Abstract
The selective elimination system blocks the accumulation of meiosis-specific mRNAs during the mitotic cell cycle in fission yeast. These mRNAs harbour a region, the determinant of selective removal (DSR), which is recognized by a YTH-family RNA-binding protein, Mmi1. Mmi1 directs target transcripts to destruction in association with nuclear exosomes. Hence, the interaction between DSR and Mmi1 is crucial to discriminate mitosis from meiosis. Here, we show that Mmi1 interacts with repeats of the hexanucleotide U(U/C)AAAC that are enriched in the DSR. Disruption of this ‘DSR core motif’ in a target mRNA inhibits its elimination. Tandem repeats of the motif can function as an artificial DSR. Mmi1 binds to it in vitro. Thus, a core motif cluster is responsible for the DSR activity. Furthermore, certain variant hexanucleotide motifs can augment the function of the DSR core motif. Notably, meiRNA, which composes the nuclear Mei2 dot required to suppress Mmi1 activity during meiosis, carries numerous copies of the core/augmenting motifs on its tail and is indeed degraded by the Mmi1/exosome system, indicating its likely role as decoy bait for Mmi1.
Keywords: mRNA degradation, RNA-binding protein, exosome, meiosis, Mmi1
2. Introduction
The gene-expression profile differs greatly between mitotic and meiotic cells. In the fission yeast Schizosaccharomyces pombe, hundreds of transcripts are newly induced or upregulated during meiosis [1], and various types of post-transcriptional regulation, in addition to transcriptional regulation, are involved in these changes [2–5]. Mmi1, which belongs to the RNA-binding protein family YTH [6], plays a pivotal role in a post-transcriptional event termed selective elimination of meiosis-specific mRNAs [3,7]. Mmi1 recognizes a group of meiosis-specific transcripts that are expressed inappropriately in mitotic cells and removes them in cooperation with nuclear exosomes [3,8]. The target transcripts carry a region known as the DSR (determinant of selective removal), to which Mmi1 binds. If this elimination system does not operate, cells cannot continue robust mitotic proliferation owing to an accumulation of deleterious meiosis-specific transcripts [3].
During meiosis, however, the DSR/Mmi1-dependent elimination system itself becomes deleterious. Mei2, the master regulator of meiosis in fission yeast, plays a key role in circumventing this. In meiotic prophase, Mei2 forms a chromosome-associated dot structure together with non-coding RNA (meiRNA) transcribed from the sme2 gene, at this gene locus on chromosome II [9–11]. The Mei2 dot sequesters Mmi1 and inhibits its function, so that meiosis-specific mRNAs may be readily and stably expressed [3]. The DSRs in four meiosis-specific genes, namely mei4, rec8, ssm4 and spo5, were precisely mapped, and the region necessary for proper DSR function was delimited in each case [3]. However, no extensive sequence homology or common features were apparent among these DSRs.
RNA-binding proteins of the YTH family appear to be conserved widely among eukaryotes [6]. Rat YT521-B, the founding member of this family, has been shown to interact with several splicing factors and to alter alternative splice sites in a dose-dependent manner [12,13]. YT521-B was demonstrated to bind to short RNA motifs with high degeneracy [14]. To understand the selective elimination system more profoundly, we set out in our current study to identify the nucleotide motifs that are essential for DSR function using both a computational method known as ‘motif sampling analysis’ [15] and a genetic method involving mutational analysis. Here, we demonstrate that clustering of certain hexanucleotide motifs is responsible for the DSR function. Furthermore, we show that the 3′-tail of the sme2 transcript is rich in these hexanucleotide motifs, indicating that this RNA may serve as decoy bait to lure Mmi1.
3. Results
3.1. Identification of a conserved motif U(U/C)AAAC in the determinant of selective removal
To determine whether a common sequence existed in the DSR of different transcripts, we performed motif sampling analysis of this region in mei4, rec8, ssm4 and spo5 mRNAs, which we have defined previously [3]. We followed the method described previously by Thijs et al. [15]. Although the DSR regions shared no extensive homology among these mRNAs, the hexanucleotide sequences UUAAAC and UCAAAC (electronic supplementary material, figure S1a) were identified as the most over-represented motifs among these molecules. As shown in electronic supplementary material, figure S1b, the mei4, rec8, ssm4 and spo5 DSRs carried seven, four, two and five copies of the DSR core motif U(U/C)AAAC in the respective DSR regions assigned previously to them (orange and yellow arrowheads). Additional copies of this core motif were found to be scattered throughout the mRNA sequences (black and grey arrowheads in electronic supplementary material, figure S1b), which might also contribute to DSR function. It was noted that the rec8 mRNA carried a second cluster of this core motif within its ORF, which we had not identified in our previous analysis. We confirmed that this region could confer the DSR activity, although it was weaker than the original one (data not shown). More precise arrangements of the core motif in each DSR are depicted in electronic supplementary material, figure S1c.
3.2. Substitution mutations affecting the core motifs block the function of determinant of selective removal
In parallel with the aforementioned computer analysis, we wished to pinpoint the region that was critical for spo5 DSR function. We performed deletion analysis of a previously described reporter gene [3] comprising the constitutive adh1 promoter, the GFP ORF and the spo5 DSR. This chimeric gene generated few GFP-spo5DSR transcripts in mitotically growing cells (electronic supplementary material, figure S2a, lane 3), whereas a control construct carrying no DSR region produced GFP transcripts abundantly (lane 1). However, in cells undergoing meiosis via the function of active Mei2 [9], the chimeric gene generated considerable levels of GFP-spo5DSR transcripts (lane 4). We introduced a series of deletions into the spo5 DSR region and found that removal of 21 nucleotides between positions 1778 and 1798 (electronic supplementary material, figure S1) rendered it non-functional (data not shown). We next introduced arbitrary substitutions into this region. One of these mutations, which we designated spo5DSR-M10 (electronic supplementary material, figure S2b), markedly impaired the DSR activity, as a result of which the mitotic cells accumulated GFP-spo5DSR-M10 transcripts, although at a slightly lower abundance than meiotic cells (electronic supplementary material, figure S2a, lane 5 versus lane 6). When matched with the DSR core motif described above, the M10 mutation was found to involve the complete loss of two contiguous copies of this motif (electronic supplementary material, figure S2b).
We next prepared a mutant strain in which all of the seven DSR core motifs in the mei4 DSR were mutated without affecting the protein coding capacity (see §5). The resultant mei4-m7 strain showed a weak growth defect (data not shown). In this strain, mei4 transcripts, but not ssm4 transcripts, were visibly expressed under the nutrient conditions, even at a greater level than in the hypomorphic mmi1-48 mutant (figure 1a) [3]. This confirms that the core motif is central to the functional activity of the DSR region, and also explains the observed growth defect of the strain, because ectopic expression of mei4 is deleterious for vegetative cell growth [3,16,17].
Figure 1.

The DSR core motif is central to DSR function. (a) Expression of mei4 and ssm4 mRNAs in the mei4-m7 cells in which DSR core motifs in the mei4 DSR were abrogated. Transcripts of mei4 and ssm4 were analysed by northern blotting in JY450 (wild-type, lane 1), JT221 (mmi1-48, lane 2) and JT925 (mei4-m7, lane 3). rRNAs stained with ethidium bromide are shown in the bottom panel as loading controls. (b) Tandem repeats of the DSR core motif can reconstitute DSR function. Expression of GFP mRNA was examined from the reporter chimeric gene with no copy of the TTAAAC core motif (JT634, lanes 1 and 2), with five copies (JT629, lanes 3 and 4), with six copies (JT630, lanes 5 and 6), with seven copies (JT631, lanes 7 and 8), with eight copies (JT632, lanes 9 and 10) or with eight copies of a mutated motif GTAAAC (JT633, lanes 11 and 12). +N lanes represent cells growing mitotically, whereas −N lanes represent cells undergoing meiosis, starved of nitrogen for 4 h. rRNAs stained with ethidium bromide are shown in the bottom panel as loading controls. (c) Expression of GFP mRNA from the reporter chimeric gene with eight copies of the TTAAAC core motif in JT923 (mmi1-ts3, lanes 1 and 2) and JT916 (wild-type, lanes 3 and 4) cells. Each strain was grown at 25°C and sampled before (lanes 1 and 3) and after (lanes 2 and 4) the shift to 36°C for 2 h. rRNAs stained with ethidium bromide are shown in the bottom panel as loading controls. (d) Electrophoretic mobility shift assay (EMSA) for the binding of GST-Mmi1 and GST-Mmi1 lacking the YTH domain (ΔYTH) to 1–4 tandem repeats of the DSR core motif (UUAAAC) fused to the GFP ORF transcript.
3.3. Tandem repeats of the core motif exhibit determinant of selective removal activity
The abovementioned observations led us to examine whether repeats of the DSR core motif exhibited DSR functional activity on their own. We constructed a set of yeast strains containing a chimeric gene composed of the adh1 promoter, the GFP ORF and a DNA fragment carrying 1–8 copies of the DSR core motif (TTAAAC). The repeat motif sequences were separated by intervals containing six-base restriction enzyme recognition sequences (see §5). The quantity of transcripts generated from the chimeric gene in each S. pombe strain was measured in both mitotic and meiotic cells. As shown in figure 1b, the chimeric gene carrying up to five copies of TTAAAC exerted no significant DSR effect (lanes 3 and 4). In contrast, the constructs carrying more than five copies of TTAAAC failed to accumulate transcripts in mitotic cells, suggesting that a tandem array of six or more copies of the core motif can function as a DSR (figure 1b, lanes 5, 7 and 9). A control strain carrying eight copies of a mutated motif GTAAAC expressed abundant transcripts in mitotic cells (lane 11). Transcripts of the construct carrying eight copies of TTAAAC were accumulated visibly when the activity of Mmi1 was weakened by the temperature-sensitive mutation (figure 1c), indicating that they are indeed eliminated through the Mmi1-dependent degradation machinery.
3.4. Physical interaction of the determinant of selective removal core motif with Mmi1
We previously showed that Mmi1 could directly bind to the DSR of mei4, rec8, ssm4 and spo5 transcripts in vitro [3]. We thus tested in our current study whether the core motif could bind Mmi1 using a non-radioisotope electrophoretic mobility shift assay (EMSA; see §5). A single copy of the DSR core motif, fused to the GFP ORF transcript, could be trapped considerably by Mmi1, and the binding was more effective if the transcript carried multiple copies of the motif, whereas Mmi1 lacking its YTH domain did not bind to it (figure 1d and electronic supplementary material, figure S3a). This indicates that the core motif is indeed a pivotal element of the DSR during recognition by the YTH domain. If a mutated motif (GUAAAC) was used, the binding was not detected, even with a transcript carrying eight copies of it (electronic supplementary material, figure S3a).
We next quantitatively determined the affinity between Mmi1 and four tandem repeats of the DSR core motif fused to the GFP ORF, by titrating the amount of Mmi1, as was done with YT521-B previously [14] (see §5). The Kd value was calculated to be 65 nM (electronic supplementary material, figure S3b). This affinity was 400 times greater than that of YT521-B for its targets (26 µM). This big difference of affinity might reflect the distinct function of the two proteins, YT521-B being a splicing regulator and Mmi1 being an inducer of mRNA degradation.
3.5. meiRNA carries ample copies of the motif on its tail
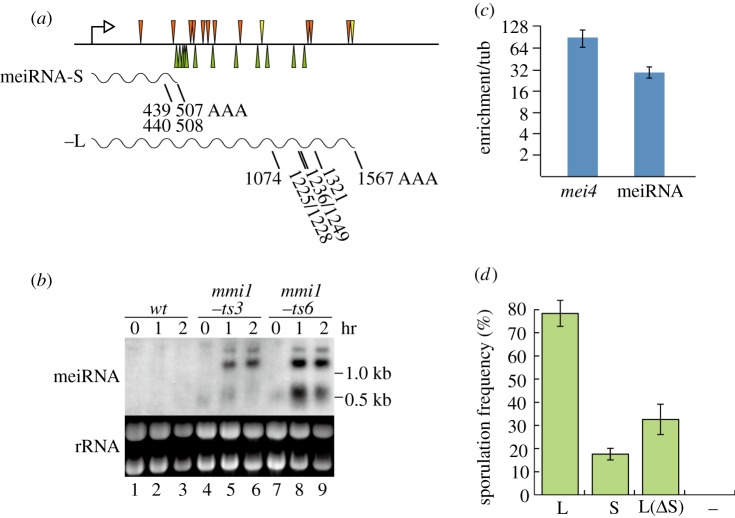
Given the above findings, we performed a genome-wide search for genes carrying repeats of the DSR core motif. This resulted in the identification of a number of candidate genes, most of which turned out to be meiosis-specific. They include mug4 (five copies), mug8 (seven copies), mug10 (six copies) and mug45 (10 copies), and their ectopic expression in the mmi1-ts mutants was indeed confirmed by northern blotting (data not shown). One highly inspiring outcome of this search, among others, was that the 3′-end region of the sme2 gene, which encodes meiRNA required to block the selective removal of DSR-containing mRNAs during meiosis [3,18], is abundantly occupied by the motif (figure 2a).
Figure 2.
meiRNA is a target of Mmi1. (a) Location of the DSR core motif within meiRNA. The UUAAAC sequence is indicated by an orange arrowhead and UCAAAC is indicated by a yellow arrowhead. The augmenting motifs (UAAAAC, UGAAAC and GUAAAC) are indicated by green arrowheads. We determined the precise 3′-ends of long transcripts (meiRNA-L) by RT–PCR. (b) Expression of meiRNA encoded by the sme2 gene in the mmi1 mutants. Transcripts of the sme2 gene (meiRNA) were analysed by northern blotting in cells of JY450 (wild-type, lanes 1–3), JV579 (mmi1-ts3, lanes 4–6) and JV582 (mmi1-ts6, lanes 7–9). Each strain was grown at 25°C, shifted to 36°C and sampled at the indicated time points. (c) Association of meiRNA with Mmi1. RNA-IP was performed for meiRNA and a positive control mei4 mRNA using anti-Mmi1 antibodies. Enrichment over tub1 mRNA is displayed for each. Error bars indicate standard deviations for four independent measurements. (d) Recovery of sporulation in the sme2Δ mutant by artificial expression of meiRNA-L, meiRNA-S and meiRNA-L without the meiRNA-S portion. Derivatives of the sme2 gene encoding either meiRNA-L (L; 1-1562), its 5′ portion (S; 1–508) or its 3′ portion (L(ΔS); 509–1562) were expressed from the vector pREP1 in homothallic sme2Δ cells (JT926), and their sporulation frequency was measured after incubation on SSA medium at 30°C for 3 days. Error bars indicate standard deviations (three measurements for each; total n > 800). The control transformant carrying pREP1 is indicated by (–).
The sme2Δ cells, which cannot develop the Mei2 dot structure to sequester Mmi1, eventually arrest prior to meiosis I [3,10]. In our original characterization, we noticed that meiRNA was transcribed from the sme2 gene mainly as short polyadenylated transcripts of about 0.5 kb in length (439/440 and 507/508 nucleotides excluding poly(A)). We also detected minor doublet bands of about 1.2 kb in meiotic cells, but we interpreted them as probable read-through products that overlapped with the 0.5 kb transcripts [18]. The results described above urged us to recharacterize sme2 transcripts. In the following analyses, we refer to the short canonical meiRNA as meiRNA-S and the long sme2 transcripts collectively as meiRNA-L.
We set out to determine polyadenylation sites of meiRNA-L, recovered from meiotic cells, by 3′-RACE (rapid amplification of cDNA ends). As summarized in figure 2a, this analysis revealed that meiRNA-L is polyadenylated at seven sites at least, which correspond to the transcript length of 1.1–1.6 kb. These meiRNA-L species carried 9–13 copies of the DSR core motif (figure 2a). We have previously shown that meiRNA-S has a moderate affinity for Mmi1 and is likely to sequester this protein in cooperation with Mei2, which also has an affinity for Mmi1 [3]. Expression of meiRNA-L was detectable only in cells undergoing meiosis [18], like DSR-containing meiotic transcripts, but meiRNA itself was dispensable for the progression of meiosis once the activity of Mmi1 was reduced [3]. These observations strongly suggested that meiRNA-L might function as a decoy bait to lure Mmi1. In agreement with this idea, expression of meiRNA-L was elevated enormously in the mmi1-ts mutants shifted to the restrictive temperature on nutrient medium: transcripts of approximately 1.2 and 1.5 kb in length were detected clearly, in addition to 0.5 kb meiRNA-S (figure 2b).
We then tested whether Mmi1 could bind to meiRNA in fission yeast cells by RNA immunoprecipitation (RNA-IP) experiments, as described in §5. Similar to the positive control mei4 mRNA, meiRNA was enriched in the immunoprecipitated Mmi1 complex (figure 2c). These results altogether indicate that meiRNA-L is a substrate of the Mmi1-dependent selective elimination, which may compete with DSR-containing meiosis-specific mRNAs. The significance of this competitor function of meiRNA in the induction of meiosis was supported by the observation that expression of meiRNA-L missing the meiRNA-S sequence could partially suppress sme2Δ (figure 2d).
3.6. Expression of artificial bait can suppress loss of meiRNA
The role of meiRNA as a competitor of the DSR-containing transcripts for selective degradation well explains our previous observation that the high expression of any DSR-containing RNA could suppress meiotic deficiency and recover sporulation in the sme2Δ strain to some extent [3]. Excess DSR-containing RNA is likely to lure Mmi1 during meiotic prophase, as meiRNA-L does, and hence to alleviate degradation of meiosis-specific mRNAs. To confirm this possibility, we mutated core motifs in the mei4 DSR and rec8 DSR, carrying seven and four copies of the motif, respectively, and tested their sme2Δ suppression activity (electronic supplementary material, figure S4). Overexpression of the DSR sequence fused to the GFP ORF from the strong nmt1 promoter suppressed the sme2Δ phenotype efficiently (mei4 DSR, about 50%; rec8 DSR, about 25%; electronic supplementary material, figure S4a,b). We then introduced mutations to motifs 4, 5 and 6 of mei4 DSR and 1–4 of rec8 DSR (see §5). Mutations that damaged a single motif in the mei4 DSR decreased the suppression efficiency to a varying extent, with the M6 mutation showing the largest decrease (electronic supplementary material, figure S4a). Thus, each motif appeared to contribute to the suppression activity to a different degree, depending on its position in the DSR. Significantly, a combination of these mutations further reduced the suppression activity, confirming that the DSR core motif was a key determinant of the suppression efficiency. We obtained the same conclusions from analyses of the rec8 DSR (electronic supplementary material, figure S4b).
3.7. Evaluation of the determinant of selective removal activity of various hexanucleotide sequences
Taking advantage of the relative ease of the sme2Δ suppression assay, we evaluated the DSR activity of various hexanucleotide sequences. Transcripts carrying more than five copies of the core motif UUAAAC suppressed the meiotic arrest of the sme2Δ mutant (figure 3a), in good agreement with the results obtained by the degradation assay (figure 1b). The suppression efficiency increased in a copy number-dependent manner (figure 3a). If the motif was altered to GUAAAC, the suppression was abrogated.
Figure 3.

Evaluation of the DSR activity of hexanucleotide repeats. (a) Sporulation frequency of JZ464 (sme2Δ) cells expressing GFP fused with tandem repeats of the DSR core motif (TTAAAC) or a mutant form (GTAAAC) from the multi-copy plasmid pRGT1. The number of repeats in each construct is depicted by ×N. The control strain carrying pRGT1 is indicated by (–). The sporulation frequency was determined by microscopic observations after incubation on SSA medium at 30°C for 3 days. Error bars indicate standard deviations (three measurements for each; total n > 400). (b) Sporulation frequency of JZ464 cells expressing GFP fused with tandem repeats of the DSR core motif (TCAAAC), assayed as in (a). (c) Sporulation frequency of JZ464 cells expressing GFP fused with tandem repeats of a variant motif (TAAAAC), assayed as in (a).
The DSR activity of repeats of UCAAAC, UAAAAC and UGAAAC was similarly evaluated. UCAAAC, which we defined as the other core motif sequence, revealed weak DSR activity in seven copies, and fairly strong activity when eight or more copies were aligned (figure 3b). UCAAAC is thus slightly less effective than UUAAAC. Two variant sequences, namely UAAAAC and UGAAAC, are also frequently seen in the DSR region, as typically illustrated for meiRNA in figure 2a. However, UAAAAC revealed very low DSR activity, if any, when eight or nine copies were aligned (figure 3c). UGAAAC showed no detectable DSR activity even when 10 copies were aligned (data not shown). Therefore, we tentatively discriminate these sequences from the core motif.
3.8. Hexanucleotides that augment the function of the determinant of selective removal core motif
Even though unable to exert significant DSR activity by themselves, the frequent occurrence of UAAAAC and UGAAAC in the DSR regions implied that they may somehow contribute to the DSR function. Natural DSRs often carry less than six copies of the core motif. We thus tested whether these hexanucleotides might augment the function of the core motif, using the test system described below. We first integrated five copies of the DSR core motif (TTAAAC), which by themselves did not confer sufficient DSR activity, at the end of the GFP ORF on pRGT1. We next placed a single hexanucleotide sequence to be tested after these five repeats. The DSR activity of the resulting composite array was then measured via the sme2Δ suppression assay. If the last copy was TTAAAC (i.e. if there were six copies of TTAAAC) the array could function as an effective DSR (figure 4a; also see figure 3a). The addition of TCAAAC also generated considerable DSR activity, as expected. Notably, TAAAAC and TGAAAC could also recover DSR activity significantly (figure 4a).
Figure 4.

DSR-augmenting motifs. (a) Sporulation frequency of JZ464 (sme2Δ) expressing GFP fused with an array of five copies of the DSR core motif (TTAAAC) plus one copy of a variant motif from a multi-copy plasmid pRGT1. Variants carrying an alteration at the second position were examined. The control strain carrying no additional variant is indicated by ×5, and that carrying only pRGT1 is indicated by (–). Error bars indicate standard deviations (three measurements for each; total n > 400). (b) Variants carrying G in the first to sixth positions were examined as in (a). The control strain carrying pRGT1 is indicated by (–). (c) Sporulation frequency of JZ464 cells expressing GFP fused with six copies of the DSR core motif (TTAAAC) at its 5′- or 3′-ends, or in the midst of the ORF. 2×gfp and 3×gfp represent constructs that have one or two extra copies of the GFP ORF at the 3′-end, respectively. Error bars indicate standard deviations (three measurements for each; total n > 400).
Variants in which the third, fourth, fifth or sixth position of the TTAAAC motif was substituted by G exhibited no sme2Δ suppression when they were linked to the 5×TTAAAC array, indicating that these altered motifs possessed no DSR-enhancing activity (figure 4b). Interestingly, however, a first position G substitution (GTAAAC) could induce weak suppression activity (figure 4b), although tandem repeats of eight copies of GUAAAC could not reconstitute DSR function (figures 1b and 3a).
The above observations indicated that the UAAAAC, UGAAAC and GUAAAC sequences could contribute to the DSR activity if combined with the canonical core motif U(U/C)AAAC. We thus refer to these sequences as ‘DSR-augmenting motifs’. Given these results, we revisited the sequences of the mei4, rec8, ssm4 and spo5 genes, and determined the number of both core and augmenting motifs in each respective DSR region (table 1). The ssm4 DSR carries only two copies of the core motif but harbours five copies of the augmenting motif (hence seven copies in total). In contrast, the spo5 DSR contains five copies of the core motif but only one copy of the augmenting motif. There are 13 copies of the core motif and 12 copies of the augmenting motif on the longest meiRNA-L (table 1).
Table 1.
The copy number of DSR core/augmenting motifs in each DSR.
| core motif |
augmenting motif |
total | ||||
|---|---|---|---|---|---|---|
| UUAAAC | UCAAAC | UAAAAC | UGAAAC | GUAAAC | ||
| mei4 DSR | 3 | 4 | 0 | 1 | 0 | 8 |
| rec8 DSR | 4 | 0 | 1 | 1 | 0 | 6 |
| ssm4 DSR | 2 | 0 | 1 | 4 | 0 | 7 |
| spo5 DSR | 2 | 3 | 1 | 0 | 0 | 6 |
| meiRNA-L | 11 | 2 | 5 | 7 | 0 | 25 |
3.9. Overall positional effects upon determinant of selective removal activity
We investigated whether the activity of a DSR might be dependent on its positioning within the target transcript. We placed an artificial DSR containing six copies of the core motif (TTAAAC) at various positions within pRGT1: immediately after the GFP ORF (3′), just before the first codon (5′) or in the midst of the ORF (middle; 167 bp downstream of the first codon; figure 4c). The DSR activity levels were again estimated by measuring the suppression of the sme2Δ phenotype. As shown in figure 4c, the 3′ insertion was the most effective in this regard, followed by the 5′ insertion, whereas the middle insertion was found to be the least effective. When the distance between the 5′ positioned DSR and the tail of the transcript was extended by insertion of one to two more copies of the GFP ORF, the suppression efficiency dropped with the increasing distance (figure 4c; 5′ 2×gfp and 3×gfp). The middle DSR also appeared to lose activity if another copy of the GFP ORF was inserted (figure 4c; middle 2×gfp).
The above results may indicate that, in general, a DSR region is more effective when positioned closer to the 3′-end of a transcript, as long as the DNA sequence surrounding it is identical. However, it is also true that the activity of a DSR is much affected by the context in which it is situated.
4. Discussion
In our present study, we establish that two hexanucleotide sequences, UUAAAC and UCAAAC, represent the DSR core motif, and play a principal role in exerting DSR activity and function. Our data also show that three further hexanucleotide sequences, UAAAAC, UGAAAC and GUAAAC, are DSR-augmenting motifs that play an assisting functional role, although GUAAAC does not seem to be a preferred choice in nature (table 1). Our analyses have suggested that the cooperation of six or more copies of the core/augmenting motifs is essential to yield full DSR function in vivo. A single copy of UUAAAC cannot do so, although it does bind Mmi1 effectively in vitro. As the emergence of a single hexanucleotide sequence is not a statistically rare event, it seems likely that the cell has developed additional regulatory processes that restrict DSR activity to a substantial number of repeats of these sequences.
It has been shown recently that Mmi1 induces facultative heterochromatin formation at the mei4 and ssm4 loci [19]. However, heterochromatin formation has not been detected at the rec8 and spo5 loci, although all four loci encode the targets of Mmi1. Red1, a component of the DSR/Mmi1-dependent elimination system [20], might have a role in discriminating these loci, because it is recruited only to the former and is essential for the assembly of heterochromatin at these loci [19]. Therefore, it is plausible that certain sequence motifs, in addition to the hexanucleotide motifs, may also contribute to the DSR function and give individual traits to each DSR.
One factor that may influence DSR activity but has not been fully explored is the space between neighbouring copies of the core motif. As shown in electronic supplementary material, figure S1c, this region is mostly 30–35 bases in length in naturally occurring DSRs, but is also variable. For example, in the mei4 DSR region, the shortest spacer region is 10 bases and the longest is 57 bases. Moreover, the spo5 DSR contains two core motif copies that are contiguous. We set the spacer region at 6 bases in our current analysis of artificial DSRs as this enabled ease of manipulation, and the obtained results suggested that constructed DSRs are functionally comparable with naturally occurring DSRs. Hence, the spacing of the DSR core/augmenting motifs may not be critically important for activity, but it remains to be seen how this affects the function of the entire array of motifs.
In a previous study from our laboratory [3], we defined some of the DSR regions within the ORFs of specific genes, which were hundreds of bases in length. This was surprising to us at the time as it was uncertain how these regions within certain genes would not compromise their coding capacity. Given that we have now mapped the essential element underlying DSR activity to a six-base motif, this appears not to be an issue any longer. UYAAAC can be translated in three reading frames, and an analysis of all of the ‘coding’ DSR core motifs shown in electronic supplementary material, figure S1b indicates that the core motif is most frequently translated as codons UYA-AAC (15 of 26), followed by UY-AAA-C (9 of 26). U-YAA-AC was found to be relatively rare, but it is unclear if this has any significance as UAA is a stop codon. Taken together, we suspect that it may not be extremely difficult for a useful coding sequence to exist that is strewn with DSR core/augmenting motifs. The use of the augmenting motifs, which are less potent than the core motif, may be rationalized by the selective pressure for certain amino acid sequences in the gene product.
Our analysis of the DSR core/augmenting motifs led to the finding that meiRNA is a target of Mmi1. We have shown that meiRNA and its binding partner Mei2 form an intranuclear dot structure during meiotic prophase, and sequester Mmi1 so that meiotic mRNAs carrying DSR may be stably expressed [3,10]. The short transcripts from sme2, namely meiRNA-S, appear to be able to fulfil this function. Given the remarkable structure of meiRNA-L, however, it is now evident that competition between this RNA and DSR-containing mRNAs as substrates for the Mmi1-dependent destruction system also contributes substantially to the suppression of the Mmi1 function during meiosis. How these two kinds of mechanisms are mutually integrated remains currently elusive. So far, in our analysis all the detected transcripts from sme2 appear to be polyadenylated at the 3′-end. While this is consistent with previous observations that the Mmi1/exosome-dependent RNA degradation is closely associated with conventional polyadenylation [8,21], it is puzzling whether each meiRNA species is transcribed as a distinct polyadenylated RNA; alternatively, meiRNA-S may represent digestion products of meiRNA-L, which somehow undergo repolyadenylation. How highly expressed meiRNA lures Mmi1 to the nuclear dot structure and dampens it offers a challenging question.
5. Material and methods
5.1. Fission yeast strains, genetic analysis and growth media
The S. pombe strains used in this study are listed in table 2. We constructed strains JT629, JT630, JT631, JT632, JT633 and JT634 by inserting a chimeric gene carrying the adh promoter, the GFP ORF and a varying number of the DSR core motif or its variant at the lys1 locus on chromosome I. JT925 was constructed by transforming a mei4::ura4+ strain with a mutated mei4 fragment in which seven DSR core motifs were altered without affecting the protein-coding capacity (M1, AGCAAT; M2, TAAAGC; M3, CTGAAT; M4, AGCAAT; M5, CTGAAT; M6, TGAAGC; M7, AGCAAT). The general genetic procedures used for the analyses of the S. pombe strains have been previously described [22]. Yeast transformations were performed using a lithium acetate method [23]. Growth medium included complete medium YE, minimal medium SD and MM [24] and synthetic sporulation medium SSA [25].
Table 2.
Schizosaccharomyces pombe strains used in this study.
| strain | genotype |
|---|---|
| JT219 | h− mmi1-48-kanR ade6-216 leu1 |
| JT221 | h90 mmi1-48-kanR ade6-216 leu1 |
| JT629 | h+/h− lys1+/lys1::adh-GFP-(5xDSR core)-KanR ade6-M216/ade6-M210 leu1/leu1 |
| JT630 | h+/h− lys1+/lys1::adh-GFP-(6xDSR core)-KanR ade6-M216/ade6-M210 leu1/leu1 |
| JT631 | h+/h− lys1+/lys1::adh-GFP-(7xDSR core)-KanR ade6-M216/ade6-M210 leu1/leu1 |
| JT632 | h+/h− lys1+/lys1::adh-GFP-(8xDSR core)-KanR ade6-M216/ade6-M210 leu1/leu1 |
| JT633 | h+/h− lys1+/lys1/lys1::adh-GFP-(8xmutated DSR core)-KanR ade6-M216/ade6-M210 leu1/leu1 |
| JT634 | h+/h− lys1+/lys1/lys1::adh-GFP-KanR ade6-M216/ade6-M210 leu1/leu1 |
| JT916 | h− lys1::adh-GFP-(8xDSR core)-KanR ade6-M216 leu1 |
| JT923 | h− lys1::adh-GFP-(8xDSR core)-KanR mmi1-ts3-bsdR ade6-M216 leu1 |
| JT925 | h90 mei4-m7 ade6-M216 leu1 |
| JT926 | h90 sme2(1-1065):: ura4+ ade6-M216 leu1 ura4-D18 |
| JV558 | h− mmi1-kanR ade6-M216 leu1 |
| JV564 | h− mmi1-ts3-kanR ade6-M216 leu1 |
| JV567 | h− mmi1-ts6-kanR ade6-M216 leu1 |
| JV579 | h90 mmi1-ts3-kanR ade6-M216 leu1 |
| JV582 | h90 mmi1-ts6-kanR ade6-M216 leu1 |
| JX383 | h− leu1::nmt41-mei2-SATA-ura4+ ade6-M210 ura4-D18 |
| JY362 | h+/h− ade6-M216/ade6-M210 leu1/leu1 |
| JY450 | h90 ade6-M216 leu1 |
| JZ464 | h90 sme2::ura4+ ade6-M216 leu1 ura4-D18 |
5.2. Plasmid construction
The DSR regions of the mei4 and rec8 genes were cloned into a GFP-fusion vector pRGT1, and that of spo5 was cloned into pAGT1. pRGT1 is a derivative of the S. pombe expression vector pREP1 [26], which harbours the ORF of a mutant version of GFP (Ser65-Thr). pAGT1 is a variant of pRGT1 in which the inducible nmt1 promoter is replaced by the constitutive adh1 promoter. The introduction of mutations into the DSR core motif (M1–M7 in mei4 DSR shown above; and m1, ACTAGT; m2, CCATGG; m3, AGATCT; and m4, CATATG in rec8 DSR) was achieved either with a standard mutagenesis protocol [27] or as indicated in the instructions for the PrimeSTAR Mutagenesis Basal Kit (Takara Bio). Plasmids carrying tandem repeats of the DSR core motifs were constructed using the oligonucleotides 5′-GGATCCTTAAACAGATCT-3′ and 5′-GGATCCTTAAACGAATTCTTAAACAGATCT-3′ (the core motif is italicized). These molecules were sequentially inserted into pRGT1. To construct a mutant version of the DSR core motif, the TTAAAC sequence in these oligonucleotides was changed to GTAAAC.
5.3. Northern and western blotting analysis
Northern blotting analysis was performed as described previously [28] using a DNA probe for the GFP sequence or for the gene indicated. Ten micrograms of total cellular RNA was used for each sample. Western blotting analysis was performed using a general method with anti-Mei2 antibodies.
5.4. Non-radioisotope electrophoretic mobility shift assay
Digoxigenin (DIG)-labelled RNA was prepared from PCR products according to the manufacturer's instructions (Roche). The RNA binding reaction was performed using 0.4 nM of DIG-labelled RNA and 40 nM of bacterially purified glutathione S-transferase (GST)-Mmi1 or GST-Mmi1 lacking the YTH domain (ΔYTH; carrying residues 1–292) in 10 µl of a modified KNET buffer: 20 mM KCl, 80 mM NaCl, 2 mM ethylene glycol bis-(2-aminoethylether) tetraacetic acid (EGTA), 50 mM Tris–HCl (pH 7.5), 0.05% NP-40, 1 mM MgCl2, 2 mM dithiothreitol, 10% glycerol and RNase Inhibitor (Roche) [18]. Samples were preincubated at room temperature with 50 µg of carrier E. coli tRNA for 20 min. The labelled RNA was then added and incubation proceeded for another 20 min. Samples were analysed by polyacrylamide gel electrophoresis and electroblotted to GeneScreen Plus membrane (NEN) using 0.5X tris-borate-EDTA (TBE) buffer. Signals were detected using a DIG Luminescent Detection Kit (Roche). Determination of the affinity of Mmi1 for the DSR core motif was done according to Zhang et al. [14]. In brief, EMSA was performed as described above, with 0.4 nM of DIG-labelled RNA carrying the GFP ORF, and four copies of the DSR core motif and 5–400 nM of GST-Mmi1. Bound and free RNA were quantified using LAS-1000plus (GE Healthcare). The Kd was calculated from the plot ln[RNAbound]/[RNAfree] versus ln[proteinfree].
5.5. RNA-immunoprecipitation
Detailed conditions for RNA-IP are described by Hiriart et al. [29]. Immunoprecipitated RNA was isolated by phenol–chloroform extraction and reverse-transcribed according to the manufacturer's instructions (Superscript II, Invitrogen). Quantitative analysis of the relative RNA levels was performed using real-time PCR (Roche) and the MESA Green qPCR Master mix (Eurogentec). A pre-amplification step, done as described [30,31], was implemented owing to the low abundance of sme2 RNAs using the following programme: 15 min incubation at 95°C, followed by 14 cycles of 95°C for 15 s, 60°C for 30 s and 70°C for 30 s. Four microlitres of a 1/400 final dilution of the preamplification reaction were used for the second quantitative PCR with a programme of 15 min incubation at 95°C, and followed by 40 cycles of 95°C for 15 s, 60°C for 30 s and 70°C for 30 s. Primers used are: tub1 forward (5′-GTACTGGCCCATACCGTGAT-3′), tub1 reverse (5′-CGAATGGAAGACGAGAAAGC-3′), mei4 forward (5′-AAAAGCGACCTTCAAGCAAA-3′), mei4 reverse (5′-TTGCATCGTTTGAGACTTCG-3′), meiRNA forward (5′-TGGTCATTCAAAAAGCTGGA-3′) and meiRNA reverse (5′-CTTGGGGGTTGGTTTAACTG-3′).
Supplementary Material
6. Acknowledgements
We thank Mai Sato and Atsushi Ueda for their help in the early stages of this study, and Juan Mata for helpful discussion. The major part of this work, done in Tokyo, was supported by Grants-in-Aid for Scientific Research (C) to A.Y. and (S) to M. Y. from Japan Society for the Promotion of Science. Y.S. is a recipient of a JSPS Research Fellowship for Young Scientists (DC). It was also supported in part by the Global COE Programme (Integrative Life Science Based on the Study of Biosignaling Mechanisms) of the Ministry of Education, Culture, Sports, Science and Technology of Japan (MEXT). Research conducted in Grenoble was supported by the International Human Frontier Science Programme Organization (HFSPO), the ARC, the Institut National de la Santé Et de la Recherche Médicale (INSERM) Avenir programme and a European Research Council (ERC) Starting Grant. Research conducted in Kobe was supported by Grants-in-Aid from MEXT to D.-Q.D. and Y.H. During preparation of this manuscript, a paper came out in which the authors noted the abundant occurrence of U(U/C/G)AAAC in the DSR regions and assumed it as a putative binding site of Mmi1, albeit they showed no substantial evidence for the binding [32].
References
- 1.Mata J, Lyne R, Burns G, Bahler J. 2002. The transcriptional program of meiosis and sporulation in fission yeast. Nat. Genet. 32, 143–147 10.1038/ng951 (doi:10.1038/ng951) [DOI] [PubMed] [Google Scholar]
- 2.Surosky RT, Esposito RE. 1992. Early meiotic transcripts are highly unstable in Saccharomyces cerevisiae. Mol. Cell. Biol. 12, 3948–3958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harigaya Y, et al. 2006. Selective elimination of messenger RNA prevents an incidence of untimely meiosis. Nature 442, 45–50 10.1038/nature04881 (doi:10.1038/nature04881) [DOI] [PubMed] [Google Scholar]
- 4.Moldon A, Malapeira J, Gabrielli N, Gogol M, Gomez-Escoda B, Ivanova T, Seidel C, Ayte J. 2008. Promoter-driven splicing regulation in fission yeast. Nature 455, 997–1000 10.1038/nature07325 (doi:10.1038/nature07325) [DOI] [PubMed] [Google Scholar]
- 5.McPheeters DS, Cremona N, Sunder S, Chen HM, Averbeck N, Leatherwood J, Wise JA. 2009. A complex gene regulatory mechanism that operates at the nexus of multiple RNA processing decisions. Nat. Struct. Mol. Biol. 16, 255–264 10.1038/nsmb.1556 (doi:10.1038/nsmb.1556) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stoilov P, Rafalska I, Stamm S. 2002. YTH: a new domain in nuclear proteins. Trends Biochem. Sci. 27, 495–497 10.1016/S0968-0004(02)02189-8 (doi:10.1016/S0968-0004(02)02189-8) [DOI] [PubMed] [Google Scholar]
- 7.Harigaya Y, Yamamoto M. 2007. Molecular mechanisms underlying the mitosis–meiosis decision. Chromosome Res. 15, 523–537 10.1007/s10577-007-1151-0 (doi:10.1007/s10577-007-1151-0) [DOI] [PubMed] [Google Scholar]
- 8.Yamanaka S, Yamashita A, Harigaya Y, Iwata R, Yamamoto M. 2010. Importance of polyadenylation in the selective elimination of meiotic mRNAs in growing S. pombe cells. EMBO J. 29, 2173–2181 10.1038/emboj.2010.108 (doi:10.1038/emboj.2010.108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Watanabe Y, Yabana S, Chikashige Y, Hiraoka Y, Yamamoto M. 1997. Phosphorylation of RNA-binding protein controls cell cycle switch from mitotic to meiotic in fission yeast. Nature 386, 187–190 10.1038/386187a0 (doi:10.1038/386187a0) [DOI] [PubMed] [Google Scholar]
- 10.Yamashita A, Watanabe Y, Nukina N, Yamamoto M. 1998. RNA-assisted nuclear transport of the meiotic regulator Mei2p in fission yeast. Cell 95, 115–123 10.1016/S0092-8674(00)81787-0 (doi:10.1016/S0092-8674(00)81787-0) [DOI] [PubMed] [Google Scholar]
- 11.Shimada T, Yamashita A, Yamamoto M. 2003. The fission yeast meiotic regulator Mei2p forms a dot structure in the horse-tail nucleus in association with the sme2 locus on chromosome II. Mol. Biol. Cell 14, 2461–2469 10.1091/mbc.E02-11-0738 (doi:10.1091/mbc.E02-11-0738) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Imai Y, Matsuo N, Ogawa S, Tohyama M, Takagi T. 1998. Cloning of a gene, YT521, for a novel RNA splicing-related protein induced by hypoxia/reoxygenation. Brain Res. Mol. Brain Res. 53, 33–40 10.1016/S0169-328X(97)00262-3 (doi:10.1016/S0169-328X(97)00262-3) [DOI] [PubMed] [Google Scholar]
- 13.Hartmann AM, Nayler O, Schwaiger FW, Obermeier A, Stamm S. 1999. The interaction and colocalization of Sam68 with the splicing-associated factor YT521-B in nuclear dots is regulated by the Src family kinase p59(fyn). Mol. Biol. Cell 10, 3909–3926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Z, et al. 2010. The YTH domain is a novel RNA binding domain. J. Biol. Chem. 285, 14 701–14 710 10.1074/jbc.M110.104711 (doi:10.1074/jbc.M110.104711) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thijs G, Marchal K, Lescot M, Rombauts S, De Moor B, Rouze P, Moreau Y. 2002. A Gibbs sampling method to detect overrepresented motifs in the upstream regions of coexpressed genes. J. Comput. Biol. 9, 447–464 10.1089/10665270252935566 (doi:10.1089/10665270252935566) [DOI] [PubMed] [Google Scholar]
- 16.Horie S, Watanabe Y, Tanaka K, Nishiwaki S, Fujioka H, Abe H, Yamamoto M, Shimoda C. 1998. The Schizosaccharomyces pombe mei4+ gene encodes a meiosis-specific transcription factor containing a forkhead DNA-binding domain. Mol. Cell Biol. 18, 2118–2129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mata J, Wilbrey A, Bahler J. 2007. Transcriptional regulatory network for sexual differentiation in fission yeast. Genome Biol. 8, R217. 10.1186/gb-2007-8-10-r217 (doi:10.1186/gb-2007-8-10-r217) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Watanabe Y, Yamamoto M. 1994. S. pombe mei2 encodes an RNA-binding protein essential for premeiotic DNA synthesis and meiosis I, which cooperates with a novel RNA species meiRNA. Cell 78, 487–498 10.1016/0092-8674(94)90426-X (doi:10.1016/0092-8674(94)90426-X) [DOI] [PubMed] [Google Scholar]
- 19.Zofall M, Yamanaka S, Reyes-Turcu FE, Zhang K, Rubin C, Grewal SI. 2012. RNA elimination machinery targeting meiotic mRNAs promotes facultative heterochromatin formation. Science 335, 96–100 10.1126/science.1211651 (doi:10.1126/science.1211651) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sugiyama T, Sugioka-Sugiyama R. 2011. Red1 promotes the elimination of meiosis-specific mRNAs in vegetatively growing fission yeast. EMBO J. 30, 1027–1039 10.1038/emboj.2011.32 (doi:10.1038/emboj.2011.32). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.St-Andre O, Lemieux C, Perreault A, Lackner DH, Bahler J, Bachand F. 2010. Negative regulation of meiotic gene expression by the nuclear poly(a)-binding protein in fission yeast. J. Biol. Chem. 285, 27 859–27 868 10.1074/jbc.M110.150748 (doi:10.1074/jbc.M110.150748). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gutz H, Heslot H, Leupold U, Loprieno N. 1974. Schizosaccharomyces pombe. In Handbook of genetics (ed. King R. D.), pp. 395–446 New York: Plenum Publishing Corporation [Google Scholar]
- 23.Okazaki K, Okazaki N, Kume K, Jinno S, Tanaka K, Okayama H. 1990. High-frequency transformation method and library transducing vectors for cloning mammalian cDNAs by trans-complementation of Schizosaccharomyces pombe. Nucleic Acids Res. 18, 6485–6489 10.1093/nar/18.22.6485 (doi:10.1093/nar/18.22.6485) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moreno S, Klar A, Nurse P. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194, 795–823 10.1016/0076-6879(91)94059-L (doi:10.1016/0076-6879(91)94059-L) [DOI] [PubMed] [Google Scholar]
- 25.Egel R, Egel-Mitani M. 1974. Premeiotic DNA synthesis in fission yeast. Exp. Cell Res. 88, 127–134 10.1016/0014-4827(74)90626-0 (doi:10.1016/0014-4827(74)90626-0) [DOI] [PubMed] [Google Scholar]
- 26.Maundrell K. 1993. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene 123, 127–130 10.1016/0378-1119(93)90551-D (doi:10.1016/0378-1119(93)90551-D) [DOI] [PubMed] [Google Scholar]
- 27.Kunkel TA. 1985. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc. Natl Acad. Sci. USA 82, 488–492 10.1073/pnas.82.2.488 (doi:10.1073/pnas.82.2.488) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamashita A, Watanabe Y, Yamamoto M. 1997. Microtubule-associated coiled-coil protein Ssm4 is involved in the meiotic development in fission yeast. Genes Cells 2, 155–166 10.1046/j.1365-2443.1997.1100307.x (doi:10.1046/j.1365-2443.1997.1100307.x) [DOI] [PubMed] [Google Scholar]
- 29.Hiriart E, et al. Submitted Mmi1 RNA surveillance machinery directs RNAi complex RITS to specific meiotic genes in fission yeast. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lao K, Xu NL, Yeung V, Chen C, Livak KJ, Straus NA. 2006. Multiplexing RT-PCR for the detection of multiple miRNA species in small samples. Biochem. Biophys. Res. Commun. 343, 85–89 10.1016/j.bbrc.2006.02.106 (doi:10.1016/j.bbrc.2006.02.106) [DOI] [PubMed] [Google Scholar]
- 31.Smith MJ, Pascal CE, Grauvogel Z, Habicht C, Seeb JE, Seeb LW. 2011. Multiplex preamplification PCR and microsatellite validation enables accurate single nucleotide polymorphism genotyping of historical fish scales. Mol. Ecol. Resourc. 11(Suppl. 1), 268–277 10.1111/j.1755-0998.2010.02965.x (doi:10.1111/j.1755-0998.2010.02965.x) [DOI] [PubMed] [Google Scholar]
- 32.Chen H-M, Futcher B, Leatherwood J. 2011. The fission yeast RNA binding protein MMi1 regulates meiotic genes by controlling intron specific splicing and polydenylation coupled RNA turnover. PLoS ONE 6, e26804. 10.1371/pone.0026804 (doi:10.1371/pone.0026804) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.