Abstract
Passive transfer of broadly neutralizing HIV antibodies can prevent infection, which suggests that vaccines that elicit such antibodies would be protective. Thus far, however, few broadly neutralizing HIV antibodies that occur naturally have been characterized. To determine whether these antibodies are part of a larger group of related molecules, we cloned 576 new HIV antibodies from four unrelated individuals. All four individuals produced expanded clones of potent broadly neutralizing CD4-binding-site antibodies that mimic binding to CD4. Despite extensive hypermutation, the new antibodies shared a consensus sequence of 68 immunoglobulin H (IgH) chain amino acids and arise independently from two related IgH genes. Comparison of the crystal structure of one of the antibodies to the broadly neutralizing antibody VRC01 revealed conservation of the contacts to the HIV spike.
Two to three years after infection, some HIV-infected patients develop serum antibodies that can neutralize a broad spectrum of HIV viruses (1–4). Among the naturally occurring monoclonal antibodies, VRC01, an antibody cloned from memory B cells that targets the CD4-binding site (CD4bs) on the HIV envelope spike, is unusual in its potency and breadth (5, 6). As do other HIV antibodies obtained by the single-cell antigen-capture method (7), VRC01 shows high levels of somatic mutations that are essential for its activity (6–8). This high frequency of mutation is a potential impediment to antibody identification, because the mutated sequences may no longer be complementary to the primers used for immunoglobulin gene amplification (9). To avert this potential problem, we developed a new primer set specifically designed to address this problem (the 5′ primer is set farther upstream to avoid the mutated region) (fig. S1A and table S1) (10). The new strategy was tested by sorting single B cells from a patient with high titers of broadly neutralizing antibodies (Pt 8) (table S2) that bind to an HIV gp120 core glycoprotein stabilized in the CD4-bound conformation and lacking the variable (V loops 1 to 3) (2CC core) ( (fig. S1B) (11, 12). In contrast to the resurfaced protein used to clone VRC01, which was designed to focus on antibodies to the CD4bs, the 2CC core should capture additional antibodies including those specific to the CD4-induced co-receptor–binding site (CD4i) (11, 12).
In side-by-side comparisons, the new primer set increased recovery of immunoglobulin H (IgH) chains when compared with the original primer set (fig. S2, A and B) (9). As expected, the antibodies obtained with the new primer set were more mutated (fig. S2, A and C) (average 35.7 versus 19.8, P = 0.0013, and maximum 85 versus 50 for IgH ) and included clones not found with the original primer set (fig. S2, A and B). Moreover, the new primers also recovered VRC01-related antibodies from cDNA samples isolated from single cells that had been sorted with the original YU2-gp140 trimer probe used to develop the single-cell antibody-cloning method (7, 13) (fig. S3, A and B).
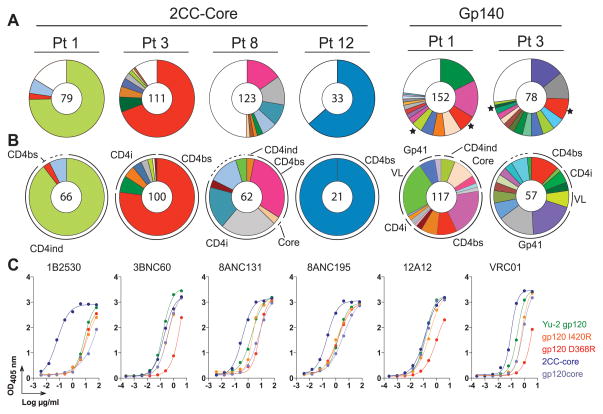
Four unrelated HIV-infected individuals showing high titers of broadly neutralizing antibodies were examined by using the 2CC core (table S2 and fig. S4, A and B). Two of these individuals, Pt 1 and Pt 3, had been studied previously, but their cloned antibodies could not account for their serologic activity (7). From single sorted B cells representing 200 different B cell clones that were diversified by mutation in germinal centers, 576 antibodies were obtained from a starting population of 1.5 × 105 IgG+ memory B cells (Fig. 1A and table S3). Representative members of each expanded B cell clone were tested for binding to gp120 and were all positive (Fig. 1, B and C; fig. S5; and table S3). The site of antibody binding on the envelope spike was mapped by using mutant proteins that interfere with either the CD4bs [gp120(D368R)] (14–16), or the CD4i site [gp120(I420R)] (17). [These mutants have substitutions, respectively, Arg for Asp at position 368 and Arg for Ile at position 420.] NIH45-46, which is a VRC01 variant, and antibodies 3BNC60, 8ANC131, and 12A12 (antibodies selected on the basis of neutralizing activity, see below) (Fig. 1C, fig. S5, and table S3) showed binding patterns similar to VRC01’s. Others, including 1B2530 and 8ANC195, could not be classified precisely solely on the basis of enzyme-linked immunosorbent assay (ELISA). As expected from earlier studies on HIV envelope–specific antibodies (8), 65% of the antibodies isolated by using the 2CC core were polyreactive (fig. S6) compared with 22.7% polyreactivity in healthy control memory B cells (18) and 17.3% in gp140-negative B cells from HIV-positive controls (8). Somatic hypermutation was likely required for development of high-affinity antigen binding and polyreactivity because reversion of four representative antibodies to the corresponding germ line led to complete loss of binding to YU2-gp140 (13) (fig. S6B and fig. S7, A to C).
Fig. 1.
The 2CC core captures CD4bs antibodies. (A and B) Top line indicates bait used for sorting, and below is the patient number. The number in the center of the pies denotes the number of antibodies; slices are unique clones and are proportional to clone size. Membership of an antibody in a B cell clone is determined by sequence analysis, in particular CDR3s and shared V and J genes of paired heavy- and light-chain genes (table S3). (A) Pie charts show a summary of detected B cell clones irrespective of their binding epitopes. (★) Star indicates clones found in both YU2-gp140 and 2CC core–sorted cells. (B) Pie charts show the distribution of antibodies binding to CD4bs, CD4i, CD4ind. (equally affected by D368R and I420R), and core (not affected by either D368R or I420R). Dashed lines indicate antibodies that were cloned but could not be produced. (C) Representative ELISAs on YU2-gp120, mutants, and 2CC core.
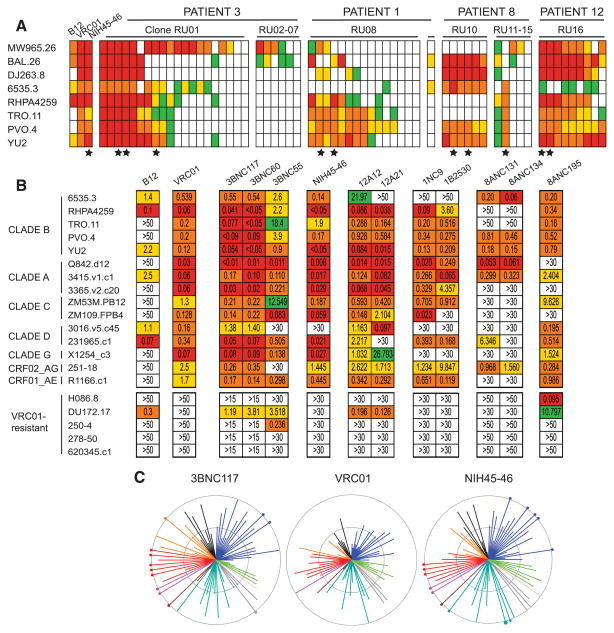
HIV-neutralizing activity was measured in vitro by using an initial panel of eight viruses including three tier 1 clade A, B, and C, and five tier 2 clade B envelope (Env) pseudovirus variants (19, 20). The neutralizing activity of the antibodies was compared with VRC01 and purified serum IgG from the donors (Fig. 2A, fig. S4, and table S4). A selection of 11 representative antibodies showing high levels of neutralizing activity (Fig. 2A and table S5) were further tested on a panel of 15 additional tier 2 Env pseudovirus variants (Fig. 2B and table S6), including five viruses that are resistant to VRC01 (Fig. 2B and table S6). Of all of the antibodies tested, 88% showed some neutralizing activity, and six clones (RU01, 08, 10, 12, 16, and NIH45-46) contained antibodies that were highly potent and broad (Fig. 2 and tables S4 and S6). These clones were also the most abundant among those captured by 2CC core in each of the four patients studied (Fig. 1A and table S3). Five antibodies representing four different broadly neutralizing founder B cell clones [clone RU01 (3BNC117 and 3BNC55), clone RU16 (12A12), clone RU12 (8ANC195), and NIH45-46] (table S5), were tested against an expanded panel of 118 tier 2 viral isolates from all known clades, including 32 transmitted founder viruses (Fig. 2C and table S7) [VRC01 had previously been tested on 82 of these (6).] The most impressive of the new antibodies, 3BNC117, belonging to a clone with 85 members (RU01), showed an 80% inhibitory concentration (IC80) on a combined group of 95 tier 2 viruses of 1.4 μg/ml (median and geometric mean IC80 values 0.3μg/ml, table S7B). When compared with the previously published VRC01 neutralization data, only 17 of the viruses tested were more sensitive to VRC01 than 3BNC117, and 3BNC117 showed greater breadth (Fig. 2, B and C, and tables S4, S6, and S7) (6). NIH45-46, a new variant of VRC01, was more potent than VRC01 on 62 of the viruses tested but still less potent than 3BNC117 (Fig. 2, B and C, and tables S4, S6, and S7) (6). Together, the new antibodies neutralized 96% of the 118 viruses tested (table S7A). Finally, the best antibodies were highly hypermutated, and this was essential for their breadth and potency (fig. S7D and tables S4, S6, and S7).
Fig. 2.
Antibodies captured by 2CC core have broad and potent HIV-neutralizing activity. (A) HIV-neutralizing activity assayed on a limited panel of viruses. Top line indicates the donor number (table S2). Antibody clones are grouped, and individual clonal relatives are represented by a column grouped as in table S4. Each row represents one virus as indicated on the left. Colors indicate concentration of clones at the median inhibitory concentration (IC50): red, ≤0.1 μg/ml; orange, 0.1 to 1 μg/ml; yellow, 1 to 10 μg/ml; green, ≥10 μg/ml; and white, not neutralized at any concentration tested. (★) Star indicates the representatives selected for the extended virus panel (table S5). (B) HIV-neutralizing activity assayed on an extended panel of viruses (tables S5 and S6). (C) Neutralization summary graph comparing the published IC50 values of VRC01 (6) with NIH45-46 and 3BNC117 (tables S5 and S7). Each color represents a different HIV clade (black corresponds to CRF01AE, blue to clade B, green to clade G, pink to clade D, gray to clade CD, emerald to clade C, brown to clade ACD, magenta to clade AC, red to clade A, and orange to CRF02AG). Length of lines and size of circles are inversely proportional to IC50. The distance between the outer and inner circle, as well as from the inner circle to the center of the spiders, in each span is two logs in IC50 concentration.
Our cloning strategy captures antibodies produced by antigen-binding memory B cells in the blood, but circulating antibodies are not produced by these cells; they originate instead from plasma cells in the bone marrow (21). To determine whether the antibodies cloned from memory B cells are also found in the bone marrow plasma cell compartment, we purified plasma cells from paired bone marrow samples from patients 3 and 8 (table S2 and fig. S8A) and used polymerase chain reaction (PCR) to specifically amplify IgVH genes from the clones RU01 and RU10 from memory B cells in these individuals (Fig. 2A, fig. S8A, and table S3). Members of these clones and large numbers of additional variants were readily identified in the respective plasma cell samples (fig. S8, B and C). We also verified that antibodies from clones RU01, RU08, and RU10 (Fig. 2A and table S3) are found in serum by mass spectrometry (fig. S9 and table S8).
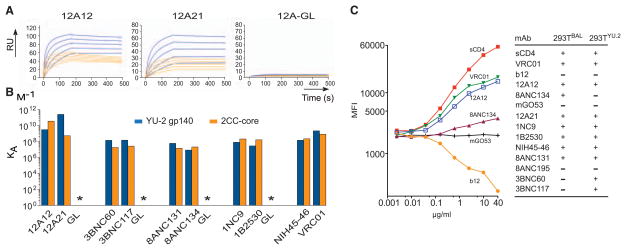
To determine whether antibody affinity to gp120 is related to neutralizing activity, we compared the binding of the highly active antibodies, selected clonal relatives, and germ line–reverted progenitors by using surface plasmon resonance (SPR) (Fig. 3, A and B; figs. S7B and S10; and table S9). The top neutralizing antibodies (tables S4 to S7) showed affinities (KA) ranging from ≃107 to 1012 (M−1) to YU2-gp140 trimers (13) and ≃107 to 1011 (M−1) to the 2CC core (11) (Fig. 3, A and B, and tables S4, S6 and S9). Consistent with their decreased neutralizing potency and breadth, 3BNC66, 3BNC156, and 3BNC55 displayed lower affinities to the YU2-gp140 trimer than 3BNC117, but surprisingly, the affinities of these antibodies to 2CC core did not correlate with their neutralizing activity (Fig. 2, fig. S10, and tables S4 and S9). Finally, we were unable to detect binding by any of the germ line–reverted antibodies tested (Fig. 3B, fig. S7B, and table S9). We conclude that the antibodies captured by the 2CC core tend to show higher affinity to the YU2-gp140 trimer than to the 2CC core.
Fig. 3.
Binding properties of antibodies. (A) Representative SPR sensor-grams for binding to YU2-gp140 and 2CC core by 12A12, 12A21 (Fig. 2B and table S5), and 12A germ line (GL)–reverted antibodies. (B) Graph shows KA for representative antibodies (tables S5 and S9). (C) Graph shows mean fluorescence intensity of CD4i antibody 3-67 (7) binding to Bal-expressing 293T cells after incubation with the indicated antibodies. Table indicates whether or not an antibody induces CD4i site accessibility (fig. S11).
When VRC01 binds to the HIV spike, it produces large conformational changes that mimic CD4 binding and expose the CD4i site (6). By contrast, the broadly neutralizing antibody b12 (22) and many other known CD4bs antibodies do not (23). To determine whether the ability to mimic CD4 is a shared feature of the most potent antibodies (tables S4 to S7), we expressed two different HIV spikes (24) on the surface of human embryonic kidney–293 (HEK 293T) cells and measured CD4i-antibody binding in the presence or absence of CD4 or CD4bs antibodies (Fig. 3C). With one exception, all of the antibodies tested resemble CD4 and VRC01 in that they facilitate CD4i-antibody binding to one or both viral spikes (Fig. 3C and fig. S11). The only antibody tested that did not share this characteristic, 8ANC195, was not a traditional CD4bs antibody in that it was equally sensitive to the D368R and I420R mutations (Fig. 1C and table S3), and it differed from the others in its neutralization pattern (Fig. 2, A and B; and tables S4, S6, and S7).
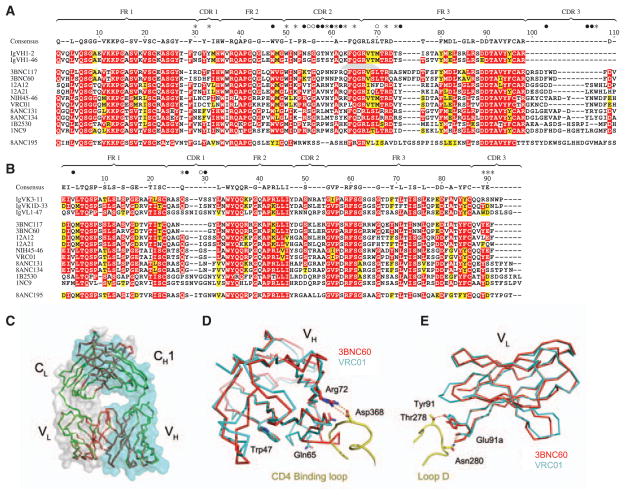
To determine whether highly active CD4bs antibodies share common sequence features, we aligned the 10 best antibodies (table S5): two variants each from independently derived antibody clones arising in each of the four patients studied and from NIH45 (Fig. 4, A and B; table S2; and fig. S3) (6). Comparison of the IgVH regions revealed a conserved consensus sequence covering 68 IgVH residues (Fig. 4A). It is noteworthy that the IgVH consensus contains seven VRC01-gp120 contact residues, including Arg 71VRC01, which mimics the key interaction of Arg59CD4 and Asp368gp120 (Fig. 4A) (5, 25). All 10 antibodies arise from only two closely related germline IgVH genes that conserve six contact residues (Fig. 4A, fig. S12A, and table S3). The codons of the consensus residues are highly somatically mutated in the 10 selected antibodies; however, the ratio of replacement to silent mutations in the consensus residues ranges from 0.7 to 1.7, whereas it is 3.5 to 22 in the nonconsensus residues, which together indicate that conservation of the consensus is strongly selected (fig. S13 and table S10). The light chain of VRC01 makes fewer contacts with gp120 (5). Consistent with its more limited role, comparison of the light-chain sequences of the same antibodies uncovers a less-extensive consensus covering 53 residues from the variable region of immunoglobulin light chain (VL residues), which includes three VRC01-gp120 contact residues (Fig. 4B and fig. S3B). Finally, like the heavy chains, the light chains arise from a limited set of germ-line genes: Two are derived from IgK1D-33, two from IgK3-11, and one from IgL1-47 (Fig. 4B, fig. S12B, and table S3) (5). Antibody 8ANC195, which differed from the others in several important respects (Figs. 1C, 2B, and 3C; fig. S11; and tables S4, S6, and S7), did not entirely conform to the consensus and did not arise from related heavy or light chains (Fig. 4, A and B; fig. S14; and table S3). Thus, there is significant sequence convergence among highly active agonistic CD4bs antibodies (HAADs).
Fig. 4.
Sequence and structural conservation of HAADs. (A) Amino acid alignment of 10 selected HAADs (table S5), their germline genes, and 8ANC195. Residues are numbered according to the 3BNC60 structure. Framework (FR) and CDR regions are indicated. Red shading shows amino acid identity; yellow shows biochemical similarity. The consensus is defined by 70% similarity between the 10 selected HAADs. The consensus sequence is shown above; dashes in this sequence indicate nonconserved residues. Contact residues between VRC01 and gp120 are shown above the consensus as closed circles for main-and side-chain interactions, open circles main chain only, and stars side chains only (5). (B) As in (A) for light chains. (C, D, and E) Crystal structure of 3BNC60 Fab. (C) Superimposed Cα traces of the two Fab molecules in the 3BNC60 asymmetric unit are shown in green and red. Semitransparent surfaces are used to outline the heavy (cyan) and light (gray) chains. (D) Superimposition of the 3BNC60 VH (red, Cα trace) and VRC01 VH (cyan, Cα trace) shown with a ribbon representation of the CD4-binding loop. The salt bridge between Arg71VRC01 and Asp368gp120 is shown as dashed lines. (E) Superimposition of the 3BNC60 VL (red, Cα trace). Hydrogen bonds between VRC01 and gp120 are shown as dashed lines.
To determine whether the structure of the antibodies in different patients is also conserved, we solved the crystal structure of the 3BNC60 Fab to 2.65 Å resolution and compared it with VRC01’s (26) (Fig. 4, C to E). Superimposition of domains in the variable region of immunoglobulin heavy chain (VH domains) from 3BNC60 and VRC01 in the VRC01-gp120 cocrystal structure (5) yields a root mean square deviation (RMSD) of 1.3 Å (calculated for 111 Cα atoms) with major differences confined to CDR2 residues 58 to 65 (3BNC60 numbering). Superimposing the structures suggests conservation of the recognition interface with gp120 (Fig. 4D). For example, Arg723BNC60 adopts a similar conformation as Arg71VRC01, which mimics an important salt bridge normally formed between Arg59CD4 and Asp368gp120 (5, 25). Gln653BNC60, which corresponds to Gln64VRC01, is within the residue segment (residues 58 to 65) that differs in structure from VRC01. The conformation of this region of 3BNC60, which is involved in a lattice contact in the crystals, is likely to change upon binding gp120, as it would clash with the CD4-binding loop on gp120. Superimposing the 3BNC60 and VRC01 VL domains yields a RMSD of 0.9Å (calculated for 95 Cα atoms) and shows that some of gp120-contacting residues are structurally conserved (Fig. 4E); Tyr913BNC60 and Glu903BNC60 adopt similar conformations as Tyr91VRC01 and Glu96VRC01, which engage loop D of gp120 via polar interactions (Fig. 4E). The VH and VL domains of the two Fabs show similar interdomain interactions and overall orientation, as super-imposition of the VH and VL domains of 3BNC60 and VRC01 yields an RMSD of 1.4 Å (calculated for 206 Cα atoms). Overall, these structural comparisons suggest that 3BNC60 binds gp120 with the same architecture as observed for the binding of VRC01 (5).
Our experiments define a class of agonistic CD4bs antibodies, HAADs, that shares IgVH and IgVL consensus sequences including 10 of the contact residues between VRC01 and the HIV spike (Fig. 4, A and B). In five different donors, these antibodies originate from only two closely related IgVH and three IgVL genes (Fig 4, A and B). Although we cannot exclude the possibility that by using the 2CC core as bait for single-cell sorting might introduce a selection bias, gp140 sorts performed on two of the donors did not reveal additional broadly neutralizing antibodies (tables S3, E and F, and S4), which suggests that a significant fraction of the CD4bs-directed repertoire was captured in our experiments. Thus, the repertoire of antibodies that are available to solve the problem of broad and potent HIV neutralization by binding to the CD4bs appears to be somewhat restricted. Despite this restriction, HAADs account for a significant fraction of the memory B cell, and plasma cell compartments, and the circulating IgGs in the patients tested. Therefore, HAADs are not rare, and once elicited, they contribute to the circulating antibody pool in patients that neutralize the virus broadly. However, HAADs are not found in all patients with high titers of broad neutralizing activity (1, 27, 28). Moreover, broad neutralization can be achieved by targeting epitopes other than the CD4bs (29–34) or by combinations of antibodies to different epitopes (7, 35), or even combination of different CD4bs antibodies (table S7).
The high levels of neutralizing activity displayed by HAADs appear to be correlated with binding to a specific surface, which is similar, but not identical, to that contacted by CD4 (5, 36). Why a specific type of binding that mimics CD4 appears to be required is not known (6); however, it has been suggested that binding of soluble CD4, or CD4 mimetic compounds, to the HIV envelope trimer leads to the formation of a metastable intermediate that decays rapidly and loses the ability to fuse (37). We speculate that HAAD binding mimics soluble CD4 and destabilizes the trimer. Irrespective of their mechanism of action, HAADs might contribute to viremic control in a subset of HIV-infected individuals and may be useful in HIV prevention, or possibly even therapy, because of the low concentrations required for viral neutralization.
Supplementary Material
Acknowledgments
The data reported in this paper are tabulated in the supporting online material, and the coordinates for the 3BNC60 Fab as well as the antibody sequences have been deposited in the Protein Data Bank and EMBL-EBI database [accession codes: 3RPI (3BNC60 coordinates), HE584535-HE584554 (antibody sequences). We thank the HIV-positive donors for their support. We thank M. Jankovic for discussions and help with cloning of HIV envelope constructs; R. Wyatt for the plasmid of YU2-gp140; J. Mascola for the plasmid of 2CC-core and YU2-gp120 mutants; and M. Connors for access to patient NIH45; C. Gaebler, M. Warncke, P. M. Marcovecchio, and H. Gao for help with protein expression; K. Moss for patient coordination. We would like to thank Francine McCutchan, George Shaw, Beatrice Hahn, Joshua Baalwa, David Montefiori, Feng Gao, Michael Thomson, Julie Overbaugh, Ronald Swanstrom, Lynn Morris, Jerome Kim, Linqi Zhang, Dennis Ellenberger, and Carolyn Williamson for contributing the HIV-1 Envelope plasmids used in our neutralization panel. We thank M. Simek, F. Priddy, and all the study participants and research staff at each of the International AIDS Vaccine Initiative (IAVI) Protocol G project; clinical and site team members; and all of the Protocol G clinical investigators, specifically, G. Miiro, A. Pozniak, D. McPhee, O. Manigart, E. Karita, A. Inwoley, W. Jaoko, J. DeHovitz, L.-G. Bekker, P. Pitisuttithum, R. Paris, J. Serwanga, and S. Allen for samples from one broadly neutralizing serum donor from Protocol G. We thank L. Walker for providing data on the neutralizing specificity of the Protocol G donor. In connection with this work, M.C.N. and J.F.S. have a pending patent application with the U.S. Patent and Trademark Office, patent number U.S. 61/486,960, entitled “Human Immunodeficiency Virus Neutralizing Antibodies and Methods of Use Thereof.” The reagents are available with a Materials Transfer Agreement. The work was supported by NIH grants, P01 AI081677, RR00862, and RR022220, and by the Bill & Melinda Gates Foundation’s Collaboration for AIDS Vaccine Discovery/Comprehensive Antibody–Vaccine Immune Monitoring Consortium, grant numbers 38619s and 38660. We thank the Molecular Observatory at Caltech (supported by the Gordon and Betty Moore Foundation) and the Stanford Synchrotron Radiation Lightsource. F.K. was supported by the German Research Foundation (Deutsche Forschungsgemeinschaft, KL 2389/1-1). M.C.N., P.J.B, and B.D.W. are Howard Hughes Medical Institute Investigators.
Footnotes
References and Notes
- 1.Walker LM, et al. PLoS Pathog. 2010;6:e1001028. doi: 10.1371/journal.ppat.1001028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Doria-Rose NA, et al. J Virol. 2010;84:1631. doi: 10.1128/JVI.01482-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gray ES, et al. J Virol. 2011;85:4828. doi: 10.1128/JVI.00198-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mikell I, et al. PLoS Pathog. 2011;7:e1001251. doi: 10.1371/journal.ppat.1001251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou T, et al. Science. 2010;329:811. doi: 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu X, et al. Science. 2010;329:856. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scheid JF, et al. Nature. 2009;458:636. doi: 10.1038/nature07930. [DOI] [PubMed] [Google Scholar]
- 8.Mouquet H, et al. Nature. 2010;467:591. doi: 10.1038/nature09385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wardemann H, et al. Science. 2003;301:1374. doi: 10.1126/science.1086907. [DOI] [PubMed] [Google Scholar]
- 10.Nussenzweig A. M C Nussenzweig, Cell. 2010;141:27. doi: 10.1016/j.cell.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dey B, et al. PLoS Pathog. 2009;5:e1000445. doi: 10.1371/journal.ppat.1000445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen B, et al. Nature. 2005;433:834. doi: 10.1038/nature03327. [DOI] [PubMed] [Google Scholar]
- 13.Yang X. M Farzan, R Wyatt, J Sodroski, J Virol. 2000;74:5716. doi: 10.1128/jvi.74.12.5716-5725.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pantophlet R, et al. J Virol. 2003;77:642. doi: 10.1128/JVI.77.1.642-658.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Olshevsky U, et al. J Virol. 1990;64:5701. doi: 10.1128/jvi.64.12.5701-5707.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thali M, et al. J Virol. 1991;65:6188. doi: 10.1128/jvi.65.11.6188-6193.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thali M, et al. J Virol. 1993;67:3978. doi: 10.1128/jvi.67.7.3978-3988.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tiller T, et al. Immunity. 2007;26:205. doi: 10.1016/j.immuni.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seaman MS, et al. J Virol. 2010;84:1439. doi: 10.1128/JVI.02108-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li M, et al. J Virol. 2005;79:10108. doi: 10.1128/JVI.79.16.10108-10125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dörner T, Radbruch A. Immunity. 2007;27:384. doi: 10.1016/j.immuni.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 22.Burton DR, et al. Proc Natl Acad Sci USA. 1991;88:10134. doi: 10.1073/pnas.88.22.10134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moore JP. J Sodroski, J Virol. 1996;70:1863. doi: 10.1128/jvi.70.3.1863-1872.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pietzsch J, et al. J Exp Med. 2010;207:1995. doi: 10.1084/jem.20101176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kwong PD, et al. Nature. 1998;393:648. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Materials and methods are available as supporting material on Science Online.
- 27.Li Y, et al. Nat Med. 2007;13:1032. doi: 10.1038/nm1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dhillon AK, et al. J Virol. 2007;81:6548. doi: 10.1128/JVI.02749-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walker LM, et al. Science. 2009;326:285. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trkola A, et al. J Virol. 1996;70:1100. doi: 10.1128/jvi.70.2.1100-1108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muster T, et al. J Virol. 1993;67:6642. doi: 10.1128/jvi.67.11.6642-6647.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zwick MB, et al. J Virol. 2001;75:10892. doi: 10.1128/JVI.75.22.10892-10905.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Purtscher M, et al. AIDS Res Hum Retroviruses. 1994;10:1651. doi: 10.1089/aid.1994.10.1651. [DOI] [PubMed] [Google Scholar]
- 34.Buchacher A, et al. AIDS Res Hum Retroviruses. 1994;10:359. doi: 10.1089/aid.1994.10.359. [DOI] [PubMed] [Google Scholar]
- 35.Zwick MB, et al. J Virol. 2001;75:12198. doi: 10.1128/JVI.75.24.12198-12208.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McClure MO, et al. Nature. 1987;330:487. doi: 10.1038/330487a0. [DOI] [PubMed] [Google Scholar]
- 37.Haim H, et al. PLoS Pathog. 2009;5:e1000360. doi: 10.1371/journal.ppat.1000360. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.