Abstract
Condensin is required for chromosome dynamics and diverse DNA metabolism. How condensin works, however, is not well understood. Condensin contains two structural maintenance of chromosomes (SMC) subunits with the terminal globular domains connected to coiled-coil that is interrupted by the central hinge. Heterotrimeric non-SMC subunits regulate SMC. We identified a novel fission yeast SMC hinge mutant, cut14-Y1, which displayed defects in DNA damage repair and chromosome segregation. It contains an amino acid substitution at a conserved hinge residue of Cut14/SMC2, resulting in diminished DNA binding and annealing. A replication protein A mutant, ssb1-418, greatly alleviated the repair and mitotic defects of cut14-Y1. Ssb1 protein formed nucleolar foci in cut14-Y1 cells, but the number of foci was diminished in cut14-Y1 ssb1-418 double mutants. Consistent with the above results, Ssb1 protein bound to single-strand DNA was removed by condensin or the SMC dimer through DNA reannealing in vitro. Similarly, RNA hybridized to DNA may be removed by the SMC dimer. Thus, condensin may wind up DNA strands to unload chromosomal components after DNA repair and prior to mitosis. We show that 16 suppressor mutations of cut14-Y1 were all mapped within the hinge domain, which surrounded the original L543 mutation site.
Keywords: structural maintenance of chromosomes, DNA damage, mitosis, DNA metabolism, condensation
2. Introduction
Condensin is a hetero-pentameric protein complex in eukaryotes that consists of two structural maintenance of chromosomes (SMC) subunits and three regulatory non-SMC subunits [1–6]. The terminal globular domains of the SMC subunits contain the Walker A and B ATPase motifs [7,8], and are connected to a coiled-coil domain that is interrupted by a central hinge, while one of the non-SMC subunits is phosphorylated by Aurora B kinase [9–12] during mitosis [13,14]. The diverse roles of condensin in chromosome dynamics, including mitotic chromosome condensation and segregation, DNA metabolism and development, are well documented [15–19], but the molecular mechanism of how it functions is not well understood. Condensin and the SMC2/4 dimer possess DNA reannealing activity, an activity not found in cohesin [20,21], suggesting that DNA reannealing activity might be required for condensin function. However, the physiological significance of the reannealing activity remains an enigma. In this study, we show that condensin antagonizes replication protein A (RPA) [22–25] activity by removing it from DNA in vitro and in vivo, which suggests that the DNA reannealing activity of condensin may facilitate the removal of proteins from chromosomes after DNA repair or prior to chromosome segregation.
3. Results
3.1. Isolation of a structural maintenance of chromosomes mutant hypersensitive to DNA damage
About 1300 temperature-sensitive (ts) haploid fission yeast Schizosaccharomyces pombe strains were constructed, and screened for cytological defects at 36°C (the restrictive temperature) that resembled previously isolated condensation-defective mutants. Identification of a novel condensation-defective cut14-Y1 mutant is described in the legend of figure 1 and electronic supplementary material, figure S1. This mutant is also highly sensitive to DNA damage at 26°C, the permissive temperature. The hydroxyurea (HU) and ultraviolet (UV) ray-sensitive phenotype co-segregated with the ts phenotype. We decided to focus on this mutant, which is the first condensin SMC mutant sensitive to DNA damage to our knowledge. Previously, cnd2-1, the Barren-like non-SMC subunit mutant of S. pombe, was shown to be sensitive to DNA damage [31].
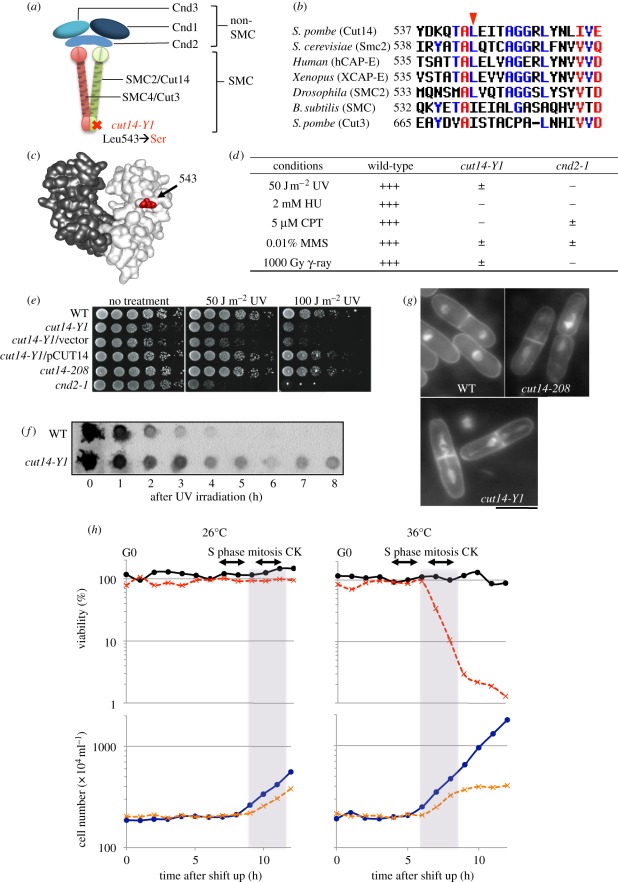
Figure 1.
Mutation site and phenotypes of condensin SMC mutant cut14-Y1. The identification of cut14-Y1: four mutant strains among 1300 ts strains examined exhibited condensation defects. Gene cloning, genetic analysis and gene sequencing established that the mutations resided in three distinct genes involved in chromosome condensation. Strain 393 was a DNA topoisomerase II top2 [26] mutant, strain 640 was a cut15 [27] (homologue of importin alpha) mutant and the remaining Y1 and 541 strains were cut14 [28,29] (SMC2 homologue) mutants. Since the cut14-Y1 strain was hypersensitive to DNA damage at 26°C (the permissive temperature), we examined whether the damage-sensitive phenotype was linked with the ts phenotype. Tetrad dissection demonstrated that the HU (hydroxyurea) and UV (ultraviolet) ray-sensitive phenotype co-segregated with the ts phenotype (electronic supplementary material, figure S1). (a) Heteropentameric condensin complex. The cut14-Y1 allele consists of a L543S substitution in the hinge. (b) Amino acid sequences of the SMC hinge that surround the mutation site (red arrowhead). (c) The mutation site (red) is shown within the three-dimensional structure of the mouse hinge domain [30]. (d) Summary of the DNA damage phenotypes of cut14-Y1 together with the previously reported response of cnd2-1 [31]. +++, normal growth; ±, very slow growth; −, no growth. (e) Wild-type (WT), cut14-Y1 and other strains were spot tested after UV irradiation at 26°C. (f) After UV irradiation (100 J m−2) at 26°C, extracts of the WT and cut14-Y1 cells harvested at intervals were immunoblotted using anti-thymine dimer antibodies. (g) The mitotic segregation defect of cut14-Y1 and cut14-208. DAPI was used to stain DNA. Scale bar, 10 µm. (h) WT and cut14-Y1 cells were first arrested at the pre-replicative G0 phase in nitrogen-deficient medium (EMM2-N) [32] at 26°C for 24 h, and then shifted to a nitrogen-replenished medium (EMM2) at 26°C (left) or at 36°C (right) for 12 h to measure cell viability (plated at 26°C) and cell number. The timing of S phase, mitosis and cytokinesis (CK) were determined by FACScan and DAPI-staining, respectively. Aliquots of the cultures were taken at 1 h intervals after replenishment, and 300 cells of each genotype were plated on three YPD plates for each time point. The plates were incubated at 26°C for 5 days, and the colony numbers were counted. Circles with solid line, WT; crosses with dashed line, cut14-Y1.
3.2. cut14-Y1 is a hinge mutant
The cut14-Y1 mutant contained a single amino acid substitution in the hinge region (L543 to S543; figure 1a). Sequence comparisons showed that this amino acid residue is conserved in SMC2 of other organisms, and similar in Cut3/SMC4 (from L to I; figure 1b). While the L543 residue is not conserved in the hinge of Escherichia coli MukB, this residue is similar in Bacillus subtilis SMC (from L to I). Therefore, MukB is probably distinct from that of condensin [33], while B. subtilis condensin contains the hinge region, which may be similar to that of eukaryote condensins [34,35]. The L543 residue (red colour in figure 1c) is located in the middle of the hinge region and not in the dimerization domain.
3.3. DNA damage repair is defective in cut14-Y1 at 26°C
The response of cut14-Y1 to various DNA damage agents at 26°C is summarized in figure 1d (data are shown in figure 1e and electronic supplementary material, figure S2). cut14-Y1 cells were sensitive to 50 J m−2 UV irradiation, 2 mM HU, 5 µM camptothecin (CPT), 0.01 per cent methylmethanesulphonate (MMS) and 1000 Gyγ-irradiation, and were more sensitive to CPT than cnd2-1. Defects in excision repair were assessed by using an antibody to detect thymine dimers produced by UV for 0–8 h at 26°C. In cut14-Y1 cells, repair after UV exposure (100 J m−2) occurred initially, but was considerably delayed later (figure 1f). The damage phenotype of cut14-Y1 differed greatly from a previously isolated mutant, cut14-208, which contains a mutation in the coiled-coil region and is not sensitive to damage [20,31].
3.4. Lethal mitosis of cut14-Y1 occurs at 36°C without delay
A culture of asynchronous cut14-Y1 mutant cells grown at 26°C was shifted to 36°C. One to two hours after the temperature shift, mutant cells stained with 4′,6-diamidino-2-phenylindole (DAPI) revealed mitotic defects (approx. 100% after 3 h), including abnormal chromosome condensation, segregation and cytokinesis (CK; figure 1g). The mitotic defects of cut14-208 were indistinguishable from those of cut14-Y1.
Wild-type (WT) and cut14-Y1 cells synchronously released from the quiescent G0 phase (figure 1h; see the legend for experimental details). The viability of cut14-Y1 cells at 36°C was identical to that of WT (approx. 100%) during S phase (4 h) and G2 phase, but decreased when cells entered mitosis and CK after 6–8 h at 36°C (dotted red line), suggesting that the lethality occurred at the restrictive temperature after mitotic entry. Potentially lethal defects that occurred before mitosis at 36°C could be rescued if cells were shifted back to 26°C. At 26°C, the viability of cut14-Y1 was high (100%) and the doubling time was slightly longer than that of the WT. At 36°C, the increase in cut14-Y1 cell number occurred only once, as cells were dead after the first mitosis (aberrant ‘cut’ cells were counted as two cells). As at 26°C, the timing of CK at 36°C, the restrictive temperature, was only slightly delayed compared with that of WT, suggesting that neither the DNA damage nor mitotic checkpoint were activated in cut14-Y1.
3.5. cut14-Y1 genetically interacts with many DNA metabolism mutations
To understand the function of the condensin hinge domain, synthetic genetic interactions were examined by pair-wise crosses between cut14-Y1 and 34 mutants defective in cell cycle, mitosis, DNA repair, replication, recombination or the DNA damage checkpoint. Results were classified into three groups (A, B and C), as shown in figure 2a and electronic supplementary material, figure S3. Group A mutants, when combined with cut14-Y1, failed to produce double mutants by tetrad analysis, suggesting that group A mutants were synthetic lethal with cut14-Y1. When cut14-Y1 was crossed with group B mutants, viable double mutants were produced, but the defects (ts, HU and UV) were additive (electronic supplementary material, figure S4). Group C mutants formed viable double mutants with cut14-Y1, and the ts and HU sensitivity were indistinguishable from that of the cut14-Y1 single mutant. Hence, we observed genetic interactions with group A and B mutants, with the greatest synthetic interactions observed with group A mutants, whereas group C mutants did not interact with cut14-Y1. Group A included mutants, many of which interact with single-stranded (ss) DNA or ssDNA-associated RPA [36,37]. Group B consisted of DNA checkpoint mutants [38], suggesting that the hinge of Cut14 might affect a damage checkpoint function. Most of the group C mutants were related to cell cycle, although two, Δuvde and Δrqh1, were involved in DNA repair (electronic supplementary material, figure S3).
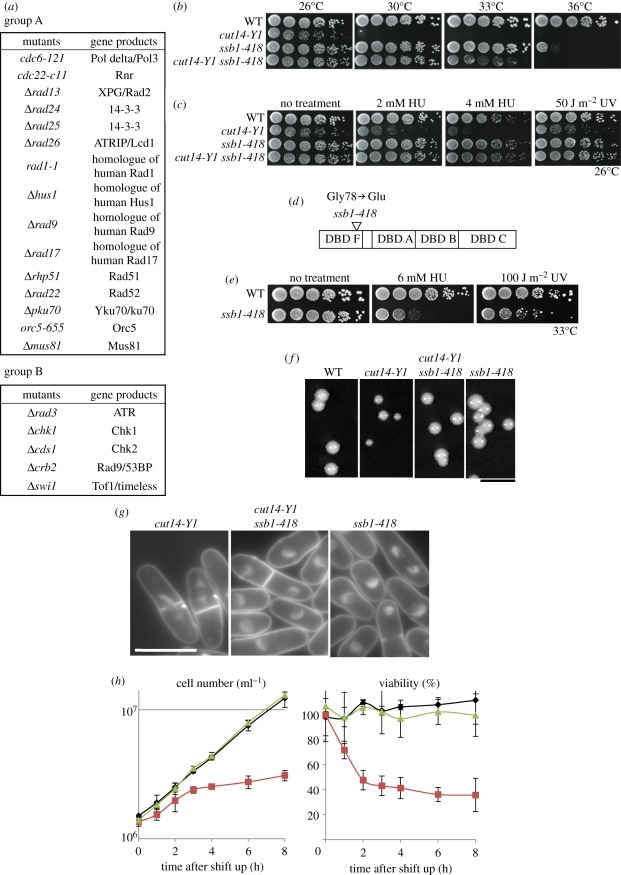
Figure 2.
cut14-Y1 defects are additive with many DNA metabolic and checkpoint mutants, and are rescued by ssb1-418. (a) Strains crossed with cut14-Y1 to obtain the double mutants are shown. Crosses with group A (15 strains) did not yield viable double mutants, while those with group B (5 strains) produced double mutants with additive defects. Crosses with group C (14 strains) produced double mutants that did not display any additive defect (see also electronic supplementary material, figure S3). Group A included mutants that are involved with the 9-1-1 complex (rad9, rad1 and hus1), double strand break repair (rhp51, rad22, mus81 and ku70), replication (cdc6, cdc22 and orc5), 14-3-3 (rad24 and rad25), ssDNA nuclease (rad13) and DNA damage checkpoint (rad17 and rad26), many of which interact with single-stranded (ss) DNA or ssDNA-associated RPA [36,37]. Group B included five DNA checkpoint mutants (rad3, chk1, cds1, crb2 [38] and swi1 [39]), suggesting that the hinge of Cut14 might have a checkpoint function. Most of the group C mutants were related to replication, cell cycle and mitosis, although two were involved in DNA repair (uvde and rqh1 [40]). (b) ssb1-418 showed a striking synthetic rescue of the cut14-Y1 ts phenotype at 30°C and 33°C. (c) ssb1-418 also rescued the 2−4 mM HU and 50 J m−2 UV sensitivity of cut14-Y1 at 26°C. (d) The ssb1-418 strain contains a G78E amino acid substitution in the DBD F. (e) Single ssb1-418 mutants were sensitive to HU and UV at the semi-permissive temperature (33°C). (f) The colony formation of single and double mutants was examined at 26°C. Scale bar, 5 mm. The colony size of cut14-Y1 single mutants was smaller than that of the cut14-Y1 ssb1-418 double mutant at 26°C. The doubling time of single cut14-Y1 was 4.5 h at 26°C, while that of the double mutant and the WT was 3.5 h at 26°C. (g,h) In YPD liquid medium at 33°C, the cut14-Y1 single mutant lost viability and displayed frequent mitotic defects, while the double mutant and ssb1-418 grew normally. The asynchronous cultures of cut14-Y1, ssb1-418 and the double mutant in the YPD liquid medium were shifted from 26°C to 30°C. The cell number (per millilitre) was counted by the cell counter. The viability was measured at each time point by plating 300 cells spread on YPD plates, incubated at 26°C for 5 days, and resulting colonies were counted. Scale bar, 10 µm. (h) Black diamonds, ssb1-418; red squares, cut14-Y1; green triangles, cut14-Y1 ssb1-418.
3.6. Synthetic rescue of cut14-Y1 by ssb1-418
After examining more mutants involved in DNA and RNA metabolism by tetrad dissection, we found that one mutant, ssb1-418, showed a striking synthetic rescue of the cut14-Y1 ts phenotype at 33°C, and also the HU and UV sensitivities at 26°C (figure 2b,c). ssb1-418 is a mutant of Ssb1, the largest subunit of heterotrimeric RPA.
ssb1-418 was isolated as a ts mutant that was suppressed by a plasmid carrying the ssb1+ gene. The mutation site, determined by genetic crossing and gene sequencing, consisted of an E substitution at G78, and is located in the amino-terminal ssDNA-binding domain called DBD F, which ensures binding to short stretches of ssDNA [41] (figure 2d). ssb1-418 was sensitive to HU (6 mM) and UV (100 J m−2) at 33°C, a semi-permissive temperature (figure 2e), but not at 26–30°C, suggesting that DNA repair is impaired in ssb1-418 at higher temperatures.
As shown in figure 2f, the colony size of cut14-Y1 is small, while the colony size of the cut14-Y1 ssb1-418 double mutant was similar to that of the WT at 26°C. The cut14-Y1 single mutant frequently exhibited aberrant chromosome segregation, but mitosis in the double mutant appeared mostly normal at 30°C (figure 2g). In liquid culture, the double mutant grew exponentially after a temperature shift from 26°C to 30°C (green line, figure 2h), while the cut14-Y1 single mutant arrested after one round of division and quickly lost viability at 30°C (red lines in figure 2h). Thus, rescue occurred at the level of mitotic chromosome segregation and cell viability.
3.7. Ssb1 nuclear foci are observed in cut14-Y1 cells, but not in the double mutant
Synthetic rescue suggested that Ssb1 and the Cut14 hinge domain antagonize each other, and that their functional balance might be important. While Ssb1 is known to stabilize DNA strand separation, the Cut14 hinge promotes DNA annealing. Therefore, Ssb1 and condensin may coordinate the dynamics of ssDNA stabilization and destabilization in chromatin. To test whether Ssb1 might anomalously remain in mitotic chromatin in the cut14-Y1 mutant, localization of Ssb1 was assessed by immunofluorescence microscopy using polyclonal antibodies against S. pombe Ssb1 [42].
Anti-Ssb1 antibody revealed that the nuclear Ssb1 signals were clearly observed in S phase cells. In those S phase cells, Ssb1 appeared as punctate signals in 100 per cent of cells (arrowheads in figure 3a). In cut14-Y1 mutant cells or the other mutants, ssb1-418 and ssb1-418 cut14-Y1, at the permissive temperature, the Ssb1 dots in S phase were similarly observed in 100 per cent of cells (arrowheads) and disappeared in G2 phase. These Ssb1 signals in S phase are distinct from the foci observed in cut14-Y1 (see below).
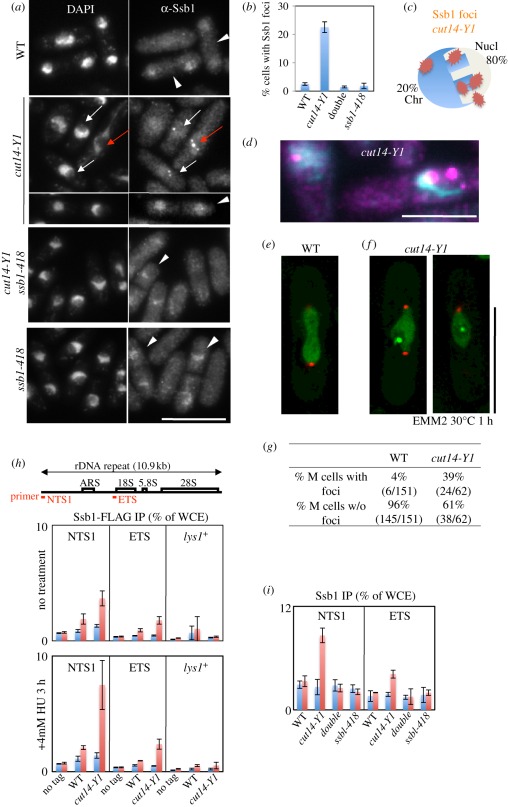
Figure 3.
Intense Ssb1-YFP foci formed in the nucleolus of cut14-Y1, but not in the double mutant. (a) Intense nuclear foci were detected by anti-Ssb1 antibody (DNA stained by DAPI). Immunofluorescence micrographs are shown for WT, single cut14-Y1, ssb1-418 and the double mutant cells that were shifted from 26°C to 30°C for 1 h. The Ssb1 foci were observed in both mitotic (red arrows) and interphase (white arrows) cut14-Y1 cells. Such foci were scarce in WT, ssb1-418 and cut14-Y1 ssb1-418 (double mutant) cells. The nuclear Ssb1 signals were clearly observed in S phase cells (white arrowheads). The S phase occurs in binucleated septated cells, allowing S phase cells to be easily distinguished. In those S phase cells, nuclear accumulation of Ssb1 appeared as punctate signals, distinct from the foci observed in cut14-Y1. Scale bar, 10 µm. (b) Quantitative data (percentage cells with Ssb1 foci) are shown. Three hundred cells were observed for each strain. (c) Anti-Ssb1 antibodies revealed that the intense foci were mostly (approx. 80%) located in the nucleolar region of the cut14-Y1 single mutant. The S. pombe interphase nucleus consists of the hemispherical chromatin region (Chr) and the remaining nucleolar (Nucl) region [43]. (d) The enlarged, merged micrograph of cut14-Y1 cells. Blue, DAPI; purple, Ssb1. Scale bar, 5 µm. (e,f) Live cell images of WT (e) and cut14-Y1 mutant (f) cells that express both Ssb1-YFP (green) and Sad1-mCherry (SPB, red). Scale bar, 10 µm. See electronic supplementary material, movies S1–S4. (g) The frequency (%) of cells showing Ssb1-YFP foci in WT and cut14-Y1 mitotic mutant cells were determined after the shift to 30°C (the restrictive temperature) from 26°C for 1 h in EMM2. (h) Chromatin immunoprecipitation (ChIP) experiment using anti-FLAG antibodies. The ssb1+ gene was chromosomally tagged with FLAG in the WT and cut14-Y1 strains, and expressed under the native promoter at 26°C in the absence (upper panel) or presence (lower panel) of 4 mM HU for 3 h. Two rDNA probes and one negative control (lys1+) probe were used. Blue and red columns indicate ChIP without and with antibodies against FLAG. An untagged (no tag) strain was used as the negative control. (i) ChIP experiment using anti-Ssb1 antibodies for the four strains cultured at 26°C. (h,i) Blue bars, −antibodies; red bars, +antibodies.
Intense nuclear foci, as detected by anti-Ssb1 antibody (DNA stained by DAPI), were frequently observed in both mitotic (red arrow, figure 3a) and interphase (short white arrows) cut14-Y1 cells. Such foci were scarce in WT, ssb1-418 and cut14-Y1 ssb1-418 (double mutant) cells. We measured the frequency of cells exhibiting these nuclear foci and, as shown in figure 3b, 22 per cent of the cut14-Y1 cells contained such foci, whereas the foci were infrequent (less than 2%) in the other three strains. This result suggested that foci formation in cut14-Y1 required the presence of WT Ssb1.
We then examined the localization of these intense foci of Ssb1 in cut14-Y1 mutant cells. The majority (80%) of foci were located in the rDNA nucleolar region, while the remaining foci (20%) resided at the periphery of the nucleus (figure 3c; the illustration shows the location of Ssb1 foci in cut14-Y1). Figure 3d shows multiple nucleolar foci in the right cell and one peri-nuclear focus in the left cell.
3.8. Live cell analysis of Ssb1-YFP foci in cut14-Y1 mitotic cells
To further investigate the results above, we constructed strains carrying chromosomally integrated Ssb1-YFP that was expressed from its native promoter, and observed Ssb1-YFP signals in WT or cut14-Y1 living cells. Ssb1-YFP was also located in the mitotic nucleoli in WT and in cut14-Y1 (figure 3e,f). In the electronic supplementary material movies, the dynamics of Ssb1-YFP signals in mitosis followed by S phase from WT and cut14-Y1 cells are shown. The YFP signals in WT were smoothly located in the nuclear chromatin region during mitosis (figure 3e) and became punctate during S phase (electronic supplementary material, movie S1). In cut14-Y1 cells, intense Ssb1 foci were observed in the nucleolar region of aberrantly elongated mitotic chromosome (figure 3f and electronic supplementary material, movies S2–S4).
After analysing a large number of movies, Ssb1-YFP foci were found in 39 per cent (24/62) of mitotic cut14-Y1 cells, but only in 4 per cent (6/151) of the WT mitotic cells (figure 3g). Movies clearly showed that the cut14-Y1 mutant cells containing foci were not delayed prior to the entry into mitosis, suggesting that the mitotic checkpoint was not activated (see below).
3.9. Ssb1 accumulated in the rDNA region of cut14-Y1, but not in the double mutant
Chromatin immunoprecipitation (ChIP) of rDNA non-coding regions was performed using chromosomally integrated Ssb1-FLAG expressed from its native promoter. Antibodies against FLAG were used for ChIP of cells cultured in the presence or absence of 4 mM HU. As shown in figure 3h, more Ssb1 bound to rDNA (primer NTS1) in cut14-Y1 cells cultured at 26°C compared with WT. In the presence of HU, the level of Ssb1 bound to rDNA increased in cut14-Y1, indicating that more Ssb1 was bound to rDNA in cut14-Y1.
To analyse the level of Ssb1-418 mutant protein that bound to rDNA, polyclonal antibodies against Ssb1 were used. As shown in figure 3i, the level of Ssb1 mutant protein bound to rDNA was diminished in the cut14-Y1 ssb1-418 double mutant when cultured at 26°C. This defect of the Ssb1-418 protein may explain why the ssb1-418 mutant suppresses the phenotypes of cut14-Y1.
3.10. Ssb1-YFP foci are also observed in cnd2-1 cells but not in mitotic cells
Non-SMC mutant cnd2-1 was also DNA damage sensitive [31]. However, the phenotypes of cnd2-1 were not rescued by ssb1-418 (figure 4a). The reason for this failure of suppression might be due to the loss of the Ssb1-YFP foci in mitotic cnd2-1 cells (described below). When the temperature of the cnd2-1 culture was shifted from 26°C to 36°C, the septation index (SI) declined rapidly owing to activation of the Cds1-dependent checkpoint [31], whereas no decrease of the SI occurred in cut14-Y1 (figure 4b). The results (explained in the legend) suggest that at 36°C, the DNA checkpoint was activated in cnd2-1, but not in cut14-Y1, and the cnd2-1 cells that entered mitosis after the delay did not contain Ssb1-YFP foci. However, the Ssb1-YFP foci were found in about 20 per cent of interphase cells of cnd2-1 (figure 4c).
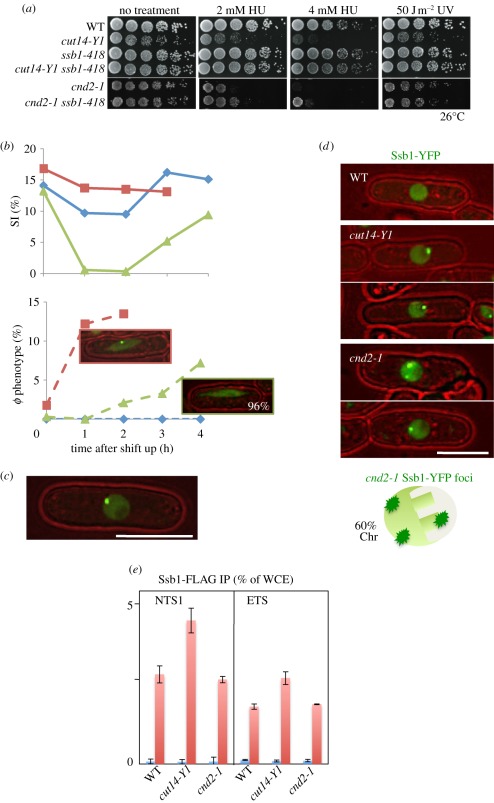
Figure 4.
The checkpoint response and nuclear localization of Ssb1-YFP differ in cnd2-1 and cut14-Y1. (a) Whereas the DNA damage sensitivities of cut14-Y1 were greatly rescued by ssb1-418 mutation, those of cnd2-1 were not. (b) WT, cut14-Y1 and cnd2-1 were cultured at 36°C for 0–4 h, and the percentage septation index (SI) and the number of cells displaying aberrant chromosome (φ phenotype) were measured. The frequency of aberrant mitotic chromosomes sharply increased in cut14-Y1 (blue diamonds with solid line, WT; red squares with solid line, cut14-Y1; green triangles with solid line, cnd2-1; top), while the appearance of such mitotic cells was delayed in cnd2-1 (blue diamonds with dashed line, WT; red squares with dashed line, cut14-Y1; green triangles with dashed line, cnd2-1; bottom). Notably, the aberrant mitotic chromosomes in cnd2-1 did not contain Ssb1 foci (96%). (c) The intense foci of Ssb1-YFP were observed in cnd2-1 cells. Note that the YFP dot (the focus) is located in the nuclear periphery chromatin region. (d) Distinct nuclear localization of the Ssb1-YFP signals in cnd2-1 and cut14-Y1. The WT cell nucleus did not show the foci of Ssb1-YFP. Two cut14-Y1 cells display the intense Ssb1-YFP foci, which are located in the nucleolar region. Two cnd2-1 cells also show the intense Ssb1-YFP foci, which are located in the non-nucleolar nuclear chromatin region. Sixty per cent of cells examined showed the nuclear chromatin localization of Ssb1-YFP foci in cnd2-1 mutant cells. (e) ChIP experiment of Ssb1-FLAG for the rDNA probe NTS1 using WT, cut14-Y1 and cnd2-1 strains. The procedures are the same employed in figure 3h. Blue bars, −antibodies; red bars, +antibodies.
The Ssb1-YFP foci were located in the non-nucleolar chromatin region of cnd2-1 mutant cells. Examples of these cells are shown in figure 4d. Consistently, ChIP of Ssb1-FLAG shows that Ssb1-FLAG is not enriched in the rDNA in cnd2-1 cells (figure 4e), in sharp contrast to the result in cut14-Y1. Taken together, Ssb1 probably binds to non-nucleolar chromosomal regions in cnd2-1, leading to the Cds1-dependent DNA damage checkpoint delay.
3.11. Purified condensin and structural maintenance of chromosomes dimer preferentially interact with ssDNA
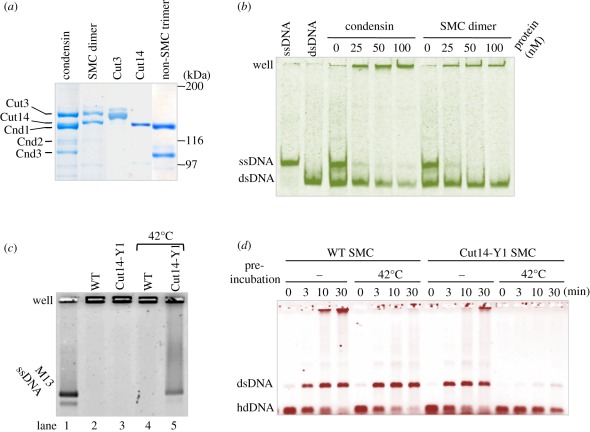
To examine DNA–protein interactions, we purified condensin protein complexes from S. pombe cells where all the condensin subunits were simultaneously overexpressed [20,21]. Protein preparations were run on a sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE) gel and stained with Coomasie brilliant blue (figure 5a).
Figure 5.
Interaction of isolated condensin and SMC dimer with different DNAs. (a) SDS-PAGE patterns of holocondensin (Cut3-Cut14-Cnd1-Cnd2-Cnd3), the SMC dimer (Cut3-Cut14) and the non-SMC trimer (Cnd1-Cnd2-Cnd3), together with single Cut3 and Cut14 as controls, stained with Coomasie brilliant blue. The procedures of isolation were previously described, and the degree of purity for these preparations was similar to those previously reported [20,21]. The Cut14 and Cnd1 overlap, and the Cnd2 band is diffuse and less intense than the other non-SMC subunits, probably owing to phosphorylation and/or degradation [9]. Limited proteolysis of Cut3 has been reported [20]. (b) Condensin and SMC dimer were incubated with a mixture of ssDNA and dsDNA, then analysed on a 10% non-denaturing acrylamide gel in the absence of SDS. DNA used was tagged with fluorescent FITC. (c) WT and mutant SMC dimer were incubated with M13 ssDNA with or without the pre-heat treatment at 42°C for 10 min, then analysed on a 0.7% native agarose gel in the absence of SDS. The mutant dimer was obtained by simultaneous overexpression of Cut3 and Cut14-Y1, and purified by affinity chromatography, stained with SYBR Gold. (d) WT and mutant SMC dimers were incubated with hdDNA with or without pre-heat treatment of the SMC dimers (see text), stained with ethidium bromide.
Holocondensin and the SMC dimer were incubated with short ssDNA (86 nucleotide long) and dsDNA (86 bp) for 10 min at 30°C. Although both bound to ssDNA and dsDNA, they bound preferentially to ssDNA, as seen in the acrylamide native gel (figure 5b). The band intensity of ssDNA decreased greatly when increasing amounts of condensin or SMC dimer were added. The SMC dimer appeared to have a stronger affinity for ssDNA than holocondensin. The associated protein–DNA complex did not produce a band shift, but instead remained in the well owing to the aggregate formation of DNA–protein [21].
3.12. Weak DNA binding and reannealing by the mutant structural maintenance of chromosomes dimer protein
We then examined whether the mutant Cut14-Y1 protein was similarly capable of binding DNA, by using a Cut3-Cut14-Y1 heterodimer. M13 phage ssDNA (7.2 kb long) was purchased (NEB) and used for the DNA–protein binding experiments. As seen in figure 5c, M13 ssDNA (producing two bands) moved to the top of the native agarose gel, which had no sodium dodecyl sulphate (SDS) added, when mixed with the WT and mutant SMC dimer (lanes 2 and 3). If the protein preparations were pre-treated with heat (42°C for 10 min [20]), M13 ssDNA associated with the WT SMC complex still remained at the top of gel (lane 4), whereas the mutant Cut3-Cut14-Y1 dimer produced smeared bands from the position of the M13 ssDNA band to the gel top (lane 5), suggesting that the complex formation of the mutant dimer on ssDNA was greatly diminished after heat treatment.
We then compared the DNA reannealing activity of WT and mutant SMC dimer with or without heat pre-treatment. As shown in figure 5d, WT and mutant (Cut14-Y1) SMC dimers with (42°C) or without (−) pre-heat treatment were incubated with heat-denatured (hd) DNA (single cut linear 3.0 kb blue script), then the reactions of hdDNA with SMC at 30°C for 0–30 min were stopped by the addition of 0.2 per cent SDS. At 0 min, only hdDNA was observed. The WT SMC dimer promoted annealing to produce dsDNA. Upon heat pre-treatment of the WT SMC dimer, a dsDNA band was also formed. Without heat treatment, the mutant SMC dimer could form dsDNA, but completely failed to reanneal hdDNA upon heat treatment.
3.13. DNA reannealing promotes the release of replication protein A bound to heat-denatured ssDNA
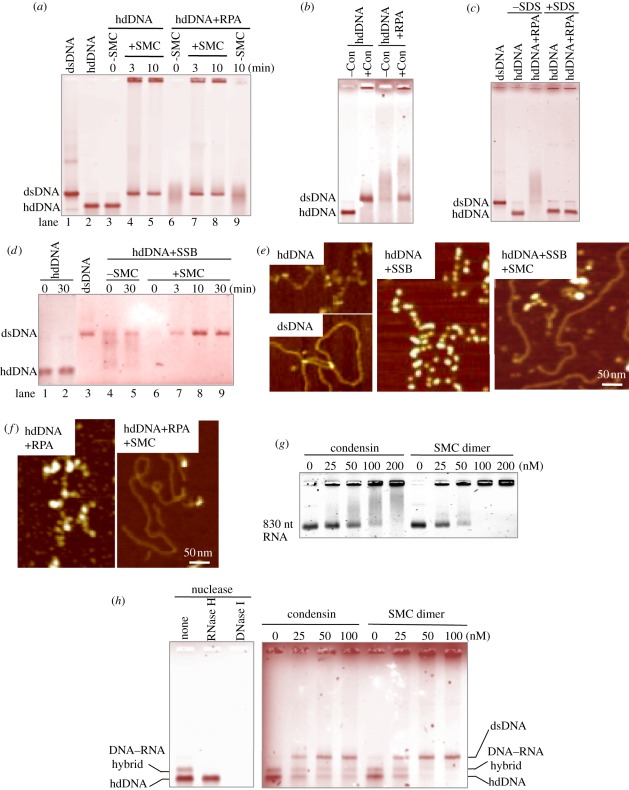
To determine whether the SMC dimer promoted reannealing in vitro when complementary ssDNAs were previously coated with purified heterotrimeric RPA [42], hdDNAs were first mixed with S. pombe heterotrimeric RPA (producing diffuse bands in the native gel) and then briefly incubated with the SMC heterodimer, yielding a duplex dsDNA band that formed within 3 min (figure 6a). Holocondensin also reannealed RPA-coated hdDNA, but less efficiently than the SMC dimer (figure 6b).
Figure 6.
Condensin SMC-mediated elimination of RPA from hdDNA. (a) SMC dimer promotes reannealing of RPA-coated hdDNA. Lanes 1,2: control ds and hdDNA; 3–5: naked hdDNA (heat denatured and then rapidly cooled) was incubated with or without SMC for 0, 3 or 10 min; 6–9: hdDNA pre-coated with RPA was further incubated with (lanes 6–8) or without (lane 9) the SMC dimers. After incubation, samples were analysed on a 0.7% native agarose gel (without SDS). (b) Holocondensin also produced dsDNA from RPA-coated hdDNA. Native agarose gel was used. (c) hdDNA incubated with RPA complex was analysed in the absence or presence of SDS. See text. (d) Lanes 1,2: hdDNA incubated alone for 0 or 30 min; 3: dsDNA; 4–9: hdDNA pre-coated with SSB for 5 min at 30°C, and further incubated for 30 min without (lanes 4,5) or with SMC for 0–30 min (lanes 6–9). The reaction mixtures were analysed by native agarose gel electrophoresis. (e) AFM images hdDNA (top left), dsDNA (bottom left), hdDNA coated with SSB (middle). SMC was added and incubated with SSB-coated hdDNA for 30 min (right). (f) AFM images of hdDNA coated with S. pombe RPA (left); SMC dimer was added and incubated with RPA-coated hdDNA for 30 min (right). (g) Condensin and SMC dimer binding to RNA that was made in electronic supplementary material, figure S5. The samples were analysed using a 4% native agarose (NuSieve) gel in the absence of SDS. (h) (left) The mixture of hdDNA and DNA–RNA hybrid was digested with DNase I or RNase H. The hybrid band was selectively digested with RNase H. (right) Condensin and SMC dimers (0–100 nM) were incubated with the mixture, and SDS was used to stop the reactions. The samples were analysed using a 0.7% agarose gel. Staining with (a–d,h) ethidium bromide and (g) SYBR Gold.
In order to firmly confirm that the diffuse bands produced by hdDNA and RPA were indeed owing to complex formation between RPA and ssDNA, and not with dsDNA, the complex formed by hdDNA (40 ng) and RPA on ice for 10 min was treated with 1 per cent SDS (+ SDS; figure 6c). When the mixture was examined in the absence of SDS (−SDS), smeared bands were observed, but not in the presence of SDS. No dsDNA formation thus occurred in this experiment.
To visualize the removal of RPA from hdDNA, atomic force microscopy (AFM) was used [4,21] (figure 6d–f). Highly purified bacterial single-strand DNA binding protein (SSB; purchased from Promega)-coated hdDNA was incubated with the S. pombe SMC dimer. Within 10 min, a sharp dsDNA band was formed (figure 6d). AFM images of hdDNA before and after the addition of SSB, followed by the addition of the SMC dimer, are shown in figure 6e. Beaded ssDNA coated with SSB was observed for hdDNA mixed with SSB (middle), while dsDNA was plentifully produced 30 min after the incubation with SMC dimer (right; control hdDNA and dsDNA images, left top and bottom, respectively).
Purified S. pombe RPA was also examined by AFM (figure 6f, left). Coated ssDNA similar to that produced by bacterial SSB was observed. Thirty minutes after the incubation with SMC dimer, dsDNA was plentifully observed (figure 6f, right). Taken together, our data show that condensin SMC promotes DNA reannealing and releases bacterial SSB or S. pombe RPA bound to complementary ssDNA.
3.14. Interactions of condensin structural maintenance of chromosomes to RNA and RNA–DNA hybrid
To substantiate these findings in a broader physiological context, condensin binding to RNA was examined. As shown in figure 6g, both condensin and the SMC dimer were bound to 830 nt RNA (electronic supplementary material, figure S5) and formed a complex that did not enter the gel, though the SMC dimer was bound to RNA more efficiently. To our knowledge, this is the first demonstration of the interaction of RNA with condensin SMC.
An RNA–DNA hybrid was then constructed by mixing transcribed RNA with hdDNA (described in electronic supplementary material, figure S5) and then adding the SMC dimer or condensin. RNase H was used to verify the hybrid. The SMC dimer reduced the amount of the RNA–DNA hybrid and increased the level of dsDNA, suggesting that condensin SMC might interact with RNA–DNA hybrid and remove RNA, followed by the formation of dsDNA (figure 6h; SDS was added before electrophoresis). Another interpretation might be possible, such that condensin SMC might interact with the RNA–DNA hybrid to form the higher-order aggregate.
3.15. Ssb1-418 mutant protein exhibits diminished ssDNA binding
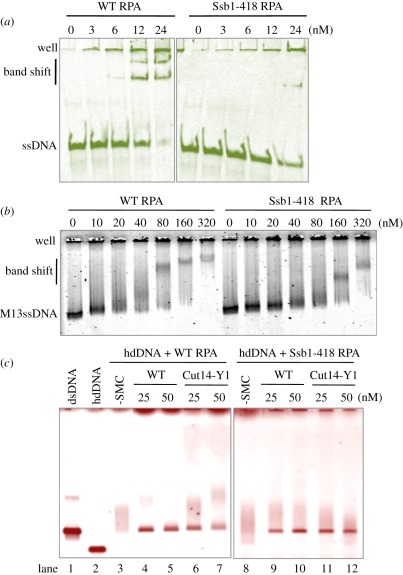
To further examine why ssb1-418 rescued the phenotypes of cut14-Y1, we characterized the properties of the mutant Ssb1 protein. ssDNA binding was tested using short and long ssDNAs. The heterotrimeric RPA containing the Ssb1-418 protein was purified and mixed with ssDNAs. Binding of the mutant RPA to short 86 nt ssDNA was greatly diminished (figure 7a), whereas the binding to long 7.2 kb M13 ssDNA was only slightly diminished (figure 7b) in comparison with WT RPA.
Figure 7.
Interaction of mutant RPA with DNA and the mutant SMC dimer. (a,b) Interaction of WT and mutant RPA complexes with (a) short and (b) long ssDNA. The heterotrimeric RPA that contained the Ssb1-418 mutant protein was purified and mixed with (a) short 86 nt ssDNA and (b) long M13 ssDNA, followed by (a) native acrylamide and (b) native agarose gel electrophoresis (in the absence of SDS). Binding of the mutant RPA to short 86 nt ssDNA was greatly diminished, whereas the binding to M13 ssDNA was only slightly diminished. (c) WT and mutant RPA (80 nM) were bound to heat-denatured hdDNA for 5 min on ice, followed by the addition of WT and mutant SMC dimer-containing Cut14-Y1 (0, 25, 50 nM) for the reannealing reaction at 30°C for 30 min. Resulting reaction mixtures were analysed using 0.7% native agarose gels and stained with ethidium bromide. Diffuse bands represented hdDNA coated with RPA, which formed with the WT and mutant RPA. The ability of mutant SMC dimer (Cut14-Y1) for reannealing was diminished for hdDNA precoated with the WT RPA, whereas the reannealing went equally well when the mutant RPA previously coated hdDNA. Staining with (a) FITC, (b) SYBR Gold and (c) ethidium bromide.
We then examined whether cut14-Y1 mutant SMC-mediated RPA removal from ssDNA differed when the WT or mutant RPA was employed. As shown in figure 7c (lane 3–7), the WT SMC dimer was more efficient than the mutant SMC dimer for the removal of WT RPA from ssDNA by reannealing. In contrast, when the mutant RPA was employed (lane 8–12), the removal of the mutant RPA from ssDNA was equally efficient by reannealing promoted by the WT and mutant SMC dimer. These results may explain the genetical rescue of the double mutant cut14-Y1 ssb1-418.
3.16. Intragenic suppressor mutations of cut14-Y1
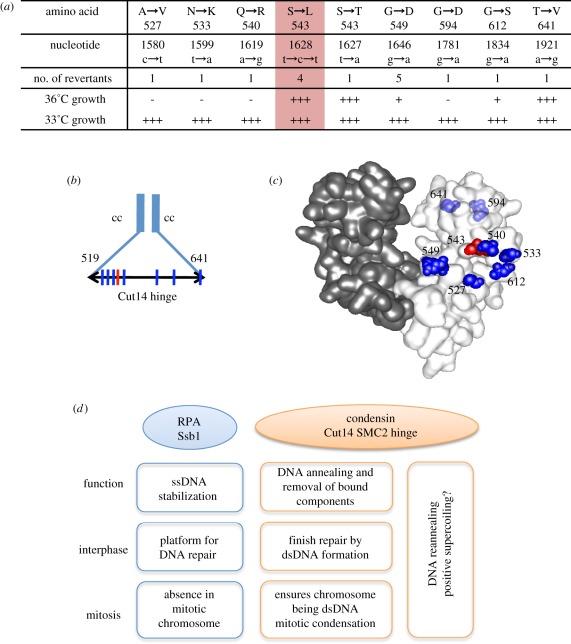
We attempted to isolate extragenic suppressors of cut14-Y1, and obtained 16 Ts+ (at 33°C) suppressors of cut14-Y1. Surprisingly, the mutations all mapped to within the cut14 locus and were all located within the hinge domain (519–641aa; figure 8a,b). Four were true revertants (S543L) and formed colonies at 36°C. One suppressor also contained a substitution at the same residue (S543T) but, curiously, behaved like the WT. Eleven contained a second site mutation in addition to the original L543S mutation. Based on the three-dimensional structure [30], the additional mutation sites (blue) surrounded the L543 site, but resided largely out of the dimerization domain (red; figure 8c). These suppressor mutations appeared to restore the DNA binding and reannealing activity of the hinge. The hinge may thus be a flexible structure in which deleterious mutations can be compensated by the presence of the additional mutations within the hinge domain.
Figure 8.
The hinge of SMC2/Cut14 is a functional entity. (a) Mapping of pseudo-revertants of cut14-Y1 that formed colonies at 33°C. The four mutants are true revertants (S543L, red column) that formed the normal colonies at 36°C. See text. (b) The mutation sites are shown by the vertical lines. The hinge region is between the residues 519 and 641. cc, coiled-coil. (c) Location of the mutation sites in the three-dimensional structure of the mouse condensin hinge [30]. The amino acid residue number is adapted for the S. pombe Cut14. The original cut14-Y1 mutation site is indicated by the red colour, while the second suppressing mutation sites are shown by blue. The residues 594 and 641 situating behind are faded. (d) A diagram depicting the relationship between the condensin SMC Cut14 hinge and Ssb1. Condensin preferentially binds to ssDNA [30,34] and promotes annealing to complementary ssDNA in vitro, and appears to oppose the action of RPA. RPA acts as a platform for various proteins involved in DNA metabolism, such as damage repair and replication through ssDNA stabilization [44–46]. The role of condensin in damage repair remains unclear, but we propose that it may be required for completing/exiting repair processes by removing RPA and forming dsDNA through reannealing.
4. Discussion
Understanding the mechanisms of how condensin functions in mitosis and DNA repair is of great importance. We identified a fission yeast ts mutant cut14-Y1 that contains a point mutation in the hinge domain of SMC2, a subunit of condensin. This mutant displayed severely reduced viability during mitosis at elevated temperatures and after exposure to DNA damage agents at the permissive temperature. Repair of UV-induced DNA damage is defective. Other phenotypes include a mitotic segregation defect and an inability to reanneal heat-denatured complementary DNAs (hdDNA). Further genetic analysis showed that a mutant of the large subunit of RPA (ssb1-418) rescued almost all the cut14-Y1 phenotypes observed.
Strikingly, we found that nucleolar RPA foci formation and Ssb1 binding to rDNA in cut14-Y1 were largely reduced in the cut14-Y1 ssb1-418 double mutant. In vitro assays demonstrated that RPA was removed by SMC-promoted reannealing of hdDNA. We propose that condensin may compete with RPA-induced DNA strand separation by re-annealing separate ssDNA strands after DNA repair and prior to mitosis in order to unload chromosomal components. We speculate that the regulation of the non-SMC trimer by Aurora kinase B [9–11] or its interaction with histone H2A [47] during mitosis may be important for the activation of its reannealing activity. In vitro, the non-SMC trimer has been shown to inhibit DNA reannealing [21]. Non-SMC trimer may thus be negatively regulated by Aurora kinase B during mitosis.
We previously showed that the non-SMC condensin cnd2-1 mutant was sensitive to DNA damage and exhibited cell cycle delays owing to DNA repair [31], but it remained possible that Cnd2 might have a role unrelated to condensin. Our present data provide additional convincing evidence supporting the idea that condensin is required for the repair of DNA damage. We also show that the condensin Cut14/SMC2 hinge may be required for damage checkpoint activation. Consistently, the damage checkpoint activation occurs in cnd2-1 that contains the WT SMC2 hinge.
Our in vitro results showed that the SMC dimer and holocondensin promote the removal of DNA-bound RPA through reannealing. This is consistent with our in vivo results demonstrating that the Ssb1 foci observed by immunofluorescence and live cell imaging are produced in the cut14-Y1 hinge mutant cells, which contain aberrant mitotic chromosomes. The striking rescue of cut14-Y1 by ssb1-418 suggested that the weakened Ssb1 activity alleviated the defects generated by the Cut14 hinge mutation. Indeed, results from ChIP and in vitro reannealing assays suggested that RPA containing the mutant Ssb1-418 subunit may be removed by mutant condensin. As shown in figure 8d, we speculate that Ssb1 and Cut14 coordinate to regulate the dynamics of ssDNA stabilization.
The damage repair defect in cnd2-1 activates damage checkpoint in a Cds1-dependent manner, and delays mitotic entry [31], whereas the damage in cut14-Y1 is slowly repaired and does not seem to activate the checkpoint. It is possible that the SMC2 hinge may act in parallel with the damage checkpoint. Ssb1 formed intense nucleolar foci in cut14-Y1, which may not be recognized by the DNA damage checkpoint and may also not be removed prior to mitosis owing to the condensin defect. By contrast, cnd2-1 formed similarly intense Ssb1 foci, but the foci were located in non-nucleolar regions. Additionally, the Ssb1-YFP foci did not remain in the aberrant chromosomes of mitotic cells after the delay. This striking difference is consistent with the failure of the rescue of cnd2-1 by ssb1-418.
The regulation of the positive supercoiling activity [48] of condensin by protein kinases such as Cdk1 [49], Aurora B and polo kinases [50], and its relationship to DNA topoisomerase II [51], have been extensively studied. These studies emphasized the role of condensin in chromatin packing, though the mechanism of how the complex functions has not been elucidated. In this study, we present evidence suggesting that condensin may have a function in clearing DNA [19]. The transition from fuzzy chromatin in the interphase nucleus to compacted mitotic chromosomes may require the removal of interphase protein components such as RPA. The SMC hinge appears to be critical for this transition. Cti1/C1D, which directly interacts with the condensin Cut3/SMC4 hinge and rescues cnd2-1 [52], is required for RNA degradation [53]. Previous structural and mutational analyses showed that the hinge is required for DNA binding and dimerization [5,30,35]. It has been suggested that the cohesin hinge may open upon DNA binding [54–56]. Based on our results, the hinge appears to be important for binding with DNA and RNA, DNA annealing, damage repair, elimination of protein and RNA from DNA, and chromosome condensation. Thus, uncovering how the condensin hinge functions may be key to understanding the many roles of condensin.
RPA and condensin appear to antagonize each other, but a balance in their activities may be important for coordinating the dynamics of strand separation. Prior to mitosis, RPA may need to be excluded from chromosomes, and condensin may remove RPA or possibly other components such as transcribed RNA to ensure that mitotic chromosomes are properly formed and maintained. The promotion of DNA reannealing by condensin SMC involves the winding up of DNA strands, which is similar to the formation of positive supercoiling (figure 8d). Condensin may supercoil DNA strands to unload chromosomal components after DNA repair and prior to mitosis. Our present finding that condensin winds up DNA to clear the DNA-bound components may provide a physiological meaning of SMC-mediated DNA reannealing. Instead, unwinding the double-stranded DNA leads to the formation of negative supercoil, which results in the association of transcription machinery and replication apparatus to unwound chromosomal DNA regions. This hypothesis will require further investigation.
5. Material and methods
5.1. Isolation of cut14-Y1 and ssb1-418
These two strains were isolated by screening the S. pombe ts strain collection made by random mutagenesis using procedures similar to those described previously [57]. cut14-Y1 was isolated owing to its cytological defect in mitotic condensation, while ssb1-418 was rescued by plasmids carrying the ssb1+ gene. Gene cloning and tetrad dissection confirmed the tight genetic linkage of the cut14-Y1 ts phenotype to the cut14 locus, whereas ssb1-418 is caused by a mutation in the ssb1 locus. The mutation sites of both cut14-Y1 and ssb1-418 (G78E) were determined by nucleotide sequencing of the mutant genes isolated by PCR.
5.2. Media, strains and plasmids
Yeast extract, polypeptone, d-glucose (YPD), sporulation agar (SPA) sporulation and minimal Edinburgh minimal medium 2 (EMM2) media were used for culturing S. pombe cells. The pre-replicative G0 cells made under nitrogen starvation conditions were replenished by the addition of 0.5 per cent NH4Cl [32]. The chromosomally integrated strain of YFP-tagged Ssb1 strain expressed under the native promoter was made as described [28]. The Sad1-mCherry was used as the spindle pole body (SPB) marker [58]. Plasmids used in this study are shown in electronic supplementary material, table S1.
5.3. Visualization of mutation sites in the three-dimensional structure
The images of mutation sites in the three-dimensional structure were made using molecular structure visualization software (MolFeat; FiatLux). The mouse SMC2-4 structure [30] was obtained from the RCSB Protein Data Bank (http://www.pdb.org/pdb/home/home.do).
5.4. DNA damage sensitivity, ultraviolet irradiation and thymine dimer detection
The procedures for UV irradiation, HU and other drug sensitivity measurements, and the detection of thymine dimers, were previously described [31].
5.5. Immunofluorescence microscopy and chip
The procedures for DAPI staining and immunofluorescence microscopy were previously described [58]. The anti-Ssb1 antibody was previously made and characterized [42]. The ChIP method was performed as previously described [58], with slight modifications. Immunoprecipitation was performed using anti-FLAG M2 antibody (Sigma-Aldrich) or anti-Ssb1 antibody. Real-time PCR was performed on the Exicycler (Bioneer). The PCR primers used were previously described [58].
5.6. Live cell analysis
Live cell analysis (figure 3e–g; electronic supplementary material, movies S1–S4) was performed as previously described [58]. In brief, S. pombe cells were cultured at 26°C in EMM2 medium and were shifted to 30°C for the appropriate duration. Cells were transferred to a glass-bottomed dish (IWAKI Glass) coated with concanavalin A (Wako) before being examined under the microscope (DeltaVision; Applied Precision). Time-lapse images were recorded by three-dimensional microscopy using the DeltaVision system. For imaging of the Ssb1-YFP with Sad1-mCherry, three optical sections were collected at 0.5 min intervals at 30°C. The vertical separations between these sections were 0.5 µm. Image projection and deconvolution were performed using an imaging workstation (SoftWoRx; Applied Precision). Video images (electronic supplementary material, movies S1–S4) were taken at 0.5 min intervals. The display speed is 3 frames s−1.
5.7. Protein purification and DNA annealing assay
Schizosaccharomyces pombe single condensin subunits, heteropentameric holocondensin, dimer or trimer subcomplexes [20,21] and the heterotrimeric RPA complex [42] were purified as described. Bacterial SSB was purchased from Promega. DNA annealing assay (in the absence of RPA or SSB) was performed as described [20]: the reactions were stopped by the addition of SDS (final 0.2%). For the protein (RPA or SSB) elimination assay, 40 nM E. coli SSB or 40 nM S. pombe heterotrimeric RPA was used for the precoating of hdDNA (linearized and heat-denatured Bluescript plasmid), and incubated with hdDNA on ice for 10 min. Condensin or the SMC dimer was then added. The annealing reaction was terminated with loading dye in the absence of SDS followed by electrophoresis. For the RNA elimination assay, 830 nt RNA was made by in vitro transcription T7 (TaKaRa). See electronic supplementary material, figure S5.
5.8. Gel shift assay
Synthetic 86 nt ssDNA [59] labelled with fluorescein isothiocyanate (FITC; Sigma-Aldrich) was incubated with a series of concentrations of condensin, the SMC dimer or individual subunits in 20 µl of binding buffer (20 mM Tris–HCl at pH 7.5, 50 mM NaCl, 2 mM MgCl2, 10 per cent glycerol, 1 mM dithiothreitol) for 10 min at 30°C. The mixtures were then analysed in 10 per cent non-denaturing polyacrylamide gels made in 0.5 × TBE (44.5 mM Tris-borate, 1 mM EDTA) buffer, followed by fluorescent imaging (Typhoon9200). The same conditions were used for M13 ssDNA, but the samples were analysed in 0.7 per cent native agarose gels, followed by SYBR Gold staining (Molecular Probes). For RNA-binding assay, 4 per cent agarose (the NuSieve 3:1, TaKaRa) gel was used.
5.9. Atomic force microscopy imaging
6. Acknowledgements
We are grateful to M. Foiani, D. Branzei, P. Russell, F. Esashi, K. Takeda, S. Saitoh and T. Hirano for their help, comments and encouragement. A.Y. thanks Prof. T. Matsumoto for hospitality in his laboratory. The present work was supported by research grants from the Japan Science and Technology Corporation (JST, CREST) and Initial Research Project for OIST, and from the Ministry of Education, Science, Culture and Sports (COE). The authors declare no competing financial interests.
References
- 1.Wood AJ, Severson AF, Meyer BJ. 2010. Condensin and cohesin complexity: the expanding repertoire of functions. Nat. Rev. Genet. 11, 391–404 10.1038/nrg2794 (doi:10.1038/nrg2794) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hudson DF, Marshall KM, Earnshaw WC. 2009. Condensin: architect of mitotic chromosomes. Chromosome Res. 17, 131–144 10.1007/s10577-008-9009-7 (doi:10.1007/s10577-008-9009-7) [DOI] [PubMed] [Google Scholar]
- 3.Anderson DE, Losada A, Erickson HP, Hirano T. 2002. Condensin and cohesin display different arm conformations with characteristic hinge angles. J. Cell Biol. 156, 419–424 10.1083/jcb.200111002 (doi:10.1083/jcb.200111002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yoshimura SH, Hizume K, Murakami A, Sutani T, Takeyasu K, Yanagida M. 2002. Condensin architecture and interaction with DNA: regulatory non-SMC subunits bind to the head of SMC heterodimer. Curr. Biol. 12, 508–513 10.1016/S0960-9822(02)00719-4 (doi:10.1016/S0960-9822(02)00719-4) [DOI] [PubMed] [Google Scholar]
- 5.Hirano M, Hirano T. 2002. Hinge-mediated dimerization of SMC protein is essential for its dynamic interaction with DNA. EMBO J. 21, 5733–5744 10.1093/emboj/cdf575 (doi:10.1093/emboj/cdf575) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cuylen S, Haering CH. 2011. Deciphering condensin action during chromosome segregation. Trends Cell Biol. 21, 552–559 10.1016/j.tcb.2011.06.003 (doi:10.1016/j.tcb.2011.06.003) [DOI] [PubMed] [Google Scholar]
- 7.Hirano T. 2006. At the heart of the chromosome: SMC proteins in action. Nat. Rev. Mol. Cell Biol. 7, 311–322 10.1038/nrm1909 (doi:10.1038/nrm1909) [DOI] [PubMed] [Google Scholar]
- 8.Nasmyth K, Haering CH. 2005. The structure and function of SMC and kleisin complexes. Annu. Rev. Biochem. 74, 595–648 10.1146/annurev.biochem.74.082803.133219 (doi:10.1146/annurev.biochem.74.082803.133219) [DOI] [PubMed] [Google Scholar]
- 9.Nakazawa N, Mehrotra R, Ebe M, Yanagida M. 2011. Condensin phosphorylated by the Aurora-B-like kinase Ark1 is continuously required until telophase in a mode distinct from Top2. J. Cell Sci. 124, 1795–1807 10.1242/jcs.078733 (doi:10.1242/jcs.078733) [DOI] [PubMed] [Google Scholar]
- 10.Lavoie BD, Hogan E, Koshland D. 2004. In vivo requirements for rDNA chromosome condensation reveal two cell-cycle-regulated pathways for mitotic chromosome folding. Genes Dev. 18, 76–87 10.1101/gad.1150404 (doi:10.1101/gad.1150404) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lipp JJ, Hirota T, Poser I, Peters JM. 2007. Aurora B controls the association of condensin I but not condensin II with mitotic chromosomes. J. Cell Sci. 120, 1245–1255 10.1242/jcs.03425 (doi:10.1242/jcs.03425) [DOI] [PubMed] [Google Scholar]
- 12.Takemoto A, Murayama A, Katano M, Urano T, Furukawa K, Yokoyama S, Yanagisawa J, Hanaoka F, Kimura K. 2007. Analysis of the role of Aurora B on the chromosomal targeting of condensin I. Nucleic Acids Res. 35, 2403–2412 10.1093/nar/gkm157 (doi:10.1093/nar/gkm157) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giet R, Glover DM. 2001. Drosophila aurora B kinase is required for histone H3 phosphorylation and condensin recruitment during chromosome condensation and to organize the central spindle during cytokinesis. J. Cell Biol. 152, 669–682 10.1083/jcb.152.4.669 (doi:10.1083/jcb.152.4.669) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ono T, Fang Y, Spector DL, Hirano T. 2004. Spatial and temporal regulation of Condensins I and II in mitotic chromosome assembly in human cells. Mol. Biol. Cell 15, 3296–3308 10.1091/mbc.E04-03-0242 (doi:10.1091/mbc.E04-03-0242) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koshland D, Strunnikov A. 1996. Mitotic chromosome condensation. Annu. Rev. Cell Dev. Biol. 12, 305–333 10.1146/annurev.cellbio.12.1.305 (doi:10.1146/annurev.cellbio.12.1.305) [DOI] [PubMed] [Google Scholar]
- 16.Hirano T. 2005. Condensins: organizing and segregating the genome. Curr. Biol. 15, R265–R275 10.1016/j.cub.2005.03.037 (doi:10.1016/j.cub.2005.03.037) [DOI] [PubMed] [Google Scholar]
- 17.Jessberger R, Frei C, Gasser SM. 1998. Chromosome dynamics: the SMC protein family. Curr. Opin. Genet Dev. 8, 254–259 10.1016/S0959-437X(98)80149-4 (doi:10.1016/S0959-437X(98)80149-4) [DOI] [PubMed] [Google Scholar]
- 18.Meyer BJ. 2010. Targeting X chromosomes for repression. Curr. Opin. Genet. Dev. 20, 179–189 10.1016/j.gde.2010.03.008 (doi:10.1016/j.gde.2010.03.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yanagida M. 2009. Clearing the way for mitosis: is cohesin a target? Nat. Rev. Mol. Cell Biol. 10, 489–496 10.1038/nrm2712 (doi:10.1038/nrm2712) [DOI] [PubMed] [Google Scholar]
- 20.Sutani T, Yanagida M. 1997. DNA renaturation activity of the SMC complex implicated in chromosome condensation. Nature 388, 798–801 10.1038/42062 (doi:10.1038/42062) [DOI] [PubMed] [Google Scholar]
- 21.Sakai A, Hizume K, Sutani T, Takeyasu K, Yanagida M. 2003. Condensin but not cohesin SMC heterodimer induces DNA reannealing through protein-protein assembly. EMBO J. 22, 2764–2775 10.1093/emboj/cdg247 (doi:10.1093/emboj/cdg247) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Flynn RL, Zou L. 2010. Oligonucleotide/oligosaccharide-binding fold proteins: a growing family of genome guardians. Crit. Rev. Biochem. Mol. Biol. 45, 266–275 10.3109/10409238.2010.488216 (doi:10.3109/10409238.2010.488216) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fanning E, Klimovich V, Nager AR. 2006. A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res. 34, 4126–4137 10.1093/nar/gkl550 (doi:10.1093/nar/gkl550) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wold MS. 1997. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem. 66, 61–92 10.1146/annurev.biochem.66.1.61 (doi:10.1146/annurev.biochem.66.1.61) [DOI] [PubMed] [Google Scholar]
- 25.Ciccia A, Elledge SJ. 2010. The DNA damage response: making it safe to play with knives. Mol. Cell 40, 179–204 10.1016/j.molcel.2010.09.019 (doi:10.1016/j.molcel.2010.09.019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Uemura T, Ohkura H, Adachi Y, Morino K, Shiozaki K, Yanagida M. 1987. DNA topoisomerase II is required for condensation and separation of mitotic chromosomes in S. pombe. Cell 50, 917–925 10.1016/0092-8674(87)90518-6 (doi:10.1016/0092-8674(87)90518-6) [DOI] [PubMed] [Google Scholar]
- 27.Matsusaka T, Imamoto N, Yoneda Y, Yanagida M. 1998. Mutations in fission yeast Cut15, an importin alpha homolog, lead to mitotic progression without chromosome condensation. Curr. Biol. 8, 1031–1034 10.1016/S0960-9822(07)00425-3 (doi:10.1016/S0960-9822(07)00425-3) [DOI] [PubMed] [Google Scholar]
- 28.Sutani T, Yuasa T, Tomonaga T, Dohmae N, Takio K, Yanagida M. 1999. Fission yeast condensin complex: essential roles of non-SMC subunits for condensation and Cdc2 phosphorylation of Cut3/SMC4. Genes Dev. 13, 2271–2283 10.1101/gad.13.17.2271 (doi:10.1101/gad.13.17.2271) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saka Y, Sutani T, Yamashita Y, Saitoh S, Takeuchi M, Nakaseko Y, Yanagida M. 1994. Fission yeast cut3 and cut14, members of a ubiquitous protein family, are required for chromosome condensation and segregation in mitosis. EMBO J. 13, 4938–4952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Griese JJ, Witte G, Hopfner KP. 2010. Structure and DNA binding activity of the mouse condensin hinge domain highlight common and diverse features of SMC proteins. Nucleic Acids Res. 38, 3454–3465 10.1093/nar/gkq038 (doi:10.1093/nar/gkq038) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aono N, Sutani T, Tomonaga T, Mochida S, Yanagida M. 2002. Cnd2 has dual roles in mitotic condensation and interphase. Nature 417, 197–202 10.1038/417197a (doi:10.1038/417197a) [DOI] [PubMed] [Google Scholar]
- 32.Su SS, Tanaka Y, Samejima I, Tanaka K, Yanagida M. 1996. A nitrogen starvation-induced dormant G0 state in fission yeast: the establishment from uncommitted G1 state and its delay for return to proliferation. J. Cell Sci. 109, 1347–1357 [DOI] [PubMed] [Google Scholar]
- 33.Ku B, Lim JH, Shin HC, Shin SY, Oh BH. 2010. Crystal structure of the MukB hinge domain with coiled-coil stretches and its functional implications. Proteins 78, 1483–1490 [DOI] [PubMed] [Google Scholar]
- 34.Hirano M, Hirano T. 1998. ATP-dependent aggregation of single-stranded DNA by a bacterial SMC homodimer. EMBO J. 17, 7139–7148 10.1093/emboj/17.23.7139 (doi:10.1093/emboj/17.23.7139) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hirano M, Hirano T. 2006. Opening closed arms: long-distance activation of SMC ATPase by hinge–DNA interactions. Mol. Cell 21, 175–186 10.1016/j.molcel.2005.11.026 (doi:10.1016/j.molcel.2005.11.026) [DOI] [PubMed] [Google Scholar]
- 36.Binz SK, Sheehan AM, Wold MS. 2004. Replication protein A phosphorylation and the cellular response to DNA damage. DNA Repair (Amst) 3, 1015–1024 10.1016/j.dnarep.2004.03.028 (doi:10.1016/j.dnarep.2004.03.028) [DOI] [PubMed] [Google Scholar]
- 37.Broderick S, Rehmet K, Concannon C, Nasheuer HP. 2010. Eukaryotic single-stranded DNA binding proteins: central factors in genome stability. Subcell. Biochem. 50, 143–163 10.1007/978-90-481-3471-7_8 (doi:10.1007/978-90-481-3471-7_8) [DOI] [PubMed] [Google Scholar]
- 38.McGowan CH, Russell P. 2004. The DNA damage response: sensing and signaling. Curr. Opin. Cell. Biol. 16, 629–633 10.1016/j.ceb.2004.09.005 (doi:10.1016/j.ceb.2004.09.005) [DOI] [PubMed] [Google Scholar]
- 39.Noguchi E, Noguchi C, Du LL, Russell P. 2003. Swi1 prevents replication fork collapse and controls checkpoint kinase Cds1. Mol. Cell. Biol. 23, 7861–7874 10.1128/MCB.23.21.7861-7874.2003 (doi:10.1128/MCB.23.21.7861-7874.2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yasui A, McCready SJ. 1998. Alternative repair pathways for UV-induced DNA damage. Bioessays 20, 291–297 (doi:10.1002/(SICI)1521-1878(199804)20:4<291::AID-BIES5>3.0.CO;2-T) [DOI] [PubMed] [Google Scholar]
- 41.Binz SK, Wold MS. 2008. Regulatory functions of the N-terminal domain of the 70-kDa subunit of replication protein A (RPA). J. Biol. Chem. 283, 21 559–21 570 10.1074/jbc.M802450200 (doi:10.1074/jbc.M802450200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haruta N, Kurokawa Y, Murayama Y, Akamatsu Y, Unzai S, Tsutsui Y, Iwasaki H. 2006. The Swi5-Sfr1 complex stimulates Rhp51/Rad51- and Dmc1-mediated DNA strand exchange in vitro. Nat. Struct. Mol. Biol. 13, 823–830 10.1038/nsmb1136 (doi:10.1038/nsmb1136) [DOI] [PubMed] [Google Scholar]
- 43.Uzawa S, Yanagida M. 1992. Visualization of centromeric and nucleolar DNA in fission yeast by fluorescence in situhybridization. J. Cell. Sci. 101, 267–275 [DOI] [PubMed] [Google Scholar]
- 44.Flynn RL, Zou L. 2011. ATR: a master conductor of cellular responses to DNA replication stress. Trends Biochem. Sci. 36, 133–140 10.1016/j.tibs.2010.09.005 (doi:10.1016/j.tibs.2010.09.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zou L, Elledge SJ. 2003. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300, 1542–1548 10.1126/science.1083430 (doi:10.1126/science.1083430) [DOI] [PubMed] [Google Scholar]
- 46.Falck J, Coates J, Jackson SP. 2005. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature 434, 605–611 10.1038/nature03442 (doi:10.1038/nature03442) [DOI] [PubMed] [Google Scholar]
- 47.Tada K, Susumu H, Sakuno T, Watanabe Y. 2011. Condensin association with histone H2A shapes mitotic chromosomes. Nature 474, 477–483 10.1038/nature10179 (doi:10.1038/nature10179) [DOI] [PubMed] [Google Scholar]
- 48.Kimura K, Hirano T. 1997. ATP-dependent positive supercoiling of DNA by 13S condensin: a biochemical implication for chromosome condensation. Cell 90, 625–634 10.1016/S0092-8674(00)80524-3 (doi:10.1016/S0092-8674(00)80524-3) [DOI] [PubMed] [Google Scholar]
- 49.Kimura K, Hirano M, Kobayashi R, Hirano T. 1998. Phosphorylation and activation of 13S condensin by Cdc2 in vitro. Science 282, 487–490 10.1126/science.282.5388.487 (doi:10.1126/science.282.5388.487) [DOI] [PubMed] [Google Scholar]
- 50.St-Pierre J, Douziech M, Bazile F, Pascariu M, Bonneil E, Sauvé V, Ratsima H, D'Amours D. 2009. Polo kinase regulates mitotic chromosome condensation by hyperactivation of condensin DNA supercoiling activity. Mol. Cell 34, 416–426 10.1016/j.molcel.2009.04.013 (doi:10.1016/j.molcel.2009.04.013) [DOI] [PubMed] [Google Scholar]
- 51.Baxter J, Sen N, Martinez VL, De Carandini ME, Schvartzman JB, Diffley JFX, Aragon L. 2011. Positive supercoiling of mitotic DNA drives decatenation by topoisomerase II in eukaryotes. Science 331, 1328–1332 10.1126/science.1201538 (doi:10.1126/science.1201538) [DOI] [PubMed] [Google Scholar]
- 52.Chen ES, Sutani T, Yanagida M. 2004. Cti1/C1D interacts with condensin SMC hinge and supports the DNA repair function of condensin. Proc. Natl Acad. Sci. USA 101, 8078–8083 10.1073/pnas.0307976101 (doi:10.1073/pnas.0307976101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mitchell P. 2010. Rrp47 and the function of the Sas10/C1D domain. Biochem. Soc. Trans. 38, 1088–1092 10.1042/BST0381088 (doi:10.1042/BST0381088) [DOI] [PubMed] [Google Scholar]
- 54.Haering CH, Lowe J, Hochwagen A, Nasmyth K. 2002. Molecular architecture of SMC proteins and the yeast cohesin complex. Mol. Cell 9, 773–788 10.1016/S1097-2765(02)00515-4 (doi:10.1016/S1097-2765(02)00515-4) [DOI] [PubMed] [Google Scholar]
- 55.Gruber S, Arumugam P, Katou Y, Kuglitsch D, Helmhart W, Shirahige K, Nasmyth K. 2006. Evidence that loading of cohesin onto chromosomes involves opening of its SMC hinge. Cell 127, 523–537 10.1016/j.cell.2006.08.048 (doi:10.1016/j.cell.2006.08.048) [DOI] [PubMed] [Google Scholar]
- 56.Shintomi K, Hirano T. 2007. How are cohesin rings opened and closed? Trends Biochem. Sci. 32, 154–157 10.1016/j.tibs.2007.02.002 (doi:10.1016/j.tibs.2007.02.002) [DOI] [PubMed] [Google Scholar]
- 57.Hayashi T, Fujita Y, Iwasaki O, Adachi Y, Takahashi K, Yanagida M. 2004. Mis16 and Mis18 are required for CENP-A loading and histone deacetylation at centromeres. Cell 118, 715–729 10.1016/j.cell.2004.09.002 (doi:10.1016/j.cell.2004.09.002) [DOI] [PubMed] [Google Scholar]
- 58.Nakazawa N, Nakamura T, Kokubu A, Ebe M, Nagao K, Yanagida M. 2008. Dissection of the essential steps for condensin accumulation at kinetochores and rDNAs during fission yeast mitosis. J. Cell. Biol. 180, 1115–1131 10.1083/jcb.200708170 (doi:10.1083/jcb.200708170) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yusufzai T, Kadonaga JT. 2008. HARP is an ATP-driven annealing helicase. Science 322, 748–750 10.1126/science.1161233 (doi:10.1126/science.1161233) [DOI] [PMC free article] [PubMed] [Google Scholar]