Abstract
Protein farnesyltransferase (FTase) inhibitors, generally called “FTIs,” block the farnesylation of prelamin A, inhibiting the biogenesis of mature lamin A and leading to an accumulation of prelamin A within cells. A recent report found that a GGTI, an inhibitor of protein geranylgeranyltransferase-I (GGTase-I), caused an exaggerated accumulation of prelamin A in the presence of low amounts of an FTI. This finding was interpreted as indicating that prelamin A can be alternately prenylated by GGTase-I and that inhibiting both protein prenyltransferases leads to more prelamin A accumulation than blocking FTase alone. Here, we tested an alternative hypothesis—GGTIs are not specific for GGTase-I, and they lead to prelamin A accumulation by inhibiting ZMPSTE24 (a zinc metalloprotease that converts farnesyl–prelamin A to mature lamin A). In our studies, commonly used GGTIs caused prelamin A accumulation in human fibroblasts, but the prelamin A in GGTI-treated cells exhibited a more rapid electrophoretic mobility than prelamin A from FTI-treated cells. The latter finding suggested that the prelamin A in GGTI-treated cells might be farnesylated (which would be consistent with the notion that GGTIs inhibit ZMPSTE24). Indeed, metabolic labeling studies revealed that the prelamin A in GGTI-treated fibroblasts is farnesylated. Moreover, biochemical assays of ZMPSTE24 activity showed that ZMPSTE24 is potently inhibited by a GGTI. Our studies show that GGTIs inhibit ZMPSTE24, leading to an accumulation of farnesyl–prelamin A. Thus, caution is required when interpreting the effects of GGTIs on prelamin A processing.
Keywords: protein farnesyltransferase, protein geranylgeranyltransferase, nuclear lamins, lopinavir, prelamin A protease
Prelamin A and other proteins containing a carboxyl-terminal CaaX motif undergo a series of posttranslational modifications, beginning with protein prenylation (1–4). First, a 15-carbon farnesyl or a 20-carbon geranylgeranyl lipid is added to the thiol group of the cysteine (the “C” of the CaaX motif) by a pair of protein prenyltransferases, protein farnesyltransferase (FTase) or protein geranylgeranyltransferase I (GGTase-I) (1, 2). Generally, the cysteine is geranylgeranylated if the “X” is a leucine or phenylalanine; otherwise, it is farnesylated (1, 2). Prelamin A terminates with –CSIM and is farnesylated (5–7). Next, the last three amino acids of the protein (i.e., the “aaX” of the CaaX motif) are clipped off by a prenylprotein-specific endoprotease (8). For most proteins, this step is carried out by RCE1 (8), but in the case of prelamin A, this step can be carried out by both zinc metalloprotease Ste24p ortholog (ZMPSTE24) and RCE1 (9, 10). Finally, the newly exposed isoprenylcysteine is methylated by ICMT (11–13). After these CaaX modifications are complete, prelamin A undergoes a second endoproteolytic processing step (14); the last 15 amino acids of the protein, including the farnesylcysteine methyl ester, are clipped off by ZMPSTE24, releasing mature lamin A (9, 15–18).
The endoprotease and the methylation reactions are carried out by enzymes that are specific for prenylated proteins (12, 19–22). Thus, when protein farnesylation is inhibited with a protein farnesyltransferase inhibitor (FTI), lamin A biogenesis is blocked, leading to an accumulation of nonfarnesylated prelamin A (5). The production of mature lamin A also can be blocked by lopinavir, an HIV protease inhibitor that inhibits ZMPSTE24 (9, 17). In the presence of lopinavir, the farnesylated form of prelamin A accumulates within cells (23, 24).
The recognition that a carboxyl-terminal CaaX motif triggers the posttranslational modification of proteins with a lipid (11) was a significant development for the field. Soon thereafter, Chen et al. (25) used “CaaX peptide chromatography” to purify and clone the mammalian protein prenyltransferases. Also, CaaX box peptidomimetic inhibitors of the protein prenyltransferases were generated (26, 27). Peptidomimetic inhibitors of FTase block the prenylation of CaaX proteins that are normally farnesylated, for example, the RAS proteins, HDJ-2, and prelamin A (5, 26); peptidomimetic inhibitors of GGTase-I (GGTIs) block the prenylation of proteins that are normally geranylgeranylated, for example, Rap1a (28). The pharmaceutical industry has been keenly interested in both FTase and GGTase-I inhibitors as anticancer agents (29, 30).
Some farnesylated CaaX proteins, notably KRAS, can be alternately prenylated by GGTase-I when FTase is blocked with an FTI (31). More recently, Varela et al. (32) suggested that alternate prenylation might also apply to prelamin A. In their experiments, they did not observe prelamin A in cells incubated with an FTI, but they did find substantial prelamin A accumulation when cells were treated with both an FTI and a GGTI. They interpreted these findings as suggesting that prelamin A is alternately prenylated by GGTase-I in the presence of an FTI. They concluded that the GGTI/FTI combination was particularly effective in inhibiting the prenylation of prelamin A, accounting for prelamin A accumulation in cells (32). However, the possibility remained that GGTIs actually lead to prelamin A accumulation by inhibiting other enzymes in the lamin A biogenesis pathway.
The notion that a GGTI might inhibit the maturation of prelamin A to lamin A by inhibiting other enzymes in the pathway is not farfetched. Most GGTase-I inhibitors are peptidomimetic compounds that structurally resemble a CaaX motif, and it would not be particularly surprising if these compounds were to affect other enzymes in the pathway that are specific for prenylated CaaX proteins. Our suspicions that ZMPSTE24 might be affected by peptidomimetic GGTIs were heightened by the fact that the substrate specificity of Ste24p, the yeast ortholog of ZMPSTE24, against prenylated peptides is affected by the sequence of the CaaX motif (33, 34). Mammalian ZMPSTE24 shares the same CaaX motif specificities (18). Given that ZMPSTE24 activity is influenced by the sequence of the substrate's CaaX motif, it is reasonable to hypothesize that some CaaX peptidomimetics (i.e., inhibitors of GGTase-I) might affect the activity of ZMPSTE24.
In the current study, we tested the hypothesis that peptidomimetic GGTIs lead to increased amounts of prelamin A in cells by blocking ZMPSTE24.
METHODS
Growth of human fibroblasts and inhibitors of the protein prenyltransferases
Human fibroblasts were obtained from ATCC. In some experiments, we used ZMPSTE24-deficient human fibroblasts from a patient with restrictive dermopathy (35). Two selective FTIs, ABT-100 and PB-43, were used at concentrations of 2–5 μM. PB-43 was obtained from Dr. Michael Gelb (University of Washington), and ABT-100 was obtained from Dr. David Frost (Abbott Laboratories). Three peptidomimetic inhibitors of GGTase-I (GGTI-298, GGTI-2147, and GGTI-2133) were obtained from Sigma-Aldrich or Calbiochem.
Urea-soluble extracts of fibroblasts were prepared as previously described (36). Extracts were size-fractionated on 4–12% gradient polyacrylamide Bis-Tris gels (Invitrogen), and the proteins transferred to nitrocellulose for Western blotting. Antibody dilutions were 1:400 for anti-lamin A/C goat IgG (sc-6215, Santa Cruz Biotechnology); 1:800 for goat anti-prelamin A (sc-6214, Santa Cruz Biotechnology); 1:1000 for anti-actin goat IgG (sc-1616, Santa Cruz Biotechnology); and 1:5000 for IRDye 800 anti-goat IgG (Rockland) or horseradish peroxidase (HRP)–conjugated anti-goat IgG (Santa Cruz Biotechnology). The IRdye-labeled antibodies were detected with an Odyssey infrared imaging scanner (LI-COR Biosciences), and the HRP-labeled antibodies were detected by Enhanced Chemiluminescence methods and exposure to X-ray film.
Hepatocyte-specific Pggt1b and Fntb knockout cells
Primary hepatocytes from mice that do not express GGTase-I in the liver (Pggt1bfl/flAlbCre+) or FTase in the liver (Fntbfl/flAlbCre+) were isolated by collagenase digestion and differential centrifugation (37). The cells were seeded in collagen-coated plates and allowed to adhere for 4 h before preparing cell extracts. Quantitative RT-PCR studies (37) showed that that Pggt1bfl/flAlbCre+ and Fntbfl/flAlbCre+ hepatocytes had a near-complete absence of Pggt1b and Fntb transcripts, respectively.
Metabolic labeling of farnesylated proteins in fibroblasts
To assess protein farnesylation, fibroblasts were metabolically labeled for 48 h with an analog of farnesol, 8-anilinogeraniol (AG) (50 μM in DMSO) (38). AG is converted to anilinogeranyl diphosphate (AGPP) and used as a substrate by FTase. The incorporation of AG into prelamin A can be detected by Western blotting with an AG-specific monoclonal antibody (1:5000) (38) and an IRDye 680–labeled anti-mouse IgG (1:2500) (LI-COR) (23, 37, 39).
Detecting prelamin A in fibroblasts by immunocytochemistry
Human fibroblasts were grown on coverslips and grown in the presence of an FTI (ABT-100) or a GGTI (GGTI-298). Cells were incubated with a rat monoclonal antibody against prelamin A (7G11; 1:50 dilution) or a mouse monoclonal antibody against Lap2β (BD Biosciences; 1:400 dilution) for 2 h. After washing, cells were incubated with a 1:200 dilution of Alexa Fluor 568–conjugated anti-rat antibody or Alexa Fluor 488–conjugated anti-mouse antibody (Invitrogen). DNA was stained with DAPI to visualize nuclei. Images were obtained with a Leica SP2 1P-FCS confocal microscope with a 63× objective.
Endoprotease-coupled methylation assays and other enzymatic assays
Endoprotease-coupled methylation assays of ZMPSTE24 activity (23, 24, 40) were performed by mixing 5 μg of membranes from Δste24Δrce1 yeast that overexpressed mouse ZMPSTE24, 8 μg of membranes from Δste24Δrce1 yeast overexpressing Ste14p, a farnesylated a-factor peptide ([YIIKGVFWDPA(farnesyl)CVIA], 5 μM; EZBioLab, Westfield, IN), and 20 μM S-adenosyl-L-[methyl-14C]methionine (55 Ci/mol; GE Healthcare) in 100 mM Tris-HCl, pH 7.5, in a final volume of 60 μl. Reactions were performed in the presence and absence of GGTI-298 and lopinavir. After incubating the reactions at 30°C for 30 min, the reactions were stopped with 50 μl of 1 M NaOH/1% SDS. The reactions were then spotted on a pleated filter paper, and [14C]methanol was allowed to diffuse into scintillation fluid for 2.5 h. The amount of radioactivity released was quantified by scintillation counting (11).
RESULTS
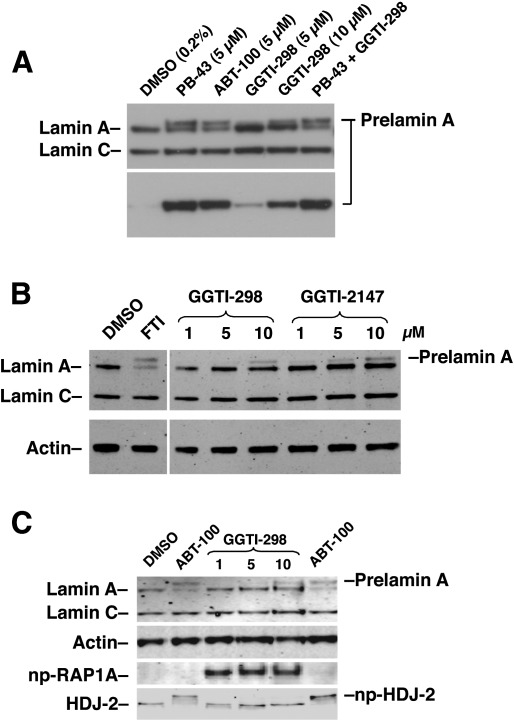
We tested whether commonly used GGTIs would lead to an accumulation of prelamin A in human fibroblasts. In our initial experiments, we observed an accumulation of prelamin A in cells treated with GGTI-298, a peptidomimetic inhibitor of GGTase-I. Indeed, the amount of prelamin A accumulation with 10 μM GGTI-298 was close to the amount of prelamin A that accumulates with selective FTIs (PB-43 and ABT-100) (Fig. 1A). When GGTI-298 was combined with an FTI (PB-43), the amount of prelamin A in cells did not increase beyond that in cells treated with FTI alone (Fig. 1A). We also observed prelamin A accumulation with another peptidomimetic GGTI, GGTI-2147 (Fig. 1B). We considered the possibility that the prelamin A accumulation in GGTI-treated cells was due to the fact that these compounds inhibited FTase; however, this was not the case. An FTI (ABT-100), but not a GGTI (GGTI-298), led to an accumulation of nonfarnesylated HDJ-2 (a CaaX protein that is normally farnesylated by FTase) (Fig. 1C). Also, the GGTI, but not the FTI, led to the accumulation of nonprenylated RAP1A (a CaaX protein that is normally geranylgeranylated by GGTase-I).
Fig. 1.
Treatment of human fibroblasts with a GGTI leads to an accumulation of prelamin A. (A) Western blot of protein extracts from human fibroblasts treated with an FTI (PB-43 or ABT-100) or a GGTI (GGTI-298) at the concentrations indicated, or with both PB-43 (5 μM) and GGTI-298 (10 μM). Prelamin A accumulation was detected with both a lamin A/C antibody (top panel) and a prelamin A–specific antibody (bottom panel). (B) Western blots, with antibodies against lamin A/C and actin, of protein extracts from human fibroblasts treated with an FTI or two different GGTIs (GGTI-298 or GGTI-2147) in increasing concentrations. (C) Western blots of protein extracts from fibroblasts treated with an FTI (ABT-100) or a GGTI (GGTI-298). As expected, only the FTI, and not the GGTI, inhibited the farnesylation of HDJ-2 (np-HDJ-2). Also, only the GGTI, and not the FTI, led to an accumulation of the nonprenylated version of RAP1A (np-RAP1A). Both the FTI and GGTI-298 led to prelamin A accumulation. In all three Western blots, the prelamin A in the GGTI-treated cells migrated slightly more rapidly than the prelamin A in FTI-treated cells.
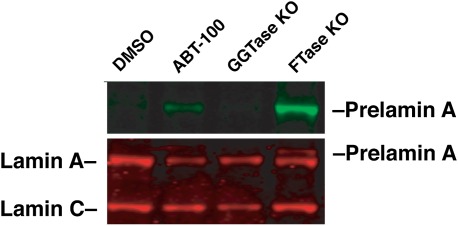
Finding an accumulation of prelamin A in both FTI- and GGTI-treated cells raised the possibility that prelamin A might normally undergo prenylation by both FTase and GGTase-I. To test this possibility, prelamin A levels were assessed in both FTase-deficient hepatocytes and GGTase-I–deficient hepatocytes [isolated from liver-specific Pggt1b knockout mice (Pggt1bfl/flAlbCre+) and liver-specific Fntb knockout mice (Fntbfl/flAlbCre+), respectively] (37). As expected, we observed a substantial amount of prelamin A accumulation in FTase-deficient liver cells. However, when GGTase-I–deficient cells were examined, no prelamin A was detected (Fig. 2). These results implied that the accumulation of prelamin A in GGTI-treated cells could not be caused by an inhibition of GGTase-I activity and, instead, must be due to an effect of the GGTI on other enzymes in the lamin A biogenesis pathway.
Fig. 2.
Inactivation of Pggt1b in mouse hepatocytes does not lead to prelamin A accumulation. We used a Pggt1b conditional knockout allele (Pggt1bfl) and an albumin-Cre (AlbCre) transgene to generate mice lacking Pggt1b in the liver (Pggt1bfl/flAlbCre+, labeled “GGTase KO”) (37). Hepatocytes were isolated from these mice, and cell extracts were blotted with antibodies against lamin A/C and prelamin A. Wild-type hepatocytes treated with DMSO or an FTI (ABT-100) and Fntb-deficient hepatocytes (labeled “FTase KO,” prepared from the liver of an Fntbfl/flAlbCre+ mouse) were included as controls. As expected, inactivation of Fntb resulted in the accumulation of prelamin A in cells, as judged by a Western blot with antibodies against lamins A/C (red) and prelamin A (green). In contrast, no prelamin A was detected in extracts from GGTase KO hepatocytes. KO, knockout.
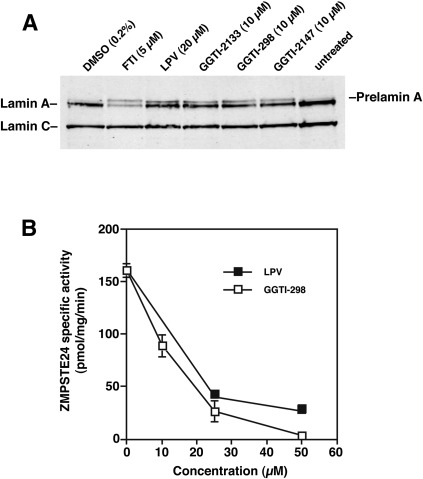
The prelamin A in GGTI-treated cells displayed a slightly faster electrophoretic mobility than nonfarnesylated prelamin A in FTI-treated fibroblasts (Fig. 1AndashC). Previously, we showed that lopinavir, a potent inhibitor of ZMPSTE24, leads to an accumulation of farnesyl–prelamin A in cells (23, 24), and we further showed that the farnesyl–prelamin A in lopinavir-treated cells has a faster electrophoretic mobility than nonfarnesylated prelamin A in FTI-treated cells (23, 24). To characterize the prelamin A in GGTI-treated cells, we compared the electrophoretic mobilities of prelamin A in GGTI-, FTI-, and lopinavir-treated cells (Fig. 3A). The electrophoretic mobility of prelamin A in GGTI- and lopinavir-treated cells was similar, and both migrated faster than prelamin A in FTI-treated cells (Fig. 3A).
Fig. 3.
GGTI-298 inhibits the enzymatic activity of ZMPSTE24. (A) Western blot of protein extracts from human fibroblasts treated with an FTI (ABT-100), the HIV-protease inhibitor lopinavir, or three different GGTIs (GGTI-2133, GGTI-298, and GGTI-2147). Lopinavir blocked ZMPSTE24 and led to an accumulation of farnesylated prelamin A (23, 24). The electrophoretic migration of the farnesyl–prelamin A in lopinavir-treated cells was more rapid than that of nonfarnesylated prelamin A in FTI-treated cells. The prelamin A in GGTI-treated cells was also more rapid, comigrating with the prelamin A in lopinavir-treated cells. (B) ZMPSTE24 activity was measured with a coupled endoprotease/methylation assay (23, 24, 40). The assay measured the ability of yeast membranes expressing mouse ZMPSTE24 to cleave a yeast a-factor substrate, rendering it susceptible to methylation. ZMPSTE24-mediated cleavage of the a-factor substrate in the presence or absence of increasing amounts of an HIV-PI (lopinavir) or a GGTI (GGTI-298) was assessed, and the results were expressed as the percentage activity with membranes incubated in DMSO alone.
The more rapid electrophoretic mobility of prelamin A in GGTI-treated cells suggested that a GGTI might inhibit ZMPSTE24 activity. Indeed, this was the case. Like lopinavir, a GGTI potently inhibited the enzymatic activity of recombinant ZMPSTE24 against a farnesylated CaaX peptide substrate (Fig. 3B).
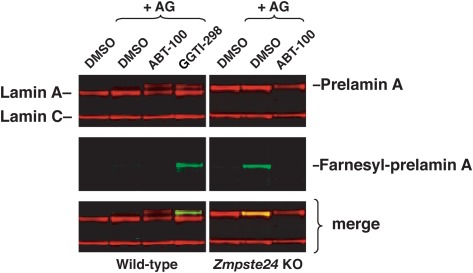
The inhibition of ZMPSTE24 by a GGTI would be expected to lead to an accumulation of farnesyl–prelamin A (23, 24). To determine whether the prelamin A in GGTI-treated cells is farnesylated, we performed metabolic labeling experiments with a farnesol analog, 8-anilinogeraniol (AG) (38), in the presence or absence of either an FTI or a GGTI. After entering cells, AG is converted to anilinogeranyl diphosphate (AGPP) and used as a substrate by FTase for protein prenylation. The incorporation of AG into prelamin A can then be detected by Western blotting with an AG-specific monoclonal antibody (23, 38, 41, 42). The AG-specific antibody detected trace amounts of prenylated prelamin A in wild-type human fibroblasts (43); the accumulation of this small amount of prenylated prelamin A was eliminated by incubation with an FTI (Fig. 4). The prelamin A that accumulates in GGTI-treated cells is farnesylated, as judged by AG incorporation into the prelamin A and consistent with its ability to inhibit ZMPSTE24. In control experiments, we found, as expected, that the prelamin A in ZMPSTE24-deficient cells is farnesylated and that this farnesylation can be blocked with an FTI (Fig. 4).
Fig. 4.
The prelamin A in GGTI-treated cells is farnesylated. Human fibroblasts were treated with a farnesol analog, 8-anilinogeraniol (AG), along with either an FTI (ABT-100) or GGTI (GGTI-298). After entering cells, the AG analog is incorporated into anilinogeranyl diphosphate (AGPP), which is used as a substrate by protein farnesyltransferase (FTase) for prelamin A modification. The incorporation of AG into prelamin A can be detected with an AG-specific antibody (38). Trace amounts of farnesyl–prelamin A were observed in untreated wild-type human fibroblasts but not in the FTI-treated cells. Large amounts of farnesyl–prelamin A were observed in GGTI-298–treated wild-type fibroblasts. Zmspte24-deficient mouse embryonic fibroblasts were included as a control; the prelamin A in these cells was farnesylated, as expected.
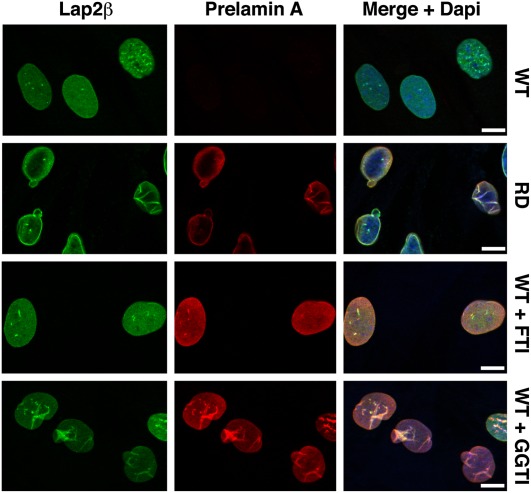
The nonfarnesylated prelamin A in mevinolin-treated fibroblasts is typically found in the nucleoplasm (44), but much of the farnesyl–prelamin A in ZMPSTE24-deficient fibroblasts is found at the nuclear rim or in folds of the nuclear membrane (35, 45), likely reflecting the role of the hydrophobic farnesyl lipid in targeting prelamin A to the nuclear membrane (46). Thus, we predicted that we would find most of the nonfarnesylated prelamin A in FTI-treated fibroblasts in the nucleoplasm and much of the farnesyl–prelamin A in GGTI-treated fibroblasts at the nuclear rim (or in nuclear membrane folds). Indeed, confocal immunofluorescence microscopy revealed an accumulation of prelamin A in nuclear membrane folds in GGTI-treated fibroblasts, whereas nearly all of the prelamin A in FTI-treated fibroblasts was in the nucleoplasm (Fig. 5). As expected, farnesylated prelamin A in human ZMPSTE24-deficient fibroblasts was at the nuclear rim or in nuclear membrane folds (Fig. 5) (35).
Fig. 5.
Confocal immunofluorescence microscopy of FTI- and GGTI-treated wild-type human fibroblasts with antibodies against Lap2β (green) and prelamin A (red). The prelamin A antibody recognizes both farnesylated and nonfarnesylated prelamin A. DNA was stained with DAPI (blue) to visualize nuclei. Restrictive dermopathy (RD) human fibroblasts, which do not express ZMPSTE24 and therefore accumulate farnesyl–prelamin A, were included as a control. Prelamin A was efficiently processed to mature lamin A in wild-type (WT) cells; thus, no prelamin A was detected. In contrast, there was substantial accumulation of farnesyl–prelamin A in RD cells, located mainly along the nuclear membrane and in folds of the nuclear membrane. When wild-type cells were treated with an FTI, there was an accumulation of nonfarnesylated prelamin A, located throughout the nucleoplasm. In GGTI-treated cells, a large percentage of the farnesyl–prelamin A was located in folds of the nuclear membrane. Scale bar, 5 μm.
DISCUSSION
In the current study, we show that commonly used inhibitors of GGTase-I lead to an accumulation of prelamin A by inhibiting ZMPSTE24. Four lines of evidence support this view. First, the prelamin A in GGTI-treated cells displayed a faster electrophoretic mobility than the prelamin A in FTI-treated cells and comigrated with the farnesyl–prelamin A in cells treated with lopinavir, a ZMPSTE24 inhibitor. Second, a GGTI inhibited the enzymatic activity of ZMPSTE24 in a biochemical assay with potency similar to that of lopinavir. Third, metabolic labeling studies showed that the prelamin A that accumulates in GGTI-treated cells is indeed farnesylated. Fourth, the prelamin A in FTI- and GGTI-treated cells exhibited different localization patterns within the cell nucleus. The nonfarnesylated prelamin A in FTI-treated fibroblasts was located mainly in the nucleoplasm, but a significant amount of the farnesyl–prelamin A in GGTI-treated cells was targeted to the nuclear membrane. At the drug concentrations that we used, the GGTI did not inhibit FTase. Only the FTI, and not the GGTI, was capable of inhibiting the farnesylation of HDJ-2, whereas only the GGTI led to the accumulation of nonprenylated RAP1A.
The ability of GGTIs to inhibit ZMPSTE24 cannot be considered unexpected, given what is known about the structure of GGTIs and the substrate specificities of ZMPSTE24. Most GGTIs, including all of the compounds used in this study, are peptidomimetics that were specifically designed to resemble a CaaX motif. Also, the enzymatic activity of the yeast ortholog of ZMPSTE24, Ste24p, is highly sensitive to the structure of the substrate's CaaX motif. Ste24p efficiently cleaves the –aaX from an a-factor substrate terminating in CAMQ, but it is incapable of cleaving an otherwise identical a-factor substrate terminating in CTLM (33, 34). Mammalian ZMPSTE24 shares these CaaX-box specificities (18). Given that the enzymatic activity of ZMPSTE24 depends on the structure of the CaaX motif, the finding that some CaaX peptidomimetics inhibit ZMPSTE24 is not particularly surprising.
The ability of GGTIs to inhibit ZMPSTE24 may explain the GGTI-induced accumulation of prelamin A in the experiments by Varela et al. (32). In their experiments, a GGTI, when added to cells with an FTI, led to substantial amounts of prelamin A accumulation. When they treated cells with an FTI alone, little or no prelamin A was apparent, a peculiar finding that raises questions about the activity of their FTI and the level of FTase inhibition that was achieved. In any case, our studies showing that GGTIs inhibit ZMPSTE24 provide a plausible explanation for the prelamin A accumulation when a GGTI is added to cells. Nevertheless the possibility that prelamin A undergoes alternate prenylation remains intriguing. In the end, a definitive understanding of prelamin A alternate prenylation will require direct biochemical assays and genetic studies in which the CaaX prenyltransferases are inactivated with genetic rather than pharmacologic approaches. Some studies with tissue-specific knockout mice have already been reported (37, 47), and they suggest that alternate prenylation of prelamin A, if it occurs in vivo, may not be particularly efficient.
Our studies showing that GGTIs inhibit ZMPSTE24 are relevant to efforts to develop GGTIs as anticancer agents. GGTIs inhibit the growth of tumor cell lines in culture, including KRAS–transformed cells, and have been shown to be efficacious in human tumor cell xenograft models (48). Also, inactivation of Pggt1b (the gene encoding the β-chain of GGTase-I) inhibits tumor cell growth in vivo and ameliorates disease phenotypes in a mouse model of KRAS–induced lung cancer (49). These observations have prompted interest in GGTase-I inhibitors as anti-cancer therapies, and clinical trials are now underway. In light of our current findings, companies developing GGTIs as anticancer agents will likely want to test whether their compounds inhibit ZMPSTE24 and elicit an accumulation of farnesyl–prelamin A. In both humans and mice, ZMPSTE24 deficiency, and the accompanying accumulation of farnesyl–prelamin A, leads to severe progeroid disorders (9, 17). Thus, it is conceivable that GGTI-mediated ZMPSTE24 inhibition could lead to unwanted complications.
Acknowledgments
The authors thank Drs. David Frost from Abbott Pharmaceuticals for ABT-100 and Michael Gelb from the University of Washington for PB-43.
Footnotes
Abbreviations:
- FTase
- protein farnesyltransferase
- FTI
- protein farnesyltransferase inhibitor
- GGTase-I
- protein geranylgeranyltransferase-I
- GGTI
- protein geranylgeranyltransferase inhibitor
- ZMPSTE24
- zinc metalloprotease Ste24p ortholog
This work was supported by National Institutes of Health Grants HL-76839, HL-86683, HL-089781, AG-035626, and GM-66152; March of Dimes Grant 6-FY2007-1012; the Ellison Medical Foundation Senior Scholar Program; and Beginning Grant-in-Aid 0865262F from the American Heart Association, Western States Affiliate. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health or other granting agencies.
REFERENCES
- 1.Zhang F. L., Casey P. J. 1996. Protein prenylation: molecular mechanisms and functional consequences. Annu. Rev. Biochem. 65: 241–269 [DOI] [PubMed] [Google Scholar]
- 2.Casey P. J., Seabra M. C. 1996. Protein prenyltransferases. J. Biol. Chem. 271: 5289–5292 [DOI] [PubMed] [Google Scholar]
- 3.Schafer W. R., Rine J. 1992. Protein prenylation: genes, enzymes, targets, and functions. Annu. Rev. Genet. 26: 209–237 [DOI] [PubMed] [Google Scholar]
- 4.Clarke S. 1992. Protein isoprenylation and methylation at carboxyl-terminal cysteine residues. Annu. Rev. Biochem. 61: 355–386 [DOI] [PubMed] [Google Scholar]
- 5.Dalton M. B., Fantle K. S., Bechtold H. A., DeMaio L., Evans R. M., Krystosek A., Sinensky M. 1995. The farnesyl protein transferase inhibitor BZA-5B blocks farnesylation of nuclear lamins and p21ras but does not affect their function or localization. Cancer Res. 55: 3295–3304 [PubMed] [Google Scholar]
- 6.Dalton M., Sinensky M. 1995. Expression systems for nuclear lamin proteins: farnesylation in assembly of nuclear lamina. Methods Enzymol. 250: 134–148 [DOI] [PubMed] [Google Scholar]
- 7.Sinensky M., Fantle K., Trujillo M., McLain T., Kupfer A., Dalton M. 1994. The processing pathway of prelamin A. J. Cell Sci. 107: 61–67 [DOI] [PubMed] [Google Scholar]
- 8.Young S. G., Ambroziak P., Kim E., Clarke S. 2000. Postisoprenylation protein processing: CXXX (CaaX) endoproteases and isoprenylcysteine carboxyl methyltransferase. In The Enzymes. Vol. 21. F. Tamanoi and D. S. Sigman, editors. Academic Press, San Diego 155–213 [Google Scholar]
- 9.Bergo M. O., Gavino B., Ross J., Schmidt W. K., Hong C., Kendall L. V., Mohr A., Meta M., Genant H., Jiang Y., et al. 2002. Zmpste24 deficiency in mice causes spontaneous bone fractures, muscle weakness, and a prelamin A processing defect. Proc. Natl. Acad. Sci. USA. 99: 13049–13054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bergo M. O., Young S. G. 2004. Zmpste24 (mammalian farnesylated protein-converting enzyme 1). In Handbook of Proteolytic Enzymes. 2nd edition. Vol. 2A. J. Barrett, N. D. Rawlings, and J. F. Woessner, editors. Academic Press, London p.677–682 [Google Scholar]
- 11.Clarke S., Vogel J. P., Deschenes R. J., Stock J. 1988. Posttranslational modification of the Ha-ras oncogene protein: evidence for a third class of protein carboxyl methyltransferases. Proc. Natl. Acad. Sci. USA. 85: 4643–4647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dai Q., Choy E., Chiu V., Romano J., Slivka S. R., Steitz S. A., Michaelis S., Philips M. R. 1998. Mammalian prenylcysteine carboxyl methyltransferase is in the endoplasmic reticulum. J. Biol. Chem. 273: 15030–15034 [DOI] [PubMed] [Google Scholar]
- 13.Xie H., Yamane H. K., Stephenson R. C., Ong O. C., Fung B. K.-K., Clarke S. 1990. Analysis of prenylated carboxyl-terminal cysteine methyl esters in proteins. Methods. 1: 276–282 [Google Scholar]
- 14.Kilic F., Dalton M. B., Burrell S. K., Mayer J. P., Patterson S. D., Sinensky M. 1997. In vitro assay and characterization of the farnesylation-dependent prelamin A endoprotease. J. Biol. Chem. 272: 5298–5304 [DOI] [PubMed] [Google Scholar]
- 15.Corrigan D. E., Kuxzczak D., Rusinol A. E., Thewke D. P., Hrycyna C. A., Michaelis S., Sinensky M. S. 2005. Prelamin A endoproteolytic processing in vitro by recombinant Zmpste24. Biochem. J. 387: 129–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Young S. G., Fong L. G., Michaelis S. 2005. Prelamin A, Zmpste24, misshapen cell nuclei, and progeria—new evidence suggesting that protein farnesylation could be important for disease pathogenesis. J. Lipid Res. 46: 2531–2558 [DOI] [PubMed] [Google Scholar]
- 17.Pendás A. M., Zhou Z., Cadiñanos J., Freije J. M. P., Wang J., Hultenby K., Astudillo A., Wernerson A., Rodríguez F., Tryggvason K., et al. 2002. Defective prelamin A processing and muscular and adipocyte alterations in Zmpste24 metalloproteinase-deficient mice. Nat. Genet. 31: 94–99 [DOI] [PubMed] [Google Scholar]
- 18.Leung G. K., Schmidt W. K., Bergo M. O., Gavino B., Wong D. H., Tam A., Ashby M. N., Michaelis S., Young S. G. 2001. Biochemical studies of Zmpste24-deficient mice. J. Biol. Chem. 276: 29051–29058 [DOI] [PubMed] [Google Scholar]
- 19.Hrycyna C. A., Sapperstein S. K., Clarke S., Michaelis S. 1991. The Saccharomyces cerevisiae STE14 gene encodes a methyltransferase that mediates C-terminal methylation of a-factor and Ras proteins. EMBO J. 10: 1699–1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hrycyna C. A., Clarke S. 1990. Farnesyl cysteine C-terminal methyltransferase activity is dependent upon the STE14 gene product in Saccharomyces cerevisiae. Mol. Cell. Biol. 10: 5071–5076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Otto J. C., Kim E., Young S. G., Casey P. J. 1999. Cloning and characterization of a mammalian prenyl protein-specific protease. J. Biol. Chem. 274: 8379–8382 [DOI] [PubMed] [Google Scholar]
- 22.Kim E., Ambroziak P., Otto J. C., Taylor B., Ashby M., Shannon K., Casey P. J., Young S. G. 1999. Disruption of the mouse Rce1 gene results in defective Ras processing and mislocalization of Ras within cells. J. Biol. Chem. 274: 8383–8390 [DOI] [PubMed] [Google Scholar]
- 23.Coffinier C., Hudon S. E., Lee R., Farber E. A., Nobumori C., Miner J. H., Andres D. A., Spielmann H. P., Hrycyna C. A., Fong L. G., et al. 2008. A potent HIV protease inhibitor, darunavir, does not inhibit ZMPSTE24 or lead to an accumulation of farnesyl-prelamin A in cells. J. Biol. Chem. 283: 9797–9804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coffinier C., Hudon S. E., Farber E. A., Chang S. Y., Hrycyna C. A., Young S. G., Fong L. G. 2007. HIV protease inhibitors block the zinc metalloproteinase ZMPSTE24 and lead to an accumulation of prelamin A in cells. Proc. Natl. Acad. Sci. USA. 104: 13432–13437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen W-J., Andres D. A., Goldstein J. L., Russell D. W., Brown M. S. 1991. cDNA cloning and expression of the peptide-binding b subunit of rat p21ras farnesyltransferase, the counterpart of yeast DPR1/RAM1. Cell. 66: 327–334 [DOI] [PubMed] [Google Scholar]
- 26.James G. L., Goldstein J. L., Brown M. S., Rawson T. E., Somers T. C., McDowell R. S., Crowley C. W., Lucas B. K., Levinson A. D., Marsters J. C., Jr 1993. Benzodiazepine peptidomimetics: potent inhibitors of Ras farnesylation in animal cells. Science. 260: 1937–1942 [DOI] [PubMed] [Google Scholar]
- 27.Reiss Y., Goldstein J. L., Seabra M. C., Casey P. J., Brown M. S. 1990. Inhibition of purified p21ras farnesyl:protein transferase by Cys-AAX tetrapeptides. Cell. 62: 81–88 [DOI] [PubMed] [Google Scholar]
- 28.Sun J., Ohkanda J., Coppola D., Yin H., Kothare M., Busciglio B., Hamilton A. D., Sebti S. M. 2003. Geranylgeranyltransferase I inhibitor GGTI-2154 induces breast carcinoma apoptosis and tumor regression in H-Ras transgenic mice. Cancer Res. 63: 8922–8929 [PubMed] [Google Scholar]
- 29.Basso A. D., Kirschmeier P., Bishop W. R. 2006. Thematic review series: lipid posttranslational modifications. Farnesyl transferase inhibitors. J. Lipid Res. 47: 15–31 [DOI] [PubMed] [Google Scholar]
- 30.Berndt N., Hamilton A. D., Sebti S. M. 2011. Targeting protein prenylation for cancer therapy. Nat. Rev. Cancer. 11: 775–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Whyte D. B., Kirschmeier P., Hockenberry T. N., Nunez-Oliva I., James L., Catino J. J., Bishop W. R., Pai J-K. 1997. K- and N-Ras are geranylgeranylated in cells treated with farnesyl protein transferase inhibitors. J. Biol. Chem. 272: 14459–14464 [DOI] [PubMed] [Google Scholar]
- 32.Varela I., Pereira S., Ugalde A. P., Navarro C. L., Suarez M. F., Cau P., Cadinanos J., Osorio F. G., Foray N., Cobo J., et al. 2008. Combined treatment with statins and aminobisphosphonates extends longevity in a mouse model of human premature aging. Nat. Med. 14: 767–772 [DOI] [PubMed] [Google Scholar]
- 33.Boyartchuk V. L., Ashby M. N., Rine J. 1997. Modulation of Ras and a-factor function by carboxyl-terminal proteolysis. Science. 275: 1796–1800 [DOI] [PubMed] [Google Scholar]
- 34.Trueblood C. E., Boyartchuk V. L., Picologlou E. A., Rozema D., Poulter C. D., Rine J. 2000. The CaaX proteases, Afc1p and Rce1p, have overlapping but distinct substrate specificities. Mol. Cell. Biol. 20: 4381–4392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Toth J. I., Yang S. H., Qiao X., Beigneux A. P., Gelb M. H., Moulson C. L., Miner J. H., Young S. G., Fong L. G. 2005. Blocking protein farnesyltransferase improves nuclear shape in fibroblasts from humans with progeroid syndromes. Proc. Natl. Acad. Sci. USA. 102: 12873–12878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fong L. G., Ng J. K., Meta M., Cote N., Yang S. H., Stewart C. L., Sullivan T., Burghardt A., Majumdar S., Reue K., et al. 2004. Heterozygosity for Lmna deficiency eliminates the progeria-like phenotypes in Zmpste24-deficient mice. Proc. Natl. Acad. Sci. USA. 101: 18111–18116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang S. H., Chang S. Y., Tu Y., Lawson G. W., Bergo M. O., Fong L. G., Young S. G. 2012. Severe hepatocellular disease in mice lacking one or both CaaX prenyltransferases. J. Lipid Res. 53: 77–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Troutman J. M., Roberts M. J., Andres D. A., Spielmann H. P. 2005. Tools to analyze protein farnesylation in cells. Bioconjug. Chem. 16: 1209–1217 [DOI] [PubMed] [Google Scholar]
- 39.Davies B. S., Barnes R. H., 2nd, Tu Y., Ren S., Andres D. A., Spielmann H. P., Lammerding J., Wang Y., Young S. G., Fong L. G. 2010. An accumulation of non-farnesylated prelamin A causes cardiomyopathy but not progeria. Hum. Mol. Genet. 19: 2682–2694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hudon S. E., Coffinier C., Michaelis S., Fong L. G., Young S. G., Hrycyna C. A. 2008. HIV-protease inhibitors block the enzymatic activity of purified Ste24p. Biochem. Biophys. Res. Commun. 374: 365–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang S. H., Andres D. A., Spielmann H. P., Young S. G., Fong L. G. 2008. Progerin elicits disease whether or not it is farnesylated. J. Clin. Invest. 118: 3291–3300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang S. H., Chang S. Y., Andres D. A., Spielmann H. P., Young S. G., Fong L. G. 2010. Assessing the efficacy of protein farnesyltransferase inhibitors in mouse models of progeria. J. Lipid Res. 51: 400–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fong L. G., Vickers T. A., Farber E. A., Choi C., Yun U. J., Hu Y., Yang S. H., Coffinier C., Lee R., Yin L., et al. 2009. Activating the synthesis of progerin, the mutant prelamin A in Hutchinson-Gilford progeria syndrome, with antisense oligonucleotides. Hum. Mol. Genet. 18: 2462–2471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lutz R. J., Trujillo M. A., Denham K. S., Wenger L., Sinensky M. 1992. Nucleoplasmic localization of prelamin A: implications for prenylation-dependent lamin A assembly into the nuclear lamina. Proc. Natl. Acad. Sci. USA. 89: 3000–3004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang S. H., Bergo M. O., Toth J. I., Qiao X., Hu Y., Sandoval S., Meta M., Bendale P., Gelb M. H., Young S. G., et al. 2005. Blocking protein farnesyltransferase improves nuclear blebbing in mouse fibroblasts with a targeted Hutchinson-Gilford progeria syndrome mutation. Proc. Natl. Acad. Sci. USA. 102: 10291–10296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hennekes H., Nigg E. A. 1994. The role of isoprenylation in membrane attachment of nuclear lamins. A single point mutation prevents proteolytic cleavage of the lamin A precursor and confers membrane binding properties. J. Cell Sci. 107: 1019–1029 [DOI] [PubMed] [Google Scholar]
- 47.Lee R., Chang S. Y., Trinh H., Tu Y., White A. C., Davies B. S., Bergo M. O., Fong L. G., Lowry W. E., Young S. G. 2010. Genetic studies on the functional relevance of the protein prenyltransferases in skin keratinocytes. Hum. Mol. Genet. 19: 1603–1617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lobell R. B., Omer C. A., Abrams M. T., Bhimnathwala H. G., Brucker M. J., Buser C. A., Davide J. P., deSolms S. J., Dinsmore C. J., Ellis-Hutchings M. S., et al. 2001. Evaluation of farnesyl:protein transferase and geranylgeranyl:protein transferase inhibitor combinations in preclinical models. Cancer Res. 61: 8758–8768 [PubMed] [Google Scholar]
- 49.Sjogren A. K., Andersson K. M., Liu M., Cutts B. A., Karlsson C., Wahlstrom A. M., Dalin M., Weinbaum C., Casey P. J., Tarkowski A., et al. 2007. GGTase-I deficiency reduces tumor formation and improves survival in mice with K-RAS-induced lung cancer. J. Clin. Invest. 117: 1294–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]