are specific to each group are identified as the smoking-mediated differential components. 5) From the differential components, genes that had common coexpression relations with MET, EGF, KRAS, TP53, E2F1, and E2F4 were pinpointed. The identified signature was then validated for prognostic (n=427) and diagnostic (n=164) prediction in 5 independent patient cohorts. The prognostic performance of the identified gene signature was also evaluated by comparing it with clinical covariates. Furthermore, the smoking-mediated gene coexpression patterns were confirmed with curated interactions published in the literature.
2. Materials and Methods
2.1. Implication induction algorithm for pair-wise coexpression network construction
An implication network is a directed graph with variables as nodes, and adjacent nodes are connected with arch representing implications. The first induction algorithm for an implication network was proposed by Liu et al. [27,28] based on binomial distribution, which is suitable for binary datasets. An alternative network induction algorithm was proposed by Guo et al. [22] based on prediction logic [29], which is applicable for more general applications, including multinomial datasets and multi-classification problems. Prediction logic reveals the implication relationships among variables in a dataset and evaluates propositions in formal logic by integrating formal logic theory and statistics. The most important aspect of prediction logic is the conceptual value of prediction analysis in constructing and evaluating useful statements, particularly in complex multinomial problems with moderate sample sizes. This feature is vital for clinical applications, in which many clinical parameters are multinomial and the patient sample size is small.
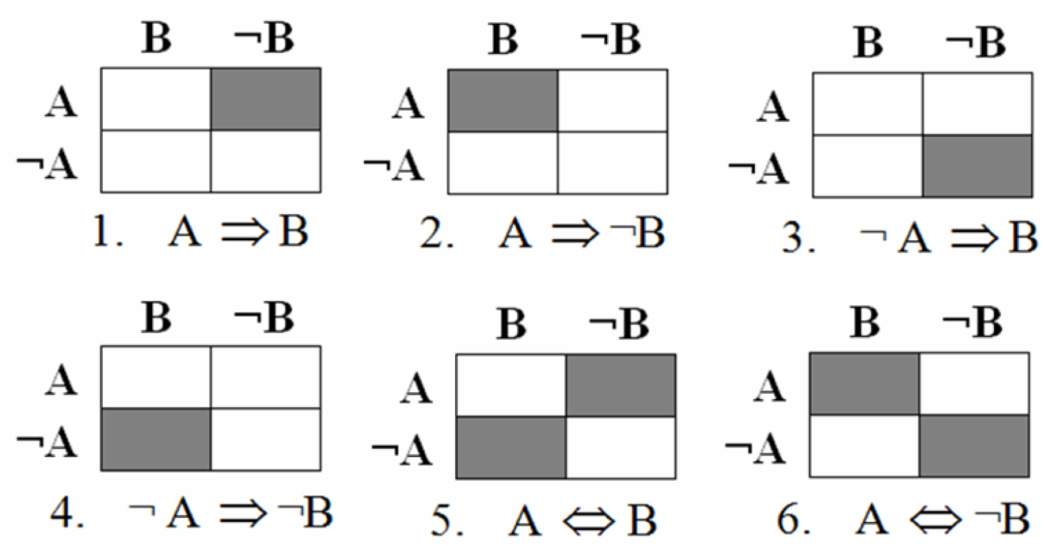
We used prediction logic based on formal logic rules relating two dichotomous variables to induce the implication network. The six most important implication rules relating two dichotomous variables are shown in Fig. 1, where each table is a contingency table and the shaded cells represent the errors for the corresponding implication rule. For example, A ∧ ¬B is the error cell for the implication rule A ⇒ B, NA ∧ ¬B represents the number of error occurrences. In the biological context, A ⇒ B: upregulation of gene A causes upregulation of gene B; A ⇒ ¬B: upregulation of gene A causes downregulation of gene B; ¬A ⇒ B: down-regulation of gene A causes upregulation of gene B; ¬A ⇒ ¬B: down-regulation of gene A causes down-regulation of gene B; A ⇔ B: upregulation of gene A causes upregulation of gene B; and upregulation of gene B causes upregulation of gene A; A ⇔ ¬B: upregulation of gene A causes down-regulation of gene B; and down-regulation of gene B causes upregulation of gene A.
Figure 1. Six implication rules relating two dichotomous variables.
A modified U-Optimality method [29] (Fig. 2) was used to derive the implication relation between each pair of variables in the dataset. In the algorithm, Up is the scope of the implication rule, representing the portion of the data covered by the implication relation, and ∇p is the precision of the implication rule, representing the prediction success of the corresponding implication relation. An implication rule has high precision when the number of error occurrences is a small portion of the data covered by the implication rule. The minimum scope and precision required by the implication rule are indicated respectively by Umin and ∇min, which must be positive for a valid implication relation. The induction algorithm derives an implication rule if it has the maximum scope, Up and it satisfies the constraint that its scope, Up and precision, ∇p are greater than the required minimum values, Umin and ∇min. To simplify the computations of the maximization problem, the ∇ij value of every error cell must be greater than that of the non-error cells for the corresponding implication rule [22].
Figure 2. Implication induction algorithm for constructing coexpression networks.
For a single error cell, where Nij is the number of error occurrences, the scope, Up and precision, ∇p are defined as:
.
For multiple error cells, they are defined as:
where ωij = 1 for error cells; otherwise, ωij = 0.
This implication induction algorithm is general for discrete datasets. With the expansion of the contingency table Mij (Fig. 2), implication rules can be induced for multinomial datasets, where error cells are those with top precision (∇ij) values and satisfying all the constraints. The proposition can then be induced according to the error set.
The complexity of the induction algorithm is O(Nv2), where N is the sample size and v is the number of variables in the dataset (i.e. nodes in the implication networks) [22]. The difference between this algorithm and that of Hildebrand et al. [29] is that minimum requirements for deriving an implication rule were set for both scope (Up) and precision (∇p), instead of for precision alone.
2.2. Microarray profiles and patient samples
Four sets of published microarray gene expression profiles were used in this study. The first set contains 442 lung adenocarcinoma patient samples in the Director’s Challenge Study [30]. The second set contains 130 adenocarcinoma and squamous cell lung cancer samples published by Raponi et al. [8]. The third set contains 111 NSCLC samples published by Bild et al. [31]. The fourth set contains 164 airway epithelial cells from current and former smokers published by Spira et al. [2]. Gene expression profiles from these studies were quantified with Affymetrix HG-U133A, except for the set from Bild et al. [31], which was quantified with Affymetrix HG-U133 Plus 2. The data used in the analyses was quantile-normalized and log2 transformed with dChip [32].
3. Results and Discussion
3.1. Identification of a smoking-associated gene signature for prognosis in lung adenocarcinoma
In this study, the UM and HLM cohorts from the Director’s Challenge Study [30] formed the training set (n = 256), whereas MSK and DFCI cohorts formed the test set (n = 186). Genes with missing values in at least half of the samples were removed, which left 19,866 genes for the analysis.
Genes associated with lung cancer survival were first selected from the entire genome. A total of 2,310 genes were significantly associated with overall survival (P < 0.05, univariate Cox model) in the training data. Second, from this set of 2,310 genes, 217 genes showed significant differential expression (P < 0.05, unpaired t-tests) in smokers versus non-smokers in the training data. These 217 survival and smoking-associated genes as well as 6 major signaling proteins, including EGF, TP53, MET, KRAS, E2F1, and E2F4, were included in the network analysis. These signaling pathways are included in human NSCLC disease mechanisms delineated by the KEGG Pathway Database1. KRAS is involved in many signaling transduction pathways and its mutation is related to many human cancer types. TP53 regulates cell cycle and functions as a tumor suppressor gene. EGF is a growth factor and regulates cell growth, proliferation and differentiation by binding to its receptor EGFR. MET is an oncogene and plays an important role in embryonic development and wound healing. E2F1 and E2F4 are members of the E2F family of transcription factors. The E2F family is essential for the control of cell cycle and action of tumor suppressor proteins. These signaling proteins are selected based on their reported clinical relevance in non-small cell lung cancer. Because tumors utilize different signaling pathways, we hypothesize that including a diverse set of pathways would perform more uniformly across heterogeneous tumor sets. These 6 hallmarks were not significantly associated with survival nor differentially expressed in smokers.
To construct implication networks, expression profiles in each patient were partitioned into binary values using the mean expression profile of each gene as the cutoff. If the expression of a gene in a patient sample was greater than the mean in the cohort, this gene was denoted as up-regulated in this tumor sample; otherwise, it was denoted as down-regulated in the tumor sample. Patient samples in the training set were separated into two groups: smokers (patients who smoked in the past or who are currently smoking) and non-smokers (patients who never smoked). For each patient group, a coexpression network among the 223 genes was constructed using the implication induction algorithm. Between each pair of the 223 genes, possible significant (P < 0.05; z-tests) coexpression relations were derived in the smoker group and the non-smoker group, respectively, constituting smoking-mediated gene coexpression networks for lung cancer. By comparing the implication rules between each pair of nodes in the two networks, differential network components were identified. These differential components are implication relations (co-expressions) that were present in the smoker group but missing in the non-smoker group, or conversely, those present in the non-smoker group but absent in the smoker group.
From the differential components associated with the smoker group and the non-smoker group, genes having direct co-expressions with the 6 lung cancer hallmarks were identified (detailed gene list provided in Supplementary File) From the non-smoker group, certain genes had direct coexpression with some of the 6 hallmarks but no gene had direct coexpression with all the 6 lung cancer hallmarks. From the smoker group, 6 genes were identified having direct coexpression with all the 6 lung cancer hallmarks. This constituted the smoking-associated 6-gene signature for lung cancer prognosis (Table 1).
Table 1.
The identified smoking associated 6-gene signature.
| Probe | Gene symbol |
Gene title | Molecular function (GO) |
|---|---|---|---|
| 200705_s_at | EEF1B2 | Eukaryotic translation elongation factor 1 beta 2 | Translation elongation factor activity; protein binding |
| 203788_s_at | SEMA3C | Sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C | Receptor activity; semaphorin receptor binding |
| 206183_s_at | HERC3 | HECT domain and RCC1-like domain-containing protein 3 | Ligase activity; acid-amino acid ligase activity |
| 209230_s_at | NUPR1 | Nuclear protein, transcriptional regulator, 1 | N/A |
| 210669_at | TFAP2A | Transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) | DNA and protein binding; transcription factor activity; transcription coactivator activity; protein dimerization activity |
| 213456_at | SOSTDC1 | Sclerostin domain containing 1 | Protein binding |
3.2. Prognostic evaluation of the 6-gene signature in lung adenocarcinoma
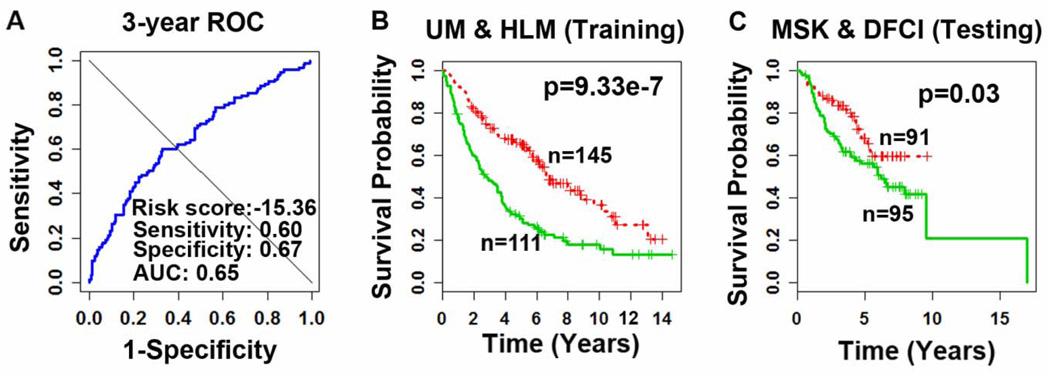
We sought to investigate if the gene signature identified could provide accurate prognostication of survival in NSCLC patients. The 6 hallmarks were not fitted in the model as they were not significantly associated with survival. On the training cohort, the original continuous expression profiles of the 6 probes were fitted into a Cox proportional hazard model as covariates. A survival risk score was generated for each patient in the training set. To identify the best patient stratification scheme, various cutoff values of the risk scores from the training set were evaluated. The cutoff value that gave the shortest distance to the point of perfect prediction, i.e. point [0,1] of the 3-year ROC curve (Fig. 3A), produced the best patient stratification in the training set (Fig. 3B). Therefore, the training model and this cutoff value were applied to the test set without re-estimating the parameters (Fig. 3C). In both training and test sets, this classification scheme generated significant patient stratifications (log-rank P < 0.03, Kaplan-Meier analyses).
Figure 3. Prognostication of survival using the 6-gene signature in lung adenocarcinoma.
On the training cohort from the Director’s Challenge Study (Shedden, 2008), the risk score giving the best prediction on the 3-year ROC curve was identified as the cutoff for patient stratification (A). This cutoff value generated significant (P < 0.03) patient stratification in the training (B) and test (C) cohorts in Kaplan-Meier analyses. Log-rank tests were used to assess the significance of difference between survival probability of the two prognostic groups
To evaluate the statistical significance of the 6-gene signature identified from the proposed network analysis, a random set of 6 genes from the 217 survival and smoking-associated genes were selected and constructed as a classifier using the same approach with the Cox proportional hazard model. Results showed that the identified signature gave significantly (P < 0.05) better lung cancer prognosis compared with 1000 random signatures.
3.3. Smoking association and smoking cessation
To evaluate the smoking association of the identified gene signature, we evaluated the performance of the prognostic signature on smokers in the studied cohorts. Results showed that the signature generated significant prognostic stratifications in smokers from both training and test cohorts (log-rank P < 0.04, Kaplan-Meier analysis) (Fig. 4), but not in non-smokers (log-rank P < 0.83, Kaplan-Meier analyses, results not shown). In addition, gene expression-defined high- and low-risk groups showed significant association with smoking (P < 0.00008, Chi-square tests) and smoking cessation (P < 0.02, Chi-square tests) (Table 2). Specifically, smokers were significantly associated with the high-risk group compared with non-smokers, and current smokers had a stronger association with the high-risk group compared with former smokers (Table 2).
Figure 4. Survival prediction in smoking lung cancer patients by the 6-gene signature.
The prognostic classifier stratified smoking patients into two prognostic groups with significantly distinct survival (P < 0.04) in both the training (A) and test (B) cohorts in the Director’s Challenge Study.
Table 2.
Association between smoking status and gene expression-defined prognostic risk groups.
| Low-risk | High-risk | Chi-square tests | |
|---|---|---|---|
| Smoker | 138 | 162 | Smoking association |
| Non-smoker | 38 | 11 | χ2 = 15.53 (P =8.10e-5) |
| Current smoker | 8 | 24 | Smoking cessation |
| Former smoker | 130 | 138 | χ2 = 5.45 (P = 0.02) |
3.4. Prognostic validation on squamous cell lung cancer
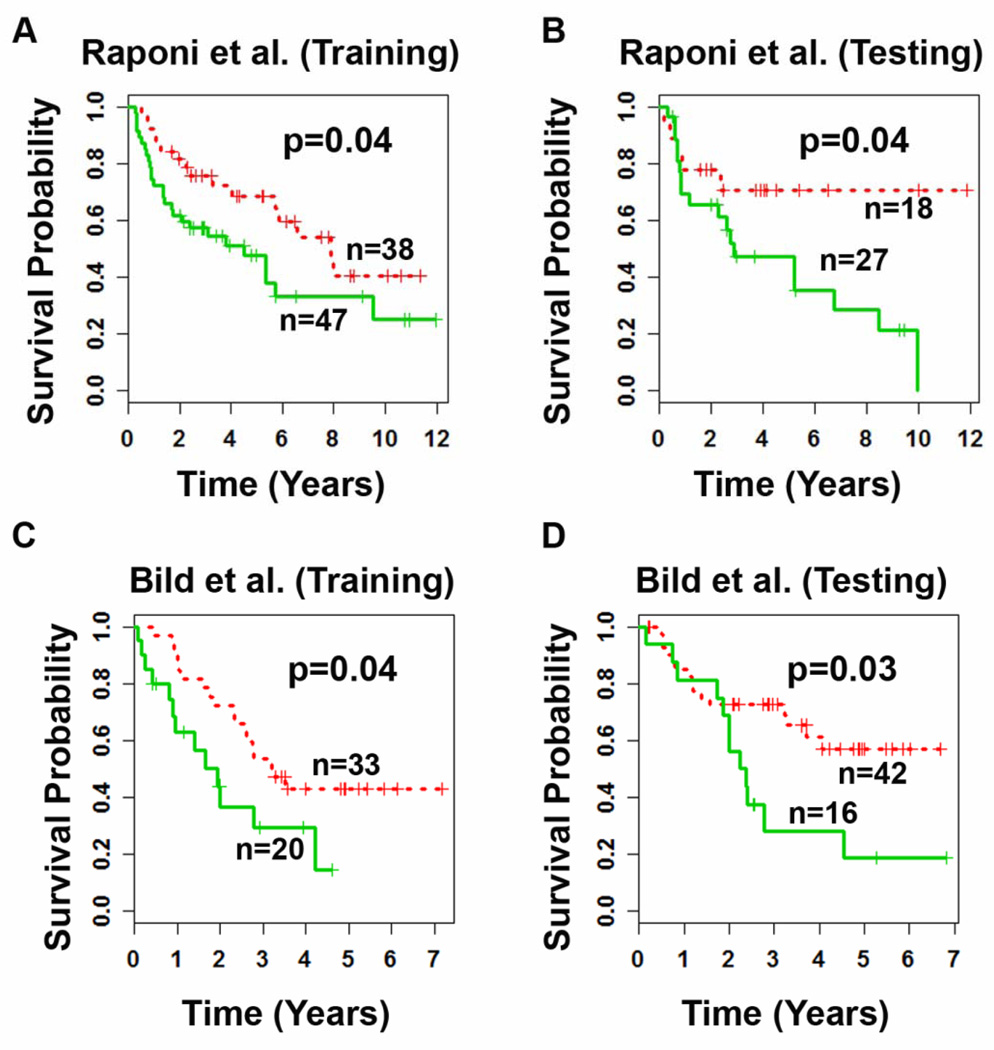
The prognostic performance of the 6-gene signature was further evaluated on the Raponi [8] and Bild [31] cohorts which include squamous cell lung cancer. For a rigorous evaluation, patient samples in the studied cohorts were randomly partitioned into separate training and test sets. A prognostic classifier was constructed on the training set using a Cox proportional hazard model and validated on the test set without re-estimating parameters. In the training set from both cohorts, the cutoff value that gave the shortest distance to the point of perfect prediction of the 3-year ROC curve produced the best patient stratification. In both training and test sets, the 6-gene signature stratified patients into two prognostic groups with distinct survival (log-rank P < 0.04; Fig. 5). These results indicate that the identified smoking-associated 6-gene signature could be used for prognosis for NSCLC patients.
Figure 5. Prognostic performance of the smoking-associated signature on other histological subtypes of NSCLC.
In Kaplan-Meier analyses, significant (P < 0.04) stratifications were obtained in the randomly partitioned training and test cohorts of patients with squamous cell lung cancer (A, B) and patients with lung adenocarcinoma or squamous cell lung cancer (C, D).
3.5. Prognosis evaluation with clinical covariates
To validate the prognostic power of the identified 6-gene signature, the constructed expression-defined prognostic model was evaluated with common lung cancer prognostic factors, including gender, age, cancer stage, and tumor differentiation in smokers in the test cohort. The prognostic outcome predicted by the gene expression model was used as a covariate in the multivariate Cox analysis.
Results from the multivariate Cox proportional analysis showed that cancer stage was the only factor significantly (P < 0.002) associated with elevated risk of lung cancer death when the model was fitted without the 6-gene prognostic model (Table 3). When the 6-gene prognostic model was added to the multivariate Cox model, the gene model demonstrated a strong association with the risk of lung cancer death (hazard ratio = 1.89, 95% CI: [1.04, 3.43]), and cancer stage remained significant (Table 3). The hazard ratio of the 6-gene prognostic model was higher than other cancer prognostic factors except for cancer stage, with no significant difference between cancer stage and the gene model. The results demonstrate that the 6-gene prognostic model is a more significant prognostic factor than some commonly used clinical parameters.
Table 3.
Multivariate Cox analyses of the gene expression-defined prognostication and major clinical covariates in smoking lung cancer patients in the test cohort.
| Variable* | P-value | Hazard ratio (95% CI)ψ | |
|---|---|---|---|
| Analysis without 6-gene prognostic prediction | |||
| Gender (Male) | 0.55 | 1.17 | (0.70, 1.95) |
| Age at diagnosis (>60) | 0.35 | 1.31 | (0.74, 2.29) |
| Tumor differentiation | |||
| Moderately differentiated | 0.30 | 0.63 | (0.26, 1.51) |
| Poorly differentiated | 0.89 | 1.06 | (0.47, 2.38) |
| Cancer Stage | |||
| Stage II | 1.54E-03 | 2.60 | (1.44, 4.71) |
| Stage III | 5.53E-05 | 4.48 | (2.16, 9.29) |
| Analysis with 6-gene prognostic prediction | |||
| Gender (Male) | 0.42 | 1.24 | (0.74, 2.08) |
| Age at diagnosis (>60) | 0.52 | 1.20 | (0.68, 2.13) |
| Tumor differentiation | |||
| Moderately differentiated | 0.39 | 0.68 | (0.28, 1.64) |
| Poorly differentiated | 0.89 | 0.94 | (0.42, 2.15) |
| Cancer Stage | |||
| Stage II | 7.30E-04 | 2.83 | (1.55, 5.19) |
| Stage III | 1.51E-05 | 5.36 | (2.50, 11.46) |
| 6-gene prognostic prediction | 0.04 | 1.89 | (1.04, 3.43) |
Gender was a binary variable (0 for female and 1 for male); age at diagnosis was a binary variable (0 for < 60 years old and 1 otherwise); tumor grade was categorical variable of 3 categories (Well [as the reference group], Moderately, and Poorly differentiated); cancer stage was categorical variable of 3 categories (Stage I [as the reference group], Stage II, and Stage III).
denotes confidence interval.
3.6. Early detection of lung cancer
We further evaluated whether the 6-gene signature could be used for lung cancer diagnosis in smokers. The smoking cohort from Spira et al. [2] was randomly separated into a training set (n = 77) and two independent test sets (n = 52 and n = 35). With the Naïve Bayes classification algorithm implemented in software package WEKA [33], the classifier could accurately identify lung cancer patients from normal patients with an overall accuracy of 73% and 69% in the two test sets (Table 4). Furthermore, the classifier’s performance was significantly (P < 0.005) better than that of random signatures with the same size using the same classifier in 1000 tests, on the same training and test sets. These results indicate that the 6-gene signature could be potentially used in diagnostic screening of lung cancer risk in smokers.
Table 4.
Prediction of lung cancer risk in smokers using the 6-gene signature with the Naïve Bayes algorithm.
| Sensitivity (lung cancer) |
Specificity (normal) |
Overall accuracy* | |
|---|---|---|---|
| Training (10-fold CV) | 71% (25/35) | 62% (26/42) | 66% (51/77) |
| Test 1 | 76% (19/25) | 65% (19/27) | 73% (38/52) |
| Test 2 | 72% (13/18) | 65% (11/17) | 69% (24/35) |
The 6-gene signature gave significantly (P<0.005) accurate performance in all three data sets when compared with 1000 random sets of 6 genes using the same algorithm.
3.7. Confirmation of smoking-mediated gene coexpression relations
The coexpression relations derived by the implication network were also evaluated. Differential network components among the signature genes and the 6 signaling hallmarks present in both training and test sets were retrieved to represent smoking-mediated gene coexpression patterns in lung cancer patients. There were 9 common coexpression relations specifically associated with smokers (Fig. 6A), and 3 specifically associated with non-smokers (Fig. 6B) in both training and testing cohorts.
Figure 6. Smoking-mediated coexpression networks.
Gene coexpression patterns specific to smokers (A) and non-smokers (B) derived by the implication network model (P < 0.05; z-tests) commonly present in both training and test sets. The biological interpretation of the implication relations is described in (C). The stability of smoking-mediated networks as evaluated with random subsets of patients from the training cohort in 100 iterations (D).
The biological relevance of the derived coexpression relations was confirmed by retrieving curated interactions related to these genes using bioinformatics tools in Ingenuity Pathway Analysis2 (IPA, Ingenuity Systems®), Pathway Studio, and the signaling pathway database STRING8. Among 12 coexpression relations derived from the implication networks, 1 interaction specific to smokers and 1 interaction specific to non-smokers were confirmed (Fig. 6).
The stability of the smoking mediated coexpression networks (Fig. 6A and 6B) was evaluated with different subsets of patient samples from the training set in 100 iterations (Fig. 6D). The stability is defined as the portion of smoking-mediated coexpression relations obtained from the original data that are retrieved by using only a random subset of the training data and the full test data. Results show that the implication network algorithm is stable as most of the coexpression relations (about 60%) could be derived using as few as half of the training samples (Fig. 6D).
In addition, we also evaluated the precision and false discovery rate (FDR) of the coexpression relations derived in the smoking-mediated coexpression networks (Fig 6A and 6B). Five gene set collections (positional, curated, motif, computational, and Gene Oncology) and canonical pathway databases from the MSigDB3 were used to evaluate the biological relevance of computationally derived coexpression relations. A coexpression relation was considered a true positive (TP) if the pair of genes belongs to the same gene set or pathway in any investigated database. If a pair of genes does not share any gene set or pathway, the coexpression relation was considered a false positive (FP). A coexpression relation was labeled as non-discriminatory (ND) if at least one gene in the pair is not annotated in a database [34]. Coexpression relations labeled as ND were excluded in the evaluation as they were not confirmed. With precision defined as TP/(TP + FP), the precision of the smoking-mediated coexpression networks (Fig. 6A and 6B) was 100% (7 relations were labeled as TP and no relation was labeled as FP; Fig. 6C). Null distribution was generated in 1,000 random permutations of the class labels in the test cohort. The precision of the smoking-mediated coexpression networks is significant at P<0.001, with no TP generated in the random tests. With FDR defined as the average of FP/(TP+FP) in 1,000 permutations, the FDR of the smoking-mediated coexpression networks is 0.0099. These results indicate that implication networks can reveal biologically relevant gene associations.
3.8 Comparison with gene association networks based on Pearson’s correlation
Large-scale gene coexpression networks have been used in biomarker discovery and disease classification, based on the observation that functionally related genes are frequently coexpressed across multiple datasets and different organisms [35–37]. These studies construct pair-wise gene coexpression networks by using correlation coefficients computed from gene expression profiles. Such networks indicate the distance or similarity between each pair of gene expression profiles but do not provide the direction or causal relations in the gene regulatory patterns. A new algorithm is needed to efficiently construct genome-scale coexpression networks and provide a convenient predictive structure of gene regulation. Prediction logic provides a convenient and more predictive structure association than correlation coefficients [29]. Boolean implications networks constructed with a similar algorithm have been used to infer gene regulations [20,21].
For comparison with implication networks, we used Pearson’s correlation coefficient to construct gene association networks for smoker and non-smoker groups in both training and testing sets using the same methodology (Figure 7). The implications networks derived more gene association rules than the networks based on Pearson’s correlation coefficients. We then evaluated the precision and FDR of the interactions specific to smoker and non-smoker groups that were present in both training and testing sets (Figure 7C). Both networks have the same precision of 0.96 and FDR of 0.04 in the evaluation with MSigDB. These results indicate that implication networks could retrieve more biologically relevant gene associations without any loss of precision and increase of FDR when compared with gene association networks based on Pearson’s correlation coefficients. Furthermore, we examined the smoking-specific and non-smoking-specific gene association networks based on Pearson’s correlation coefficients in the training set. No gene was coexpressed with all 6 lung cancer hallmarks based on the Pearson’s correlation. In other words, using gene association networks based on Pearson’s correlation coefficients, we would not be able to identify any gene with concurrent coexpression with the 6 signaling pathways using the proposed methodology. Together, these results demonstrate the advantage of implication networks based on prediction logic in biomarker discovery.
Figure 7. Comparison with gene association networks based on Pearson’s correlation coefficients.
Number of gene associations derived with implication networks and Pearson’s correlation coefficients on the training set (A), testing set (B) and common gene associations in both training and testing sets (C). Genet: implications networks.
4. Conclusions
This study presents an implication network-based approach to the identification of a smoking-associated 6-gene signature that was co-expressed with major NSCLC signaling pathways. The identified 6-gene signature could accurately estimate disease-specific survival in NSCLC patients and could potentially be used for screening of lung cancer risk in smokers. The gene expression-defined prognostication also showed strong association with smoking and smoking cessation. This gene signature is a more accurate prognostic factor than some commonly used clinical parameters such as age, gender, and tumor differentiation, and is comparable with cancer stage in terms of hazard ratio. Some of the computationally derived coexpression patterns have been experimentally verified in previous studies.
Our previous studies have demonstrated that implication network-based methodology is efficient in modeling disease-mediated genome-scale coexpression networks for biomarker identification [23,24]. In this study, the methodology was applied to a more focused set of genes related to smoking and lung cancer survival. The results from this study demonstrate that combined analysis of smoking mediated coexpression networks and crosstalk with lung cancer signaling pathways could identify important biomarkers and elucidate mechanistic and possibly synergistic processes underlying oncogenesis and metastasis in lung cancer. The gene coexpression relations derived with implication networks have been validated with biological experiments (results not shown).
Supplementary Material
Acknowledgements
We are grateful for Rebecca Raese for editing the manuscript. We thank Changchang Xiao for processing the microarray data. This project is supported by NIH R01LM009500 (PI: Guo) and NIH/NCRR P20RR16440 and Supplement (PD: Guo). Software license and training for Ingenuity Pathway Analysis and Pathway Studio is supported by NIH/NCRR P2016477.
Footnotes
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics: 2009. CA Cancer J. Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Spira A, Beane JE, Shah V, Steiling K, Liu G, Schembri F, et al. Airway epithelial gene expression in the diagnostic evaluation of smokers with suspect lung cancer. Nat Med. 2007;13:361–366. doi: 10.1038/nm1556. [DOI] [PubMed] [Google Scholar]
- 3.Massion PP, Zou Y, Chen H, Jiang A, Coulson P, Amos CI, et al. Smoking-related genomic signatures in non-small cell lung cancer. Am. J. Respir. Crit Care Med. 2008;178:1164–1172. doi: 10.1164/rccm.200801-142OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Woenckhaus M, Klein-Hitpass L, Grepmeier U, Merk J, Pfeifer M, Wild P, et al. Smoking and cancer-related gene expression in bronchial epithelium and non-small-cell lung cancers. J. Pathol. 2006;210:192–204. doi: 10.1002/path.2039. [DOI] [PubMed] [Google Scholar]
- 5.Guo NL, Tosun K, Horn K. Impact and interactions between smoking and traditional prognostic factors in lung cancer progression. Lung Cancer. 2009;66:386–392. doi: 10.1016/j.lungcan.2009.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beer DG, Kardia SL, Huang CC, Giordano TJ, Levin AM, Misek DE, et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat. Med. 2002;8:816–824. doi: 10.1038/nm733. [DOI] [PubMed] [Google Scholar]
- 7.Chen HY, Yu SL, Chen CH, Chang GC, Chen CY, Yuan A, et al. A five-gene signature and clinical outcome in non-small-cell lung cancer. N. Engl. J. Med. 2007;356:11–20. doi: 10.1056/NEJMoa060096. [DOI] [PubMed] [Google Scholar]
- 8.Raponi M, Zhang Y, Yu J, Chen G, Lee G, Taylor JM, et al. Gene expression signatures for predicting prognosis of squamous cell and adenocarcinomas of the lung. Cancer Res. 2006;66:7466–7472. doi: 10.1158/0008-5472.CAN-06-1191. [DOI] [PubMed] [Google Scholar]
- 9.Chuang HY, Lee E, Liu YT, Lee D, Ideker T. Network-based classification of breast cancer metastasis. Mol. Syst. Biol. 2007;3:140. doi: 10.1038/msb4100180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muller FJ, Laurent LC, Kostka D, Ulitsky I, Williams R, Lu C, et al. Regulatory networks define phenotypic classes of human stem cell lines. Nature. 2008;455:401–405. doi: 10.1038/nature07213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taylor IW, Linding R, Warde-Farley D, Liu Y, Pesquita C, Faria D, et al. Dynamic modularity in protein interaction networks predicts breast cancer outcome. Nat Biotechnol. 2009;27:199–204. doi: 10.1038/nbt.1522. [DOI] [PubMed] [Google Scholar]
- 12.Emilsson V, Thorleifsson G, Zhang B, Leonardson AS, Zink F, Zhu J, et al. Genetics of gene expression and its effect on disease. Nature. 2008;452:423–428. doi: 10.1038/nature06758. [DOI] [PubMed] [Google Scholar]
- 13.Csermely P, Agoston V, Pongor S. The efficiency of multi-target drugs: the network approach might help drug design. Trends Pharmacol. Sci. 2005;26:178–182. doi: 10.1016/j.tips.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 14.Yildirim MA, Goh KI, Cusick ME, Barabasi AL, Vidal M. Drug-target network. Nat. Biotechnol. 2007;25:1119–1126. doi: 10.1038/nbt1338. [DOI] [PubMed] [Google Scholar]
- 15.Calvano SE, Xiao W, Richards DR, Felciano RM, Baker HV, Cho RJ, et al. A network-based analysis of systemic inflammation in humans. Nature. 2005;437:1032–1037. doi: 10.1038/nature03985. [DOI] [PubMed] [Google Scholar]
- 16.Jansen R, Yu H, Greenbaum D, Kluger Y, Krogan NJ, Chung S, et al. A Bayesian networks approach for predicting protein-protein interactions from genomic data. Science. 2003;302:449–453. doi: 10.1126/science.1087361. [DOI] [PubMed] [Google Scholar]
- 17.Sahoo D, Dill DL, Gentles AJ, Tibshirani R, Plevritis SK. Boolean implication networks derived from large scale, whole genome microarray datasets. Genome Biol. 2008;9:R157. doi: 10.1186/gb-2008-9-10-r157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sachs K, Perez O, Pe'er D, Lauffenburger DA, Nolan GP. Causal protein-signaling networks derived from multiparameter single-cell data. Science. 2005;308:523–529. doi: 10.1126/science.1105809. [DOI] [PubMed] [Google Scholar]
- 19.Milo R, Shen-Orr S, Itzkovitz S, Kashtan N, Chklovskii D, Alon U. Network motifs: simple building blocks of complex networks. Science. 2002;298:824–827. doi: 10.1126/science.298.5594.824. [DOI] [PubMed] [Google Scholar]
- 20.Sahoo D, Dill DL, Gentles AJ, Tibshirani R, Plevritis SK. Boolean implication networks derived from large scale, whole genome microarray datasets. Genome Biol. 2008;9:R157. doi: 10.1186/gb-2008-9-10-r157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sahoo D, Seita J, Bhattacharya D, Inlay MA, Weissman IL, Plevritis SK, et al. MiDReG: a method of mining developmentally regulated genes using Boolean implications. Proc. Natl. Acad. Sci. U. S. A. 2010;107:5732–5737. doi: 10.1073/pnas.0913635107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo L, Cukic B, Singh H. Predicting Fault Prone Modules by the Dempster-Shafer Belief Networks. Proceedings of 18th IEEE International Conference on Automated Software Engineering (ASE'03) 2003:249–252. doi: 10.1109/ASE.2003.1240314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo NL, Wan YW, Bose S, Denvir J, Kashon ML, Andrew ME. A novel network model identified a 13-gene lung cancer prognostic signature. Int. J. Comput. Biol. Drug Des. 2011;4:19–39. doi: 10.1504/IJCBDD.2011.038655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wan YW, Bose S, Denvir J, Guo NL. A Novel Network Model for Molecular Prognosis. Proc. ACM International Conference on Bioinformatics and Computational Biology. 2010:342–345. doi: 10.1145/1854776.1854825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mani KM, Lefebvre C, Wang K, Lim WK, Basso K, la-Favera R, et al. A systems biology approach to prediction of oncogenes and molecular perturbation targets in B-cell lymphomas. Mol. Syst. Biol. 2008;4:169. doi: 10.1038/msb.2008.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cui Q, Ma Y, Jaramillo M, Bari H, Awan A, Yang S, et al. A map of human cancer signaling. Mol. Syst. Biol. 2007;3:152. doi: 10.1038/msb4100200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu J, Desmarais MC. A Method of Learning Implication Networks from Empirical Data: Algorithm and Monte-Carlo Simulation-Based Validation. IEEE Transactions on Knowledge and Data Engineering. 1997;9:990–1004. [Google Scholar]
- 28.Liu J, Maluf D, Desmarais MC. A New Uncertainty Measure for Belief Networks with Applications to Optimal Evidential Inferencing. IEEE Transactions on Knowledge and Data Engineering. 2001;13:416–425. [Google Scholar]
- 29.Hildebrand DK, Laing JD, Rosenthal H. Prediction Analysis of Cross Classifications. John Wiley & Sons; 1977. [Google Scholar]
- 30.Shedden K, Taylor JM, Enkemann SA, Tsao MS, Yeatman TJ, Gerald WL, et al. Gene expression-based survival prediction in lung adenocarcinoma: a multi-site, blinded validation study. Nat. Med. 2008;14:822–827. doi: 10.1038/nm.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 32.Li C. Automating dChip: toward reproducible sharing of microarray data analysis. BMC. Bioinformatics. 2008;9:231. doi: 10.1186/1471-2105-9-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Witten IH, Frank E. Data Mining: Practical Machine Learning Tools and Techniques (2nd Edition) Morgan Kaufmann; 2005. [Google Scholar]
- 34.Ucar D, Neuhaus I, Ross-MacDonald P, Tilford C, Parthasarathy S, Siemers N, et al. Construction of a reference gene association network from multiple profiling data: application to data analysis. Bioinformatics. 2007;23:2716–2724. doi: 10.1093/bioinformatics/btm423. [DOI] [PubMed] [Google Scholar]
- 35.Choi JK, Yu U, Yoo OJ, Kim S. Differential coexpression analysis using microarray data and its application to human cancer. Bioinformatics. 2005;21:4348–4355. doi: 10.1093/bioinformatics/bti722. [DOI] [PubMed] [Google Scholar]
- 36.Elo LL, Jarvenpaa H, Oresic M, Lahesmaa R, Aittokallio T. Systematic construction of gene coexpression networks with applications to human T helper cell differentiation process. Bioinformatics. 2007 doi: 10.1093/bioinformatics/btm309. [DOI] [PubMed] [Google Scholar]
- 37.Liu CC, Chen WS, Lin CC, Liu HC, Chen HY, Yang PC, et al. Topology-based cancer classification and related pathway mining using microarray data. Nucleic Acids Res. 2006;34:4069–4080. doi: 10.1093/nar/gkl583. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.