Abstract
The current consensus for the eukaryote tree of life consists of several large assemblages (supergroups) that are hypothesized to describe the existing diversity. Phylogenomic analyses have shed light on the evolutionary relationships within and between supergroups as well as placed newly sequenced enigmatic species close to known lineages. Yet, a few eukaryote species remain of unknown origin and could represent key evolutionary forms for inferring ancient genomic and cellular characteristics of eukaryotes. Here, we investigate the evolutionary origin of the poorly studied protist Collodictyon (subphylum Diphyllatia) by sequencing a cDNA library as well as the 18S and 28S ribosomal DNA (rDNA) genes. Phylogenomic trees inferred from 124 genes placed Collodictyon close to the bifurcation of the “unikont” and “bikont” groups, either alone or as sister to the potentially contentious excavate Malawimonas. Phylogenies based on rDNA genes confirmed that Collodictyon is closely related to another genus, Diphylleia, and revealed a very low diversity in environmental DNA samples. The early and distinct origin of Collodictyon suggests that it constitutes a new lineage in the global eukaryote phylogeny. Collodictyon shares cellular characteristics with Excavata and Amoebozoa, such as ventral feeding groove supported by microtubular structures and the ability to form thin and broad pseudopods. These may therefore be ancient morphological features among eukaryotes. Overall, this shows that Collodictyon is a key lineage to understand early eukaryote evolution.
Keywords: 18S and 28S rDNA; Collodictyon; Diphyllatia; tree of life; phylogenomics; cDNA, pyrosequencing
Introduction
Over the last few years, molecular sequence data have addressed some of the most intriguing questions about the eukaryote tree of life. Phylogenomic analyses have confirmed the existence of several major eukaryote groups (supergroups) as well as shown various levels of evidences for the relationships among them (Burki et al. 2007; Parfrey et al. 2010). Recently, two new large assemblages, SAR (Stramenopila, Alveolata, and Rhizaria) and CCTH (Cryptophyta, Centrohelida, Telonemia, and Haptophyta), were proposed to encompass a large fraction of the eukaryote diversity, together with the other supergroups Opisthokonta, Amoebozoa, Archaeplastida, and Excavata (Patron et al. 2007; Burki et al. 2009). Solid phylogenomic evidence supports the monophyly of Amoebozoa, Opisthokonta, Archaeplastida, and SAR (Rodriguez-Ezpeleta et al. 2007; Burki et al. 2009; Minge et al. 2009), but the monophyly of Excavata and CCTH (also called Hacrobia; Okamoto et al. 2009) remains controversial, often dependent on the selection of taxa and gene data set (Burki et al. 2009; Hampl et al. 2009; Baurain et al. 2010). Despite several attempts, the evolutionary relationships between these supergroups are still uncertain because of the ancient and complex genome histories (Simpson and Roger 2004; Parfrey et al. 2006; Roger and Simpson 2009).
Identification of sister lineages to these supergroups is crucial for resolving the eukaryote tree and understanding the early history of eukaryotes. If these key lineages exist, they may be found among the few species that harbor distinct morphological features but are of unknown evolutionary origin in single-gene phylogenies (Patterson 1999; Shalchian-Tabrizi et al. 2006; Kim et al. 2011). Indications that such enigmatic species can be placed in the eukaryote tree come from recent phylogenomic analyses. For instance, Ministeria (Opisthokonta), Breviata (Amoebozoa) and Telonemia, Centroheliozoa, and Picobiliphyta have been shown to constitute deep lineages within their respective supergroups (Shalchian-Tabrizi, Minge, et al. 2008; Burki et al. 2009; Minge et al. 2009; Yoon et al. 2011).
Here, we investigate a member of such a key lineage, Collodictyon, which was first described in 1865 (Carter 1865), but its cellular structure and outer morphology were analyzed only recently (Klaveness 1995; Brugerolle et al. 2002). Collodictyon was originally proposed to be closely related to Diphylleia and Sulcomonas and classified in the family Diphylleidae (Cavalier-Smith 1993; the synonymous family Collodictyonidae in Brugerolle et al. 2002) and subphylum Diphyllatia (Cavalier-Smith 2003). Collodictyon is an omnivorous amoeba-flagellate with a mix of cellular features that makes it unique among eukaryotes. The cell has an egg- or heart-like outline without walls or any other external ornamentation in spite of a highly vacuolated cytoplasm (Rhodes 1917; Klaveness 1995). It possesses four equally long flagella and mitochondria with unconventional tubular-shaped cristae. An important character of Collodictyon is a broad ventral feeding groove dividing the cell longitudinally. This groove is supported by both left and right microtubular roots along the entire length of the lips, similar to comparable structures in other eukaryotes such as in Excavata (Simpson 2003). It also forms pseudopods typical of Amoebozoa at the base of the groove, which are actively used for catching prey.
Despite its interesting morphological features, it remains unclear whether Collodictyon is closely related to either Excavata or Amoebozoa or to any of the other supergroups because no molecular data are available. Furthermore, the position of the closely related Diphylleia is totally unresolved in 18S ribosomal DNA (rDNA) phylogenies (Brugerolle et al. 2002; Shalchian-Tabrizi et al. 2006). In order to explore the origin of Collodictyon, we established a culture of Collodictyon triciliatum, sequenced the 18S and 28S rDNA genes, and carried out a deep survey of a cDNA library with 454 pyrosequencing. About 300,000 sequence reads were generated and used to assemble an alignment of 124 genes (27,638 amino acid characters) that covered a taxon-rich sampling of eukaryotes (79 species). To further understand the evolutionary history of this lineage, we also screened the cDNA library for the dihydrofolate reductase (DHFR) and thymidylate synthase (TS) genes and extended the DHFR gene by 3′ Rapid Amplification of cDNA Ends (RACE) and polymerase chain reaction (PCR).
Materials and Methods
Culturing, Harvesting, and cDNA Library Construction
Collodictyon triciliatum was isolated from Lake Årungen, Norway, and cultured on a modified Guillard and Lorenzen medium (Guillard and Lorenzen 1972). Collodictyon triciliatum was inoculated in a culture of the cryptomonad Plagioselmis nannoplanktica (Klaveness 1995; Shalchian-Tabrizi, Bråte, et al. 2008). cDNA libraries were constructed by Vertis Biotechnology AG (Freising, Germany) according to their random-primed cDNA protocol: Total RNA was extracted with mirVana RNA isolation kit (Ambion, Austin, TX), and poly(A) + RNA was isolated from the total RNA. First-strand cDNA synthesis was performed with randomized primers, and second-strand cDNA was synthesized using Gubler and Hoffman protocol (Gubler and Hoffman 1983). Double-stranded DNA (dsDNA) was blunted, and 454 GSFLX adapters A and B were ligated to its 5′ and 3′ ends. dsDNA carrying both adapters was selected and amplified with PCR (24 cycles). Differently expressed genes were normalized with a method developed by Vertis Biotechnology AG. cDNA in the size range of 250–600 bp was eluted from a preparative agarose gel and sequenced by the Norwegian ultra-high throughput sequencing service unit at the University of Oslo and Macrogen Inc (South Korea) yielding a total of 300,000 sequence reads.
Sequence Analysis
All the 454 pyrosequencing reads were assembled into contigs using Newbler v2.5 (Margulies et al. 2005) with default parameters. We retrieved contigs larger than 200 bp with significant similarity to genes recently used in a multigene phylogeny (Burki et al. 2010). The translated contigs were screened by BlastP using our single-gene sequences as queries, and the homologous copies (e value < 1 × 10−20) were added to the single-gene data set. These new sequences were automatically aligned by Mafft with the linsi algorithm (Katoh et al. 2002), and ambiguously aligned positions were removed using Gblocks (Castresana 2000) with half of the gapped positions allowed, the minimum number of sequences for a conserved and a flank position set to 50% of the number of taxa, the maximum of contiguous nonconserved positions set to 12, and the minimum length of a block set to 5. The orthology and possible contamination in each single-gene alignment were assessed by maximum likelihood (ML) reconstructions with 100 bootstrap replicates using RAxML v7.2.6 under the PROTCATLGF substitution model (Stamatakis 2006), followed by visual evaluation of the resulting individual trees. For several single genes (i.e., prmt8, tubb, rpsa, suclg1, tcp1-beta, hsp90, ubc, and crfg), the PROTGAMMALGF model was used in addition to the PROTCATLGF model for better identification of the orthology. We used published global eukaryotic trees such as in Rodriguez-Ezpeleta et al. (2007) and Burki et al. (2009) as framework to identify and remove the sequences that showed unexpected grouping and were supported with more than 70% bootstrap in the single genes trees. In order to identify hidden paralogs in the data, we added more taxa in the single-gene phylogenetic analyses than in analyses of the supermatrix. Deletion of long-branch taxa (i.e., Trichomonas, Giardia, and Spironucleus) was done in a subsample of the single-gene alignments, but it did not change the phylogeny or the bootstrap values significantly. Hence, although inclusion of fast-evolving species could potentially introduce systematic errors in the trees, these types of taxa seemed not to strongly impact our paralog identification. Importantly, we included gene sequences from the cryptomonad Guillardia theta in all alignments in order to phylogenetically distinguish sequences from Collodictyon and its prey (P. nannoplanktica). This left in total 124 single-gene alignments containing Collodictyon sequences that were used for further analyses. The concatenation of the 124 single genes was done by Scafos (Roure et al. 2007) and amounted to 27,638 amino acid positions with average missing characters 34.4% (For detail, see supplementary table S2, Supplementary Material online). The sequences generated here were submitted to GenBank with accession number JN618831–JN618979. The single-gene trees and alignments as well as the concatenated alignment are available at http://www.mn.uio.no/bio/english/people/aca/kamran/.
Phylogeny of rDNA and Multigene Alignments
Reconstructions of ML phylogenies from 18S and 28S rDNA sequence alignments were done using RAxML v7.2.6. The best tree was determined after 100 heuristic searches starting from different random trees under the general time reversible (GTR) + GAMMA + I model. Bootstrap analyses were performed with 100 pseudoreplicates using the same model as in the initial tree search. Bayesian analyses were done with MrBayes v3.1.2 (Huelsenbeck and Ronquist 2001) under the GTR + GAMMA + I + COV evolutionary model that accounts for covarion substitution pattern across the sequences. Two independent runs, each starting from a random tree for Markov chain Monte Carlo (MCMC) chains, were run for 6,000,000 (18S rDNA) and 4,000,000 (18S + 28S rDNA) generations and sampled every 100 generations. Posterior probabilities and average branch lengths were calculated from the consensus of trees sampled after burn-in set to 3,000,000 (18S rDNA) and 1,000,000 (18S + 28S rDNA) generations. Chains were considered to be convergent when the average split frequency was lower than 0.01.
Several concatenated protein alignments with different taxonomic compositions were constructed to investigate the influence of species sampling and missing data on the phylogeny of Collodictyon. Phylogenies were inferred by ML and Bayesian approaches, as implemented in RAxML v7.2.6 and Phylobayes v3.2 (Lartillot and Philippe 2004), respectively. Following both the Akaike information criterion and the likelihood ratio test computed with ProtTest 3.0 (Darriba et al. 2011), the optimal model LG + GAMMA + F available in RAxML v.7.2.6 was chosen to infer ML trees. The best ML topology was determined in heuristic searches from ten random starting trees. Due to computational burden, statistical support was evaluated with 100 bootstrap replicates under the PROTCATLGF model that approximates the gamma distribution for site-rate variation (Stamatakis et al. 2008). Bayesian inferences were done with the CAT site-heterogeneous mixture model. Two independent MCMC chains in PhyloBayes starting from random trees were run for 24,000 cycles with trees being sampled every cycle. Consensus topology and posterior probability (PP) values were calculated from saved trees after burn-in. Convergence between the two chains was ascertained by examining the difference in frequency for all their bipartitions (maxdiff < 0.15). In addition, a bootstrap analysis under the CAT model was performed on 100 pseudoreplicates generated by Seqboot (Phylip package; Felsenstein 2001). For each replicate, two Phylobayes MCMC chains were run for 5,000 cycles with a conservative burn-in of 2,000 cycles. Manual verification of 10% randomly chosen replicates showed that the burn-in was optimal between 1,000 and 2,000 cycles. Consense (Phylip package) was used to calculate the bootstrap support based on these 100 Bayesian consensus trees.
Testing Robustness of Trees by Removal of Fast-Evolving Sites
We applied the AIR package (Kumar et al. 2009; Yang 2007) to estimate evolutionary rates of sites under the Whelan and Goldman + GAMMA model. The ML topology constructed from a sample of 76 taxa (i.e., removal of two Malawimonas species and Collodictyon) was used as starting tree for the estimate of site rates. The rationale for choosing this topology was to ensure that the site rates were calculated independently of the evolutionary affinity between these two lineages and their positions in the tree. The sites were then removed in 5% intervals (i.e., removal of the 5%, 10%, 15%, 20%, 25%, 30%, 35%, 40%, 45%, and 50% fastest evolving sites) from a full alignment that contained the two Malawimonas species and Collodictyon (i.e., 79 taxa) and an alignment where only the two Malawimonas species were removed (i.e., 77 taxa). The bootstrap values (BP) for the nodes defining the supergroups as well as for the position of Collodictyon and Malawimonas were inferred from each of these processed alignments by RAxML v7.2.6 under the PROTCATLGF model (with 100 bootstrap replicates). These trimmed alignments were then used for the estimation of amino acid composition (see supplementary materials and methods, Supplementary Material online). All bioinformatics analyses were done on the Bioportal at the University of Oslo (www.bioportal.uio.no; Kumar et al. 2009).
Topology Comparisons
Topology testing was performed using the approximately unbiased (AU) test (Shimodaira 2002). For each tested tree, site likelihoods were calculated using RAxML v7.2.6 with the PROTGAMMALGF model, and the AU test was performed using CONSEL (Shimodaira and Hasegawa 2001).
3′ RACE and Sequencing of the DHFR-TS Genes
All assembled contigs were used as queries in BLAST search against the nonredundant protein sequences database available at NCBI. Three contigs (contig15348, contig15349, and contig06264) showed a significant similarity to the DHFR gene (e value < 1 × 10−10). In order to verify that these contigs belong to Collodictyon and not the prey, we designed forward and reverse primers, then different combinations of primers were used to amplify genomic DNA from three cultures: 1) P. nannoplanktica (PN), 2) P. nannoplanktica + C. triciliatum (PN + CT), and 3) Chlorella pyreuoidosa + C. triciliatum (CP + CT). Bands were observed on the agarose gel solely when using forward primer in contig15348 and reverse primer in contig15349 for PCR amplification from PN + CT and CP + CT cultures. Both sequences were identical and matched the 3′-end region of contig15348 and the 5′-end region of contig15349. Since identical sequences were only obtained in the cultures containing Collodictyon, it confirmed that these two contigs corresponded to the Collodictyon gene, not the Plagioselmis or Chlorella one. Total RNA was isolated from PN + CT cultures with the RNAqueous-Micro Kit (Ambion, Austin, TX) following the standard protocol. The 3′ RACE system from Invitrogen (Carlsbad, CA) was performed to obtain the full-length 3′-end of the DHFR cDNA. Two specific forward primers (DHFR1F: 5′-CGAGTGCGTTGAATGATTCGTCAAA-3′ and DHFR2F: 5′-CTCAATGTTATTGTCAGCAGCACT-3′), together with a universal reverse primer (AUAP: 5′-GGCCACGCGTCGACTAGTAC-3′), were used in a two-step protocol to improve the specificity of the amplification process. The PCR products were sequenced to validate whether the DHFR gene and the TS gene were fused or not (GenBank accession number: JN618830).
Results and Discussion
Collodictyon Is an Ancient and Distinct Eukaryote Lineage
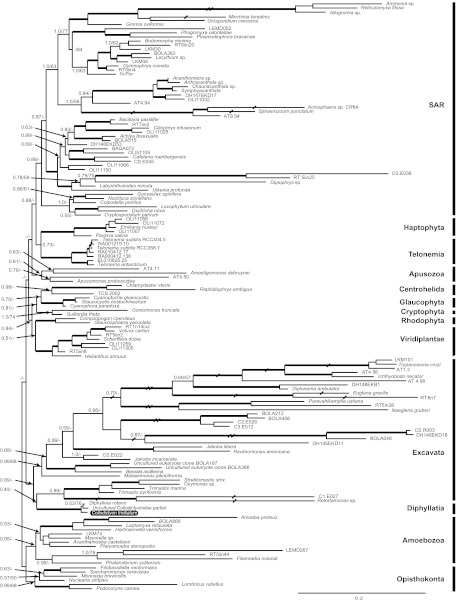
In order to clarify the origin of Collodictyon, we first obtained the 18S rDNA sequence for C. triciliatum. Phylogenetic analysis recovered most of the eukaryote supergroups as monophyletic clades, except CCTH and Archaeplastida, congruent with several recent reports (fig. 1; Burki et al. 2007, 2008; Yoon et al. 2008; Hampl et al. 2009). More interestingly, this phylogeny robustly supported Collodictyon and Diphylleia as sister lineages with 100% bootstrap support (BP) and 1.00 posterior probabilities (PP), confirming that these two species indeed are closely related. In an attempt to enrich the species diversity for this group and estimate their potential abundance and diversity in nature, we searched for Collodictyon-like 18S rDNA sequences by blastn against the environmental database in NCBI. Twenty of the top Blast hits were used for phylogenetic analysis, but only a single partial sequence grouped with Diphylleia (results not shown), suggesting a low diversity and abundance of the Diphyllatia in the environment. This partial sequence was included in the 18S phylogeny (fig. 1).
FIG. 1.
18S rDNA phylogeny of the Diphyllatia species Collodictyon triciliatum (highlighted by black box) and Diphylleia rotans. The topology was reconstructed by MrBayes v3.1.2 under the GTR + GAMMA + I + covarion model. Posterior probabilities (PP) and ML bootstrap supports (BP, inferred by RAxML v7.1.2 under GTR + GAMMA + I model) are shown at the nodes. Thick lines indicate PP > 0.90 and BP > 80%. Dashes “-” indicate PP < 0.5 or BP < 50%. A few long branches are shortened by 50% (/) or 75% (//).
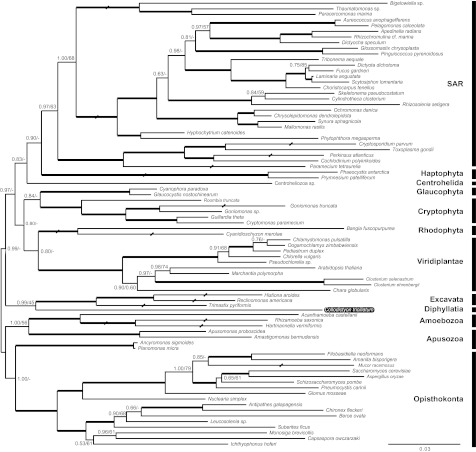
To improve the rDNA tree, we also sequenced the 28S rDNA gene for Collodictyon and reconstructed a combined 18S + 28S rDNA phylogeny (fig. 2). This tree showed Collodictyon as a deep lineage with possible affinity to Excavata with 45% BP and 0.99 PP. Interestingly, our data did not show any affiliation to Apusozoa, even though this group has been proposed to be closely related to Collodictyon (Cavalier-Smith 2003). Instead, the 18S + 28S rDNA tree suggested Apusomonas to be sister to Amoebozoa (56% BP and 1.00 PP), although Ancyromonas grouped with the Opisthokonta (<50% BP and 1.00 PP).
FIG. 2.
18S + 28S rDNA phylogeny of Collodictyon triciliatum (highlighted by black box) reconstructed with MrBayes v3.1.2 under the GTR + GAMMA+I + covarion model. Numbers at nodes are PP and ML bootstrap values (BP, inferred by RAxML v7.2.6 under the GTR + GAMMA + I model). Thick lines show PP > 0.9 and BP > 80%. Nodes marked with symbol “-” indicate BP < 50% or PP < 0.5. Some branches are shortened by half in order to save space (marked with “/”).
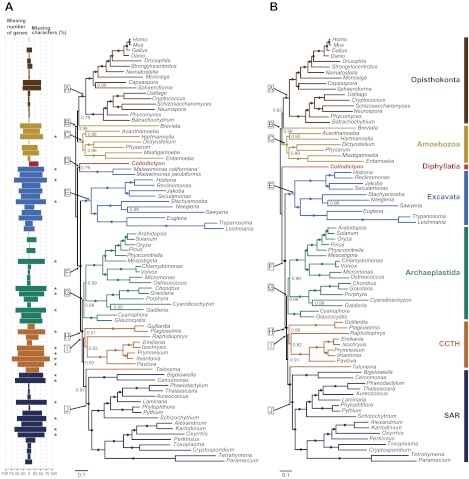
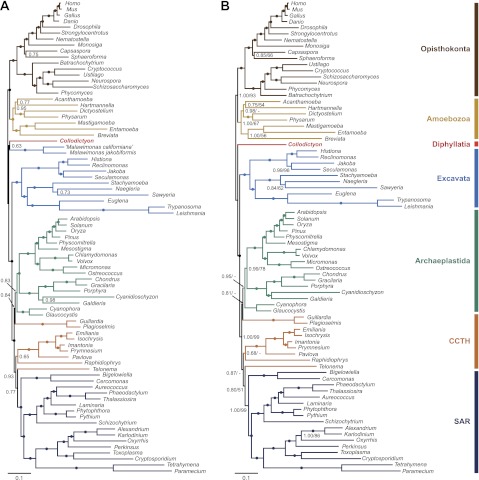
Because our 18S and 18S + 28S rDNA trees suggested that Collodictyon might have diverged very early in eukaryote evolution and that these two genes alone were not sufficient to infer ancient relationships, we sought to increase the phylogenetic signal by constructing an alignment of 124 protein-coding genes and 79 taxa. Phylogenomic trees inferred with both Bayesian and ML methods consistently recovered most eukaryote supergroups as in recent studies (Rodriguez-Ezpeleta et al. 2007; Burki et al. 2009; Hampl et al. 2009), generally with high statistical support (table 1). Differing from published phylogenies (Burki et al. 2009; Minge et al. 2009), the Bayesian inference (fig. 3A) did not recover Breviata as sister to Amoebozoa and Telonema did not branch within CCTH, but these were instead placed as a sister to Opisthokonta (0.75 PP) and SAR (0.91 PP). Of much interest, our analyses showed that Collodictyon branched outside any of the major lineages (fig. 3A and supplementary fig. S1A, Supplementary Material online), more specifically at the bifurcation of the so-called “unikonts” (Amoebozoa and Opisthokonta) and “bikonts” (Archaeplastida, SAR, Excavata, CCTH; the terms unikonts and bikonts are used here for simplicity and do not refer to their original description; Stechmann and Cavalier-Smith 2002; Roger and Simpson 2009). Although Collodictyon did not fall within any of the supergroups, an affinity to another enigmatic genus Malawimonas was recovered with 0.79 PP and 86% BP.
Table 1.
Maximum likelihood bootstrap values (ML) and bayesian posterior probabilities (Bayes) of the Eukaryote Supergroups in the Phylogenomic Trees.
| 79 Taxa |
74 Taxaa |
77 Taxab |
72 Taxac |
||||||||
| All Sites |
20% Removedd |
All Sites | All Sites |
20% Removedd |
All Sites | ||||||
| Nodee | Groups | ML | Bayes | ML | Bayes | ML | ML | Bayes | ML | Bayes | ML |
| A | Opisthokonta | 100 | 1.00 | 100 | 1.00 | 100 | 100 | 1.00 | 100 | 1.00 | 100 |
| B | Unikonts | 79 | 0.99 | 99 | 1.00 | 87 | 57 | 0.99 | 96 | 1.00 | 58 |
| C | Amoebozoa | 86 | 1.00f | 100 | 1.00 | 100 | 84 | 1.00f | 100 | 1.00 | 100 |
| D | Collodictyon + Malawimonas | 86 | 0.79 | 98 | 0.63 | 94 | NA | NA | NA | NA | NA |
| E | Excavata | 100 | 1.00 | 100 | 1.00 | 100 | 100 | 1.00 | 100 | 1.00 | 100 |
| F | Bikonts | 98 | 1.00 | 98 | 1.00 | 95 | 98 | 1.00 | 100 | 1.00 | 93 |
| G | Archaeplastida | - | 0.98 | 63 | 0.84 | - | - | 0.99 | 71 | 0.95 | - |
| H | Archaeplastida + CCTH + SAR | - | 1.00 | 81 | 1.00 | - | - | 1.00 | 88 | 1.00 | - |
| I | CCTH | - | * | 54 | * | - | 50 | * | 60 | * | - |
| J | SAR | 98 | 1.00 | 100 | 1.00 | 96 | 99 | 1.00 | 100 | 1.00 | 96 |
NOTE.—“-” indicate bootstrap values < 50% or PP < 0.5; “*” indicate that CCTH (Cryptophyta, Centrohelida, Telonemia, and Haptophyta) is not monophyletic.
Five taxa (Leishmania, Trypanosoma, Sawyeria, Entamoeba, and Breviata) were removed.
Two Malawimonas taxa were removed.
Two Malawimonas taxa and five taxa (Leishmania, Trypanosoma, Sawyeria, Entamoeba, and Breviata) were removed.
Removal of the 20% fastest evolving sites from the alignment.
The capital letters correspond to supergroups marked in figure 3.
Breviata is sister to Opisthokonta (fig 3).
FIG. 3.
Phylogenomic position of Collodictyon inferred from 124 genes under the CAT mixture model in PhyloBayes v3.2. Branches that received 1.00 PP are marked by filled circles. The branch length of Entamoeba is shortened by 50% to save space. (A) Tree topology constructed with 79 taxa from the saved 18,000 trees after discarding the first 6,000 cycles as burn-in (maxdiff = 0.137). Missing data for each taxon is shown as a color barplot (left bar: missing number of genes; right bar: missing percentage of characters). Bars marked by “*” indicate the missing percentage of characters is over 60% of the full-length alignment. (B) Tree topology constructed with 77 taxa (i.e., two Malawimonas excluded) from the saved 16,000 trees after discarding first 8,000 cycles as burn-in (maxdiff = 0.083). CCTH is the abbreviation of Cryptophyta, Centrohelida, Telonemia, and Haptophyta. Additional statistical support values for the main nodes in the tree marked by capital letters in boxes are listed in table 1.
To test whether the deep position of Collodictyon was stable or instead sensitive to taxonomic sampling, we performed several taxon removal experiments, but Collodictyon was consistently recovered in the same position. Most interestingly, the position of Collodictyon in the global eukaryote phylogeny remained identical when Malawimonas was removed from our alignment (fig. 3B and supplementary fig. S1B, Supplementary Material online). It was still placed close to the split between unikonts and bikonts, suggesting that this position was not caused by erroneous attraction to Malawimonas or other Excavata species (i.e., Trimastix; see supplementary fig. S2, Supplementary Material online). The high statistical support for the bikont group recovered with this reduced data set strongly excluded Collodictyon from being member of this assemblage (bikonts: BP = 98% and PP = 1.00). On the other hand, removing Malawimonas lowered the bootstrap support for the unikonts (BP = 57% and PP = 0.99; table 1), pointing to a possible attraction between Collodictyon and this other major group. In order to evaluate the potential impact of missing data on the position of Collodictyon, we removed taxa with more than 60% missing characters (fig. 3A). The phylogenies inferred from this data set showed Collodictyon in the same position, which indicated that taxa with low sequence coverage did not affect the construction of Collodictyon phylogeny (supplementary figs. S3 and S4, Supplementary Material online). Finally, we tested the possibility of Collodictyon branching within unikonts or bikonts using similar taxonomic sampling as reported by Hampl et al. 2009 and Rodriguez-Ezpeleta et al. 2007 (i.e., Leishmania, Trypanosoma, Sawyeria, Entamoeba, and Breviata removed). Again, no alternative position was observed for Collodictyon (see table 1 and supplementary fig. S5, Supplementary Material online).
All phylogenetic analyses described above were done based on a “concatenated model,” without considering the evolutionary tempo and mode of each protein composing the concatenated alignment. We therefore assessed the impact of using a “separate model” that takes into account the evolutionary specificity of each gene (see supplementary materials and methods, Supplementary Material online). The topologies inferred from the separate model again recovered Collodictyon in the same position near the bifurcation of unikonts and bikonts, either alone or as sister to Malawimonas (supplementary fig. S1 and S5, Supplementary Material online). Furthermore, the separate model generated similar bootstrap support values as the concatenated model (see supplementary table S1, Supplementary Material online), altogether demonstrating that the phylogenetic position of Collodictyon is not an artifact caused by oversimplification of the concatenated model.
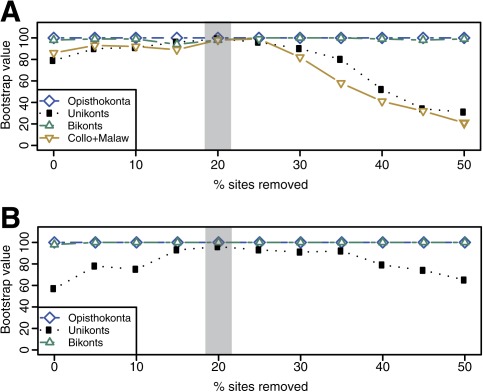
To further investigate the evolutionary origin of Collodictyon, we attempted to increase the phylogenetic versus nonphylogenetic signal ratio by removing the fastest evolving sites, which have been shown to bear the highest degree of homoplasy (Brinkmann and Philippe 1999). Because our analyses suggested that Collodictyon is excluded from the known eukaryote supergroups, we successively monitored the statistical support for unikonts and bikonts. Most notably, the bootstrap support for unikonts increased as the fastest evolving sites were removed, reaching a peak value of 96% after removing 20% of sites (table 1 and fig. 4B), whereas the bikonts remained highly supported (BP > 95%) during this experiment. Moreover, a Bayesian phylogeny constructed with the alignment removing the 20% fastest evolving sites showed strong evidence for excluding Collodictyon from unikonts (PP = 1.00; CAT-BP = 93%) or bikonts (PP = 1.00; CAT-BP = 100%) (fig. 5 and table 1). Cross-validation test showed that the CAT model fits our data better than the LG model with a score averaged over 10 replicates of 2451.36 ± 132.9 (all replicates favored the “CAT” model). The global phylogeny inferred from the CAT model should be favored, although both models recovered the same position of Collodictyon (fig. 5B and supplementary fig. S6B, Supplementary Material online). Hence, after the removal of the noisiest positions in our alignment, Collodictyon was robustly placed close to the bifurcation of unikonts and bikonts.
Fig. 4.
Changes in bootstrap support for key nodes in the inferred trees as fast-evolving sites were removed. Site rates were estimated from an alignment without two Malawimonas and Collodictyon species (76 taxa). Sites were then removed in 5% increments from alignments consisting of (A) 79 taxa (including Collodictyon and Malawimonas) and (B) 77 taxa (including Collodictyon). ML Bootstrap values (BP) for Collodictyon + Malawimonas, unikonts, bikonts, and Opisthokonta (used as a reference) were calculated under the PROTCATLGF model in RAxML v7.2.6. BP values shaded by gray rectangles are listed in table 1 and supplementary figure S6 (Supplementary Material online).
Fig. 5.
Bayesian phylogeny of Collodictyon constructed from 124 genes after removal of the fastest evolving sites. The consensus topology was calculated under the CAT model from 18,000 saved trees after discarding the first 6,000 cycles as burn-in. Branches showing 1.00 PP are marked by filled circles. The branch length of Entamoeba is shortened by 50% to save space. (A) Tree topology inferred from the trimmed alignment with the 20% fastest evolving sites removed (marked by gray rectangles in fig. 4A). Chains were considered to have converged (maxdiff = 0.104). (B) Tree topology inferred from the trimmed alignment (i.e., two Malawimonas excluded) with the 20% fastest evolving sites removed (marked by gray rectangles in fig. 4B). Chains were considered to have converged (maxdiff = 0.065). Numbers at the nodes in (B) indicate PP/bootstrap values calculated from from 100 pseudoreplicates with Phylobayes under CAT mixture model. Dashes “-” indicate bootstrap supports < 50%. CCTH is the abbreviation of Cryptophyta, Centrohelida, Telonemia, and Haptophyta. Additional statistical support values for the supergroups are shown in table 1.
Consistent with the phylogenetic analyses mentioned above, the AU test based on the data set without the 20% fastest evolving sites rejected topologies where Collodictyon was placed within unikonts or bikonts. The same results hold true for the bikonts when the full-length alignment was used, but the possibility of Collodictyon branching within unikonts, that is, sister to Amoebozoa (P = 0.372) or Opisthokonta (P = 0.076), could not be discarded at the 5% level of significance (table 2). These two alternative trees were evaluated by comparing with the optimal likelihood topology (supplementary fig. S1B, Supplementary Material online) under a covarion model in ProCov (Wang et al. 2009). The alternative topologies obtained substantially lower likelihood values (ΔlnL = −31 and ΔlnL = −15) than the optimal topology. Nevertheless, in order to examine other possible affinities of Collodictyon within Amoebozoa or Opisthokonta, 24 topologies where Collodictyon branched with basal lineages of unikonts were compared. Strikingly, all of them were rejected (P < 0.05), thus weakening the suspicion of a closer relationship between Collodictyon and unikonts (supplementary fig. S7, Supplementary Material online).
Table 2.
AU Test of Tree Topologies.
|
P Valueb |
|||
| Rank | Tree Topology Based on a Sample of 79 Taxaa | Allc | 20%d |
| 1 | (((Opst,Amoe),(Mala,Coll)),(Exca,(Plan,(SAR,((Cryp,Hapt),TelRap))))) | 0.982 | 0.995 |
| 2 | (((Opst,Amoe),Coll),((Exca,Mala),(Plan,(SAR,((Cryp,Hapt),TelRap))))) | 0.064 | 0.045 |
| 3 | (((Amoe,(Mala,Coll)),Opst),(Exca,(Plan,(SAR,((Cryp,Hapt),TelRap))))) | 0.067 | 0.006 |
| 4 | (((Opst,(Mala,Coll)),Amoe),(Exca,(Plan,(SAR,((Cryp,Hapt),TelRap))))) | 0.009 | 0.005 |
| 5 | (((Opst,Mala),(Coll,Amoe)),(Exca,(Plan,(SAR,((Cryp,Hapt),TelRap))))) | 0.007 | 0.010 |
| 6 | (((Opst,Coll),(Mala,Amoe)),(Exca,(Plan,(SAR,((Cryp,Hapt),TelRap))))) | 0.006 | 0.006 |
| 7 | ((((Opst,Amoe),Coll),Mala),(Exca,(Plan,(SAR,((Cryp,Hapt),TelRap))))) | 0.005 | 0.014 |
| 8 | (((Opst,(Coll,Amoe)),Mala),(Exca,(Plan,(SAR,((Cryp,Hapt),TelRap))))) | 0.003 | 2 × 10−04 |
| 9 | ((((Opst,Coll),Amoe),Mala),(Exca,(Plan,(SAR,((Cryp,Hapt),TelRap))))) | 0.002 | 0.001 |
| 10 | (((Opst,Amoe),Mala),((Exca,Coll),(Plan,(SAR,((Cryp,Hapt),TelRap))))) | 3 × 10−04 | 7 × 10−05 |
| 11 | (((Opst,Amoe),Mala),(Exca,((Plan,Coll),(SAR,((Cryp,Hapt),TelRap))))) | 2 × 10−04 | 8 × 10−05 |
| 12 | (((Opst,Amoe),Mala),(Exca,(Plan,(SAR,((Cryp,Hapt),(Coll,TelRap)))))) | 1 × 10−04 | 0.001 |
| 13 | (((Opst,Amoe),Mala),(Exca,(Plan,(SAR,(((Cryp,Coll),Hapt),TelRap))))) | 8 × 10−05 | 8 × 10−06 |
| 14 | (((Opst,Amoe),Mala),(Exca,(Plan,(SAR,((Cryp,(Coll,Hapt)),TelRap))))) | 2 × 10−05 | 6 × 10−07 |
| 15 | ((((Opst,Amoe),Coll),Mala),(Exca,(Plan,(SAR,((Cryp,Hapt),TelRap))))) | 1 × 10−05 | 2 × 10−07 |
| 16 | ((Opst,Amoe),((Exca,(Mala,Coll)),(Plan,(SAR,((Cryp,Hapt),TelRap))))) | 6 × 10−15 | 5 × 10−14 |
| 17 | ((Opst,Amoe),((Exca,Mala),Coll)),(Plan,(SAR,((Cryp,Hapt),TelRap))))) | 4 × 10−12 | 3 × 10−11 |
| P Valueb | |||
| Rank | Tree Topology Based on a Sample of 77 Taxaa (i.e., two Malawimonas excluded) | Allc | 20%e |
| 1 | (((Opst,Amoe),Coll),(Exca,(Plan,(SAR,((Cryp,Hapt),TelRap))))) | 0.532 | 0.614 |
| 2 | (((Opst,Amoe),Coll),(Exca,(SAR,(Plan,((Cryp,Hapt),TelRap))))) | 0.630 | 0.529 |
| 3 | (((Opst,Coll),Amoe),(Exca,(Plan,(SAR,((Cryp,Hapt),TelRap))))) | 0.071 | 0.046 |
| 4 | (((Opst,Coll),Amoe),(Exca,(SAR,(Plan,((Cryp,Hapt),TelRap))))) | 0.076 | 0.045 |
| 5 | ((Opst,(Amoe,Coll)),(Exca,(Plan,(SAR,((Cryp,Hapt),TelRap)))) | 0.284 | 0.040 |
| 6 | ((Opst,(Amoe,Coll)),(Exca,(SAR,(Plan,((Cryp,Hapt),TelRap)))) | 0.372 | 0.037 |
| 7 | ((Opst,Amoe),((Exca,Coll),(Plan,(SAR,((Cryp,Hapt),TelRap))))) | 0.001 | 0.001 |
| 8 | ((Opst,Amoe),((Exca,Coll),(SAR,(Plan,((Cryp,Hapt),TelRap))))) | 0.003 | 2 × 10−04 |
| 9 | ((Opst,Amoe),(Exca,((Plan,Coll),(SAR,((Cryp,Hapt),TelRap))))) | 2 × 10−06 | 4 × 10−07 |
| 10 | ((Opst,Amoe),(Exca,((SAR,Coll),(Plan,((Cryp,Hapt),TelRap))))) | 7 × 10−06 | 3 × 10−08 |
| 11 | ((Opst,Amoe),(Exca,(Plan,(SAR,(((Cryp,Coll),Hapt),TelRap))))) | 6 × 10−07 | 1 × 10−04 |
| 12 | ((Opst,Amoe),(Exca,(SAR,(Plan,(((Cryp,Coll),Hapt),TelRap))))) | 2 × 10−05 | 6 × 10−05 |
| 13 | ((Opst,Amoe),(Exca,(Plan,(SAR,((Cryp,(Coll,Hapt)),TelRap))))) | 6 × 10−09 | 7 × 10−39 |
| 14 | ((Opst,Amoe),(Exca,(SAR,(Plan,((Cryp,(Coll,Hapt)),TelRap))))) | 8 × 10−05 | 2 × 10−47 |
| 15 | ((Opst,Amoe),(Exca,(Plan,(SAR,((Cryp,Hapt),(Coll,TelRap)))))) | 1 × 10−74 | 5 × 10−51 |
| 16 | ((Opst,Amoe),(Exca,(SAR,(Plan,((Cryp,Hapt),(Coll,TelRap)))))) | 1 × 10−69 | 8 × 10−54 |
| 17 | ((Opst,Amoe),(Exca,(Plan,((SAR,Coll),((Cryp,Hapt),TelRap))))) | 2 × 10−63 | 7 × 10−40 |
| 18 | ((Opst,Amoe),(Exca,(SAR,((Plan,Coll),((Cryp,Hapt),TelRap))))) | 2 × 10−51 | 2 × 10−43 |
The abbreviation of major groups: Opst, Opisthokonta; Amoe, Amoebozoa; Exca, Excavata; Plan, Archaeplastida; SAR, Stramenopila + Alveolata + Rhizaria; Cryp, Guillardia + Plagioselmis; Hapt, Haptophyta; TelRap, Telonemia + Raphidiophrys; Mala, Malawimonas; and Coll, Collodictyon.
P values in which the topologies cannot be rejected at the 5% level of significance were underlined.
P values were calculated from the original alignment (i.e., no sites removed).
P values were calculated from the trimmed alignment with removal of the 20% fastest evolving sites (marked by gray rectangles in fig. 4A).
P values were calculated from the trimmed alignment (i.e., two Malawimonas excluded) with removal of the 20% fastest evolving sites (marked by gray rectangles in fig. 4B).
Relationship between Collodictyon and Malawimonas
Malawimonas has proven to be particularly challenging to place in the eukaryote tree, even with very large alignments, but it has typically been associated with Excavata based on its ultrastructure (Simpson 2003). In our analyses, Malawimonas generally branched outside of Excavata (fig. 3A, supplementary figs. S1A and S3A and S3C, Supplementary Material online), in agreement with previous observations (Rodriguez-Ezpeleta et al. 2007; Hampl et al. 2009). Because Malawimonas grouped with Collodictyon and not with Excavata in our Bayesian and ML trees, we took a closer look at this relationship by applying several strategies. One model violation that is known to cause tree reconstruction artifacts is bias in the amino acid (AA) composition. Interestingly, our heatmap analyses showed a weak deviation from amino acid homogeneity that could partially account for the grouping of Collodictyon and Malawimonas, together with a few other taxa (supplementary fig. S8 and table S3, Supplementary Material online). Removing up to 20% of the fastest evolving sites seemed not to overcome the amino acid compositional bias (supplementary fig. S8, Supplementary Material online). However, recoding the amino acids into functional categories (Hrdy et al. 2004) still recovered the grouping of Malawimonas and Collodictyon (supplementary fig. S9, Supplementary Material online), suggesting that the bias may not significantly affect the phylogeny.
Despite this apparent close relationship between them, it is important to note that the Bayesian tree inferred under the better fitted CAT model from the alignment after removing the 20% fastest evolving sites only weakly recovered Collodictyon and Malawimonas as a group (PP = 0.63; fig. 5A and table 1). Moreover, when Collodictyon and five other taxa (i.e., Leishmania, Trypanosoma, Sawyeria, Entamoeba, and Breviata) were removed from the data set, Malawimonas grouped as sister to Excavata in our ML tree (BP = 60%; supplementary fig. S5B, Supplementary Material online), in agreement with recent examination of the Excavata phylogeny (Rodriguez-Ezpeleta et al. 2007; Hampl et al. 2009). In addition, the alternative position of Malawimonas within Excavata was not rejected by the AU test (P = 0.064; table 2), altogether suggesting that the position of Malawimonas was not stable and highly sensitive to taxonomic sampling. Hence, although the grouping of Collodictyon and Malawimonas remains unclear after our analyses, the unstable position of Malawimonas and low support in Bayesian analyses applying the CAT model indicates that these two lineages may belong to different groups of eukaryotes.
Collodictyon Is Placed Near the “Unikont–Bikont” Bifurcation
Our phylogenetic inferences suggest that Collodictyon diverged near the unikont—bikont bifurcation. Although the root of the eukaryote tree is controversial and no clear evidence exists for its position, a lineage that is not included within either unikonts or bikonts is likely of early origin. The poor diversity of known Diphyllatia (Collodictyon and Diphylleia) is striking in this respect as one would expect to find more related lineages along its branch, but it remains to see if Diphyllatia in fact represent a larger group: they could be closely related to other groups that are yet to be sequenced or discovered. Regardless of these possible sister groups, interpretations of the evolutionary origin of Collodictyon are largely dependent on the position of the root of the eukaryote tree.
Two rare genomic changes have suggested an ancient split between the unikonts and bikonts; the bikonts have been shown to share a fusion of the dihydrofolate reductase (DHFR) and thymidylate synthase (TS) genes, whereas all unikonts appear to have a unique glycine insertion to myosin class II paralogues (Stechmann and Cavalier-Smith 2002; Richards and Cavalier-Smith 2005). At face value, investigating these characters in Collodictyon should be very informative. However, the bikont species Amastigomonas, bearing the fused DHFR-TS genes, is unexpectedly placed within unikonts (Kim et al. 2006; Derelle and Lang 2011), a result also recovered by our 18S + 28S rDNA tree (fig. 2). This seriously questioned the validity of this genomic marker as a synapomorphy for the bikonts (Roger and Simpson 2009). Nevertheless, we identified a fragment of the DHFR gene in our cDNA library and extended it by 3′ RACE. Annotation of the sequence by searches against the Pfam database revealed a fused TS and DHFR domain. The obtained sequence was furthermore confirmed to be from Collodictyon and not the cryptomonad prey by both successful amplification and sequencing of the gene from the culture grown with green algal prey (Chlorella) and phylogenetic analysis of the DHFR domain (for details, see supplementary fig. S10, Supplementary Material online). In contrast, the myosin class II synapomorphy for unikonts could not be found within our cDNA data set. The broad distribution of the fused DHFR-TS gene within bikonts and its presence in Collodictyon might indicate that Collodictyon is more closely related to bikonts than unikonts. On the other hand, if the eukaryote root falls instead within bikonts, as it was recently proposed (Rogozin et al. 2009; Cavalier-Smith 2010), Collodictyon would then branch as a sister lineage to Amoebozoa and Opisthokonta. Regardless of the position of the root, the phylogeny shows that Collodictyon is an early diverging lineage and therefore useful for inferring the evolution of eukaryote morphology. Features of Collodictyon, such as the ventral feeding groove and the ability to form broad and thin pseudopods from the ventral groove resemble defining features of the Excavata and Amoebozoa. The question is whether these structures are homologous to those in Collodictyon, in which case Collodictyon has a unique combination of ancient morphological characteristics.
Conclusion
Collodictyon is one of the few remaining species that have had no clear affiliation in the eukaryote tree of life (Brugerolle et al. 2002; Shalchian-Tabrizi et al. 2006; Roger and Simpson 2009). Our results suggest that Collodictyon, together with Diphylleia, belongs to a distinct branch that originated very early in the evolution of eukaryotes. Apusozoa seems not to be closely related to Collodictyon but rather belong to two different lineages among unikonts (see also Derelle and Lang 2011). Further attention to this and other enigmatic lineages such as Palpitomonas (Yabuki et al. 2010) as well as short branching Amoebozoa and Excavata will help clarify the relationships at the base of the eukaryote tree. Another major question that remains to be addressed is how large the diversity of the Diphyllatia subphylum is. Strikingly, only one Collodictyon-like sequence could be identified from all environmental sequences in public databases, showing that the diversity in this ancient group needs further exploration.
Supplementary Material
Supplementary figures S1–S10, tables S1–S4, and materials and methods are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
We thank the 454-sequencing lab and Bioportal at University of Oslo (UiO) for sequencing and bioinformatics services. We thank John M. Archibald for granting access to the Guillardia theta genome sequences generated by the Joint Genome Institute (JGI). We would also like to thank Mark van der Giezen and Jeff Silbermann for allowing us to use expressed sequence tag sequences of Breviata. In addition, Anders K. Krabberød gave us a great help to validate Collodictyon DHFR-TS sequences. F.B. is currently supported by a prospective researcher postdoctoral fellowship from the Swiss National Science Foundation and by a grant to the Centre for Microbial Diversity and Evolution from Tula Foundation. P.J.K. is a fellow of the Canadian Institute of Advanced Research. This work has been supported by research grants from the University of Oslo to K.S.-T. and D.K. as well PhD fellowship for S.Z. and J.B.
References
- Baurain D, Brinkmann H, Petersen J, Rodriguez-Ezpeleta N, Stechmann A, Demoulin V, Roger AJ, Burger G, Lang BF, Philippe H. Phylogenomic evidence for separate acquisition of plastids in cryptophytes, haptophytes, and stramenopiles. Mol Biol Evol. 2010;27:1698–1709. doi: 10.1093/molbev/msq059. [DOI] [PubMed] [Google Scholar]
- Brinkmann H, Philippe H. Archaea sister group of Bacteria? Indications from tree reconstruction artifacts in ancient phylogenies. Mol Biol Evol. 1999;16:817–825. doi: 10.1093/oxfordjournals.molbev.a026166. [DOI] [PubMed] [Google Scholar]
- Brugerolle G, Bricheux G, Philippe H, Coffe G. Collodictyon triciliatum and Diphylleia rotans (=Aulacomonas submarina) form a new family of flagellates (Collodictyonidae) with tubular mitochondrial cristae that is phylogenetically distant from other flagellate groups. Protist. 2002;153:59–70. doi: 10.1078/1434-4610-00083. [DOI] [PubMed] [Google Scholar]
- Burki F, Inagaki Y, Bråte J, et al. (14 co-authors) Large-scale phylogenomic analyses reveal that two enigmatic protist lineages, Telonemia and Centroheliozoa, are related to photosynthetic chromalveolates. Genome Biol Evol. 2009;1:231–238. doi: 10.1093/gbe/evp022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burki F, Kudryavtsev A, Matz MV, Aglyamova GV, Bulman S, Fiers M, Keeling PJ, Pawlowski J. Evolution of Rhizaria: new insights from phylogenomic analysis of uncultivated protists. BMC Evol Biol. 2010;10:377. doi: 10.1186/1471-2148-10-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burki F, Shalchian-Tabrizi K, Minge M, Skjaeveland A, Nikolaev SI, Jakobsen KS, Pawlowski J. Phylogenomics reshuffles the eukaryotic supergroups. PLoS One. 2007;2:e790. doi: 10.1371/journal.pone.0000790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burki F, Shalchian-Tabrizi K, Pawlowski J. Phylogenomics reveals a new ‘megagroup’ including most photosynthetic eukaryotes. Biol Lett. 2008;4:366–369. doi: 10.1098/rsbl.2008.0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter HJ. On the fresh- and salt-water Rhizopoda of England and India. Ann Mag Nat Hist. 1865;15:227–293. [Google Scholar]
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. Kingdom protozoa and its 18 phyla. Microbiol Rev. 1993;57:953–994. doi: 10.1128/mr.57.4.953-994.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T. The excavate protozoan phyla Metamonada Grasse emend. (Anaeromonadea, Parabasalia, Carpediemonas, Eopharyngia) and Loukozoa emend. (Jakobea, Malawimonas): their evolutionary affinities and new higher taxa. Int J Syst Evol Microbiol. 2003;53:1741–1758. doi: 10.1099/ijs.0.02548-0. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. Kingdoms Protozoa and Chromista and the eozoan root of the eukaryotic tree. Biol Lett. 2010;6:342–345. doi: 10.1098/rsbl.2009.0948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics. 2011;27:1164–1165. doi: 10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derelle R, Lang BF. doi: 10.1093/molbev/msr295. 2011. Rooting the eukaryotic tree with mitochondrial and bacterial proteins. Mol Biol Evol. Advance Access published December 1, 2011. doi:10.1093/molbev/msr295. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP (phylogeny inference package). 3.6. 2001. Distributed by the author. Seattle (WA): Department of Genetics, University of Washington. [Google Scholar]
- Gubler U, Hoffman BJ. A simple and very efficient method for generating cDNA libraries. Gene. 1983;25:263–269. doi: 10.1016/0378-1119(83)90230-5. [DOI] [PubMed] [Google Scholar]
- Guillard RRL, Lorenzen CJ. Yellow-green algae with chlorophyllide c. J Phycol. 1972;8:10–14. [Google Scholar]
- Hampl V, Hug LA, Leigh JW, Dacks JB, Lang BF, Simpson AGB, Roger AJ. Phylogenetic analyses support the monophyly of Excavata and resolve relationships among eukaryotic “supergroups”. Proc Natl Acad Sci U S A. 2009;106:3859–3864. doi: 10.1073/pnas.0807880106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrdy I, Hirt RP, Dolezal P, Bardonova L, Foster PG, Tachezy J, Embley TM. Trichomonas hydrogenosomes contain the NADH dehydrogenase module of mitochondrial complex I. Nature. 2004;432:618–622. doi: 10.1038/nature03149. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Harrison JW, Sudek S, Jones MD, Wilcox HM, Richards TA, Worden AZ, Archibald JM. Newly identified and diverse plastid-bearing branch on the eukaryotic tree of life. Proc Natl Acad Sci U S A. 2011;108:1496–1500. doi: 10.1073/pnas.1013337108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Simpson AG, Graham LE. Evolutionary relationships of apusomonads inferred from taxon-rich analyses of 6 nuclear encoded genes. Mol Biol Evol. 2006;23:2455–2466. doi: 10.1093/molbev/msl120. [DOI] [PubMed] [Google Scholar]
- Klaveness D. Collodictyon triciliatum H.J. Carter (1865)—a common but fixation-sensitive algivorous flagellate from the limnopelagial. Nord J Freshw Res. 1995;70:3–11. [Google Scholar]
- Kumar S, Skjaeveland A, Orr RJ, Enger P, Ruden T, Mevik BH, Burki F, Botnen A, Shalchian-Tabrizi K. AIR: a batch-oriented web program package for construction of supermatrices ready for phylogenomic analyses. BMC Bioinformatics. 2009;10:357. doi: 10.1186/1471-2105-10-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lartillot N, Philippe H. A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol Biol Evol. 2004;21:1095–1109. doi: 10.1093/molbev/msh112. [DOI] [PubMed] [Google Scholar]
- Margulies M, Egholm M, Altman WE, et al. (56 co-authors) Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minge MA, Silberman JD, Orr RJ, Cavalier-Smith T, Shalchian-Tabrizi K, Burki F, Skjaeveland A, Jakobsen KS. Evolutionary position of breviate amoebae and the primary eukaryote divergence. Proc R Soc B Biol Sci. 2009;276:597–604. doi: 10.1098/rspb.2008.1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto N, Chantangsi C, Horak A, Leander BS, Keeling PJ. Molecular phylogeny and description of the novel katablepharid Roombia truncata gen. et sp. nov., and establishment of the Hacrobia taxon nov. PLoS One. 2009;4:e7080. doi: 10.1371/journal.pone.0007080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parfrey LW, Barbero E, Lasser E, Dunthorn M, Bhattacharya D, Patterson DJ, Katz LA. Evaluating support for the current classification of eukaryotic diversity. PLoS Genet. 2006;2:e220. doi: 10.1371/journal.pgen.0020220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parfrey LW, Grant J, Tekle YI, Lasek-Nesselquist E, Morrison HG, Sogin ML, Patterson DJ, Katz LA. Broadly sampled multigene analyses yield a well-resolved eukaryotic tree of life. Syst Biol. 2010;59:518–533. doi: 10.1093/sysbio/syq037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patron NJ, Inagaki Y, Keeling PJ. Multiple gene phylogenies support the monophyly of cryptomonad and haptophyte host lineages. Curr Biol. 2007;17:887–891. doi: 10.1016/j.cub.2007.03.069. [DOI] [PubMed] [Google Scholar]
- Patterson DJ. The diversity of eukaryotes. Am Nat. 1999;154:S96–S124. doi: 10.1086/303287. [DOI] [PubMed] [Google Scholar]
- Rhodes RC. Binary fission in Collodictyon triciliatum carter. The faculty of the college of letters and science. Berkeley (CA): University of California; 1917. p. 74. [Google Scholar]
- Richards TA, Cavalier-Smith T. Myosin domain evolution and the primary divergence of eukaryotes. Nature. 2005;436:1113–1118. doi: 10.1038/nature03949. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Ezpeleta N, Brinkmann H, Burger G, Roger AJ, Gray MW, Philippe H, Lang BF. Toward resolving the eukaryotic tree: the phylogenetic positions of jakobids and cercozoans. Curr Biol. 2007;17:1420–1425. doi: 10.1016/j.cub.2007.07.036. [DOI] [PubMed] [Google Scholar]
- Roger AJ, Simpson AG. Evolution: revisiting the root of the eukaryote tree. Curr Biol. 2009;19:R165–R167. doi: 10.1016/j.cub.2008.12.032. [DOI] [PubMed] [Google Scholar]
- Rogozin IB, Basu MK, Csuros M, Koonin EV. Analysis of rare genomic changes does not support the unikont-bikont phylogeny and suggests cyanobacterial symbiosis as the point of primary radiation of eukaryotes. Genome Biol Evol. 2009;1:99–113. doi: 10.1093/gbe/evp011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roure B, Rodriguez-Ezpeleta N, Philippe H. SCaFoS: a tool for selection, concatenation and fusion of sequences for phylogenomics. BMC Evol Biol. 2007;7(Suppl 1):S2. doi: 10.1186/1471-2148-7-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalchian-Tabrizi K, Bråte J, Logares R, Klaveness D, Berney C, Jakobsen KS. Diversification of unicellular eukaryotes: cryptomonad colonizations of marine and fresh waters inferred from revised 18S rRNA phylogeny. Environ Microbiol. 2008;10:2635–2644. doi: 10.1111/j.1462-2920.2008.01685.x. [DOI] [PubMed] [Google Scholar]
- Shalchian-Tabrizi K, Eikrem W, Klaveness D, et al. (12 co-authors) Telonemia, a new protist phylum with affinity to chromist lineages. Proc R Soc B Biol Sci. 2006;273:1833–1842. doi: 10.1098/rspb.2006.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalchian-Tabrizi K, Minge MA, Espelund M, Orr R, Ruden T, Jakobsen KS, Cavalier-Smith T. Multigene phylogeny of choanozoa and the origin of animals. PLoS One. 2008;3:e2098. doi: 10.1371/journal.pone.0002098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimodaira H. An approximately unbiased test of phylogenetic tree selection. Syst Biol. 2002;51:492–508. doi: 10.1080/10635150290069913. [DOI] [PubMed] [Google Scholar]
- Shimodaira H, Hasegawa M. CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinformatics. 2001;17:1246–1247. doi: 10.1093/bioinformatics/17.12.1246. [DOI] [PubMed] [Google Scholar]
- Simpson AGB. Cytoskeletal organization, phylogenetic affinities and systematics in the contentious taxon Excavata (Eukaryota) Int J Syst Evol Microbiol. 2003;53:1759–1777. doi: 10.1099/ijs.0.02578-0. [DOI] [PubMed] [Google Scholar]
- Simpson AGB, Roger AJ. The real ‘kingdoms’ of eukaryotes. Curr Biol. 2004;14:R693–R696. doi: 10.1016/j.cub.2004.08.038. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML Web servers. Syst Biol. 2008;57:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- Stechmann A, Cavalier-Smith T. Rooting the eukaryote tree by using a derived gene fusion. Science. 2002;297:89–91. doi: 10.1126/science.1071196. [DOI] [PubMed] [Google Scholar]
- Wang HC, Susko E, Roger AJ. PROCOV: maximum likelihood estimation of protein phylogeny under covarion models and site-specific covarion pattern analysis. BMC Evol Biol. 2009;9:225. doi: 10.1186/1471-2148-9-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabuki A, Inagaki Y, Ishida K. Palpitomonas bilix gen. et sp. nov.: a novel deep-branching heterotroph possibly related to Archaeplastida or Hacrobia. Protist. 2010;161:523–538. doi: 10.1016/j.protis.2010.03.001. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Yoon HS, Grant J, Tekle YI, Wu M, Chaon BC, Cole JC, Logsdon JM, Jr, Patterson DJ, Bhattacharya D, Katz LA. Broadly sampled multigene trees of eukaryotes. BMC Evol Biol. 2008;8:14. doi: 10.1186/1471-2148-8-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon HS, Price DC, Stepanauskas R, Rajah VD, Sieracki ME, Wilson WH, Yang EC, Duffy S, Bhattacharya D. Single-cell genomics reveals organismal interactions in uncultivated marine protists. Science. 2011;332:714–717. doi: 10.1126/science.1203163. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.