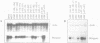Abstract
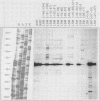
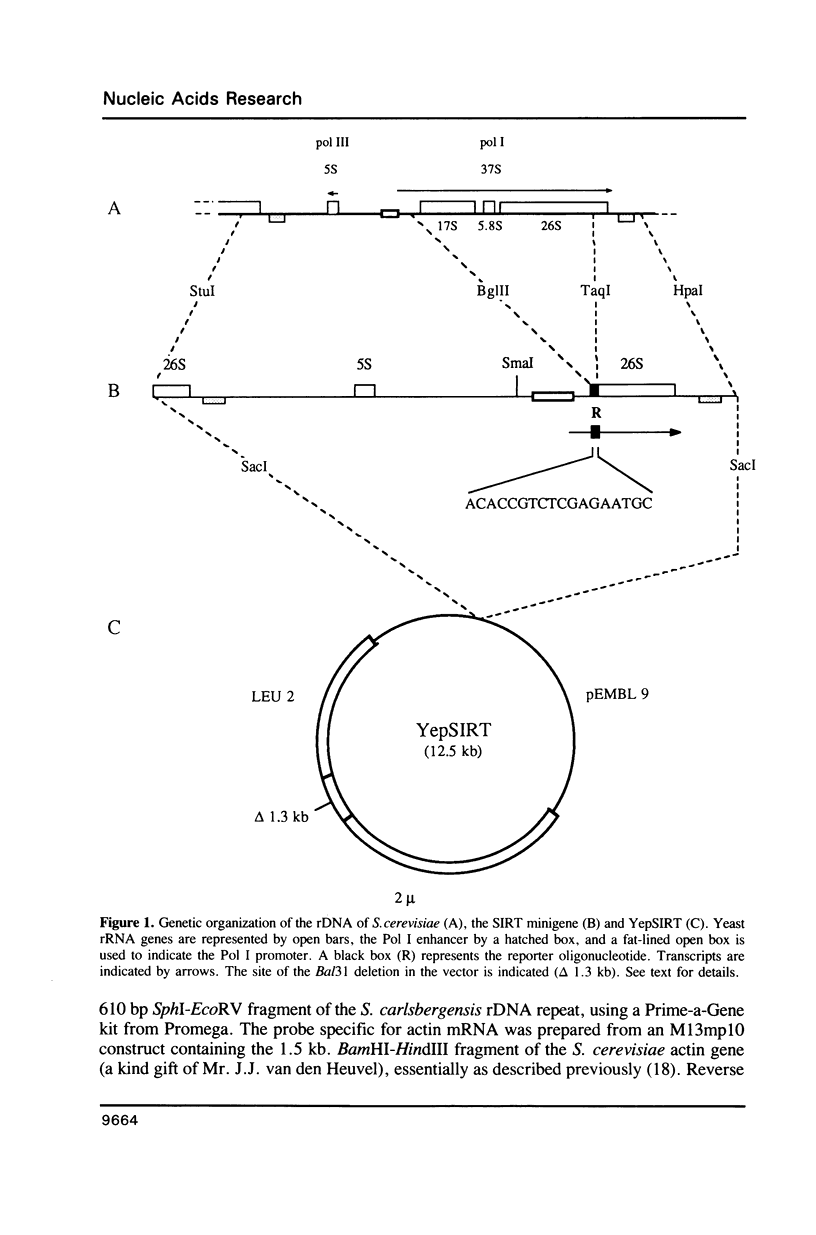
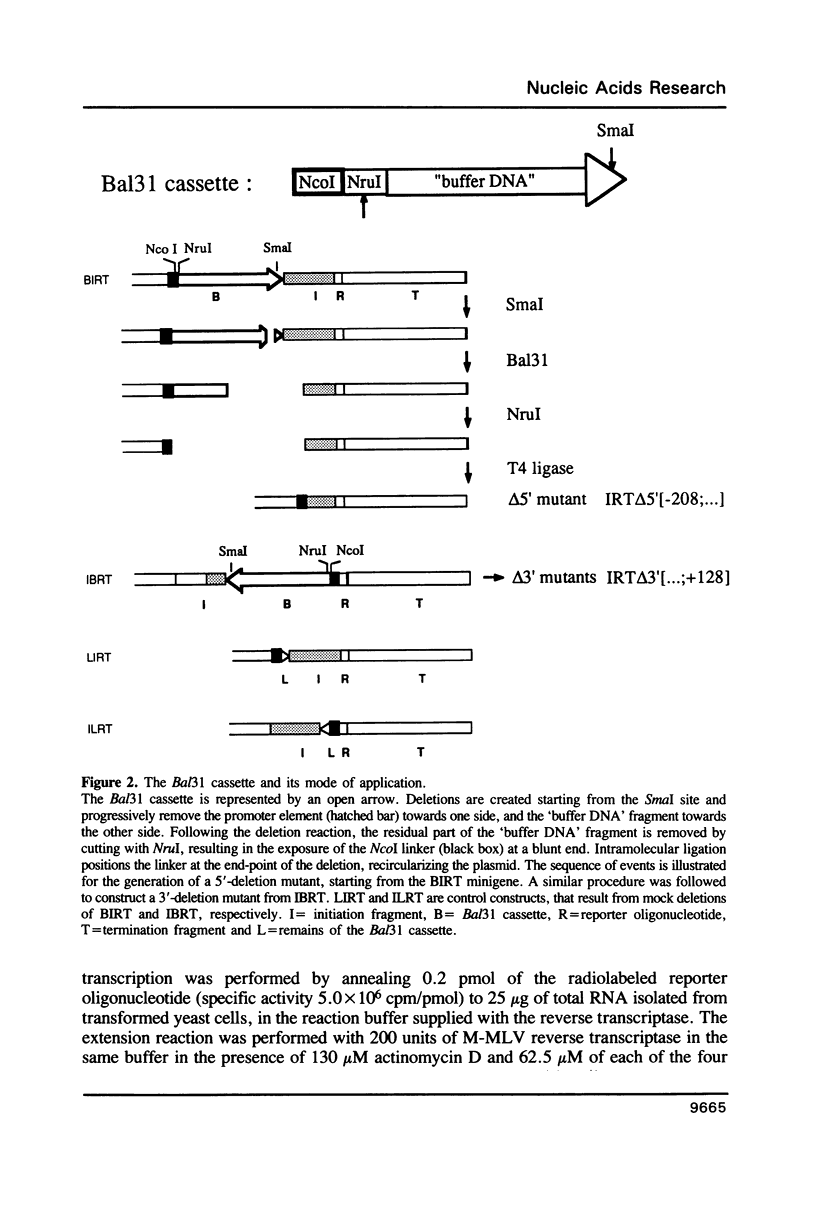
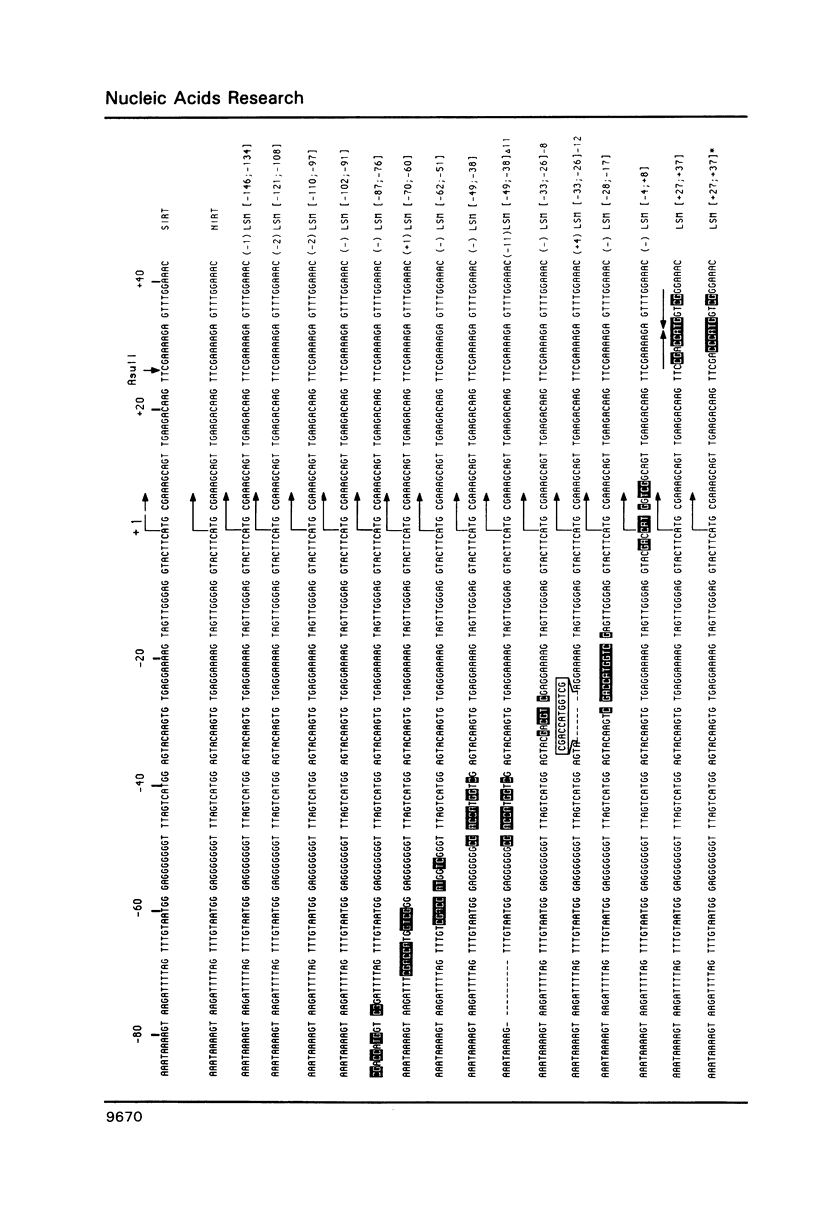
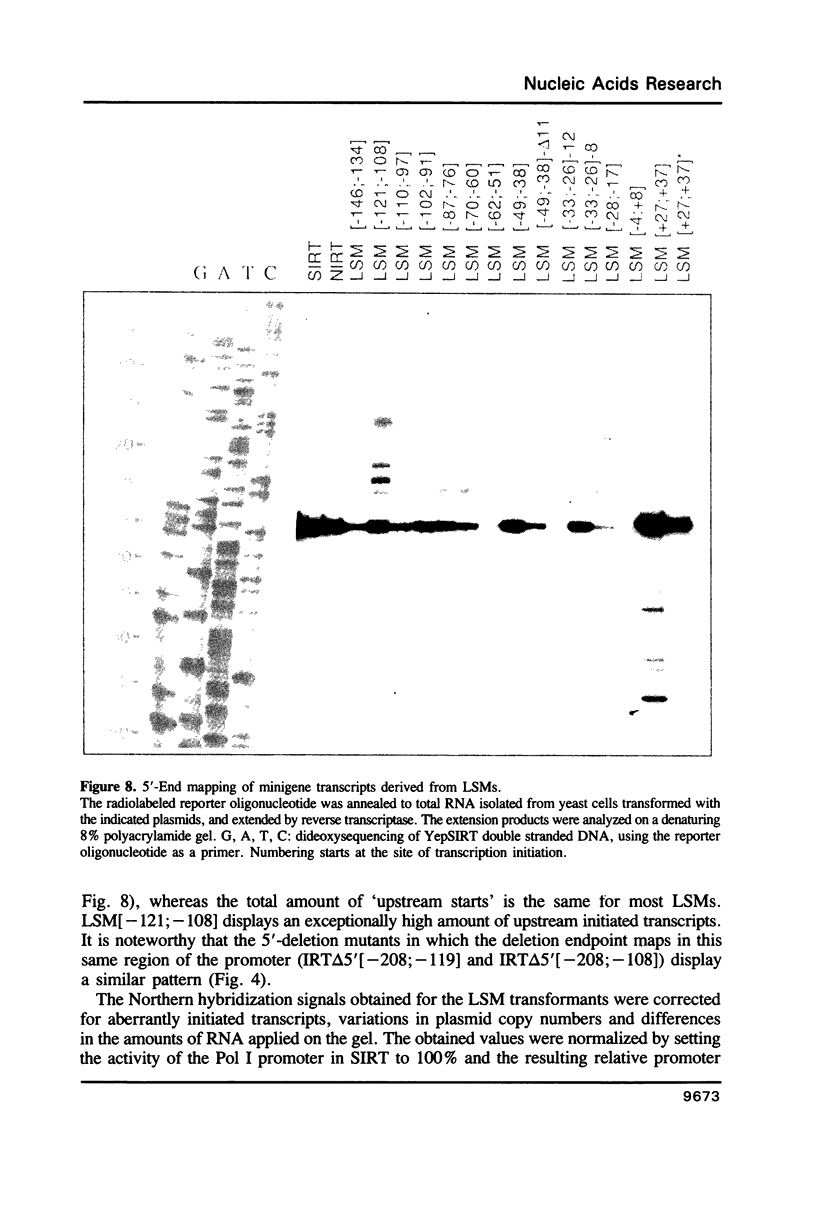
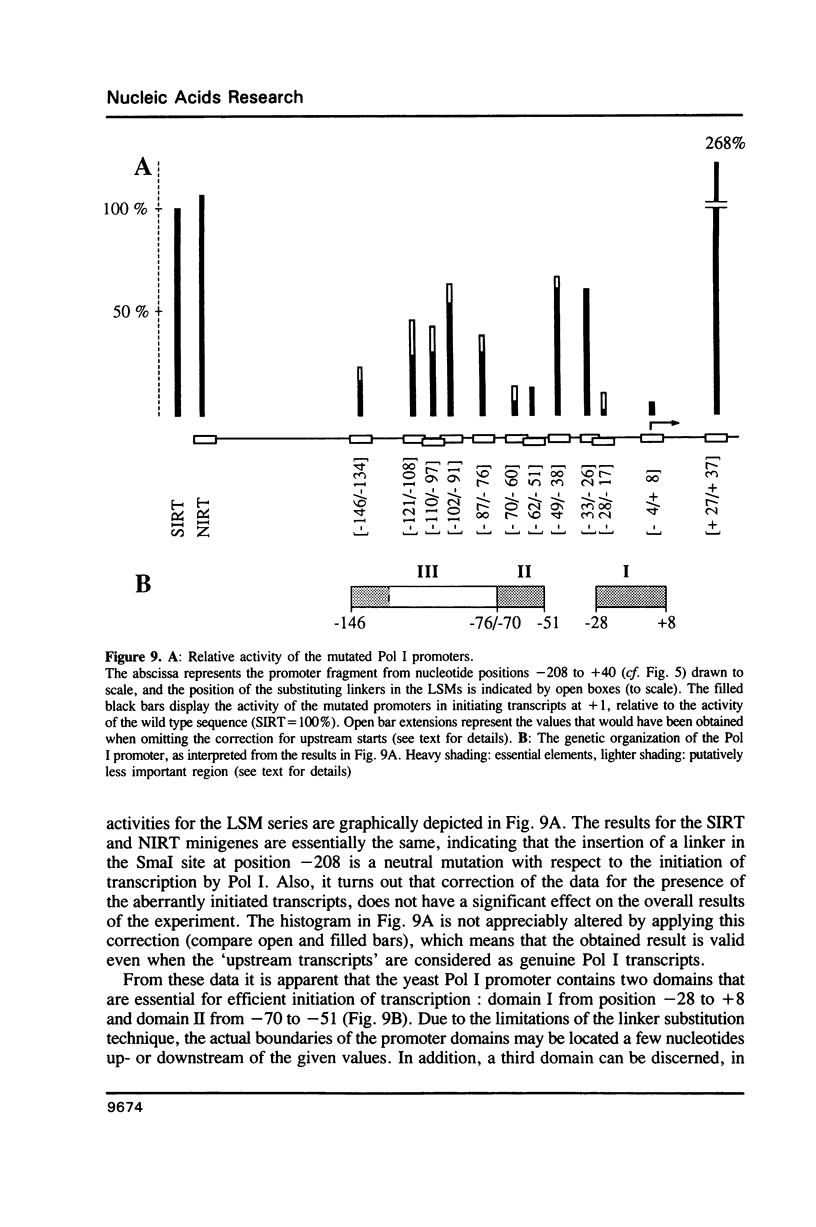
To define the RNA polymerase I promoter in the rDNA of Saccharomyces cerevisiae more precisely, we have constructed a series of 5'- and 3'-deletion mutants in a novel, plasmid-borne rDNA minigene, that also contains the transcriptional enhancer. Our data show that the Pol I promoter, in this context, extends from position -155 to +27, with 5'-deletions up to -134 and 3'-deletions up to -2 removing essential sequence information. To investigate the internal organization of the yeast Pol I promoter, linker scanning mutants were constructed, that traverse the Pol I promoter region and comprise between 5 and 12 clustered point mutations. Analysis of minigene transcription in yeast cells transformed with these plasmids demonstrates that the pol I promoter consists of three domains. Mutations in Domain I (from position -28 to +8) and Domain II (-70 to -51) drastically reduce promoter activity, whereas clustered point mutations in Domain III (starts at position -146 and presumably extends to position -76) appear to have less effect. Furthermore, the insertion of 4 nt between Domains I and II diminishes minigene transcription, indicating that the relative positions of these domains is essential.
Full text
PDF












Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baldari C., Cesareni G. Plasmids pEMBLY: new single-stranded shuttle vectors for the recovery and analysis of yeast DNA sequences. Gene. 1985;35(1-2):27–32. doi: 10.1016/0378-1119(85)90154-4. [DOI] [PubMed] [Google Scholar]
- Bell G. I., DeGennaro L. J., Gelfand D. H., Bishop R. J., Valenzuela P., Rutter W. J. Ribosomal RNA genes of Saccharomyces cerevisiae. I. Physical map of the repeating unit and location of the regions coding for 5 S, 5.8 S, 18 S, and 25 S ribosomal RNAs. J Biol Chem. 1977 Nov 25;252(22):8118–8125. [PubMed] [Google Scholar]
- Dunaway M., Reeder R. H. DNase I footprinting shows three protected regions in the promoter of the rRNA genes of Xenopus laevis. Mol Cell Biol. 1985 Feb;5(2):313–319. doi: 10.1128/mcb.5.2.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elion E. A., Warner J. R. An RNA polymerase I enhancer in Saccharomyces cerevisiae. Mol Cell Biol. 1986 Jun;6(6):2089–2097. doi: 10.1128/mcb.6.6.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elion E. A., Warner J. R. The major promoter element of rRNA transcription in yeast lies 2 kb upstream. Cell. 1984 Dec;39(3 Pt 2):663–673. doi: 10.1016/0092-8674(84)90473-2. [DOI] [PubMed] [Google Scholar]
- Haltiner M. M., Smale S. T., Tjian R. Two distinct promoter elements in the human rRNA gene identified by linker scanning mutagenesis. Mol Cell Biol. 1986 Jan;6(1):227–235. doi: 10.1128/mcb.6.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haltiner M., Kempe T., Tjian R. A novel strategy for constructing clustered point mutations. Nucleic Acids Res. 1985 Feb 11;13(3):1015–1025. doi: 10.1093/nar/13.3.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M. H., Learned R. M., Tjian R. Analysis of clustered point mutations in the human ribosomal RNA gene promoter by transient expression in vivo. Proc Natl Acad Sci U S A. 1988 Feb;85(3):669–673. doi: 10.1073/pnas.85.3.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempers-Veenstra A. E., Musters W., Dekker A. F., Klootwijk J., Planta R. J. Deletion mapping of the yeast Pol I promoter. Curr Genet. 1985;10(4):253–260. doi: 10.1007/BF00365621. [DOI] [PubMed] [Google Scholar]
- Kempers-Veenstra A. E., Oliemans J., Offenberg H., Dekker A. F., Piper P. W., Planta R. J., Klootwijk J. 3'-End formation of transcripts from the yeast rRNA operon. EMBO J. 1986 Oct;5(10):2703–2710. doi: 10.1002/j.1460-2075.1986.tb04554.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempers-Veenstra A. E., van Heerikhuizen H., Musters W., Klootwijk J., Planta R. J. Transcription of an artificial ribosomal RNA gene in yeast. EMBO J. 1984 Jun;3(6):1377–1382. doi: 10.1002/j.1460-2075.1984.tb01980.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kief D. R., Warner J. R. Coordinate control of syntheses of ribosomal ribonucleic acid and ribosomal proteins during nutritional shift-up in Saccharomyces cerevisiae. Mol Cell Biol. 1981 Nov;1(11):1007–1015. doi: 10.1128/mcb.1.11.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kownin P., Bateman E., Paule M. R. Eukaryotic RNA polymerase I promoter binding is directed by protein contacts with transcription initiation factor and is DNA sequence-independent. Cell. 1987 Aug 28;50(5):693–699. doi: 10.1016/0092-8674(87)90327-8. [DOI] [PubMed] [Google Scholar]
- Mestel R., Yip M., Holland J. P., Wang E., Kang J., Holland M. J. Sequences within the spacer region of yeast rRNA cistrons that stimulate 35S rRNA synthesis in vivo mediate RNA polymerase I-dependent promoter and terminator activities. Mol Cell Biol. 1989 Mar;9(3):1243–1254. doi: 10.1128/mcb.9.3.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller K. G., Tower J., Sollner-Webb B. A complex control region of the mouse rRNA gene directs accurate initiation by RNA polymerase I. Mol Cell Biol. 1985 Mar;5(3):554–562. doi: 10.1128/mcb.5.3.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishima Y., Financsek I., Kominami R., Muramatsu M. Fractionation and reconstitution of factors required for accurate transcription of mammalian ribosomal RNA genes: identification of a species-dependent initiation factor. Nucleic Acids Res. 1982 Nov 11;10(21):6659–6670. doi: 10.1093/nar/10.21.6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss T. Transcription of cloned Xenopus laevis ribosomal DNA microinjected into Xenopus oocytes, and the identification of an RNA polymerase I promoter. Cell. 1982 Oct;30(3):835–842. doi: 10.1016/0092-8674(82)90288-4. [DOI] [PubMed] [Google Scholar]
- Musters W., Venema J., van der Linden G., van Heerikhuizen H., Klootwijk J., Planta R. J. A system for the analysis of yeast ribosomal DNA mutations. Mol Cell Biol. 1989 Feb;9(2):551–559. doi: 10.1128/mcb.9.2.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petes T. D. Yeast ribosomal DNA genes are located on chromosome XII. Proc Natl Acad Sci U S A. 1979 Jan;76(1):410–414. doi: 10.1073/pnas.76.1.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quincey R. V., Arnold R. E. Transcription of a yeast ribosomal RNA minigene in Saccharomyces cerevisiae. Biochem J. 1984 Dec 1;224(2):497–503. doi: 10.1042/bj2240497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeder R. H., Pennock D., McStay B., Roan J., Tolentino E., Walker P. Linker scanner mutagenesis of the Xenopus laevis ribosomal gene promoter. Nucleic Acids Res. 1987 Sep 25;15(18):7429–7441. doi: 10.1093/nar/15.18.7429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubtsov P. M., Musakhanov M. M., Zakharyev V. M., Krayev A. S., Skryabin K. G., Bayev A. A. The structure of the yeast ribosomal RNA genes. I. The complete nucleotide sequence of the 18S ribosomal RNA gene from Saccharomyces cerevisiae. Nucleic Acids Res. 1980 Dec 11;8(23):5779–5794. doi: 10.1093/nar/8.23.5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skryabin K. G., Eldarov M. A., Larionov V. L., Bayev A. A., Klootwijk J., de Regt V. C., Veldman G. M., Planta R. J., Georgiev O. I., Hadjiolov A. A. Structure and function of the nontranscribed spacer regions of yeast rDNA. Nucleic Acids Res. 1984 Mar 26;12(6):2955–2968. doi: 10.1093/nar/12.6.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollner-Webb B., Tower J. Transcription of cloned eukaryotic ribosomal RNA genes. Annu Rev Biochem. 1986;55:801–830. doi: 10.1146/annurev.bi.55.070186.004101. [DOI] [PubMed] [Google Scholar]
- Sollner-Webb B., Wilkinson J. A., Roan J., Reeder R. H. Nested control regions promote Xenopus ribosomal RNA synthesis by RNA polymerase I. Cell. 1983 Nov;35(1):199–206. doi: 10.1016/0092-8674(83)90222-2. [DOI] [PubMed] [Google Scholar]
- Swanson M. E., Holland M. J. RNA polymerase I-dependent selective transcription of yeast ribosomal DNA. Identification of a new cellular ribosomal RNA precursor. J Biol Chem. 1983 Mar 10;258(5):3242–3250. [PubMed] [Google Scholar]
- Swanson M. E., Yip M., Holland M. J. Characterization of an RNA polymerase I-dependent promoter within the spacer region of yeast ribosomal cistrons. J Biol Chem. 1985 Aug 15;260(17):9905–9915. [PubMed] [Google Scholar]
- Veldman G. M., Klootwijk J., de Regt V. C., Planta R. J., Branlant C., Krol A., Ebel J. P. The primary and secondary structure of yeast 26S rRNA. Nucleic Acids Res. 1981 Dec 21;9(24):6935–6952. doi: 10.1093/nar/9.24.6935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbeet M. P., Klootwijk J., van Heerikhuizen H., Fontijn R. D., Vreugdenhil E., Planta R. J. A conserved sequence element is present around the transcription initiation site for RNA polymerase A in Saccharomycetoideae. Nucleic Acids Res. 1984 Jan 25;12(2):1137–1148. doi: 10.1093/nar/12.2.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windle J. J., Sollner-Webb B. Two distant and precisely positioned domains promote transcription of Xenopus laevis rRNA genes: analysis with linker-scanning mutants. Mol Cell Biol. 1986 Dec;6(12):4585–4593. doi: 10.1128/mcb.6.12.4585. [DOI] [PMC free article] [PubMed] [Google Scholar]