Summary
How does a common RNA polymerase II apparatus generate a complex pattern of transcripts in response to many gene-specific transcription factors and in accordance with cell’s state? Cai et al. (2012) reveal that the process involves coordinated conformational changes in Pol II and Mediator.
In this issue of Structure, Cai et al. describe a 16 Å cryo-electron microscopy (cryo-EM) structure of the yeast pre-initiation complex (PIC) in a minimal functional form (mPIC) consisting of RNA polymerase II (Pol II), three general transcription factors (GTFs), TBP, TFIIB and TFIIF, a 53-bp DNA core promoter, and the Head module of the Mediator complex. Strikingly, the authors observe large-scale conformational changes involving structural domains of both polymerase and the Mediator Head module. These structural changes appear to be synergistic, or at least coordinated, among the domains involved.
The dodecameric Pol II is the central enzyme that catalyzes transcription of pre-mRNAs and noncoding RNAs, and it does it by assembling with five GTFs (TBP, TFIIB, TFIIF, TFIIE and TFIIH) onto core promoter DNAs to form a Pol II basal apparatus. Basal activity of the Pol II apparatus is tightly controlled, and this regulation acts as the end point of signaling pathways. Regulated transcription generates patterns of transcripts (transcriptome) that are each characteristic of the cell-cycle stage and different environmental context. Extensive past work has revealed extreme complexity in the mechanism underpinning eukaryotic regulation of transcription. Even the first step, from the binding of DNA sequence(s) by a signal-modulated transcription factor to initiating transcription from the promoter, is not direct as it is in bacterial systems. Rather, it involves yet another complex called Mediator (Kornberg, 2005) that has a bewildering composition of protein subunits (25 in yeast), many of which show intimate relationship to transcription. Mediator fulfills the task of regulating transcription in a gene- and stimulus-specific way by conveying signals from transcription factors bound at distal regulatory cis elements to the Pol II basal apparatus located at the core promoter. Mediator functions via a number of mechanisms: it interacts with Pol II to form the Pol II holoenzyme; it facilitates recruitment of Pol II and additional factors and assembly of the PIC; it may enhance the release of early elongation complex from promoter proximal regions; it participates in the re-initiation from pre-assembled promoter scaffold, and it can counter the actions of negative regulators such as Gdown1 and NELF. Thus, Mediator provides a physical interface that contacts the Pol II apparatus on one side, and hundreds of eukaryotic gene-specific transcription factors on the other. The question then arises: what is the mechanism that enables Mediator to live up to its name and mediate communication between one common receiver and the diverse signal couriers?
The answer is emerging from structural analyses of various Mediator complexes. Studies in both yeast and human systems have established a conserved Mediator architecture comprising Head, Middle, Tail and CKD8 modules. Further results have converged on “malleability” as the most characteristic property of Mediator’s structure. For instance, human Mediator undergoes specific large-scale conformational shifts in response to binding of different types of transcription activators to its Tail module (Taatjes, 2010). Cai et al. have previously shown that both Head and Tail modules in yeast Mediator adopt a range of conformations in solution. The Middle module of yeast Mediator ‘morphs’ from the more compact unbound form into an arch that grasps polymerase in the holoenzyme complex (Cai et al., 2009).
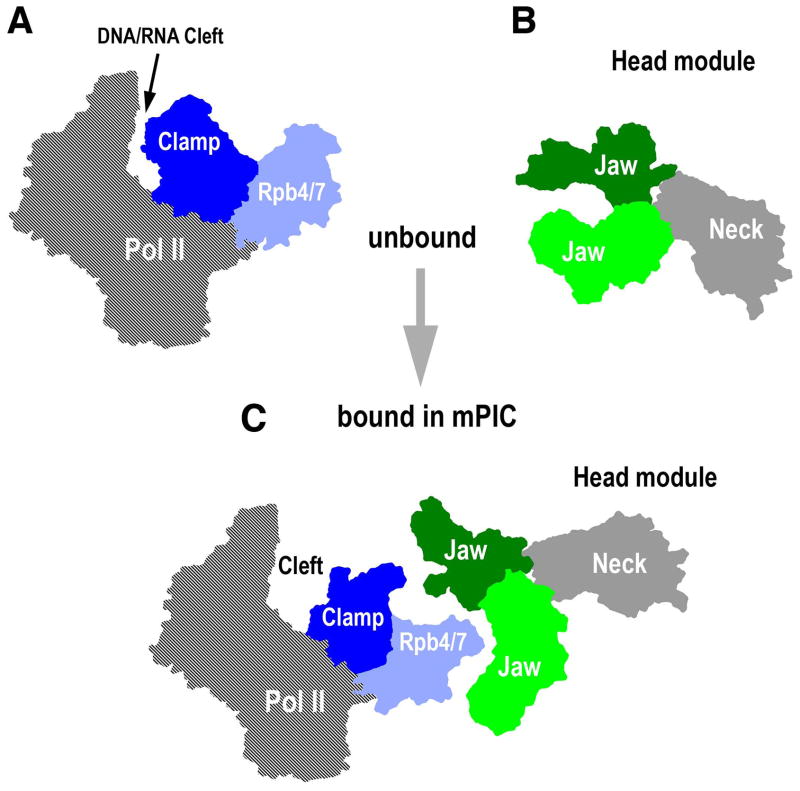
The same group now presents further EM results that reinforce the idea of structural malleability in Mediator. They show that the Mediator Head-Pol II interaction within the mPIC leads to concerted large-scale structural rearrangements in Mediator and Pol II. In mPIC, the Head module jaws embrace the Rpb4/Rpb7 subcomplex of Pol II, while the rest of the Head structure extends toward the back, Rpb3 side of Pol II. In the resulting structure, the Clamp domain and Rpb4/7 of Pol II are repositioned, with an outward rotation of the Clamp resulting in a DNA/RNA cleft that is considerably more open than in any of the previous Pol II structures (Fig. 1). In this complementary union of the Head and Pol II, all three domains of the Head module also undergo large rearrangements from their conformations in the isolated Head (Imasaki et al., 2011). As such, the Pol II and Head structures seem to undergo ‘mutual remodeling’ as they form a stable complex.
Fig. 1.
Mutual remodeling of Pol II and the Mediator Head in the mPIC. (A) Architecture of unbound Pol II as found in its crystal structure. The relevant domains are marked. (B) Domain arrangement of unbound Head module in its crystal structure (PDB ID: 3RJ1). (C) The rearranged domains of Pol II and the Head module when they interact in the mPIC.
Motions of the Clamp were originally discerned in the initial Pol II crystallographic work in which the Clamp was predicted to swivel around its hinge-like base (Fu et al., 1999). That conformational change was understood as a crucial event that could control DNA/RNA engagement during various stages of transcription (Gnatt, 2002), with closed Clamp refractory to DNA and RNA binding, and half-open Clamp accommodating to DNA/RNA hybrid binding. Now, Cai et al. (2012) present activity data demonstrating enhanced basal transcription in the presence of the Head protein, bolstering Clamp opening as a potential mechanism for regulating DNA engagement. Their results lend support for a direct role of the Clamp in promoter DNA engagement and suggest a mechanistic basis for the long-established stimulation of basal transcription by Mediator. Furthermore, their mPIC EM map includes density arising from TFIIF which was biochemically incorporated during mPIC assembly, and both TFIIB and promoter DNA, allowing a model building for the PIC. This work helps decipher Mediator’s enigmatic mode of operation by revealing mutual structural remodeling between Mediator and Pol II in the context of PIC, and provides direct evidence in support of the proposal that contacts between Mediator and Pol II may promote conformational changes in the polymerase that facilitate promoter engagement (Conaway and Conaway, 2009).
This detection of coordinated rearrangement between the jaws and neck domains of the Head module invites speculation that conformational rearrangements may be correlated amongst all Mediator modules. It has emerged from studies of the human Mediator (Taatjes, 2010), that a binding event in the Tail may initiate a ‘wave’ of remodeling that propagates through the Middle module to the Head, reaching Pol II basal machinery and changing its promoter engagement potential, and/or affinities toward cofactors. Such an allosteric mechanism may also exist in the yeast Mediator, which would be consistent with the observation that yeast Tail subunits, which are main binding targets of activators, influence the PIC assembly process that is physically facilitated by the Head module. Conceivably, these allosteric shifts would be mediated by the intrinsically disordered peptide segments that are abundantly encoded within many of the Mediator subunits (Toth-Petroczy et al., 2008). Conformational signals would be ultimately parsed through triggering points on Pol II such as its Rpb4/7 and Clamp domains.
Do the conformations that Pol II and the Mediator Head adopt in the mPIC complex exist freely in solution in agreement with “conformation selection” theory (Boehr et al., 2009)? This theory postulates the pre-existence of ensembles of different conformations for a given protein and a redistribution of the ensembles upon binding. Free Pol II displays at least three Clamp conformations: closed on a lipid layer, open in the uncomplexed Pol II (Fu et al., 1999), and half-open in Pol II-DNA/RNA ternary complex (Gnatt, 2002). The open and half-open Clamp conformations have also been observed for human Pol II (Kostek et al., 2006). The Head module displays a number of different conformations in solution as well (Cai et al., 2010). These observations suggest pre-existence of heterogeneous conformations in the free Pol II and Head populations, which supports the idea of conformational selection mechanism, as opposed to induced-fit that would govern changes in static Pol II and Head conformations. However, since the new conformations, the wide-open Pol II Clamp and rotated Head jaws and neck, have not been seen in their free forms yet, it is premature to rule either one of the mechanisms out. Interestingly, the cryo-EM map obtained by Cai et al. reveals that other Pol II domains (e.g. Rpb8, Rpb1-Foot) might also have shifted as a result of interacting with the Mediator Head, and their analysis suggests that mPIC might adopt a number of slightly different conformations in solution.
Overall, it appears that Pol II-Mediator complex possesses conformational variability that enables it to respond to a large array of inputs from gene-specific activators and repressors. With the binding of different activators leading to the different Mediator conformations (Taatjes, 2010), it is not difficult to envisage an allosteric propagation of conformational changes throughout the entire complex to yield a correspondingly remodeled Pol II structure (Tsai and Nussinov, 2011), potentiated for transcriptional activities on genes targeted by the activators.
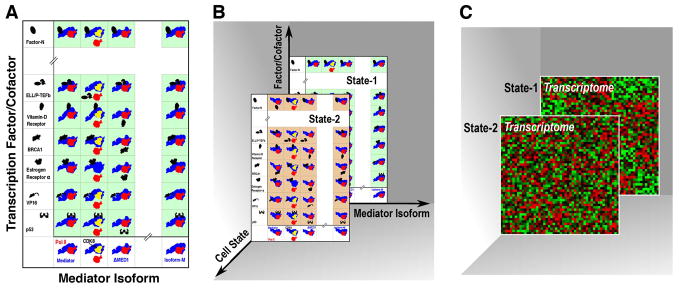
To help organize these structural scenarios, we suggest a two-dimensional array of discrete Pol II holoenzyme structures with the Mediator isoforms as one variable (horizontal axis) and the conditions according to the binding with transcription factors/cofactors as the second variable (vertical axis). Each array position would correspond to a specific holoenzyme conformation induced (or selected) as Mediator interacts with a transcription factor under a given cellular condition (Fig. 2A). Current knowledge suggests three holoenzyme isoforms: bound with the CDK8 kinase module; lacking the Mediator subunit Med1 (indicated in Fig. 2A–B); and associated with Med26 which helps the recruitment of elongation factors. The vertical dimension would be more densely populated due to the sheer number of activators, repressors and cofactors in the cell. Of note, a particular set of activators may preferentially target one form of the holoenzyme over another (Taatjes and Tjian, 2004), adding to the structural diversity of the conformational array (Fig. 2A). It is not difficult to see that a change in cellular context would lead to a change in posttranslational modifications, stability, concentration and/or subcellular localization of the entire collection of transcription factors and cofactors, which would affect their affinities for DNA recognition sites and/or Mediator. As interactions with transcription factors change, the many holoenzyme conformations (Fig. 2B, green array) would correspondingly vary, resulting in a changed conformational array (Fig. 2B, brown array). As such, cellular states would contribute to a third dimension to produce a 3-D grid of discrete holoenzyme conformations, with closely related states displaying more subtle conformational differences. Conceivably, each cell state-specific array of Pol II holoenzyme conformations (Fig. 2B, State-1 or -2) would contribute to the specific transcriptome of that state (Fig. 2C, State-1 or -2). As a challenge to structural biology methods, it remains to be seen how many positions of the holoenzyme conformation grid can be experimentally sampled.
Fig. 2.
A conformational grid for the Pol II holoenzyme. (A) Variations in both the subunit composition of Mediator (horizontal dimension) and the binding of transcription factor/cofactor (vertical) give rise to an array of discrete holoenzyme conformations, under a given cell state (State-1, green array). Pol II, red; Mediator, dark blue. The CDK8 module (yellow) is depicted following the S. pombe complex. (B) In the third dimension, a change in cell state leads to modifications of various transcription factors and cofactors, resulting in a different array of conformations (State-2, brown array). (C) Transcriptomes (represented as gene-chip patterns) of State-1 and State-2 are generated corresponding to the cell state-specific conformation arrays, the green and brown array in B, respectively.
Acknowledgments
We thank Vaughn Jackson (Medical College of Wisconsin) for comments. The authors acknowledge research support from the NIH (GM100997 to JF).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Boehr DD, Nussinov R, Wright PE. Nat Chem Biol. 2009;5:789–796. doi: 10.1038/nchembio.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai G, Imasaki T, Takagi Y, Asturias FJ. Structure. 2009;17:559–567. doi: 10.1016/j.str.2009.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai G, Imasaki T, Yamada K, Cardelli F, Takagi Y, Asturias FJ. Nat Struct Mol Biol. 2010;17:273–279. doi: 10.1038/nsmb.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conaway RC, Conaway JW. Structure. 2009;17:485–486. doi: 10.1016/j.str.2009.03.003. [DOI] [PubMed] [Google Scholar]
- Fu J, Gnatt AL, Bushnell DA, Jensen GJ, Thompson NE, Burgess RR, David PR, Kornberg RD. Cell. 1999;98:799–810. doi: 10.1016/s0092-8674(00)81514-7. [DOI] [PubMed] [Google Scholar]
- Gnatt A. Biochim Biophys Acta. 2002;1577:175–190. doi: 10.1016/s0167-4781(02)00451-7. [DOI] [PubMed] [Google Scholar]
- Imasaki T, Calero G, Cai G, Tsai KL, Yamada K, Cardelli F, Erdjument-Bromage H, Tempst P, Berger I, Kornberg GL, Asturias FJ, Kornberg RD, Takagi Y. Nature. 2011;475:240–243. doi: 10.1038/nature10162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg RD. Trends Biochem Sci. 2005;30:235–239. doi: 10.1016/j.tibs.2005.03.011. [DOI] [PubMed] [Google Scholar]
- Kostek SA, Grob P, De Carlo S, Lipscomb JS, Garczarek F, Nogales E. Structure. 2006;14:1691–1700. doi: 10.1016/j.str.2006.09.011. [DOI] [PubMed] [Google Scholar]
- Taatjes DJ. Trends Biochem Sci. 2010;35:315–322. doi: 10.1016/j.tibs.2010.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth-Petroczy A, Oldfield CJ, Simon I, Takagi Y, Dunker AK, Uversky VN, Fuxreiter M. PLoS Comput Biol. 2008;4:e1000243. doi: 10.1371/journal.pcbi.1000243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai CJ, Nussinov R. Biochem J. 2011;439:15–25. doi: 10.1042/BJ20110972. [DOI] [PMC free article] [PubMed] [Google Scholar]