Abstract
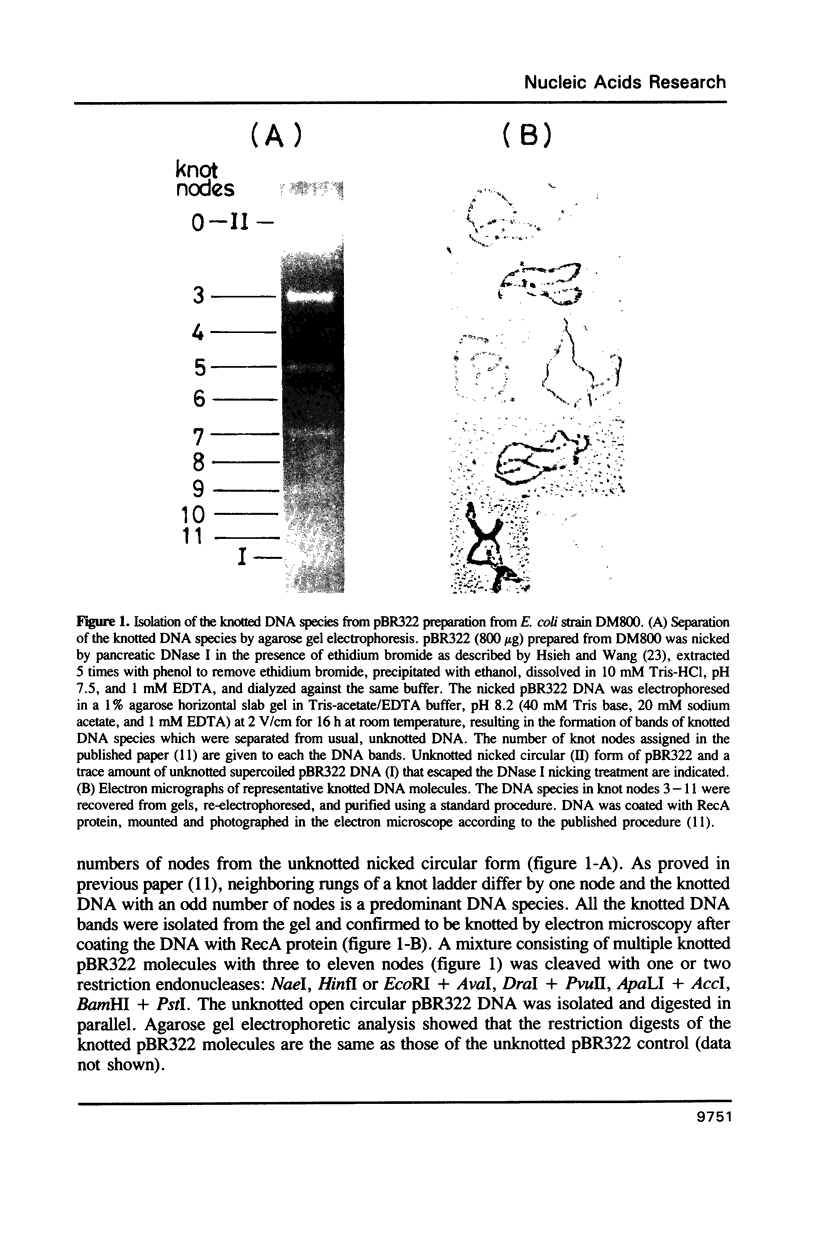
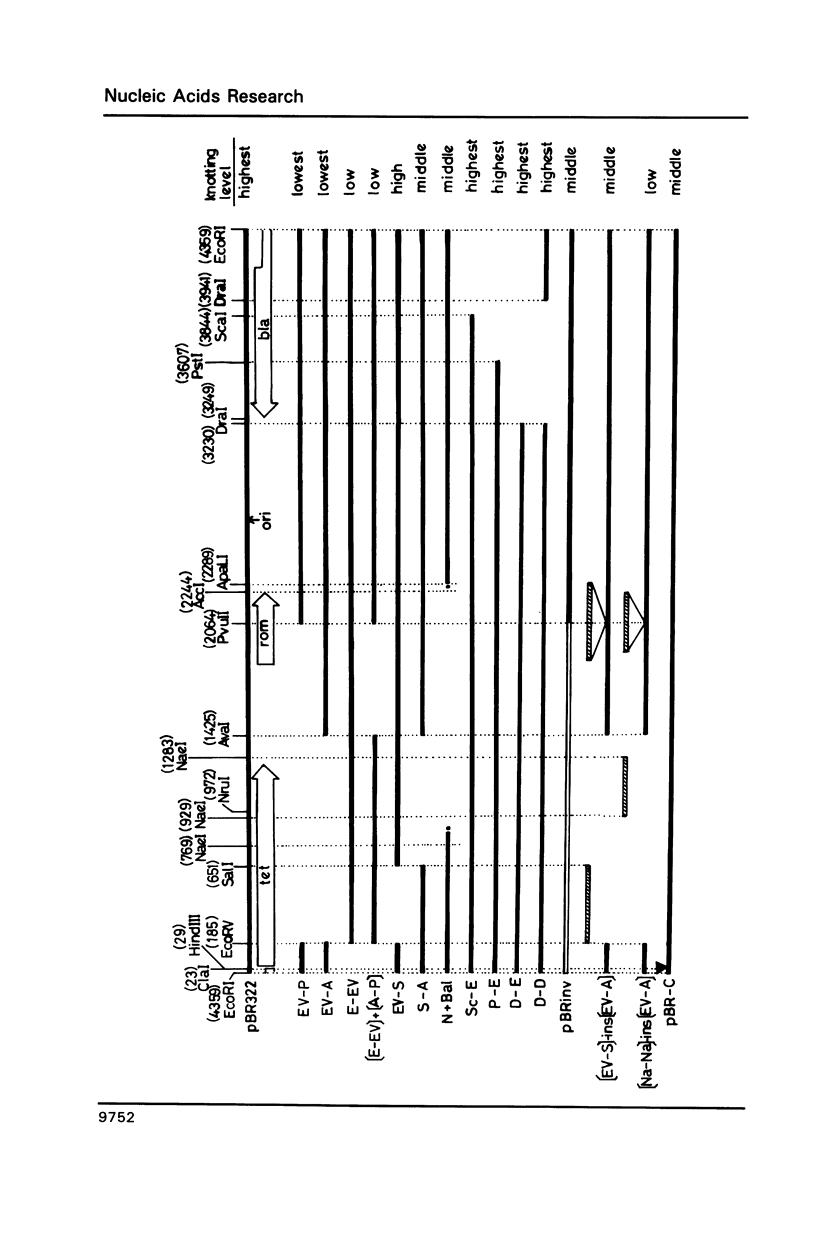
Plasmid pBR322 DNA isolated from Escherichia coli DNA topoisomerase I deletion mutant DM800 is estimated to contain about 10% of the knotted forms (Shishido et al., 1987). These knotted DNA species were shown to have the same primary structure as usual, unknotted pBR322 DNA. Analysis of the knotting level of deletion, insertion and sequence-rearranged derivatives of pBR322 in DM800 showed that the presence of the region on pBR322 encoding resistance to tetracycline (tet) is required for high levels of plasmid knotting. When the entire tet region is present in a native orientation, the level of knotting is highest. Inactivating the tet promoter is manifested by a middle level of knotting. For deletion derivatives lacking various portions of the tet region, the level of knotting ranges from lowest to high depending on the site and length of the tet gene remaining. Inverting the orientation of tet region on the pBR322 genome results in a middle level of knotting. Deleting the ampicillin-resistance (bla)gene outside of its second promoter does not affect the level of knotting, if the entire tet gene remains. A possible mechanism of regulation of plasmid knotting is discussed.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brosius J., Cate R. L., Perlmutter A. P. Precise location of two promoters for the beta-lactamase gene of pBR322. S1 mapping of ribonucleic acid isolated from Escherichia coli or synthesized in vitro. J Biol Chem. 1982 Aug 10;257(15):9205–9210. [PubMed] [Google Scholar]
- Dean F. B., Stasiak A., Koller T., Cozzarelli N. R. Duplex DNA knots produced by Escherichia coli topoisomerase I. Structure and requirements for formation. J Biol Chem. 1985 Apr 25;260(8):4975–4983. [PubMed] [Google Scholar]
- DiNardo S., Voelkel K. A., Sternglanz R., Reynolds A. E., Wright A. Escherichia coli DNA topoisomerase I mutants have compensatory mutations in DNA gyrase genes. Cell. 1982 Nov;31(1):43–51. doi: 10.1016/0092-8674(82)90403-2. [DOI] [PubMed] [Google Scholar]
- Griffith J. D., Nash H. A. Genetic rearrangement of DNA induces knots with a unique topology: implications for the mechanism of synapsis and crossing-over. Proc Natl Acad Sci U S A. 1985 May;82(10):3124–3128. doi: 10.1073/pnas.82.10.3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harley C. B., Lawrie J., Betlach M., Crea R., Boyer H. W., Hedgpeth J. Transcription initiation at the tet promoter and effect of mutations. Nucleic Acids Res. 1988 Aug 11;16(15):7269–7285. doi: 10.1093/nar/16.15.7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh T. S., Wang J. C. Thermodynamic properties of superhelical DNAs. Biochemistry. 1975 Feb 11;14(3):527–535. doi: 10.1021/bi00674a011. [DOI] [PubMed] [Google Scholar]
- Hsieh T. Knotting of the circular duplex DNA by type II DNA topoisomerase from Drosophila melanogaster. J Biol Chem. 1983 Jul 10;258(13):8413–8420. [PubMed] [Google Scholar]
- Kreuzer K. N., Cozzarelli N. R. Formation and resolution of DNA catenanes by DNA gyrase. Cell. 1980 May;20(1):245–254. doi: 10.1016/0092-8674(80)90252-4. [DOI] [PubMed] [Google Scholar]
- Liu L. F., Liu C. C., Alberts B. M. Type II DNA topoisomerases: enzymes that can unknot a topologically knotted DNA molecule via a reversible double-strand break. Cell. 1980 Mar;19(3):697–707. doi: 10.1016/s0092-8674(80)80046-8. [DOI] [PubMed] [Google Scholar]
- Liu L. F., Perkocha L., Calendar R., Wang J. C. Knotted DNA from bacteriophage capsids. Proc Natl Acad Sci U S A. 1981 Sep;78(9):5498–5502. doi: 10.1073/pnas.78.9.5498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L. F., Wang J. C. Supercoiling of the DNA template during transcription. Proc Natl Acad Sci U S A. 1987 Oct;84(20):7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodge J. K., Kazic T., Berg D. E. Formation of supercoiling domains in plasmid pBR322. J Bacteriol. 1989 Apr;171(4):2181–2187. doi: 10.1128/jb.171.4.2181-2187.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peden K. W. Revised sequence of the tetracycline-resistance gene of pBR322. Gene. 1983 May-Jun;22(2-3):277–280. doi: 10.1016/0378-1119(83)90112-9. [DOI] [PubMed] [Google Scholar]
- Pruss G. J. DNA topoisomerase I mutants. Increased heterogeneity in linking number and other replicon-dependent changes in DNA supercoiling. J Mol Biol. 1985 Sep 5;185(1):51–63. doi: 10.1016/0022-2836(85)90182-2. [DOI] [PubMed] [Google Scholar]
- Pruss G. J., Drlica K. Topoisomerase I mutants: the gene on pBR322 that encodes resistance to tetracycline affects plasmid DNA supercoiling. Proc Natl Acad Sci U S A. 1986 Dec;83(23):8952–8956. doi: 10.1073/pnas.83.23.8952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shishido K., Komiyama N., Ikawa S. Increased production of a knotted form of plasmid pBR322 DNA in Escherichia coli DNA topoisomerase mutants. J Mol Biol. 1987 May 5;195(1):215–218. doi: 10.1016/0022-2836(87)90338-x. [DOI] [PubMed] [Google Scholar]
- Spengler S. J., Stasiak A., Cozzarelli N. R. The stereostructure of knots and catenanes produced by phage lambda integrative recombination: implications for mechanism and DNA structure. Cell. 1985 Aug;42(1):325–334. doi: 10.1016/s0092-8674(85)80128-8. [DOI] [PubMed] [Google Scholar]
- Sternglanz R., DiNardo S., Voelkel K. A., Nishimura Y., Hirota Y., Becherer K., Zumstein L., Wang J. C. Mutations in the gene coding for Escherichia coli DNA topoisomerase I affect transcription and transposition. Proc Natl Acad Sci U S A. 1981 May;78(5):2747–2751. doi: 10.1073/pnas.78.5.2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomizawa J., Som T. Control of ColE1 plasmid replication: enhancement of binding of RNA I to the primer transcript by the Rom protein. Cell. 1984 Oct;38(3):871–878. doi: 10.1016/0092-8674(84)90282-4. [DOI] [PubMed] [Google Scholar]
- Tsao Y. P., Wu H. Y., Liu L. F. Transcription-driven supercoiling of DNA: direct biochemical evidence from in vitro studies. Cell. 1989 Jan 13;56(1):111–118. doi: 10.1016/0092-8674(89)90989-6. [DOI] [PubMed] [Google Scholar]
- Wasserman S. A., Dungan J. M., Cozzarelli N. R. Discovery of a predicted DNA knot substantiates a model for site-specific recombination. Science. 1985 Jul 12;229(4709):171–174. doi: 10.1126/science.2990045. [DOI] [PubMed] [Google Scholar]
- Watson N. A new revision of the sequence of plasmid pBR322. Gene. 1988 Oct 30;70(2):399–403. doi: 10.1016/0378-1119(88)90212-0. [DOI] [PubMed] [Google Scholar]
- White J. H., Cozzarelli N. R. A simple topological method for describing stereoisomers of DNA catenanes and knots. Proc Natl Acad Sci U S A. 1984 Jun;81(11):3322–3326. doi: 10.1073/pnas.81.11.3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. Y., Shyy S. H., Wang J. C., Liu L. F. Transcription generates positively and negatively supercoiled domains in the template. Cell. 1988 May 6;53(3):433–440. doi: 10.1016/0092-8674(88)90163-8. [DOI] [PubMed] [Google Scholar]