Abstract
Both genome-wide genetic and epigenetic alterations are fundamentally important for the development of cancers, but the interdependence of these aberrations is poorly understood. Glioblastomas and other cancers with the CpG island methylator phenotype (CIMP) constitute a subset of tumours with extensive epigenomic aberrations and a distinct biology1–3. Glioma CIMP (G-CIMP) is a powerful determinant of tumour pathogenicity, but the molecular basis of G-CIMP remains unresolved. Here we show that mutation of a single gene, isocitrate dehydrogenase 1 (IDH1), establishes G-CIMP by remodelling the methylome. This remodelling results in reorganization of the methylome and transcriptome. Examination of the epigenome of a large set of intermediate-grade gliomas demonstrates a distinct G-CIMP phenotype that is highly dependent on the presence of IDH mutation. Introduction of mutant IDH1 into primary human astrocytes alters specific histone marks, induces extensive DNA hypermethylation, and reshapes the methylome in a fashion that mirrors the changes observed in G-CIMP-positive lower-grade gliomas. Furthermore, the epigenomic alterations resulting from mutant IDH1 activate key gene expression programs, characterize G-CIMP-positive proneural glioblastomas but not other glioblastomas, and are predictive of improved survival. Our findings demonstrate that IDH mutation is the molecular basis of CIMP in gliomas, provide a framework for understanding oncogenesis in these gliomas, and highlight the interplay between genomic and epigenomic changes in human cancers.
The isocitrate dehydrogenase genes IDH1 and IDH2 are mutated in >70%of lower-grade gliomas (grades II and III), in some glioblastomas4,5, and in leukaemias and several other cancers6,7. The most common IDH1 mutations in glioma (>95%) result in an amino acid substitution at arginine 132 (R132), which resides in the enzyme’s active site. Mutation of IDH imparts the ability to produce 2-hydroxyglutarate (2-HG), a potential oncometabolite8–10. Alterations in the methylation landscape have been shown to have important roles during oncogenesis11. CIMP has emerged as a distinct molecular subclass of tumours in a number of human malignancies, including glioblastoma1–3. This phenotype is associated with extensive, coordinated hypermethylation at specific loci1,2,12,13. In glioblastomas, G-CIMP is associated with the proneural subgroup of tumours and IDH mutation1. Exactly how mutant IDH promotes tumorigenesis and causes G-CIMP—or CIMP in any type of human cancer—is unknown.
To determine whether IDH1 mutation directly causes G-CIMP, we used immortalized primary human astrocytes14 and constructed isogenic cells expressing either mutant IDH1 (R132H), wild-type IDH1, or neither. These astrocytes are well characterized14–17. Introduction of wild-type IDH1 and the R132H IDH1 mutant resulted in equal expression of protein (modest threefold increase) (Fig. 1a). Expression of mutant but not wild-type IDH1 in human astrocytes resulted in the production of 2-HG (Fig. 1b). To determine whether mutant IDH1 altered the methylation landscape, we analysed genomic DNA from these cells using the Illumina InfiniumHumanMethylation450 platform. The platform provides genome-wide coverage and is both well validated and highly reproducible18,19.
Figure 1. Introduction of mutant IDH1 into human astrocytes remodels the methylome.
a, Expression of wild-type and mutant IDH1 (R132H) in immortalized human astrocytes (passage 5). b, Overexpression of mutant IDH1 but not wild-type(WT) IDH1 in human astrocytes leads to production of 2-HG8. Error bars show 1 standard deviation (s.d.) (n = 2). c, Self-organizing map (SOM) analysis of methylome data for wild-type IDH1-expressing, mutant IDH1-expressing (R132H), and parental (control) cell lines shows changes in the methylome in mutant IDH1-expressing and wild-type IDH1-expressing astrocytes, compared to parental cells. Mosaic patterns are pseudo-coloured SOMs from different time points (P indicates passage number). Tile colours indicate methylation level of centroids. d, Hierarchical clustering showing divergence of the methylome of IDH1-expressing astrocytes from that of parental astrocytes. MUT, mutant; PAR, parental. e, Heatmap showing the 10,678 most significant differentially methylated probes (ANOVA) in IDH1 mutant astrocytes and parental astrocytes (passages 2 and 40). Colour scale indicates β values. f, Kinetics of differential methylation in mutant and wild-type-expressing astrocytes. Error bars indicate inter-quartile range (n = 2).
Previous data demonstrated that de novo DNA methylation in in vitro models occurs over extended periods, requiring time to ‘lock in’ epigenomic changes12,20. We thus analysed the methylomes of astrocytes expressing mutant or wild-type IDH1 over successive passages (up to 50). Analysis using self-organizing maps demonstrated that mutant IDH1 progressively remodelled the glial methylome over time (Fig. 1c, d), an effect that was not seen in control astrocytes. Expression of mutant IDH1 caused a marked increase in hypermethylation at a large number of genes, although there was a small group of hypomethylated genes as well (Fig. 1e and Supplementary Fig. 1a and Supplementary Table 1). Surprisingly, expression of wild-type IDH1 also reshaped the methylome but in a manner that differed from effects due to expression of mutant IDH1 (Fig. 1f). Expression of wild-type IDH1 caused hypomethylation at specific loci, suggesting that both the production of 2-HG and the levels of α-ketoglutarate can affect the methylome. Unsupervised hierarchical clustering of the methylome data showed that the hypermethylated genes included both genes that underwent de novo methylation as well as genes that originally possessed low levels of methylation but subsequently acquired high levels of methylation (Fig. 1e). Control astrocytes did not undergo these methylome changes (Fig. 1c, d). Mutant IDH1-induced remodelling of the methylome was progressive and reproducible, and resulted in significant changes in gene expression (Fig. 1f and Supplementary Fig. 1a, Supplementary Tables 2 and 3).
We sought to define the methylation targets of mutant IDH in astrocytes. Of the 44,334 CpG sites that were differentially methylated in mutant IDH-expressing cells, 30,988 sites were hypermethylated (3,141 unique genes with promoter CpG island methylation changes; Supplementary Table 1). Transcriptional module mapping showed that the genes undergoing methylation changes were highly enriched for polycomb complex 2 (PRC2)-targeted loci (Supplementary Fig. 1b and Supplementary Table 4)12,21. These observations demonstrate that mutant IDH1 is sufficient to reshape the epigenome by altering the global methylation landscape.
Lower-grade gliomas (LGGs; World Health Organization grades II and III) and secondary glioblastomas are biologically distinct from primary or de novo glioblastomas22. Present knowledge of G-CIMP is based on the examination of primary glioblastomas in which IDH mutations are infrequent1,4,5. To determine the impact of IDH mutation on the methylation landscape in primary LGGs, we generated a high-resolution, genome-wide set of LGG methylome data from patients with complete clinical follow-up using the same Infinium 450K platform as described earlier (72 WHO grade II and III gliomas; Fig. 2 and Supplementary Table 5). We first performed consensus clustering (Fig. 2a and Supplementary Fig. 2a) and unsupervised hierarchical clustering (Fig. 2b and Supplementary Fig. 2b) to identify LGG subgroups. We identified two robust DNA methylation clusters, one encompassing tumours with markedly high methylation levels (cluster 2) and another without the hypermethylated loci (cluster 1). Cluster 2 tumours demonstrated a characteristic DNA methylation profile with high-coordinate cancer-specific methylation at a subset of loci, concordant with the G-CIMP phenotype defined in glioblastomas (Supplementary Fig. 2b and Supplementary Table 6)1. The composition of the G-CIMP group in these LGGs was confirmed by two independent clustering methods (K-means consensus and two-dimensional hierarchical clustering) (Fig. 2a, b). Probes defining CIMP in LGGs included those in CpG islands and shores (Supplementary Fig. 2c, d) and were enriched for PRC2-target genes (Supplementary Table 7). Global expression profiles showed that G-CIMP+ tumours possessed markedly different transcriptional profiles than G-CIMP− tumours (Supplementary Tables 8 and 9). EpiTYPER (Sequenom) mass spectrometry was used to validate the methylation status of loci in both the astrocyte model and in the tumours (Supplementary Fig. 2e–g)23.
Figure 2. Global epigenetic analysis of LGGs reveals dependence of G-CIMP on IDH mutation.
a, Identification of G-CIMP by K-means consensus clustering of LGG samples. Unsupervised clustering was performed with the most variant probes (9,711 probes, top 2%). Tumours are listed in the same order along the x and y axes. G-CIMP status is indicated by the black and white bars. Consensus index values range from 0 to 1, with 0 being dissimilar (white) and 1 being similar (red). K = 2 is identified by the Lorenz curve. b, Two-dimensional (2D) hierarchical clustering of the same probes as in a identified the same two clusters. Each row represents a tumour and each column represents a probe. CIMP and IDH mutation status are indicated by the colour code. The level of DNA methylation (β value) for each probe is represented by colour scale (red, methylated; blue, non-methylated). Only cancer-specific events were used27. c, Kaplan–Meier survival curve of Memorial Sloan-Kettering Cancer Center (MSKCC) patients (n = 72) with LGG (grade II and III). d, Receiver operating characteristic (ROC) curve comparing the sensitivity and specificity of G-CIMP status compared with MGMT methylation or MGMT expression status, in LGGs. Areas under the curve are noted in the inset. G-CIMP, MGMT methylation and MGMT expression were determined as described in Methods.
To determine the mutational status of IDH1and IDH2, we sequenced the entire coding sequence of the two genes in all the samples above (Fig. 2b). Ninety-eight per cent (49/50) of the G-CIMP+ tumours possessed either an IDH1 mutation or IDH2 mutation. Notably, none of the G-CIMP− tumours possessed mutant IDH (Supplementary Fig. 2h). These genomic data show that G-CIMP is highly dependent on the presence of IDH mutation and, in LGGs that are CIMP−, IDH mutations do not occur (0%). Currently, the methylation status of O-6-methylguanine DNA methyltransferase (MGMT) is a widely used molecular biomarker for glioblastoma prognosis and response to temozolomide24. In LGGs, G-CIMP associated with markedly better clinical endpoints (Fig. 2c and Supplementary Figs 3– 6, Supplementary Tables 10 and 11). Importantly, G-CIMP was significantly superior to MGMT methylation or MGMT messenger RNA expression as a predictor of survival (Fig. 2d).
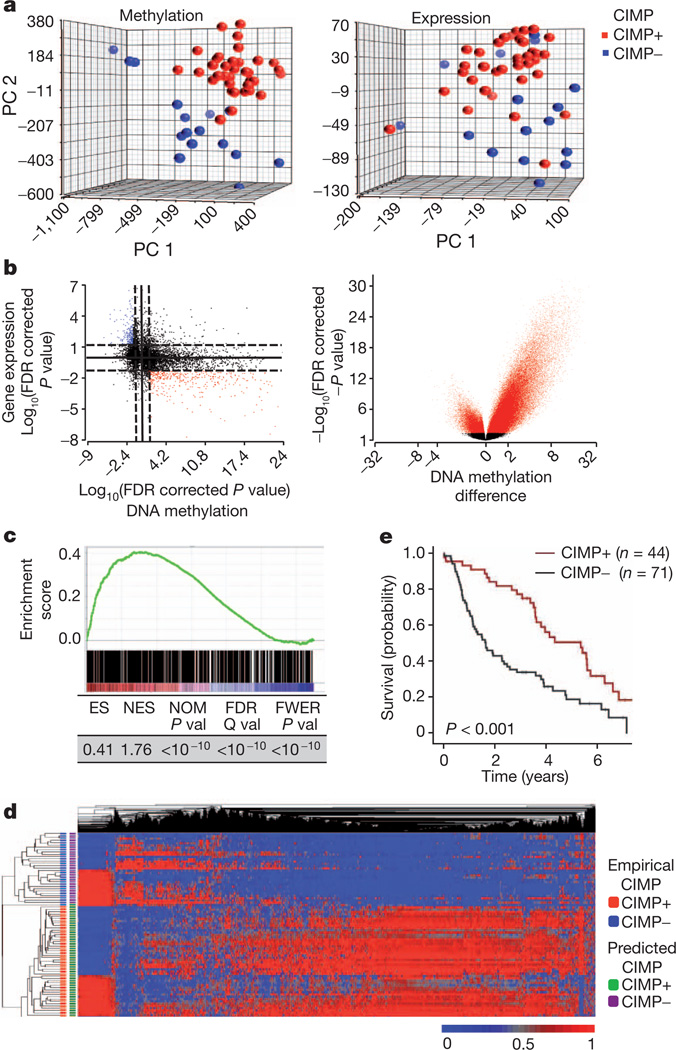
We next sought to define the nature of the methylome differences between IDH mutant and wild-type tumours and characterize the effects of these differences on the LGG transcriptome. Figure 3a shows a principal component analysis (PCA) of methylome and expression data from our tumours. PCA shows that G-CIMP+ and G-CIMP− LGGs methylome subgroups correlate with marked transcriptome differences (Fig. 3a). Of the 140,016 sites that were differentially methylated between IDH mutant and wild-type tumours, 121,660 were hypermethylated (Supplementary Table 6). There were 2,611 unique genes with alterations in promoter CpG islands represented in this group. Consistent with the results in Fig. 2b, a volcano plot showing differentially methylated genes between G-CIMP+ and G-CIMP− tumours was highly asymmetric (Fig. 3b). A starburst plot showing the relationship between DNA methylation and expression is shown in Fig. 3b. Integration of the normalized gene expression and DNA methylation gene sets identified 429 genes with both significant hypermethylation and downregulation and 176 genes that were hypomethylated and upregulated in G-CIMP+ LGGs (Supplementary Table 12). Among these genes are those known to be involved in glioma initiation and outcome, including CDKN2C and GAP43 (refs 25, 26).
Figure 3. IDH1 mutation directly generates the methylation patterns present in G-CIMP tumours.
a, The methylomes and transcriptomes of LGGs are distinct. PCA plot of LGG tumours for all methylation probes (left) and expression probes (right) (n = 52). PC, principal component. b, Starburst plot (left) for comparison of DNA methylation and gene expression. The log10 (FDR-corrected P value) is plotted for β value forDNAmethylation (x axis) and gene expression (y axis) for each gene. Black dotted line shows the FDR-adjusted P value of 0.05. Red points indicate downregulated and hypermethylated genes in G-CIMP+ LGGs versus G-CIMP− LGGs. Blue points show hypomethylated and upregulated genes. Volcano (right) plot of all CpG loci analysed for G-CIMP association. The β-value difference in DNA methylation between G-CIMP+ and G-CIMP− tumours is plotted along the x axis. The P value between G-CIMP+ and G-CIMP− tumours is plotted on the y axis (−log10 scale). Red indicates significantly different probes. c, Concordance between hypermethylated sites in mutant IDH1-expressing astrocytes and G-CIMP+ LGGs. GSEA shows significant enrichment between 730 hypermethylated unique CpG sites identified in IDH1 mutant astrocytes (ANOVA between passage 2 and 40) and those present in G-CIMP+ gliomas. GSEA correlation shown in colour scale. ES, enrichment score; FDR, false discovery rate; FEWR, familywise error rate; NES, normalized enrichment score; NOM, nominal P value. d, Differential methylation in IDH mutant astrocytes correctly classifies G-CIMP in the human LGGs. Two-dimensional-unsupervised hierarchical clustering of 81 human gliomas with top variant probes (n = 10,000) frommutant IDH1 astrocytes. Tumours are shown on the y axis, probes along the x axis. Methylation (β value) for each probe is represented with the colour scale. G-CIMP classification as determined by the astrocyte-derived data is denoted by the colour bars at the left. e, Kaplan–Meier survival curve of 115 patients with grade II or grade III gliomas in the Rembrandt Database grouped by CIMP status. P value calculated by log rank.
As a critical experiment to prove causality between IDH1 mutation and G-CIMP, we performed an in-depth comparison of methylation marks and gene expression alterations between human astrocytes expressing mutant IDH1 and the LGGs with endogenous IDH1 mutation. We first focused on the comparison of methylation marks and found that both sets of methylome alterations targeted similar loci. Gene set enrichment analysis (GSEA) of the mutant IDH1-induced methylation changes in the isogenic astrocyte system (Fig. 1) and the G-CIMP genes demonstrated very significant enrichment and concordance (Fig. 3c and Supplementary Table 13 and Supplementary Fig. 7). Importantly, the genes that were methylated after mutant IDH1 expression correctly classified LGG tumours into CIMP+ or CIMP− groups with very high accuracy (Fig. 3d and Supplementary Table 14). To confirm the impact of these alterations on glioma pathobiology, we used the transcriptomic footprint of mutant IDH to generate an expression signature (mutant IDH repression signature) composed of the most significantly methylated and downregulated genes in both the isogenic astrocyte system and the G-CIMP gene set (17 genes; Supplementary Table 15). As expected, this signature classified an independent LGG cohort (Rembrandt) into two distinct subgroups (Fig. 3e and Supplementary Figs 8–10 and Supplementary Table 16). Together, our findings show that introduction of mutant IDH reprograms the epigenome and generates the foundations of G-CIMP.
IDH mutation is highly enriched in the CIMP+, proneural subgroup of glioblastomas. Using data from The Cancer Genome Atlas (TCGA), we applied the mutant IDH repression signature as a classifier to the transcriptomes of all four subgroups of glioblastomas27. The signature segregated IDH mutant and wild-type proneural glioblastomas into two distinct subgroups associated with very different prognoses, but did not do so in other glioblastoma subgroups (Supplementary Fig. 11a, b). These data demonstrate that mutant IDH-induced epigenomic alterations have profound biological implications within the proneural class of glioblastomas that are specific for this subclass. Comparison of gene expression programs that occur in astrocytes expressing mutant IDH1 to those in LGG tumours that harbour the IDH mutation showed remarkable similarity (Fig. 4a and Supplementary Fig. 12). Moreover, introduction of mutant but not wild-type IDH1 into astrocytes resulted in the upregulation of nestin (and other genes associated with stem cell identity) at the time of DNA methylation increase and the adoption of a neurosphere/stem-like phenotype (Fig. 4b and Supplementary Fig. 13)28. These data suggest that mutant IDH1 functions by interfering with differentiation state.
Figure 4. Functional implications of IDH1-mutation-induced alterations in the glioma epigenome.
a, Concordance of transcriptional programs regulated by mutant IDH1 in astrocytes and G-CIMP in LGGs. P value for significance is shown along the x axis. Yellow lines indicate threshold of significance (P = 0.05). b, Mutant IDH1 results in the expression of markers of self-renewal and stem cell identity. Left, mutant IDH1 results in expression of nestin. P indicates passage number. Right, expression of mutant IDH1 promotes the adoption of a neurosphere phenotype. Astrocytes (passage 15) that express IDHR132H or IDH1 wild type were used in the neurosphere assay. Error bars indicate 1 s.d. **P < 0.01 (t-test). c, Alterations in histone marks in IDH1-mutant-expressing human astrocytes. Left, western blot results are shown using the indicated antibodies. Astrocytes are from passage 27. Right, ChIP of the indicated histone marks for representative hypermethylated genes. Error bars indicate 1 s.d. *P < 0.05. d, Mutant IDH1 inhibits the production of 5hmC in human astrocytes. Left, mutant IDH inhibits TET2-dependent 5hmC production in astrocytes. Parental astrocytes were infected with lentivirus directing the expression of TET2 catalytic domain and green fluorescent protein (GFP) ± mutant IDH1. FACS analyses are shown for 5hmC. Right, astrocytes expressing IDH R132H (passage 10) have less 5hmC than astrocytes that do not express the IDH mutant.
Our data show that IDH1 mutation is the mechanistic cause of G-CIMP. To gain further insight, we determined the effects of mutant IDH1 on histone alterations in our astrocyte system. Figure 4c (left) shows that expression of the IDH1 mutant increases levels of H3K9me2, H3K27me3 and H3K36me3, consistent with previous findings29. Chromatin immunoprecipitation (ChIP) experiments examining representative genes that undergo hypermethylation show H3K9 and H3K27 methylation are both enriched in cells expressing mutant IDH1 (Fig. 4c, right). As both of these marks can promote DNA methylation, alterations in histone marks may contribute to the accumulation of DNA methylation.
Next, we determined the effects the mutation had onTET2-dependent 5-hydroxymethylcytosine (5hmC) levels. We used a well-established assay9,29 and first confirmed that we were able to detect TET-dependent alterations in 5hmC (Supplementary Fig. 14). We found that expression of the IDH1 mutant in astrocytes resulted in a significant decrease in 5hmC (Fig. 4d, right). Expression of TET2 in the astrocytes produced 5hmC, which was inhibited by mutant but not wild-type IDH1 (Fig. 4d, left). Because TET-mediated production of 5hmC is a primary mode of DNA demethylation30, inhibition of this activity in the IDH1-mutant-expressing astrocytes may be a mechanistic basis for accumulation of DNA methylation, ultimately leading to a CIMP pattern.
IDH mutation and the CIMP phenotype are two very common features in cancer, the underlying mechanisms for which are obscure. The fundamental questions regarding these features are (1) how the IDH mutation contributes to oncogenesis, and (2) what the root cause of CIMP is. Our data address these important questions by demonstrating that IDH mutation is the cause of CIMP and leads to the CIMP phenotype by stably reshaping the epigenome. This remodelling involves modulating patterns of methylation on a genome-wide scale, changing transcriptional programs and altering the differentiation state. Our observations suggest that the activity of IDH may form the basis of an ‘epigenomic rheostat’, linking alterations in cellular metabolism to the epigenetic state. In summary, these data provide a mechanistic framework for how IDH mutation leads to oncogenesis and the molecular basis of CIMP in gliomas. We believe our observations have critical implications for the understanding of gliomas and the development of novel therapies for this disease.
METHODS
Cell culture
Immortalized human astrocytes were a gift from R. O. Pieper (University of California, San Francisco) and were prepared as previously described14. Cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) plus 10% fetal bovine serum (FBS; Invitrogen). Expression of IDH was accomplished by cloning wild-type or mutant IDH1 (R132H) into the vector pLNCX2. These constructs were used to construct retroviruses used for infection of target cells. The retroviral packaging cell line GP-293 was seeded in 10-cm-diameter dishes and (at 30–50% confluency) was transfected using Lipofectamine (Invitrogen) with pVSV-G (Clontech) and pLNCX2-IDH1 wild type or IDH1 R132H. Retroviral particles were collected, filtered through a 0.45-µm syringe filter and polybrene was added (8 µgml−1 final concentration) to infect the human astrocytes for 12 h. Stable transfectants were selected with G418 and pooled populations of G418-resistant cells expressing either wild-type IDH1 or IDH1 R132H were confirmed by western blot analysis with anti-IDH1 antibody (rabbit anti-IDH1; Cell Signaling). All experiments were performed in duplicate.
Tumours
All tumours (n = 81) were obtained following surgical resection at the MSKCC as part of routine clinical care, and snap frozen. Tumours were obtained in accordance with Institutional Review Board policies at the MSKCC. Each sample was examined histologically by a neuropathologist. Before analysis, tumours were sectioned and microdissected. Genomic DNA or RNA was extracted using the DNeasy kit (Qiagen) or Triazol (Invitrogen) as per the manufacturer’s instructions. Data from TCGA tumours (n = 173) are publically available27. For the LGG validation set, expression data sets of 115 patients with grade II and grade III gliomas were identified from the NCI Repository for Molecular Brain Neoplasia Data (Rembrandt; http://rembrandt.nci.nih.gov).
Sample preparation
DNA from wild-type IDH1, R132H IDH1 and parental astrocytes was extracted with the Puregene Cell and Tissue Kit (Qiagen) at various passages (passages 2, 5, 10, 15, 20, 25, 30, 40 and 50) and RNA was extracted with Trizol (Invitrogen) according to the manufacturer’s directions. All experiments with the astrocytes were performed in duplicate, each with two corresponding technical (microarray) replicates. Genomic DNA and RNA from human tumours were extracted from frozen primary tumours for the methylation and expression studies. Frozen samples were snap frozen in liquid nitrogen and stored at −80 °C. Each sample was examined histologically with haematoxylin-and-eosin-stained sections by a neuropathologist and representative sections were microdissected from the slides. Genomic DNA was extracted with the Qiagen DNeasy Blood and Tissue Kit using the manufacturer’s instructions. RNA was extracted with Qiagen RNeasy Lipid Tissue Mini Kit using the manufacturer’s instructions. Nucleic acid quality was determined with the Agilent 2100 Bioanalyzer.
Genomic analysis
Expression analysis of astrocytes and tumours was performed using the Affymetrix U133 2.0 microarray (Affymetrix). Genome-wide methylation analysis was performed using the Infinium HumanMethylation450 bead array (Illumina). Processing of the arrays was per themanufacturer’s protocol. Methylation data were extracted using GenomeStudio software (Illumina). Methylation values for each site are expressed as a β value, representing a continuous measurement from 0 (completely unmethylated) to 1 (completely methylated). This value is based on following calculation: β value = (signal intensity of methylation-detection probe)/(signal intensity of methylation-detection probe + signal intensity of non-methylation detection probe).
Data analysis
For methylation analysis, Illumina data were imported into Partek software. β Values were logit-transformed and adjusted for batch effects before analysis. Analysis of variance (ANOVA) with false discovery correction (FDR) was used to identify genes that were differentially methylated between the astrocytes expressing wild-type IDH1, mutant IDH1, and control astrocytes. Significant changes were defined as genes having an FDR-corrected P value < 0.05. In human tumours, unsupervised consensus clustering of the β values was performed with K-means clustering (Kmax = 5) with Euclidean distance and average linkage over 1,000 resampling iterations with random restart on the top 2% of the most variant probes (9,750 probes) using Gene Pattern v.2.031. This identified an optimal number of K = 2 groups. This was repeated using unsupervised hierarchical clustering using Pearson dissimilarity. The cluster of samples that exhibited a large degree of hypermethylation was identified as CIMP+, and the remaining group CIMP−. ANOVA with FDR correction was used to identify genes that were differentially methylated between the CIMP groups. Significant changes were defined as genes having an FDR-corrected P value < 0.05.
For gene expression analysis of astrocytes, Affymetrix CEL files were imported into the R statistical software (v.2.13.0; http://www.R-project.org). Normalization was performed with the AffyPLM package in BioConductor (v.2.4), using RMA background correction, quantile normalization, and the Tukey biweight summary method. Differential expression was detected using the limma package and P values were adjusted for multiple testing using the FDR approach. A probe set is considered differentially expressed if the FDR-adjusted P value < 0.05. For gene expression analysis of the human tumours, the Affymetrix data were imported into the Partek Genomics Suite (Partek) as Affymetrix CEL files. The data were RMA normalized and median-scaled for analysis. ANOVA followed by FDR was used to identify genes that were differentially expressed between the CIMP groups. To derive the 17-gene mutant IDH1 repression signature, we identified 605 unique genes that had either statistically significant hypermethylation at promoter-associated CpG islands and decreased gene expression, or had hypomethylation at promoter-associated CpG Islands and increased gene expression in CIMP+ versus CIMP− tumours. We identified common genes in a comparison of this gene set with that derived from mutant IDH1-expressing astrocytes versus wild type. Differential methylation in the cell lines was defined as an FDR-adjusted P value < 0.05, and differential expression was defined as a P value < 0.05 with concordant fold change of at least 1.5 fold.
The 17-gene expression signature was used to predict CIMP in the Rembrandt data set. Unsupervised hierarchical clustering using Pearson dissimilarity identified two unique clusters that were categorized as ‘predicted CIMP+’ and ‘predicted CIMP−’. The 17-gene expression signature was also used to identify subgroups from the TCGA GBM data set of 173 patients27. Unsupervised consensus clustering using Pearson dissimilarity was performed on each of the subclasses identified by the TCGA to identify clusters32. Rembrandt data sets were obtained at http://caintegrator-info.nci.nih.gov/rembrandt.
Functional analysis of gene lists was performed using the PANTHER database and categories with adjusted P values (Benjamini–Hochberg) < 0.05 were considered as significantly over-represented in our gene lists33. Concepts module mapping was performed as follows. The hypermethylation signature identified from our analysis of differentially methylated genes in IDH1 mutants compared to IDH1 wild-type was imported into Oncomine (http://www.oncomine.org) to identify associations with molecular concepts signatures derived from independent cancer profiling studies. Statistically significant concordances of our methylation gene signature with the pre-defined concepts were identified and Q value was calculated as previously described34.
Methylation data of parental, wild-type IDH1-expressing astrocytes, and mutant IDH1-expressing astrocytes were clustered using self-organizing maps and visualized with the Gene Expression Dynamics Inspector (GEDI; v.2.1). For GEDI analysis, methylation data were normalized as a group across all passages and genotypes. Further hierarchical clustering (average-linkage) of GEDI map centroids was performed in R using the hclust library in the stats package. GSEA was performed using GSEA software v.2.0 and MSigDB database v.2.5. We assessed the significance of the curated gene sets (MSigDB collection c2) with the following parameters: number of permutations = 1,000 and permutation_type = phenotype, with an FDR Q-value cut-off of 5% (ref. 35).
G-CIMP was compared to MGMT methylation and MGMT expression in 52 LGG samples in the MSKCC cohort. We identified the Illumina 450K methylation probe ID (cg12981137) that corresponded to the MGMT MSP primer sequence as identified previously36 and the Affymetrix probe ID (204880_at) that corresponds to MGMT expression.
Clinical and pathological characteristics between cohorts were compared using the χ2 test. Overall survival was calculated from the date of surgery to death from any cause. Patients were censored at the time they were last known to be alive. Overall survival was assessed using the Kaplan–Meier method and the log-rank test was used for comparison between groups. Multivariate analysis was performed using a Cox proportional hazards model to assess the independent effect of prognostic variables on outcome, and using binary logistic regression to predict the probability of occurrence of CIMP+. An ROC curve was generated to graph the sensitivity and specificity of CIMP, MGMT methylation, and MGMT expression to predict survival ≥3 years. MGMT methylation and MGMT expression was considered continuous variables, and CIMP a categorical variable (defined by unsupervised hierarchical analysis as described above). Patients that were alive and had less than 3 years of follow-up were excluded from this analysis. Data was analysed using SPSS software (IBM SPSS statistics version 19.0).
Quantitative DNA methylation analysis using mass spectrometry
DNA methylation analysis was performed using the EpiTYPER system (Sequenom). The EpiTYPER assay is a tool for the detection and quantitative analysis of DNA methylation using base-specific cleavage of bisulphite-treated DNA and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS)23. For primer sequences, target chromosomal sequence, and EpiTYPER-specific tags, see Supplementary Table 17. SpectroCHIPs were analysed using a Bruker Biflex III MALDI-TOF mass spectrometer (SpectroREADER, Sequenom). Results were analysed using the EpiTYPER Analyzer software, and manually inspected for spectra quality and peak quantification. CIMP positivity was defined as a mean methylated allelic frequency of >50% or a twofold increase over normal breast tissue and the CIMP– state.
PCR amplification and sequencing
Exonic regions for the IDH1 and IDH2 genes (NCBI Human Genome Build 36.1) were broken into amplicons of 500 bp or less, and specific primers were designed using Primer3. Standard M13 tails were added to the primers to facilitate Sanger sequencing. PCR reactions were carried out in 384-well plates in a Duncan DT-24 water bath thermal cycler with 10 ng of whole-genome amplified DNA (REPLI-g Midi, Qiagen) as a template, using a touchdown PCR protocol with KAPA Fast HotStart (Kapa Biosystems). The touchdown PCR method consisted of: 1 cycle of 95 °C for 5 min; 3 cycles of 95 °C for 30 s, 64 °C for 15 s, 72 °C for 30 s; 3 cycles of 95 °C for 30 s, 62 °C for 15 s, 72 °C for 30 s; 3 cycles of 95 °C for 30 s, 60 °C for 15 s, 72 °C for 30 s; 37 cycles of 95 °C for 30 s, 58 °C for 15 s, 72 °C for 30 s; 1 cycle of 70 °C for 5 min. Templates were purified using AMPure (Agencourt Biosciences). The purified PCR reactions were split into two and sequenced bidirectionally with M13 forward and reverse primer and the Big Dye Terminator Kit v.3.1 (Applied Biosystems) at Agencourt Biosciences. Dye terminators were removed using the CleanSEQ kit (Agencourt Biosciences), and sequence reactions were run on ABI PRISM 3730xl sequencing apparatus (Applied Biosystems). Sanger sequencing of IDH1 and IDH2 produced an average coverage of 96.1% of coding sequence nucleotides across all samples.
Mutation detection
Passing reads were assembled against reference sequences, containing all coding exons including 5 kb upstream and downstream of the gene, using command line Consed 16.037. Assemblies were passed on to Polyphred 6.02b38, which generated a list of putative candidate mutations, and to Polyscan 3.039, which generated a second list of putative mutations. The lists were merged together into a combined report, and the putative mutation calls were normalized to ‘+’ genomic coordinates and annotated using the Genomic Mutation Consequence Calculator40. The resulting list of annotated putative mutations was loaded into a Postgres database along with select assembly details for each mutation call (assembly position, coverage, and methods supporting mutation call). To reduce the number of false positives generated by the mutation detection software packages, only point mutations that were supported by at least one bi-directional read pair and at least one sample mutation called by Polyphred were considered, and only the putative mutations that were annotated as having non-synonymous coding effects, occurred within 1 bp of an exon boundary, or had a conservation score > 0.699 were included in the final candidate list. Indels were manually reviewed and included in the candidate list if found to hit an exon. All putative mutations were confirmed by a second PCR and sequencing reaction, in parallel with amplification and sequencing of matched normal tissue DNA.
ChIP
Cells were fixed with 1% formaldehyde for 10 min at room temperature (21 °C) and formaldehyde was inactivated by the addition of 125 mM glycine. ChIP assays were performed using a protocol recommended by the manufacturer of a commercially available ChIP assay kit (17-371, Millipore). Chromatin extracts were immunoprecipitated using anti-H3K9me3 (Ab8898, Abcam) or anti-H3K27me3 (07-449, Millipore) antibodies. After washing, ChIPed DNA was eluted from the beads and analysed on an Eppendorf Realplex using SYBR Green (Applied Biosystems). Relative occupancy values were calculated by determining ratios of the amount of immunoprecipitated DNA to that of the input sample (2% of total).
Flow cytometry and 5hmC assay
HEK 293T cells were transiently transfected with Flag–TET2 inpCMV6-ENTRY vector with Lipofectamine 2000 (GIBCO). For two-colour flow cytometry, 106 cells were washed with ice-cold PBS, permeabilized and fixed using BD Cytoperm/Cytofix solution (BD, PharMingen), and incubated with anti-Flag (1:200, Sigma) and anti-5hmC (1:400, Active Motif #39770) antibodies for 30 min at room temperature. Cells were washed with PBS and incubated with secondary antibodies conjugated with Alexa Fluor 488 or Cy5 (Invitrogen) for 30 min in the dark. For single-colour flow cytometry, parental and IDH1 mutant astrocytes were stained using anti-5hmC (1:400) followed by Alexa Fluor 488 secondary antibody. Cells were washed in PBS and analysed using the FACScan flow cytometer (Becton Dickinson). FACS data were analysed using FLowJo Software (TreeStar).
Neurosphere assay
IDH1 (R123H)-expressing astrocytes and parental controls were grown in media permissive of neural stem cell growth as previously described41,42. Briefly, immortalized human astrocyte (IHA) cells stably expressing wild-type or R132H mutant IDH1 at passage 15 were seeded in 6-well plates at 200,000 cells per well. The next day, proliferation medium (DMEM plus 10% FCS) was replaced with neural stem cell medium made from serum-free DMEM supplemented with B27 and N2 supplements (all from Invitrogen), bFGF, EGF and PDGFAA (all at 20 ngml−1, all from PeproTech). Medium was replaced every 2–3 days. Neurospheres were quantified using microscopy. Experiments were performed in triplicate.
Supplementary Material
Acknowledgements
We thank A. Kayserian, K. Huberman, I. Dolgalev and S. Thomas for technical expertise. We thank C. Sawyers and E. Holland for helpful discussions. This grant was supported in part by the National Institutes of Health (R01CA154767-01) (T.A.C.), the National Cancer Institute (U54-CA143798) (I.K.M.), an Advanced Clinical Research Award in Glioma from the American Society of Clinical Oncology (I.K.M.), the Doris Duke Charitable Fund (I.K.M., T.A.C.), a National Brain Tumor Society Systems Biology Research Grant (I.K.M.), the MSKCC Brain Tumor Center (T.A.C.), the Louis Gerstner Foundation (T.A.C.), the STARR Cancer Consortium (T.A.C.) and the Sontag Foundation (T.A.C., I.K.M.).
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions T.A.C., S.T., A.G. and I.K.M. designed the experiments. S.T., A.G., F.F., D.R., A.H., L.A.W., C.C., E.Y., C.L., P.S.W., A.V., J.T.H., A.W.M.F. and L.G.T.M. performed the experiments. S.T., J.T.H., A.G., F.F., A.K., A.H., E.Y., A.V., P.S.W., C.B.T., T.A.C. and I.K.M. analysed the data. D.R., O.G., R.L. and I.K.M. contributed new reagents. T.A.C., S.T., I.K.M. and A.G. wrote the paper.
Author Information Data sets have been deposited in the Gene Expression Omnibus under accession number GSE30339.
The authors declare competing financial interests: details accompany the full-text HTML version of the paper at www.nature.com/nature.
References
- 1.Noushmehr H, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17:510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fang F, et al. Breast cancer methylomes establish an epigenomic foundation for metastasis. Sci. Transl. Med. 2011;3:75ra25. doi: 10.1126/scitranslmed.3001875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Toyota M, et al. CpG island methylator phenotype in colorectal cancer. Proc. Natl Acad. Sci. USA. 1999;96:8681–8686. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yan H, et al. IDH1 and IDH2 mutations in gliomas. N. Engl. J. Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parsons DW, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sjoblom T, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 7.Mardis ER, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N. Engl. J. Med. 2009;361:1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dang L, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Figueroa ME, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ward PS, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting α-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17:225–234. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ohm JE, et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nature Genet. 2007;39:237–242. doi: 10.1038/ng1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng YW, et al. CpG island methylator phenotype associates with low-degree chromosomal abnormalities in colorectal cancer. Clin. Cancer Res. 2008;14:6005–6013. doi: 10.1158/1078-0432.CCR-08-0216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sonoda Y, et al. Formation of intracranial tumors by genetically modified human astrocytes defines four pathways critical in the development of human anaplastic astrocytoma. Cancer Res. 2001;61:4956–4960. [PubMed] [Google Scholar]
- 15.Vivanco I, et al. Identification of the JNK signaling pathway as a functional target of the tumor suppressor PTEN. Cancer Cell. 2007;11:555–569. doi: 10.1016/j.ccr.2007.04.021. [DOI] [PubMed] [Google Scholar]
- 16.Sonoda Y, et al. Akt pathway activation converts anaplastic astrocytoma to glioblastoma multiforme in a human astrocyte model of glioma. Cancer Res. 2001;61:6674–6678. [PubMed] [Google Scholar]
- 17.Pieper RO. Defined human cellular systems in the study of glioma development. Front. Biosci. 2003;8:s19–s27. doi: 10.2741/983. [DOI] [PubMed] [Google Scholar]
- 18.Bibikova M, et al. High density DNA methylation array with single CpG site resolution. Genomics. 2011;98:288–295. doi: 10.1016/j.ygeno.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 19.Sandoval J, et al. Validation of a DNA methylation microarray for 450,000 CpG sites in the human genome. Epigenetics. 2011;6:692–702. doi: 10.4161/epi.6.6.16196. [DOI] [PubMed] [Google Scholar]
- 20.Mutskov V, Felsenfeld G. Silencing of transgene transcription precedes methylation of promoter DNA and histone H3 lysine 9. EMBO J. 2004;23:138–149. doi: 10.1038/sj.emboj.7600013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bracken AP, et al. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 2006;20:1123–1136. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Furnari FB, et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21:2683–2710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- 23.Docherty SJ, et al. Bisulfite-based epityping on pooled genomic DNA provides an accurate estimate of average group DNA methylation. Epigenetics Chromatin. 2009;2:3. doi: 10.1186/1756-8935-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hegi ME, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N. Engl. J. Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 25.Solomon DA, et al. Identification of p18INK4c as a tumor suppressor gene in glioblastoma multiforme. Cancer Res. 2008;68:2564–2569. doi: 10.1158/0008-5472.CAN-07-6388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Etcheverry A, et al. DNA methylation in glioblastoma: impact on gene expression and clinical outcome. BMC Genomics. 2010;11:701. doi: 10.1186/1471-2164-11-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.The Cancer Genome Atlas Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lendahl U, Zimmerman LB, McKay RD. CNS stem cells express a new class of intermediate filament protein. Cell. 1990;60:585–595. doi: 10.1016/0092-8674(90)90662-x. [DOI] [PubMed] [Google Scholar]
- 29.Xu W, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He YF, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- 31.Reich M, et al. GenePattern 2.0. Nature Genet. 2006;38:500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 32.Verhaak RG, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thomas PD, et al. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003;13:2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rhodes DR, et al. Molecular concepts analysis links tumors, pathways, mechanisms, and drugs. Neoplasia. 2007;9:443–454. doi: 10.1593/neo.07292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vlassenbroeck I, et al. Validation of real-time methylation-specific PCR to determine O6-methylguanine-DNA methyltransferase gene promoter methylation in glioma. J. Mol. Diagn. 2008;10:332–337. doi: 10.2353/jmoldx.2008.070169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gordon D, Abajian C, Green P. Consed: a graphical tool for sequence finishing. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- 38.Nickerson DA, Tobe VO, Taylor SL. PolyPhred: automating the detection and genotyping of single nucleotide substitutions using fluorescence-based resequencing. Nucleic Acids Res. 1997;25:2745–2751. doi: 10.1093/nar/25.14.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen K, et al. PolyScan: an automatic indel and SNP detection approach to the analysis of human resequencing data. Genome Res. 2007;17:659–666. doi: 10.1101/gr.6151507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Major JE. Genomic mutation consequence calculator. Bioinformatics. 2007;23:3091–3092. doi: 10.1093/bioinformatics/btm339. [DOI] [PubMed] [Google Scholar]
- 41.Galli R, et al. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res. 2004;64:7011–7021. doi: 10.1158/0008-5472.CAN-04-1364. [DOI] [PubMed] [Google Scholar]
- 42.De Filippis L, et al. A novel, immortal, and multipotent human neural stem cell line generating functional neurons and oligodendrocytes. Stem Cells. 2007;25:2312–2321. doi: 10.1634/stemcells.2007-0040. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.