Abstract
Autophagy is a lysosomal degradation pathway that degrades damaged or superfluous cell components into basic biomolecules, which are then recycled back into the cytosol. In this respect, autophagy drives a flow of biomolecules in a continuous degradation-regeneration cycle. Autophagy is generally considered a pro-survival mechanism protecting cells under stress or poor nutrient conditions. Current research clearly shows that autophagy fulfills numerous functions in vital biological processes. It is implicated in development, differentiation, innate and adaptive immunity, ageing and cell death. In addition, accumulating evidence demonstrates interesting links between autophagy and several human diseases and tumor development. Therefore, autophagy seems to be an important player in the life and death of cells and organisms. Despite the mounting knowledge about autophagy, the mechanisms through which the autophagic machinery regulates these diverse processes are not entirely understood. In this review, we give a comprehensive overview of the autophagic signaling pathway, its role in general cellular processes and its connection to cell death. In addition, we present a brief overview of the possible contribution of defective autophagic signaling to disease.
Keywords: autophagy, autophagosome, vesicle, degradation, cell death
Introduction
Cells have mechanisms that clear out superfluous, misfolded, damaged and harmful proteins: the ubiquitin-proteasome degradation pathway and autophagy. In contrast to the proteasome, autophagy has a nearly unlimited degradative capacity. It can target large protein aggregates and entire organelles. It is not restricted to protein degradation but breaks down lipids, DNA and RNA. In this way, autophagy provides new pools of amino acids, fatty acids and nucleosides for anabolic processes and drives a continuous flow of materials in the cell in a degradation-regeneration cycle 1. The executors of autophagic degradation are the lysosomes, which enclose acidic hydrolases, such as peptidases, lipases and nucleases, which degrade large molecules into their basic units. Although all autophagic routes end up in the lysosomal compartment (vacuole in yeast), many ways are used to reach the lysosomes.
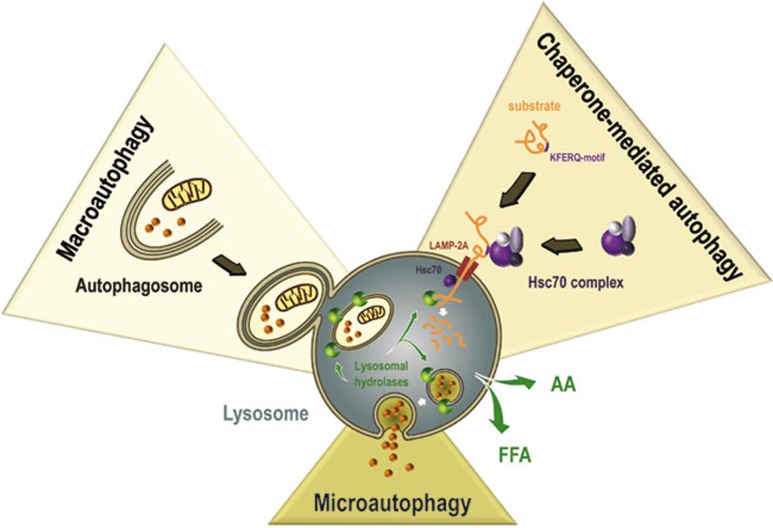
Three types of autophagy mechanisms have been recognized in mammalian cells: chaperone-mediated autophagy, microautophagy and macroautophagy 2 (Figure 1). Chaperone-mediated autophagy sequesters proteins that expose a KFERQ-like motif. Mediated by heat shock cognate 70 and its co-chaperones, these proteins are directly targeted to the lysosomes. Degradation then occurs upon interaction with and internalization through the lysosomal receptor lysosomal-associated membrane protein 2 (LAMP-2A) 3. During microautophagy, the lysosomal membrane invaginates to engulf portions of the cytoplasm,which are broken down once they are entirely enclosed. Macroautophagy differs from the other types of autophagy because it involves specialized vacuoles for cargo transportation. These vacuoles, called autophagosomes, result from de novo synthesis of autophagic membranes (phagophores), which upon closure form vesicles with a double membrane. Macroautophagy is evolutionarily well conserved and occurs in all eukaryotes. Because mouse models only exist for macroautophagy so far, extensive research has been dedicated to the understanding of this type of autophagy. This research has brought to light the clear relevance of macroautophagy to human disease. Therefore, in this review, we will focus on macroautophagy, and for the sake of simplicity, we will refer to it as autophagy.
Figure 1.
Schematic representation of the different types of autophagy. Chaperone-mediated autophagy sequesters proteins harboring a KFERQ-like motif that, mediated by the Hsc70 complex, are directly targeted to the lysosomes for degradation. During microautophagy the lysosomal membrane invaginates to engulf portions of the cytoplasm, which are consequently broken down once entirely enclosed. During macroautophagy specialized vacuoles are formed for cargo transportation. These vacuoles, called autophagosomes, are double membrane bound and deliver proteins, lipids and organelles to the lysosome.
Autophagy is primarily a non-selective, bulk degradation pathway, but the importance of more selective forms of autophagy is becoming increasingly apparent. Mitophagy, pexophagy, reticulophagy, nucleophagy, lipophagy and xenophagy refer to the selective removal of mitochondria, peroxisomes, endoplasmic reticulum (ER), nuclei, lipids and intruding microorganisms, respectively. Moreover, autophagy can sequester selective protein targets, such as ubiquitinated protein aggregates or key effectors of important signaling pathways 4, 5, 6. The importance of autophagic signaling to homeostasis has been shown by the study of autophagy-defective systems. Autophagy primarily fulfills a pro-survival role during adaptation to unfavorable growth conditions or following cellular stress. Accumulating data also demonstrate its involvement in general processes such as development, differentiation, immune homeostasis, defense against pathogens, ageing and cell death. Therefore, interest in autophagy has experienced exponential growth during the last decade. Yet many questions concerning its specific role in these diverse cellular and (patho)physiological processes remain unanswered, and our knowledge about its molecular signaling is far from complete.
Molecular signaling of autophagy
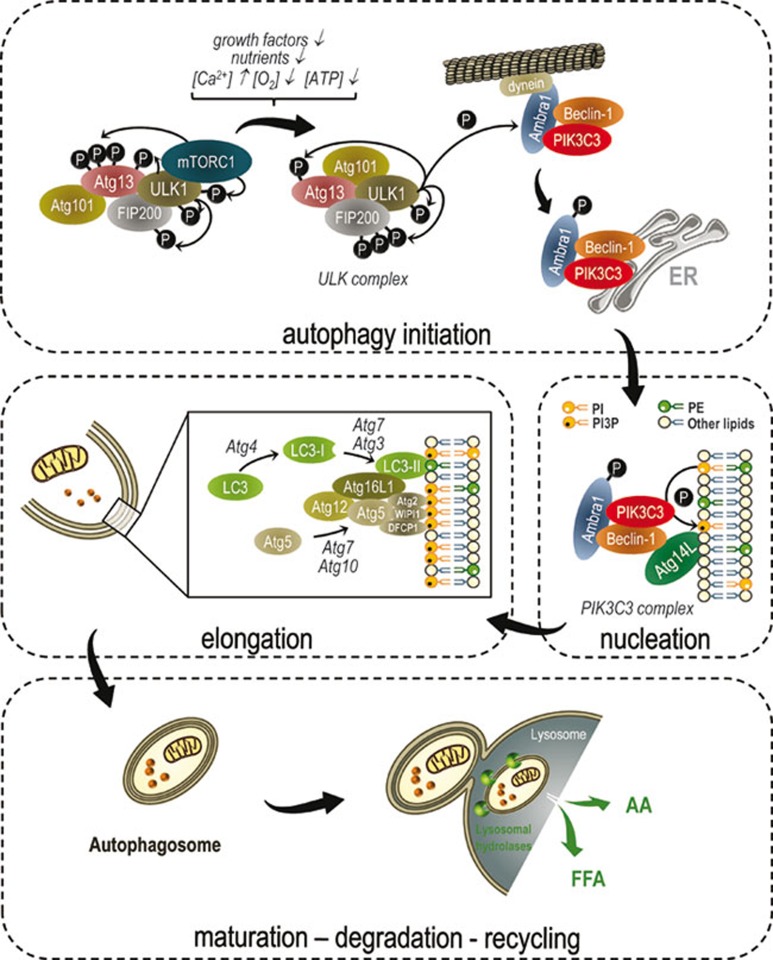
Autophagy induction is tightly controlled by complex regulatory mechanisms involving diverse input signals, including nutrients, growth factors, hormones, intracellular Ca2+-concentrations, adenosine triphosphate (ATP) levels, hypoxia, accumulation of misfolded proteins and many more (Figure 2). Many signals converge at the level of the mammalian target of rapamycin complex 1 (mTORC1). mTORC1 consists of mTOR, regulatory associated protein of mTOR (raptor), DEP-domain-containing mTOR-interacting protein (Deptor), proline-rich AKT substrate 40 kDa (PRAS40) and G-protein β-subunit-like protein (GβL) 7. mTORC1 regulates a variety of cellular responses, such as cell growth, proliferation, protein synthesis and autophagy. When amino acids and growth factors are present, class I phosphatidylinositol-3-kinase (PIK3C1) activates mTORC1, which suppresses autophagic signaling. Active mTORC1 inhibits autophagy by binding and phosphorylating uncoordinated-51 (unc-51)-like kinase 1 or 2 (ULK1 or ULK2) and Atg13 within the ULK complex 8, 9, 10. This complex is composed of ULK1 or ULK2, Atg13, focal adhesion kinase family interacting protein of 200 kDa (FIP200), and Atg101 10, 11, 12. Consequently, repression of mTORC1 by nutrient deprivation or rapamycin treatment is commonly used to activate autophagy. When mTORC1 is inactivated, it dissociates from the ULK complex, promoting ULK activity and FIP200 hyperphosphorylation 10. The exact role of the ULK complex has long been elusive. However, recent data demonstrate its involvement in the proper localization of another crucial autophagy-inducing complex, the phosphatidylinositol-3-kinase class-III (PIK3C3) complex 13. In nutrient-rich conditions, the PIK3C3 complex connects to the cytoskeleton. This interaction is mediated by the activating molecule in Beclin-1-regulated autophagy 1 (Ambra1), which binds both the PIK3C3 complex and the microtubule-associated dynein motor complex 13. During starvation, ULK1 phosphorylates Ambra1, which releases Ambra1 and the PIK3C3 complex from the microtubules and enables the PIK3C3 complex to relocalize to the ER, which is regarded as the major organelle contributing to autophagosome formation 13. Also in yeast, the Atg1 (ortholog of ULK1) complex is implicated in regulating the localization of the vacuolar protein sorting 34 (homolog of PIK3C3) complex, as Atg13 targets the latter to the site of autophagosome formation, called the phagophore assembly site (PAS) or pre-autophagosomal structure 14. In addition, ULK1 has an important role in regulating the trafficking of mAtg9 (Atg9), a multi-spanning transmembrane protein that cycles between the trans Golgi network (TGN) and endosomal compartment in basal conditions 15. During starvation, mAtg9 is recruited to the growing autophagosome, where it might supply the lipids needed for membrane elongation 16. This mAtg9 trafficking was found to involve ULK1-mediated activation of the myosin II motor protein 17.
Figure 2.
Molecular signaling of macroautophagy. During cellular and metabolic stress the mTOR is inactivated, which allows ULK complex activation. ULK complex activation involves ULK1-dependent phosphorylation of Atg13, FIP200 and ULK1 itself. These phosphorylation events are required for autophagy initiation. ULK1 also phosphorylates Ambra1, which interacts with the PIK3C3 complex at the microtubule. Phosphorylation of Ambra1 results in the release of Ambra1 and the PIK3C3 complex from the microtubule and their translocation to the ER, the major site of autophagosome formation. During autophagosome nucleation, the PIK3C3 complex generates PI3P, which interacts with DFCP1, Atg2 and WIPI1 and recruits other Atg proteins involved in membrane elongation. To this end, two ubiquitin-like processes are carried out. Atg7 and Atg10 mediate Atg5-Atg12 complex formation. This Atg5-Atg12 complex subsequently binds to Atg16L1, generating the Atg16L1 complex. LC3 is cleaved by the cysteine protease, Atg4, to LC3-I. Mediated by Atg7, Atg10 and the Atg16L1 complex, LC3-I is conjugated to phosphatidylethanolamine (PE), generating LC3-II. After the completion of autophagosome formation, the outer autophagosomal membrane fuses with the lysosome, releasing the autophagic cargo into the lysosomal lumen. The inner autophagosomal membrane and its content become degraded and the resulting amino acids (AA), free fatty acids (FFA) etc. are released back into the cytosol. Figures were produced using Servier Medical Art (http://www.servier.com).
Following autophagy induction, autophagosomes are formed. Autophagosome biosynthesis can be divided into three main processes: vesicle nucleation, elongation and maturation (Figure 2) 18. PIK3C3, p150, Ambra1 and Bcl-2-interacting protein (Beclin-1) form the core of the PIK3C3 complex, which can further bind either UV-radiation resistance associated gene (UVRAG) or Atg14L 19. Within this complex, Beclin-1 (Atg6) constitutes a platform for the binding of several interactors regulating the kinase activity of PIK3C3 20. PIK3C3 generates phosphatidyl-inositol-3-phosphate (PI3P), which recruits additional autophagy-related (Atg) proteins to the site of autophagosome formation, a process called autophagosome nucleation. Some of these PI3P-binding autophagic proteins have recently been identified: WD repeat domain phosphoinositide interacting 1 (WIPI1) and 2 (human orthologs of Atg18), mAtg2 (Atg2) and double-FYVE containing protein-1 (DFCP1) 21. However, further studies are needed to clarify their precise functions in autophagosome biogenesis. Two ubiquitin-like conjugation systems are implicated in autophagosome membrane expansion, shaping and sealing 18. First, Atg12 is activated by the E1-like enzyme, Atg7. Atg12 is then transferred to Atg10, an E2-like enzyme, and then covalently linked to Atg5. This Atg12-Atg5 conjugate further interacts with Atg16L1 to form a trimer, which consequently homodimerizes to form a large multimeric complex 22, 23. This Atg16L1 complex is transiently associated with the outer autophagosomal membrane of the growing autophagosome, and has been suggested to determine its curvature. In addition, the Atg16L1 complex acts as an E3 ligase, allowing the second conjugation reaction to be completed 24. This second ubiquitin-like conjugation pathway involves LC3 (microtubule-associated light chain-3) lipidation. LC3 (homolog of Atg8) is cleaved by Atg4, a cysteine protease, and this exposes a glycine at the C-terminal end 25. Following the combined action of Atg7, Atg3 (E2-like enzyme) and the Atg16L1 complex (E3-ligase), LC3 is finally conjugated to phosphatidylethanolamine (PE) to produce LC3-PE (also called LC3-II). In contrast to LC3 (or LC3-I), LC3-II specifically localizes to the autophagosomal membranes and so it is suited to serving as an autophagy-specific marker 26. Upon vesicle completion, Atg4 removes LC3-II from the outer autophagosomal membrane in a process called deconjugation. However, LC3-II remains associated with the inner autophagosomal membrane and thus becomes partially degraded after fusion with the lysosome.
Once autophagosome formation is completed, autophagosomes can fuse with early or late endosomes to become amphisomes. Fusion with early or late endosomes delivers cargo and components of the membrane fusion machinery and lowers the pH before delivery of the lysosomal hydrolases. Autophagosomes can also be directly targeted to the lysosomes along the microtubules 27. The outer autophagosomal membrane fuses with the lysosomes, releasing the inner autophagosomal membrane and its contents (the autophagic body) into the lysosomal lumen. Several proteins are involved in these fusion events, including LAMP2, the UVRAG-C-Vps tethering complex, Rab (Ras-related GTP-binding protein), HOPS (homotypic fusion and protein sorting), SNAREs (soluble N-ethylmalemide-sensitive factor attachment protein receptor), AAA ATPases, LC3, FYCO1 and the ESCRT (endosomal sorting complex required for transport) machinery 28. Also “kiss-and-run” fusions often take place, during which the autophagic cargo is transferred from the autophagosome to the lysosome before both vesicles segregate 29. Once the autophagic body is taken up by the lysosome, it disintegrates and its cargo is degraded by lysosomal hydrolases and lipases. Subsequently, lysosomal efflux transporters, e.g., Atg22 in yeast (mammalian ortholog not yet identified), mediate the release of the resulting amino acids, fatty acids and nucleosides back into the cytosol 30.
Although much progress has been made in the research on autophagosome biogenesis, several key questions remain on the cellular topology of autophagosome formation and on the origin of the lipids that constitute the autophagic membrane. In yeast, autophagosome formation starts at the PAS, which is located near the vacuole but is still of unknown origin. In mammals, autophagosome formation occurs at dispersed locations in the cytosol. Most studies designate the ER as the main source of autophagic membranes 31. However, several others also demonstrate the emergence of autophagic vesicles from mitochondria, TGN, plasma membrane and nucleus 16, 31, 32, 33, 34. Whether all these organelles collectively participate in autophagosome formation in the same autophagy process, or whether regulation mechanisms determine which membrane source is used, needs further investigation.
Autophagy in quality control
Although autophagy is especially known for its pro-survival function during metabolic stress, it is constitutively active in virtually all tissues of eukaryotic organisms and exerts an important housekeeping role. At basal levels, autophagy maintains cellular homeostasis and provides an intracellular quality control mechanism. To this end, autophagy targets misfolded proteins and damaged organelles for degradation, preventing the accumulation of aberrant cellular components, which can disturb cellular function. Accordingly, protein aggregates and inclusion bodies are found in autophagy-deficient hepatocytes (Atg7−/−), cardiomyocytes (Atg5−/−) and neural cells (Atg5−/− or Atg7−/−) 35, 36, 37, 38. Moreover, liver-specific Atg7-knockout mice develop hepatomegaly and neuron-specific Atg5 or Atg7 deficient mice suffer from neurodegeneration 35, 37, 38.
One of the key events in the removal of misfolded proteins is ubiquitination 6. Misfolded proteins are recognized by molecular chaperones of the heat shock protein family and are targeted for ubiquitination by E3-ligases, such as CHIP (carboxyl terminus of the heat shock cognate protein 70-interacting protein) and Parkin 39, 40. Often these ubiquitinated misfolded proteins polymerize and form protein aggregates and inclusion bodies, which cannot be removed by the proteasome, but are cleared by autophagy. Two ubiquitin-binding proteins, p62/SQSTM1 (sequestosome 1) and NBR1 (neighbor of BRCA1 gene 1), are involved in the formation of these inclusion bodies and have been implicated in the selective removal of aggregation-prone proteins 41, 42. In addition to an ubiquitin-binding domain, both proteins contain an LC3-interacting region (WxxL), which enables them to target their substrates to the growing autophagosome 42, 43. Interestingly, when the rate of formation exceeds that of clearance, protein aggregates are delivered to microtubule-organizing center (MTOC)-associated aggresomes, a process requiring histone deacetylase 6 (HDAC6) 44. HDAC6 binds ubiquitin and directly interacts with dynein motor proteins to transport aggregated proteins to the MTOC along the microtubules 45. At the MTOC, lysosomes are enriched, which allows for efficient autophagic clearance of protein aggregates.
As mentioned above, autophagy also removes damaged organelles, including mitochondria and ER. For example, loss of mitochondrial potential can lead to the selective degradation of mitochondria in a process called mitophagy 46. Although the mechanisms involved in selective organelle removal are not well understood, specific autophagy-receptors seem to play a role. In yeast, two mitochondria-associated proteins, Uth1p and Aup1, mark mitochondria for sequestration in autophagosomes 47, 48. Similarly, in mammals, BNIP3L (BCL2/adenovirus E1B 19 kDa protein-interacting protein 3-like)/Nix is indispensable for mitochondria elimination during reticulocyte maturation 49. Recent data also suggest the importance of ubiquitination for organelle removal by autophagy 50. Parkin, an E3 ubiquitin ligase, was found to localize at depolarized mitochondria, to ubiquitinate outer mitochondrial membrane proteins and to induce mitophagy 51, 52. Parkin requires PINK1 (PTEN-induced putative kinase 1)-dependent phosphorylation for its recruitment to damaged mitochondria 53. Interestingly, PINK1 is stabilized by its interaction with Parkin and enhances basal and starvation-induced autophagy through binding with Beclin-1 54, 55.
Autophagy in development and differentiation
Studies using autophagy-defective yeast, ameba, fungi, worms, flies and mice have demonstrated an important role for autophagy during development 56. In lower eukaryotes, autophagy is often associated with tissue remodeling or metamorphosis, while in mammals it is mainly activated during pre-implantation development. Unicellular organisms, such as yeast (S. cerevisiae and S. pombe) and ameba (D. discoideum) activate adaptive processes in nutrient poor conditions that result in sporulation or the generation of fruiting bodies, respectively. During these processes, autophagy recycles amino acids needed for de novo protein synthesis and increases the organism's chance of survival 57, 58. Also C. elegans uses autophagy to adapt to stressful conditions 59. In times of nutrient starvation, increased temperature or high population density, C. elegans enters dauer diapause, a third larval stage that can survive unfavorable conditions. Several key autophagy proteins, e.g., unc-51 (homolog of yeast Atg1), Atg7, LGG-1 (homolog of Atg8), Atg18 and Bec-1, are required for normal dauer morphogenesis, and their silencing causes various defects 59, 60. Furthermore, during metamorphosis of D. melanogaster, autophagy contributes to degradation of larval midgut and salivary gland cells so that they can be replaced by adult tissues, although the role of autophagy in midgut cell death is debated 61, 62, 63. In addition, autophagy in D. melanogaster is involved in synaptic development and regulates neuromuscular junction formation 64.
In mammals, autophagy is strongly activated during the early stages of embryogenesis and seems to be crucial for the pre-implantation development of mouse embryos 65. After fertilization, autophagy is triggered to enable degradation of maternal mRNAs, proteins and sperm mitochondria in the oocyte. Autophagy-defective oocytes derived from oocyte-specific Atg5-knockout mice fail to develop beyond the four- and eight-cell stages when fertilized by Atg5-null sperm 65. Furthermore, protein synthesis rates are reduced in autophagy-deficient embryos. How autophagy governs later stages of development is not completely clear. However, the importance of Atg proteins during mammalian development is emphasized by the early embryonic death of mice deficient in Beclin-1, FIP200 or Ambra1 66, 67, 68, 69, 70. These mice suffer from early developmental abnormalities such as defective pro-amniotic cavity formation, neuronal tube formation and heart and liver development. Yet, many other knock-out mice (Atg3, Atg4C, Atg5, Atg7, Atg9, Atg16L1 and ULK1) do not exhibit severe embryonic defects and survive the entire embryonic period 37, 38, 71, 72, 73, 74, 75. This disparity of phenotypes between different Atg proteins might be explained by the pleiotropy of some autophagic proteins that also have essential non-autophagic functions. Alternatively, redundancy in functions or pathways could compensate for the loss of certain Atg proteins, which is probably the case for ULK1 75. Nevertheless, except for ULK1 and Atg4C, all other Atg-knockout mice that are born die within 1 day due to nutrient deficit and energy depletion following disruption of the trans-placental nutrient supply 71, 75, 76.
Autophagy is activated during different stages of embryogenesis, and thereafter it is involved in differentiation of specialized tissues. Many studies were performed using conditional knock-out mice to investigate the consequences of autophagy deficiency on cellular and tissue differentiation. These studies clearly demonstrate the importance of autophagy during differentiation of multiple cell types. Cellular differentiation often requires extensive morphological changes and intracellular remodeling. Maturation of erythrocytes and T-lymphocytes, for example, requires a reduction in mitochondrial content, which is made possible by autophagy 77, 78. Lack of key autophagy components (e.g., ULK1 or Atg7) results in mitochondrial accumulation and, in the case of Atg7, severely alters blood cell counts 75, 77, 78. Similarly, autophagy is involved in lipid droplet formation during adipogenesis, the efficiency of which is drastically reduced when autophagy is perturbed, for example by Atg5 or Atg7 deficiency 79, 80, 81. Further, autophagy impairment inhibits neuronal differentiation of neuroblastoma cells and affects glioma stem/progenitor cell differentiation 82, 83.
Autophagy in immunity
Increasing data show that autophagy plays a considerable role in the control of immunity and inflammation. At distinct levels, autophagy contributes to both innate and adaptive immunity as a regulator and effector 84. One autophagy effector function in innate immunity is xenophagy, which refers to the autophagy-dependent elimination of intracellular pathogens, including bacteria (S. pyogenes 85, S. flexneri 86, M. tuberculosis 87, S. typhimurium 88 and L. monocytogenes 89), parasites (T. gondii 90) and viruses (Herpes simplex virus 91). In this way, autophagy restricts pathogen replication and survival, and inhibits pathogen-induced cell death in both plants and mice 92, 93. Interestingly, NDP52 (nuclear dot protein 52 kDa) and p62 (also called /SQSTM1), both of which contain LC3- and ubiquitin-binding domains, have been identified as autophagy receptors targeting intracellular ubiquitin-coated pathogens to the autophagic pathway 94, 95. The importance of p62 in innate immunity has been emphasized by the recent finding that p62 is a downstream target of innate defense regulator-1, an antimicrobial peptide that suppresses infection and inflammation 96. Some pathogens, however, have evolved mechanisms to escape autophagy-mediated capture or degradation 84. The existence of virulence factors that counter autophagy indicates a fundamental role for autophagy in innate immunity. ICP34.5, which is expressed by Herpes simplex virus, effectively inhibits autophagy induction by binding host Beclin-1 97. Also, human immunodeficiency virus (HIV) protein Nef interacts with Beclin-1 and thereby inhibits autophagosome maturation to protect HIV from degradation 98. Other viruses, such as gamma-herpes virus and human cytomegalovirus, possess B-cell lymphoma-2 (Bcl-2)-like proteins or activate the mTOR signaling pathway to render the cells less sensitive to autophagy induction 99, 100, 101.
Interestingly, when Toll-like (TLR) and NOD-like (NLR) receptors recognize pathogen-associated molecular patterns, they not only regulate pro-inflammatory cytokine production but can also stimulate autophagy. Following activation of TLR-1, -3, -4, -5, -6 or -7, myeloid differentiation primary response gene 88 (MyD88) and TIR-domain-containing adapter-inducing interferon-β (TRIF) recruit Beclin-1 into the TLR signaling complex and thereby reduce the autophagy-inhibiting interaction between Beclin-1 and Bcl-2 102. In addition, TNF receptor-associated factor 6 (TRAF6)-mediated ubiquitination of Beclin-1 was shown to be required for TLR4-triggered autophagy in macrophages 103. The NLRs (NOD1 (nucleotide-binding oligomerization domain-containing protein 1) and NOD2) direct Atg16L1 to the site of bacterial entry at the plasma membrane, a process critical for elimination of bacteria by autophagy 104. Autophagy is also induced by many immune signals, such as the T helper (Th1) cytokines interferon (IFN)-γ and tumor necrosis factor-α, and by CD40 signaling. In contrast, Th2 cytokines, such as interleukin (IL)-4 and IL-13, inhibit autophagy 105. Conversely, cytokine production can be regulated by autophagy, as shown by the increased production of potent anti-viral factors upon RIG-I receptor activation in the absence of autophagy 106, enhanced IFN-β production in dsDNA-stimulated Atg9a-deleted mouse embryonic fibroblast cells 73, and increased IL-1β and IL-18 production in autophagy-deficient macrophages (Atg16−/−, LC3B−/− and Beclin-1+/−) 74, 107. In relation to this, it has been reported that inhibition of autophagy by Beclin-1 or Atg5 knockdown results in reactive oxygen species (ROS) accumulation and sensitization of NLRP3 inflammasome activation 108. In addition, autophagy has been reported to target pro-IL-1β for lysosomal degradation 109.
Autophagy also participates in adaptive immunity. First, it is important in the regulation of T- and B-cell survival and differentiation and in Paneth cell homeostasis 78, 110, 111, 112, 113. Second, autophagy contributes to major histocompatibility complex (MHC) class I and II antigen presentation. Molecules captured by autophagosomes are delivered to the endosomal compartment, where antigens are proteolytically processed and loaded onto the MHC class I and II molecules for presentation to CD8+ and CD4+ T-cells, respectively 114. For example, during herpes simplex virus (HSV-1) infection, autophagy facilitates presentation of HSV-1 antigens on MHC class I molecules, and mice with Atg5-deficient dendritic cells show impaired CD4+ T-cell priming upon HSV-1 infection 115, 116. Also, immunization with cells undergoing autophagy enhances the efficiency of cross-priming of antigen-specific CD8+ T-cells 117. Furthermore, autophagy functions in delivery of cytoplasmic viral nucleic acids to endosomal TLRs, which results in type I IFN production and IFN-dependent immune responses 118. Finally, autophagy modulates self-tolerance and controls T-cell selection in the mouse thymus 119. Consequently, Atg5 deficiency in thymic epithelium causes colitis and multi-organ lymphoid infiltration.
Autophagy in cell death processes
Autophagy is a cytoprotective mechanism that enables the cell to survive unfavorable growth conditions. It can prevent cell death by apoptosis, necrosis, necroptosis or pyroptosis 120. In unfavorable settings, autophagy is crucial for reducing cellular stress, whether this implies energy shortage, ROS production, genomic instability, accumulation of misfolded proteins, organelle damage or microbial infection. However, accumulating evidence also demonstrates a tight interconnection of autophagy with several cell death pathways and reveals an active contribution of autophagy to cell death. When autophagy has a functional role in the death process, the cell death process is designated “autophagic cell death” (ACD) 121. Although many studies failed to demonstrate a causal link between autophagy and cell death, an increasing number of reports suggest a contribution of Atg proteins to the execution of cell death. The interplay between autophagy and cell death is complex and diverse. Depending on the cell type, environment and stimulus, autophagy and cell death mechanisms can have inhibitory, additive or even synergistic effects.
Apoptotic cell death
As mentioned above, autophagy can prevent apoptotic cell death. The crucial role for autophagy in cell survival is apparent from studies using Atg-knockout mice. Mice deficient in Atg3, Atg5, Atg7, Atg9 or Atg16L1 fail to induce autophagy and die on the day of birth due to starvation following disruption of the trans-placental nutrient supply 122. Also, mice with neuron-specific Atg5 or Atg7 knockout suffer from neurodegeneration and apoptotic death of neurons, and T-cell-specific Atg5 deficiency results in increased apoptosis in peripheral T-cells upon T-cell activation 35, 37, 111. Furthermore, autophagy promotes epithelial cell survival during anoikis 123. Apparently, autophagy prevents apoptotic cell death in several ways. During starvation for example, autophagy inhibits apoptosis by breakdown of cellular components to recycle essential metabolites and to replenish the energy reserves 124. Moreover, autophagy degrades protein aggregates and targets damaged organelles, such as mitochondria, preventing ROS accumulation and cell death. In addition, recent reports demonstrate selective removal of apoptotic effectors by autophagosomes. In this regard, TRAIL-induced autophagy continuously targets active caspase-8 for lysosomal degradation, thereby inhibiting cell death by apoptosis 4.
Conversely, autophagy can also contribute to apoptosis. In this case, depletion of key autophagy regulators and pharmacological interference with autophagy prevent apoptosis. For example, autophagy precedes apoptotic cell death of salivary glands in D. melanogaster and inhibition of apoptosis only partially prevents cell loss 62, 125. Also, in a model for HIV infection in which T cell expressing cluster of differentiation 4 (CD4)/CXCR4 interact with cells expressing HIV-1-encoded envelope glycoproteins, autophagy precedes apoptosis 126. When autophagy is inhibited using 3-methyladenine (3-MA) or by knockdown of Beclin-1 or Atg7, caspase activity is reduced and apoptosis is attenuated. Moreover, autophagy contributes to damage-induced apoptosis as Atg5 knockdown causes a dramatic decrease in cell death 127. The mechanisms of how the autophagy machinery drives apoptotic cell death are still under intense investigation. Interestingly, during late D. melanogaster oogenesis, autophagy controls DNA fragmentation and apoptosis in nurse cells by removal of dBruce, the D. melanogaster inhibitor of apoptosis 128. Furthermore, several Atg proteins were shown to trigger cell death and to engage the apoptotic pathway (Table 1). Atg5 was shown to directly bind Fas-associated protein with death domain (FADD), thereby triggering IFN-γ-induced cell death 129; Atg7 mediates lysosome dysfunction-induced apoptosis in neural cells 130; and Beclin-1 stimulates apoptosis in response to the Bcl-2 inhibitor, obatoclax 131. Moreover, a recent study uncovered the existence of an Atg12-Atg3 conjugation, which sensitizes cells to death downstream of mitochondrial pathways 132. Also fragments resulting from proteolytic cleavage of typical Atg proteins can enhance apoptotic cell death. Truncated Atg5, Atg4D and Beclin-1 localize at the mitochondria and trigger apoptosis, for example by release of pro-apoptotic factors from mitochondria 133, 134, 135, 136. Finally, autophagy can also assist in the clearance of apoptotic cells. It has been suggested that autophagy maintains the ATP levels needed for exposure of phosphatidylserine (“eat-me” signal) and secretion of lysophosphatidylcholine (“come-and-get-me” signals) 68.
Table 1. List of Atg proteins exerting a pro-apoptotic function.
| Protein | Prot event | Role in apoptosis | Reference |
|---|---|---|---|
| Atg4D | Casp-3 (Atg4D-C) | Targets mitochondria Induces apoptosis | 136 |
| Atg5 | Uncleaved | Triggers IFN-γ-induced cell death Binds FADD | 129 |
| Calp-I, -II (Atg5-N) | Induces apoptosis Binds Bcl-xL Induces cyt c release | 133 | |
| Beclin-1 | Uncleaved | Stimulates obatoclax-induced apoptosis Triggers caspase-3 activation | 131 |
| Casp-3, -8, -9 (Beclin-1-C) | Enhances IL-3 withdrawal-induced cell death Targets mitochondria Induces Omi and cyt c release | 135 | |
| Atg7 | Uncleaved | Mediates bafilomycin-induced apoptosis Mediates chloroquine-induced apoptosis Triggers p53 activation Triggers caspase-3 activation | 130 |
| Atg12-Atg3 | Uncleaved | Enhances mitochondrial apoptosis Decreases Bcl-xL levels | 132 |
Prot event: proteolytic event, -C: C-terminal fragment, -N: N-terminal fragment
The interplay between apoptosis and autophagy is also evident from the extensive molecular crosstalk between autophagy-related and apoptosis-related proteins. Several apoptotic proteins regulate the autophagic process. Conversely, typical autophagy-related proteins can modify apoptotic signaling. One of the most obvious interactions between apoptosis and autophagy effectors occurs within the Bcl-2-Beclin-1 complex. Bcl-2 is a central apoptosis inhibitor that also effectively impedes autophagy 137. Bcl-2 directly binds Beclin-1 via its BH1 and Bcl-2 homology domain 3 (BH3) domains, and this inhibits the autophagic activity of Beclin-1 92. B-cell lymphoma extra long (Bcl-xL) and myeloid cell leukemia sequence-1 (Mcl-1), two other anti-apoptotic Bcl-2 family members, also inhibit autophagy by binding Beclin-1. In fact, Beclin-1 has been identified as a new BH3-only protein 138. However, binding of Beclin-1 to Bcl-2 does not seem to affect the anti-apoptotic role of Bcl-2 139. Bcl-2 has also been shown to inhibit autophagy induction through its interaction with Ambra1 at the mitochondria. Bcl-2-Ambra1 complex formation prevents Ambra1 from promoting Beclin-1 activity 140. Further, FLICE-inhibitory protein (FLIP) can inhibit autophagy by interacting directly with Atg3. By binding and inactivating Atg3, FLIP abrogates LC3 lipidation, which is essential for autophagosome formation 141. In contrast, other typical apoptosis proteins stimulate autophagy induction. For example, several BH3-only proteins, such as Bcl-2-associated death promoter protein (Bad), tBid (truncated BH3 interacting domain death agonist) and BNIP3, abrogate the Bcl-2-Beclin-1 interaction by competing with Beclin-1 for Bcl-2 binding. Furthermore, other apoptotic modulators (death-associated protein kinase, c-jun N-terminal kinase, TRAF6 and A20) mediate Beclin-1 or Bcl-2 post-translational modifications that enable dissociation of Beclin-1 from Bcl-2 103, 142, 143. Surprisingly, caspases were also implicated in autophagy-mediated survival. Indeed, caspase-9 promotes autophagic flux probably by regulating lysosomal pH and cathepsin activity 144. Another interesting finding is the involvement of p53, a well-known pro-apoptotic tumor suppressor, in autophagy regulation. Depending on its subcellular localization, p53 either decreases or stimulates autophagic signaling 145. When located in the cytoplasm, p53 can repress autophagy. p53 inactivation probably induces ER stress, a potent activator of autophagy, as IRE1α (inositol requiring 1) deficiency prevents induction of autophagy following p53 neutralization 146. Conversely, nuclear p53 signals to autophagy via transactivation of DRAM (Damage-regulated autophagy modulator), a lysosomal protein that induces autophagy upon DNA damage 127. In addition, p53 can inhibit mTOR through activation of AMP-activated protein kinase (AMPK) 147. p62 also constitutes a molecular link between autophagic and apoptotic signaling. During autophagy, p62 binds ubiquitinated targets and is involved in their aggregation and delivery to the autophagosome through its interaction with LC3 42. p62 is also implicated in the activation of caspase-8 when cell death is triggered 148. Following cullin-3-dependent ubiquitination of caspase-8, interaction between p62 and caspase-8 results in caspase-8 aggregation, which enhances caspase-8 activity and enables the autoproteolytic release of caspase-8 into the cytosol to trigger apoptosis 148.
Non-apoptotic cell death
Also non-apoptotic modes of cell death, such as necrosis, necroptosis, pyroptosis and NETosis, are linked to autophagy. In this regard, autophagy can protect apoptosis-deficient cells from necrosis following metabolic stress caused by ischemia 149. Accordingly, autophagy-deficiency in apoptosis-defective tumor cells activates necrotic cell death and promotes necrosis-related inflammation and tumor growth 149. Autophagy also protects macrophages from caspase-1-dependent cell death and against pyroptosis induced by Shigella infection 150. Similarly, malfunction of autophagy due to absence of Atg7 or Atg16L1 increases pyroptosis upon LPS treatment through enhancement of caspase-1 activity and IL-1β and IL-18 production 74. In contrast, autophagy activity can also provoke non-apoptotic cell death. For example, in C. elegans, autophagy is required for necrotic neuron destruction, and both 3-MA and knockdown of key Atg proteins (Bec-1, Igg-1, Atg18) reduce necrotic cell death 151, 152. Often, autophagy-associated necrotic cell death occurs when apoptosis is defective. In L929 cells, zVAD-fmk-induced necrotic cell death is triggered by ROS-mediated membrane lipid peroxidation resulting from autophagy-dependent degradation of catalase, one of the major ROS scavenging enzymes 153, 154. Similarly, triggers of cell death induce cytotoxic autophagy in Bax/Bak double knockout cells 155, 156, 157. Also, although autophagy is required for T cell homeostasis, in proliferating T cells lacking FADD or caspase-8 it is strongly increased and triggers receptor-interacting protein 1 (RIP1)-dependent necroptotic cell death 112. Interestingly, this mechanism of cell death can be used to re-sensitize glucocorticoid-resistant leukemia cells to dexamethasone after treatment with obatoclax (GX15-070), a Bcl-2 antagonist (see below) 158. Of note, p62 was shown to interact with RIP1, which regulates nuclear factor κB activation in a TRADD/RIP/p62/aPKCs/IKKbeta signaling cascade 159. Finally, autophagy enables the neutrophil extracellular trap cell death (NETosis) induced by phorbol-myristate acetate (PMA). NETosis is a specialized form of neutrophil cell death in which microbes are trapped and killed within neutrophil extracellular traps (NETs); these traps are chromatin structures loaded with concentrated anti-microbial molecules 160. During PMA-induced NETosis, autophagy prevents caspase activation and triggers chromatin decondensation, which is required for NET formation and concurrent cell death 160.
Autophagy and ageing
Aged cells are characterized by the accumulation of altered or damaged DNA, proteins, lipids and organelles, increase in oxidative stress and defects in several biological processes. One of the most intriguing properties of autophagy is its ability to counteract the ageing process and to promote longevity in different animal species. Indeed, environmental conditions or use of agents that promote the induction of autophagy have been shown to improve cellular fitness and survival 161. In C. elegans and D. melanogaster, for example, caloric restriction (not involving malnutrition) enhances autophagic activity and increases the mean lifespan 162, 163. In addition, other autophagy-promoting treatments, e.g., rapamycin, resveratrol and spermidine, enhance cell survival, fitness and/or lifespan. Conversely, knockdown or deficiency of key autophagy components reverses these effects 164, 165, 166. How resveratrol and spermidine regulate autophagy needs further investigation, but it seems that hypoacetylation might be involved. Both caloric restriction and resveratrol require sirtuin-1, a deacetylase, to promote lifespan extension 167, 168. Interestingly, sirtuin-1, apart from targeting histones, also targets key Atg proteins, such as Atg5, Atg7 and Atg8, and its overexpression induces autophagy 169, 170. Similarly, spermidine triggers epigenetic deacetylation through inhibition of acetyltransferases and, in addition, prevents oxidative stress and necrosis 171. Strikingly, the altered epigenetic acetylation status did not affect Atg7, which points to a regulatory mechanism that protects Atg genes from hypoacetylation 171. Resveratrol-induced autophagy might also involve the induction of ER stress as resveratrol has been shown to induce several ER stress markers, e.g., eukaryotic translation initiation factor 2α (eIF2α) phosphorylation, X-box-binding protein 1 splicing and CHOP (C/EBP homologous protein) expression 172. In line with the notion that autophagy is important in slowing down the ageing process, a decline in formation and elimination of autophagosomes is often observed in ageing cells 173. Thus, diminished autophagy increases the accumulation of harmful protein aggregates and damaged organelles. This results in increased ROS, higher risk of mutations in DNA and defects in biological processes, which eventually affect lysosomal and autophagic functionality and give rise to a vicious pro-aging cycle 174.
Autophagy in tumorigenesis and cancer treatment
Because of its major role in cell survival during unfavorable conditions, it is not surprising that autophagy protects tumor cells from cellular and metabolic stress. This implies that autophagy has an oncogenic function. However, much data also favor a tumor suppressive role for autophagy. Seemingly, autophagy plays a dual role in cancer 175. One current idea is that, depending on cell type, context or stage of tumor development, autophagy exerts one of the two opposing functions: autophagy, as an intracellular quality control mechanism, prevents malignant transformation and cancer progression, while its pro-survival role during non-optimal growth conditions provides the established tumor with an adaptation mechanism that promotes its survival 175. Taking into account this apparent dual role, the design of cancer treatments that modulate autophagy becomes quite complex 176. Recent data suggest that therapeutic interventions enhancing or decreasing autophagy, depending on the context, can be beneficial to cancer patients, as described in more detail in the paragraphs below 176, 177, 178, 179.
Autophagy has tumor-promoting properties and most likely provides growth advantages to tumor cells residing in a nutrient poor and hypoxic environment 175. Indeed, autophagy increases the viability of cells exposed to deprivation of serum, amino acids or growth factors 124, 180, 181. In this respect, recent reports nicely demonstrate the autophagy dependency of tumors for growth and survival 181, 182. Autophagy supports Ras-mediated tumorigenesis by maintaining mitochondrial metabolic function and energy levels 181 and is required for tumorigenic growth of pancreatic cancers probably by constraining oxidative damage and preserving metabolic homeostasis 182. In addition, autophagy can protect cells from anoikis upon detachment from the extracellular matrix, which suggests a role for autophagy in metastasis 123. In line with a tumor-protecting role for autophagy, tumor cells induce autophagy in response to most anticancer therapies and utilize autophagy to overcome the therapeutically induced stress and cellular damage 176, 177, 183. In these cases, autophagy inhibition might enhance the efficacy of anticancer treatments. Inhibition of autophagy by use of chloroquine or Atg5 knockdown has been shown to enhance tumor death induced by p53 or alkylating drugs 184. In addition, use of 3-MA or Atg7 knockdown in 5-fluorouracil-treated cancer cells enhances apoptosis induction 185. These data demonstrate a potential role for autophagy inhibitors in combination with conventional therapies in sensitizing apoptosis induction in human cancers.
However, there is generally a strong association between molecules that are implicated in autophagy induction, e.g., PTEN (phosphatase and tensin homolog), p53, tuberous sclerosis protein 1 (TSC1), TSC2 and Bax-interacting factor-1, and tumor suppression 186. More importantly, several Atg proteins, including Beclin-1, UVRAG, Atg5 and Atg4C, have tumor-suppressing effects 67, 71, 133, 187 (Table 2). Indeed, monoallelic deletions of Beclin-1 and UVRAG are frequently found in human cancers, and both Beclin-1 and UVRAG overexpression inhibit proliferation and tumorigenesis of tumor cell lines 187, 188, 189. Similarly, forced expression of Atg5 effectively reduces tumor growth of breast cancer cells, and Atg4C−/− mice are more susceptible to fibrosarcoma development following treatment with chemical carcinogens 71, 133. The mechanisms by which autophagy protects against tumorigenesis are not entirely clear. In favor of a tumor-suppressing role, it has been shown that dysfunctional autophagy in apoptosis-defective tumors stimulates necrotic cell death and inflammation, which promotes tumor growth 149. Additionally, autophagy plays an important role in limiting the accumulation of genomic damage, which favors oncogene activation and tumor initiation 190, 191. In this regard, autophagy can eliminate damaged or harmful organelles, such as mitochondria, that produce ROS and cause oxidative stress that would otherwise result in a higher rate of DNA damage. Moreover, protein aggregates, imbalanced energy homeostasis and enhanced proliferation could contribute to genomic instability when autophagy is defective 67, 192, 193. In addition, a recent report demonstrates enhanced proliferation of epithelial cells through Ras-dependent degradation of Beclin-1 following detachment from the extracellular matrix 193. As autophagy is able to contribute to cell death in apoptosis-deficient cells, pharmacological intervention aiming at enhancing autophagic function could be of high therapeutic interest. For example, in acute lymphoblastic leukemia cells, the BH3 mimetic, obatoclax, abrogates glucocorticoid resistance by promoting Beclin-1-mediated autophagy and inducing necroptotic cell death 158. In addition, knockdown of transglutaminase 2 (TG2) or Bcl-2, both of which are important autophagy suppressors strongly expressed in many tumors, was shown to induce ACD in cancer cells 194, 195.
Table 2. List of Atg protein mutations found in distinct cancer types.
| Protein | Alteration | Cancer type | Reference |
|---|---|---|---|
| Atg2B | Frameshift mutation | Gastric cancer Colorectal cancer | 215 |
| Atg5 | Frameshift mutation | Gastric cancer Colorectal cancer | 215 |
| Atg9b | Frameshift mutation | Gastric cancer Colorectal cancer | 215 |
| Beclin-1 | Mono-allelic deletion | Breast cancer Ovarian cancer Prostate cancer | 187, 189 |
| CpG hypermethylation | Breast cancer | 216 | |
| Loss-of-heterozygosity | Breast cancer | 216 | |
| Missense mutations | Gastric cancer Colorectal cancer Breast cancer | 217 | |
| FIP200 | Compound heterozygous deletions Loss-of-heterozyosity | Breast cancer | 218 |
| UVRAG | Mono-allelic deletion | Colon cancer | 219 |
| Frameshift mutation | Colon cancer Gastric cancer | 220, 221 |
Autophagy in other diseases
As autophagy plays a role in many important biological processes, defects in autophagy signaling could be associated with many diseases. Autophagy has been linked to neuronal disorders, liver and heart diseases, infectious diseases, cancer, diabetes type II, cystic fibrosis and many more 186. A considerable part of these diseases are associated with accumulation of proteins that are prone to aggregation due to mutation or misfolding. Protein aggregates can be sequestered and subsequently degraded by autophagosomes 196. When their removal is perturbed, e.g., when autophagy is defective, they can be detrimental 197. Therefore, autophagy can prevent the emergence of neurodegenerative diseases. Indeed, autophagy protects against aggregation-prone mutant proteins in spinocerebellar ataxia, mutated forms of α-synuclein in Parkinson's disease, mutant Huntingtin in Huntington's disease, tau mutants that cause frontotemporal dementia, pathogenic intraneuronal amyloid beta in Alzheimer disease brain and polyglucosan inclusion bodies in Lafora disease 162, 196, 198, 199, 200, 201, 202. Interestingly, most of these neurodegenerative diseases are associated with decreased Beclin-1 levels, which might account for the impaired autophagic clearance. In this respect, Beclin-1 expression in human brain was found to decrease with age 203. Moreover, caspase-mediated cleavage of Beclin-1 was observed in brain tissues of Alzheimer's patients 204.
Cystic fibrosis (CF), which is caused by mutations in cystic fibrosis transmembrane conductance regulator, is associated with impaired autophagy and the accumulation of protein aggregates in CF lung epithelia. In this condition, ROS accumulation causes TG2-dependent crosslinking of Beclin-1 and the sequestration of the PIK3C3 complex in perinuclear aggregates, which prevents the PIK3C3 complex from fulfilling its autophagic function at its proper locations 205.
Also certain myopathies are associated with impaired autophagic clearance of aggregation-prone proteins or damaged organelles. Muscular dystrophies linked to collagen VI deficiency, for example, are characterized by the accumulation of dysfunctional organelles and spontaneous apoptosis of muscle fibers caused by defective autophagy induction 206. Reduced levels of Beclin-1 and BNIP3 were observed in muscle biopsies from subjects with Bethlem myopathy or Ullrich congenital muscular dystrophy 206. Strikingly, forced activation of autophagy by genetic, dietary and pharmacological approaches restored myofiber survival and ameliorated the dystrophic phenotype of collagen VI-knockout mice 206. Pompe disease and Danon disease involve primary defects in the lysosomal proteins, LAMP-2 and acid alpha-glucosidase, respectively, and they are associated with extensive accumulation of autophagosomes in muscles. In addition, the skeletal muscles of patients with X-linked myopathy show excessive autophagy 207. Autophagy has also been implicated in certain heart diseases 208. LAMP-2-deficient mice suffer from cardiomyopathy, and cardiac-specific Atg5 deficiency results in increased accumulation of polyubiquitinated proteins, ER stress and apoptosis 36, 209. Moreover, autophagic activity protects from hemodynamic stress such as pressure overload and hypertrophy in failing hearts 36. However, autophagy seems detrimental for the heart during ischemia/reperfusion, possibly due to excessive activation of autophagy and consequent cell death 210, 211.
As mentioned above, autophagy also protects against human liver diseases. α1 anti-trypsin deficiency is a liver disorder associated with chronic inflammation and carcinogenesis. The Z mutation in α1 anti-trypsin causes protein misfolding and polymerization and the formation of intrahepatic inclusions, which are degraded by autophagy. Carbamazepine, an anti-epileptic drug, induces autophagy, which clears out inclusions of α1 anti-trypsin in mice and reduces hepatic fibrosis 212, 213. In addition, autophagy might be involved in the development of type II diabetes 214. Pancreatic β-cells of mice with β-cell-specific Atg7 knockout accumulated large ubiquitinated inclusion bodies and dysfunctional mitochondria and exhibited hallmarks of ER stress, possibly due to impaired autophagy. Moreover, these mice showed increased β-cell death, hyperglycemia, reduced insulin production and higher insulin resistance 214. Finally, recent studies have implicated autophagy in the predisposition to Crohn's disease (CD), a major form of chronic inflammatory bowel disease. Both Atg16L1 and IRGM1 (immunity-related GTPase family M member 1), two autophagy-related genes, have been identified as susceptibility loci for CD. The CD-associated Atg16L1*300A polymorphism results in impaired autophagic activity, reduced autophagic clearance of bacteria and Paneth cell granule abnormalities 113. Interestingly, the CD-related frameshift mutation in NOD2 (L1007insC) encodes a mutant NOD2 protein incapable of recruiting Atg16L1 to the site of bacterial entry, which leads to inefficient clearance of bacteria by autophagy 104.
Conclusions and perspectives
Over recent years, our knowledge about the autophagic process has greatly expanded. Many autophagy-related proteins have been identified and functionally characterized in yeast and mammals. In addition, we are starting to unveil the physiological roles of autophagy. But despite these recent advances, many questions remain largely unanswered. How is the specificity in the many types of autophagy regulated? Which organelles contribute to autophagosome biogenesis and what determines the origin of the membranes? What is the redundancy between the Atg proteins and which pleiotropic functions do they fulfill? Does the effect of caloric restriction on lifespan occur mainly through autophagy induction and how can this be therapeutically exploited to treat obesity? What are the downstream targets of autophagy induction during dietary restriction? How do micro-autophagy and chaperone-mediated autophagy contribute to human disease? Which parameters determine whether autophagy is pro-survival or pro-death, tumor promoting or tumor suppressive? Hopefully, future studies will increase our understanding of the mechanisms underlying autophagy, its functions in multiple biological processes and the regulatory signaling pathways that control its activation. This knowledge will be of great importance in validating autophagy as an effective target for the treatment of various diseases, including cancer.
Acknowledgments
This research has been supported by Flanders Institute for Biotechnology (VIB), by European grants (FP6 ApopTrain, MRTN-CT-035624; FP7 EC RTD Integrated Project, Apo-Sys, FP7-200767; Euregional PACT II), Belgian grants (Interuniversity Attraction Poles, IAP 6/18), Flemish grants (Fonds Wetenschappelijk Onderzoek Vlaanderen, FWO G.0875.11 and FWO G.0973.11), and Ghent University grants (MRP, GROUP-ID). PV is holder of a Methusalem grant (BOF09/01M00709) from the Flemish Government. The doctoral fellowship of EW has been supported by Ghent University grants (BOF 2001-GOA 12050502 en BOF 2005-GOA 01GC0205) and by the Methusalem project BOF09/01M00709. SL and TV are postdoctoral fellows with the FWO. Research in PA's laboratory is supported by FWO G.0661.09 and G.0728.10, the Catholic University of Leuven (GOA/11/009) and the Stichting tegen Kanker. We thank Dr Amin Bredan (DMBR-VIB, Ghent) for editing the manuscript.
Glossary
- 3-MA
(3-methyladenine)
- ACD
(autophagic cell death)
- Ambra1
(activating molecule in Beclin-1-reguated autophagy)
- AMPK
(AMP-activated protein kinase)
- Atg
(autophagy related)
- ATP
(adenosine triphosphate)
- Bad
(Bcl-2-associated death promoter protein)
- Bcl-2
(B-cell lymphoma-2)
- Bcl-xL
(B-cell lymphoma extra long)
- Beclin-1
(Bcl-2-interacting protein)
- BH3
(Bcl-2 homology domain 3)
- BNIP3
(BCL2/adenovirus E1B 19 kDa protein-interacting protein 3)
- CD
(Crohn's disease)
- CD4
(cluster of differentiation 4)
- Deptor
(DEP-domain-containing mTOR-interacting protein)
- DFCP1
(double FYVE-containing protein-1)
- eIF2α
(eukaryotic translation initiation factor 2α)
- DRAM
(damage-regulated autophagy modulator)
- ER
(endoplasmic reticulum)
- ESCRT
(endosomal sorting complex required for transport)
- FADD
(Fas-associated protein with death domain)
- FIP200
(focal adhesion kinase family interacting protein of 200 kDa)
- FLIP
(FLICE-inhibitory protein)
- FYCO1
(FYVE and coiled-coil domain containing 1)
- GβL
(G-protein β-subunit-like protein)
- HDAC6
(histone deacetylase 6)
- HIV
(human immunodeficiency virus)
- HOPS
(homotypic fusion and protein sorting)
- HSV
(herpes simplex virus)
- IL-1β
(interleukin-1β)
- IRE1α
(inositol requiring 1)
- IRGM1
(immunity-related GTPase family M member 1)
- IFN
(interferon)
- LAMP-2
(lysosomal-associated membrane protein 2)
- LC3
(microtubule-associated light chain-3)
- Mcl-1
(myeloid cell leukemia sequence-1)
- MHC
(major histocompatibility complex)
- MTOC
(microtubule-organizing centre)
- mTORC1
(mammalian target of rapamycin complex 1)
- MyD88
(myeloid differentiation primary response gene 88)
- NBR1
(neighbor of BRCA1 gene 1)
- NDP52
(nuclear dot protein 52kDa)
- NLR
(NOD-like receptor)
- NOD1
(nucleotide-binding oligomerization domain-containing protein 1)
- PAS
(phagophore assembly site)
- PE
(phosphatidylethanolamine)
- PIK3C3
(phosphatidyl-inositol-3-kinase class III)
- PINK1
(PTEN-induced putative kinase 1)
- PI3P
(phosphatidyl-inositol-3-phosphate)
- PMA
(phorbol-myristate acetate)
- PRAS40
(proline-rich AKT substrate 40 kDa)
- PTEN
(phosphatase and tensin homolog)
- Rab
(Ras-related GTP-binding protein)
- ROS
(reactive oxygen species)
- RIP1
(receptor-interacting protein 1)
- SQSTM1
(sequestosome 1)
- SNARE
(soluble N-ethylmalemide-sensitive factor attachment protein receptor)
- tBid
(truncated BH3 interacting domain death agonist)
- TG2
(transglutaminase 2)
- TGN
(trans Golgi network)
- Th1
(T helper)
- TLR
(Toll-like receptor)
- TRAF6
(TNF receptor associated factor 6)
- TRIF
(TIR-domain-containing adapter-inducing interferon-β)
- TSC1
(tuberous sclerosis protein 1)
- UCMD
(Ullrich congenital muscular dystrophy)
- ULK
(unc-51-like kinase)
- unc-51
(uncoordinated-51)
- UVRAG
(UV-radiation resistance associated gene)
- WIPI1
(WD repeat domain phosphoinositide interacting 1)
References
- Rabinowitz JD, White E. Autophagy and metabolism. Science. 2010;330:1344–1348. doi: 10.1126/science.1193497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuervo AM. Autophagy: many paths to the same end. Mol Cell Biochem. 2004;263:55–72. doi: 10.1023/B:MCBI.0000041848.57020.57. [DOI] [PubMed] [Google Scholar]
- Kaushik S, Bandyopadhyay U, Sridhar S, et al. Chaperone-mediated autophagy at a glance. J Cell Sci. 2011;124:495–499. doi: 10.1242/jcs.073874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou W, Han J, Lu C, Goldstein LA, Rabinowich H. Autophagic degradation of active caspase-8: a crosstalk mechanism between autophagy and apoptosis. Autophagy. 2010;6:891–900. doi: 10.4161/auto.6.7.13038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C, Cao W, Bao L, et al. Autophagy negatively regulates Wnt signalling by promoting Dishevelled degradation. Nat Cell Biol. 2010;12:781–790. doi: 10.1038/ncb2082. [DOI] [PubMed] [Google Scholar]
- Kirkin V, McEwan DG, Novak I, Dikic I. A role for ubiquitin in selective autophagy. Mol Cell. 2009;34:259–269. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- Sengupta S, Peterson TR, Sabatini DM. Regulation of the mTOR complex 1 pathway by nutrients, growth factors, and stress. Mol Cell. 2010;40:310–322. doi: 10.1016/j.molcel.2010.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa N, Hara T, Kaizuka T, et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell. 2009;20:1981–1991. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganley IG, Lam du H, Wang J, Ding X, Chen S, Jiang X. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem. 2009;284:12297–12305. doi: 10.1074/jbc.M900573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung CH, Jun CB, Ro SH, et al. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell. 2009;20:1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer CA, Kaliappan A, Dennis PB. A novel, human Atg13 binding protein, Atg101, interacts with ULK1 and is essential for macroautophagy. Autophagy. 2009;5:649–662. doi: 10.4161/auto.5.5.8249. [DOI] [PubMed] [Google Scholar]
- Hosokawa N, Sasaki T, Iemura S, Natsume T, Hara T, Mizushima N. Atg101, a novel mammalian autophagy protein interacting with Atg13. Autophagy. 2009;5:973–979. doi: 10.4161/auto.5.7.9296. [DOI] [PubMed] [Google Scholar]
- Di Bartolomeo S, Corazzari M, Nazio F, et al. The dynamic interaction of AMBRA1 with the dynein motor complex regulates mammalian autophagy. J Cell Biol. 2010;191:155–168. doi: 10.1083/jcb.201002100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obara K, Sekito T, Ohsumi Y. Assortment of phosphatidylinositol 3-kinase complexes—Atg14p directs association of complex I to the pre-autophagosomal structure in Saccharomyces cerevisiae. Mol Biol Cell. 2006;17:1527–1539. doi: 10.1091/mbc.E05-09-0841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webber JL, Tooze SA. New insights into the function of Atg9. FEBS Lett. 2010;584:1319–1326. doi: 10.1016/j.febslet.2010.01.020. [DOI] [PubMed] [Google Scholar]
- Young AR, Chan EY, Hu XW, et al. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J Cell Sci. 2006;119:3888–3900. doi: 10.1242/jcs.03172. [DOI] [PubMed] [Google Scholar]
- Tang HW, Wang YB, Wang SL, Wu MH, Lin SY, Chen GC. Atg1-mediated myosin II activation regulates autophagosome formation during starvation-induced autophagy. EMBO J. 2011;30:636–651. doi: 10.1038/emboj.2010.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanida I. Autophagosome formation and molecular mechanism of autophagy. Antioxid Redox Signal. 2011;14:2201–2214. doi: 10.1089/ars.2010.3482. [DOI] [PubMed] [Google Scholar]
- He C, Levine B. The Beclin 1 interactome. Curr Opin Cell Biol. 2010;22:140–149. doi: 10.1016/j.ceb.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funderburk SF, Wang QJ, Yue Z. The Beclin 1-VPS34 complex—at the crossroads of autophagy and beyond. Trends Cell Biol. 2010;20:355–362. doi: 10.1016/j.tcb.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polson HE, de Lartigue J, Rigden DJ, et al. Mammalian Atg18 (WIPI2) localizes to omegasome-anchored phagophores and positively regulates LC3 lipidation. Autophagy. 2010;6:506–522. doi: 10.4161/auto.6.4.11863. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Kuma A, Kobayashi Y, et al. Mouse Apg16L, a novel WD-repeat protein, targets to the autophagic isolation membrane with the Apg12-Apg5 conjugate. J Cell Sci. 2003;116:1679–1688. doi: 10.1242/jcs.00381. [DOI] [PubMed] [Google Scholar]
- Kuma A, Mizushima N, Ishihara N, Ohsumi Y. Formation of the approximately 350-kDa Apg12-Apg5.Apg16 multimeric complex, mediated by Apg16 oligomerization, is essential for autophagy in yeast. J Biol Chem. 2002;277:18619–18625. doi: 10.1074/jbc.M111889200. [DOI] [PubMed] [Google Scholar]
- Fujita N, Itoh T, Omori H, Fukuda M, Noda T, Yoshimori T. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol Biol Cell. 2008;19:2092–2100. doi: 10.1091/mbc.E07-12-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanida I, Sou YS, Ezaki J, Minematsu-Ikeguchi N, Ueno T, Kominami E. HsAtg4B/HsApg4B/autophagin-1 cleaves the carboxyl termini of three human Atg8 homologues and delipidates microtubule-associated protein light chain 3- and GABAA receptor-associated protein-phospholipid conjugates. J Biol Chem. 2004;279:36268–36276. doi: 10.1074/jbc.M401461200. [DOI] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Yamamoto A, Oshitani-Okamoto S, Ohsumi Y, Yoshimori T. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J Cell Sci. 2004;117:2805–2812. doi: 10.1242/jcs.01131. [DOI] [PubMed] [Google Scholar]
- Kochl R, Hu XW, Chan EY, Tooze SA. Microtubules facilitate autophagosome formation and fusion of autophagosomes with endosomes. Traffic. 2006;7:129–145. doi: 10.1111/j.1600-0854.2005.00368.x. [DOI] [PubMed] [Google Scholar]
- Tong J, Yan X, Yu L. The late stage of autophagy: cellular events and molecular regulation. Protein Cell. 2010;1:907–915. doi: 10.1007/s13238-010-0121-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahreiss L, Menzies FM, Rubinsztein DC. The itinerary of autophagosomes: from peripheral formation to kiss-and-run fusion with lysosomes. Traffic. 2008;9:574–587. doi: 10.1111/j.1600-0854.2008.00701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Huang J, Geng J, Nair U, Klionsky DJ. Atg22 recycles amino acids to link the degradative and recycling functions of autophagy. Mol Biol Cell. 2006;17:5094–5104. doi: 10.1091/mbc.E06-06-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axe EL, Walker SA, Manifava M, et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hailey DW, Rambold AS, Satpute-Krishnan P, et al. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 2010;141:656–667. doi: 10.1016/j.cell.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravikumar B, Moreau K, Jahreiss L, Puri C, Rubinsztein DC. Plasma membrane contributes to the formation of pre-autophagosomal structures. Nat cell biol. 2010;12:747–757. doi: 10.1038/ncb2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English L, Chemali M, Desjardins M. Nuclear membrane-derived autophagy, a novel process that participates in the presentation of endogenous viral antigens during HSV-1 infection. Autophagy. 2009;5:1026–1029. doi: 10.4161/auto.5.7.9163. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Chiba T, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- Nakai A, Yamaguchi O, Takeda T, et al. The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nat Med. 2007;13:619–624. doi: 10.1038/nm1574. [DOI] [PubMed] [Google Scholar]
- Hara T, Nakamura K, Matsui M, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Ueno T, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai Y, Soda M, Hatakeyama S, et al. CHIP is associated with Parkin, a gene responsible for familial Parkinson's disease, and enhances its ubiquitin ligase activity. Mol Cell. 2002;10:55–67. doi: 10.1016/s1097-2765(02)00583-x. [DOI] [PubMed] [Google Scholar]
- Goldberg AL. Protein degradation and protection against misfolded or damaged proteins. Nature. 2003;426:895–899. doi: 10.1038/nature02263. [DOI] [PubMed] [Google Scholar]
- Kirkin V, Lamark T, Sou YS, et al. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell. 2009;33:505–516. doi: 10.1016/j.molcel.2009.01.020. [DOI] [PubMed] [Google Scholar]
- Pankiv S, Clausen TH, Lamark T, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Koike M, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- Pandey UB, Nie Z, Batlevi Y, et al. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature. 2007;447:859–863. doi: 10.1038/nature05853. [DOI] [PubMed] [Google Scholar]
- Kawaguchi Y, Kovacs JJ, McLaurin A, Vance JM, Ito A, Yao TP. The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell. 2003;115:727–738. doi: 10.1016/s0092-8674(03)00939-5. [DOI] [PubMed] [Google Scholar]
- Wang K, Klionsky DJ. Mitochondria removal by autophagy. Autophagy. 2011;7:297–300. doi: 10.4161/auto.7.3.14502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissova I, Deffieu M, Manon S, Camougrand N. Uth1p is involved in the autophagic degradation of mitochondria. J Biol Chem. 2004;279:39068–39074. doi: 10.1074/jbc.M406960200. [DOI] [PubMed] [Google Scholar]
- Tal R, Winter G, Ecker N, Klionsky DJ, Abeliovich H. Aup1p, a yeast mitochondrial protein phosphatase homolog, is required for efficient stationary phase mitophagy and cell survival. J Biol Chem. 2007;282:5617–5624. doi: 10.1074/jbc.M605940200. [DOI] [PubMed] [Google Scholar]
- Schweers RL, Zhang J, Randall MS, et al. NIX is required for programmed mitochondrial clearance during reticulocyte maturation. Proc Natl Acad Sci USA. 2007;104:19500–19505. doi: 10.1073/pnas.0708818104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra DP, Youle RJ. Targeting Mitochondrial Dysfunction: Role for PINK1 and Parkin in Mitochondrial Quality Control. Antioxid Redox Signal. 2011;14:1929–1938. doi: 10.1089/ars.2010.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vives-Bauza C, Zhou C, Huang Y, et al. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc Natl Acad Sci USA. 2010;107:378–383. doi: 10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda N, Tanaka K. Uncovering the roles of PINK1 and parkin in mitophagy. Autophagy. 2010;6:952–954. doi: 10.4161/auto.6.7.13039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Park J, Kim S, et al. PINK1 controls mitochondrial localization of Parkin through direct phosphorylation. Biochem Biophys Res Commun. 2008;377:975–980. doi: 10.1016/j.bbrc.2008.10.104. [DOI] [PubMed] [Google Scholar]
- Shiba K, Arai T, Sato S, et al. Parkin stabilizes PINK1 through direct interaction. Biochem Biophys Res Commun. 2009;383:331–335. doi: 10.1016/j.bbrc.2009.04.006. [DOI] [PubMed] [Google Scholar]
- Michiorri S, Gelmetti V, Giarda E, et al. The Parkinson-associated protein PINK1 interacts with Beclin1 and promotes autophagy. Cell Death Differ. 2010;17:962–974. doi: 10.1038/cdd.2009.200. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Levine B. Autophagy in mammalian development and differentiation. Nat Cell Biol. 2010;12:823–830. doi: 10.1038/ncb0910-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukaiyama H, Nakase M, Nakamura T, Kakinuma Y, Takegawa K. Autophagy in the fission yeast Schizosaccharomyces pombe. FEBS Lett. 2010;584:1327–1334. doi: 10.1016/j.febslet.2009.12.037. [DOI] [PubMed] [Google Scholar]
- Otto GP, Wu MY, Kazgan N, Anderson OR, Kessin RH. Macroautophagy is required for multicellular development of the social amoeba Dictyostelium discoideum. J Biol Chem. 2003;278:17636–17645. doi: 10.1074/jbc.M212467200. [DOI] [PubMed] [Google Scholar]
- Melendez A, Talloczy Z, Seaman M, Eskelinen EL, Hall DH, Levine B. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science. 2003;301:1387–1391. doi: 10.1126/science.1087782. [DOI] [PubMed] [Google Scholar]
- Takacs-Vellai K, Vellai T, Puoti A, et al. Inactivation of the autophagy gene bec-1 triggers apoptotic cell death in C. elegans. Curr Biol. 2005;15:1513–1517. doi: 10.1016/j.cub.2005.07.035. [DOI] [PubMed] [Google Scholar]
- Lee CY, Cooksey BA, Baehrecke EH. Steroid regulation of midgut cell death during Drosophila development. Dev Biol. 2002;250:101–111. doi: 10.1006/dbio.2002.0784. [DOI] [PubMed] [Google Scholar]
- Berry DL, Baehrecke EH. Growth arrest and autophagy are required for salivary gland cell degradation in Drosophila. Cell. 2007;131:1137–1148. doi: 10.1016/j.cell.2007.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhasz G, Erdi B, Sass M, Neufeld TP. Atg7-dependent autophagy promotes neuronal health, stress tolerance, and longevity but is dispensable for metamorphosis in Drosophila. Genes Dev. 2007;21:3061–3066. doi: 10.1101/gad.1600707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W, Ganetzky B. Autophagy promotes synapse development in Drosophila. J Cell Biol. 2009;187:71–79. doi: 10.1083/jcb.200907109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto S, Kuma A, Murakami M, Kishi C, Yamamoto A, Mizushima N. Autophagy is essential for preimplantation development of mouse embryos. Science. 2008;321:117–120. doi: 10.1126/science.1154822. [DOI] [PubMed] [Google Scholar]
- Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci USA. 2003;100:15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu X, Yu J, Bhagat G, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112:1809–1820. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu X, Zou Z, Sun Q, et al. Autophagy gene-dependent clearance of apoptotic cells during embryonic development. Cell. 2007;128:931–946. doi: 10.1016/j.cell.2006.12.044. [DOI] [PubMed] [Google Scholar]
- Hara T, Takamura A, Kishi C, et al. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol. 2008;181:497–510. doi: 10.1083/jcb.200712064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fimia GM, Stoykova A, Romagnoli A, et al. Ambra1 regulates autophagy and development of the nervous system. Nature. 2007;447:1121–1125. doi: 10.1038/nature05925. [DOI] [PubMed] [Google Scholar]
- Marino G, Salvador-Montoliu N, Fueyo A, Knecht E, Mizushima N, Lopez-Otin C. Tissue-specific autophagy alterations and increased tumorigenesis in mice deficient in Atg4C/autophagin-3. J Biol Chem. 2007;282:18573–18583. doi: 10.1074/jbc.M701194200. [DOI] [PubMed] [Google Scholar]
- Sou YS, Waguri S, Iwata J, et al. The Atg8 conjugation system is indispensable for proper development of autophagic isolation membranes in mice. Mol Biol Cell. 2008;19:4762–4775. doi: 10.1091/mbc.E08-03-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh T, Fujita N, Hayashi T, et al. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc Natl Acad Sci USA. 2009;106:20842–20846. doi: 10.1073/pnas.0911267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh T, Fujita N, Jang MH, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature. 2008;456:264–268. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- Kundu M, Lindsten T, Yang CY, et al. Ulk1 plays a critical role in the autophagic clearance of mitochondria and ribosomes during reticulocyte maturation. Blood. 2008;112:1493–1502. doi: 10.1182/blood-2008-02-137398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuma A, Hatano M, Matsui M, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- Mortensen M, Simon AK. Nonredundant role of Atg7 in mitochondrial clearance during erythroid development. Autophagy. 2010;6:423–425. doi: 10.4161/auto.6.3.11528. [DOI] [PubMed] [Google Scholar]
- Pua HH, Guo J, Komatsu M, He YW. Autophagy is essential for mitochondrial clearance in mature T lymphocytes. J Immunol. 2009;182:4046–4055. doi: 10.4049/jimmunol.0801143. [DOI] [PubMed] [Google Scholar]
- Baerga R, Zhang Y, Chen PH, Goldman S, Jin S. Targeted deletion of autophagy-related 5 (atg5) impairs adipogenesis in a cellular model and in mice. Autophagy. 2009;5:1118–1130. doi: 10.4161/auto.5.8.9991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman SJ, Zhang Y, Jin S. Autophagic degradation of mitochondria in white adipose tissue differentiation. Antioxid Redox Signal. 2011;14:1971–1978. doi: 10.1089/ars.2010.3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Goldman S, Baerga R, Zhao Y, Komatsu M, Jin S. Adipose-specific deletion of autophagy-related gene 7 (atg7) in mice reveals a role in adipogenesis. Proc Natl Acad Sci USA. 2009;106:19860–19865. doi: 10.1073/pnas.0906048106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Huang Q, Yang J, et al. Autophagy impairment inhibits differentiation of glioma stem/progenitor cells. Brain Res. 2010;1313:250–258. doi: 10.1016/j.brainres.2009.12.004. [DOI] [PubMed] [Google Scholar]
- Zeng M, Zhou JN. Roles of autophagy and mTOR signaling in neuronal differentiation of mouse neuroblastoma cells. Cell Signal. 2008;20:659–665. doi: 10.1016/j.cellsig.2007.11.015. [DOI] [PubMed] [Google Scholar]
- Deretic V, Levine B. Autophagy, immunity, and microbial adaptations. Cell Host Microbe. 2009;5:527–549. doi: 10.1016/j.chom.2009.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa I, Amano A, Mizushima N, et al. Autophagy defends cells against invading group A Streptococcus. Science. 2004;306:1037–1040. doi: 10.1126/science.1103966. [DOI] [PubMed] [Google Scholar]
- Ogawa M, Yoshimori T, Suzuki T, Sagara H, Mizushima N, Sasakawa C. Escape of intracellular Shigella from autophagy. Science. 2005;307:727–731. doi: 10.1126/science.1106036. [DOI] [PubMed] [Google Scholar]
- Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119:753–766. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- Birmingham CL, Smith AC, Bakowski MA, Yoshimori T, Brumell JH. Autophagy controls Salmonella infection in response to damage to the Salmonella-containing vacuole. J Biol Chem. 2006;281:11374–11383. doi: 10.1074/jbc.M509157200. [DOI] [PubMed] [Google Scholar]
- Yano T, Mita S, Ohmori H, et al. Autophagic control of listeria through intracellular innate immune recognition in drosophila. Nat Immunol. 2008;9:908–916. doi: 10.1038/ni.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade RM, Wessendarp M, Gubbels MJ, Striepen B, Subauste CS. CD40 induces macrophage anti-Toxoplasma gondii activity by triggering autophagy-dependent fusion of pathogen-containing vacuoles and lysosomes. J Clin Invest. 2006;116:2366–2377. doi: 10.1172/JCI28796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talloczy Z, Virgin HWt, Levine B. PKR-dependent autophagic degradation of herpes simplex virus type 1. Autophagy. 2006;2:24–29. doi: 10.4161/auto.2176. [DOI] [PubMed] [Google Scholar]
- Liang XH, Kleeman LK, Jiang HH, et al. Protection against fatal Sindbis virus encephalitis by beclin, a novel Bcl-2-interacting protein. J Virol. 1998;72:8586–8596. doi: 10.1128/jvi.72.11.8586-8596.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Czymmek K, Talloczy Z, Levine B, Dinesh-Kumar SP. Autophagy regulates programmed cell death during the plant innate immune response. Cell. 2005;121:567–577. doi: 10.1016/j.cell.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Thurston TL, Ryzhakov G, Bloor S, von Muhlinen N, Randow F. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat Immunol. 2009;10:1215–1221. doi: 10.1038/ni.1800. [DOI] [PubMed] [Google Scholar]
- Zheng YT, Shahnazari S, Brech A, Lamark T, Johansen T, Brumell JH. The adaptor protein p62/SQSTM1 targets invading bacteria to the autophagy pathway. J Immunol. 2009;183:5909–5916. doi: 10.4049/jimmunol.0900441. [DOI] [PubMed] [Google Scholar]
- Yu HB, Kielczewska A, Rozek A, et al. Sequestosome-1/p62 is the key intracellular target of innate defense regulator peptide. J Biol Chem. 2009;284:36007–36011. doi: 10.1074/jbc.C109.073627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orvedahl A, Alexander D, Talloczy Z, et al. HSV-1 ICP34.5 Confers Neurovirulence by Targeting the Beclin 1 Autophagy Protein. Cell Host Microbe. 2007;1:23–35. doi: 10.1016/j.chom.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Kyei GB, Dinkins C, Davis AS, et al. Autophagy pathway intersects with HIV-1 biosynthesis and regulates viral yields in macrophages. J Cell Biol. 2009;186:255–268. doi: 10.1083/jcb.200903070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattingre S, Tassa A, Qu X, et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Sinha S, Colbert CL, Becker N, Wei Y, Levine B. Molecular basis of the regulation of Beclin 1-dependent autophagy by the gamma-herpesvirus 68 Bcl-2 homolog M11. Autophagy. 2008;4:989–997. doi: 10.4161/auto.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaumorcel M, Souquere S, Pierron G, Codogno P, Esclatine A. Human cytomegalovirus controls a new autophagy-dependent cellular antiviral defense mechanism. Autophagy. 2008;4:46–53. doi: 10.4161/auto.5184. [DOI] [PubMed] [Google Scholar]
- Shi CS, Kehrl JH. MyD88 and Trif target Beclin 1 to trigger autophagy in macrophages. J Biol Chem. 2008;283:33175–33182. doi: 10.1074/jbc.M804478200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi CS, Kehrl JH. TRAF6 and A20 regulate lysine 63-linked ubiquitination of Beclin-1 to control TLR4-induced autophagy. Sci Signal. 2010;3:ra42. doi: 10.1126/scisignal.2000751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travassos LH, Carneiro LA, Ramjeet M, et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat Immunol. 2010;11:55–62. doi: 10.1038/ni.1823. [DOI] [PubMed] [Google Scholar]
- Harris J, De Haro SA, Master SS, et al. T helper 2 cytokines inhibit autophagic control of intracellular Mycobacterium tuberculosis. Immunity. 2007;27:505–517. doi: 10.1016/j.immuni.2007.07.022. [DOI] [PubMed] [Google Scholar]
- Tal MC, Sasai M, Lee HK, Yordy B, Shadel GS, Iwasaki A. Absence of autophagy results in reactive oxygen species-dependent amplification of RLR signaling. Proc Natl Acad Sci USA. 2009;106:2770–2775. doi: 10.1073/pnas.0807694106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahira K, Haspel JA, Rathinam VA, et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat Immunol. 2011;12:222–30. doi: 10.1038/ni.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Yazdi AS, Menu P, Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469:221–225. doi: 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]
- Harris J, Hartman M, Roche C, et al. Autophagy controls IL-1beta secretion by targeting pro-IL-1beta for degradation. J Biol Chem. 2011;286:9587–9597. doi: 10.1074/jbc.M110.202911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller BC, Zhao Z, Stephenson LM, et al. The autophagy gene ATG5 plays an essential role in B lymphocyte development. Autophagy. 2008;4:309–314. doi: 10.4161/auto.5474. [DOI] [PubMed] [Google Scholar]
- Pua HH, Dzhagalov I, Chuck M, Mizushima N, He YW. A critical role for the autophagy gene Atg5 in T cell survival and proliferation. J Exp Med. 2007;204:25–31. doi: 10.1084/jem.20061303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell BD, Leverrier S, Weist BM, et al. FADD and caspase-8 control the outcome of autophagic signaling in proliferating T cells. Proc Natl Acad Sci USA. 2008;105:16677–16682. doi: 10.1073/pnas.0808597105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadwell K, Liu JY, Brown SL, et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259–263. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotzer VL, Blum JS. Autophagy and adaptive immunity. Immunology. 2010;131:9–17. doi: 10.1111/j.1365-2567.2010.03321.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English L, Chemali M, Duron J, et al. Autophagy enhances the presentation of endogenous viral antigens on MHC class I molecules during HSV-1 infection. Nat Immunol. 2009;10:480–487. doi: 10.1038/ni.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HK, Mattei LM, Steinberg BE, et al. In vivo requirement for Atg5 in antigen presentation by dendritic cells. Immunity. 2010;32:227–239. doi: 10.1016/j.immuni.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhl M, Kepp O, Jusforgues-Saklani H, Vicencio JM, Kroemer G, Albert ML. Autophagy within the antigen donor cell facilitates efficient antigen cross-priming of virus-specific CD8+ T cells. Cell Death Differ. 2009;16:991–1005. doi: 10.1038/cdd.2009.8. [DOI] [PubMed] [Google Scholar]
- Lee HK, Lund JM, Ramanathan B, Mizushima N, Iwasaki A. Autophagy-dependent viral recognition by plasmacytoid dendritic cells. Science. 2007;315:1398–1401. doi: 10.1126/science.1136880. [DOI] [PubMed] [Google Scholar]
- Nedjic J, Aichinger M, Emmerich J, Mizushima N, Klein L. Autophagy in thymic epithelium shapes the T-cell repertoire and is essential for tolerance. Nature. 2008;455:396–400. doi: 10.1038/nature07208. [DOI] [PubMed] [Google Scholar]
- Duprez L, Wirawan E, Vanden Berghe T, Vandenabeele P. Major cell death pathways at a glance. Microbes Infect. 2009;11:1050–1062. doi: 10.1016/j.micinf.2009.08.013. [DOI] [PubMed] [Google Scholar]
- Kroemer G, Galluzzi L, Vandenabeele P, et al. Classification of cell death: recommendations of the Nomenclature Committee on Cell Death 2009. Cell Death Differ. 2009;16:3–11. doi: 10.1038/cdd.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuma A, Mizushima N. Physiological role of autophagy as an intracellular recycling system: with an emphasis on nutrient metabolism. Semin Cell Dev Biol. 2010;21:683–690. doi: 10.1016/j.semcdb.2010.03.002. [DOI] [PubMed] [Google Scholar]
- Fung C, Lock R, Gao S, Salas E, Debnath J. Induction of autophagy during extracellular matrix detachment promotes cell survival. Mol Biol Cell. 2008;19:797–806. doi: 10.1091/mbc.E07-10-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum JJ, Bauer DE, Kong M, et al. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell. 2005;120:237–248. doi: 10.1016/j.cell.2004.11.046. [DOI] [PubMed] [Google Scholar]
- Martin DN, Baehrecke EH. Caspases function in autophagic programmed cell death in Drosophila. Development. 2004;131:275–284. doi: 10.1242/dev.00933. [DOI] [PubMed] [Google Scholar]
- Espert L, Denizot M, Grimaldi M, et al. Autophagy is involved in T cell death after binding of HIV-1 envelope proteins to CXCR4. J Clin Invest. 2006;116:2161–2172. doi: 10.1172/JCI26185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crighton D, Wilkinson S, O'Prey J, et al. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell. 2006;126:121–134. doi: 10.1016/j.cell.2006.05.034. [DOI] [PubMed] [Google Scholar]
- Nezis IP, Shravage BV, Sagona AP, et al. Autophagic degradation of dBruce controls DNA fragmentation in nurse cells during late Drosophila melanogaster oogenesis. J Cell Biol. 2010;190:523–531. doi: 10.1083/jcb.201002035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyo JO, Jang MH, Kwon YK, et al. Essential roles of Atg5 and FADD in autophagic cell death: dissection of autophagic cell death into vacuole formation and cell death. J Biol Chem. 2005;280:20722–20729. doi: 10.1074/jbc.M413934200. [DOI] [PubMed] [Google Scholar]
- Walls KC, Ghosh AP, Franklin AV, et al. Lysosome dysfunction triggers Atg7-dependent neural apoptosis. J Biol Chem. 2010;285:10497–10507. doi: 10.1074/jbc.M110.103747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidari N, Hicks MA, Harada H. GX015-070 (obatoclax) overcomes glucocorticoid resistance in acute lymphoblastic leukemia through induction of apoptosis and autophagy. Cell Death Dis. 2010;1:e76. doi: 10.1038/cddis.2010.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radoshevich L, Murrow L, Chen N, et al. ATG12 conjugation to ATG3 regulates mitochondrial homeostasis and cell death. Cell. 2010;142:590–600. doi: 10.1016/j.cell.2010.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousefi S, Perozzo R, Schmid I, et al. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat Cell Biol. 2006;8:1124–1132. doi: 10.1038/ncb1482. [DOI] [PubMed] [Google Scholar]
- Xia HG, Zhang L, Chen G, et al. Control of basal autophagy by calpain1 mediated cleavage of ATG5. Autophagy. 2010;6:61–66. doi: 10.4161/auto.6.1.10326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirawan E, Vandewalle L, Kersse K, et al. Caspase-mediated cleavage of Beclin-1 inactivates Beclin-1-induced autophagy and enhances apoptosis by promoting the release of proapoptotic factors from mitochondria. Cell Death Dis. 2010;1:e18. doi: 10.1038/cddis.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betin VM, Lane JD. Caspase cleavage of Atg4D stimulates GABARAP-L1 processing and triggers mitochondrial targeting and apoptosis. J Cell Sci. 2009;122:2554–2566. doi: 10.1242/jcs.046250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Sinha S, Kroemer G. Bcl-2 family members: dual regulators of apoptosis and autophagy. Autophagy. 2008;4:600–606. doi: 10.4161/auto.6260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberstein A, Jeffrey PD, Shi Y. Crystal structure of the Bcl-XL-Beclin 1 peptide complex: Beclin 1 is a novel BH3-only protein. J Biol Chem. 2007;282:13123–13132. doi: 10.1074/jbc.M700492200. [DOI] [PubMed] [Google Scholar]
- Ciechomska IA, Goemans GC, Skepper JN, Tolkovsky AM. Bcl-2 complexed with Beclin-1 maintains full anti-apoptotic function. Oncogene. 2009;28:2128–2141. doi: 10.1038/onc.2009.60. [DOI] [PubMed] [Google Scholar]
- Strappazzon F, Vietri-Rudan M, Campello S, et al. Mitochondrial BCL-2 inhibits AMBRA1-induced autophagy. EMBO J. 2011;30:1195–1208. doi: 10.1038/emboj.2011.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Li Q, Lee JY, et al. FLIP-mediated autophagy regulation in cell death control. Nat Cell Biol. 2009;11:1355–1362. doi: 10.1038/ncb1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalckvar E, Berissi H, Eisenstein M, Kimchi A. Phosphorylation of Beclin 1 by DAP-kinase promotes autophagy by weakening its interactions with Bcl-2 and Bcl-XL. Autophagy. 2009;5:720–722. doi: 10.4161/auto.5.5.8625. [DOI] [PubMed] [Google Scholar]
- Wei Y, Pattingre S, Sinha S, Bassik M, Levine B. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol Cell. 2008;30:678–688. doi: 10.1016/j.molcel.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong HS, Choi HY, Lee ER, et al. Involvement of caspase-9 in autophagy-mediated cell survival pathway. Biochim Biophys Acta. 2011;1813:80–90. doi: 10.1016/j.bbamcr.2010.09.016. [DOI] [PubMed] [Google Scholar]
- Tasdemir E, Chiara Maiuri M, Morselli E, et al. A dual role of p53 in the control of autophagy. Autophagy. 2008;4:810–814. doi: 10.4161/auto.6486. [DOI] [PubMed] [Google Scholar]
- Tasdemir E, Maiuri MC, Galluzzi L, et al. Regulation of autophagy by cytoplasmic p53. Nat cell biol. 2008;10:676–687. doi: 10.1038/ncb1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z, Zhang H, Levine AJ, Jin S. The coordinate regulation of the p53 and mTOR pathways in cells. Proc Natl Acad Sci USA. 2005;102:8204–8209. doi: 10.1073/pnas.0502857102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z, Li Y, Pitti R, et al. Cullin3-based polyubiquitination and p62-dependent aggregation of caspase-8 mediate extrinsic apoptosis signaling. Cell. 2009;137:721–735. doi: 10.1016/j.cell.2009.03.015. [DOI] [PubMed] [Google Scholar]
- Degenhardt K, Mathew R, Beaudoin B, et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 2006;10:51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Franchi L, Toma C, et al. Differential regulation of caspase-1 activation, pyroptosis, and autophagy via Ipaf and ASC in Shigella-infected macrophages. PLoS Pathog. 2007;3:e111. doi: 10.1371/journal.ppat.0030111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samara C, Syntichaki P, Tavernarakis N. Autophagy is required for necrotic cell death in Caenorhabditis elegans. Cell Death Differ. 2008;15:105–112. doi: 10.1038/sj.cdd.4402231. [DOI] [PubMed] [Google Scholar]
- Castino R, Bellio N, Follo C, Murphy D, Isidoro C. Inhibition of PI3k class III-dependent autophagy prevents apoptosis and necrosis by oxidative stress in dopaminergic neuroblastoma cells. Toxicol Sci. 2010;117:152–162. doi: 10.1093/toxsci/kfq170. [DOI] [PubMed] [Google Scholar]
- Yu L, Alva A, Su H, et al. Regulation of an ATG7-beclin 1 program of autophagic cell death by caspase-8. Science. 2004;304:1500–1502. doi: 10.1126/science.1096645. [DOI] [PubMed] [Google Scholar]
- Yu L, Wan F, Dutta S, et al. Autophagic programmed cell death by selective catalase degradation. Proc Natl Acad Sci USA. 2006;103:4952–4957. doi: 10.1073/pnas.0511288103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu S, Kanaseki T, Mizushima N, et al. Role of Bcl-2 family proteins in a non-apoptotic programmed cell death dependent on autophagy genes. Nat Cell Biol. 2004;6:1221–1228. doi: 10.1038/ncb1192. [DOI] [PubMed] [Google Scholar]
- Ullman E, Fan Y, Stawowczyk M, Chen HM, Yue Z, Zong WX. Autophagy promotes necrosis in apoptosis-deficient cells in response to ER stress. Cell Death Differ. 2008;15:422–425. doi: 10.1038/sj.cdd.4402234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buytaert E, Callewaert G, Vandenheede JR, Agostinis P. Deficiency in apoptotic effectors Bax and Bak reveals an autophagic cell death pathway initiated by photodamage to the endoplasmic reticulum. Autophagy. 2006;2:238–240. doi: 10.4161/auto.2730. [DOI] [PubMed] [Google Scholar]
- Bonapace L, Bornhauser BC, Schmitz M, et al. Induction of autophagy-dependent necroptosis is required for childhood acute lymphoblastic leukemia cells to overcome glucocorticoid resistance. J Clin Invest. 2010;120:1310–1323. doi: 10.1172/JCI39987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz L, Sanchez P, Lallena MJ, Diaz-Meco MT, Moscat J. The interaction of p62 with RIP links the atypical PKCs to NF-kappaB activation. EMBO J. 1999;18:3044–3053. doi: 10.1093/emboj/18.11.3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remijsen Q, Berghe TV, Wirawan E, et al. Neutrophil extracellular trap cell death requires both autophagy and superoxide generation. Cell Res. 2011;21:290–304. doi: 10.1038/cr.2010.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeo F, Tavernarakis N, Kroemer G. Can autophagy promote longevity. Nat Cell Biol. 2010;12:842–846. doi: 10.1038/ncb0910-842. [DOI] [PubMed] [Google Scholar]
- Jia K, Hart AC, Levine B. Autophagy genes protect against disease caused by polyglutamine expansion proteins in Caenorhabditis elegans. Autophagy. 2007;3:21–25. doi: 10.4161/auto.3528. [DOI] [PubMed] [Google Scholar]
- Mair W, Goymer P, Pletcher SD, Partridge L. Demography of dietary restriction and death in Drosophila. Science. 2003;301:1731–1733. doi: 10.1126/science.1086016. [DOI] [PubMed] [Google Scholar]
- Alvers AL, Wood MS, Hu D, Kaywell AC, Dunn WA, Jr, Aris JP. Autophagy is required for extension of yeast chronological life span by rapamycin. Autophagy. 2009;5:847–849. doi: 10.4161/auto.8824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison DE, Strong R, Sharp ZD, et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–395. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morselli E, Galluzzi L, Kepp O, et al. Autophagy mediates pharmacological lifespan extension by spermidine and resveratrol. Aging. 2009;1:961–970. doi: 10.18632/aging.100110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood JG, Rogina B, Lavu S, et al. Sirtuin activators mimic caloric restriction and delay ageing in metazoans. Nature. 2004;430:686–689. doi: 10.1038/nature02789. [DOI] [PubMed] [Google Scholar]
- Howitz KT, Bitterman KJ, Cohen HY, et al. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature. 2003;425:191–196. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- Lee IH, Cao L, Mostoslavsky R, et al. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc Natl Acad Sci USA. 2008;105:3374–3379. doi: 10.1073/pnas.0712145105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morselli E, Maiuri MC, Markaki M, et al. The life span-prolonging effect of sirtuin-1 is mediated by autophagy. Autophagy. 2010;6:186–188. doi: 10.4161/auto.6.1.10817. [DOI] [PubMed] [Google Scholar]
- Eisenberg T, Knauer H, Schauer A, et al. Induction of autophagy by spermidine promotes longevity. Nat Cell Biol. 2009;11:1305–1314. doi: 10.1038/ncb1975. [DOI] [PubMed] [Google Scholar]
- Park JW, Woo KJ, Lee JT, et al. Resveratrol induces pro-apoptotic endoplasmic reticulum stress in human colon cancer cells. Oncol Rep. 2007;18:1269–1273. [PubMed] [Google Scholar]
- Terman A. The effect of age on formation and elimination of autophagic vacuoles in mouse hepatocytes. Gerontology. 1995;41 Suppl 2:319–326. doi: 10.1159/000213753. [DOI] [PubMed] [Google Scholar]
- Cuervo AM, Bergamini E, Brunk UT, Droge W, Ffrench M, Terman A. Autophagy and aging: the importance of maintaining “clean” cells. Autophagy. 2005;1:131–140. doi: 10.4161/auto.1.3.2017. [DOI] [PubMed] [Google Scholar]
- Mathew R, Karantza-Wadsworth V, White E. Role of autophagy in cancer. Nat Rev Cancer. 2007;7:961–967. doi: 10.1038/nrc2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalby KN, Tekedereli I, Lopez-Berestein G, Ozpolat B. Targeting the prodeath and prosurvival functions of autophagy as novel therapeutic strategies in cancer. Autophagy. 2010;6:322–329. doi: 10.4161/auto.6.3.11625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maycotte P, Thorburn A. Autophagy and cancer therapy. Cancer Biol Ther. 2011;11:127–137. doi: 10.4161/cbt.11.2.14627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaravadi RK, Lippincott-Schwartz J, Yin XM, et al. Principles and current strategies for targeting autophagy for cancer treatment. Clin Cancer Res. 2011;17:654–666. doi: 10.1158/1078-0432.CCR-10-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White E, DiPaola RS. The double-edged sword of autophagy modulation in cancer. Clin Cancer Res. 2009;15:5308–5316. doi: 10.1158/1078-0432.CCR-07-5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boya P, Gonzalez-Polo RA, Casares N, et al. Inhibition of macroautophagy triggers apoptosis. Mol Cell Biol. 2005;25:1025–1040. doi: 10.1128/MCB.25.3.1025-1040.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JY, Chen HY, Mathew R, et al. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 2011;25:460–470. doi: 10.1101/gad.2016311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Wang X, Contino G, et al. Pancreatic cancers require autophagy for tumor growth. Genes Dev. 2011;25:717–729. doi: 10.1101/gad.2016111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewaele M, Maes H, Agostinis P. ROS-mediated mechanisms of autophagy stimulation and their relevance in cancer therapy. Autophagy. 2010;6:838–854. doi: 10.4161/auto.6.7.12113. [DOI] [PubMed] [Google Scholar]
- Amaravadi RK, Yu D, Lum JJ, et al. Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. J Clin Invest. 2007;117:326–336. doi: 10.1172/JCI28833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Hou N, Faried A, Tsutsumi S, Kuwano H. Inhibition of autophagy augments 5-fluorouracil chemotherapy in human colon cancer in vitro and in vivo model. Eur J Cancer. 2010;46:1900–1909. doi: 10.1016/j.ejca.2010.02.021. [DOI] [PubMed] [Google Scholar]
- Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang XH, Jackson S, Seaman M, et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- Liang C, Feng P, Ku B, et al. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat Cell Biol. 2006;8:688–699. doi: 10.1038/ncb1426. [DOI] [PubMed] [Google Scholar]
- Aita VM, Liang XH, Murty VV, et al. Cloning and genomic organization of beclin 1, a candidate tumor suppressor gene on chromosome 17q21. Genomics. 1999;59:59–65. doi: 10.1006/geno.1999.5851. [DOI] [PubMed] [Google Scholar]
- Karantza-Wadsworth V, Patel S, Kravchuk O, et al. Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis. Genes Dev. 2007;21:1621–1635. doi: 10.1101/gad.1565707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew R, Kongara S, Beaudoin B, et al. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev. 2007;21:1367–1381. doi: 10.1101/gad.1545107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koneri K, Goi T, Hirono Y, Katayama K, Yamaguchi A. Beclin 1 gene inhibits tumor growth in colon cancer cell lines. Anticancer Res. 2007;27:1453–1457. [PubMed] [Google Scholar]
- Yoo BH, Wu X, Li Y, et al. Oncogenic ras-induced down-regulation of autophagy mediator Beclin-1 is required for malignant transformation of intestinal epithelial cells. J Biol Chem. 2010;285:5438–5449. doi: 10.1074/jbc.M109.046789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akar U, Ozpolat B, Mehta K, Fok J, Kondo Y, Lopez-Berestein G. Tissue transglutaminase inhibits autophagy in pancreatic cancer cells. Mol Cancer Res. 2007;5:241–249. doi: 10.1158/1541-7786.MCR-06-0229. [DOI] [PubMed] [Google Scholar]
- Akar U, Chaves-Reyez A, Barria M, et al. Silencing of Bcl-2 expression by small interfering RNA induces autophagic cell death in MCF-7 breast cancer cells. Autophagy. 2008;4:669–679. doi: 10.4161/auto.6083. [DOI] [PubMed] [Google Scholar]
- Williams A, Jahreiss L, Sarkar S, et al. Aggregate-prone proteins are cleared from the cytosol by autophagy: therapeutic implications. Curr Top Dev Biol. 2006;76:89–101. doi: 10.1016/S0070-2153(06)76003-3. [DOI] [PubMed] [Google Scholar]
- Knaevelsrud H, Simonsen A. Fighting disease by selective autophagy of aggregate-prone proteins. FEBS Lett. 2010;584:2635–2645. doi: 10.1016/j.febslet.2010.04.041. [DOI] [PubMed] [Google Scholar]
- Spencer B, Potkar R, Trejo M, et al. Beclin 1 gene transfer activates autophagy and ameliorates the neurodegenerative pathology in alpha-synuclein models of Parkinson's and Lewy body diseases. J Neurosci. 2009;29:13578–13588. doi: 10.1523/JNEUROSCI.4390-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickford F, Masliah E, Britschgi M, et al. The autophagy-related protein beclin 1 shows reduced expression in early Alzheimer disease and regulates amyloid beta accumulation in mice. J Clin Invest. 2008;118:2190–2199. doi: 10.1172/JCI33585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger PA, Pickford F, Sun CH, Lucin KM, Masliah E, Wyss-Coray T. Regulation of amyloid precursor protein processing by the Beclin 1 complex. PLoS One. 2010;5:e11102. doi: 10.1371/journal.pone.0011102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roze E, Bonnet C, Betuing S, Caboche J. Huntington's disease. Adv Exp Med Biol. 2010;685:45–63. [PubMed] [Google Scholar]
- Aguado C, Sarkar S, Korolchuk VI, et al. Laforin, the most common protein mutated in Lafora disease, regulates autophagy. Hum Mol Genet. 2010;19:2867–2876. doi: 10.1093/hmg/ddq190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata M, Lu T, Furuya T, et al. Regulation of intracellular accumulation of mutant Huntingtin by Beclin 1. J Biol Chem. 2006;281:14474–14485. doi: 10.1074/jbc.M600364200. [DOI] [PubMed] [Google Scholar]
- Rohn TT, Wirawan E, Brown RJ, Harris JR, Masliah E, Vandenabeele P. Depletion of Beclin-1 due to proteolytic cleavage by caspases in the Alzheimer's disease brain. Neurobiol Dis. 2011;43:68–78. doi: 10.1016/j.nbd.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luciani A, Villella VR, Esposito S, et al. Defective CFTR induces aggresome formation and lung inflammation in cystic fibrosis through ROS-mediated autophagy inhibition. Nat Cell Biol. 2010;12:863–875. doi: 10.1038/ncb2090. [DOI] [PubMed] [Google Scholar]
- Grumati P, Coletto L, Sabatelli P, et al. Autophagy is defective in collagen VI muscular dystrophies, and its reactivation rescues myofiber degeneration. Nat Med. 2010;16:1313–1320. doi: 10.1038/nm.2247. [DOI] [PubMed] [Google Scholar]
- Malicdan MC, Noguchi S, Nonaka I, Saftig P, Nishino I. Lysosomal myopathies: an excessive build-up in autophagosomes is too much to handle. Neuromuscul Disord. 2008;18:521–529. doi: 10.1016/j.nmd.2008.04.010. [DOI] [PubMed] [Google Scholar]
- Nishida K, Kyoi S, Yamaguchi O, Sadoshima J, Otsu K. The role of autophagy in the heart. Cell Death Differ. 2009;16:31–38. doi: 10.1038/cdd.2008.163. [DOI] [PubMed] [Google Scholar]
- Nishino I, Fu J, Tanji K, et al. Primary LAMP-2 deficiency causes X-linked vacuolar cardiomyopathy and myopathy (Danon disease) Nature. 2000;406:906–910. doi: 10.1038/35022604. [DOI] [PubMed] [Google Scholar]
- Valentim L, Laurence KM, Townsend PA, et al. Urocortin inhibits Beclin1-mediated autophagic cell death in cardiac myocytes exposed to ischaemia/reperfusion injury. J Mol Cell Cardiol. 2006;40:846–852. doi: 10.1016/j.yjmcc.2006.03.428. [DOI] [PubMed] [Google Scholar]
- Matsui Y, Takagi H, Qu X, et al. Distinct roles of autophagy in the heart during ischemia and reperfusion: roles of AMP-activated protein kinase and Beclin 1 in mediating autophagy. Circ Res. 2007;100:914–922. doi: 10.1161/01.RES.0000261924.76669.36. [DOI] [PubMed] [Google Scholar]
- Marciniak SJ, Lomas DA. Alpha1-antitrypsin deficiency and autophagy. N Engl J Med. 2010;363:1863–1864. doi: 10.1056/NEJMcibr1008007. [DOI] [PubMed] [Google Scholar]
- Hidvegi T, Ewing M, Hale P, et al. An autophagy-enhancing drug promotes degradation of mutant alpha1-antitrypsin Z and reduces hepatic fibrosis. Science. 2010;329:229–232. doi: 10.1126/science.1190354. [DOI] [PubMed] [Google Scholar]
- Jung HS, Lee MS. Role of autophagy in diabetes and mitochondria. Ann N Y Acad Sci. 2010;1201:79–83. doi: 10.1111/j.1749-6632.2010.05614.x. [DOI] [PubMed] [Google Scholar]
- Kang MR, Kim MS, Oh JE, et al. Frameshift mutations of autophagy-related genes ATG2B, ATG5, ATG9B and ATG12 in gastric and colorectal cancers with microsatellite instability. J Pathol. 2009;217:702–706. doi: 10.1002/path.2509. [DOI] [PubMed] [Google Scholar]
- Li Z, Chen B, Wu Y, Jin F, Xia Y, Liu X. Genetic and epigenetic silencing of the beclin 1 gene in sporadic breast tumors. BMC Cancer. 2010;10:98. doi: 10.1186/1471-2407-10-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JW, Jeong EG, Lee SH, Yoo NJ. Somatic mutations of BECN1, an autophagy-related gene, in human cancers. APMIS. 2007;115:750–756. doi: 10.1111/j.1600-0463.2007.apm_640.x. [DOI] [PubMed] [Google Scholar]
- Chano T, Kontani K, Teramoto K, Okabe H, Ikegawa S. Truncating mutations of RB1CC1 in human breast cancer. Nat Genet. 2002;31:285–288. doi: 10.1038/ng911. [DOI] [PubMed] [Google Scholar]
- Knaevelsrud H, Ahlquist T, Merok MA, et al. UVRAG mutations associated with microsatellite unstable colon cancer do not affect autophagy. Autophagy. 2010;6:863–870. doi: 10.4161/auto.6.7.13033. [DOI] [PubMed] [Google Scholar]
- Ionov Y, Nowak N, Perucho M, Markowitz S, Cowell JK. Manipulation of nonsense mediated decay identifies gene mutations in colon cancer cells with microsatellite instability. Oncogene. 2004;23:639–645. doi: 10.1038/sj.onc.1207178. [DOI] [PubMed] [Google Scholar]
- Kim MS, Jeong EG, Ahn CH, Kim SS, Lee SH, Yoo NJ. Frameshift mutation of UVRAG, an autophagy-related gene, in gastric carcinomas with microsatellite instability. Hum Pathol. 2008;39:1059–1063. doi: 10.1016/j.humpath.2007.11.013. [DOI] [PubMed] [Google Scholar]