Abstract
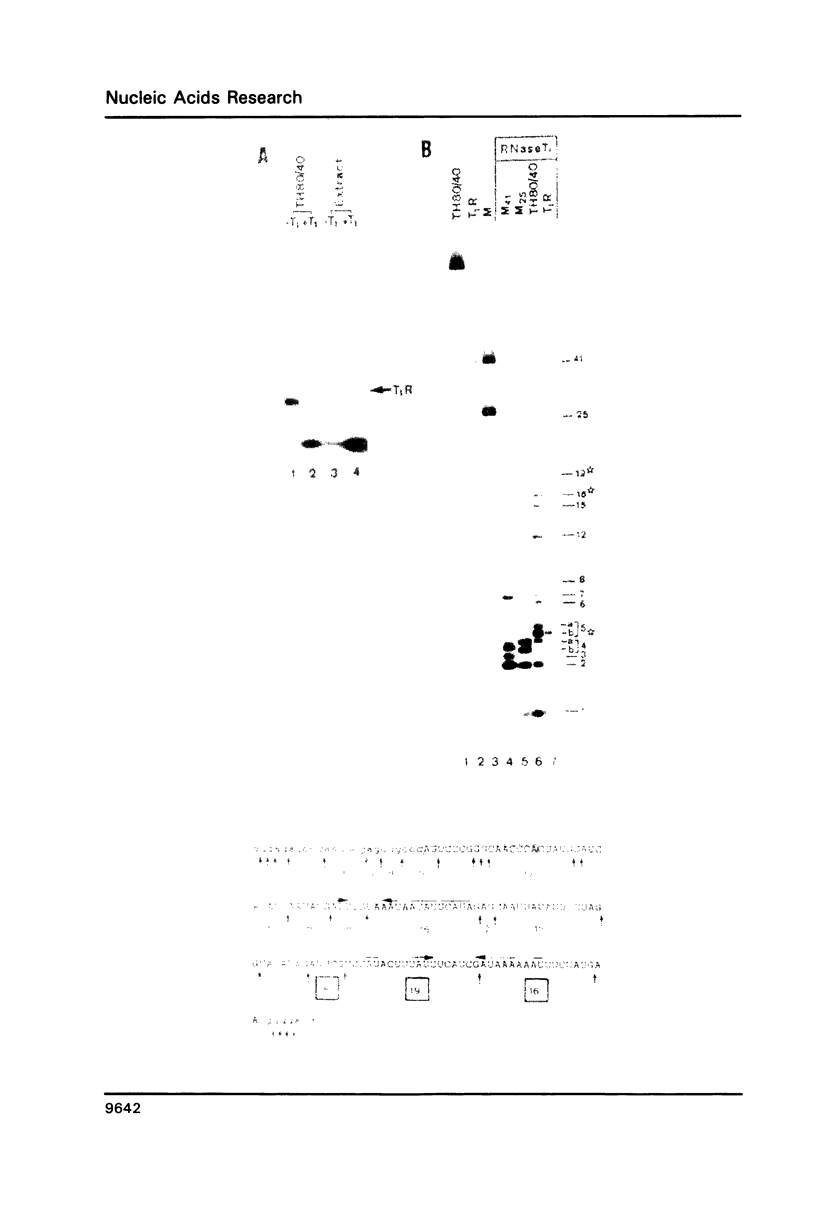
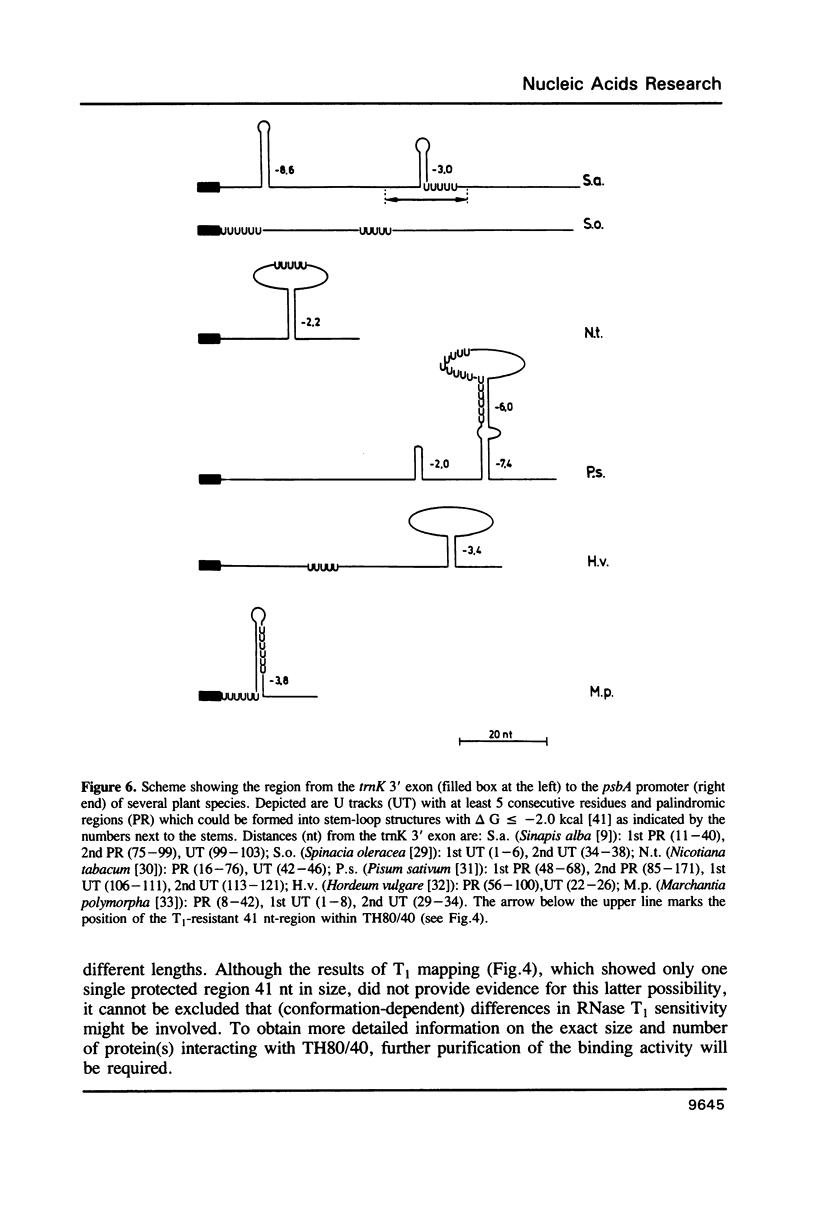
The 3' flanking region of the chloroplast trnK gene for tRNALys of mustard contains a palindromic sequence previously implicated with transcription termination and/or processing of the precursor RNA. Here we have investigated whether RNA sequences from the trnK 3' region are capable of interacting with chloroplast proteins in vitro. We find specific binding to an RNA region which is located further downstream from the palindromic sequence. The approximate length and position of this 3' binding region is reflected by a 41 nt spanning RNA segment which is protected against RNase T1 digestion by chloroplast protein(s). Competition experiments and sequence analyses suggest that U residues play an essential role in the RNA-protein interaction. Only a small number of proteins, possibly one single species, is in contact with the trnK 3' RNA.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Belasco J. G., Higgins C. F. Mechanisms of mRNA decay in bacteria: a perspective. Gene. 1988 Dec 10;72(1-2):15–23. doi: 10.1016/0378-1119(88)90123-0. [DOI] [PubMed] [Google Scholar]
- Birnstiel M. L., Busslinger M., Strub K. Transcription termination and 3' processing: the end is in site! Cell. 1985 Jun;41(2):349–359. doi: 10.1016/s0092-8674(85)80007-6. [DOI] [PubMed] [Google Scholar]
- Bogenhagen D. F., Brown D. D. Nucleotide sequences in Xenopus 5S DNA required for transcription termination. Cell. 1981 Apr;24(1):261–270. doi: 10.1016/0092-8674(81)90522-5. [DOI] [PubMed] [Google Scholar]
- Bottomley W., Smith H. J., Bogorad L. RNA polymerases of maize: partial purification and properties of the chloroplast enzyme. Proc Natl Acad Sci U S A. 1971 Oct;68(10):2412–2416. doi: 10.1073/pnas.68.10.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L. J., Orozco E. M., Jr Recognition of prokaryotic transcription terminators by spinach chloroplast RNA polymerase. Nucleic Acids Res. 1988 Sep 12;16(17):8411–8431. doi: 10.1093/nar/16.17.8411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efimov V. A., Andreeva A. V., Reverdatto S. V., Jung R., Chakhmakhcheva O. G. Nucleotide sequence of the barley chloroplast psbA gene for the QB protein of photosystem II. Nucleic Acids Res. 1988 Jun 24;16(12):5685–5685. doi: 10.1093/nar/16.12.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried M., Crothers D. M. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981 Dec 11;9(23):6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman D. I., Imperiale M. J., Adhya S. L. RNA 3' end formation in the control of gene expression. Annu Rev Genet. 1987;21:453–488. doi: 10.1146/annurev.ge.21.120187.002321. [DOI] [PubMed] [Google Scholar]
- Garner M. M., Revzin A. A gel electrophoresis method for quantifying the binding of proteins to specific DNA regions: application to components of the Escherichia coli lactose operon regulatory system. Nucleic Acids Res. 1981 Jul 10;9(13):3047–3060. doi: 10.1093/nar/9.13.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb E., Steitz J. A. Function of the mammalian La protein: evidence for its action in transcription termination by RNA polymerase III. EMBO J. 1989 Mar;8(3):851–861. doi: 10.1002/j.1460-2075.1989.tb03446.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb E., Steitz J. A. The RNA binding protein La influences both the accuracy and the efficiency of RNA polymerase III transcription in vitro. EMBO J. 1989 Mar;8(3):841–850. doi: 10.1002/j.1460-2075.1989.tb03445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg J. R. Ultraviolet light-induced crosslinking of mRNA to proteins. Nucleic Acids Res. 1979 Feb;6(2):715–732. doi: 10.1093/nar/6.2.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruissem W. Chloroplast gene expression: how plants turn their plastids on. Cell. 1989 Jan 27;56(2):161–170. doi: 10.1016/0092-8674(89)90889-1. [DOI] [PubMed] [Google Scholar]
- Gruissem W., Greenberg B. M., Zurawski G., Prescott D. M., Hallick R. B. Biosynthesis of chloroplast transfer RNA in a spinach chloroplast transcription system. Cell. 1983 Dec;35(3 Pt 2):815–828. doi: 10.1016/0092-8674(83)90114-9. [DOI] [PubMed] [Google Scholar]
- Higgins C. F., McLaren R. S., Newbury S. F. Repetitive extragenic palindromic sequences, mRNA stability and gene expression: evolution by gene conversion? A review. Gene. 1988 Dec 10;72(1-2):3–14. doi: 10.1016/0378-1119(88)90122-9. [DOI] [PubMed] [Google Scholar]
- Konarska M. M., Sharp P. A. Electrophoretic separation of complexes involved in the splicing of precursors to mRNAs. Cell. 1986 Sep 12;46(6):845–855. doi: 10.1016/0092-8674(86)90066-8. [DOI] [PubMed] [Google Scholar]
- Krieg P. A., Melton D. A. Functional messenger RNAs are produced by SP6 in vitro transcription of cloned cDNAs. Nucleic Acids Res. 1984 Sep 25;12(18):7057–7070. doi: 10.1093/nar/12.18.7057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn A., Grummt I. 3'-end formation of mouse pre-rRNA involves both transcription termination and a specific processing reaction. Genes Dev. 1989 Feb;3(2):224–231. doi: 10.1101/gad.3.2.224. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Leibold E. A., Munro H. N. Cytoplasmic protein binds in vitro to a highly conserved sequence in the 5' untranslated region of ferritin heavy- and light-subunit mRNAs. Proc Natl Acad Sci U S A. 1988 Apr;85(7):2171–2175. doi: 10.1073/pnas.85.7.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link G., Langridge U. Structure of the chloroplast gene for the precursor of the Mr 32,000 photosystem II protein from mustard (Sinapis alba L.). Nucleic Acids Res. 1984 Jan 25;12(2):945–958. doi: 10.1093/nar/12.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattaj I. W. A binding consensus: RNA-protein interactions in splicing, snRNPs, and sex. Cell. 1989 Apr 7;57(1):1–3. doi: 10.1016/0092-8674(89)90164-5. [DOI] [PubMed] [Google Scholar]
- Neuhaus H., Link G. The chloroplast tRNALys(UUU) gene from mustard (Sinapis alba) contains a class II intron potentially coding for a maturase-related polypeptide. Curr Genet. 1987;11(4):251–257. doi: 10.1007/BF00355398. [DOI] [PubMed] [Google Scholar]
- Pelletier J., Sonenberg N. Photochemical cross-linking of cap binding proteins to eucaryotic mRNAs: effect of mRNA 5' secondary structure. Mol Cell Biol. 1985 Nov;5(11):3222–3230. doi: 10.1128/mcb.5.11.3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt T. Transcription termination and the regulation of gene expression. Annu Rev Biochem. 1986;55:339–372. doi: 10.1146/annurev.bi.55.070186.002011. [DOI] [PubMed] [Google Scholar]
- Queen C., Korn L. J. A comprehensive sequence analysis program for the IBM personal computer. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):581–599. doi: 10.1093/nar/12.1part2.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern D. B., Gruissem W. Control of plastid gene expression: 3' inverted repeats act as mRNA processing and stabilizing elements, but do not terminate transcription. Cell. 1987 Dec 24;51(6):1145–1157. doi: 10.1016/0092-8674(87)90600-3. [DOI] [PubMed] [Google Scholar]
- Sugita M., Shinozaki K., Sugiura M. Tobacco chloroplast tRNA(UUU) gene contains a 2.5-kilobase-pair intron: An open reading frame and a conserved boundary sequence in the intron. Proc Natl Acad Sci U S A. 1985 Jun;82(11):3557–3561. doi: 10.1073/pnas.82.11.3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Wang M. J., Davis N. W., Gegenheimer P. Novel mechanisms for maturation of chloroplast transfer RNA precursors. EMBO J. 1988 Jun;7(6):1567–1574. doi: 10.1002/j.1460-2075.1988.tb02981.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis I., Frendewey D., Nichols M., Hottinger-Werlen A., Schaack J., Söll D. A single base change in the intron of a serine tRNA affects the rate of RNase P cleavage in vitro and suppressor activity in vivo in Saccharomyces cerevisiae. J Biol Chem. 1986 May 5;261(13):5878–5885. [PubMed] [Google Scholar]
- Wilusz J., Shenk T. A 64 kd nuclear protein binds to RNA segments that include the AAUAAA polyadenylation motif. Cell. 1988 Jan 29;52(2):221–228. doi: 10.1016/0092-8674(88)90510-7. [DOI] [PubMed] [Google Scholar]
- Zurawski G., Bohnert H. J., Whitfeld P. R., Bottomley W. Nucleotide sequence of the gene for the M(r) 32,000 thylakoid membrane protein from Spinacia oleracea and Nicotiana debneyi predicts a totally conserved primary translation product of M(r) 38,950. Proc Natl Acad Sci U S A. 1982 Dec;79(24):7699–7703. doi: 10.1073/pnas.79.24.7699. [DOI] [PMC free article] [PubMed] [Google Scholar]






