Abstract
Postnatal intestinal ontogenesis in an animal model of diabesity may recapitulate morphological and transduction features of diabesity-induced intestinal dysplasia and its amelioration by endogenous (n-3) polyunsaturated fatty acids (PUFA). Proliferation, differentiation, and transduction aspects of intestinal ontogenesis have been studied here in obese, insulin-resistant db/db mice, in fat-1 transgene coding for desaturation of (n-6) PUFA into (n-3) PUFA, in db/db crossed with fat-1 mice, and in control mice. Diabesity resulted in increased colonic proliferation and dedifferentiation of epithelial colonocytes and goblet cells, with increased colonic β-catenin and hepatocyte nuclear factor (HNF)-4α transcriptional activities accompanied by enrichment in HNF-4α–bound (n-6) PUFA. In contrast, in fat-1 mice, colonic proliferation was restrained, accompanied by differentiation of crypt stem cells into epithelial colonocytes and goblet cells and by decrease in colonic β-catenin and HNF-4α transcriptional activities, with concomitant enrichment in HNF-4α-bound (n-3) PUFA at the expense of (n-6) PUFA. Colonic proliferation and differentiation, the profile of β-catenin and HNF-4α-responsive genes, and the composition of HNF-4α-bound PUFA of db/db mice reverted to wild-type by introducing the fat-1 gene into the db/db context. Suppression of intestinal HNF-4α activity by (n-3) PUFA may ameliorate diabesity-induced intestinal ontogenesis and offer an effective preventive modality for colorectal cancer.
Keywords: obesity, diabetes, epithelial cell growth, epithelial cell differentiation, colorectal neoplasia
The colon consists of a self-renewing epithelium that turns over every three to five days. Thus, slowly dividing, stationary, pluripotent stem cells at the base of the crypt give rise to rapidly proliferating cells that differentiate into postmitotic columnar absorptive colonocytes, mucin-secreting goblet cells, and enteroendocrine cells as they migrate from the crypt base to the surface, followed finally by their shedding into the intestinal lumen (reviewed in Ref. 1). Several signaling pathways, notably β-catenin/Tcf and hepatocyte nuclear factor (HNF)-4α, modulate and control the proliferation and differentiation of adult colonocytes, whereas their perturbation via mutation or epigenetic modification may ultimately result in colorectal cancer (CRC). Moreover, cell dedifferentiation correlates with key tumor features, such as tumor progression rates, invasiveness, drug resistance, and metastatic potential (2). Hence, perturbation of programming commitment during ontogenesis may mimic primary features of intestinal oncogenesis. In contrast to humans, major intestinal maturation processes of rodents take place only following birth (3), allowing for searching intestinal ontogenesis as a function of variable conditions reported to promote or suppress CRC.
The metabolic syndrome (MetS) and its individual diseases (upper-body obesity, type 2 diabetes, dyslipidemia, and hypertension) are associated with an increased risk for CRC, and CRC incidence seems to be associated with the number of MetS components present at baseline (reviewed in Refs. 4, 5). Thus, the relative risk (RR) of CRC increases 2-fold with increase in waist circumference of MetS subjects (6), whereas the age- and sex-adjusted RR of CRC for an hemoglobin (Hb)A1c ≥ 7% amounts to 2.94, compared with 1.13 for HbA1c of 5.0–5.9%. Overall, individuals with MetS have a 75% increased risk for CRC, after controlling for age, race, gender, smoking, and alcohol use.
Based on epidemiological and experimental evidence, total fat consumption and in particular dietary fatty acid composition may modulate colon tumorigenesis (7). Thus, saturated fat promotes CRC, whereas (n-3) polyunsaturated fatty acids (PUFA) are protective. Beneficial effects of (n-3) PUFA in CRC have been proposed to reflect the following: i) synthesis of less inflammatory 3-series prostaglandins/thromboxanes and 5-series leukotrienes derived from (n-3) PUFA, at the expense of 2-series prostaglandins/thromboxanes and 4-series leukotrienes derived from (n-6) PUFA (reviewed in Ref. 8); ii) modulation of membrane fluidity by enrichment with (n-3) PUFA, resulting in modulating receptor binding, cell interaction, or transduction pathways (8); iii) suppression of the transcriptional activity of HNF-4α by (n-3) PUFA acting as HNF-4α ligand antagonists (9); and iv) dephosphorylation and activation of GSK-3β by (n-3) PUFA, resulting in β-catenin degradation and disruption of the WNT pathway (10). The HNF-4α and WNT pathways seem to be tightly associated (9).
HNF-4α is a member of the super family of nuclear receptors (11) expressed in liver, intestine, pancreas, and kidney, where it is required for tissue-specific expression of many of their respective traits (12–15). HNF-4α global deletion in the mouse is lethal, resulting in defective visceral endoderm and its differentiation into functional colon (12–14, 16–18). We previously reported the transcriptional activity of HNF-4α to be modulated by the CoA-thioesters of long-chain fatty acids (LCFA) as a function of their chain length and degree of saturation (19). Thus, saturated LCFA were reported to activate (n-6) PUFA to be essentially neutral and to activate (n-3) PUFA (e.g., C20:5, C22:6, and C18:3) to act as potent suppressors of HNF-4α transcriptional activity. Indeed, HNF-4α was found to catalyze the hydrolysis of LCFA-CoA by its intrinsic thioesterase activity, followed by intramolecular channeling of the free acid product into the free acid pocket of its ligand binding domain (LBD) (20). However, binding of LCFA to endogenous HNF-4α within a physiological setup still remained to be verified.
Studying intestinal ontogenesis in an animal model for MetS as function of the (n-6)/(n-3) PUFA status may recapitulate morphological and transduction aspects of MetS-induced CRC and its amelioration by (n-3) PUFA. Morphological and transduction aspects of intestinal ontogenic programs that are significantly modulated by MetS characteristics and/or by (n-3) PUFA have been studied here using the following animal models: i) obese, hypertriglyceridemic, hyperinsulinemic, insulin-resistant BKS.Cg-Leprdb/Leprdb/OlaHsdb/db mice (BKS.Cg) mutated in the leptin receptor; ii) fat-1 transgenic mice that express the Caenorhabditis elegans desaturase (fat-1 gene), leading to the endogenous formation of n-3PUFA from n-6 PUFA and higher tissue content of n-3 PUFA compared with wild-type littermates (21); iii) first-generation of BKS.Cg mice crossed with fat-1 mice that express the fat-1 gene in the BKS.Cg db/db context (f×B); and iv) wild-type C57BL mice that serve as control and background mice for both BKS.Cg and fat-1 mice.
MATERIALS AND METHODS
Animals
Heterozygous BKS.Cg-Leprdb/Leprdb/OlaHsd (BKS.Cg) mice (Harlan) were bred to yield homozygous mice as verified by their fasting blood glucose levels (Optimum Xceed/Plus; Abbot, UK), serum leptin (Quantikine Mouse Leptin Immunoassay kit; R and D Systems, Minneapolis, MN), and insulin levels (Mercodia ultrasensitive mouse insulin ELISA kit; Uppsala, Sweden). Fat-1 breeders were bred in-house. Progeny of heterozygous fat-1 mice (fat-1) were genotyped by PCR of tail samples (see below). First-generation BKS.Cg/fat-1–crossed mice (f×B) were bred in-house and were genotyped as described. All mice were from the same genetic background (i.e., C57BL). This is the reason why we used C57BL as control mice. All mice were kept in plastic cages with wire tops in a light-temperature–controlled specific pathogen free (SPF) room, and they were maintained on commercial chow (Teklad Sterilizable Rodent Diet; Harlan). All mice were caged in the same animal facility and all were littermates. Animal care and experimental procedures were in accordance with the accredited animal ethics committee of the Hebrew University.
Analysis of (n-3) PUFA desaturase by PCR and tissue fatty acid analysis
The presence of the fat-1 gene was confirmed both by PCR genotyping (F: CCACGCATCCAATTACCAAC and R: GATGGCATTGCTTCCCAATC) and fatty acid analysis of tissue lipids. Extraction of tail and colonic fatty acids was performed as described by Kang et al. (22). Briefly, 1 cm of each mouse tail or colon was snap-frozen in liquid nitrogen. Frozen tissues were separately ground to rough powder using a crater and pestle, followed by adding 1 ml of GC-grade hexane and 1 ml of 14% boron trifluoride in methanol (BF3) reagent. The mixture was flushed with nitrogen for about 30 s, incubated for 1 h at 100°C, and then cooled on ice for 5–10 min. After adding 1 ml of H2O, the extract was vortexed and centrifuged at 18,000 g for 1 min. The upper phase, containing the methylated fatty acids, was concentrated under nitrogen. Fatty acid methyl esters (FAME) were analyzed using an Agilent gas chromatograph (7890A) equipped with a flame ionization detector (FID) and capillary column (Agilent DB-23, 60 m, 0.25 mm, 0.25 µm). Samples were injected in pulsed splitless mode (pulsed pressure of 30 psi for 0.65 min). Conditions of analysis were the following: flow of carrier gas (hydrogen), 1 ml/min; temperature of injector, 270 C; temperature of detector, 250 C; temperature of column oven, 150 C (1 min) then ramped at 5 C/min to 230 C (held for 10 min). The concentrations of eicosapentaenoic acid [EPA; (n-3) 20:5], α-linolenic acid [(n-3) 18:3]), arachidonic acid [(n-6) 20:4], and linoleic acid [LA; (n-6) 18:2] were evaluated using authentic standards (Sigma-Aldrich, Rehovot, Israel).
HNF-4alpha-bound fatty acid analysis
Nuclear extracts were prepared as described by Yuan et al. (23), with some modifications. Briefly, pooled colon samples of 2–3 mice were rinsed in PBS, homogenized in buffer A [10 mM HEPES (pH 7.8), 0.32 M sucrose, 0.3% (v/v) Triton X-100, 25 m MKCl, 0.15 mM spermine, 0.5 mM spermidine, 1 mM EGTA, 1 mM EDTA, 1 mM DTT, 0.5 mM PMSF] containing protease and phosphatase inhibitors (Sigma), and centrifuged at 3,500 g for 10 min. The nuclear pellet was washed twice in buffer A and twice in a low-salt buffer [buffer A devoid of sucrose and TritonX-100 but with added 20% (v/v) glycerol]. The washed pellet was resuspended in a high-salt buffer (low-salt buffer with 0.5 M KCl), extracted by gentle agitation for 60 min, and centrifuged at 14,000 g for 20 min. The supernatant (nuclear extract) was removed, aliquoted, snap-frozen in liquid nitrogen, and stored at −80°C till further use. Nuclear extract aliquots were immunoprecipitated with HNF-4α antibody as previously described (23). Immunoprecipitates were extracted with 150 μl methanol, 900 μl chloroform, and 1% formic acid, followed by adding 150 μl water. The extract was sonicated for 20 s, centrifuged at low speed for 15 min, and then the lower organic phase containing fatty acids was filtered through a 0.2 μ filter, dried under nitrogen, dissolved in 50 μl of isopropanol, and analyzed by LC-MS using the Accela LC system coupled to LTQ Orbitrap Discovery hybrid FT mass spectrometer (Thermo Fisher Scientific) equipped with an electrospray ionization ion source (40 C, flow at 300 µl/min, and volume injected at 2.20 µl). LC conditions consisted of variable proportions of water plus 0.05% acetic acid (solvent A) / acetonitrile plus 0.05% acetic acid (solvent B) / isopropyl alcohol (solvent C) with gradual increase in solvent C at the expense of solvent B. The mass spectrometer was operated in negative ionization mode; ion source spray voltage, 3.5 kV; capillary temperature, 250°C; capillary voltage, –35 V; disabled source fragmentation; sheath gas rate (30); and auxiliary gas rate (10). Mass spectra were acquired in the m/z 150–2,000 Da range. Data were analyzed using Xcalibur software (Thermo Fisher Scientific).
Immunohistochemistry: PAS assay and morphologic measurements
Colonic tissues were fixed and then embedded in paraffin. Sections (5 μm) on Superfrost Plus microscope slides (D-38116; Menzel, Braunschweig, Germany) were exposed to the following primary antibodies: anti-proliferating cell nuclear antigen (PCNA) (M0879; Dako, Glostrup, Denmark) (1:200 dilution); anti-E-cadherin (610181; BD Biosciences) (1:100 dilution); monoclonal mouse anti-human HNF-4α (ab41898; Abcam, Cambridge, UK) (1:200 dilution); monoclonal mouse anti-human β-catenin (1:100 dilution) (Santa Cruz Biotechnology, CA); polyclonal rabbit anti-aquaporin-8 (AQP8) (H-85; Santa Cruz Biotechnology) (1:300 dilution); and anti-MUC2 antibody (ab11197; Abcam) (1:200 dilution). The secondary antibodies used were goat anti-mouse or goat anti-rabbit (Jackson Laboratory) (1:200 dilution). PCNA and HNF-4α staining was quantified using ImageJ software (National Institutes of Health, Bethesda, MD).
Crypt depth was measured in distal colonic tissues sections (5 μm), fixed, embedded in paraffin, and stained with hematoxylin and eosin (HE). Crypts were measured using ImageJ software (National Institutes of Health) that had been calibrated with a micrometer slide image. The observer was blinded to genotype while images were captured and measurements were performed. The criteria for selecting which crypts to measure included a clearly seen and continuous cell column on each side of the crypt and a completely visible crypt lumen and opening. Many crypts, from multiple sections prepared from four to five animals per group, were measured to evaluate a total of 40–50 crypts per group. Mature goblet cells were phenotyped by the intensity of periodic acid-Shiff (PAS) staining as previously described (24). Gut mucosal goblet cell number was performed in PAS-stained distal colon samples. Goblet cells were identified by classical morphology, and total goblet cell number was determined from the crypt base to the luminal surface of 10–15 well-oriented crypt units in each section and in 10–15 well-oriented colonic crypt units from the crypt base to the luminal surface. The average number of goblet cells per colonic unit counted in colonic segments and calculated for each animal and group means were determined using ImageJ.
Mucin content was estimated by using integrated morphometric analysis based on pixel density of differential color staining of pink PAS-positive cytoplasm corrected for blue nuclear staining over five microscopic fields at 40× magnification on at least three mice per genotype. This was done by using the MetaMorph computer program (Universal Imaging, Downingtown, PA) in a comparable manner to a previous study (25).
Immunofluorescence and confocal microscopy
Colonic sections (5 µm) after deparafinization were exposed to the primary antibodies (β-catenin and E-cadherin). To block unspecific staining, cells were incubated for 1 h at room temperature with 5% (v/v) donkey serum in Tris-Buffered Saline Tween (TBST). Cells were sequentially stained with E-cadherin primary antibody (dilution 1:100) or β-catenin primary antibody (dilution 1:100) in a humidity chamber overnight at 4 C. Cover slips were washed three times for 30 min with TBST solution and then incubated with Alexa Fluor 488 goat anti-rabbit IgG secondary antibody (dilution 1:500) in a humidity chamber for two h at room temperature. Cover slips were washed four times with TBST and then mounted upside down with mounting solution [70% (v/v)] mixed with mounting solution with DAPI [30% (v/v)] on glass slides. Cells were observed under Leica-CTR4000 confocal microscope at 63× magnification using immersion oil.
RNA extraction and quantitative real-time PCR
Tissue RNA was extracted using TRI Reagent (Sigma) as previously described (26). RNA concentration and purity were validated by verifying the 260/280 and 230/280 ratios (Nanodrop ND-1000 V.3.1.0; NanoDrop Technologies Inc, DE). Two micrograms of DNase-treated RNA were reverse-transcribed using Superscript First-Strand kit (Invitrogen, CA), followed by subjecting the cDNA to quantitative real-time PCR using the SYBR Green Master kit (Applied Biosystems, Foster City, CA) and the ABI Prism 7300 Sequence Detection System. Primers were designed using Primer Express v.2.0 (Applied Biosystems) and their specificity was validated by respective dissociation curves. Primers were validated over four orders of magnitude and analyzed using 7300 System software. Primers consisted of HNF-4α [F: 5′-ACGTGCTGCTCCTAGGCAAT-3′, R: 5′-CTAGCTCTGGACAGTGCCGAG-3′]; PCNA [F: 5′-TTTGAGGCACGCCTGATCC-3′, R: 5′-GGAGACGTGAGACGAGTCCAT-3′]; c-myc [F: 5′-TGGACACGCTGACGAAAGT-3′, R:5′-AGGCGAAGCAGCTCTATTTCT-3′]; axin2 [F: 5′-CCATGACGGACAGTAGCGTA-3′, R: 5′-GCCATTGGCCTTCACACT-3′]; Tcf4 [F: 5′-GAAAAGTTCCTCCGGGTTTG-3′, R: 5′-GAGAGTTCCCTGGCTGTGTC-3′]; muc4 [F: 5′-GAGAGTTCCCTGGCTGTGTC-3′, R: 5′-GGGACATGGGTGTCTGTGTT-3′]; Apo B [F: 5′-GCCCATTGTGGACAAGTTGATC-3′, R: 5′-CCAGGACTTGGAGGTCTTGGA-3′]; Apo A4 [F: 5′-GACTACTTCAGCCAAAACAGTTGGA-3′, R: 5′-AAGCTGCCTTTCAGGTTCTCCT-3′]; KLF4 [F: 5′-AGAGGAGCCCAAGCCAAAGAGG-3′, R: 5′-CCACAGCCGTCCCAGTCACAGT-3′]; and Math1 [F: 5′-AGATCTACATCAACGCTCTGTC-3′, R: 5′-ACTGGCCTCATCAGAGTCACTG-3′]. Genes were normalized to mouse ribosomal protein S18 (RPS18) gene [F: 5′-GTAACCCGTTGAACCCCATT-3′, R: 5′-CCATCCAATCGGTAGTAGCG-3′]. The fold change in target gene expression was calculated by the comparative CT method, also referred to as the 2−ΔΔct method (Applied Biosystems). Transcript values for respective mice groups represent mean ± SE of independent triplicates, in which each biological triplicate was analyzed in technical triplicates.
Protein extraction and Western blot analysis
E-cadherin, HNF-4α, and aquaporin-8 were detected in colonic tissues lysates as previously described (9).
Statistical analysis
Data is presented as the mean ± SE, using t-test and one-way ANOVA (ANOVA). Differences were considered significant at probability levels of P < 0.05.
RESULTS
Strain phenotypes
In line with the db/db phenotype, body weight gain of BKS.Cg mice was significantly increased compared with the C57BL wild-type or fat-1 strain, whereas it reverted to the control value in the f×B cross (not shown). The increase in body weight gain of BKS.Cg mice was not accompanied by increase in tail length (not shown), indicating that the higher body weight gain of BKS.Cg mice reflects increased adiposity. Also, in line with the db/db genotype, blood glucose, insulin, and leptin levels of three- and six-month-old BKS.Cg mice were significantly higher than those of the C57BL wild-type or fat-1 strain and were partially reverted to the control value in the f×B cross (Table 1).
TABLE 1.
Blood leptin, glucose, and insulin levels of respective strains
| C57BL | fat-1 | BKS.Cg | f×B | |
| Leptin (ng/ml) | 6.3 ± 0.6 | 5.4 ± 0.5 | 31.1 ± 1.0a | 16.3 ± 0.9ab |
| Fasting glucose (mg/dl) | 59.8 ± 9.5 | 53.4 ± 8.1 | 195 ± 17a | 113 ± 0.3ab |
| Fasting insulin (µU/ml) | 68 ± 9 | 61 ± 8 | 110 ± 8a | 90 ± 7ab |
Blood leptin, glucose, and insulin of six-month-old C57BL wild-type, fat-1, BKS.Cg, and f×B mouse strains were determined as described in Materials and Methods. Mean ± SE (n = 10 for each strain).
Significant compared with wild-type (P < 0.005).
Significance of f×B strain compared with BKS.Cg (P < 0.001).
Table 2 summarizes the PUFA composition of three-month-old mice of the four strains. In line with previous reports (27, 28), PUFA composition determined in colonic tissue was similar to other organs, including tails (not shown). The (n-6) PUFA content is shown to decrease by about 50% in the fat-1 and f×B strains compared with the wild-type and BKS.Cg strains, with concomitant 2-fold increase in (n-3) PUFA content, resulting in 5-fold increase in (n-3)/(n-6) PUFA ratio in the fat-1 and f×B mice. The relative PUFA composition remained unaffected during the first six months of maturation, in spite of a progressive increase in the absolute concentrations of (n-3) PUFA (not shown), indicating that Table 2 reflects the PUFA composition of the four strains irrespective of age. Similar changes have been previously reported for various organs in fat-1 mice (27).
TABLE 2.
Colonic composition of (n-3)/(n-6) PUFA of respective strains
| C57BL | fat-1 | BKS.Cg | f×B | |
| Arachidonic acid (n-6, 20:4) | 2.18 ± 0.19 | 0.30 ± 0.03a | 7.05 ± 0.12a | 0.15 ± 0.07ab |
| Linoleic acid (n-6, 18:2) | 10.20 ± 1.85 | 10.40 ± 0.87 | 13.92 ± 1.03a | 8.06 ± 1.81b |
| Eicosapentaenoic acid (n-3, 20:5) | 0.20 ± 0.05 | 2.00 ± 0.22a | 0.28 ± 0.06 | 1.42 ± 0.31ab |
| α-Linolenic acid (n-3, 18:3) | 1.14 ± 0.08 | 3.40 ± 0.12a | 1.23 ± 0.33 | 2.40 ± 0.34ab |
| Total (n-3) | 1.34 ± 0.03 | 5.4 ± 0.04a | 1.51 ± 0.02 | 3.82 ± 0.05ab |
| Total (n-6) | 12.38 ± 1.18 | 10.7 ± 0.14 | 20.97 ± 2.11a | 8.21 ± 0.86b |
| (n-6)/(n-3) | 9.24 ± 1.13 | 1.98 ± 0.10a | 13.9 ± 2.01a | 2.15 ± 0.41ab |
Percentage of (n-3) and (n-6) PUFA in total fatty acid extract six-month-old wild-type, fat-1, BKS.Cg, and f×B colonic tissue was determined as described in Materials and Methods. Mean ± SE (n = 10 for each strain).
Significant compared with the wild-type strain (P < 0.01).
Significance of f×B strain compared with BKS.Cg (P < 0.001).
Colonic ontogenesis
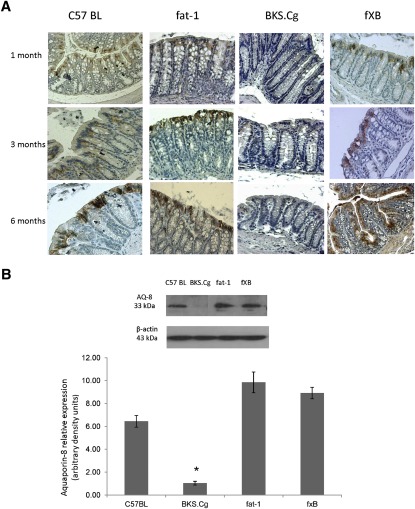
The time course of intestinal ontogenesis of the four strains has been evaluated in terms of colonic markers that represent the proliferation (PCNA) and differentiation of crypt stem cells into epithelial colonocytes (aquaporin-8) (29) and goblet cells (PAS) (30) and MUC-2 expression. The differentiated colonic epithelium plays a major role in salt and water transport and contributes to body fluid homeostasis. The colonic epithelium is a tight tissue characterized by high electrical resistance; therefore, water flux occurs mainly by transporters and/or aquaporins. Aquaporin-8 plays a major role in water movement through the colon by acting on the apical side of the superficial cells (29). As shown in Fig. 1A, intestinal aquaporin-8 was minimally expressed in BKS.Cg strain during the ages assessed (one, three, and six months). In contrast, intestinal aquaporin-8 was fully expressed in the fat-1 strain at all ages, and in f×B strains that express the fat-1 gene, it gradually increased with age, implying gradual modulation of differentiation by (n-3) PUFA. Wild-type C57BL mice also expressed median aquaporin-8 levels at all ages. Immunohistochemistry data for intestinal aquaporin-8 were further verified by Western blot analyses of colonic tissue. As shown in Fig. 1B, colonic aquaporin-8 expression of three-month-old BKS.Cg mice was ∼6-fold decreased compared with wild-type mice, implying delayed colonic ontogenesis in obese diabetic mice. In contrast, intestinal aquaporin-8 content increased 1.5-fold in fat-1 mice compared with wild-type, implying modulation of aquaporin-8 expression by (n-3) PUFA. Moreover, aquaporin-8 expression of the f×B strain increased∼5-fold compared with the BKS.Cg strain, indicating that (n-3) PUFA may counteract the delayed expression of intestinal aquaporin-8 in obese diabetic mice. Similar differences in intestinal aquaporin-8 expression were observed in one- and six-month-old mice of the four strains (not shown).
Fig. 1.
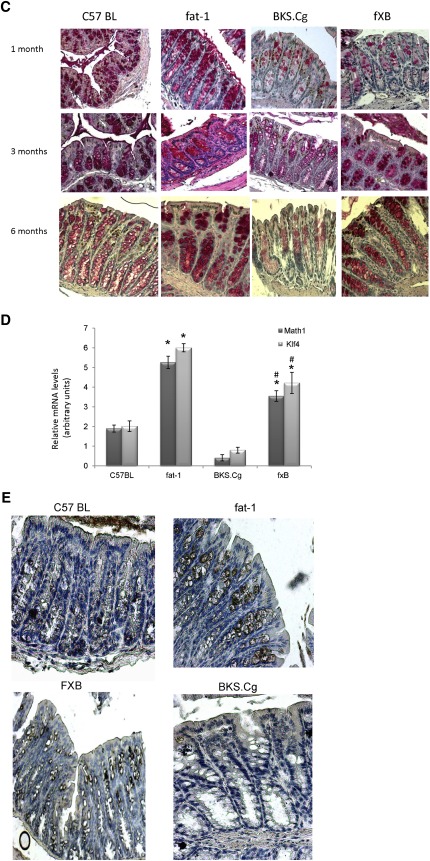
Colonic differentiation-associated ontogenesis. A: Representative colonic aquaporin-8 immunostaining patterns (magnification 200×) of wild-type, BKS.Cg, fat-1, and f×B strains at 1,3, and 6 months of age. n = 6 for each strain and age. B: Colonic aquaporin-8 content normalized to β-actin of 3-month-old mice of the four strains. SDS-PAGE analysis as described in Materials and Methods. Mean ± SE of six independent determinations. *Significant compared with the wild-type strain, fat-1, and f×B (P < 0.001). Inset: representative blots. C: Colonic PAS ontogenesis. Representative colonic PAS staining (magnification 200×) of wild-type, BKS.Cg, fat-1, and f×B strains at 1, 3, and 6 months of age. n = 6 for each strain and age. D: Math-1 and Klf-4 transcripts were determined by qRT-PCR as described in Materials and Methods. Mean ± SE (n = 3 for each 1-month-old mice strain). *Significant compared with the wild-type strain (P < 0.001). #Significance of BKS.Cg strain compared with f×B (P < 0.01). E: Representative colonic muc-2 immunohistograms (magnification 200×) of 3-month-old wild-type, BKS.Cg, fat-1, and f×B strains.
PAS staining served as marker for intestinal ontogenesis into differentiated goblet cells of the four strains concerned. Indeed, PAS staining (Fig. 1B) was significantly age-dependently suppressed in BKS.Cg mice, whereas it was highly expressed in fat-1 mice. In f×B strains, PAS staining was gradually increased with age, indicating again that the fat-1 gene plays a role in colonic intestinal differentiation.
Morphologic measurements performed in HE- and PAS-stained distal colon sections allowed measuring colonic crypt depth, counting goblet cells, and estimating mucin content following measurement of PAS staining intensity. As indicated in Table 3, crypt depths were significantly larger in BKS.Cg mice than in any other strain. Crossing fat-1 with BKS.Cg to obtain f×B resulted in colonic crypt depths similar to control mice.
TABLE 3.
Morphological assessments in the different mice strains
| C57BL | fat-1 | BKS.Cg | f×B | |
| Crypt depth (µm) | 151.9 ± 4.0 | 152.8 ± 4.0 | 171.6 ± 13.0a | 159.9 ± 5.0b |
| Number of goblet cells/crypt | 31 ± 3.2 | 41 ± 5.4a | 24.1 ± 4.4a | 39 ± 4.2ab |
| Number of colonocytes/crypt | 65 ± 5.2 | 63 ± 5.5 | 81.7 ± 9.7a | 69 ± 7.2b |
| Relative mucin content | 1.8 ± 0.2 | 2.8 ± 0.3a | 1.1 ± 0.2a | 2.2 ± 0.3ab |
Colon crypt depths and number of cells per crypt were determined in HE-stained distal colonic sections as described in Materials and Methods. PAS staining of colonic crypts allowed measurement of number of goblet cells. The extent of PAS staining was assessed using Metamorph software as described in Materials and Methods. Mean ± SE (at least n = 3 for each mice strain).
Significant compared with wild-type (P < 0.001).
Significance of f×B strain compared with BKS.Cg (P < 0.01).
Goblet cells are glandular epithelial cells scattered in the colonic epithelium. The cytoplasm of these cells is filled with granules containing mucins, which are secreted into the intestinal lumen. Goblet cells derive from intestinal stem and progenitor cells located in the lower part of the crypt. Their differentiation is controlled by several transcription factors, especially by the basic helix-loop-helix transcription factor Hath1, the zinc-finger transcription factor KLF4, and others (31). For example, Math1 null mice (Math1 is the mouse homolog of human Hath1) are not able to develop goblet cells, whereas the columnar cells are intact (32). Hath1 seems to be involved in an early stage of goblet cell differentiation; KLF4 appears to play a crucial role, especially in the terminal stage of goblet cell differentiation (33). We determined here transcript levels of KLF4 and Math1 and demonstrated that their expression is significantly higher in fat-1 mice, resulting in more goblet cells and filled with more PAS-positive staining (i.e., appearance of more neutral mucin). The secretory mucin MUC-2, which is the predominant structural component of the intestinal mucus layer, is exclusively and abundantly expressed by goblet cells in the colon (34). After synthesis, MUC-2 is secreted into the lumen and forms a protective mucus gel layer that acts as a selective barrier to protect the epithelium from mechanical stress and noxious agents. Expression of MUC-2 is considered a phenotypic marker of colonic differentiation, which can be inversely correlated with the severity of inflammation in inflammatory bowel disease (35). In the normal colon, MUC-2 is the predominant secreted mucin expressed by goblet cells. MUC-2 expression here was detected by immunohistochemistry. As seen in Fig. 1E, microscopic images of the intestinal mucosa show that there is increased staining for MUC-2 in the epithelium of fat-1 mice compared with wild-type C57BL mice, consistent with the PAS finding presented in Fig. 1C. In contrast, in BKS.Cg mice, very low MUC-2 staining is detected. Introduction of fat-1 genotype to BKS.Cg mice also induced upregulation of MUC-2 expression. Together, these data suggest that fat-1 genotype may affect goblet cell differentiation.
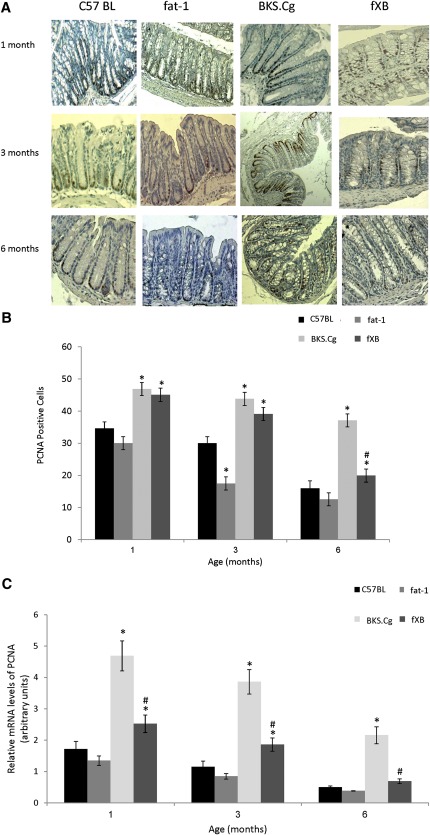
PCNA-positive cells are mostly confined to the differentiating crypt compartment, whereas their expansion beyond the crypt base marks enhanced proliferation (36). As shown in Fig. 2A, B, PCNA-positive cells of the wild-type and fat-1 strains are highly expressed in one-month-old animals, and then their number gradually decreases by about 50% in six-month-old mice. In contrast, the proliferation index of one-month-old BKS.Cg mice is significantly higher and remains significantly increased throughout the maturation process, implying an expansion of the crypt compartment where PCNA is expressed. In contrast to BKS.Cg mice, the increased proliferation index of one-month-old f×B mice gradually decreased upon maturation, reaching the six-month level of wild-type or fat-1 mice, implying that endogenous (n-3) PUFA may restrain colonocyte proliferation in the obese diabetic context. The expansion of the PCNA-positive compartment in obese diabetic mice and its shrinkage in the f×B cross-strain has been further confirmed by quantifying the colonic PCNA transcript (Fig. 2C), implying that the PCNA profile of the respective strains was controlled at the transcript level.
Fig. 2.
Colonic proliferation-associated ontogenesis. A: Representative colonic PCNA immunostaining patterns (magnification 200×) of wild-type, fat-1, BKS.Cg, and f×B strains at 1, 3, and 6 months of age. n = 6 for each strain and age. B: PCNA staining index was expressed as the mean percentage of the colonic area showing positive staining relative to total colonic area of four different individual sections. PCNA-positive cells. Mean ± SE (n = 6 for each strain and age). *Significant compared with the corresponding age-matched wild-type strain (P < 0.001). #Significance of f×B strain compared with BKS.Cg (P < 0.01). C: PCNA transcript levels determined by qRT-PCR as described in Materials and Methods. Mean ± SE (n = 6 for each strain and age). *Significant compared with the corresponding age-matched wild-type strain (P < 0.001). #Significance of f×B strain compared with BKS.Cg (P < 0.01).
Ontogenesis and colonic transduction pathways
Colonic ontogenesis is controlled by several transduction pathways, including HNF-4α and β-catenin/Tfc. Hence, differences in ontogenic profiles of the four strains concerned have been further pursued in terms of colonic β-catenin, E-cadherin, and HNF-4α expression.
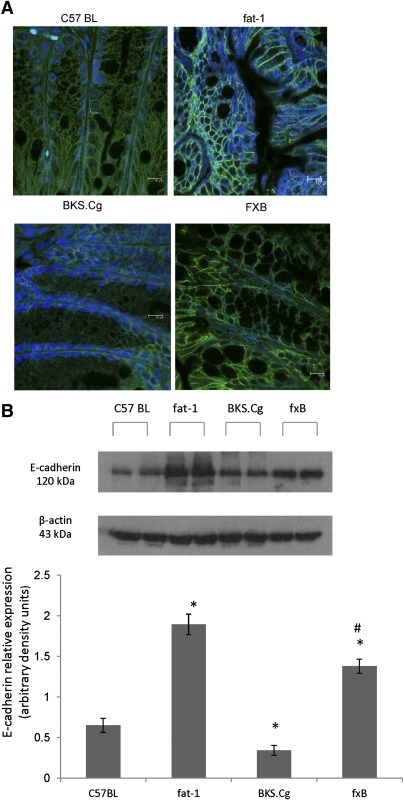
As shown in Fig. 3A, expression of E-cadherin as measured by immunofluorescence in colonic tissue of three-month-old wild-type C57BL mice was clearly evident in membrane boundaries of colonic epithelial cells. Nevertheless, expression of colonic E-cadherin was significantly upregulated in the fat-1 and f×B strains that express the fat-1 gene, implying modulation of its expression by (n-3) PUFA. Similar differences in intestinal E-cadherin expression were observed in one- and six-month-old mice of the four strains (not shown). Immunofluorescence data for colonic E-cadherin were further verified by Western blot analyses of colonic tissue. As shown in Fig. 3B, colonic E-cadherin expression of three-month-old BKS.Cg mice was 2-fold decreased compared with wild-type mice, implying delayed colonic ontogenesis in obese diabetic mice. In contrast, intestinal E-cadherin content increased 4-fold in fat-1 mice, implying modulation of E-cadherin expression by (n-3) PUFA. Moreover, E-cadherin expression of the f×B strain increased 4-fold compared with the BKS.Cg strain, indicating that (n-3) PUFA may counteract the delayed expression of intestinal E-cadherin in obese diabetic mice. Similar differences in intestinal E-cadherin expression were observed in one- and six-month-old mice of the four strains (not shown).
Fig. 3.
Colonic E-cadherin expression. A: E-cadherin expression in three-month-old wild-type, fat-1, BKS.Cg, and f×B strains. Pictures shown are single optical sections in the X-Y axis. Blue nuclei (DAPI); green (E-cadherin); Bar = 10 µm. B: Colonic E-cadherin content normalized to β-actin of 3-month-old mice of the four strains. SDS-PAGE analysis as described in Materials and Methods. Mean ± SE of six independent determinations.*Significant compared with the wild-type strain (P < 0.01). #Significance of f×B strain compared with BKS.Cg (P < 0.001). Inset: representative blots.
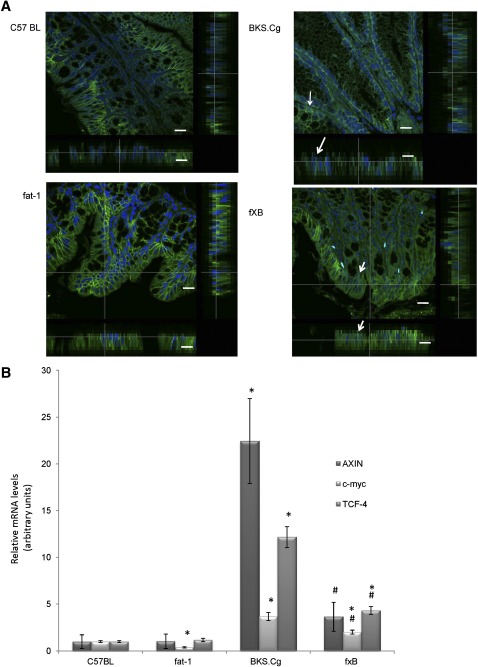
As shown in Fig. 4A, expression of β-catenin in colonic tissue of three-month-old C57BL wild-type mice was localized to the cell membrane and cytoplasm. In fat-1 and f×B animals, β-catenin staining was confined predominantly to the cell membrane of colonocytes. Nonetheless, no dramatic changes in β-catenin immunostaining intensity among these three mice strains groups were observed. However β-catenin of BKS.Cg colonic tissue demonstrated that a large proportion β-catenin was translocated to the nucleus. Analyses of lateral images of cells reconstituted from confocal Z-sectioned images (β-catenin, green; nuclei, blue DAPI) demonstrated that this view captured β-catenin accumulation in the central nuclear region, whereas β-catenin levels in cytoplasmatic or membrane regions (arrows) were reduced. Introducing the fat-1 gene into the obese diabetic context resulted in a robust decrease in β-catenin expression followed by its confinement to the plasma membrane (Fig. 4A), indicating that β-catenin cellular distribution is tightly controlled by colonic (n-3) PUFA. Diabesity-induced increase in nuclear β-catenin and its suppression by (n-3) PUFA has been further verified by probing β-catenin/Tcf-responsive transcripts (e.g., Axin-2, Tcf-4, and c-myc; Fig. 4B).
Fig. 4.
Colonic β-catenin expression and β-catenin-responsive transcripts. A: The distribution of β-catenin was analyzed by immunofluorescence in wild-type, BKS.Cg, fat-1, and f×B strains (n = 6 of each strain). Pictures shown are single optical sections in the X-Y axis. Corresponding orthogonal sections in the X-Z and Y-Z axis are shown beside and at the sides. Arrows in the merged picture indicate where nuclei (DAPI) and β-catenin colocalize (evidenced by green-blue mixed color). Bar = 10 µm. β-catenin–responsive transcripts. B: Axin, Tcf, and c-myc transcripts were determined by qRT-PCR as described in Materials and Methods. Mean ± SE (n = 3 for each 1-month-old mice strain). *Significant compared with the wild-type strain (P < 0.001). #Significance of f×B strain compared with BKS.Cg (P < 0.01).
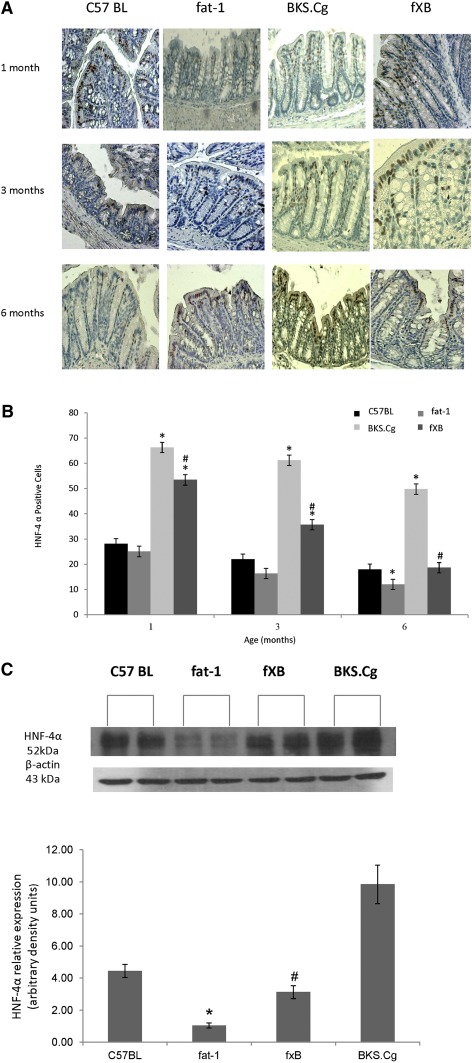
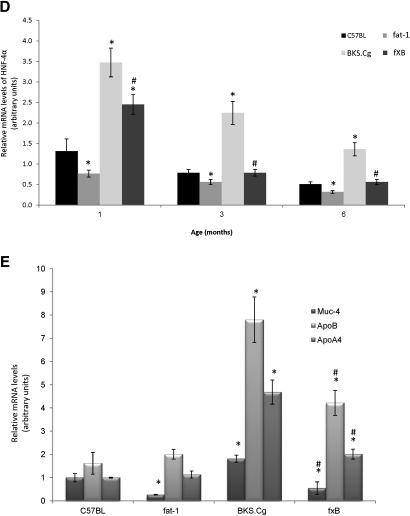
In line with β-catenin expression, the expression and distribution of HNF-4α along the crypt-villus axis varied among the mice strains. Thus, colonic HNF-4α expression in wild-type, fat-1, and f×B strains was mainly confined to the differentiation compartment of the crypt, whereas it extended to the proliferation compartment in the BKS.Cg strain (Fig. 5A). In contrast to the wild-type and fat-1 strains, HNF-4α-positive cells were highly expressed in the BKS.Cg strain at all maturation stages, whereas they approached the wild-type level in the f×B strain (Fig. 5B). Immunohistochemistry data for HNF-4α were further verified by Western blot analyses of colonic tissue. As shown in Fig. 5C, colonic HNF-4α expression of three-month-old BKS.Cg mice increased ∼2.5-fold compared with wild-type mice. In contrast, HNF-4α expression decreased 4-fold in fat-1 mice compared with wild-type, implying modulation of HNF-4α expression by (n-3) PUFA. Moreover, HNF-4α expression of the f×B strain decreased ∼5-fold compared with the BKS.Cg strain, indicating that (n-3) PUFA may counteract the increased expression of intestinal HNF-4α in obese diabetic mice. Similar differences in intestinal HNF-4α expression were observed in one- and six-month-old mice of the four strains (not shown). The expansion of HNF-4α-positive cells in obese diabetic mice and their progressive decrease in the f×B cross-strain was further confirmed by quantifying the colonic HNF-4α transcript (Fig. 5D) as well as the HNF-4α-responsive transcripts (namely, muc-4, ApoA4, and ApoB) (Fig. 5E), implying suppression of diabesity-induced HNF-4α transcriptional activity by (n-3) PUFA.
Fig. 5.
Colonic HNF-4α expression and HNF-4α-responsive transcripts. A: Representative colonic HNF-4α immunostaining patterns (magnification 200×) of wild-type, BKS.Cg, fat-1, and f×B strains at 1, 3, and 6 months of age. n = 6 for each strain and age. B: HNF-4α staining index was expressed as the mean percentage of the colonic area showing positive staining relative to total colonic area of four different individual sections. HNF-4α-positive cells: Mean ± SE (n = 10 for each strain and age). *Significant compared with the corresponding age-matched wild-type strain (P < 0.001). #Significance of f×B strain compared with BKS.Cg (P < 0.01). C: Colonic HNF-4α content normalized to β-actin of three-month-old mice of the four strains. SDS-PAGE analysis as described in Materials and Methods. Mean ± SE of six independent determinations. *Significant compared with the wild-type strain, fat-1, and f×B (P < 0.001). #Significance of f×B strain compared with BKS.Cg (P < 0.01). Inset: representative blots. (D) HNF-4α transcript levels determined by qRT-PCR as described in Materials and Methods. Mean ± SE (n = 6 for each strain and age). *Significant compared with the corresponding age-matched wild-type strain (P < 0.001). #Significance of f×B strain compared with BKS.Cg (P < 0.01). D: HNF-4α-responsive transcripts Muc-4, ApoB, and ApoA4 were determined by qRT-PCR as described in Materials and Methods. Mean ± SE (n = 3 for each one-month-old mice strain). *Significant compared with the wild-type strain (P< 0.001). #Significance of f×B strain compared with BKS.Cg (P < 0.01).
As the transcriptional activity of HNF-4α has previously been reported to be modulated by LCFA as function of chain length and degree of saturation being suppressed by (n-3) PUFA (19), diabesity-induced increase in the expression of HNF-4α-responsive genes and their suppression by (n-3) PUFA was been further pursued in terms of the composition of (n-6) and (n-3) PUFA bound to HNF-4α of the strains. As shown in Table 4, wild-type HNF-4α-bound PUFA consisted of about 65% of (n-6) LA, complemented by 35% of dietary (n-3) α-linolenic acid. In contrast to wild-type, BKS.Cg colonic HNF-4α consisted exclusively of (n-6) LA with only traces of (n-3) PUFA. In contrast to BKS.Cg, PUFA composition of fat-1 colonic HNF-4α was dominated by (n-3) α-linolenic acid at the expense of a robust decrease in (n-6) PUFA. Also, in contrast to HNF-4α being essentially devoid of EPA and DHA in BKS.Cg, HNF-4α-bound EPA in fat-1 amounted to 7% of total PUFA. Most importantly, introducing the fat-1 gene into the BKS.Cg context resulted in reverting HNF-4α-PUFA composition to the wild-type profile. It is worth noting that the respective PUFA compositions of HNF-4α reflect specific HNF-4α-PUFA affinity characteristics, rather than the endogenous PUFA composition of the respective strains. Indeed, a 1.5-fold increase in BKS.Cg (n-6)/(n-3) endogenous PUFA ratio compared with wild-type (Table 2) resulted in a 240-fold increase in BKS.Cg (n-6)/(n-3) HNF-4α-PUFA (Table 3). Similarly, a 7-fold decrease in fat-1 or f×B (n-6)/(n-3) endogenous PUFA ratio compared with BKS.Cg (Table 2) resulted in a greater than 500-fold decrease in(n-6)/(n-3) HNF-4α-PUFA in fat-1 or f×B mice (Table 3). Hence, the expression of HNF-4α–responsive genes in the respective strains may reflect the extent of HNF-4α expression combined with modulation of its transcriptional activity by the respective PUFA ligands.
TABLE 4.
Composition of (n-3)/(n-6) PUFA bound to colonic HNF-4α of respective strains
| C57BL | fat-1 | BKS.Cg | f×B | |
| Linoleic acid (n-6, 18:2) | 64.9 ± 7.0 | 25.8 ± 5.0a | 97.4 ± 13.0a | 46.9 ± 5.0ab |
| Arachidonic acid (n-6, 20:4) | 0.11 ± 0.06 | 0.01 ± 0.01a | 0.52 ± 0.40a | 0.32 ± 0.10b |
| α-Linolenic acid (n-3, 18:3) | 33.5 ± 2.0 | 65.9 ± 5.0a | 0.18 ± 0.02a | 43.4 ± 6.0ab |
| Eicosapentaenoic acid (n-3, 20:5) | 1.01 ± 0.60 | 7.6 ± 0.7a | 0.006 ± 0.002a | 1.01 ± 0.05b |
| Docosahexaenoic acid (n-3, 22:6) | 0.16 ± 0.01 | 0.20 ± 0.03a | 0.004 ± 0.003a | 0.16 ± 0.01b |
| Total (n-3) | 34.70 ± 2.12 | 73.70 ± 6.11a | 0.19 ± 0.07a | 44.6 ± 6.12ab |
| Total (n-6) | 65.51 ± 7.37 | 25.80 ± 6.12a | 98.13 ± 11.21a | 47.2 ± 5.11ab |
| (n-6)/(n-3) | 1.88 ± 0.02 | 0.35 ± 0.04a | >500a | 1.06 ± 0.01ab |
Colonic nuclear HNF-4α of wild-type, fat-1, BKS.Cg, and f×B strains was quantitatively purified by immunoprecipitation as described in Materials and Methods. Percentage composition of (n-3) and (n-6) PUFA bound to colonic HNF-4α was determined by LC/MS as described in Materials and Methods. Mean ± SE (n = 3 for each strain).
Significant compared with wild-type (P < 0.01).
Significance of f×B strain compared with BKS.Cg (P < 0.001).
DISCUSSION
The profile of colonic ontogenesis of db/db mice reported here may indicate that diabesity is associated with an increase in colonic proliferation (PCNA), with concomitant dedifferentiation of epithelial colonocytes (aquaporin-8) and goblet cells (PAS). The colon is a tight epithelium characterized by high electrical resistance and considerable water flux; therefore, water flux may occur mainly by transporters and/or aquaporins. Accordingly, the water transporter aquaporin-8 plays a pivotal role for aquaporin-mediated water transport in colonic superficial cells, an activity associated with full-cell differentiation. The colonic epithelium is a continually renewing tissue, containing cells at different stages of proliferation and differentiation aligned in an orderly pattern along the crypt continuum. The sizes of colon proliferative and maturational compartments are maintained within precise boundaries by several cell homeostasis-related mechanisms. When these mechanisms are affected, the sizes of the crypt compartments are altered. In the present study, crypt depths and total number of colonocytes were significantly larger in BKS.Cg mice than in any other mice strain. Crossing fat-1 with BKS.Cg to obtain f×B resulted in colonic crypt depths similar to control mice. PCNA staining supports the finding that the proliferative compartment is larger in BKS.Cg mice than in any other mice strain, whereas the maturational compartment as detected by aquaporine-8 staining was significantly downregulated.
Goblet cells are glandular epithelial cells scattered among the absorptive cells in the epithelium. The cytoplasm of these cells is filled with granula containing mucins, which are secreted into the intestinal lumen. These mucins form the mucus layer, acting as a barrier between the luminal contents and the epithelial surface (37). Terminal differentiation means a higher number of goblet cells. Accordingly, in fat-1 mice, we detected a higher number of goblet cells, whereas in BKS.Cg mice, a lower number of these cells were detected, indicating that (n-3) PUFA may induce differentiation toward this cell lineage. Cross f×B mice demonstrated a higher number of goblet cells compared with BKS.Cg, supporting this finding. Goblet cells derive from intestinal stem and progenitor cells located in the lower part of the crypt. Their differentiation is controlled by several transcription factors, especially the basic helix-loop-helix transcription factor Hath1, the zinc-finger transcription factor KLF4, and others. We demonstrated here that fat-1 mice express higher transcripts of KLF4 and Math-1 (the mouse homolog of human Hath1), indicating that these transcription factors may be controlled by (n-3) PUFA. Goblet cells in fat-1 mice not only existed at higher numbers but also contained more mucin (as detected by intensity of PAS staining; see Table 3).We also demonstrated that goblet cells express higher amounts of the mucin Muc-2, an epithelial mucin expressed in the intestine, with particularly abundant expression in the colon (38). Katz et al. (33) demonstrated altered expression of the goblet cell marker Muc2 in Klf4−/− mice, indicating that Muc2 expression is controlled by KLF4; as the extent of KLF4 transcription also depends on (n3) PUFA, Muc-2 expression in goblet cells depends on n-3 PUFA levels. Muc2 appears to be the most prominent colonic mucin in the mouse, and throughout the intestine, it is expressed exclusively in pregoblet and goblet cells.
In contrast to the reported decrease in liver HNF-4α (39), diabesity results in increase in colonic HNF-4α expression with concomitant increase in HNF-4α-responsive genes, implying tissue-specific expression of HNF-4α in response to hyperinsulinemia. Diabesity further results in increased nuclear translocation of β-catenin, with increased expression of β-catenin/Tcf-responsive genes. In contrast to diabesity, the fat-1 transgene is characterized by restrained colonic proliferation, accompanied by differentiation of crypt stem cells into epithelial colonocytes and goblet cells. The fat-1 profile is further associated with decrease in colonic HNF-4α expression and in nuclear β-catenin, with concomitant decrease in their responsive genes. Most importantly, morphological and transcriptional characteristics of diabesity may be reverted to wild-type by introducing the fat-1 gene into the diabesity context, implying that enrichment with (n-3) PUFA is effective in ameliorating diabesity-induced colorectal ontogenesis. The mode of action of hyperinsulinemia/insulin resistance in promoting intestinal HNF-4α and β-catenin expression in diabesity remains to be investigated. Negative autoregulation of HNF-4α transcription by HNF-4α (40) may limit the diabesity-induced increase in intestinal HNF-4α protein and transcript. Amelioration by (n-3) PUFA may be accounted for by the decrease in HNF-4α expression, accompanied by the direct inhibition of HNF-4α transcriptional activity by binding of (n-3) PUFA to the HNF-4α LCFA pocket (19, 20). Amelioration by (n-3) PUFA may further be complemented by β-catenin degradation due to (n-3) PUFA activation of GSK3 (10, 41).
Positive correlation between colonic HNF-4α/β-catenin transcriptional activities and colonic proliferation and dedifferentiation apparently counters previous reports in which conditional intestinal HNF-4α knockout within a period following epithelial cellular determination was found to have only minor morphological and functional consequences compared with wild-type (16, 42) or claimed to result in increased proliferation of crypt stem cells with increased activity of the β-catenin/Tcf system (43). Although the disparity between the knockout studies concerned remains to be resolved, it is worth noting that the knockout data may imply an obligatory role of HNF-4α in modulating intestinal ontogenesis but may not predict the functional impact of fatty acyl ligands in modulating the transcriptional activity of prevalent HNF-4α. Indeed, as nuclear-receptor/ligand pairs may have distinct ligand-dependent and ligand-independent activities, the global effect of knocking out a nuclear receptor may not simulate ligand-dependent suppression of its activity in the context of a defined promoter. Thus, increased epithelial proliferation due to HNF-4α knockout may concur with decreased epithelial proliferation due to HNF-4α suppression by its (n-3) PUFA ligand antagonists.
The functional impact and pattern of ligand binding to HNF-4α as a function of the LCFA composition of respective animal models reported here seems to finally resolve a long-time issue initiated upon our previous claim that HNF-4α may serve as target for LCFA and that its transcriptional activity may be modulated as function of chain length and degree of unsaturation of respective fatty acyl-CoA ligands (19). That initial claim was followed by crystallographic and modeling studies using the bacterially expressed LBD of HNF-4α (lacking the N-terminal as well as the C-terminal F-domain of HNF-4α), which identified LCFA tightly bound to the LBD rather than their respective CoA thioesters (44–46). However, the F-domain of HNF-4α appears to have a crucial impact on the ligand binding affinity, ligand specificity, and secondary structure of HNF-4α, implying that characteristics of F-truncated mutants may not reflect the properties of full-length HNF-4α (47). Moreover, the E-F domain of HNF-4α was found to bind LCFA-CoA (but not the respective free acids) to catalyze the hydrolysis of bound fatty acyl-CoA by its intrinsic thioesterase activity, followed by intramolecular channeling of the free fatty acid product into the free acid pocket of HNF-4α E-domain (20). These studies have highlighted the need to identify endogenous ligands that bind to HNF-4α in a physiological setup and to verify their functional impact. An initial attempt in that direction has been recently accomplished upon identifying (n-6) LA bound to nuclear HNF-4α overexpressed in COS-7 cells or to hepatic HNF-4α of fed mice (23). Of note, HNF-4α-bound [H1]LA could be exchanged with endogenous [H2]LA but not in vitro, presumably implying a requirement for the acyl-CoA serving as the immediate ligand. The present study further advances these findings by underscoring the specific capacity of HNF-4α to respond to a variety of LCFA in a variable physiological setup while pointing to its functional impact. Thus, diabesity resulted in shifting the composition of HNF-4α-bound PUFA in the (n-6) direction with only traces of (n-3) PUFA, whereas HNF-4α-bound PUFA in colonic fat-1 was mostly dominated by (n-3) α-linolenic acid at the expense of robust decrease in (n-6) PUFA. Similarly, colonic HNF-4α-bound EPA amounted to 7% of total PUFA in fat-1 mice, whereas it was essentially devoid in BKS.Cg HNF-4α. Moreover, in line with our initial claim (19), binding of (n-3) PUFA was associated with decrease in the expression of HNF-4α-responsive genes. Most importantly, introducing the fat-1 gene into the diabesity context resulted in reverting the HNF-4α-PUFA composition to the wild-type profile, with concomitant suppression of its transcriptional activity. These findings imply that HNF-4α may indeed serve as target for LCFA, which may modulate its transcriptional activity as function of chain length and degree of unsaturation.
As colonic ontogenic programs may recapitulate morphological and transduction aspects of CRC, the findings reported here may offer a molecular rational for the reported increase in CRC prevalence in MetS/diabesity subjects and its amelioration by (n-3) PUFA (4, 48, 49). Indeed, the intestinal ontogenic program of diabesity and the transformation sequel of CRC are both characterized by increased epithelial proliferation and dedifferentiation. Moreover, the apparent linkage between the intestinal ontogenic program of diabesity and the increase in intestinal HNF-4α activity may indicate that overactivity of HNF-4α may promote CRC. Also, the apparent linkage between the intestinal ontogenic program of fat-1 mice and the decrease in intestinal HNF-4α activity may indicate that suppression of HNF-4α transcriptional activity may alleviate CRC. These predictions seem to be corroborated by the following reported findings: i) HNF-4α transcript and protein levels increased 2- to 3-fold in human CRC samples compared with their paired margin resections (50); ii) silencing HNF-4α expression of HT29 CRC cells by SiHNF-4α resulted in suppressing their proliferation and in activating E-cadherin expression (9); iii) LCFA analogs of the MEDICA series that act as HNF-4α ligand antagonists inhibited proliferation of HT29 and Caco2 CRC cells and suppressed growth of HT29 CRC xenografts (9); and iv) postnatal HNF-4α conditional deletion resulted in robust decrease in polyp multiplicity in ApcMin mice (50).
Altogether, these findings may indicate that suppression of intestinal HNF-4α activity by (n-3) PUFA may ameliorate diabesity-induced intestinal ontogenesis and offer an effective modality for preventing the development of CRC.
Footnotes
Abbreviations:
- CRC
- colorectal cancer
- EPA
- eicosapentaenoic acid
- HE
- hematoxylin and eosin
- HNF-4α
- hepatocyte nuclear factor 4α
- LA
- linoleic acid
- LBD
- ligand binding domain
- LCFA
- long-chain fatty acid
- MetS
- metabolic syndrome
- PCNA
- proliferating cell nuclear antigen
- RR
- relative risk
This work was supported by Israel Science Foundation Grant 134/06.
REFERENCES
- 1.Radtke F., Clevers H. 2005. Self-renewal and cancer of the gut: two sides of a coin. Science. 307: 1904–1909 [DOI] [PubMed] [Google Scholar]
- 2.de Lau W., Barker N., Clevers H. 2007. WNT signaling in the normal intestine and colorectal cancer. Front. Biosci. 12: 471–491 [DOI] [PubMed] [Google Scholar]
- 3.Drozdowski L. A., Clandinin T., Thomson A. B. 2010. Ontogeny, growth and development of the small intestine: understanding pediatric gastroenterology. World J. Gastroenterol. 16: 787–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giovannucci E. 2007. Metabolic syndrome, hyperinsulinemia, and colon cancer: a review. Am. J. Clin. Nutr. 86: s836–s842 [DOI] [PubMed] [Google Scholar]
- 5.Pais R., Silaghi H., Silaghi A. C., Rusu M. L., Dumitrascu D. L. 2009. Metabolic syndrome and risk of subsequent colorectal cancer. World J. Gastroenterol. 15: 5141–5148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cowey S., Hardy R. W. 2006. The metabolic syndrome: a high-risk state for cancer? Am. J. Pathol. 169: 1505–1522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fujise T., Iwakiri R., Kakimoto T., Shiraishi R., Sakata Y., Wu B., Tsunada S., Ootani A., Fujimoto K. 2007. Long-term feeding of various fat diets modulates azoxymethane-induced colon carcinogenesis through Wnt/beta-catenin signaling in rats. Am. J. Physiol. Gastrointest. Liver Physiol. 292: G1150–G1156 [DOI] [PubMed] [Google Scholar]
- 8.Berquin I. M., Edwards I. J., Chen Y. Q. 2008. Multi-targeted therapy of cancer by omega-3 fatty acids. Cancer Lett. 269: 363–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwartz B., Algamas-Dimantov A., Hertz R., Nataf J., Kerman A., Peri I., Bar-Tana J. 2009. Inhibition of colorectal cancer by targeting hepatocyte nuclear factor-4alpha. Int. J. Cancer. 124: 1081–1089 [DOI] [PubMed] [Google Scholar]
- 10.Ng S. S., Mahmoudi T., Danenberg E., Bejaoui I., de Lau W., Korswagen H. C., Schutte M., Clevers H. 2009. Phosphatidylinositol 3-kinase signaling does not activate the wnt cascade. J. Biol. Chem. 284: 35308–35313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Benoit G., Cooney A., Giguere V., Ingraham H., Lazar M., Muscat G., Perlmann T., Renaud J. P., Schwabe J., Sladek F., et al. 2006. International Union of Pharmacology. LXVI. Orphan nuclear receptors. Pharmacol. Rev. 58: 798–836 [DOI] [PubMed] [Google Scholar]
- 12.Duncan S. A., Manova K., Chen W. S., Hoodless P., Weinstein D. C., Bachvarova R. F., Darnell J. E., Jr 1994. Expression of transcription factor HNF-4 in the extraembryonic endoderm, gut, and nephrogenic tissue of the developing mouse embryo: HNF-4 is a marker for primary endoderm in the implanting blastocyst. Proc. Natl. Acad. Sci. USA. 91: 7598–7602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garrison W. D., Battle M. A., Yang C., Kaestner K. H., Sladek F. M., Duncan S. A. 2006. Hepatocyte nuclear factor 4alpha is essential for embryonic development of the mouse colon. Gastroenterology. 130: 1207–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maestro M. A., Cardalda C., Boj S. F., Luco R. F., Servitja J. M., Ferrer J. 2007. Distinct roles of HNF1beta, HNF1alpha, and HNF4alpha in regulating pancreas development, beta-cell function and growth. Endocr. Dev. 12: 33–45 [DOI] [PubMed] [Google Scholar]
- 15.Watt A. J., Garrison W. D., Duncan S. A. 2003. HNF4: a central regulator of hepatocyte differentiation and function. Hepatology. 37: 1249–1253 [DOI] [PubMed] [Google Scholar]
- 16.Babeu J. P., Darsigny M., Lussier C. R., Boudreau F. 2009. Hepatocyte nuclear factor 4alpha contributes to an intestinal epithelial phenotype in vitro and plays a partial role in mouse intestinal epithelium differentiation. Am. J. Physiol. Gastrointest. Liver Physiol. 297: G124–G134 [DOI] [PubMed] [Google Scholar]
- 17.Chen W. S., Manova K., Weinstein D. C., Duncan S. A., Plump A. S., Prezioso V. R., Bachvarova R. F., Darnell J. E., Jr 1994. Disruption of the HNF-4 gene, expressed in visceral endoderm, leads to cell death in embryonic ectoderm and impaired gastrulation of mouse embryos. Genes Dev. 8: 2466–2477 [DOI] [PubMed] [Google Scholar]
- 18.Wang Z., Burke P. A. 2008. Modulation of hepatocyte nuclear factor-4alpha function by the peroxisome-proliferator-activated receptor-gamma co-activator-1alpha in the acute-phase response. Biochem. J. 415: 289–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hertz R., Magenheim J., Berman I., Bar-Tana J. 1998. Fatty acyl-CoA thioesters are ligands of hepatic nuclear factor-4alpha. Nature. 392: 512–516 [DOI] [PubMed] [Google Scholar]
- 20.Hertz R., Kalderon B., Byk T., Berman I., Za'tara G., Mayer R., Bar-Tana J. 2005. Thioesterase activity and acyl-CoA/fatty acid cross-talk of hepatocyte nuclear factor-4{alpha}. J. Biol. Chem. 280: 24451–24461 [DOI] [PubMed] [Google Scholar]
- 21.Kang J. X., Wang J., Wu L., Kang Z. B. 2004. Transgenic mice: fat-1 mice convert n-6 to n-3 fatty acids. Nature. 427: 504. [DOI] [PubMed] [Google Scholar]
- 22.Kang J. X., Wang J. 2005. A simplified method for analysis of polyunsaturated fatty acids. BMC Biochem. 6: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yuan X., Ta T. C., Lin M., Evans J. R., Dong Y., Bolotin E., Sherman M. A., Forman B. M., Sladek F. M. 2009. Identification of an endogenous ligand bound to a native orphan nuclear receptor. PLoS ONE. 4: e5609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bergstrom K. S., Guttman J. A., Rumi M., Ma C., Bouzari S., Khan M. A., Gibson D. L., Vogl A. W., Vallance B. A. 2008. Modulation of intestinal goblet cell function during infection by an attaching and effacing bacterial pathogen. Infect. Immun. 76: 796–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Malaterre J., Carpinelli M., Ernst M., Alexander W., Cooke M., Sutton S., Dworkin S., Heath J. K., Frampton J., McArthur G., et al. 2007. c-Myb is required for progenitor cell homeostasis in colonic crypts. Proc. Natl. Acad. Sci. USA. 104: 3829–3834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lavi I., Levinson D., Peri I., Nimri L., Hadar Y., Schwartz B. 2010. Orally administered glucans from the edible mushroom Pleurotus pulmonarius reduce acute inflammation in dextran sulfate sodium-induced experimental colitis. Br. J. Nutr. 103: 393–402 [DOI] [PubMed] [Google Scholar]
- 27.Li F., Marchette L. D., Brush R. S., Elliott M. H., Davis K. R., Anderson A. G., Anderson R. E. 2010. High levels of retinal docosahexaenoic acid do not protect photoreceptor degeneration in VPP transgenic mice. Mol. Vis. 16: 1669–1679 [PMC free article] [PubMed] [Google Scholar]
- 28.Wan J. B., Huang L. L., Rong R., Tan R., Wang J., Kang J. X. 2010. Endogenously decreasing tissue n-6/n-3 fatty acid ratio reduces atherosclerotic lesions in apolipoprotein E-deficient mice by inhibiting systemic and vascular inflammation. Arterioscler. Thromb. Vasc. Biol. 30: 2487–2494 [DOI] [PubMed] [Google Scholar]
- 29.Laforenza U., Cova E., Gastaldi G., Tritto S., Grazioli M., LaRusso N. F., Splinter P. L., D'Adamo P., Tosco M., Ventura U. 2005. Aquaporin-8 is involved in water transport in isolated superficial colonocytes from rat proximal colon. J. Nutr. 135: 2329–2336 [DOI] [PubMed] [Google Scholar]
- 30.Stringer A. M., Gibson R. J., Logan R. M., Bowen J. M., Yeoh A. S., Laurence J., Keefe D. M. 2009. Irinotecan-induced mucositis is associated with changes in intestinal mucins. Cancer Chemother. Pharmacol. 64: 123–132 [DOI] [PubMed] [Google Scholar]
- 31.Gersemann M., Stange E. F., Wehkamp J. 2011. From intestinal stem cells to inflammatory bowel diseases. World J. Gastroenterol. 17: 3198–3203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang Q., Bermingham N. A., Finegold M. J., Zoghbi H. Y. 2001. Requirement of Math1 for secretory cell lineage commitment in the mouse intestine. Science. 294: 2155–2158 [DOI] [PubMed] [Google Scholar]
- 33.Katz J. P., Perreault N., Goldstein B. G., Lee C. S., Labosky P. A., Yang V. W., Kaestner K. H. 2002. The zinc-finger transcription factor Klf4 is required for terminal differentiation of goblet cells in the colon. Development. 129: 2619–2628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tytgat K. M., Buller H. A., Opdam F. J., Kim Y. S., Einerhand A. W., Dekker J. 1994. Biosynthesis of human colonic mucin: Muc2 is the prominent secretory mucin. Gastroenterology. 107: 1352–1363 [DOI] [PubMed] [Google Scholar]
- 35.Allen A., Hutton D. A., Pearson J. P. 1998. The MUC2 gene product: a human intestinal mucin. Int. J. Biochem. Cell Biol. 30: 797–801 [DOI] [PubMed] [Google Scholar]
- 36.Shpitz B., Bomstein Y., Mekori Y., Cohen R., Kaufman Z., Grankin M., Bernheim J. 1997. Proliferating cell nuclear antigen as a marker of cell kinetics in aberrant crypt foci, hyperplastic polyps, adenomas, and adenocarcinomas of the human colon. Am. J. Surg. 174: 425–430 [DOI] [PubMed] [Google Scholar]
- 37.Shirazi T., Longman R. J., Corfield A. P., Probert C. S. 2000. Mucins and inflammatory bowel disease. Postgrad. Med. J. 76: 473–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Klinken B. J., Einerhand A. W., Duits L. A., Makkink M. K., Tytgat K. M., Renes I. B., Verburg M., Buller H. A., Dekker J. 1999. Gastrointestinal expression and partial cDNA cloning of murine Muc2. Am. J. Physiol. 276: G115–G124 [DOI] [PubMed] [Google Scholar]
- 39.Xie X., Liao H., Dang H., Pang W., Guan Y., Wang X., Shyy J. Y., Zhu Y., Sladek F. M. 2009. Down-regulation of hepatic HNF4alpha gene expression during hyperinsulinemia via SREBPs. Mol. Endocrinol. 23: 434–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Magenheim J., Hertz R., Berman I., Nousbeck J., Bar-Tana J. 2005. Negative autoregulation of HNF-4alpha gene expression by HNF-4alpha1. Biochem. J. 388: 325–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lim K., Han C., Dai Y., Shen M., Wu T. 2009. Omega-3 polyunsaturated fatty acids inhibit hepatocellular carcinoma cell growth through blocking beta-catenin and cyclooxygenase-2. Mol. Cancer Ther. 8: 3046–3055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ahn S. H., Shah Y. M., Inoue J., Morimura K., Kim I., Yim S., Lambert G., Kurotani R., Nagashima K., Gonzalez F. J., et al. 2008. Hepatocyte nuclear factor 4alpha in the intestinal epithelial cells protects against inflammatory bowel disease. Inflamm. Bowel Dis. 14: 908–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cattin A. L., Le Beyec J., Barreau F., Saint-Just S., Houllier A., Gonzalez F. J., Robine S., Pincon-Raymond M., Cardot P., Lacasa M., et al. 2009. Hepatocyte nuclear factor 4alpha, a key factor for homeostasis, cell architecture, and barrier function of the adult intestinal epithelium. Mol. Cell. Biol. 29: 6294–6308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bogan A. A., Dallas-Yang Q., Ruse M. D., Jr, Maeda Y., Jiang G., Nepomuceno L., Scanlan T. S., Cohen F. E., Sladek F. M. 2000. Analysis of protein dimerization and ligand binding of orphan receptor HNF4alpha. J. Mol. Biol. 302: 831–851 [DOI] [PubMed] [Google Scholar]
- 45.Dhe-Paganon S., Duda K., Iwamoto M., Chi Y. I., Shoelson S. E. 2002. Crystal structure of the HNF4 alpha ligand binding domain in complex with endogenous fatty acid ligand. J. Biol. Chem. 277: 37973–37976 [DOI] [PubMed] [Google Scholar]
- 46.Wisely G. B., Miller A. B., Davis R. G., Thornquest A. D., Jr, Johnson R., Spitzer T., Sefler A., Shearer B., Moore J. T., Willson T. M., et al. 2002. Hepatocyte nuclear factor 4 is a transcription factor that constitutively binds fatty acids. Structure. 10: 1225–1234 [DOI] [PubMed] [Google Scholar]
- 47.Petrescu A. D., Hertz R., Bar-Tana J., Schroeder F., Kier A. B. 2002. Ligand specificity and conformational dependence of the hepatic nuclear factor-4alpha (HNF-4alpha). J. Biol. Chem. 277: 23988–23999 [DOI] [PubMed] [Google Scholar]
- 48.Nowak J., Weylandt K. H., Habbel P., Wang J., Dignass A., Glickman J. N., Kang J. X. 2007. Colitis-associated colon tumorigenesis is suppressed in transgenic mice rich in endogenous n-3 fatty acids. Carcinogenesis. 28: 1991–1995 [DOI] [PubMed] [Google Scholar]
- 49.Phinney S. D. 1996. Metabolism of exogenous and endogenous arachidonic acid in cancer. Adv. Exp. Med. Biol. 399: 87–94 [DOI] [PubMed] [Google Scholar]
- 50.Darsigny M., Babeu J. P., Dupuis A. A., Furth E. E., Seidman E. G., Levy E., Verdu E. F., Gendron F. P., Boudreau F. 2009. Loss of hepatocyte-nuclear-factor-4alpha affects colonic ion transport and causes chronic inflammation resembling inflammatory bowel disease in mice. PLoS ONE. 4: e7609. [DOI] [PMC free article] [PubMed] [Google Scholar]