Summary
More than any other methodology, transmission electron microscopy (TEM) has contributed to our understanding of the architecture and organization of cells. With current detection limits approaching atomic resolution, it will ultimately become possible to ultrastructurally image intracellular macromolecular assemblies in situ. Presently, however, methods to unambiguously identify proteins within the crowded environment of the cell’s interior are lagging behind. We describe a novel approach, metal-tagging TEM (METTEM) that allows detection of intracellular proteins in mammalian cells with high specificity, exceptional sensitivity and at molecular scale resolution. In live cells treated with gold salts, proteins bearing a small metal-binding tag will form 1-nm gold nanoclusters, readily detectable in electron micrographs. The applicability and strength of METTEM is demonstrated by a study of Rubella virus replicase and capsid proteins, which revealed virus-induced cell structures not seen before.
INTRODUCTION
As life of the cell depends on compartmentalized, spatially-coordinated activities and interactions of macromolecular assemblies, ultrastructural imaging of these biomolecules in their natural context should significantly further our understanding of cell biology. Technically this is within reach, as current detection limits of transmission electron microscopy (TEM) are approaching atomic scale resolution. Electron tomography methods, in combination with technical advances in sample preparation and preservation, already permit 3D reconstruction of intracellular structures to resolutions of 3–8 nm (Robinson et al., 2007; Volkmann, 2010). Unfortunately, however, the development of methods to unambiguously identify proteins in thin sections and in (cryo) electron tomograms has not kept pace. Ultrastructural detection of proteins still relies largely on immunogold labeling assays with specific antibodies and colloidal gold conjugates. The sensitivity and resolution of immuno-electron microscopy (IEM) are limited, however (see Supplementary Introduction), and there is a need for more robust techniques to directly and quantitatively identify individual proteins in the crowded environment of the cell’s interior. One solution would be to provide target proteins with a genetically clonable tag, an approach that could open new possibilities for TEM, potentially as far-reaching as the applications of fluorescent protein tagging in light microscopy. Attempts in this direction have as yet met with modest success. For example, proteins have been provided with fluorescent tags to generate electron-dense deposits through photoconversion that are visible by TEM (Gaietta, et al., 2002; Grabenbauer et al., 2005; Shu et al., 2011), but the resulting signals are diffuse, lacking the resolution of particulate probes (Kireev et al., 2008), and may be too weak to visualize low-abundance proteins (Wang et al., 2011). Bacterial ferritin, or rather its monomer Ftna1, has been used as a clonable tag for cryo-EM in bacteria (Wang et al., 2011). Ftna1 forms 12 nm cages, comprised of 24 monomers and containing up to 4200 atoms of iron, that are discernable by TEM as ~7 nm electron-dense particles. The sheer size of ferritin, 18-times that of GFP, poses a potential problem, however, as it might affect trafficking and biological function of target proteins. Moreover, target protein mislocation and clustering may result as a consequence of Ftna1-mediated oligomerization.
Building on seminal work by Mercogliano and DeRosier, we recently explored the possibilities of using the metal-binding protein metallothionein (MT) as a tag (Diestra et al., 2009: Mercogliano and DeRosier, 2007). MT, a metal-ion chelator, controls the storage and interchange of biologically essential metals such as zinc and copper (Chan et al., 2002) in eukaryotic cells. Isoform 1 of murine MT, a small 61-aa protein, comprises 20 cysteine residues that bind metal atoms very efficiently. When fused with a protein of interest and treated in vitro with gold salts, a single MT tag will build an electron-dense gold-thiolate cluster of ~1 nm diameter (Mercogliano and DeRosier, 2006) (Figure 1A). We provided proof of principle by demonstrating that metal-tagging transmission EM (METTEM) can be used to detect intracellular proteins in bacteria (Diestra et al., 2009). It remained uncertain, however, whether the method could be adapted for mainstream cell biology, i.e. made to work in mammalian cells. In contrast to bacteria, these cells cannot be adapted to grow in the presence of heavy metals and are intolerant to prolonged exposure to high AuCl concentrations (Diestra et al., 2009). Also, it was not known whether heavy metals will be transported into these cells with the required efficiency to allow for gold cluster formation, and to what extent resident endogenous MTs might produce background (Diestra et al., 2009). Here we demonstrate that MT can be used as a clonable tag for EM in mammalian cells. Our findings are potentially transformative as METTEM allows identification and localization of intracellular proteins with high specificity and exceptional sensitivity at molecular-scale resolution.
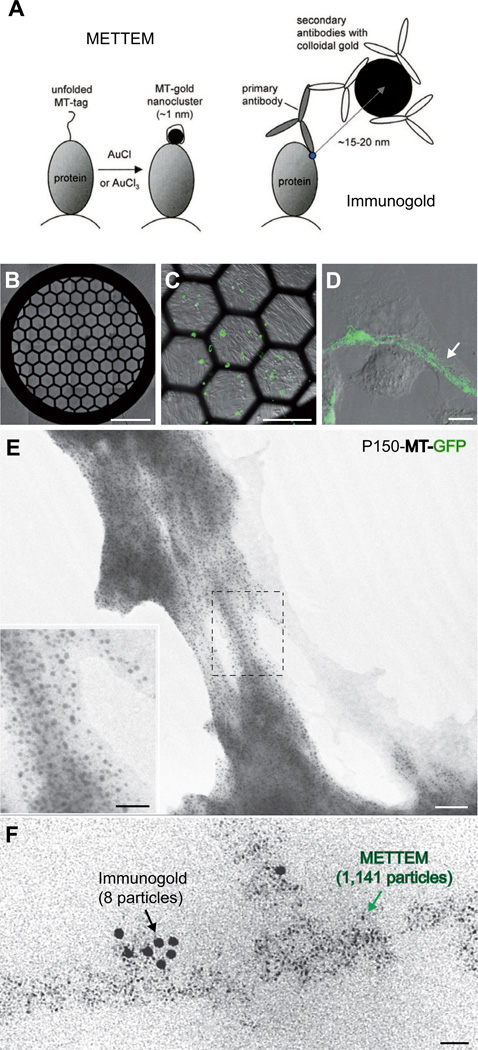
Figure 1. Detection of MT-gold-tagged intracellular proteins in mammalians cells.
(A) Cartoon summarizing the principles and differences between METTEM and IEM. (B–E) Correlative GPF-fluorescence and metal-tagging transmission electron microscopy. BHK-21 cells were cultured on EM grids and transfected with plasmid VR-P150-HA-MT-GFP (Figure S1) as to express MT-GFP-tagged RUBV P150. (B–D) Differential interference contrast and GFP-fluorescence microscopy; the white arrow indicates filament arrays formed by P150-MT-GFP. (E) P150-MT-GFP-expressing cells cultured on grid were treated with SLO and AuCl, dried on-grid and analyzed by TEM. Electron microscopy of the cell periphery showed that within minutes, intracellular green filaments had built numerous electron-dense nanoclusters. Inset, 2.2-fold enlargement of the area marked with a dashed rectangle. (F) Correlative immunogold and metal-tagging transmission electron microscopy. Cells, treated with SLO and AuCl as in (E), were embedded in acrylic resin. Ultra-thin sections were incubated with anti-GFP antibody and 10-nm colloidal gold conjugate and subjected to TEM. A typical image is presented, showing co-localization of immunogold particles and MT-induced gold nanoclusters; note, however, the difference in number of particles (8 versus 1,141, respectively). Scale bars: 750 µm for (B), 250 µm for (C), 10 µm for (D), 50 nm for (E), 25 nm for (F) and for inset in (E). See also Figure S1.
RESULTS
Rubella virus (RUBV), an enveloped, positive-stranded RNA virus in the family Togaviridae and an important human teratogenic pathogen, served as a model system. The biosynthesis and trafficking of the viral proteins that constitute the RUBV replication sites have been studied in considerable detail by fluorescent light microscopy and by IEM both in infected cells and in cells stably- or transiently-transfected with single round replicons (Fontana et al., 2007; Fontana et al., 2010). As targets, we selected the RUBV replicase subunit P150 and the capsid protein that build different types of structures in a variety of intracellular locations (Figure S1). When expressed in isolation, P150 assembles into nonfunctional cytoplasmic filament arrays (Matthews et al., 2010), but in association with replicase subunit P90 it will form biologically-active replication complexes (RCs) (Fontana et al., 2007; Tzeng et al., 2001). The available data (Fontana et al., 2007; Fontana et al., 2010) (and vide infra) suggests that P150-P90 hetero-oligomers initially accumulate at the cytoplasmic face of the plasma membrane to induce the formation of multi-layered membranous protrusions. These structures become internalized, ending up in vesicles that eventually fuse with lysosomes to form so-called cytopathic vacuoles (CPVs) (Fontana et al., 2010; Magliano et al., 1998) (Figure S2). It is in these latter virus-induced organelles, located perinuclearly and often enwrapped by rough endoplasmic reticulum, that viral RNA synthesis is believed to take place (Fontana et al., 2007; Lee et al., 1994). The RUBV capsid protein, when expressed separately, will display a disperse cytoplasmic distribution, but in the presence of P150 and P90, becomes recruited to the CPVs (Fontana et al., 2007; Tzeng and Frey, 2009).
To test the feasibility of MT-gold tagging of mammalian proteins, we cultured BHK-21 cells on EM grids and transfected them to transiently express a RUBV P150 derivative, dually-tagged with GFP and MT. The cells were treated with exotoxin streptolysin O (SLO) to permeabilize the plasma membrane (Fontana et al., 2007), incubated for 30 min with 1 mM AuCl and dried. Expression of RUBV P150-MT-GFP was confirmed by fluorescence microscopy, the protein assembling into a cytoplasmic microtubular-like fiber network (Figure 1B–1D), as seen before (Matthews et al., 2010) (see also Figure S1). Strikingly, in thin, semi-translucent peripheral areas of the cell, we detected by correlative EM a multitude of electron-dense particles of 1–2 nm in diameter, apparently in close association with these fibers (Figure 1E). Importantly, the particles were detected exclusively in transfected cells.
In ultra-thin sections of cells embedded in acrylic resin, the particles appeared more homogeneous in size, measuring ~1 nm in diameter, and they co-localized with deposits of P150-MT-GFP as shown by IEM (Figure 1F). Of note, the number of nanoparticles detected exceeded that of the colloidal gold particles associated with MT-P150-bound immune complexes by at least two orders of magnitude. Also in thin sections, these particles were observed in transfected cells only. In a series of control experiments (n = 10), untransfected and transfected cells were treated in parallel with gold salts, and EM was performed on serial sections covering the entire cell volume. Thus, more than 500 untransfected cells were analyzed, each of which tested negative. These findings firmly establish that MT-tagged proteins can be detected efficiently in SLO-permeabilized mammalian cells. The size of the particles corresponds to that of a metal nanocluster comprised of 20–40 gold atoms build by a single MT molecule (Mercogliano and DeRosier, 2006), suggesting that each gold cluster represents an individual MT-tagged protein molecule.
Conveniently, yet surprisingly enough, endogenous cellular MTs -though readily detectable in cell lysates by western blot analysis and in cryosections by IEM (Figure S3)- did not seem to induce formation of gold clusters. Possibly, this is due to the fact that MT levels are tightly controlled, such that the resident cellular MTs are already fully metallated with little or no free MT available and free Cu and Zn virtually absent in the cell (Beyersmann and Haase, 2001; Rae et al., 1999). As these metals, when bound to MT, are only partially displaced by gold (Schmitz et al., 1980), the resident cellular MTs would be unable to build gold clusters large enough to be detected by TEM. Be-it-as-it-may, our data decidedly show that also in mammalian cells MT-tagged intracellular proteins can be identified with high specificity and sensitivity. Whereas recombinant P150-MT-GFP expressed in isolation accumulates in the cytoplasm, the intact RUBV replicase, comprised of P150-P90 complexes, associates with membranes and becomes incorporated in CPVs, i.e. lysosome-derived virus-induced organelles. To assess whether METTEM would also allow detection of MT-tagged P150 in these more secluded intracellular locations, we studied P150 distribution in cells transfected with RUBV replicons. This approach also allowed us to ask whether MT-tagging would be compatible with proper multiprotein-complex formation, intracellular protein trafficking, and biological function. Cells transfected with a replicon encoding a P150 derivative, tagged with the HA epitope and MT (Figure S1), were first analyzed by confocal immunofluorescence microscopy. P150-HA-MT was detected at the cell periphery and in perinuclear foci (Figure 2A) and thus displayed an intracellular distribution indistinguishable from that of replicon-expressed wildtype P150 (Fontana et al., 2007). While GFP-tagging of P150 results in loss of RUBV RNA-dependent RNA polymerase activity, RCs comprising P150-HA-MT appeared to be fully functional. Viral RNA synthesis as indicated by the detection of dsRNA (Fontana et al., 2007) was confined to perinuclear foci, the number and distribution of which was similar to that seen for the wildtype RUBV replicon (Figures 2B and 2C). Ultra-thin sections of cells transfected with the P150-HA-MT replicon were analyzed by METTEM. The sections were left unstained as to avoid masking of the small gold nanoclusters by deposition of uranium or lead, routinely used for EM staining. In accordance with the results of immunofluorescence analysis, gold nanoclusters were detected primarily at the plasma membrane and in perinuclear vacuoles (Figures 2D–2I and S4). Where sections included large planar areas of plasma membrane, we observed randomly distributed groups of clusters (Figures S4A and S4B) associated with P150-HA-MT deposits as shown by IEM (not shown). We also found gold nanoclusters in multilayered, convoluted membranes, either protruding from the cell surface (Figures S4C–S4E) or located within intracellular vesicles near the plasma membrane (Figure S4F). In these vesicles, the membraneous structures appear to be drawn into spherical-elliptical arrangements with the P150 molecules grouping more closely together than at the cell surface (Figures S4G–S4L). Inside CVPs, large numbers of gold clusters were located very closely together suggesting that in these organelles P150-HA-MT molecules are densely packed (Figures 2F–2I), possibly as a result of membrane condensation. The combined findings provide proof of principle that proteins that assemble into nanomachines as complex as the RUBV RC (i) can be tagged with MT without affecting their intracellular trafficking and without loss of function and (ii) can be readily detected by METTEM even in secluded membranous compartments within the mammalian cell. To formally demonstrate that proteins other than RUBV P150 can also be studied by METTEM, we expressed the RUBV capsid protein, tagged with the FLAG epitope and MT. In cells stably transfected with replicon RUBrep/GFP/neo (Fontana et al., 2007) (Figure S1), MT-tagged capsid behaved similarly to untagged capsid and capsid-GFP. As shown by confocal immunofluorescence microscopy, the capsid protein accumulated in cytosol and was recruited to CPVs (Figures 3A–3C). In full accordance with these findings, METTEM detected capsid-MT molecules in the cytosol, where they showed a disperse, seemingly random distribution (Figure 3D), and inside CPVs, where they formed aggregates (Figures 3E–3G). The advantages of METTEM over indirect methods like IEM are saliently illustrated by the fact that IEM weakly marked the capsid in cytosol (Figure 3D), and completely failed to detect the protein inside CPVs (Figures 3F and 3G). In the experiments described so far, METTEM was performed on SLO-treated cells. Although permeabilization by SLO is not necessarily lethal and is in fact reversible, we did wonder whether it would be possible to perform gold-labeling under more physiological conditions, for example by exposing intact live cells to low concentrations of gold. BHK-21 cells were incubated in culture medium supplemented with gold salts (1 mM AuCl or 0.5 mM AuCl3) for different lengths of time (15 min to 3 h) and monitored for signs of toxicity by trypan blue exclusion, phase contrast light microscopy, actin immunofluorescence and electron microscopy of ultra-thin sections. No changes in gross or ultrastructural morphology or in viability were observed in cells exposed to gold salts for up to 1h. Cytopathic effects such as shrinking and partial actin depolymerization were noted only after prolonged incubation (2–3 hr). Based on these observations, incubation with AuCl or AuCl3 was restricted to 15 or 30 min, respectively, periods sufficiently brief to prevent toxicity, but long enough for intracellular MT-tagged proteins to form 1 nm gold nanoclusters. Indeed, in cells, transiently transfected with the RUBV P150-HA-MT replicon, gold clusters were detected both on the cell surface and in CPVs, just as we observed in SLO-treated cells (Figure S5). These findings suggest that in many cases, permeabilization by SLO may not be required to detect intracellular proteins by METTEM. To detect the small gold nanoclusters directly and unequivocally within the complex intracellular environment, we performed METTEM routinely on unstained ultra-thin sections of cells. Even by conventional TEM, these conditions provide sufficient contrast and detail to recognize the main cellular substructures and organelles. Ideally, however, one would want to combine detection of MT-tagged proteins by METTEM with the high contrast and detailed ultrastructural imaging attainable by staining with uranyl acetate and lead citrate. Collecting paired images of gold-labeled cells before and after staining was not a viable option. Sections of cells irradiated in the electron microscope to photograph the gold nanoclusters no longer incorporate heavy metal stains. As an alternative, comparison of stained and non-stained sequential serial sections does provide valuable information on the approximate location of the gold-tagged target proteins within a cell or organelle (Figures S6A–S6C). Still, as each section represents a different plane of a larger structure (ultra-thin sections are 30–50 nm thick, while nanoclusters have a diameter of ~1 nm), their super-position cannot be used to assess the fine-distribution of MT-tagged proteins (Figures S6D and S6E). To overcome this limitation, we analyzed stained thin sections of gold-treated cells by elastic bright field (EBF) and electron spectroscopic imaging (ESI) (Krivanek et al., 1995a). As shown in Figure 4, EBF and elemental gold imaging allowed simultaneous visualization of ultrastructural details and molecule location of P150-MT.
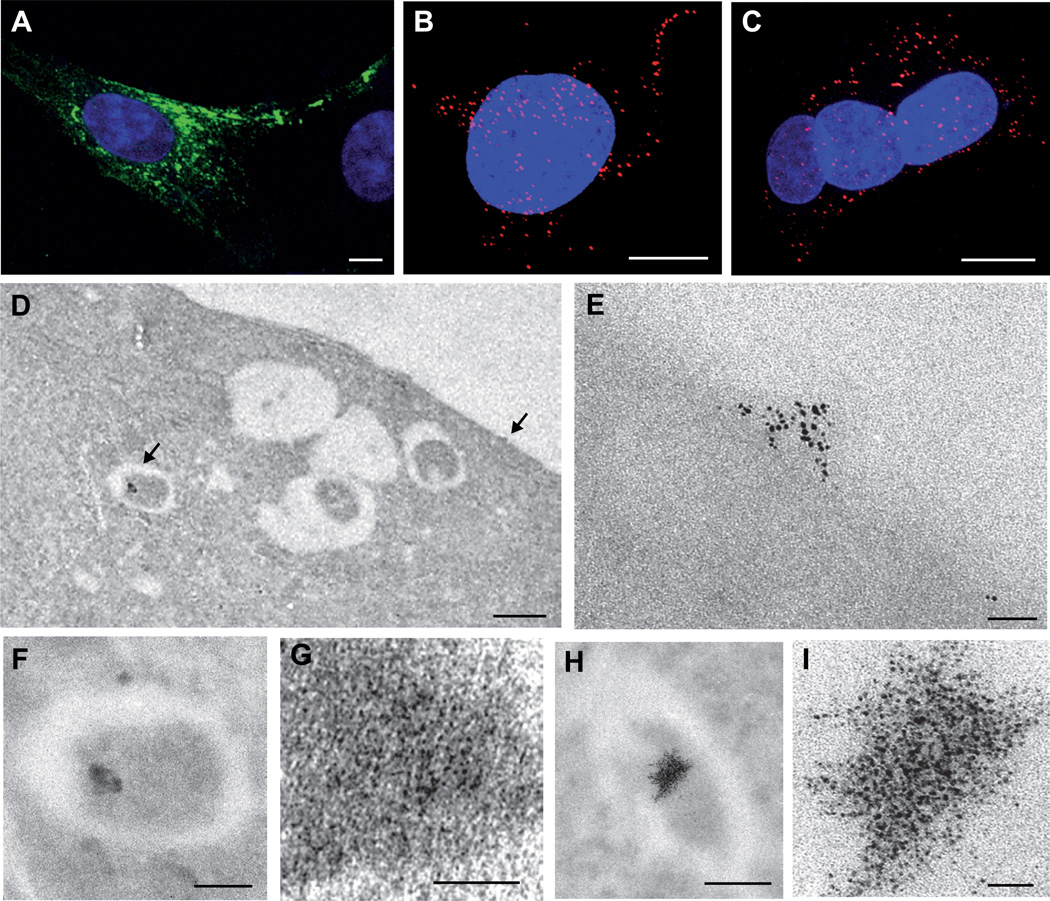
Figure 2. MT-tagging does not affect intracellular trafficking and function of RUBV P150, and allows protein detection even in secluded membranous compartments.
(A) Intracellular distribution of HA/MT-tagged RUBV P150, when expressed in the presence of RUBV P90. BHK-21 cells were transfected with replicon RUBrep-HA-MT/GFP (Figure S1), stained with anti-P150 antibodies (green) and analyzed by confocal immunofluorescence microscopy; nuclei were stained with DAPI (blue). Note that the intracellular distribution of P150-HA-MT (at the cell periphery and in perinuclear foci) is indistinguishable from that of replicon-expressed wildtype P150 (Fontana et al., 2007). (B) and (C) RUBV RCs comprising P150-HA-MT are biologically active. BHK21 cells were transfected with replicon RUBrep-HA-MT/GFP (B) or RUBrep/GFP (C) (Figure S1) to express HA-MT-tagged P150 or wild type P150, respectively. Biological activity of RUBV RCs (RNA synthesis) was assessed by confocal immunofluorescence microscopy after staining, 24 hr post transfection, with antibodies against dsRNA (red), an established marker of functional RUBV replication complexes (Fontana et al., 2007; Lee et al., 1994); nuclei were stained with DAPI (blue). (D–I) Intracellular distribution of HA/MT-tagged RUBV P150 as detected by METTEM. Cells were treated with SLO and AuCl, sectioned and visualized without heavy metal staining. (D) Low magnification EM of a RUBrep-HA-MT/GFP-transfected cell. Arrows indicate areas enlarged in (E) and (F). (E) METTEM detection of P150 at the cell surface. Typical image showing groups of gold nanoclusters at the plasma membrane. (F),(G) and (H), (I) Accumulation of P150 within CPVs. Corresponding low (F) and (H) and high magnification (G) and (I) views of perinuclear CPVs, high magnifications showing dense packing of large numbers of gold nanoclusters in a CPV subcompartment. Scale bars: 10 µm for (A) to (C), 0.5 µm for (D), 25 nm for (E), (G) and (I), 200 nm for (F) and (H). See also Figures S1–S5.
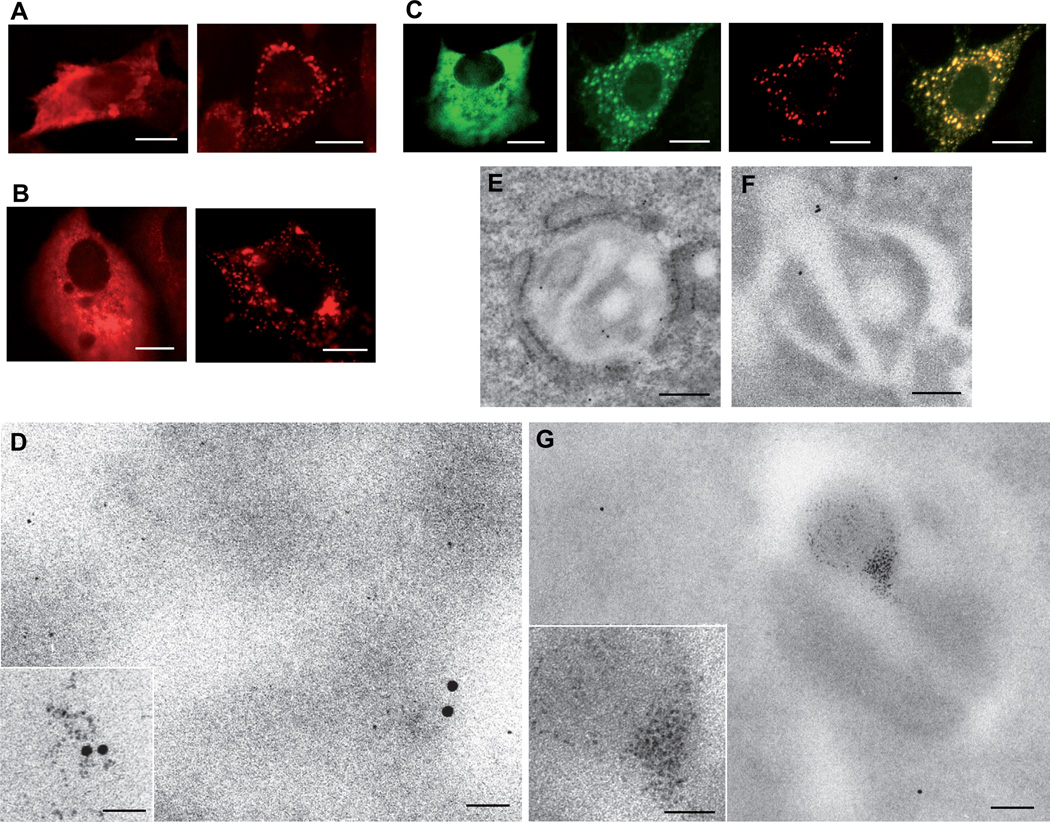
Figure 3. Fluorescence microscopy, immunogold and metal-tagging electron microscopy showing recruitment of MT-tagged RUBV capsid protein to CPVs.
(A) and (B) Intracellular distribution of RUBV FLAG-MT-tagged RUBV capsid protein (A) and wildtype capsid protein (B) expressed either in isolation (left hand panels) or in BHK21-cells, stably transfected with replicon RUBrep/GFP/Neo replicon (right hand panels). Cells were labeled with anti-FLAG antibodies (A) or anti-capsid antibodies (B) and analyzed by confocal immunofluorescence microscopy. (C) Intracellular distribution of GFP-tagged RUBV capsid protein as determined by confocal fluorescence microscopy. Distribution of capsid-GFP, when expressed in the absence of other RUBV proteins (left hand panel) or in cells, stably transfected with replicon RUBrep/C-GFP/Neo (remaining right hand panels). Capsid protein (green), RUBV P150 replicase subunit labeled with anti-P150 specific antibodies (red) and GFP-capsid/P150 co-localization (yellow). (D–G) Intracellular distribution of MT-tagged RUBV capsid protein as determined by immunogold and metal-tagging electron microscopy. RUBrep/GFP/neo-transfected cell expressing FLAG-MT-tagged RUBV capsid, were treated with SLO and AuCl (panels D and G) or left untreated (E) and (F), thin-sectioned and immunogold-labeled with antibodies against RUBV replicase protein P90 or FLAG. (D) METTEM and IEM detection of FLAG-MT-capsid molecules in the cytosol. Note the distinction between the 10-nm colloidal gold particles and the MT-induced ~1-nm gold nanoclusters. (E) and (F) Ultrathin sections showing CPVs, immunogold-labeled with anti-replicase P90 antibodies and stained with uranyl acetate (E) or with anti-FLAG antibodies and left unstained (F). (G) METTEM detection of accumulations of FLAG-MT-capsid molecules inside CPVs. The two large gold particles in the cytoplasm in the low magnification image correspond to immunogold labeling with anti-FLAG antibodies. Scale bars: 10 µm for (A) to (C), 200 nm for (E), (F) and (G), 25 nm for (D) and insets in (D) and (G). See also Figures S1–S3.
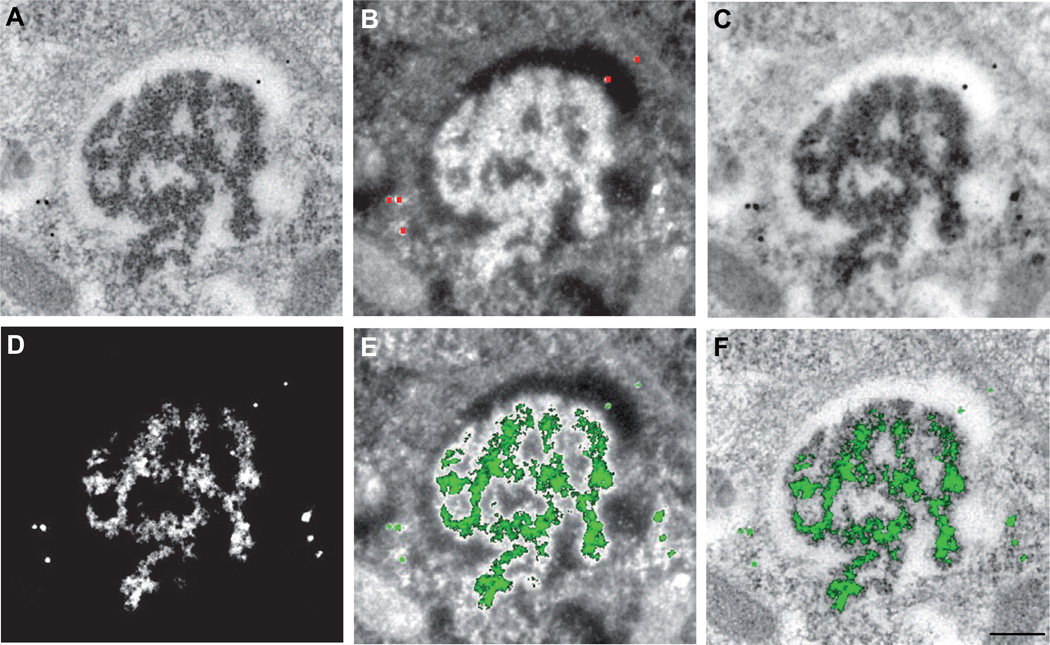
Figure 4. Elemental distribution of gold in stained sections as visualized by METTEM and Electron Spectroscopic Imaging (ESI).
Replicon-transfected cells expressing P150-HA-MT at 24 h post transfection, incubated with AuCl3 (30 min), processed for embedding and thin-sectioning, and stained with uranyl acetate and lead citrate. (A) Ultrastructure of an early CPV (Figure S2, stage 3) visualized by elastic bright field microscopy. (B) High contrast image (HCI) recorded at 250 eV. Red dots mark 10-nm colloidal gold particles added to the section as an internal gold standard. (C) Inverted HCI image. (D) Net gold elemental distribution obtained by ESI and background subtraction. (E) Gold signal (green) superimposed on the high contrast image or (F) on elastic bright field microscopy image, showing the precise distribution of MT-gold - tagged P150 molecules in the CPV subdomains. Scale bar: 200 nm. See also Figures S2, S5 and S6.
DISCUSSION
Recently, we reported METTEM detection of methallothionein-gold-tagged proteins in live bacteria (Diestra et al., 2009). Transferring this procedure to eukaryotic cells, particularly to mammalian cell lines, was not trivial, however, in view of the toxicity of gold salts, the potential interference from endogenous MTs and questions concerning the efficiency of gold uptake. We now demonstrate (i) that gold salts reach intracellular locations within minutes -a timespan too brief for cytopathology to occur- with MT-gold nanoclusters forming rapidly both in the cytosol and within organelles, (ii) that background arising from cellular MTs is negligible, and (iii) that gold nanoclusters can be readily detected in nonstained thin sections by conventional TEM, and in heavy metal-stained sections by EBF-ESI. With respect to specificity and resolution, METTEM greatly surpasses immunogold detection, its sensitivity exceeding that of IEM by orders of magnitude. An added advantage, METTEM is compatible with a variety of sample processing methods.
We illustrate the applicability and strength of the method in a series of experiments aimed to study the intracellular distribution of viral replicase and capsid proteins. Our findings extend previous observations in that they reveal the existence of hitherto unknown RUBV-induced cell structures and mini-organelles as apparent intermediates in the biogenesis of the CPVs (Fontana et al., 2007).
The convoluted multilamellar membraneous structures at the cell surface and within membrane proximals vesicles have not been described before, and this is also the first report of RUBV RCs aggregating within the CPVs. The combined data suggest a tentative model, reminiscent of the one recently proposed for the morphogenesis of alphavirus RCs (Frolova et al., 2010; Spuul et al., 2010), in which RUBV P150-P90 hetero-oligomers assemble at and associate with the cytoplasmic face of the PM to induce the formation of complex multilayered membraneous cell surface protrusions. These are taken up in large vesicles that in turn fuse with lysosomes to mature into mixed-content CPVs (Fontana et al., 2007; Fontana et al., 2010). Concurrently, the RUBV replicase complexes organize in densely packed arrays, in a process that would seem to involve progressive membrane condensation, such as to form distinct sub-compartments within the CPVs, to which also RUBV capsid proteins become recruited.
As demonstrated here, METTEM opens new avenues to study intracellular distribution and trafficking of proteins by conventional 2D TEM. As the gap between cellular ultrastructure and atomic-scale resolution closes rapidly, we envisage that methallothionein-gold tagging may also become a valuable tool to guide the docking of crystal or NMR structures of proteins into the 3D density maps produced by cryo-electron tomography of cells. Indeed, as was shown already, MT-nanoclusters can be visualized unambiguously in vitrified isolated macromolecular complexes (Mercogliano and DeRosier, 2006, 2007; Bouchet-Marquis et al, 2012) and in frozen bacteria (Diestra et al., 2009). We consider METTEM to be an important addition to the list of recent electron microscopical innovations that ultimately may allow ultrastructural imaging of intracellular macromolecular assemblies in their native state and in their natural environment.
EXPERIMENTAL PROCEDURES
Cells, constructs, antibodies and other reagents
BHK-21 cells from the American Type Culture Collection (ATCC) were grown as described (Fontana et al., 2007). For immunolabeling and Western blot analysis the following antibodies were used: rabbit antisera against P150 and P90 RUBV replicase components were previously characterized (Fontana et al., 2007); a mouse monoclonal antibody (mAb) specific for RUBV capsid and a mouse anti-FLAG mAb were purchased from Chemicon; K2 mAb specific for dsRNA was provided by Dr. N. Lukács (Corvinus University, Budapest, Hungary); rabbit anti-GFP (green fluorescent protein) was provided by Dr. David Shima (Imperial Cancer Research Foundation, London, UK); a mouse anti-MT mAb was purchased from Abcam and a mouse anti-HA mAb was from BAbCO. Fluorescent secondary antibodies conjugated with Alexa488 (green) or Alexa594 (red) were from Molecular Probes/Invitrogen. Secondary antibodies conjugated with 10 nm colloidal gold particles used for immunogold labeling were purchased from BBInternational. DAPI and gold salts (AuCl and AuCl3) were from Sigma-Aldrich. Acrylic resins Lowicryl HM23 and LR White were from Taab Laboratories and London Resin Company Ltd, respectively. SLO was purchased from Dr Sucharit Bhakdi’s laboratory (Inst. Medical Microbiology and Hygiene, Mainz, Germany).
Molecular biology
To construct the VR-p150-HA-MT-GFP plasmid (Figure S1), two PCR fragments were amplified. The mouse MT1 gene was produced by PCR from pET3d-MT (provided by Dr. Dennis Winge’s laboratory at University of Utah) using primer (5’-GTCCGCGGCCGCAGCGTAATCCGGAACATCATACGGGTAGGCACAGCACGTGCACTTGTCCGCGGCGCCTTTGCAGACAC-3’) and primer (5’GCGCGGATCCATGGACCCCAACTGCTCCTGCTCCACCGGCGG CTCC-3’) restricted with BamHI and NotI. The PinAI-BamHI fragment was amplified by primer (5’-GCGCGGATCCCGTATCGGCGCGCGCGCGGAGAGC-3’) and primer (5’-CCGGACACCGCCCACCCCGGGCGC-3’) and RUBrep/GFP template (Tzeng et al., 2001). Two fragments were ligated with PinAI-NotI-restricted VR-p150-GFP (Matthews et al., 2010) to generate VR-p150-HA-MT-GFP-ΔNotI. The NotI-NotI fragment from NotI restricted RUBrep/GFP was ligated with NotI-restricted and CIP-treated VR-p150-HA-MT-GFP-ΔNotI to make VR-p150-HA-MT-GFP. To construct RUBrep-HA-MT/GFP (Figure S1), two PCR fragments were amplified. The MT gene was produced by PCR from pET3d-MT using primer (5’-GTCCGCGGCCGCGATGGACCCCAACTGCTCCTGCTCCACCGGCGGCTCC-3’) and primer (5’GCGCGGATCCGGCACAGCACGTGCACTTGTCCGCGGCGCCTTTGCAGAC AC-3’) restricted with NotI and BamHI. The BamHI-NheI fragment was amplified by primer (5’-GCGCGGATCCTCACCGCCACCCGG CGACCCCCCG-3’) and primer (5’GCACGCGACACACGCCCACCGACG-3’) with RUBrep/GFP (Tzeng et al., 2001) template. Two fragments were ligated with NotI-NheI restricted RUBrep-HA/GFP (Tzeng et al., 2006) to generate RUBrep-HA-MT/GFP-ΔNotI. The NotI-NotI fragment from NotI restricted RUBrep/GFP was ligated with NotI-restricted and CIP-treated RUBrep-HA-MT/GFP-ΔNotI to make RUBrep-HA-MT/GFP.
To construct the pcDNA-N961-MT plasmid (Figure S1), the MT gene was produced by PCR from pET3d-MT. A PCR fragment was amplified with primer (5’CGCTCGGCGCGCCATCCTTGGCGCATCCGCATGGACCCCAACTGCTCCTGCTCCA CCGGCGGCTCC-3’) and primer (5’GCACTCTAGATCAGGCACAGCACGTGCACTTGTCCGCGGCGCCTTTGCAGACAC-3’) restricted with AcsI and XbaI, ligated with AscI-XbaI restricted pcDNA-N961 (Tzeng et al., 2006).
RUBrep/GFP/Neo and RUBrep/C-GFP/Neo replicons were previously characterized (Fontana et al., 2007). They express two heterologous genes, GFP or a C-GFP fusion under control of the subgenomic promoter followed by the neomycin resistance gene under control of a picornaviral IRES (Figure S1).
Immunofluorescence, confocal microscopy and differential interference contrast (DIC) light microscopy
Samples were prepared for immunofluorescence as previously described in detail (Fontana et al., 2007). Differential interference contrast (DIC) and confocal microscopy studies were done in a Leica TCS SP5 spectral confocal microscope. Digital photographs were processed with Adobe Photoshop, Image J, LAS and CW4000 FISH (Leica Microsystems) software packages.
Cell transfection, treatment with gold salts and processing for EM
In vitro transcription of RUBV replicons, purification of constructs and transfections were performed as described (Tzeng et al., 2001). Cell monolayers were grown on gold, hexagonal 100 mesh EM grids (Taab Laboratories) covered with collodion for direct visualization of P150-MT-GFP expression by fluorescence microscopy and EM. Alternatively, cells were grown on Thermanox plastic pieces from Nunc for oriented serial sectioning. BHK-21 cells were permeabilized with streptolysin O (SLO) as reported (Fontana et al., 2007). SLO-permeabilized cells at 9 and 24 h pt were treated (30 min) with 1 mM AuCl diluted in Dulbecco’s minimal essential medium (DMEM). Cells were washed with DMEM before fixation with 4% paraformaldehyde and 0.1% glutaraldehyde in PBS, fast-freezing in liquid ethane, freeze-substitution in methanol at −90°C and embedding in Lowicryl HM23 as described (Diestra et al., 2009). Alternatively, live cells incubated with 1 mM AuCl or 0.5 mM AuCl3 were monitored for signs of toxicity by trypan blue exclusion, phase contrast light microscopy and electron microscopy of ultra-thin sections. Live cells showed the first minor signs of alteration 1 h after addition of gold salts to the cultures. No effects in cell shape or ultrastructure were detected when live cells were incubated up to 1 h with gold salts. Based on these data, incubation times were established as 15 min for 1 mM AuCl and 15–30 min for 0.5 mM AuCl3 for live cells at 9, 24 or 48 h pt. The indicated incubation times with gold salts were insufficient to induce higher endogenous MT expression levels (Wei et al., 2008) or to alter cell ultrastructure, but sufficient to build ~1 nm gold nanoclusters associated to exogenous MT-tagged proteins, as confirmed by TEM. Ultra-thin (50–70 nm) sections were collected in 300 mesh Quantifoil holey carbon grids (R 3.5/1 Cu/ Rh, Quantifoil Micro Tools) and studied without staining in a Jeol JEM 1011 electron microscope. Some sections were stained (30 min) with saturated uranyl acetate, washed in distilled water and stained (1 min) with lead citrate following standard procedures (Fontana et al., 2007).
Endogenous MT and specificity of METTEM
Endogenous MT expression in untransfected control cells was studied by 8%, 12% and 14% SDS-PAGE and Western blot. Nuclear and cytosolic fractions were prepared and washed with 1X PBS and then resuspended in a buffer containing 20 mM HEPES (pH 7.4), 100 mM NaCl, 100 mM NaF, 1 mM sodium orthovanadate, 5 mM EDTA, 1 µM okadaic acid, 1% Triton X-100, and Complete protease inhibitor cocktail (Roche). Alternatively, whole cells were collected in lysis buffer (30 mM Tris, pH 8.5, 7M urea, 2M thiourea, 4% CHAPS and 50 mM DTT). Even in reducing and denaturing conditions, MT is visualized as multiple bands that correspond to MT multimers generated during sample preparation, as described (Aoki and Suzuki, 1991). For Western blot analysis, proteins were transferred to nitrocellulose membranes by standard procedures. Membranes were blocked with PBS with non-fat dry milk and 0.05% Tween 20 (overnight, 4°C) and incubated with anti-MT and -HA mAbs in saturation buffer (dil. 1:1000, 1 h, room temperature). After washing with the same buffer, membranes were incubated with secondary antibodies conjugated with horseradish peroxidase (1:2000 in saturation buffer). After several washes in saturation buffer, signal in membranes was visualized by chemiluminescence (ECL plus kit, Amersham).
To estimate the potential background from endogenous MT, we treated untransfected control cells with gold salts before processing for embedding, ultrathin sectioning and EM visualization. Serial sections were obtained to cover the whole cell volume. Gold nanoclusters were not detected in oriented serial sections of more than 500 cells from 10 independent experiments. In cell cultures transfected with MT-tagged plasmids or replicons, we obtained transfection efficiencies of 60–80%. In SLO- and AuCl-treated cells, correlative microscopy showed that untransfected cells in these cultures had no detectable gold nanoclusters, while surrounding transfected cells showed gold nanoclusters associated with MT-tagged proteins, as confirmed by immunogold labeling with specific antibodies.
Immunogold labeling
Cryosections of frozen cells and ultra-thin sections of cells embedded in acrylic resin were processed for immunogold labeling following standard procedures (Diestra et al., 2009; Fontana et al., 2007; Fontana et al., 2010). Primary antibodies and dilutions for immunogold assays were anti- P90 (1:100), -MT (1:50), -FLAG (1:25), and -GFP (1:100). Secondary antibodies conjugated with 10 nm colloidal gold particles were diluted 1:40.
Elastic Bright Field (EBF) and Electron Spectroscopic Imaging (ESI)
Ultra-thin sections collected on collodion-coated EM grids were analyzed in a Zeiss LIBRA 120 LaB6 electron microscope equipped with an in column EFTEM (energy filter TEM) and OSIS iTEM software for image analysis. For high contrast (Elastic Bright Field, EBF) energy selecting slit was inserted to eliminate inelastically scattered electrons. Spectrometer was set to 0 eV energy loss and blurring from chromatic aberration was removed resulting in a significant gain in image contrast. For elemental gold detection two core-loss edges are suitable with EFTEM (Krivanek et al., 1995b). The Au M4,5 edge at ΔE ion=2206 eV and ΔE max=2386 eV requires high electron doses but this is a very strong edge with practically no elemental overlaps. Samples were stable when pre-irradiated 20 min at low magnifications and gold elemental analysis was therefore done on M4,5 edge with exposure times of 50 s, illumination angle >4.0 mrad, and emission current of 11 µA. Köhler illumination was used for specimen stabilization by limiting the field of irradiation to the selected area. Evaluation of the gold elemental distribution and background subtraction were done as described (Egerton, 1996). For high contrast imaging (HCI), a special mode of ESI (Bauer, 1988), the spectrometer was set to 250 eV of energy loss. The ionization energy of the carbon K-edge is at 284 eV of energy loss. Therefore, imaging at 250 eV excludes all carbon signal. This results in excellent structural contrast in the absence of staining.
Highlights.
-
.
Metallothionein (MT) can be used in live mammalian cells as a clonable tag for EM.
-
.
Metal-tagging TEM using MT shows proteins in cells at molecular scale resolution.
-
.
METTEM exceeds sensitivity of immunogold detection by orders of magnitude.
-
.
METTEM revealed virus-induced cell structures and organelles not seen before.
Supplementary Material
ACKNOWLEDGEMENTS
Our gratitude to Sylvia Gutiérrez Erlandsson for expert support with confocal microscopy, and Catherine Mark for editorial assistance. This work was funded by grants from the Spanish Ministry of Science and Innovation (BIO2009-07255 to CR) and the US National Institutes of Health (AI21389 to TKF).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions: C.R. and R.J.D.G. designed research; C.R., E.S.C., and V.S. performed research; W.P.T. and T.K.F. contributed new reagents/analytic tools; C.R. and R.J.D.G. analyzed the data and wrote the manuscript.
Competing financial interests: C.R. and E.S.C. filed a patent application (ES201031880) in part related to this manuscript. V.S. is an employee of Carl Zeiss Nano Technology Systems.
REFERENCES
- Aoki Y, Suzuki KT. Detection of metallothionein by western blotting. Methods Enzymol. 1991;205:108–114. doi: 10.1016/0076-6879(91)05092-a. [DOI] [PubMed] [Google Scholar]
- Bauer R. Electron Spectroscopic Imaging: an advanced technique for imaging and analysis in transmission electron microscopy. Methods Microbiol. 1988;20:113–146. [Google Scholar]
- Beyersmann D, Haase H. Functions of zinc in signalling, proliferation and differentiation of mammalian cells. BioMetals. 2001;14:331–341. doi: 10.1023/a:1012905406548. [DOI] [PubMed] [Google Scholar]
- Bouchet-Marquis C, Pagratis M, Kirmse R, Hoenger A. Metallothionein as a clonable high-density marker for cryo-electron microscopy. J. Struct. Biol. 2012;177:119–127. doi: 10.1016/j.jsb.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J, Huang Z, Merrifield ME, Salgado MT, Stillman MJ. Studies of metal binding reactions in metallothioneins by spectroscopic, molecular biology, and molecular modelling techniques. Coord. Chem. Rev. 2002;233–234:319–339. [Google Scholar]
- Diestra E, Fontana J, Guichard P, Marco S, Risco C. Visualization of proteins in intact cells with a clonable tag for electron microscopy. J. Struct. Biol. 2009;165:157–168. doi: 10.1016/j.jsb.2008.11.009. [DOI] [PubMed] [Google Scholar]
- Egerton RF. Electron Energy-Loss Spectroscopy in the Electron Microscope. New York: Plenum Press; 1996. [Google Scholar]
- Fontana J, Tzeng W-P, Calderita G, Fraile-Ramos A, Frey TK, Risco C. Novel replication complex architecture in rubella replicon-transfected cells. Cell. Microbiol. 2007;9:875–890. doi: 10.1111/j.1462-5822.2006.00837.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana J, López-Iglesias C, Tzeng W-P, Frey TK, Fernández JJ, Risco C. Three-dimensional structure of Rubella virus factories. Virology. 2010;405:579–591. doi: 10.1016/j.virol.2010.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolova EI, Gorchakov R, Pereboeva L, Atasheva S, Frolov I. Functional Sindbis virus replicative complexes are formed at the plasma membrane. J. Virol. 2010;84:11679–11695. doi: 10.1128/JVI.01441-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaietta G, Deerinck TJ, Adams SR, Bouwer J, Tour O, Laird DW, Sosinsky GE, Tsien RY, Ellisman MH. Multicolor and electron microscopy imaging of connexion trafficking. Science. 2002;296:503–507. doi: 10.1126/science.1068793. [DOI] [PubMed] [Google Scholar]
- Grabenbauer M, Geerts WJ, Fernández-Rodríguez J, Hoenger A, Koster AJ, Nilsson T. Correlative microscopy and electron tomography of GFP through photooxidation. Nat. Methods. 2005;2:857–862. doi: 10.1038/nmeth806. PubMed: 16278657. [DOI] [PubMed] [Google Scholar]
- Kireev I, Lakonishok M, Liu W, Joshi VN, Powell R, Belmont AS. In vivo immunogold labelling confirms large-scale chromatin folding motifs. Nat. Methods. 2008;5:311–313. doi: 10.1038/nmeth.1196. [DOI] [PubMed] [Google Scholar]
- Krivanek OL, Friedman SL, Gubbens AJ, Kraus B. An imaging filtering for biological applications. Ultramicroscopy. 1995a;59:267–282. doi: 10.1016/0304-3991(95)00034-x. PubMed: 7571120. [DOI] [PubMed] [Google Scholar]
- Krivanek OL, Kundmann MK, Kimoto K. Spatial resolution in EFTEM elemental maps. J. Microsc. 1995b;180:277–287. [Google Scholar]
- Lee J-Y, Marshall JA, Bowden DS. Characterization of Rubella virus replication complexes using antibodies to double-stranded RNA. Virology. 1994;200:307–312. doi: 10.1006/viro.1994.1192. [DOI] [PubMed] [Google Scholar]
- Magliano D, Marshall JA, Bowden DS, Vardaxis S, Meanger J, Lee JY. Rubella virus replication complexes are virus modified lysosomes. Virology. 1998;240:57–63. doi: 10.1006/viro.1997.8906. [DOI] [PubMed] [Google Scholar]
- Matthews JD, Tzeng W-P, Frey TK. Analysis of the function of cytoplasmic fibers formed by the rubella virus nonstructural replicase proteins. Virology. 2010;406:212–227. doi: 10.1016/j.virol.2010.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercogliano CP, DeRosier DJ. Gold cluster formation using metallothionein: mass spectrometry and electron microscopy. J. Mol. Biol. 2006;355:211–223. doi: 10.1016/j.jmb.2005.10.026. [DOI] [PubMed] [Google Scholar]
- Mercogliano CP, DeRosier DJ. Concatenated metallothionein as a clonable gold label for electron microscopy. J. Struct. Biol. 2007;160:70–82. doi: 10.1016/j.jsb.2007.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rae TD, Schmidt PJ, Pufahl RA, Culotta VC, O’Halloran TV. Undetectable intracellular free copper: the requirement of a copper chaperone for superoxide dismutase. Science. 1999;284:805–808. doi: 10.1126/science.284.5415.805. [DOI] [PubMed] [Google Scholar]
- Robinson CV, Sali A, Baumeister W. The molecular sociology of the cell. Nature. 2007;450:973–982. doi: 10.1038/nature06523. [DOI] [PubMed] [Google Scholar]
- Schmitz G, Minkel DT, Gingrich D, Shaw CF., III The binding of gold (I) to metallothionein. J. Inorg. Biochem. 1980;12:293–306. doi: 10.1016/s0162-0134(00)80270-6. [DOI] [PubMed] [Google Scholar]
- Shu X, Lev-Ram V, Deerinck TJ, Qi Y, Ramko EB, Davidson MW, Jin Y, Ellisman MH, Tsien RY. A genetically encoded tag for correlated light and electron microscopy of intact cells, tissues, and organisms. PLoS Biol. 2011;9 doi: 10.1371/journal.pbio.1001041. e1001041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spuul P, Balistreri G, Kääriäinen L, Ahola T. Phosphatidylinositol 3-kinase-, actin-, and microtubule-dependent transport of semliki forest virus replication complexes from the plasma membrane to modified lysosomes. J. Virol. 2010;84:7543–7557. doi: 10.1128/JVI.00477-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzeng W-P, Chen MH, Derdeyn CA, Frey TK. Rubella virus DI RNAs and replicons: requirement for non-structural proteins acting in cis for amplification by helper virus. Virology. 2001;289:63–73. doi: 10.1006/viro.2001.1088. [DOI] [PubMed] [Google Scholar]
- Tzeng W-P, Frey TK. Functional replacement of a domain in the Rubella virus P150 replicase protein by the virus capsid protein. J. Virol. 2009;83:3549–3555. doi: 10.1128/JVI.02411-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzeng W-P, Matthews JD, Frey TK. Analysis of Rubella virus capsid protein-mediated enhancement of replicon replication and mutant rescue. J. Virol. 2006;80:3966–3974. doi: 10.1128/JVI.80.8.3966-3974.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkmann N. Methods for segmentation and interpretation of electron tomographic reconstructions. Methods Enzymol. 2010;483:31–46. doi: 10.1016/S0076-6879(10)83002-2. [DOI] [PubMed] [Google Scholar]
- Wang Q, Mercogliano CP, Löwe J. A ferritin-based label for cellular electron cryotomography. Structure. 2011;19:147–154. doi: 10.1016/j.str.2010.12.002. [DOI] [PubMed] [Google Scholar]
- Wei H, Mokhtar Desouki M, Lin S, Xiao D, Franklin RB, Feng P. Differential expression of metallothioneins (MTs) 1, 2, and 3 in response to zinc treatment in human prostate normal and malignant cells and tissues. Mol. Cancer. 2008;7:7. doi: 10.1186/1476-4598-7-7. PubMed: 18208603. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.