The single nucleotide polymorphism (SNP) rs1344706 within the second intron of ZNF804A shows robust association with schizophrenia (1-5), but does not have an obvious effect on protein coding sequence. While this could reflect linkage disequilibrium with the actual causal variant(s), re-sequencing and detailed association analyses of ZNF804A in the largest sample to date have not uncovered a variant that is more strongly associated (2). Given its intronic location, any functional effects of this SNP are likely to be at the level of transcription or splicing, a hypothesis supported by reports of association between rs13344706 and ZNF804A RNA expression (2, 3). However, additional tests of ZNF804A allelic expression carried out in one of these studies (2) suggest that the association with ZNF804A expression may not be directly attributable to this SNP, or that other variants are also involved.
As gene expression is fundamentally controlled by binding of nuclear proteins to regulatory regions in genomic DNA, we investigated the functional potential of rs13344706 by assessing its binding to nuclear proteins derived from human neural cell lines using electromobility shift assays (EMSA). Fifty base complementary 5′ biotinylated DNA oligonucleotides were synthesised along with unlabelled competitor oligonucleotides for each allele of rs1344706 (sequences in Supplementary Table S1). Equimolar amounts of complementary oligonucleotides were annealed in a thermocycler to form double-stranded DNA. Nuclear extracts were prepared from undifferentiated SH-SY5Y cells and two human neural progenitor cell lines, one derived from foetal cortical neuroepithelium and one from foetal hippocampus (described in references 6 and 7, respectively). Nuclear extract binding reactions were carried out using the LightShift EMSA Optimisation and Control Kit (Thermo Scientific). Binding specificity was assessed by addition of a 200-fold excess (4 pmols) of unlabelled oligonucleotide. Electrophoresis was carried out using 6% Novex® DNA retardation gels (Invitrogen). Gels were transferred to positively charged nylon membranes and cross-linked using a UV transilluminator. Membranes were visualised using the LightShift Chemiluminescent EMSA kit (Thermo Scientific) and image acquisition and analysis performed using a ChemiDoc-It camera system (UVP). DNA-protein bands were quantified as percentage intensity of total lane intensity. For comparison of binding between sequences containing alternative alleles of rs1344706, 2 separate incubation reactions were performed for each allele using the same volume (3μl) of the same nuclear extract for all 4 reactions, which were then assayed on a single gel. This was repeated on a further 2 occasions, using separate nuclear extracts, to yield 6 measures of binding intensity for each allele.
Incubation of SH-SY5Y nuclear extracts with double-stranded DNA containing either allele of rs1344706 gave rise to a prominent band in addition to the free DNA probe, indicative of a DNA-protein interaction (Figure 1a). The band was abolished by excess of the specific unlabelled oligonucleotide competitor containing the same allele of rs1344706, indicating that the impeded electrophoretic mobility of the labelled DNA was due to formation of a DNA-protein complex specific to these sequences. This band was also observed using nuclear extracts from the two human neural progenitor cell lines (Supplementary Figure S1). Moreover, repeat experiments demonstrated a significant allelic difference in the intensity of bound DNA, with the nuclear protein(s) binding on average 46% less oligonucleotide containing the schizophrenia-associated T-allele of rs1344706 than oligonucleotide containing the G-allele of this SNP (t-test P <0.001, Figure 1b). The greater binding potential of DNA containing the G-allele of rs1344706 was additionally demonstrated by competitor experiments in which unlabelled oligonucleotides containing the T-allele were unable to prevent protein binding to the G-allele, even when present at a 200-fold excess (Figure 1c).
Figure 1.
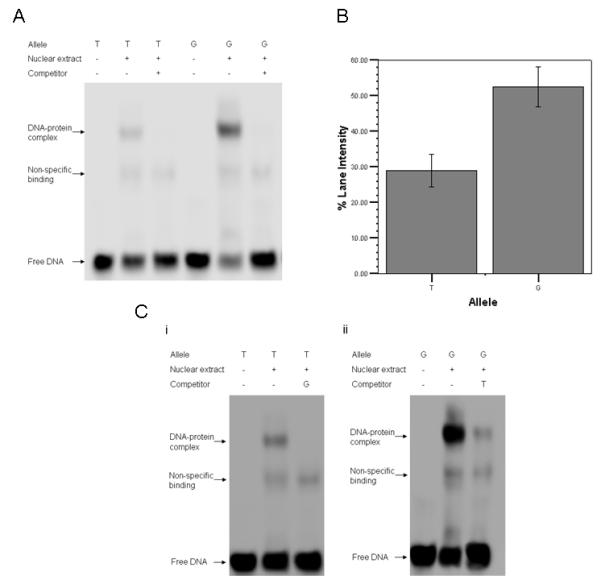
A) Binding of proteins from a single SH-SY5Y nuclear extract to alleles of rs1344706. Electromobility shift assays were used to investigate the formation of DNA-protein complexes with oligonucleotides specific to each rs1344706 allele. “+” and “−” indicate the presence or absence of nuclear extract or unlabelled competitor probe in each reaction. B) Quantification of binding of nuclear factors from SH-SY5Y cells to DNA oligonucleotides containing each rs1344706 allele. DNA-protein complexes were quantified as percentage total lane intensity. A significant reduction in binding intensity, averaging 46%, was observed for the T-containing oligonucleotide compared to the G-containing oligonucleotide (t-test P< 0.001, N= 6). Error bars represent +/− 1 S.D. There was no difference in total lane intensities between the two alleles (t-test P= 0.69). C) Competition of DNA-protein binding using unlabelled oligonucleotides containing the alternative allele of rs1344706. i.) Lane 1 = labelled T-containing oligonucleotide incubated in the absence of both nuclear extract and competitor. Lane 2 = labelled T-containing oligonucleotide incubated in the presence of nuclear extract only. Lane 3 = labelled T-containing oligonucleotide incubated in the presence of both nuclear extract and unlabelled G-containing oligonucleotide competitor at a 200-fold excess. DNA-protein complex formation at the T allele is abolished. ii.) Lane 1 = labelled G-containing oligonucleotide incubated in the absence of both nuclear extract and competitor. Lane 2 = labelled G-containing oligonucleotide incubated in the presence of SH-SY5Y nuclear extract only. Lane 3 = labelled G-containing oligonucleotide incubated in the presence of both nuclear extract and unlabelled T-containing oligonucleotide competitor. A 200-fold excess of T-containing oligonucleotides was unable to abolish DNA-protein complex formation at the G allele.
We tested whether our findings reflected binding of the nuclear proteins homez and hmx2, as previous bioinformatic analyses suggest that the G-allele of rs1344706 creates binding sites for these transcription factors (3). However, even when present at a 100-fold excess, competitor oliogonucleotides for the consensus binding sequences of these proteins failed to prevent binding to the 50 nucleotide sequence containing (the G-allele of) rs1344706 (Supplementary Figure S2), strongly suggesting that these are not the specific bound proteins. The identity of the nuclear protein(s) that binds to sequence containing rs1344706 is therefore, at present, unknown.
This study establishes rs1344706 as a functional polymorphism, with potential effects on ZNF804A expression through altered DNA-protein interactions. The identification of the bound protein and its temporal and cellular co-expression with ZNF804A would be valuable in determining the circumstances under which rs1344706 is active, as cis-effects on ZNF804A expression are known to differ between brain regions (8). Reporter gene assays of rs1344706 are also now warranted as a means of addressing the specific effect of the rs1344706 risk allele on ZNF804A expression (i.e. up- or down- regulation). While our data do not exclude the possibility of other functional variants in ZNF804A, they provide functional legitimacy for rs1344706 as a locus with direct effects on schizophrenia risk, and therefore experimental support for the view that variants promoting susceptibility to psychiatric disorders need not be in protein coding sequence.
Supplementary Material
Acknowledgements
This work was supported by the Medical Research Council, UK [Grant ID: G0802166].
Footnotes
Conflict of Interest The authors declare no conflict of interest.
References
- 1.O’Donovan MC, Craddock N, Norton N, Williams H, Peirce T, Moskvina V, et al. Nat Genet. 2008;40:1053–1055. doi: 10.1038/ng.201. [DOI] [PubMed] [Google Scholar]
- 2.Williams HJ, Norton N, Dwyer S, Moskvina V, Nikolov I, Carroll L, et al. Mol Psychiatry. 2010 e-pub ahead of print 6 April; doi: 10.1038/mp.2010.36. [Google Scholar]
- 3.Riley B, Thiselton D, Maher BS, Bigdeli T, Wormley B, McMichael GO, et al. Mol Psychiatry. 2010;15:29–37. doi: 10.1038/mp.2009.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Steinberg S, Mors O, Børglum AD, Gustafsson O, Werge T, Mortensen PB, et al. Mol Psychiatry. 2010 e-pub ahead of print 5 Jan; doi: 10.1038/mp.2009.149. [Google Scholar]
- 5.Zhang R, Lu SM, Qiu C, Liu XG, Gao CG, Guo TW, et al. Mol Psychiatry. 2010 doi: 10.1038/mp.2010.55. e-pub ahead of print 11 May; doi: 10.1038/mp.2010.55. [DOI] [PubMed] [Google Scholar]
- 6.Pollock K, Stroemer P, Patel S, Stevanato L, Hope A, Miljan E, et al. Exp Neurol. 2006;199:143–155. doi: 10.1016/j.expneurol.2005.12.011. [DOI] [PubMed] [Google Scholar]
- 7.Johansson S, Price J, Modo M. Stem Cells. 2008;26:2444–2454. doi: 10.1634/stemcells.2008-0116. [DOI] [PubMed] [Google Scholar]
- 8.Buonocore F, Hill HJ, Campbell CD, Oladimeji PB, Jeffries AR, Troakes C, et al. Hum Mol Genet. 2010;19:4490–4496. doi: 10.1093/hmg/ddq380. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


