SUMMARY
Fragile X syndrome (FXS), the leading monogenic cause of intellectual disability and autism, results from loss of function of the RNA-binding protein FMRP. Here we show that FMRP regulates the translation of neuronal nitric oxide synthase 1 (NOS1) in the developing human neocortex. Whereas NOS1 mRNA is ubiquitously expressed, NOS1 protein is transiently co-expressed with FMRP during early synaptogenesis in layer- and region-specific subpopulations of pyramidal neurons. These include mid-fetal layer 5 subcortically projecting neurons arranged into alternating columns in the prospective Broca’s area and orofacial motor cortex. Human NOS1 translation is activated by FMRP via interactions with coding region binding motifs absent from mouse Nos1 mRNA, which is expressed in mouse pyramidal neurons, but not efficiently translated. Correspondingly, neocortical NOS1 protein levels are severely reduced in developing human FXS cases but not FMRP-deficient mice. Thus, alterations in FMRP post-transcriptional regulation of NOS1 in developing neocortical circuits may contribute to cognitive dysfunction in FXS.
INTRODUCTION
The development of neural circuits is a precisely regulated process susceptible to genetic alterations that can lead to disorders affecting the most distinctively human aspects of cognition, including speech and language, theory of mind, and complex social behavior (Geschwind and Levitt, 2007; Lui et al., 2011; Ramocki and Zoghbi, 2008; State, 2010; Walsh et al., 2008). One such disorder, fragile X syndrome (FXS), is the leading inherited cause of intellectual disability and is often accompanied by autistic-like features, aggression, attention deficits, and delays in speech and language development (Abbeduto et al., 2007; Rogers et al., 2001; Willemsen et al., 2011). FXS is caused by loss of function of the FMR1 gene, which encodes an RNA-binding protein (FMRP) involved in mRNA localization, stability, and translation (Ashley et al., 1993; Bagni and Greenough, 2005; Bassell and Warren, 2008; Zalfa et al., 2007). Many FMRP mRNA targets function in synaptic development and plasticity (Brown et al., 2001; Darnell et al., 2011). Concordantly, Fmr1-deficient mice show neural deficits also found in FXS patients (The Dutch-Belgian Fragile X Consortium, 1994). However, FMRP target mRNAs and their role in human neurodevelopment are not as well understood.
The study of human FMRP function may provide insights into the molecular mechanisms and neural circuits affected in autism spectrum disorders (ASD), which are highly co-morbid with FXS (Rogers et al., 2001). ASD are a group of complex developmental syndromes characterized by impairments in social communication and language development, and repetitive behaviors. Multiple lines of evidence point to the dysfunction of neocortical circuits involved in social, emotional, and language processing in ASD (Geschwind and Levitt, 2007; State, 2010; Walsh et al., 2008). While no overt neuroanatomical alterations have been linked to the autistic brain, there is emerging evidence of abnormal organization of cortical minicolumns (Casanova et al., 2002; Peters, 2010), which are composed of vertically arranged neurons connected into a local network and thought to originate from developmental radial units (Mountcastle, 1997; Rakic, 1988). Whether the molecular mechanisms altered in ASD are associated with the development of specific human cortical circuits, including minicolumns, remains unknown.
Here, we report that FMRP binds human neuronal nitric oxide synthase 1 (NOS1, nNOS ) mRNA and increases its translation in the developing neocortex in a species-dependent manner. NOS1 produces the gaseous signaling molecule nitric oxide (NO), which plays important roles in the development and function of the nervous system (Bredt and Snyder, 1994; Garthwaite, 2008). Our study of NOS1 post-transcriptional regulation was instigated by our observation of a marked discrepancy between the mid-fetal human neocortex expression patterns of NOS1 mRNA, which is ubiquitous, and NOS1 protein, which is restricted to layer- and region-specific subpopulations of pyramidal neurons. These include layer (L) 5 subcortically projecting neurons with an alternating minicolumnar arrangement in the frontoparietal operculum (FOp). The FOp encompasses the prospective Broca’s area and orofacial motor cortex, regions involved in speech production and language comprehension (Keller et al., 2009). After our screen for RNA-binding proteins revealed that FMRP is abundantly bound to human NOS1 mRNA, we found that FMRP interacts with sequences in the NOS1 coding region that contain G-quartet motifs and leads to increased NOS1 protein expression. These motifs are absent from mouse Nos1 mRNA and replacing the G-quartet-containing region of human NOS1 with the mouse orthologous sequence abrogates FMRP-dependent activation of translation. Concordantly, neocortical NOS1 protein levels are dramatically reduced in human FXS but not Fmr1-deficient mice. Thus, we identified a species-dependent post-transcriptional regulation of human NOS1 by FMRP in specific neocortical circuits during column development and synaptogenesis, and showed it to be altered in FXS brains.
RESULTS
NOS1 Protein is Transiently Expressed in Developing Human Pyramidal Neurons
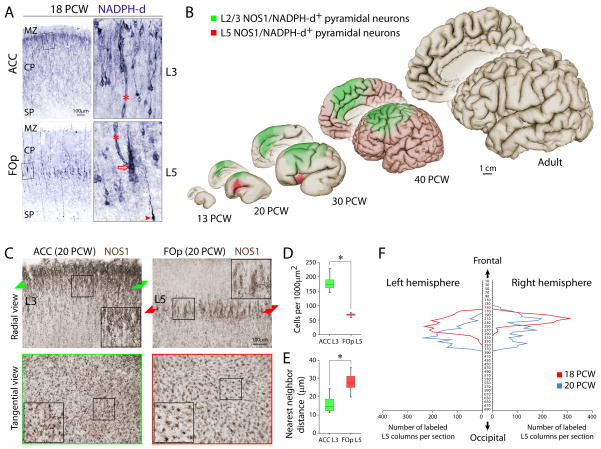
The current research stems from our unexpected observation that strong NADPH-diaphorase (NADPH-d) activity, a reliable histoenzymatic marker of NOS (Dawson et al., 1991; Hope et al., 1991), is transiently present in subpopulations of pyramidal neurons in the developing human neocortex (Sestan and Kostovic, 1994), in addition to its previously reported localization to interneurons and cortical plate (CP) neuropil (Fertuzinhos et al., 2009; Judas et al., 1999). Our comprehensive analysis of pre- and postnatal postmortem brains ranging from 8 postconceptional weeks (PCW) to adulthood identified transient expression of NOS1/NADPH-d in two layer- and region-specific populations of pyramidal neurons with a predominant localization to somata and apical dendrites (Figures 1A–C and S1A). Specifically, morphologically immature pyramidal neurons expressing NOS1 were present in the middle of the CP corresponding to the future L5 exclusively in the ventrolateral frontal cortex of the FOp and the dorsal part of the anterior insula starting around 15 PCW. One week later, NOS1+ pyramidal neurons were also found in the anterior cingulate cortex (ACC) and adjacent dorsolateral frontoparietal cortex in the upper CP corresponding to the future L2 and L3. NOS1 expression in both of these regions was also temporally regulated. The ACC L2/3 expression of NOS1 was maintained at high levels throughout the late fetal ages, and decreased during early infancy (Figures 1B and S1A and data not shown). In contrast, neocortical L5 expression of NOS1 occurred in two transient waves. First, the L5 expression was restricted to the FOp, and started at 15 PCW, peaked at 18–20 PCW, and was rapidly downregulated at 23 PCW, after which a small number of NOS1+ pyramidal neurons were present in the ventral part of the anterior insula. Second, sparse pyramidal NOS1 expression was present throughout neocortical L5 in the weeks immediately prior to birth and was progressively downregulated after birth (Figures 1B and S1A and data not shown). Thus, in developing pyramidal neurons, NOS1 expression is precisely regulated, exhibiting temporal, laminar, and regional specificity.
Figure 1. Spatio-Temporal Dynamics of NOS1 Expression in Pyramidal Neurons of the Human Neocortex.
(A) NADPH-d histochemistry at 18 PCW revealed intensely labeled pyramidal neurons in ACC L3 and FOp L5, where stained neurons were arranged into alternating vertical columns (open arrow). Interneurons (arrowhead) and blood vessels (asterisks) were also labeled. (B) Schematic summary of the spatiotemporal dynamics of NOS1/NADPH-d staining in pyramidal neurons of L2/3 (green) and L5 (red) in the developing and adult human neocortex. (C) NOS1 immunohistochemisty of radial and tangential sections at 20 PCW. NOS1+ pyramidal neurons exhibited a clear columnar organization in FOp L5 but not ACC L3. (D and E) Analysis of cell density (D) and cluster spacing (E) revealed that NOS1+ neurons in FOp L5 were significantly distinct in cytoarchitectonic organization from those in ACC L3. *P < 0.05. Error bars represent the 5th and 95th percentiles of thirty measurements. (F) Serial section analysis of NADPH-d+ L5 columns in two brains at 18 and 20 PCW. NADPH-d+ columns were present bilaterally.
Fetal L5 NOS1+ Pyramidal Neurons Form Alternating Columns
Further analysis of the mid-fetal FOp L5 NOS1+ pyramidal neurons revealed that they were arranged vertically into alternating arrays of intensely (NOS1+) and lightly (NOS1−) stained pyramidal neurons (Figures 1A and 1C) resembling previously described ontogenetic columns (Rakic, 1988). In contrast, the ACC L2/3 NOS1+ pyramidal neurons were more densely distributed (Figure 1D; FOp L5, 67.86±5.66 cells per 1000 μm2; ACC L3, 178.57±27.95 cells per 1000 μm2; P=4.12×10−5) and lacked this alternating columnar arrangement (Figure 1E; nearest neighbor distance between cell clusters: FOp L5, 27.87±5.26 μm; ACC L3, 15.75±4.68 μm; P=4.24×10−9). In contrast to mid-gestation, perinatal NOS1+ L5 neurons did not exhibit columnar organization (Figure S1A).
Because the FOp is structurally and functionally lateralized (Keller et al., 2009), we investigated whether L5 NOS1+ columns exhibited left-right asymmetry in two whole mid-fetal brains (18 and 20 PCW). Serial reconstruction confirmed the two separate domains of NOS1+ pyramidal neurons in the FOp and ACC of both hemispheres (Figures 1F and S1B) and provided approximate total numbers of FOp NOS1+ columns (18 PCW: 41,380 and 20 PCW: 45,150). Although the number of NOS1+ columns was not significantly different between the left and right hemispheres (P=0.569), the distribution of NOS1+ columns showed an asymmetric trend, peaking more rostrally in the right hemisphere, in both brains. Thus, the columnar organization of NOS1+ neurons in the mid-fetal FOp L5 is bilaterally present.
Molecular and Projectional Identity of NOS1+ Pyramidal Neurons
To molecularly characterize the identity of NOS1+ neurons, we examined their expression of neuronal subtype markers. In the mid-fetal FOp L5, markers of subcortical projection neurons, BCL11B (CTIP2) and FEZF2 (FEZL, ZFP312) (Chen et al., 2005; Kwan et al., 2008; Leone et al., 2008; Molyneaux et al., 2007), were selectively co-expressed by L5 NOS1+ neurons, forming an alternating columnar pattern identical to that of NOS1 (Figures 2A, 2B, and 2D). NOS1+ neurons also co-expressed FOXP2 (Figure 2E), a gene altered in a developmental disorder that severe affects speech and language (Lai et al., 2001). In contrast, SATB2, a marker of upper layer corticocortical pyramidal neurons (Britanova et al., 2008), was highly expressed in NOS1−, but not NOS1+, neurons (Figures 2A and 2B), suggesting that NOS1− neurons were later-born and likely migrating in between L5 NOS1+ columns to the upper layers. Consistent with this, we observed vimentin (VIM)-positive radial glial fibers in between but not within NOS1+ columns (Figure S2A). This suggests that glial-guided migration of upper layer neurons occurs via corridors formed between L5 neuronal columns. In the ACC, all L2/3 NOS1+ neurons co-expressed SATB2 (Figure S2B), confirming their upper-layer identity and distinction from the FOp NOS1+ neurons.
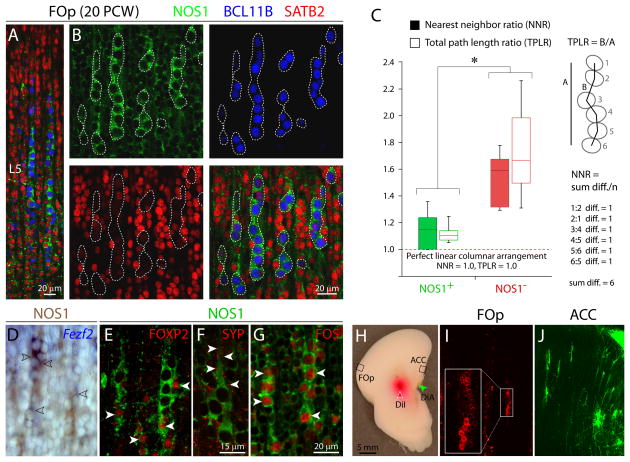
Figure 2. Molecular, Structural, and Axonal Connectivity Analyses of L5 NOS1+ Columns in the Mid-Fetal Human FOp.
(A and B) Triple immunofluorescent staining for NOS1 (green), BCL11B (blue), a marker of L5 subcortical projection neurons, and SATB2 (red), a marker of upper layer corticocortical projection neurons, in 20 PCW FOp L5. NOS1+ and BCL11B+ pyramidal neurons formed alternating columns (outlined) separated by clusters of SATB2+ neurons. (C) Analysis of columnarity in FOp L5 neurons. NOS1+ neurons were significantly more columnar in organization compared to NOS1− neurons. *P< 0.05. Error bars represent the 5th and 95th percentiles of thirty measurements. (D) NOS1 immunostaining (brown) and FEZF2 in situ hybridization (blue) of FOp L5 at 18 PCW. NOS1+ neurons co-expressed FEZF2 (open arrowheads). (E, F, and G) Immunofluorescent staining for NOS1 (green) and FOXP2, synaptophysin (SYP), or FOS (red in E, F, and G). NOS1+ neurons co-expressed FOXP2 and FOS (arrowheads in E and G), and were encircled by SYP punta (arrowheads in F). (H, I, and J) Retrograde axonal tracing at 20 PCW. Retrograde travel of Fast DiI inserted into dorsal internal capsule (red arrowhead) and Fast DiA inserted into the corpus callosum (green arrowhead) were examined after seven months in incubation. In the FOp L5 (I), DiI-labeled subcortical projection neurons formed columns similar those composed of NOS1+ neurons. In the ACC (J), DiA-labeled corticocortical projection neurons did not exhibit obvious columnar organization.
To examine whether the mid-fetal pyramidal neurons of diverse subtypes have distinct cytoarchitectonic arrangements, we first measured the nearest neighbor ratio (NNR) and total path length ratio (TPLR) (Buxhoeveden et al., 1996) of NOS1+ and NOS1− L5 neurons in the 20 PCW FOp (Figure 2C). This confirmed that NOS1+ L5 neurons were significantly closer to being perfectly columnar (1.0) compared to NOS1− L5 neurons, as determined by both NNR (NOS1+, 1.145±0.146; NOS1−, 1.528±0.202; P=2.97×10−5) and TPLR (NOS1+, 1.122±0.077; NOS1-, 1.740±0.369; P=2.41×10−3). Next, we retrogradely traced projection neurons in a postmortem 20 PCW brain. We labeled subcortical projection neurons with Fast DiI inserted into the internal capsule and corticocortical projection neurons with Fast DiA inserted into the corpus callosum (Figure 2H). DiI-labeled subcortical projection neurons in the FOp formed columns similar in organization to the NOS1+ columns (Figure 2I). In contrast, DiA-labeled callosal neurons in the ACC did not exhibit columnar organization (Figure 2J). Collectively, these results indicate that FOp NOS1+ neurons exhibit the columnar organization and molecular identity of post-migratory L5 subcortical projection neurons.
Transient NOS1 Expression in Pyramidal Neurons is Concomitant with Early Synaptogenesis
Previous studies have shown that significant neocortical synaptogenesis starts during mid-gestation (Molliver et al., 1973). Consistent with the possibility that NOS1 expression is associated with synaptogenesis, we found numerous pre-synaptic synaptophysin (SYP) puncta encircling the cell membrane of FOp NOS1+ L5 neurons at the soma and apical dendrite (Figure 2F). Our pre-embedding NOS1 immuno-EM in the 18 and 20 PCW FOp, however, revealed only sparse mature synapses in CP (Figures S2C and S2D), suggesting that the majority of SYP+ puncta on L5 NOS1+ columns were immature terminals. Concordantly, immature synapses that have not yet become electron dense or accumulated vesicles and non-synaptic contacts were observed in L5 and on NOS1+ dendrites (Figures S2E–H). Interestingly, FOS (C-FOS), a marker of recent neuronal activity, was expressed by virtually all NOS1+ columnar neurons and was mostly absent from NOS1− intercolumnar neurons in the 20 PCW FOp (Figure 2G), suggesting that NOS1+ L5 neurons may be electrically active. Together, these findings suggest that the expression of NOS1 in L5 FOp neurons is concomitant with early synaptogenesis and neuronal activity.
Cross-Species Comparison of Neocortical NOS1 Expression
To determine whether the spatiotemporal expression pattern of NOS1 exhibits species differences, we examined NADPH-d/NOS1 expression in the gyrated macaque monkey neocortex and lissencephalic mouse neocortex (Figure 3 and data not shown). In the macaque, NADPH-d+ pyramidal neurons were present in L2/3 of the ACC and adjacent frontoparietal regions, starting as early as embryonic day (E) 62, an age equivalent to human mid-gestation (Kostovic and Rakic, 1990), and persisting until the late fetal period (Figures 3B and S3 and data not shown). NADPH-d+ pyramidal neurons were present in L5 columns of the FOp and adjacent regions (Figures 3B and 3C), starting as early as E 73, peaking near E 82, and persisting until at least E 113 (Figures 3B and S3 and data not shown). Consistent with previous studies, our analyses of the mouse neocortex from E18.5 to P14, a period equivalent to human mid-fetal to early postnatal development, revealed that intense NADPH-d activity was present exclusively in interneurons and neuropil (Figure 3A and data not shown), indicating that pyramidal expression of NOS1 is species-dependent.
Figure 3. Comparative Analysis of NADPH-d Activity in Mouse, Macaque, and Human Neocortex at Equivalent Developmental Ages.
In the P4 mouse ACC and lateral frontal cortex (A), intense NADPH-d staining was restricted to interneurons, with neuropil staining in ACC L2/3. In the E73 macaque neocortex (B), intense NADPH-d activity was present in ACC L2/3 pyramidal neurons similar to those labeled in the human 18 PCW ACC (C). In the macaque FOp, NADPH-d+ L5 pyramidal neurons were arranged into vertical columns similar in organization to the human FOp columns (C). Strong interneuronal and weak neuropil NADPH-d staining was present in all cortical areas in both macaque and human neocortex.
Discordant NOS1 mRNA and Protein Expression Patterns in the Fetal Neocortex
To examine the expression pattern of NOS1 mRNA, adjacent tissue sections of the mid-fetal neocortex were analyzed with NOS1 in situ hybridization, NADPH-d, and NOS1 immunostaining (Figure 4A). Surprisingly, NOS1 mRNA was abundantly and ubiquitously present in the CP in all cortical layers and regions examined, including the great majority of pyramidal neurons that did not express NOS1 protein. This strikingly difference between the highly restricted NOS1 protein and nearly ubiquitous NOS1 mRNA expression suggests that NOS1 is post-transcriptionally regulated.
Figure 4. Discordant NOS1 mRNA and NOS1 Protein Expression Patterns in the Developing Human and Mouse Neocortex.
(A) Nissl staining, FEZF2 and NOS1 in situ hybridization, NOS1 immunohistochemistry and NADPH-d histochemistry in adjacent sections from 18 PCW FOp, ACC and dorsal lateral prefrontal cortex (PFC). NOS1 mRNA was abundantly and ubiquitously present in all cortical areas and layers examined. Intense NOS1 and NADPH-d labeling in L5 pyramidal columns (asterisks) were present exclusively in the FOp. In other cortical regions, NOS1 and NADPH-d were restricted to interneurons (open arrowheads) and neuropil. (B) Nos1 in situ hybridization, NOS1 and NADPH-d staining in adjacent sections from P3 mouse frontal neocortex. Nos1 mRNA was ubiquitously present, intense NOS1 and NADPH-d staining was exclusively present in interneurons (open arrowheads). Neuropil was weakly stained. (C) Pyramidal neurons of the Emx1 lineage were isolated from the P3 mouse neocortex by fluorescent cell sorting (FACS) and analyzed by quantitative (q) RT-PCR. Nos1 mRNA was abundantly present in pyramidal neurons. Error bars represent the 5th and 95th percentiles of four measurements.
Remarkably, Nos1 mRNA was also abundantly and ubiquitously expressed in the early postnatal mouse neocortex (Figure 4B). Pyramidal expression of mouse Nos1 mRNA was confirmed by quantitative RT-PCR of fluorescently sorted pyramidal neurons fate-mapped in mice doubly transgenic for Emx1-Cre and a CRE-responsive GFP (CAG-Cat-GFP) (Figure 4C). Therefore, whereas NOS1 mRNA is expressed in pyramidal neurons of both human and mouse neocortex, its efficient translation into NOS1 protein occurs in subpopulations of human, but not mouse, pyramidal neurons. This indicates that pyramidal NOS1 expression is driven by species-dependent post-transcriptional regulation.
NOS1 mRNA Associates with FMRP in Human Fetal Neocortex
To identify potential NOS1 mRNA-binding proteins, we used immobilized full-length human NOS1 mRNA to pull down candidate proteins from the human frontal CP at 20 and 21 PCW. To facilitate the isolation of sequence-dependent RNA-binding proteins, we used three negative control RNAs (GAPDH, EGFP, and NeoR). NOS1 mRNA interacting proteins showed a distinct enrichment at a molecular weight of approximately 75kDa (Figure 5A). To identify the protein present in this band, we analyzed our human brain transcriptome dataset (www.humanbraintranscriptome.org; Johnson et al., 2009; Kang et al., 2011) for RNA-binding proteins near 75kDa that are expressed in the mid-fetal frontal neocortex. Analysis of four candidates (FMRP, FXR1, CPEB3, and EIF2C2) by immunoblotting of pulled down proteins revealed that FMRP, but not the others, was strongly and specifically associated with NOS1 mRNA (Figures 5A and S4A). The presence of FMRP in this NOS1-enriched band was confirmed by mass-spectrometry (data not shown), which also revealed the putative presence of PABPC4, a poly-adenylate-binding protein, and HSPA8, a chaperone protein. Double immunofluorescent staining showed that FMRP was highly co-expressed in NOS1+ pyramidal neurons in the mid-fetal FOp and ACC (Figures 5B and S4B). Subcellularly, FMRP and NOS1 co-localized to the soma and apical dendrite. Interestingly, most NOS1+ interneurons in the SP and CP did not express FMRP at high levels during mid-gestation. Together, these results suggest a potential role of FMRP in the post-transcriptional regulation of NOS1 in fetal human pyramidal neurons.
Figure 5. FMRP Binds NOS1 mRNA in the Human Fetal Neocortex.
(A) Proteins eluted from an mRNA pull-down assay using lysates of a 21 PCW human neocortex were analyzed by silver staining and immunoblotting. NOS1, but not control (GAPDH, GFP, and NeoR), mRNA specifically associated with a ~75kDa protein that was immunopositive for FMRP. Asterisk indicates an artifact of gel transfer. (B) NOS1 (green), FMRP (red), and DAPI (blue) staining of 20 PCW FOp. FMRP was co-expressed by L5 columnar NOS1+ pyramidal neurons (solid arrowheads) but not most interneurons (arrow). (C and D) FMRP immunoprecipitated mRNAs from a 21 PCW human and P0 mouse CP were analyzed by quantitative RT-PCR. Relatively to control GAPDH and MAP1B mRNAs, FMRP strongly associated with NOS1 mRNA (green bar) in human but not mouse. Error bars represent the 5th and 95th percentiles of four measurements.
Species Differences in FMRP-NOS1 mRNA Association in the Developing Neocortex
To confirm the putative FMRP-NOS1 mRNA interaction, we performed RNA-binding protein immunoprecipitation (RIP) using 21 PCW frontal CP lysate. RNAs co-immunoprecipitated with FMRP were analyzed using quantitative RT-PCR (Figure 5C). Compared to rabbit immunoglobulin (IgG) control, anti-FMRP antibodies immunoprecipitated 6.8±0.8 fold more NOS1 mRNA, a level of enrichment similar to MAP1B mRNA (7.1±0.6 fold), a well known target of FMRP (Darnell et al., 2011), and significantly higher than GAPDH mRNA (1.5±0.4 fold), a negative control. In contrast, in the early postnatal mouse neocortex (Figure 5D), Nos1 mRNA was enriched only 2.7±0.3 fold by anti-FMRP immunoprecipitation, markedly lower than the 8.6±0.5 fold enrichment for Map1b mRNA and comparable to Gapdh mRNA (2.0±0.1 fold). Consistent with this, Nos1 was absent from the FMRP targets identified in a recent, comprehensive HITS-CLIP analysis of the mouse brain (Darnell et al., 2011). Thus, FMRP strongly associates with human but not mouse NOS1 mRNA in the developing neocortex, suggesting that FMRP may underlie species differences in NOS1 translation.
FMRP Binds G-Quartet-containing Sequences in the Human NOS1 Coding Region
FMRP can interact with specific mRNA sequences including G-quartet (GQ) structures (Darnell et al., 2001; Schaeffer et al., 2001) and poly-uridine stretches (Chen et al., 2003). Analysis of human NOS1 mRNA revealed three putative GQ motifs and six poly-uridine stretches (Figure 6A). RNA pull-down assays from 21 PCW frontal CP lysate revealed that FMRP had strong affinity for each of the two NOS1 coding region GQs (GQ1 and GQ2), but not GQ3 or the U-rich regions (UR1-UR6) (Figure 6B). To confirm this, we synthesized a fragment of RNA representing both GQ1 and GQ2 and performed an eletrophoretic mobility shift assay (EMSA; Figure 6C). In the presence of FMRP protein, this RNA exhibited a significant shift that was abolished by the addition of excess non-biotinylated “cold” RNA or a neutralizing FMRP antibody. To determine whether human GQ1 and GQ2 form RNA G-quadruplex structures, we used a reverse transcription termination assay (Figure 6D). Reverse transcriptase activity pauses at sites of GQ structures in a cation-dependent manner (Schaeffer et al., 2001). Reverse transcription from both GQ1 and GQ2 RNA exhibited a significant pause at the expected GQ site in the presence of potassium, which facilitates GQ formation, but not lithium, which abrogates it. Therefore, FMRP interacts with GQ-forming sequences found within the coding region of human NOS1 mRNA.
Figure 6. FMRP Binds Human NOS1 G-Quartet-Containing Sequences and Activates Human, but not Mouse, NOS1 mRNA Translation.
(A) Prediction of putative FMRP-binding GQ and U-rich (UR) motifs in the human NOS1 mRNA sequence and alignment of GQ1 and GQ2. GQ1 and GQ2 were highly conserved in primates but not rodents. (B) FMRP association with each putative binding motif was analyzed by an mRNA pull-down assay using 21 PCW human neocortex lysates. FMRP selectively associated with GQ1 and GQ2. (C) EMSA of GQ1 and GQ2. RNA containing both GQs exhibited a significant shift in mobility in the presence of FMRP. This shift was abolished by addition of excess unbiotinylated (“cold”) RNA or a neutralizing anti-FMRP antibody. (D) Reverse transcription termination assay. Reverse transcription paused at the expected GQ sites for both GQ1 and GQ2 in the presence of K+, which facilitates GQ formation, but not Li+, which disrupts it. (E) Colorimetric NOS assays in Neuro2a cells co-transfected with CAG-hFMR1 or CAG-hFMR1(I304N) and one of CAG-hNOS1, CAG-mNos1, or CAG-murinized-hNOS1. NOS activity from hNOS1, but not mNos1 or murinized hNOS1, increased dose-dependently with increasing wildtype FMRP. The I304N mutation in FMRP abolished its activation of hNOS1 translation. *P < 0.05. Error bars represent the 5th and 95th percentiles of four measurements. (F) Luciferase assays in Neuro2a cells transfected with an empty reporter construct (GL3-empty), or constructs containing the human NOS1 GQs (GL3-hNOS1(GQs)) or the orthologous sequence in mouse Nos1 (GL3-mNOS1(GQortholog)). Luciferase activity in cells transfected with GL3-hNOS1(GQs) increased dose-dependently with co-transfection of human CAG-hFMR1 (solid blue line) or mouse CAG-mFmr1 (broken blue line). Lucifase activity from the GL3-mNos1(GQortholog) decreased with increasing amounts of CAG-hFMR1 (solid red line). Error bars represent the 5th and 95th percentiles of six measurements.
Evolution of NOS1 mRNA G-Quartet-Containing Sequences
To investigate whether GQ motifs are present in other mammals, we analyzed the 21 species for which NOS1 mRNA sequence was available. Highly stable tetrads at both GQ1 and GQ2 positions were predicted only in the great apes and macaque monkey (Figure S5A). Among great apes, which otherwise have perfectly conserved GQs, only orangutan has a point mutation that leads to a less stable two-stack GQ1 quartet, but a fully conserved GQ2 quartet. In marmoset, a New World monkey, and non-primate mammals, with the exception of the guinea pig which exhibited one quartet, they are absent from both positions. Further analysis of the entire NOS1 coding region in nine placental mammals revealed a very high degree of conservation (Figure S5B), with the vast majority of substitutions being synonymous. The few non-synonymous substitutions, however, were selectively clustered in the GQ region. This marked reduction in amino acid identity in an otherwise highly conserved protein is consistent with the hypothesis that the sequences containing the GQ motifs evolved and made possible post-transcriptional regulation by FMRP, perhaps at the expense of protein integrity. Furthermore, these sequences have remained quite stable since their emergence in catarrhine primates, which is consistent with the expression of NOS1 in human and macaque pyramidal neurons.
FMRP Increases NOS1 Expression via Interaction with a G-Quartet-Containing Sequence
To test the functional consequences of FMRP on NOS1 translation, we co-transfected human expression constructs of FMRP (CAG-hFMR1) and NOS1 (CAG-hNOS1) into Neuro2a cells and quantified NOS activity (Figure 6E). With CAG-hFMR1 co-transfection, NOS1 activity was increased in a dose-dependent manner, by up to 3.6±0.9-fold (P=0.043), indicating that FMRP acts as a positive regulator of NOS1 expression. No increase in NOS1 activity occurred when a mouse Nos1 construct (CAG-mNos1) or a human FMR1 construct harboring the I304N mutation (CAG-hFMR1 (I304N)) (Siomi et al., 1994) was used, or when the GQ-containing sequence of the human NOS1 was replaced with the orthologous sequence from mouse Nos1 (CAG-murinized-hNOS1). Therefore, the FMRP-mediated increase in NOS1 expression is dependent on the species of the NOS1 sequence, the intact KH2 domain of FMRP, and the presence of GQ-containing sequences in the NOS1 mRNA. To specifically examine the GQ region, we cloned the NOS1 sequences containing GQ1 and GQ2 into the 3′ UTR of SV40-GL3 and performed luciferase assays in Neuro-2a cells (Figure 6F). The inclusion of the human NOS1 GQs (SV40-GL3-hNOS1-GQs) led to significant dose-dependent increases in luciferase activity in response to CAG-hFMR1, indicating that FMRP increases NOS1 translation via binding to these sequences. Importantly, a mouse FMRP expression construct (CAG-mFmr1) dose-dependently increased luciferase expression in a manner highly similar to CAG-hFMR1. However, when the human NOS1 GQ sequences were replaced with the orthologous region of the mouse Nos1 (SV40-GL3-mNos1-GQortholog), FMRP failed to enhance luciferase activity. These results indicate that FMRP activates NOS1 protein expression via binding to a sequence containing GQ-motifs and that this interaction exhibits species differences. Together, these data strongly support a scenario wherein FMRP activation of NOS1 translation evolved through NOS1 nucleotide substitutions that gave rise to a GQ-containing sequence targeted by FMRP.
Mouse Pyramidal Neurons Efficiently Translate Human NOS1 in an FMRP-Dependent Manner
Since mouse FMRP is able to enhance human NOS1 expression, we hypothesized that exogenous human NOS1 mRNA can be efficiently translated in mouse pyramidal neurons, likely in an Fmr1-dependent manner. To test this in vivo, we introduced a NOS1 expression construct with the CAG-Gfp reporter into mouse neocortical ventricular zone (VZ) using in utero electroporation (IUE) at embryonic day (E) 13.5 to target L5 pyramidal neurons. At P0, the majority of CAG-hNOS1-electroporated L5 pyramidal neurons expressed NOS1 protein at high levels (Figures 7A, 7B, and 7C). In contrast, those electroporated with CAG-mNos1 or CAG-murinized-hNOS1 expressed only low levels of NOS1 in a minority of neurons, indicating reduced protein expression efficiency consistent with our in vitro assays. Human and mouse NOS1 exhibited similar somatodendritic localization (Figures 7A and 7B). Therefore, our results show that mouse pyramidal neurons possess all of the cellular machinery necessary for the translation of human NOS1 protein and suggest that their diminished expression of endogenous NOS1 is a result of differences in the NOS1 mRNA sequence between human and mouse.
Figure 7. Translation of Human NOS1 in Pyramidal Neurons Requires FMRP and is Severely Reduced in Fetal FXS Neocortex.
(A, B, and C) Neocortex of wildtype or Fmr1 KO mouse electroporated in utero at E13.5 and immunostained for NOS1 (red) at P0. In wildtype neocortex, the majority of pyramidal neurons transfected with hNOS1 expressed high levels of NOS1 properly localized to the soma and apical dendrite. NOS1 protein expression from mNos1 or murinized-hNOS1 in wildtype and from hNOS1 in Fmr1 KO neocortex was dramatically reduced in comparison. The fluorescent intensity of NOS1 staining normalized to GFP (B) and the proportion of GFP+ cells expressing NOS1 (C) were quantified. *P < 0.05. Error bars represent the 5th and 95th percentiles of at least four animals. (D) Immunoblots of human fetal FXS and P0 mouse Fmr1 KO neocortex. Normalized to GAPDH levels, the neocortical expression of NOS1 protein was severely reduced in both human fetal FXS cases. In neonatal mouse neocortex, loss of Fmr1 did not alter neocortical NOS1 expression.
To assess FMRP dependence, we further electroporated Fmr1 knockout (KO) mice with CAG-hNOS1 and CAG-Gfp (Figure 7A). Both the number of GFP+ neurons expressing NOS1 and the levels of NOS1 protein decreased significantly compared to wildtype (Figures 7B and 7C). In addition, neurons cultured from E14.5 Fmr1 KO and transfected with CAG-hNOS1 also exhibited a significant reduction in NOS1 levels compared to control (Figure S6A; 43.8±7.8% reduction, P=0.0065). These data show that FMRP is required for the efficient expression of human NOS1 protein in pyramidal neurons.
Severe Reduction in NOS1 Protein Levels in Developing Human FXS but not Mouse Fmr1 KO Neocortex
To determine whether NOS1 protein levels are altered in human FXS cases, we performed immunoblotting of neocortex from confirmed mid-fetal and postnatal FXS cases (15 and 18 PCW; and 9, 22, and 85 years) and age-matched controls. Neocortical lysates normalized to GAPDH levels were immunoblotted for NOS1, FMRP, and GAPDH (Figure 7D). Remarkably, in both fetal cases of FXS, neocortical NOS1 protein levels were severely reduced compared to matched controls. Furthermore, this deficit was age-dependent, being very dramatic in the fetal cases, less so in the cases aged 9 and 22 years, and absent in the 85 years specimen (Figure S6C). These results indicate that NOS1 protein expression is greatly reduced in the developing human FXS neocortex. Notably, neocortical NOS1 levels were not affected in early post-natal Fmr1 KO mice (Figures 7D and S6B), indicating that the requirement of FMRP for NOS1 expression is species-dependent.
DISCUSSION
In this study, we demonstrate that human neocortical NOS1 expression is post-transcriptionally regulated by FMRP in a species-dependent manner. Molecular analyses revealed that FMRP binds GQ motif-containing sequences present in the coding region of human, but not mouse, NOS1 mRNA and facilitates NOS1 protein expression. Concordantly, NOS1 expression is severely reduced in the developing FXS human, but not FMRP-deficient mouse, neocortex. In the human neocortex, NOS1 and FMRP are transiently co-expressed during synaptogenesis in subpopulations of pyramidal neurons in regions involved in language and complex social behaviors. Together, these findings provide a novel candidate mechanism and insights into the potential connectional pathology of FXS and possibly ASD.
Our analyses indicate that the FMRP-NOS1 interaction emerged as result of closely clustered nucleotide substitutions within the otherwise highly conserved coding sequence of NOS1 that gave rise to the GQ-containing motifs, occurring at the potential expense of protein integrity. FMRP binding to GQ motifs has been associated with translational repression (Bechara et al., 2009; Schaeffer et al., 2001). There is, however, a precedent for positive, activity-dependent post-transcriptional regulation in PSD-95 (DLG4), which has an FMRP-binding GQ motif (Todd et al., 2003; Zalfa et al., 2007). Interestingly, NOS1 and PSD-95 are functionally related. NOS1 is anchored to the synaptic membrane via a physical interaction with PSD-95 (Brenman et al., 1996) and its enzymatic product, NO, S-nitrosylates PSD-95 (Ho et al., 2011). Although the abundant presence of NOS1 mRNA in pyramidal neurons suggests that translational regulation is involved, FMRP may also control the stability of the NOS1 transcript in a manner similar to its control of PSD-95 mRNA stability (Zalfa et al., 2007). Furthermore, the GQ motif has been shown to mediate the dendritic localization of PSD-95 (Dictenberg et al., 2008) and may also play a role in NOS1 mRNA targeting. The possibility that NOS1 and PSD-95 are similarly regulated by FMRP is consistent with their shared post-synaptic localization, physical interaction, and related functions. The binding of FMRP to GQs has been demonstrated both in vitro (Bagni and Greenough, 2005; Bassell and Warren, 2008) and in vivo (Rackham et al., 2004; Iioka et al., 2011). Recently, however, it was shown that the presence GQ motifs is not predictive of FMRP binding (Darnell et al., 2011). Therefore, the context-dependence of FMRP interactions with GQs remains to be fully elucidated and individual potential interactions should be validated empirically.
Animal models of FXS exhibit multiple phenotypes present in human FXS, indicating that many aspects of FMRP function are well conserved. Therefore, any contribution of NOS1 to the FXS phenotype would likely involve the higher cognitive functions that are absent from mouse. This possibility is supported by the co-expression of NOS1 and FMRP in projection neurons of the FOp and the ACC and adjacent dorsal frontoparietal neocortex. The FOp encompasses the future Broca’s area and its contralateral hemisphere equivalent, as well as the orofacial motor cortex, regions involved in speech production, language comprehension, and action recognition (Keller et al., 2009). The ACC is involved in decision making, attention, emotional processing, and social awareness (Devinsky et al., 1995). NOS1 expression in these regions is also temporally regulated from mid-gestation to early infancy, a developmental period critical for early synaptogenesis, dendritic spine formation, and ingrowth of cortical afferents (Kang et al., 2011). Therefore, the neuroanatomical localization and timing of the FMRP-NOS1 interaction is consistent with a putative role in the development of neocortical circuits, including those involved in linguistic and social functions likely affected in FXS and ASD.
This potential role of NOS1 in the development and function of human neural circuits is further supported by studies of a human NOS1 hypomorphic allele, which has been linked to attention deficit hyperactivity disorder (ADHD), impulsivity, and aggression (Reif et al., 2009), behavioral features often comorbid with FXS (Rogers et al., 2001). This NOS1 hypomorphism has also been linked to hypoactivity in the ACC (Reif et al., 2009), the cortical area with the most prominent mid-fetal pyramidal expression of NOS1. Functional imaging studies revealed a similar reduction in ACC activation in FXS and ADHD patients during attentional processing tasks (Bush et al., 1999; Menon et al., 2004) and in autistic children in response to a familiar face (Pierce and Redcay, 2008). The overlapping neural and behavioral deficits between NOS1 hypomorphism, FXS, and FXS comorbidities are consistent with a functional role of NOS1 in human brain circuitry related to FXS and ASD. NOS1 may also be associated with other psychiatric disorders, as sequence variations in NOS1 have been associated with schizophrenia (Cui et al., 2010; Reif et al., 2006; Shinkai et al., 2002).
Structural alteration in the organization of minicolumns has been reported in autism and other psychiatric disorders (Casanova et al., 2002). In this study, we found that within the FOp, alternating L5 fetal columns co-express FMRP and NOS1, as well as FOXP2, which is implicated in the development of language and cognition (Lai et al., 2001), two functions affected in FXS and ASD. We also showed that neurons within the same column have a shared subcortical molecular identity and connectivity. Positioned in between the columns are migratory corridors containing radial glial fibers and corticocortical projection neurons en route to the superficial layers. Thus, this fetal organization may have implications for the developmental basis of normal minicolumns (Rakic, 1988), as well as columnopathies (Casanova et al., 2002). Interestingly, the NOS1+ columnar neurons of the fetal FOp share some areal and projection properties with adult mirror neurons, which are present in macaque area F5 (Rizzolatti and Craighero, 2004), an area equivalent to the human Broca’s area, and project subcortical axons (Kraskov et al., 2009). Mirror neurons, which are activated during both the observation and execution of a particular goal-directed action, are thought to contribute to theory of mind and language abilities (Rizzolatti and Craighero, 2004), and in autistic children, the mirror neuron activity that is normally observed in the FOp is absent (Dapretto et al., 2006). Therefore, the molecular profile of NOS1+ columns, as well as their shared location and connectivity with mirror neurons, is consistent with a potential role in cognition and motor control.
The synthesis of NO, a short-lived gas that cannot be stored or transported, must be precisely regulated and amenable to rapid, localized activation. As FMRP controls both the dendritic localization and translation of target mRNAs, it is well suited to contribute to the dynamic regulation of NOS1 activity. It should be noted, however, that NOS1 mRNA may also be under additional, perhaps negative, post-transcription control, as suggested by the lack of NOS1 protein expression in the majority of NOS1 mRNA-expressing pyramidal neurons. The modulation of neuronal function by NO in the brain has been widely studied and post-synaptic NO is thought to represent a retrograde signal that promotes pre-synaptic differentiation (Bredt and Snyder, 1994; Garthwaite, 2008). Blockade of NOS1 function has been shown to disrupt synapse formation and result in spine loss (Nikonenko et al., 2008). Given the potential role of NO in synapse development, the loss of NOS1 expression in the fetal FXS brain during early synaptogenesis may contribute to this phenotype. Studies have also shown that NO mediates neuronal synchronization (O’Donnell and Grace, 1997) and can modulate protein function via S-nitrosylation (Jaffrey et al., 2001), including that of histones (Nott et al., 2008), which can mediate transcriptome changes. In future studies, it will be important to characterize the mechanisms of NO function in the developing human neocortex and their potential contribution to FXS.
EXPERIMENTAL PROCEDURES
Human brain tissue processing
The sources and methods for the collection, dissection, and fixation of control and fragile X postmortem human tissues are described in the Extended Experimental Procedures. All specimens were collected under guidelines approved by institutional review boards and anonymized prior to our receipt. Fixed tissues were vibratome- or cryostat-sectioned. For NADPH-d staining, sections were incubated in beta-NADPH, nitro blue tetrazolium, and Triton X-100. Sections were pre-incubated in hydrogen peroxide for immunohistochemisty or directly pre-blocked in blocking solution for immunofluorescent staining prior to incubation with primary antibodies followed by biotinylated or fluorophore-conjugated secondary antibodies. For immunohistochemistry, sections were further incubated in avidin-biotin-peroxidase complex andvisualized using DAB.
RNA pull-down assay and RIP
For pull-down assays, RNAs were transcribed from cDNA or PCR products, biotinylated, and captured using streptavidin beads. Human mid-fetal CP lysates were added and bound proteins were analyzed by SDS-PAGE, silver staining, and immunoblotting. For RIP, FMRP-bound mRNAs were immunoprecipitated from mid-fetal human and neonatal mouse CP lysates and analyzed using quantitative RT-PCR.
Expression assays and in utero electroporation
The generation of DNA constructs is described in the Extended Experimental Procedures. Neuro-2a cells were transfected by lipofection. Luciferase or NOS activity was assayed 48 hours after transfection and normalized to transfection efficiency controls. For electroporation, DNA was injected into the lateral ventricles of embryonic mice and transferred into ventricular zone cells by 40 V pulses. Electroporated brains were analyzed at P 0 by immunostaining.
Supplementary Material
Highlights.
NOS1 and FMRP are co-expressed in subsets of pyramidal neurons in human fetal brain
FMRP directly interacts with motifs present in human, but not mouse, NOS1 mRNA
NOS1 translation is increased by FMRP in a species-dependent manner
NOS1 protein levels are severely reduced in developing human fragile X neocortex
Acknowledgments
We thank D. Budinščak, Z. Cmuk, Z. Krsnik, S. Liu-Chen, B. Popović, B. Poulos, and B. Sajin for help with tissue acquisition; M. Nakane, W. Sessa, and I. Grkovic for antibodies and reagents; F. Cheng and M. Li for analyzing transcriptome data; E. Gulcicek for help with mass-spectrometry; and M. Brown, L. Kaczmarek, A. Louvi, and members of the Sestan laboratory for discussions and comments. E.J.H. and D.H.R. received technical support from the Pediatric Neuropathology Research Lab at UCSF (UCOP Award #142675). D.H.R. is a HHMI Investigator. A.M.M.S. is supported by a fellowship from the Portuguese Foundation for Science and Technology. This work was supported by grants from the NIH, MH081896, MH089929, NS051869 (N.S.), 1K99NS064303 (M-R. R.), Kavli Foundation, NARSAD, and James S. McDonnell Foundation Scholar Award (N.S.), and ZonMw 912-07-022 (R.W.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abbeduto L, Brady N, Kover ST. Language development and fragile X syndrome: profiles, syndrome-specificity, and within-syndrome differences. Ment Retard Dev Disabil Res Rev. 2007;13:36–46. doi: 10.1002/mrdd.20142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashley CT, Wilkinson KD, Reines D, Warren ST. FMR1 protein: conserved RNP family domains and selective RNA binding. Science. 1993;262:563–566. doi: 10.1126/science.7692601. [DOI] [PubMed] [Google Scholar]
- Bagni C, Greenough WT. From mRNP trafficking to spine dysmorphogenesis: the roots of fragile X syndrome. Nat Rev Neurosci. 2005;6:376–387. doi: 10.1038/nrn1667. [DOI] [PubMed] [Google Scholar]
- Bassell GJ, Warren ST. Fragile X syndrome: loss of local mRNA regulation alters synaptic development and function. Neuron. 2008;60:201–214. doi: 10.1016/j.neuron.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechara EG, Didiot MC, Melko M, Davidovic L, Bensaid M, Martin P, Castets M, Pognonec P, Khandjian EW, Moine H, et al. A novel function for fragile X mental retardation protein in translational activation. PLoS Biol. 2009;7:e16. doi: 10.1371/journal.pbio.1000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredt DS, Snyder SH. Nitric oxide: a physiologic messenger molecule. Annu Rev Biochem. 1994;63:175–195. doi: 10.1146/annurev.bi.63.070194.001135. [DOI] [PubMed] [Google Scholar]
- Brenman JE, Chao DS, Gee SH, McGee AW, Craven SE, Santillano DR, Wu Z, Huang F, Xia H, Peters MF, et al. Interaction of nitric oxide synthase with the postsynaptic density protein PSD-95 and alpha1-syntrophin mediated by PDZ domains. Cell. 1996;84:757–767. doi: 10.1016/s0092-8674(00)81053-3. [DOI] [PubMed] [Google Scholar]
- Britanova O, de Juan Romero C, Cheung A, Kwan KY, Schwark M, Gyorgy A, Vogel T, Akopov S, Mitkovski M, Agoston D, et al. Satb2 is a postmitotic determinant for upper-layer neuron specification in the neocortex. Neuron. 2008;57:378–392. doi: 10.1016/j.neuron.2007.12.028. [DOI] [PubMed] [Google Scholar]
- Brown V, Jin P, Ceman S, Darnell JC, O’Donnell WT, Tenenbaum SA, Jin X, Feng Y, Wilkinson KD, Keene JD, et al. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell. 2001;107:477–487. doi: 10.1016/s0092-8674(01)00568-2. [DOI] [PubMed] [Google Scholar]
- Bush G, Frazier JA, Rauch SL, Seidman LJ, Whalen PJ, Jenike MA, Rosen BR, Biederman J. Anterior cingulate cortex dysfunction in attention-deficit/hyperactivity disorder revealed by fMRI and the counting stroop. Biol Psychiatry. 1999;45:1542–1552. doi: 10.1016/s0006-3223(99)00083-9. [DOI] [PubMed] [Google Scholar]
- Buxhoeveden D, Lefkowitz W, Loats P, Armstrong E. The linear organization of cell columns in human and nonhuman anthropoid Tpt cortex. Anat Embryol. 1996;194:23–36. doi: 10.1007/BF00196312. [DOI] [PubMed] [Google Scholar]
- Casanova MF, Buxhoeveden DP, Switala AE, Roy E. Minicolumnar pathology in autism. Neurology. 2002;58:428–432. doi: 10.1212/wnl.58.3.428. [DOI] [PubMed] [Google Scholar]
- Chen J, Rasin M, Kwan K, Sestan N. Zfp312 is required for subcortical axonal projections and dendritic morphology of deep-layer pyramidal neurons of the cerebral cortex. Proc Natl Acad Sci USA. 2005;102:17792–17797. doi: 10.1073/pnas.0509032102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Yun SW, Seto J, Liu W, Toth M. The fragile X mental retardation protein binds and regulates a novel class of mRNAs containing U rich target sequences. Neuroscience. 2003;120:1005–1017. doi: 10.1016/s0306-4522(03)00406-8. [DOI] [PubMed] [Google Scholar]
- Cui H, Nishiguchi N, Yanagi M, Fukutake M, Mouri K, Kitamura N, Hashimoto T, Shirakawa O, Hishimoto A. A putative cis-acting polymorphism in the NOS1 gene is associated with schizophrenia and NOS1 immunoreactivity in the postmortem brain. Schizophr Res. 2010;121:172–178. doi: 10.1016/j.schres.2010.05.003. [DOI] [PubMed] [Google Scholar]
- Dapretto M, Davies MS, Pfeifer JH, Scott AA, Sigman M, Bookheimer SY, Iacoboni M. Understanding emotions in others: mirror neuron dysfunction in children with autism spectrum disorders. Nat Neurosci. 2006;9:28–30. doi: 10.1038/nn1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Jensen KB, Jin P, Brown V, Warren ST, Darnell RB. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell. 2001;107:489–499. doi: 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- Darnell JC, Van Driesche SJ, Zhang C, Hung KY, Mele A, Fraser CE, Stone EF, Chen C, Fak JJ, Chi SW, et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146:247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson TM, Bredt DS, Fotuhi M, Hwang PM, Snyder SH. Nitric oxide synthase and neuronal NADPH diaphorase are identical in brain and peripheral tissues. Proc Natl Acad Sci USA. 1991;88:7797–7801. doi: 10.1073/pnas.88.17.7797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devinsky O, Morrell MJ, Vogt BA. Contributions of anterior cingulate cortex to behaviour. Brain. 1995;118:279–306. doi: 10.1093/brain/118.1.279. [DOI] [PubMed] [Google Scholar]
- Dictenberg JB, Swanger SA, Antar LN, Singer RH, Bassell GJ. A direct role for FMRP in activity-dependent dendritic mRNA transport links filopodial-spine morphogenesis to fragile X syndrome. Dev Cell. 2008;14:926–939. doi: 10.1016/j.devcel.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fertuzinhos S, Krsnik Z, Kawasawa Y, Rasin M, Kwan K, Chen J, Judas M, Hayashi M, Sestan N. Selective depletion of molecularly defined cortical interneurons in human holoprosencephaly with severe striatal hypoplasia. Cereb Cortex. 2009;19:2196–2207. doi: 10.1093/cercor/bhp009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garthwaite J. Concepts of neural nitric oxide-mediated transmission. Eur J Neurosci. 2008;27:2783–27802. doi: 10.1111/j.1460-9568.2008.06285.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geschwind D, Levitt P. Autism spectrum disorders: developmental disconnection syndromes. Curr Opin Neurobiol. 2007;17:103–111. doi: 10.1016/j.conb.2007.01.009. [DOI] [PubMed] [Google Scholar]
- Ho GP, Selvakumar B, Mukai J, Hester LD, Wang Y, Gogos JA, Snyder SH. S-nitrosylation and S-palmitoylation reciprocally regulate synaptic targeting of PSD-95. Neuron. 2011;71:131–141. doi: 10.1016/j.neuron.2011.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope BT, Michael GJ, Knigge KM, Vincent SR. Neuronal NADPH diaphorase is a nitric oxide synthase. Proc Natl Acad Sci USA. 1991;88:2811–2814. doi: 10.1073/pnas.88.7.2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iioka H, Loiselle D, Haystead TA, Macara IG. Efficient detection of RNA-protein interactions using tethered RNAs. Nucleic Acids Res. 2011;39:e53. doi: 10.1093/nar/gkq1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffrey SR, Erdjument-Bromage H, Ferris CD, Tempst P, Snyder SH. Protein S-nitrosylation: a physiological signal for neuronal nitric oxide. Nat Cell Biol. 2001;3:193–197. doi: 10.1038/35055104. [DOI] [PubMed] [Google Scholar]
- Johnson M, Kawasawa Y, Mason C, Krsnik Z, Coppola G, Bogdanović D, Geschwind D, Mane S, State M, Sestan N. Functional and evolutionary insights into human brain development through global transcriptome analysis. Neuron. 2009;62:494–509. doi: 10.1016/j.neuron.2009.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judas M, Sestan N, Kostović I. Nitrinergic neurons in the developing and adult human telencephalon: transient and permanent patterns of expression in comparison to other mammals. Microsc Res Tech. 1999;45:401–419. doi: 10.1002/(SICI)1097-0029(19990615)45:6<401::AID-JEMT7>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Kang HJ, Imamura Kawasawa Y, Cheng F, Zhu Y, Xu X, Li M, Sousa AMM, Pletikos M, Meyer KA, Sedmak G, et al. Spatiotemporal transcriptome of the human brain. Nature. 2011;478:483–489. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller SS, Crow T, Foundas A, Amunts K, Roberts N. Broca’s area: nomenclature, anatomy, typology and asymmetry. Brain Lang. 2009;109:29–48. doi: 10.1016/j.bandl.2008.11.005. [DOI] [PubMed] [Google Scholar]
- Kostovic I, Rakic P. Developmental history of the transient subplate zone in the visual and somatosensory cortex of the macaque monkey and human brain. J Comp Neurol. 1990;297:441–470. doi: 10.1002/cne.902970309. [DOI] [PubMed] [Google Scholar]
- Kraskov A, Dancause N, Quallo MM, Shepherd S, Lemon RN. Corticospinal neurons in macaque ventral premotor cortex with mirror properties: a potential mechanism for action suppression? Neuron. 2009;64:922–930. doi: 10.1016/j.neuron.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan K, Lam M, Krsnik Z, Imamura Kawasawa Y, Lefebvre V, Sestan N. SOX5 postmitotically regulates migration, postmigratory differentiation, and projections of subplate and deep-layer neocortical neurons. Proc Natl Acad Sci USA. 2008;105:16021–16026. doi: 10.1073/pnas.0806791105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CS, Fisher SE, Hurst JA, Vargha-Khadem F, Monaco AP. A forkhead-domain gene is mutated in a severe speech and language disorder. Nature. 2001;413:519–523. doi: 10.1038/35097076. [DOI] [PubMed] [Google Scholar]
- Leone DP, Srinivasan K, Chen B, Alcamo E, McConnell SK. The determination of projection neuron identity in the developing cerebral cortex. Curr Opin Neurobiol. 2008;18:28–35. doi: 10.1016/j.conb.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui JH, Hansen DV, Kriegstein AR. Development and evolution of the human neocortex. Cell. 2011;146:18–36. doi: 10.1016/j.cell.2011.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon V, Leroux J, White CD, Reiss AL. Frontostriatal deficits in fragile X syndrome: relation to FMR1 gene expression. Proc Natl Acad Sci USA. 2004;101:3615–3620. doi: 10.1073/pnas.0304544101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molliver ME, Kostovic I, van der Loos H. The development of synapses in cerebral cortex of the human fetus. Brain Res. 1973;50:403–407. doi: 10.1016/0006-8993(73)90741-5. [DOI] [PubMed] [Google Scholar]
- Molyneaux B, Arlotta P, Menezes J, Macklis J. Neuronal subtype specification in the cerebral cortex. Nat Rev Neurosci. 2007;8:427–437. doi: 10.1038/nrn2151. [DOI] [PubMed] [Google Scholar]
- Mountcastle VB. The columnar organization of the neocortex. Brain. 1997;120:701–722. doi: 10.1093/brain/120.4.701. [DOI] [PubMed] [Google Scholar]
- Nikonenko I, Boda B, Steen S, Knott G, Welker E, Muller D. PSD-95 promotes synaptogenesis and multiinnervated spine formation through nitric oxide signaling. J Cell Biol. 2008;183:1115–1127. doi: 10.1083/jcb.200805132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nott A, Watson PM, Robinson JD, Crepaldi L, Riccio A. S-Nitrosylation of histone deacetylase 2 induces chromatin remodelling in neurons. Nature. 2008;455:411–415. doi: 10.1038/nature07238. [DOI] [PubMed] [Google Scholar]
- O’Donnell P, Grace AA. Cortical afferents modulate striatal gap junction permeability via nitric oxide. Neuroscience. 1997;76:1–5. doi: 10.1016/s0306-4522(96)00433-2. [DOI] [PubMed] [Google Scholar]
- Peters A. The Morphology of Minicolumns. In: Blatt GJ, editor. The Neurochemical Basis of Autism. Springer; 2010. pp. 45–68. [Google Scholar]
- Pierce K, Redcay E. Fusiform function in children with an autism spectrum disorder is a matter of “who”. Biol Psychiatry. 2008;64:552–560. doi: 10.1016/j.biopsych.2008.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rackham O, Brown CM. Visualization of RNA-protein interactions in living cells: FMRP and IMP1 interact on mRNAs. EMBO J. 2004;23:3346–3355. doi: 10.1038/sj.emboj.7600341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakic P. Specification of cerebral cortical areas. Science. 1988;241:170–176. doi: 10.1126/science.3291116. [DOI] [PubMed] [Google Scholar]
- Ramocki MB, Zoghbi HY. Failure of neuronal homeostasis results in common neuropsychiatric phenotypes. Nature. 2008;455:912–918. doi: 10.1038/nature07457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reif A, Herterich S, Strobel A, Ehlis AC, Saur D, Jacob CP, Wienker T, Töpner T, Fritzen S, Walter U, et al. A neuronal nitric oxide synthase (NOS-I) haplotype associated with schizophrenia modifies prefrontal cortex function. Mol Psychiatry. 2006;11:286–300. doi: 10.1038/sj.mp.4001779. [DOI] [PubMed] [Google Scholar]
- Reif A, Jacob CP, Rujescu D, Herterich S, Lang S, Gutknecht L, Baehne CG, Strobel A, Freitag CM, Giegling I, et al. Influence of functional variant of neuronal nitric oxide synthase on impulsive behaviors in humans. Arch Gen Psychiatry. 2009;66:41–50. doi: 10.1001/archgenpsychiatry.2008.510. [DOI] [PubMed] [Google Scholar]
- Rizzolatti G, Craighero L. The mirror-neuron system. Annu Rev Neurosci. 2004;27:169–192. doi: 10.1146/annurev.neuro.27.070203.144230. [DOI] [PubMed] [Google Scholar]
- Rogers SJ, Wehner DE, Hagerman R. The behavioral phenotype in fragile X: symptoms of autism in very young children with fragile X syndrome, idiopathic autism, and other developmental disorders. J Dev Behav Pediatr. 2001;22:409–417. doi: 10.1097/00004703-200112000-00008. [DOI] [PubMed] [Google Scholar]
- Schaeffer C, Bardoni B, Mandel JL, Ehresmann B, Ehresmann C, Moine H. The fragile X mental retardation protein binds specifically to its mRNA via a purine quartet motif. EMBO J. 2001;20:4803–4813. doi: 10.1093/emboj/20.17.4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sestan N, Kostovic I. Histochemical localization of nitric oxide synthase in the CNS. Trends Neurosci. 1994;17:105–106. doi: 10.1016/0166-2236(94)90115-5. [DOI] [PubMed] [Google Scholar]
- Shinkai T, Ohmori O, Hori H, Nakamura J. Allelic association of the neuronal nitric oxide synthase (NOS1) gene with schizophrenia. Mol Psychiatry. 2002;7:560–563. doi: 10.1038/sj.mp.4001041. [DOI] [PubMed] [Google Scholar]
- Siomi H, Choi M, Siomi MC, Nussbaum RL, Dreyfuss G. Essential role for KH domains in RNA binding: impaired RNA binding by a mutation in the KH domain of FMR1 that causes fragile X syndrome. Cell. 1994;77:33–39. doi: 10.1016/0092-8674(94)90232-1. [DOI] [PubMed] [Google Scholar]
- State MW. The genetics of child psychiatric disorders: focus on autism and Tourette syndrome. Neuron. 2010;68:254–269. doi: 10.1016/j.neuron.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd PK, Mack KJ, Malter JS. The fragile X mental retardation protein is required for type-I metabotropic glutamate receptor-dependent translation of PSD-95. Proc Natl Acad Sci USA. 2003;100:14374–14378. doi: 10.1073/pnas.2336265100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Dutch-Belgian Fragile X Consortium. Bakker CE, Verheij C, Willemsen R, Vanderhelm R, Oerlemans F, Vermey M, Bygrave A, Hoogeveen AT, Oostra BA, et al. Fmr1 knockout mice: a model to study fragile X mental retardation. The Dutch-Belgian Fragile X Consortium. Cell. 1994;78:23–33. [PubMed] [Google Scholar]
- Walsh CA, Morrow EM, Rubenstein JL. Autism and brain development. Cell. 2008;135:396–400. doi: 10.1016/j.cell.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willemsen R, Levenga J, Oostra B. CGG repeat in the FMR1 gene: size matters. Clin Genet. 2011;80:214–225. doi: 10.1111/j.1399-0004.2011.01723.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalfa F, Eleuteri B, Dickson KS, Mercaldo V, De Rubeis S, di Penta A, Tabolacci E, Chiurazzi P, Neri G, Grant SG, et al. A new function for the fragile X mental retardation protein in regulation of PSD-95 mRNA stability. Nat Neurosci. 2007;10:578–587. doi: 10.1038/nn1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.