Abstract
Multi-cellular organisms need to successfully link cell growth and metabolism to environmental cues during development. Insulin receptor–target of rapamycin (InR–TOR) signalling is a highly conserved pathway that mediates this link. Herein, we describe poly, an essential gene in Drosophila that mediates InR–TOR signalling. Loss of poly results in lethality at the third instar larval stage, but only after a stage of extreme larval longevity. Analysis in Drosophila demonstrates that Poly and InR interact and that poly mutants show an overall decrease in InR–TOR signalling, as evidenced by decreased phosphorylation of Akt, S6K and 4E-BP. Metabolism is altered in poly mutants, as revealed by microarray expression analysis and a decreased triglyceride : protein ratio in mutant animals. Intriguingly, the cellular distribution of Poly is dependent on insulin stimulation in both Drosophila and human cells, moving to the nucleus with insulin treatment, consistent with a role in InR–TOR signalling. Together, these data reveal that Poly is a novel, conserved (from flies to humans) mediator of InR signalling that promotes an increase in cell growth and metabolism. Furthermore, homology to small subunits of Elongator demonstrates a novel, unexpected role for this complex in insulin signalling.
Keywords: cell growth, nucleus, signalling, insulin
2. Introduction
Multi-cellular organisms have evolved mechanisms to link cellular metabolism and growth to external environmental cues, such as nutrient and growth factor levels, in order to survive and adapt to fluctuations in the availability of these factors. The InR–TOR pathway is one of the key regulators of cellular energy homeostasis and growth [1], and this signalling pathway is evolutionarily conserved among metazoa [1,2]. Thus, studies carried out using Drosophila as a model system have played a major role in expanding our understanding of the mechanisms, as well as downstream consequences, of signalling via this pathway [3–5]. In humans, aberrations of InR–TOR signalling lead to various metabolic syndromes, including diabetes and obesity, as well as to the development of various types of cancers [6].
A cascade of phosphorylation events mediates signalling through the InR–TOR pathway. The binding of insulin to the InR leads to the phosphorylation of insulin receptor substrate (IRS) by the InR. IRS acts as a recruitment site for phosphatidylinositol 3-kinase (PI3K), which catalyses the conversion of phosphatidylinositol (4,5)-bisphosphate (PIP2) into phosphatidylinositol (3,4,5)-trisphosphate (PIP3) at the cell membrane. PIP3 in turn recruits PDK1 and Akt to the membrane, where PDK1 phosphorylates and activates Akt. Phosphorylated Akt signals inhibit the tuberous sclerosis complex (TSC, Tsc1–Tsc2) [7–9]. When TSC is inhibited, the small GTPase Rheb becomes active [10]. This then activates TOR, integrating TOR into the insulin signalling process.
TOR is a component of two different complexes: TORC1 and TORC2. Activation of TORC1 has various downstream effects contributing to an increase in cell growth and proliferation. For example, TORC1 directly phosphorylates S6K and 4E-BP, resulting in an increase in ribosome biogenesis and m7G cap-dependent translation [1]. In addition, TORC1 activation inhibits autophagy [11]. A negative feedback loop signals back to the IRS through S6K, ensuring attenuation of TOR signalling above a certain level [12]. The TORC2 complex phosphorylates and activates Akt kinase [13], resulting in the phosphorylation of the forkhead-like transcription factor FoxO. Phosphorylated FoxO is excluded from the nucleus, precluding the transcription of FoxO target genes [14–16].
A critical consequence of the activation of InR–TOR signalling is the inhibition of autophagy: a cellular response to starvation in which components of the cytoplasm are engulfed in small double-membrane-enclosed vesicles. The contents of these vesicles are degraded by the autophagic machinery, and breakdown products then serve as a nutrient source for the cell until more nutrients become available in the environment. Alterations to autophagy have been found in cancer and neurodegenerative diseases [17,18].
Herein, we report the identification of Poly as a novel mediator of the InR–TOR signalling pathway. poly is an essential gene in Drosophila that was mutated in a P-element transposon mutagenesis screen [19]. Crucially, the gene product is conserved in higher eukaryotes, including humans, showing homology to the ELP6 subunit of the Elongator complex. Loss of poly function results in lethality at the late larval stage, but only after extreme larval longevity. Many intriguing phenotypic features are observed in larvae lacking poly, including abnormal nuclear morphology in neuroblasts and the development of large melanotic masses in third instar larvae. We have combined genetic, biochemical and bioinformatic approaches to functionally characterize poly. Our data reveal that poly is a novel mediator of InR–TOR signalling and that loss of poly results in downregulation of a number of components of the InR–TOR pathway. We therefore propose that the wild-type Poly protein is a positive regulator of cell growth and metabolism.
3. Results
3.1. Characterization of the poly mutant phenotype
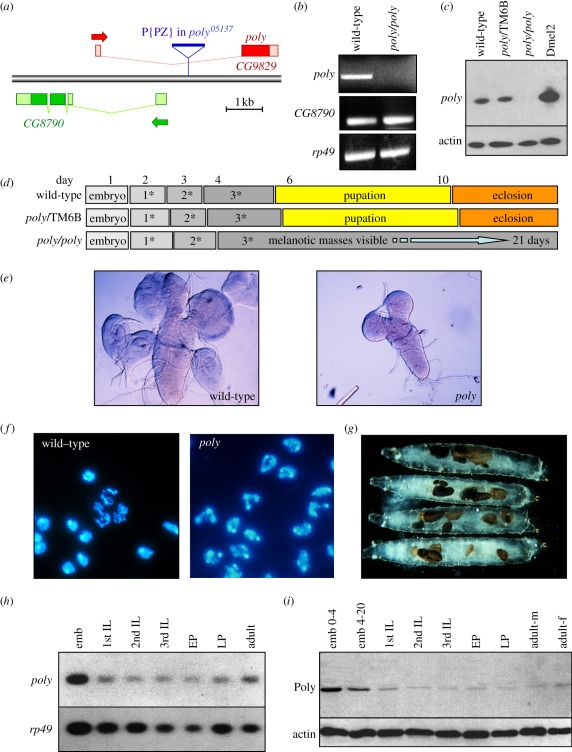
The poly mutation was isolated in a P-element mutagenesis screen that aimed to generate a large collection of single P-element-induced mutations in Drosophila [19]. The P-element insertion that led to the lethal poly05137 allele was mapped to the single intron of the CG9829 gene, localizing to 87E7-8 on the third chromosome (figure 1a). The poly05137 insertion led to an absence of poly mRNA as assessed by Northern blotting (not shown), reverse transcriptase–polymerase chain reaction (RT-PCR; figure 1b) and Poly protein as revealed by immunoblotting (figure 1c). RT-PCR verified that expression of the overlapping CG8790 gene was not affected by the P-element insertion in the poly05137 allele (figure 1b). Two independent experiments additionally corroborate lesion of the CG9829 gene as being responsible for the mutant phenotype: (i) excision of the P-element following genetic exposure to transposase completely reverted the mutant phenotype, and (ii) successful rescue of poly05137 lethality was achieved by using a hs-Gal4 driver to direct expression of a UAS-poly transgene during larval development.
Figure 1.
Characterization of the poly gene and mutant phenotype. (a) Gene map showing the P-element insertion in the first intron of poly. (b) RT-PCR from wild-type and homozygous poly third instar larvae demonstrating the absence of Poly in poly mutant animals. The level of CG8790 is unaffected in poly mutants. RP49 serves as a control. (c) Immunoblotting of extracts from wild-type, heterozygous and homozygous poly third instar larvae demonstrating the absence of Poly in poly mutant animals. Poly is also present in Dmel2-cultured Drosophila cells. (d) Developmental timing of wild-type, heterozygous and homozygous poly animals. (e) Third instar larval brain and imaginal discs dissected from wild-type and poly animals. Note the reduced brain size and absence of imaginal discs in the homozygous poly mutant. (f) DAPI staining of wild-type and poly larval neuroblasts. Note the abnormally shaped nuclei in the homozygous poly mutant. (g) Homozygous poly larvae develop melanotic masses during the third larval instar, which increase in number with time. (h,i) Expression analysis of poly mRNA and protein throughout development. Northern and immunoblotting of wild-type extracts at specific developmental stages showing Poly mRNA and protein expression levels. emb, embryo (0–4, 4–20 = age in hours); IL, instar larvae (1st, 2nd and 3rd); EP, early pupae; LP, late pupae; adult-m, adult males; adult-f, adult females.
Mutation of poly results in pleiotropic effects manifesting as a particularly striking phenotype. poly mutants appear to progress normally through embryogenesis, but larval development proceeds much more slowly. While the normal generation time is 10 days at 25°C, poly mutant larvae exhibit extreme third instar longevity—up to 21 days—before dying without pupation (figure 1d). When homozygous mutant larvae were examined, the morphology of many tissues appeared abnormal: the brain, ring gland, salivary glands and imaginal discs were reduced in size compared with heterozygous siblings and wild-type animals, suggesting cell growth and/or proliferation defects (figure 1e). Mutant larval neuroblasts were characterized by abnormally shaped nuclei, though mitotic figures, evident even in 20 day old mutant larvae, were normal in appearance. These lobulated nuclei resembled the nuclei of mammalian polymorphonuclear leucocytes, thus suggesting the name poly (figure 1f). During the lengthened third instar larval phase, melanotic masses appeared in the haemolymph of poly mutants, increasing in size and number with time (figure 1g). A second hypomorphic allele of poly has recently been reported to result in abnormal nurse-cell chromosome dispersal and oocyte polarity in the Drosophila germline, possibly owing to a proposed interaction with an mRNP complex involved in similar processes [20].
Expression analysis of poly revealed high levels of mRNA and protein in the first 4 h of embryogenesis, suggestive of maternal loading of the mRNA and possibly protein into oocytes (figure 1h,i). While the level of Poly decreased during the remainder of embryogenesis, the level was still higher than in later stages of development. Overall, poly was expressed throughout development (figure 1h,i).
3.2. Poly is conserved among higher eukaryotes and resembles the Elp6 subunit of the Elongator complex
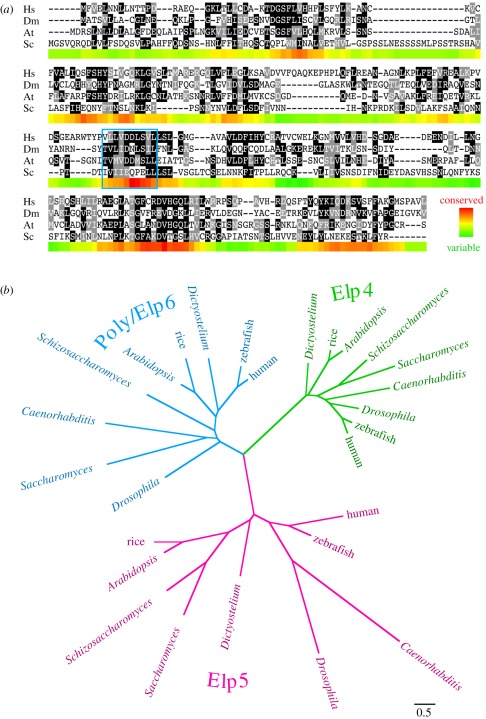
The Drosophila poly gene encodes a 251-amino-acid-long protein, lacking any motifs suggestive of specific function. Sequence similarity searches revealed that Poly is conserved among higher eukaryotes and, importantly, has a human homologue (figure 2a). The apparent orthologue of Poly in Saccharomyces cerevisiae is the Elp6 protein, part of the 6-subunit Elongator complex that, in association with the RNA polymerase II holoenzyme, is responsible for transcriptional elongation [21,22]. The Elongator holocomplex is conserved in composition from yeast to humans [23,24], with acetylation activity contributed by the Elp3 catalytic subunit [25]. Intriguingly, acetylation is directed to different substrates, depending on where in the cell the complex is (e.g. tubulin in the cytoplasm, and histone in the nucleus) [26,27]. Elongator has additionally been linked to translational control through tRNA modification in the cytoplasm. To date, only the Elp3 subunit has been studied in Drosophila, where the mutant phenotype has recently been reported to be remarkably similar to that described herein [28].
Figure 2.
Alignment and phylogenetic analysis of Poly. (a) Alignment showing Poly homologues in human (Hs; NP_001026873), Drosophila melanogaster (Dm; AAF40432), Arabidopsis thaliana (At; NP_567351) and Saccharomyces cerevisiae (Sc; NP_014043), with shading of identical and similar amino acids as rendered by the BOXSHADE program. The coloured band underneath each section of the alignment indicates sequence conservation according to the PAM250 matrix, averaged over a sliding window of 10 amino acids. The location of the Walker B motif (ordinarily associated with ATP binding) is indicated by a blue box, although no Walker A (P-loop) motif required for ATP hydrolysis is similarly conserved. (b) Phylogenetic tree showing the relationship between Poly, identified as the Drosophila orthologue of Elp6, and homologous sequences. The scale bar indicates distance according to the WAG substitution matrix.
Elp6 in budding yeast is part of an Elongator subcomplex that also contains the Elp4 and Elp5 proteins [21,22], which share sufficient homology to be aligned with one another (and can also be identified in flies and humans). Consistent with the previous identification of Elp4 and Elp6 proteins as paralogues [29], our resulting phylogeny implies that each of the three different Elongator subunits probably arose from common gene duplication events during eukaryotic evolution (figure 2b; electronic supplementary material, figure S1). Thus, Poly is an evolutionarily conserved protein that exhibits homology to the yeast Elp6 Elongator subcomplex component.
3.3. Identification of protein interactors of Poly
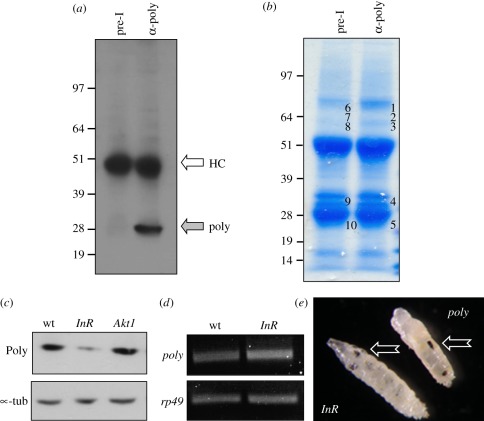
As the poly gene appeared to be a ‘pioneer’, we adopted an unbiased approach to determine the pathways and processes in which Poly might be involved. In order to identify proteins interacting physically with Poly, immunoprecipitation with an antibody generated against recombinant Poly was performed using 0–5 h Drosophila embryo extracts (figure 3a,b). Poly can only be immunoprecipitated with immune serum, and only from wild-type extracts. Samples were analysed using tandem mass spectrometry [30,31]. Immunoprecipitates with pre-immune serum served as the control. Among the five Poly-binding proteins identified with significant scores, InR scored highest and was represented by numerous peptides in two distinct bands (table 1). This result suggests that Poly and the InR might coexist within a complex, possibly playing a role in the InR signalling pathway.
Figure 3.
Poly and the insulin receptor physically interact. (a) An antibody generated to recombinant Poly immunoprecipitates Poly from wild-type Drosophila third instar larval extracts. HC represents the heavy chain recognized by the secondary antibody during detection. Poly migrates at its predicted size of 28 kDa. (b) Poly was immunoprecipitated from 0–5 h wild-type embryos using Poly antibodies and compared with an identical sample immunoprecipitated with pre-immune serum. Bands numbered 1–5 in the immunoprecipitate lane and their corresponding bands in the pre-immune lane (numbered 6–10) were excised from the Coomassie-stained gel and analysed by mass spectrometry. (c) Immunoblotting using the Poly antibody was carried out on wild-type, InR05545 and Akt104226 larval extracts. (d) RT-PCR was performed using primers specific for poly and rp49 on RNA extracted from wild-type and InR05545 larvae. (e) Melanotic masses (arrows) observed in InR05545 and poly mutant larvae.
Table 1.
Mass spectrometry identifies the insulin receptor as a physical interactor of Poly. Analysis of pre-immune and immunoprecipitation samples by tandem mass spectrometry identified interacting proteins specifically found in the immune-precipitate sample. Insulin receptor was identified in bands 2 and 3, with the highest scores. ‘exp score’ quantifies on a log scale (base 10) the expectation that the hit was achieved by chance, calculated using the program X!Tandem.
| band | exp score | unique/total peptides | protein |
|---|---|---|---|
| 1 | −3 | 1/1 | ATP-dependent RNA helicase (Rm62) |
| 2 | −13 | 4/7 | insulin-like receptor |
| 3 | −13 | 4/7 | insulin-like receptor |
| 4 | −5.3 | 3/3 | cadherin |
| 5 | −2.5 | 2/2 | transient receptor potential cation channel subfamily A (TrpA1) |
3.4. Poly is decreased in insulin receptor mutants
Considering the physical interaction of Poly with InR, we assessed the level of Poly in various InR signalling mutants. Strikingly, the level of Poly was dramatically decreased in InR05545 mutant larvae, whereas the level of Poly in Akt1 mutant larvae was unaffected (Akt1 is situated downstream of InR; figure 3c). The level of poly mRNA was unchanged in the InR mutant, suggesting that the difference in protein level might be due to instability or decreased protein synthesis of Poly in the absence of InR (figure 3d).
Further evidence for a connection between Poly and the InR was provided by phenotypic analysis of InR05545 mutant larvae (figure 3e). While this allele was reported previously to be embryonic lethal [32], we noticed that a small number of homozygous InR05545 animals reached the third instar larval stage. Strikingly, the phenotype of these InR mutant larvae resembled that observed in poly larvae with increased larval lifespan and development of melanotic masses (figure 3e). These observations demonstrate that loss of InR leads to a decrease in Poly and that InR05545 mutants exhibit phenotypic features similar to those of poly mutant larvae.
3.5. Poly mutation results in decreased signalling activity downstream of the insulin receptor
Based on the results presented so far, we hypothesized that there should be genetic interactions between poly and components of the InR signalling pathway. Consequences of such interactions could be revealed by the state of downstream effectors of the InR pathway.
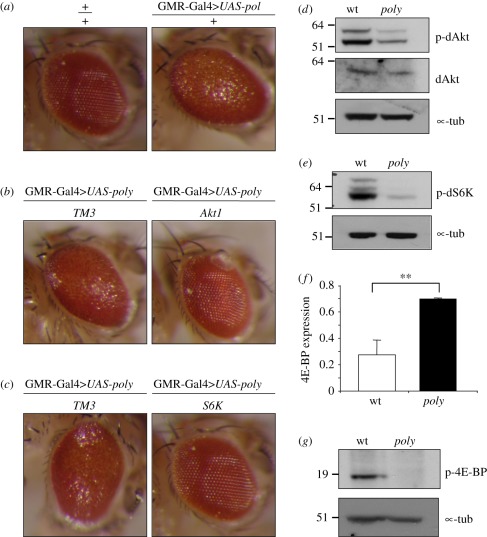
We therefore over-expressed poly in adult fly eyes, using a GMR-Gal4 construct to drive the expression of a UAS-poly transgene [33]. At 27°C, this led to a rough eye (disorganized ommatidia) phenotype (figure 4a). Disruption of InR–TOR signalling through mutation of either dAkt1 or dS6K led to a striking suppression of the poly-induced rough eye phenotype (figure 4b,c), suggesting that an intact InR–TOR signalling cascade was required for Poly to exert its effects. These experiments also critically demonstrate the cell autonomous importance of Poly to insulin signalling.
Figure 4.
Phosphorylation levels of Akt and S6K are decreased in poly larvae. (a–c) Images showing Drosophila compound eyes. (a) A rough eye phenotype is caused by UAS-poly overexpression under the control of GMR-Gal4 driver. (b,c) The rough eye phenotype caused by UAS-poly overexpression under the control of GMR-Gal4 driver is suppressed by Akt104226 and S6K07084 mutations. TM3 represents the balancer chromosome carried by control sibling progeny. (d,e) Immunoblotting of third instar larval extracts using phospho-specific antibodies to dAkt and dS6K reveals decreased phosphorylation of both kinases in poly larval extracts. (f) d4E-BP transcript levels were assessed by real-time qPCR on RNA isolated from wild-type and poly mutant larvae. d4E-BP levels were normalized to Actin5C levels. The error bars derive from reactions performed on biological triplicate samples. The double asterisk represents significant difference (p < 0.01) in 4E-BP expression levels between wild-type and poly larvae. (g) Immunoblotting of third instar larval extracts showing decreased phosphorylation of d4E-BP in poly compared with wild-type larvae.
We hypothesized that if dAkt and dS6K mutants can act as suppressors of the poly over-expression phenotype, the activity of these kinases might be altered in poly mutant animals. We therefore probed early third instar larval extracts with antibodies that recognize specifically the phosphorylated (active) forms of dAkt and dS6K. Indeed, phosphorylation of both dAkt and dS6K was decreased upon mutation of poly (figure 4d,e). These data reveal that the activity of both dAkt and dS6K kinases was decreased in poly mutant animals. In addition, the level of p-Akt at the tissue level, examined by immunofluorescence of whole mount third instar larval brains, additionally corroborates this decrease (electronic supplementary material, figure S2).
One crucial downstream effector of TOR is 4E-BP, the translation initiation factor eIF-4E-binding protein. Phosphorylation of 4E-BP by TOR leads to its dissociation from the m7G-cap-binding protein eIF-4E, thereby allowing activation of cap-dependent translation with consequent positive effects on cell growth [34]. Disruption of InR–TOR signalling causes inhibition of cap-dependent translation as a decrease in 4E-BP phosphorylation results in its binding to eIF-4E [14]. Disruption of signalling also causes increased transcription of 4E-BP owing to increased FoxO activity. Consistent with a decrease in InR–TOR signalling, the level of d4E-BP transcript was almost threefold greater in poly mutant larvae compared with control animals (figure 4f). In addition, immunoblotting revealed that d4E-BP was hypo-phosphorylated in poly mutant extracts compared with wild-type extracts (figure 4g). Taken together, these results are consistent with a decrease in cap-dependent translation upon loss of Poly function.
3.6. Autophagy is constitutively active in the fat body of poly larvae
A critical consequence of the activation of InR–TOR signalling is the inhibition of autophagy. Drosophila undergoes developmentally programmed autophagy at defined times to facilitate tissue remodelling during metamorphosis [35–38]. On the other hand, starvation-induced autophagy (in response to nutrient deprivation) takes place specifically in the larval fat body [11]. As the larval fat body serves a similar function to the vertebrate liver by acting as a nutrient storage organ, it has been commonly used to examine autophagy during both starvation-induced and developmentally regulated autophagy [11,39,40]. While the fat body from fed wild-type larvae does not normally exhibit autophagy, autophagy is evident within a short period of amino acid starvation.
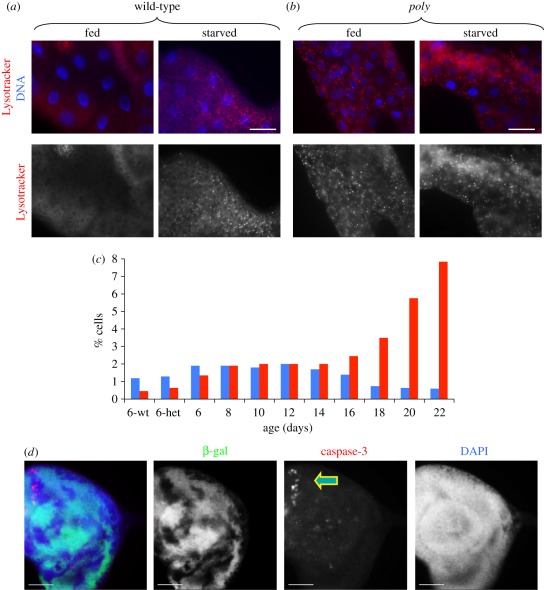
TOR directly inhibits starvation-induced autophagy, while components of InR signalling (such as InR and Akt, acting upstream of TOR) also behave as negative regulators of autophagy. We hypothesized that if poly acts in InR signalling, the state of autophagy as visualized with Lysotracker staining may be altered in the poly mutant fat body compared with wild-type fat body. As anticipated, no Lysotracker puncta were observed in fed wild-type early third instar fat body (figure 5a, fed). However, Lysotracker puncta became apparent following a 4 h amino acid starvation, owing to the activation of autophagy (figure 5a, starved). Strikingly, Lysotracker puncta were abundant even in fed poly fat bodies, demonstrating that autophagy was constitutively active in the fat body of poly mutants (figure 5b, compare fed with starved).
Figure 5.
poly mutations leads to constitutive autophagy and increased cell death. (a,b) Lysotracker (an acidic component-specific fluorescent dye) was used to monitor the state of autophagy in early third instar larval fat body. Autophagy was indicated by a punctate Lysotracker appearance. Live fat body tissues were stained with Lysotracker (red) and Hoechst 33342 to label DNA (blue). (a) In wild-type larval fat body, no autophagy is evident. As indicated by Lysotracker positive bright spots, autophagy is active upon 4 h amino acid starvation in 20 per cent sucrose solution. (b) Lysotracker puncta can be detected in fed poly fat body, as well as in starved poly fat body. (a,b) Scale bar, 50 µm. (c) Quantitation of mitosis and apoptosis in larval neuroblasts over development. While mitotic activity decreases, apoptosis increases during the extended larval stage of poly larva. Blue bars, p∼H3; red bars, TUNEL. (d) Images showing imaginal disc dissected from third instar larva of the genotype eyFLP; FRT82Bβ-Gal/FRT82Bpoly05137. Immunostaining for cleaved caspase-3 (red), β-galactosidase (green) and DNA (blue) was performed. poly mutant clones are indicated by the lack of β-Gal staining. Scale bar, 30 µm.
3.7. Poly loss of function leads to an increase in apoptotic cell death
Autophagy and apoptosis are closely related, and increased levels of autophagy can lead to apoptosis [39,41]. For example, increased autophagy resulting from the clonal over-expression of Atg1 in the wing disc resulted in elevated apoptosis, as evidenced by the appearance of cleaved caspase-3 in these clones [39].
Because loss of poly led to constitutive activation of autophagy in the fat body, we investigated whether the loss of poly also resulted in increased apoptosis. Indeed, cell death increased dramatically in third instar larval neuroblasts as poly larvae aged. This was readily seen when larval neuroblasts were stained for pS10-histone H3 and TUNEL to detect mitosis and apoptosis, respectively (figure 5c). Mitotic figures, though rare at the latest stages, were still normal in appearance (data not shown). As the generation of mosaic imaginal discs allows the side-by-side comparison of wild-type versus mutant cells, we generated poly loss-of-function clones in imaginal discs. Staining of mosaic discs revealed increased levels of cleaved caspase-3 in poly mutant clones, which were discernible by the lack of ß-galactosidase (figure 5d). Cleaved caspase-3 was not detectable in the adjacent wild-type cells.
Thus, several independent lines of evidence demonstrate that loss of poly leads to an activation of autophagy coupled with an increase in apoptotic cell death.
3.8. Metabolism is affected in poly mutant larvae
Drosophila is frequently used to study metabolic regulation as fruitflies share the majority of metabolic functions with vertebrates [42]. The larval fat body is the main organ for regulation of energy homeostasis as excess energy is stored in the form of glycogen and triglycerides (TAGs; lipids). Activation of signalling through the InR pathway promotes both anabolic metabolism and the storage of nutrients such as TAGs [43]. This is important for development, as the breakdown of larval fat body and consequent release of energy facilitates Drosophila metamorphosis.
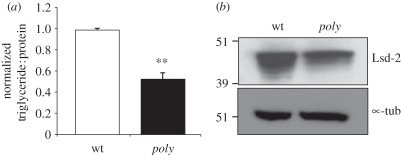
In order to identify genes that were differentially expressed in poly mutant larvae, we carried out a microarray analysis. Among the list of 106 genes downregulated in poly mutants, functional enrichment group analysis identified a strong enrichment in gene ontology (GO) terms belonging to metabolic processes, suggesting that metabolism might be affected in poly mutants (table 2). We therefore investigated whether the decrease in InR–TOR activity was also manifest at the metabolic level in the storage of TAGs in poly mutants. TAG levels were normalized to total protein to give an accurate quantification of lipids per unit mass in wild-type and poly mutant larval extracts. Consistent with a decrease in InR–TOR signalling, we found the TAG : protein ratio to be half that detected in wild-type larvae (figure 6a).
Table 2.
Microarray analysis revealed that several metabolic functions were among the genes downregulated in poly mutants. DAVID (the Database for Annotation, Visualization and Integrated Discovery) functional enrichment group analysis of the list of differentially expressed genes identified enrichment in gene ontology (GO) terms belonging to metabolic processes as among those genes downregulated in poly mutants. The data were analysed by Limma (see §5.10) and were for four biological replicates. Genes were selected for functional/pathway analysis if the adjusted (corrected for multiple testing) p-value was less than 0.05. Log (base 2) fold changes are given (log2FC). ‘rep’ indicates reported gene function, while ‘pred’ indicates predicted gene function according to the gene-specific FlyBase report.
| GO TERM | name of affected gene (log2FC) | fold-enrichment |
|---|---|---|
| GO:0006739∼NADP metabolic process | FBgn0004057 // CG12529 (−1.42) | 81.9 |
| glucose-6-phosphate dehydrogenase rep | ||
| FBgn0023477 // CG2827 (−1.27) | ||
| transaldolase rep | ||
| FBgn0004654 // CG3724 (−2.15) | ||
| phosphogluconate dehydrogenase rep | ||
| FBgn0030239 // CG17333 (−1.34) | ||
| 6-phosphogluconolactonase pred | ||
| FBgn0037607 // CG8036 (−1.39) | ||
| transketolase pred | ||
| GO:0006769∼nicotinamide metabolic process | same five genes | 64.4 |
| GO:0019362∼pyridine nucleotide metabolic process | same five genes | 60.1 |
| GO:0006006∼glucose metabolic process | same five genes, and | 16.4 |
| FBgn0002569 // CG8694 (−0.89) | ||
| maltase A2 rep |
Figure 6.
poly mutant larvae are characterized by reduced levels of TAGs and Lsd-2. (a) TAG levels were assessed in wild-type and poly mutant third instar larvae. The total TAG level was normalized to the total protein level. Error bars derive from the standard deviation of three independent experiments. The unpaired two-tailed t-test was used to compare the data from wild-type and poly larvae. The double asterisk represents a significant difference (p < 0.01) in TAG : protein ratio between control and poly mutant larvae. (b) Immunoblotting was carried out on wild-type and poly mutant larval extracts with an antibody specifically recognizing Lsd2 protein.
Lipid storage droplet-2 protein (Lsd-2), a protector against lipolysis and the Drosophila perilipin homologue, is localized on the outer membrane of lipid droplets, the main storage organelle for TAGs [44,45]. We found that the level of Lsd-2 in poly larval extracts was decreased relative to wild-type fat body, consistent with the decreased TAG : protein ratio observed in poly mutants (figure 6b).
Together, the reduction in TAG storage and Lsd-2 protein in poly mutant larvae suggest a decrease in anabolic metabolism, consistent with—and an expected consequence of—diminished InR–TOR signalling.
3.9. The Poly protein relocalizes following insulin stimulation in Drosophila haemocytes and human cultured cells
Given the physical interaction of Poly with the InR and functional links between Poly and InR–TOR signalling, we were interested to determine whether the level and/or distribution of Poly in the cell were dependent on InR activity.
Strikingly, staining for Poly was noticeably increased following insulin stimulation of larval haemocytes (cells of the innate immune system). Prior to isolation of haemocytes, larvae were starved for 3 h in 20 per cent sucrose. Haemocytes were then stimulated in culture with 200 nM insulin (electronic supplementary material, figure S3). This increased staining in Poly appears to be a rapid response as it is detectable following only 15 min of insulin stimulation, and levels of Poly remain elevated after 75 min.
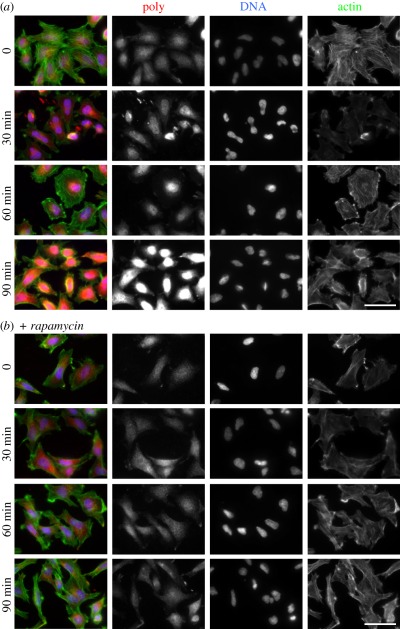
A change in the appearance of Poly following insulin stimulation was conserved in human cells. Overnight serum-starved HeLa cells were stimulated with insulin for up to 90 min. As in Drosophila haemocytes, staining for HsPoly was significantly stronger following insulin stimulation, appearing maximal at 90 min. It additionally appeared that this increase in HsPoly was concentrated in or near the nucleus, with relocalization already evident by 30 min of insulin stimulation (figure 7a). Nearly, 60 per cent of cells showed a relocalization of Poly by 60 min of insulin stimulation, with this level increasing to 100 per cent by 90 min.
Figure 7.
Insulin stimulation of HeLa cells leads to an increase in HsPoly staining, accumulated in the nuclear area. (a) HeLa cells were serum-starved overnight and then stimulated with 100 nM insulin for 30, 60 and 90 min prior to staining for HsPoly (red), DNA (blue) and F-actin (green). Staining for HsPoly indicated peri-nuclear accumulation already by 30 min, reaching a maximum following 90 min of insulin stimulation. (b) The relocalization of HsPoly was inhibited by overnight incubation of cells with 20 nM rapamycin prior to insulin treatment. Scale bars, 50 µm.
This rapid change in HsPoly behaviour was dependent on TOR signalling. Incubation of HeLa cells with rapamycin during the overnight starvation period prevented the increase in HsPoly staining following insulin stimulation (figure 7b). In rapamycin-treated cells, HsPoly remained evenly distributed throughout the cell even following 90 min of insulin treatment.
Taken together, these observations demonstrate that the increase and nuclear relocalization of HsPoly following insulin treatment occurs in a TOR-dependent manner. Overall, our results identify Poly as a novel component of InR–TOR signalling that is conserved from flies to humans.
4. Discussion
Poly is a conserved protein that plays a novel essential role in InR signalling and, crucially, promotes the effects of the TOR kinase on cell growth and metabolism.
4.1. The activity of InR signalling is decreased in poly larvae
Our data reveal an essential involvement for Poly in InR signalling, an important pathway linking nutritional status to metabolism and cell growth [3]. The two kinases, Akt and S6K, are key effectors of InR–TOR signalling. The Akt kinase is located upstream of the TSC1–TSC2 complex in this pathway and has a critical role in promoting cell growth. Phosphorylation of Akt substrates contributes to a range of cellular processes including cell growth, proliferation and survival [46]. Dysregulation of Akt is involved in various diseases, including type-2 diabetes and cancer [47,48]. S6K kinase is one of the most downstream effectors of InR–TOR signalling, and is subject to phosphorylation and activation by TOR. Activation of S6K leads to an increase in translation through its phosphorylation and activation of ribosomal protein S6 [1].
Poly acts upstream of both of these kinases in the InR signalling pathway, as levels of phosphorylated (active) dAkt and dS6K kinases were decreased in poly larvae. That Poly acts in the activation of both dAkt and dS6K is also supported by genetic data that revealed suppression of a poly-induced rough eye phenotype by mutations in dAkt and dS6K. These two kinases should only act as genetic suppressors if they are situated downstream of poly in the signalling pathway, resulting in an overall decrease in InR–TOR signalling in the absence of Poly.
The biochemical data presented herein revealed a physical interaction of Poly with InR. Intriguingly, the level of Poly was reduced in InR mutant larval extracts independently of transcription, suggesting that a fraction of Poly may be unstable and subject to degradation in the absence of InR. Indeed, there are numerous examples of such instability if one partner in a protein complex is absent [49–51]. Whether Poly exists in a discrete complex with InR, and how such complex(es) may be regulated in response to insulin stimulation, are currently under investigation. Such an interaction may well be transient and highly dynamic, as Poly appears to accumulate in the nucleus following insulin stimulation. Considering the insulin-induced, TOR-dependent nuclear enrichment of Poly in HeLa cells, it is likely that insulin-mediated signalling via Poly is conserved from flies to humans.
Together our data predict a decrease in InR–TOR signalling in the absence of Poly. As one read-out of TOR signalling, we examined autophagy, a multi-step, catabolic process used for nutrient recycling during development and starvation. TOR activity is responsible for inhibiting autophagy under cell growth-promoting conditions. Our finding that autophagy is constitutively active in the fat body of poly mutant larvae is in accord with the observation of reduced InR–TOR signalling, and consistent with an inhibition of cell growth and/or proliferation in poly mutant animals. Furthermore, the increase in apoptotic cell death in poly mutant animals and clones generated in imaginal discs might result from increased levels of autophagy upon mutation of poly.
4.2. Metabolism is disrupted in poly mutants
A major function of the InR–TOR pathway is the modulation of metabolism. For example, 4E-BP mutant animals show increased sensitivity to starvation [16]. Interestingly, it was previously shown that the loss of the tumour suppressor PTEN (responsible for the dephosphorylation of PIP3) in Drosophila nurse cells results in the accumulation of activated Akt in the cytoplasm. This activated Akt drives the formation of enlarged lipid droplets along with an increase in the expression of Drosophila Lsd-2 [52]. Lsd-2 mutants are characterized by decreased TAG levels [45]. Interestingly, autophagy and lipid metabolism were shown to be two interlinked processes, as suggested by a decrease in autophagy resulting in an increase of lipid storage in the cell [53]. Consistent with decreased InR–TOR signalling in the absence of poly, both Lsd-2 and the TAG : protein ratio were reduced in poly mutant animals. Therefore, we propose that Poly affects metabolism via its interaction with the InR–TOR pathway, acting as a positive regulator of anabolic metabolism.
4.3. Comparison of poly to other mutants and Elongator
Numerous aspects of the poly phenotype have been observed in other mutations, including that of InR and TOR. An extreme larval longevity, one of the most remarkable aspects of the poly mutant phenotype, has been observed in both Tor [54] and InR mutants (this study). The appearance of melanotic masses is frequently seen in mutations with aberrant immune responses and/or haematopoietic defects [55–57]. In poly third instar larvae, melanotic masses appearing at multiple different locations along the larval body are likely to result from either an alteration in the immune response and/or a hyper-proliferation of blood cells, or haemocytes. The observation that the few InR mutant larvae that reach the third instar stage also develop melanotic masses highlights the phenotypic similarities of poly and InR loss-of-function mutations, and further corroborates a functional relationship between poly and InR.
Given the apparent similarity of Poly to yeast Elp6, it is therefore intriguing that the recently described mutant phenotype for the Elongator Elp3 catalytic subunit in Drosophila resembles that of poly [28]. These observations are suggestive of a role for the Elongator holocomplex in insulin signalling—through histone and/or tubulin acetylation—or even translational control through tRNA modification. In addition, as human mutations in Elp3 have been linked to familial dysautonomia [58], it is likely that the future analysis of Elongator function in model organisms will be of significant preclinical value.
4.4. Model for Poly action
We have shown that Poly binds to InR (a transmembrane receptor) and modulates the activity of various downstream proteins. While the lack of clearly discernible functional motifs hampered a prediction of Poly's molecular function, extensive database searches and phylogenetic analyses identified Poly as a member of the Elp6 subfamily of Elongator proteins, with more distant homology to various members of the RecA/Rad51/DCM1 superfamily (such as KaiC and RadB); these observations are described in a distinct study from our laboratory. Both KaiC and Elp6 have been shown to significantly affect gene expression, pointing to a degree of functional conservation. Thus, we suggest an involvement for Poly during transcription (perhaps once relocated to the nucleus following insulin stimulation).
If Poly has a role during transcription, as the phylogenetic data suggest, how do we explain its binding to a transmembrane receptor? One explanation is the dynamic changes in the cellular localization of Poly, occurring in a TOR-dependent manner. Interestingly, InR phosphorylation of IRS-1 and IRS-2 (two of the human IRS homologues) not only leads to activation of PI3K signalling, but is also associated with IRS-1 translocation to the nucleus, where it activates transcription of various genes [59–61]. Given that Poly also interacts with InR, and moves to the nucleus following insulin stimulation, it is possible that changes in the localization of Poly are also ultimately in response to InR signalling. Immunofluorescence on both Drosophila haemocytes and HeLa cells demonstrated that the level and/or distribution of Poly are significantly affected following insulin stimulation. Importantly, in HeLa cells, Poly relocalization to the nucleus occurred in a rapamycin-sensitive manner. Thus, we speculate that the stimulatory effects of Poly on cell growth and metabolism may be exerted via effects on transcription. However, further analysis is required to assess whether Poly participates in an Elongator complex and, if so, in which of the myriad functions currently ascribed to Elongator.
Future research will address the detailed nature of the interaction of Poly with the InR. Does Poly interact with the InR in the absence or presence of insulin? How dynamic is this interaction? And is the level or post-translational state of Poly modified upon insulin treatment?
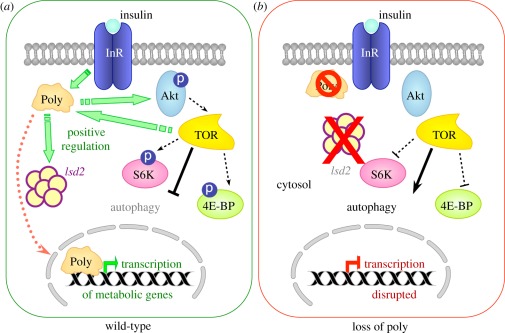
In the light of the data presented herein, we propose that Poly is a novel mediator of InR–TOR signalling in the regulation of cell metabolism and growth in Drosophila (figure 8). We suggest that the physical interaction between Poly and the InR is followed by the translocation of Poly into the nucleus (upon insulin stimulation), wherein the expression of key metabolic genes is affected, thus contributing to the promotion of cell growth and metabolism. While the detailed nature and regulation of Poly's interaction with the InR remain to be elucidated, it is highly likely that, given the conservation of Poly, this crucial interaction and function will also hold true in human cells.
Figure 8.
Model for the function of Poly in the InR–TOR signalling pathway. The interaction of Poly with (a) the insulin receptor allows it to fulfil a role as a positive regulator of InR–TOR signalling, which has the overall effect of increasing the phosphorylation and, hence, activity of numerous positive regulators of the InR–TOR pathway, such as Akt, S6K and 4E-BP, as well as inhibiting negative regulators of cell growth such as autophagy. Furthermore, Poly aids to promote anabolic metabolism resulting in an increase of TAG and Lsd-2 levels. (b) The absence of Poly leads to a decrease in the activation of positive regulators of cell growth such as Akt and S6K kinases. The 4E-BP protein becomes hypo-phosphorylated, consistent with an overall downregulation of InR–TOR signalling. Furthermore, autophagy—a negative effector of InR/TOR signalling—becomes constitutively active. Anabolic metabolism is downregulated as indicated by a decrease in TAG and Lsd-2 levels.
5. Material and methods
5.1. Drosophila culture
All fly stocks were maintained at 25°C on standard medium unless otherwise stated. Fly stocks used in this study were: Canton S (wild-type), poly05137/TM6B. The following fly lines were obtained from the Bloomington Stock Center: InR05545/TM3, Akt104226/TM3, S6K07084/TM3, Cg-Gal4.
5.2. Phylogenetic analysis
Sequences homologous to Drosophila Poly were identified by multiple iterative searches using the PSI-BLAST program [62] and the HHpred interactive server [63]. Alignments between the corresponding sequences were generated using M-Coffee [64] and manually adjusted based on predicted secondary structure according to Ali2D [65]. Identical and similar amino acids in a representative subset of aligned sequences were shaded using the BOXSHADE server (http://www.ch.embnet.org/software/BOX_form.html). Phylogenetic trees were calculated using TREE-PUZZLE [66], MrBayes [67] and the Proml program of the PHYLIP Package [68] after clustering of related sequences into smaller groups using SplitsTree4 [69]. Branch lengths were calculated by application of the WAG substitution matrix [70] using TREE-PUZZLE, modelling rate heterogeneity according to a gamma distribution with 16 rate categories, and bootstrap confidence intervals (provided in electronic supplementary material, figure S1) were estimated using the Seqboot program of the PHYLIP Package [68].
5.3. Immunoprecipitation
Fifty microlitres of 0–5 h wild-type embryos (raised at 18°C) were homogenized in 1 ml of cold lysis buffer (100 mM NaCl, 10 mM EDTA, 50 mM Tris–HCl pH 7.6, 0.1% Triton-X100, 10 µg ml−1 each of chymostatin, leupeptin, antipain and pepstatin, and 50 µg ml−1 PMSF) and briefly sonicated. Eight microlitres of rabbit pre-immune or anti-poly serum coupled to 40 µl of Dynal beads (Invitrogen) were added to pre-cleared embryo lysates overnight. Ten per cent of each sample was subjected to immunoblot analysis to verify successful immunoprecipitation of Poly, while the remaining 90 per cent of sample was resolved on precast 4–12 per cent Bis-Tris polyacrylamide gels (Novex) and stained with Colloidal Coomassie Blue (Invitrogen). Comparable molecular weight regions of interest were excised from each lane (pre-immune- and immune-precipitations) and mass spectrometry analysis performed (Dr Gerard Cagney, Dublin).
5.4. Mass spectrometry
The proteins in slices from sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS–PAGE) gels were digested in-gel with trypsin by the method of Shevchenko et al. [71]. The resulting peptide mixtures were resuspended in 1 per cent formic acid and analysed by nano-electrospray liquid chromatography mass spectrometry (nano-LC MS/MS). A high-performance liquid chromatography (HPLC) instrument (Dionex, UK) was interfaced with an LTQ ion trap mass spectrometer (ThermoFinnigan, CA). Chromatography buffer solutions (buffer A: 5% acetonitrile and 0.1% formic acid; buffer B: 80% acetonitrile and 0.1% formic acid) were used to deliver a 60 min gradient (35 min to 45% buffer B, 10 min to 90%, hold 10 min, 3 min to 5% and hold for 15 min). A flow rate of 2 µl min−1 was used at the electrospray source. One full scan was followed by 10 MS/MS events, and the duty cycle programmed to enforce dynamic exclusion for 2 min. In-house proteomics pipeline software (Proline) was used to process data. Spectra were searched using the Sequest algorithm [72] against SwissProt.2007.04.19 database restricted to Drosophila melanogaster entries. Proteins with (i) peptide prophet probability score greater than 0.99 [73] and (ii) identified by a minimum of two different peptide spectra were automatically accepted, while spectra for the minority of proteins identified by single spectra were manually checked for quality.
5.5. Larval manipulations
Prior to dissections or protein extractions, 10–15 second instar larvae were transferred to a fresh vial of food supplemented with fresh yeast paste. Manipulations were carried out when larvae reached early third instar stage. For starvation experiments, early third instar larvae were starved in 20 per cent sucrose solution for 3–4 h prior to dissection.
5.6. RNA extraction and quantitative reverse transcriptase–polymerase chain reaction
Larvae were transferred to RNase-free Eppendorf tubes (Ambion). RNA was extracted with the Qiagen RNeasy Plus Kit (Qiagen Hilden, Germany) according to manufacturer's instructions. cDNA synthesis was carried out by using the SuperScript III (Invitrogen) following the manufacturer's instructions. Real-time quantitative RT-PCR analysis was performed on the LightCycler system (Roche) using the universal probe library. The reactions were set up following manufacturer's recommendation with the LightCycler master mix kit. The relative cDNA ratio was calculated with Lightcycler software 480. Actin5C was used as control to normalize equal loading of template cDNA.
5.7. Preparation of larval protein extracts for immunoblotting
Fifteen to twenty wandering third instar larvae of the appropriate genotype were placed in 1.5 ml tubes and rinsed three times with Ephrussi–Beadle Ringer's solution (130 mM NaCl, 4.7 mM KCl, 1.9 mM CaCl2, 10 mM HEPES and pH 6.9). Next, 300µl of cold lysis buffer (Ephrussi–Beadle Ringer's solution with 10 mM EDTA, 10 mM DTT, 1 µg ml−1 of each of chymostatin, leupeptin, antipain and pepstatin [CLAP, Sigma], 1 mM phenylmethanesulphonyl fluoride [PMSF, Sigma], and 1 unit of Aprotinin [Calbiochem] was added to the tubes. The larvae were then homogenized using a motorized hand pestle starting at lowest speed and gently increasing the speed to the maximum for approximately 2 min. Then, 150 µl of hot (70°C) 3X SDS-PAGE Sample Buffer containing 10 mM DTT was added to the homogenate and the tube placed at 100°C for 10 min. Particulate matter was pelleted at 13 000 r.p.m. for 2 min, and the supernatant transferred to a fresh tube. Samples were stored at −20°C until required.
5.8. Sodium dodecyl sulphate polyacrylamide gel electrophoresis and immunoblotting
Protein samples were resolved at 170 V by SDS-PAGE on precast 4–12% Bis-Tris polyacrylamide gels (Novex) and transferred onto nitrocellulose membranes in a Trans-Blot apparatus (Biorad). Membranes were blocked in TBSTw (Tris-buffered saline (150 mM NaCl, 20 mM Tris pH 7.5) + 0.05% Tween 20) and 5% (w/v) semi-skimmed milk for 1 h at room temperature and then incubated for 1 h with primary antibody in TBSTw. After washing three times for 5 min with TBSTw, the membranes were incubated in an appropriate horseradish peroxidase-linked secondary antibody (1 : 10 000) for 1 h in TBSTw at room temperature. Finally, the membranes were washed as above in Tris-buffered saline plus 0.2% Triton X and immune-complexes detected by enhanced chemiluminescence (ECL; Amersham Biosciences).
Primary antibodies and dilutions used in immunoblotting experiments were as follows: DmPoly 504 (1 : 1000), anti-Isd2 (1 : 2000), p-Akt antibody (1 : 1000), p-S6K antibody (1 : 1000) and p-4E-BP antibody (1 : 200). The anti-Isd2 antibody was a generous gift from Ronald Kühnlein (MPI, Göttingen), while the last three were purchased from Cell Signaling Technology.
5.9. Lysotracker staining
Lysotracker staining was performed as described [74]. Fat body from fed or starved larvae was dissected in PBS, and incubated for 2 min in 100 nM LysoTracker Red DND-99 (Molecular Probes), with 1 µM Hoechst 33342 (Molecular Probes) in PBS. Fat bodies were mounted with phosphate-buffered saline (PBS) on a glass slide and visualized using an Olympus AX-70 Provis epifluorescence microscope and Hamamatsu Orca II charge coupled device (CCD) camera. Images were captured using SmartCapture 3 and processed using Adobe Photoshop.
5.10. Microarray processing and analysis
Ten to fifteen second instar wild-type Canton S and poly larvae were transferred into vials supplemented with fresh yeast paste. Early third instar larvae were transferred into TRIzol 24 h later for RNA extraction. All microarray processing was by the Flychip team at the University of Cambridge (http://www.flychip.org.uk/). RNA from wild-type Canton S and poly early third instar larvae was extracted (medium scale) using TRIzol (http://www.flychip.org.uk/protocols/gene_expression/standard_extraction.php). RNA was reverse transcribed and labelled using Klenow labelling: 5 µg of total RNA were reverse transcribed to cDNA (anchored oligo (dT)23 (Sigma), Superscript III (Invitrogen)) and second strand synthesis was then performed (Second strand buffer (Invitrogen), DNA Polymerase I (Invitrogen), RNaseH (New England Biolabs), Escherichia coli DNA ligase (GE Healthcare)) to obtain double-stranded DNA. Random primers are annealed to 500 ng of this denatured DNA template and extended by Klenow fragment using the Bioprime DNA Labeling System (Invitrogen), while fluorescent dyes Cy3-dCTP or Cy5-dCTP (GE Healthcare) are incorporated (http://www.flychip.org.uk/protocols/gene_expression/klenow_v2.php). Hybridization to FL002 microarrays: hybridization to amino-modified long oligonucleotide microarrays using a Genomic Solutions hybridization station with the Biosolutions hybridization buffer (http://www.flychip.org.uk/protocols/gene_expression/hyb_oligoMWG.php). Scanning was with the Genepix 4000B dual laser scanner at 5 µM pixel resolution (http://www.flychip.org.uk/protocols/gene_expression/scanning2.php). Spot finding and quantitation were via the Dapple package. Raw data were normalized by the vsn method in bioconductor (http://www.bioconductor.org/packages/2.8/bioc/html/vsn.html) to generate log (base 2) fold changes and average signals. Differential expression was tested using Limma, also in bioconductor (http://www.bioconductor.org/packages/2.8/bioc/html/limma.html). GO and KEGG biological pathway enrichment in the differentially expressed gene set were assessed using the DAVID functional annotation bioinformatics microarray analysis tool (http://david.abcc.ncifcrf.gov/). Microarray data were deposited in the gene expression omnibus (GEO) under the accession number GSE32637 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE32637).
5.11. Triglyceride assay on larvae
TAG assay on larvae was carried out as described by Gronke et al. [75]. Briefly, six whole larvae corresponding to each genotype were collected in 500 µl of 0.05 per cent Tween 20 and homogenized using a Polytron apparatus, followed by treatment at 70°C for 5 min. Samples were centrifuged for 1 min at 3500 r.p.m. and 350 µl of the supernatant were transferred to a new vial and centrifuged for 3 min at 2500 r.p.m. Six hundred microlitres of Thermo Infinity Trig solution (Thermo Electron, 981786) were added to 75 µl of isolated supernatant and the absorbance at 540 nm was measured following incubation for 30 min. Similarly, protein content was determined by adding 10 µl of isolated supernatant to Bradford reagent (Sigma, B6916) and reading the absorbance at 595 nm. TAG levels were normalized to corresponding protein levels.
5.12. Antibody staining of haemocytes
Third instar larvae were bled on multispot microscope slides (Hendley-Essex) using a pair of forceps and a 25-gauge needle in 20 μl of PBS. Cells were left to settle on the slide for 1 h at room temperature in a humidified chamber to allow adherence to the slide. Cells were fixed with 20 µl of 3.7 per cent paraformaldehyde in PBS for 5 min. Cells were washed with PBS for 5 min, followed by a 5 min permeabilization in PBS + 0.1% Triton X-100. Following an additional 5 min wash in PBS, blocking was performed by incubating cells in PBS + 3% BSA for 1 h. Cells were then incubated with primary antibody diluted in PBS + 3% BSA overnight at 4°C. The following day cells were washed in PBS three times and incubated at room temperature with secondary antibody diluted in PBS + 3% BSA. Following the secondary antibody, incubation cells were washed twice in PBS for 5 min. Cells were incubated with DAPI diluted in PBS (1 : 5000 dilution) for 5 min followed by a final 5 min wash in PBS. Coverslips were mounted with mowiol on top of the slide.
Primary rabbit anti-DmPoly serum was used in immunofluorescence experiments at a 1 : 1000 dilution. Alexa Fluor-488 anti-rabbit (1 : 500) and Alexa Fluor-594 conjugated phalloidin (1 : 500) were purchased from Molecular Probes.
5.13. HeLa cell culture
HeLa cells were maintained in Dulbecco's modified Eagle's medium (DMEM; Sigma) supplemented with 10 per cent foetal bovine serum (FBS; Gibco). Cells were grown to 80–90% confluence before overnight serum withdrawal. For insulin stimulation, serum-starved cells were treated with 100 nM insulin (Sigma). Cells were treated with 20 nM rapamycin (Cell Signaling Technology) during an overnight serum starvation.
5.14. Immunofluorescence on HeLa cells
HeLa cells were cultured on coverslips in six-well plates overnight. Cells were rinsed in PBS for 3 min followed by a 3 min fixation in PBS containing 4 per cent paraformaldehyde (diluted from 16% ampoules). The fixative was removed by a 2 min wash in PBS. Cells were permeabilized by a 5 min incubation in PBS + 0.5% Triton X-100 followed by a 1 h block in PBS containing 3 per cent BSA. Subsequently, cells were washed in PBS + 0.1% Triton X-100 for 5 min. Primary antibody was diluted in PBS + 0.3% BSA + 0.1% Triton X-100 and incubated on the cells for 1 h at room temperature. Cells were washed twice for 5 min followed by a 1 h secondary antibody incubation diluted in PBS + 0.3% BSA + 0.1% Triton X-100. Subsequently, 45 min washes were performed in PBS + 0.1% Triton X-100. DAPI was included in the penultimate wash at 0.1 µg ml−1 concentration.
Primary rabbit anti-HsPoly serum was used in immunofluorescence experiments at a 1 : 1000 dilution. Texas-Red conjugated goat anti-rabbit (1 : 500) secondary antibody and Alexa Fluor-488-conjugated phalloidin (1 : 500) were purchased from Molecular Probes.
Supplementary Material
Supplementary Material
Supplementary Material
6. Acknowledgements
The authors would like to express their gratitude to Dr Gerard Cagney (Conway Institute, University College Dublin) for invaluable assistance with the mass spectrometry experiments. We also thank Kasper Pedersen and the Conway Institute Mass Spectrometry Resource for experimental support, and Andreas de Stefani for bioinformatics support. Emanuela Giacometti, Josefin Fernius, Shubha Gururaja Rao, Swati Naidu and Mei Xuan Lye made valuable contributions at various stages of this project. We thank William Earnshaw and Paul Hartley for critical feedback on the manuscript. E.B., S.V., N.C. and M.M.S.H. were supported by The Wellcome Trust. B.N. was supported by the Natural Sciences and Engineering Research Council of Canada, while V.S. was an MSc student at the University of Edinburgh. D.R.D. was supported by a British Heart Foundation Centre of Research Excellence Award.
References
- 1.Wullschleger S, Loewith R, Hall MN. 2006. TOR signaling in growth and metabolism. Cell 124, 471–484 10.1016/j.cell.2006.01.016 (doi:10.1016/j.cell.2006.01.016) [DOI] [PubMed] [Google Scholar]
- 2.Grewal SS. 2009. Insulin/TOR signaling in growth and homeostasis: a view from the fly world. Int. J. Biochem. Cell Biol. 41, 1006–1010 10.1016/j.biocel.2008.10.010 (doi:10.1016/j.biocel.2008.10.010) [DOI] [PubMed] [Google Scholar]
- 3.Britton JS, et al. 2002. Drosophila's insulin/PI3-kinase pathway coordinates cellular metabolism with nutritional conditions. Dev. Cell 2, 239–249 10.1016/S1534-5807(02)00117-X (doi:10.1016/S1534-5807(02)00117-X) [DOI] [PubMed] [Google Scholar]
- 4.Hennig KM, Colombani J, Neufeld TP. 2006. TOR coordinates bulk and targeted endocytosis in the Drosophila melanogaster fat body to regulate cell growth. J. Cell Biol. 173, 963–974 10.1083/jcb.200511140 (doi:10.1083/jcb.200511140) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Puig O, et al. 2003. Control of cell number by Drosophila FOXO: downstream and feedback regulation of the insulin receptor pathway. Genes Dev. 17, 2006–2020 10.1101/gad.1098703 (doi:10.1101/gad.1098703) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manning BD. 2004. Balancing Akt with S6K: implications for both metabolic diseases and tumorigenesis. J. Cell Biol. 167, 399–403 10.1083/jcb.200408161 (doi:10.1083/jcb.200408161) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cai SL, et al. 2006. Activity of TSC2 is inhibited by AKT-mediated phosphorylation and membrane partitioning. J. Cell Biol. 173, 279–289 10.1083/jcb.200507119 (doi:10.1083/jcb.200507119) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao X, Pan D. 2001. TSC1 and TSC2 tumor suppressors antagonize insulin signaling in cell growth. Genes Dev. 15, 1383–1392 10.1101/gad.901101 (doi:10.1101/gad.901101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Potter CJ, Huang H, Xu T. 2001. Drosophila Tsc1 functions with Tsc2 to antagonize insulin signaling in regulating cell growth, cell proliferation, and organ size. Cell 105, 357–368 10.1016/S0092-8674(01)00333-6 (doi:10.1016/S0092-8674(01)00333-6) [DOI] [PubMed] [Google Scholar]
- 10.Long X, et al. 2005. Rheb binds and regulates the mTOR kinase. Curr. Biol. 15, 702–713 10.1016/j.cub.2005.02.053 (doi:10.1016/j.cub.2005.02.053) [DOI] [PubMed] [Google Scholar]
- 11.Scott RC, Schuldiner O, Neufeld TP. 2004. Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev. Cell. 7, 167–178 10.1016/j.devcel.2004.07.009 (doi:10.1016/j.devcel.2004.07.009) [DOI] [PubMed] [Google Scholar]
- 12.Radimerski T, et al. 2002. dS6K-regulated cell growth is dPKB/dPI(3)K-independent, but requires dPDK1. Nat. Cell. Biol. 4, 251–255 10.1038/ncb763 (doi:10.1038/ncb763) [DOI] [PubMed] [Google Scholar]
- 13.Sarbassov DD, et al. 2008. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 307, 1098–1101 10.1126/science.1106148 (doi:10.1126/science.1106148) [DOI] [PubMed] [Google Scholar]
- 14.Marr MT, et al. 2007. IRES-mediated functional coupling of transcription and translation amplifies insulin receptor feedback. Genes Dev. 21, 175–183 10.1101/gad.1506407 (doi:10.1101/gad.1506407) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Puig O, Tjian R. 2005. Transcriptional feedback control of insulin receptor by dFOXO/FOXO1. Genes Dev. 19, 2435–2446 10.1101/gad.1340505 (doi:10.1101/gad.1340505) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Teleman AA, Chen YW, Cohen SM. 2005. 4E-BP functions as a metabolic brake used under stress conditions but not during normal growth. Genes Dev. 19, 1844–1848 10.1101/gad.341505 (doi:10.1101/gad.341505) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mizushima N, et al. 2008. Autophagy fights disease through cellular self-digestion. Nature 451, 1069–1075 10.1038/nature06639 (doi:10.1038/nature06639) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levine B, Kroemer G. 2008. Autophagy in the pathogenesis of disease. Cell 132, 27–42 10.1016/j.cell.2007.12.018 (doi:10.1016/j.cell.2007.12.018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karpen GH, Spradling AC. 1992. Analysis of subtelomeric heterochromatin in the Drosophila minichromosome Dp1187 by single P element insertional mutagenesis. Genetics 132, 737–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klusza S, Deng WM. 2010. poly is required for nurse-cell chromosome dispersal and oocyte polarity in Drosophila. Fly (Austin) 4, 128–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krogan NJ, Greenblatt JF. 2001. Characterization of a six-subunit holo-elongator complex required for the regulated expression of a group of genes in Saccharomyces cerevisiae. Mol. Cell Biol. 21, 8203–8212 10.1128/MCB.21.23.8203-8212.2001 (doi:10.1128/MCB.21.23.8203-8212.2001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Winkler GS, et al. 2001. RNA polymerase II elongator holoenzyme is composed of two discrete subcomplexes. J. Biol. Chem. 276, 32 743–32 749 10.1074/jbc.M105303200 (doi:10.1074/jbc.M105303200) [DOI] [PubMed] [Google Scholar]
- 23.Hawkes NA, et al. 2002. Purification and characterization of the human elongator complex. J. Biol. Chem. 277, 3047–3052 10.1074/jbc.M110445200 (doi:10.1074/jbc.M110445200) [DOI] [PubMed] [Google Scholar]
- 24.Nelissen H, et al. 2005. The elongata mutants identify a functional Elongator complex in plants with a role in cell proliferation during organ growth. Proc. Natl Acad. Sci. USA. 102, 7754–7759 10.1073/pnas.0502600102 (doi:10.1073/pnas.0502600102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Winkler GS, et al. 2002. Elongator is a histone H3 and H4 acetyltransferase important for normal histone acetylation levels in vivo. Proc. Natl Acad. Sci. USA 99, 3517–3522 10.1073/pnas.022042899 (doi:10.1073/pnas.022042899) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Svejstrup JQ. 2007. Elongator complex: how many roles does it play? Curr. Opin. Cell. Biol. 19, 331–336 10.1016/j.ceb.2007.04.005 (doi:10.1016/j.ceb.2007.04.005) [DOI] [PubMed] [Google Scholar]
- 27.Versees W, De Groeve S, Van Lijsebettens M. 2010. Elongator, a conserved multitasking complex? Mol. Microbiol. 76, 1065–1069 10.1111/j.1365-2958.2010.07162.x (doi:10.1111/j.1365-2958.2010.07162.x) [DOI] [PubMed] [Google Scholar]
- 28.Walker J, et al. 2011. Role of Elongator Subunit Elp3 in Drosophila melanogaster larval development and immunity. Genetics 187, 1067–1075 10.1534/genetics.110.123893 (doi:10.1534/genetics.110.123893) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ponting CP. 2002. Novel domains and orthologues of eukaryotic transcription elongation factors. Nucleic Acids Res. 30, 3643–3652 10.1093/nar/gkf498 (doi:10.1093/nar/gkf498) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Figeys D, McBroom LD, Moran MF. 2001. Mass spectrometry for the study of protein–protein interactions. Methods 24, 230–239 10.1006/meth.2001.1184 (doi:10.1006/meth.2001.1184) [DOI] [PubMed] [Google Scholar]
- 31.Shiio Y, et al. 2002. Quantitative proteomic analysis of Myc oncoprotein function. Embo J. 21, 5088–5096 10.1093/emboj/cdf525 (doi:10.1093/emboj/cdf525) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fernandez R, et al. 1995. The Drosophila insulin receptor homolog: a gene essential for embryonic development encodes two receptor isoforms with different signaling potential. Embo J. 14, 3373–3384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brand AH, Perrimon N. 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401–415 [DOI] [PubMed] [Google Scholar]
- 34.Hay N, Sonenberg N. 2004. Upstream and downstream of mTOR. Genes Dev. 18, 1926–1945 10.1101/gad.1212704 (doi:10.1101/gad.1212704) [DOI] [PubMed] [Google Scholar]
- 35.Butterworth FM, Forrest EC. 1984. Ultrastructure of the preparative phase of cell death in the larval fat body of Drosophila melanogaster. Tissue Cell. 16, 237–250 10.1016/0040-8166(84)90047-8 (doi:10.1016/0040-8166(84)90047-8) [DOI] [PubMed] [Google Scholar]
- 36.Butterworth FM, Emerson L, Rasch EM. 1988. Maturation and degeneration of the fat body in the Drosophila larva and pupa as revealed by morphometric analysis. Tissue Cell 20, 255–268 10.1016/0040-8166(88)90047-X (doi:10.1016/0040-8166(88)90047-X) [DOI] [PubMed] [Google Scholar]
- 37.Juhasz G, et al. 2003. The Drosophila homolog of Aut1 is essential for autophagy and development. FEBS Lett. 543, 154–158 10.1016/S0014-5793(03)00431-9 (doi:10.1016/S0014-5793(03)00431-9) [DOI] [PubMed] [Google Scholar]
- 38.Rusten TE, et al. 2004. Programmed autophagy in the Drosophila fat body is induced by ecdysone through regulation of the PI3K pathway. Dev. Cell 7, 179–192 10.1016/j.devcel.2004.07.005 (doi:10.1016/j.devcel.2004.07.005) [DOI] [PubMed] [Google Scholar]
- 39.Scott RC, Juhasz G, Neufeld TP. 2007. Direct induction of autophagy by Atg1 inhibits cell growth and induces apoptotic cell death. Curr. Biol. 17, 1–11 10.1016/j.cub.2006.10.053 (doi:10.1016/j.cub.2006.10.053) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Juhasz G, et al. 2007. Gene expression profiling identifies FKBP39 as an inhibitor of autophagy in larval Drosophila fat body. Cell Death Differ. 14, 1181–1190 10.1038/sj.cdd.4402123 (doi:10.1038/sj.cdd.4402123) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Berry DL, Baehrecke EH. 2007. Growth arrest and autophagy are required for salivary gland cell degradation in Drosophila. Cell 131, 1137–1148 10.1016/j.cell.2007.10.048 (doi:10.1016/j.cell.2007.10.048) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baker KD, Thummel CS. 2007. Diabetic larvae and obese flies-emerging studies of metabolism in Drosophila. Cell Metab. 6, 257–266 10.1016/j.cmet.2007.09.002 (doi:10.1016/j.cmet.2007.09.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saltiel AR, Kahn CR. 2001. Insulin signalling and the regulation of glucose and lipid metabolism. Nature 414, 799–806 10.1038/414799a (doi:10.1038/414799a) [DOI] [PubMed] [Google Scholar]
- 44.Gronke S, et al. 2007. Dual lipolytic control of body fat storage and mobilization in Drosophila. PLoS Biol. 5, e137. 10.1371/journal.pbio.0050137 (doi:10.1371/journal.pbio.0050137) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Teixeira L, et al. 2003. Drosophila Perilipin/ADRP homologue Lsd2 regulates lipid metabolism. Mech. Dev. 120, 1071–1081 10.1016/S0925-4773(03)00158-8 (doi:10.1016/S0925-4773(03)00158-8) [DOI] [PubMed] [Google Scholar]
- 46.Manning BD, Cantley LC. 2007. AKT/PKB signaling: navigating downstream. Cell 129, 1261–1274 10.1016/j.cell.2007.06.009 (doi:10.1016/j.cell.2007.06.009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bellacosa A, et al. 2005. Activation of AKT kinases in cancer: implications for therapeutic targeting. Adv. Cancer Res. 94, 29–86 10.1016/S0065-230X(05)94002-5 (doi:10.1016/S0065-230X(05)94002-5) [DOI] [PubMed] [Google Scholar]
- 48.Engelman JA, Luo J, Cantley LC. 2006. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat. Rev. Genet. 7, 606–619 10.1038/nrg1879 (doi:10.1038/nrg1879) [DOI] [PubMed] [Google Scholar]
- 49.Ciapponi L, et al. 2004. The Drosophila Mre11/Rad50 complex is required to prevent both telomeric fusion and chromosome breakage. Curr. Biol. 14, 1360–1366 10.1016/j.cub.2004.07.019 (doi:10.1016/j.cub.2004.07.019) [DOI] [PubMed] [Google Scholar]
- 50.Savvidou E, et al. 2005. Drosophila CAP-D2 is required for condensin complex stability and resolution of sister chromatids. J. Cell. Sci. 118, 2529–2543 10.1242/jcs.02392 (doi:10.1242/jcs.02392) [DOI] [PubMed] [Google Scholar]
- 51.Vass S, et al. 2003. Depletion of Drad21/Scc1 in Drosophila cells leads to instability of the cohesin complex and disruption of mitotic progression. Curr. Biol. 13, 208–218 10.1016/S0960-9822(03)00047-2 (doi:10.1016/S0960-9822(03)00047-2) [DOI] [PubMed] [Google Scholar]
- 52.Vereshchagina N, Wilson C. 2006. Cytoplasmic activated protein kinase Akt regulates lipid-droplet accumulation in Drosophila nurse cells. Development 133, 4731–4735 10.1242/dev.02659 (doi:10.1242/dev.02659) [DOI] [PubMed] [Google Scholar]
- 53.Singh R, et al. 2009. Autophagy regulates lipid metabolism. Nature 458, 1131–1135 10.1038/nature07976 (doi:10.1038/nature07976) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang H, et al. 2000. Regulation of cellular growth by the Drosophila target of rapamycin dTOR. Genes Dev. 14, 2712–2724 10.1101/gad.835000 (doi:10.1101/gad.835000) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Braun A, Hoffmann JA, Meister M. 1998. Analysis of the Drosophila host defense in domino mutant larvae, which are devoid of hemocytes. Proc. Natl Acad. Sci. USA. 95, 14 337–14 342 10.1073/pnas.95.24.14337 (doi:10.1073/pnas.95.24.14337) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dearolf CR. 1998. Fruit fly ‘leukemia’. Biochim. Biophys. Acta 1377, M13–M23 [DOI] [PubMed] [Google Scholar]
- 57.Minakhina S, Steward R. 2006. Melanotic mutants in Drosophila: pathways and phenotypes. Genetics 174, 253–263 10.1534/genetics.106.061978 (doi:10.1534/genetics.106.061978) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nguyen L, et al. 2010. Elongator—an emerging role in neurological disorders. Trends Mol. Med. 16, 1–6 10.1016/j.molmed.2009.11.002 (doi:10.1016/j.molmed.2009.11.002) [DOI] [PubMed] [Google Scholar]
- 59.Prisco M, et al. 2002. Nuclear translocation of insulin receptor substrate-1 by the simian virus 40 T antigen and the activated type 1 insulin-like growth factor receptor. J. Biol. Chem. 277, 32 078–32 085 10.1074/jbc.M204658200 (doi:10.1074/jbc.M204658200) [DOI] [PubMed] [Google Scholar]
- 60.Sun H, et al. 2003. Insulin-like growth factor I receptor signaling and nuclear translocation of insulin receptor substrates 1 and 2. Mol. Endocrinol. 17, 472–486 10.1210/me.2002-0276 (doi:10.1210/me.2002-0276) [DOI] [PubMed] [Google Scholar]
- 61.Wu A, Chen J, Baserga R. 2008. Nuclear insulin receptor substrate-1 activates promoters of cell cycle progression genes. Oncogene 27, 397–403 10.1038/sj.onc.1210636 (doi:10.1038/sj.onc.1210636) [DOI] [PubMed] [Google Scholar]
- 62.Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 10.1093/nar/25.17.3389 (doi:10.1093/nar/25.17.3389) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Söding J, Biegert A, Lupas AN. 2005. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33, W244–W248 10.1093/nar/gki408 (doi:10.1093/nar/gki408) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wallace IM, et al. 2006. M-Coffee: combining multiple sequence alignment methods with T-Coffee. Nucleic Acids Res. 34, 1692–1699 10.1093/nar/gkl091 (doi:10.1093/nar/gkl091) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Biegert A, et al. 2006. The MPI Bioinformatics Toolkit for protein sequence analysis. Nucleic Acids Res. 34, W335–W339 10.1093/nar/gkl217 (doi:10.1093/nar/gkl217) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schmidt HA, et al. 2002. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics 18, 502–504 10.1093/bioinformatics/18.3.502 (doi:10.1093/bioinformatics/18.3.502) [DOI] [PubMed] [Google Scholar]
- 67.Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574 10.1093/bioinformatics/btg180 (doi:10.1093/bioinformatics/btg180) [DOI] [PubMed] [Google Scholar]
- 68.Felsenstein J. 2005. PHYLIP (Phylogeny Inference Package) version 3.6. Distributed by the author. Seattle, WA: Department of Genome Sciences, University of Washington [Google Scholar]
- 69.Huson DH, Bryant D. 2006. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23, 254–267 10.1093/molbev/msj030 (doi:10.1093/molbev/msj030) [DOI] [PubMed] [Google Scholar]
- 70.Whelan S, Goldman N. 2001. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 18, 691–699 [DOI] [PubMed] [Google Scholar]
- 71.Shevchenko A, et al. 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68, 850–858 10.1021/ac950914h (doi:10.1021/ac950914h) [DOI] [PubMed] [Google Scholar]
- 72.Yates JR, III, et al. 1995. Method to correlate tandem mass spectra of modified peptides to amino acid sequences in the protein database. Anal. Chem. 67, 1426–1436 10.1021/ac00104a020 (doi:10.1021/ac00104a020) [DOI] [PubMed] [Google Scholar]
- 73.Keller A, et al. 2002. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74, 5383–5392 10.1021/ac025747h (doi:10.1021/ac025747h) [DOI] [PubMed] [Google Scholar]
- 74.Juhasz G, Neufeld TP. 2008. Experimental control and characterization of autophagy in Drosophila. Methods Mol. Biol. 445, 125–133 10.1007/978-1-59745-157-4_8 (doi:10.1007/978-1-59745-157-4_8) [DOI] [PubMed] [Google Scholar]
- 75.Gronke S, et al. 2003. Control of fat storage by a Drosophila PAT domain protein. Curr. Biol. 13, 603–606 10.1016/S0960-9822(03)00175-1 (doi:10.1016/S0960-9822(03)00175-1) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.