Abstract
Recent studies have revealed that bacterial protein methylation is a widespread post-translational modification that is required for virulence in selected pathogenic bacteria. In particular, altered methylation of outer-membrane proteins has been shown to modulate the effectiveness of the host immune response. In this study, 2D gel electrophoresis combined with MALDI-TOF MS identified a Leptospira interrogans serovar Copenhageni strain Fiocruz L1-130 protein, corresponding to ORF LIC11848, which undergoes extensive and differential methylation of glutamic acid residues. Immunofluorescence microscopy implicated LIC11848 as a surface-exposed outer-membrane protein, prompting the designation OmpL32. Indirect immunofluorescence microscopy of golden Syrian hamster liver and kidney sections revealed expression of OmpL32 during colonization of these organs. Identification of methylated surface-exposed outer-membrane proteins, such as OmpL32, provides a foundation for delineating the role of this post-translational modification in leptospiral virulence.
Introduction
Pathogenic Leptospira are the causative agents of the emerging zoonotic disease leptospirosis (Levett, 2001). Rats and house mice are asymptomatic carriers of Leptospira and serve as both reservoir (Bharti et al., 2003; Ko et al., 2009; Levett, 2001) and vector for the maintenance and transmission of bacteria to humans. Transmission to humans is thought to occur via contact with water contaminated by the urine of reservoir hosts (Levett, 2001). Leptospirosis is highly prevalent in developing nations with urban slums (Ko et al., 2009; McBride et al., 2005), and is endemic in settings where farmers that conduct their work manually are exposed to animals shedding leptospiral pathogens (Bharti et al., 2003; Ko et al., 2009; Levett, 2001). Disease symptoms in humans most commonly include uveitis (Verma et al., 2008), and hepatic (Merien et al., 1998) and renal (Seguro et al., 1990) dysfunction. The most recent WHO statistics report more than 500 000 severe human leptospirosis cases per year with mortality rates greater than 10 % (WHO, 1999).
Understanding pathogenicity mechanisms surrounding the leptospiral infection process has been hampered by the lack of targeted genetic tools available to leptospiral researchers, such as signature tagged mutagenesis (STM) (Hensel et al., 1995) and transposon site hybridization (TraSH) (Sassetti et al., 2001). Random transposon mutagenesis, while not as robust and efficient as STM and TraSH, has been successfully employed to identify virulence factor-encoding genes of leptospiral pathogens (Bourhy et al., 2005). To date, five of six genes encoding virulence factors have been identified in this manner, including la0222 in Leptospira interrogans serovar Lai, which encodes the outer-membrane lipoprotein Loa22 (Ristow et al., 2007), la2613 in serovar Lai, which encodes the flagella motor switch protein FliY (Liao et al., 2009), an orthologue of serovar Lai lb186 in L. interrogans serovar Manilae, which encodes the haem oxygenase HemO (Murray et al., 2009), and two genes which encode LPS biosynthesis proteins in serovar Manilae, with one being orthologous to serovar Lai la1641 and the other having no observed serovar Lai orthologue (Murray et al., 2010). Lastly, invA, a Nudix hydrolase specific for numerous dinucleoside oligophosphate substrates, has been directly inactivated using insertion mutagenesis and demonstrated to be required for virulence of serovar Lai in hamsters (Luo et al., 2011). In addition to these genetic approaches, various protein-based approaches have been utilized to identify potential leptospiral virulence factors. Such studies have elucidated the subcellular location for various proteins (Beck et al., 2009; Cullen et al., 2005; Haake & Matsunaga, 2002; Monahan et al., 2008; Pinne & Haake, 2009; Sakolvaree et al., 2007), identified immunoreactive proteins (Artiushin et al., 2004; Guerreiro et al., 2001; Sakolvaree et al., 2007), quantified absolute protein numbers per bacterium (Malmström et al., 2009), provided evidence for differential protein expression in response to changes in microenvironments (Cullen et al., 2002; Eshghi et al., 2009; Lo et al., 2009; Matsunaga et al., 2007; Nally et al., 2001a, b, 2007; Velineni et al., 2006), contributed to the identification of the first confirmed leptospiral virulence factor, Loa22 (Nally et al., 2007), and revealed diverse and extensive post-translational modifications (PTMs) (Cao et al., 2010).
Of particular interest, a recent global proteomic analysis of L. interrogans serovar Lai identified a total of 155 methylated proteins (Cao et al., 2010), suggesting a widespread role for methylation in L. interrogans protein function and/or regulation. In the context of virulence, methylation has been shown to be an essential PTM in various bacterial pathogens. Using genetic approaches, native methylation has been shown to be required for regulation of transcription and phase variation of outer-membrane components (Deitsch et al., 2009), and for maintenance of virulence (Giacomodonato et al., 2009). Methylation of glycolipids has been shown to be essential for Mycobacterium avium virulence in mice (Krzywinska et al., 2005), while methylation of proteins has been shown to alter the antigenicity of the outer-membrane protein OmpB from Rickettsia typhi (Chao et al., 2008), and to alter both the antigenicity of and the host T cell-mediated immune response against the heparin-binding haemagglutinin from Mycobacterium tuberculosis (Parra et al., 2004; Temmerman et al., 2004). In addition, methylation has been demonstrated to be essential for functional type III secretion, and thus virulence, in Yersinia pseudotuberculosis (Garbom et al., 2004, 2007), and methylation of the surface protein OmpB in Rickettsia prowazaki has been suggested to be central to the pathogenesis of that bacterium (Chao et al., 2004, 2007). Collectively, these investigations highlight the importance of methylation of outer-membrane surface components in bacterial virulence.
The present study describes the proteomic identification of the novel L. interrogans outer-membrane protein OmpL32 (LIC11848). Immunological analyses confirmed exposure of OmpL32 on the leptospiral surface and expression of this protein during the course of bacterial infection. Proteomic analysis revealed differential and extensive glutamic acid methylation of this protein under all the tested environmental conditions. The potential implications of this PTM are discussed within the context of leptospiral virulence.
Methods
Leptospira and culture conditions.
L. interrogans serovar Copenhageni strain Fiocruz L1-130 is a clinical isolate originating from Salvador, Brazil (Ko et al., 1999). L. interrogans serovar Pomona type kennewicki strain RM-211 is an isolate from a swine abortion case (Thiermann et al., 1984). Cultures were maintained in Ellinghausen and McCullough (Ellinghausen & McCullough, 1965) as modified by Johnson and Harris (Johnson & Harris, 1967) (EMJH) medium at 29.5 °C. Bacterial enumeration and media shift experiments were performed as previously described (Eshghi et al., 2009). In these experiments, L. interrogans was grown under the following four conditions: (1) in EMJH medium at 37 °C; (2) in EMJH medium depleted of iron; (3) in EMJH medium supplemented with 10 % fetal bovine serum (FBS); or (4) in EMJH medium supplemented with 10 % FBS and depleted of iron.
2D gel electrophoresis (2DGE) and MALDI-TOF MS experiments.
L. interrogans was harvested at 8500 g and washed twice with PBS supplemented with 5 mM MgCl2. Cell pellets were lyophilized, weighed and resuspended to a concentration of 2.5 mg ml−1. 2DGE, staining of gels with colloidal Coomassie blue and MALDI-TOF MS experiments were performed as previously described (Eshghi et al., 2009). Briefly, 2D gels (pH range 3–11) containing equal amounts of Leptospira by dry weight from each of the four different growth conditions were run in triplicate. Gels originating from the four different growth conditions were compared to detect proteins displaying altered intensity between the comparative conditions, while validation of protein spot intensity within a growth condition was obtained by comparison of matched triplicate gels.
Liquid chromatography-electrospray ionization-tandem MS (LC-ESI-MS/MS).
Trypsin digests were performed with Genomic Solutions ProGest (Digilab). Briefly, gel slices were manually cut into 1 mm cubes and transferred to a Genomics Solutions ProGest perforated digestion tray. The gel pieces were de-stained (50 : 45 : 5, v/v, methanol/water/acetic acid) prior to reduction and alkylation with 10 mM DTT and 100 mM iodoacetamide, respectively (Sigma). Modified sequencing grade porcine trypsin solution (20 ng µl−1, Promega) was added to the gel slices at an enzyme : protein ratio of 1 : 50. Proteins were digested for 5 h at 37 °C prior to collection of resulting peptides and acid extraction of the gel slices (50 : 40 : 10, v/v, acetonitrile/water/formic acid). Samples were then Speed Vac-centrifuged to dry and stored at −20 °C until analysed by LC-ESI-MS/MS. An UltiMate Nano HPLC (LC Packings/Dionex) coupled to an Applied Biosystems/MDS Sciex QSTAR Pulsar I hybrid quadrupole-TOF LC-MS/MS mass spectrometer (AB Sciex) was used to perform the LC-MS/MS analyses. The nano-column was connected to a 20 µm internal diameter emitter tip (New Objective, Inc.) positioned at the orifice plate of the mass spectrometer with spray established by applying a tip voltage of 1900 V to the pre-nano column via a platinum wire nano-tee high-voltage connection.
The lyophilized samples were rehydrated in 40 µl 2 % (v/v) acetonitrile/0.1 % formic acid, and 15 µl of each sample was injected using a FAMOS Autosampler (LC Packings/Dionex) for each analysis. The samples were concentrated/desalted using a SwitchOSII loading pump (LC Packings/Dionex) with 98 % solvent A (2 %, v/v, acetonitrile, 0.1 % formic acid) at a rate of 30 µl min−1 for 10 min over an Agilent Zorbax C18-SB trap column (5×0.3 mm) (Agilent Technologies) to protect the in-house-prepared nano-analytical column Magic C18AQ resin, 150 mm×75 µm column diameter (Michrom Bioresources). After the 10 min loading period, the trap column was switched in-line with the nano-column gradient flow. The following 1 h HPLC analytical separation was performed at 300 nl min−1 [33 min linear gradient from 5–60 % solvent B (98 %, v/v, acetonitrile, 0.1 % formic acid), 2 min linear gradient from 60–75 % solvent B, 5 % solvent B over 3 min and re-equilibrated with 5 % solvent B for 12 min before the next injection].
Mass spectra analyses were acquired by collecting a 1 s TOFMS survey scan of 400–1200 m/z followed by two 2.5 s product ion scans in the 100–1500 m/z mass range. MS/MS spectra were acquired in a data-dependent manner, selecting the top two most intense eluting ions in the 400–1200 m/z range with a 2+ to 4+ charge state greater than 20 counts. Following selection for MS/MS analysis, precursor ions were excluded from selection for MS/MS analysis for 180 s. A rolling collision energy for fragmentation was selected based on the precursor ion mass using the formula 0.05 m/z+5 V. Known keratin tryptic and trypsin autolysis product masses were excluded to prevent these contaminant ions from being selected for fragmentation.
Mass spectrometer parameters used were as follows: declustering potential setting of 50, focusing potential setting of 220, curtain gas setting of 25, and CAD gas setting of 5 with nitrogen in the collision cell.
Bioinformatic analyses.
Peptide mass fingerprint (PMF) searches were conducted as previously described (Eshghi et al., 2009), with the following change: no variable modifications were selected. PMFs were further analysed for potential PTMs using the ExPASy Proteomics Server FindMod tool (Wilkins et al., 1999) (http://web.expasy.org/findmod/). Predictions were conducted with the following parameter settings: ion mode was set to [M+H]+, mass was set to monoisotopic, and mass tolerance was set to 10 p.p.m.
MS/MS data were searched in mascot script against the L. interrogans proteome in UniProtKB annotation (http://www.uniprot.org/uniprot/?query=leptospira+interrogans+serovar+copenhageni&sort=score&format=*) that was maintained in-house at the University of Victoria-Genome BC Proteomics Centre. Search parameters used were as follows: taxonomy was set to Leptospira, enzyme was set to trypsin allowing for 1 missed cleavage site, and variable modifications were set to carbamedomethyl (C), deamidated (NQ), methyl (DE) and oxidation (M). Peptide and MS/MS tolerances were set to 0.6 and 0.3 Da respectively, monoisotope mass was selected, peptide charge was set to 1+, 2+ and 3+, and ESI-QUAD-TOF was the selected instrument. Glutamic acid methylation was also confirmed via manual de novo sequencing in peaks (Bioinformatics Solutions, Inc.) (Ma et al., 2003) using the user input sequence for a given peptide spectrum. To be classified as a methylated peptide, spectral ions representing the methylated glutamic acid residue and the N- and C-terminal residues adjacent to the methylated glutamic acid residues had to be present. The only exception to this rule was when the methylated glutamic acid residue was in the second residue position from the N terminus of a given peptide, in which case spectral ions representing the glutamic acid residue and the C-terminal residue adjacent to the glutamic acid residue had to be present (b1 ions are not commonly detected in peptide spectra). Annotated spectra were exported as .svg files to easily search spectra and are available in Supplementary Figs S2–S5.
For prediction of subcellular localization, the LIC11848 amino acid sequence was analysed for the presence of a putative signal peptide sequence using the LipoP 1.0 (Rahman et al., 2008) (http://www.cbs.dtu.dk/services/LipoP/), SpLip (Setubal et al., 2006) and SignalP (Emanuelsson et al., 2007) (http://www.cbs.dtu.dk/services/SignalP/) signal peptide prediction programs. Secondary structure analyses for prediction of β-sheets was conducted using GOR (Kloczkowski et al., 2002; Sen et al., 2005) (http://pbil.ibcp.fr/htm/index.php), and homology predictions to Gram-negative outer-membrane proteins were conducted using OMPdb (Tsirigos et al., 2011) (http://aias.biol.uoa.gr/OMPdb/index.php). Transmembrane domain prediction was conducted using the hidden Markov model outer-membrane β-barrel protein prediction program PRED-TMBB (Bagos et al., 2004) (http://biophysics.biol.uoa.gr/PRED-TMBB/). B cell epitope predictions were made using the BCPreds B cell epitope prediction server (El-Manzalawy et al., 2008), using the search parameter ‘fixed length epitope prediction’ and epitope lengths of 12, 14, 20 or 22 aa (http://ailab.cs.iastate.edu/bcpreds/predict.html).
Recombinant protein expression and purification.
ORF LIC11848 was PCR-amplified from L. interrogans serovar Copenhageni strain Fiocruz L1-130 genomic DNA using the primer pair 5′-CTAGACCATATGTCCGGATCCGATCAAAAATC-3′ and 5′-GTCAGCTCGAGTTAGTAGCGGAGGGAATCC-3′ (incorporated restriction sites indicated by underlined nucleotides). The LIC11848 amplicon (693 bp in length encompassing basepairs 70–762 of the ORF) was ligated first into the cloning vector pJET1 (CloneJet, Fermentas) and digested with NdeI and XhoI, followed by ligation into a similarly digested pET28a expression vector (Novagen). The sequence and reading frame of the expression construct were verified by DNA sequencing with vector-specific primers. The LIC11848/pET28a construct was transformed into the Escherichia coli expression strain BL21 Star BL21(DE3) (Invitrogen). Bacteria were grown at 37 °C in a shaking incubator, and recombinant expression was induced at OD600 1.0 using a final concentration of 0.4 mM IPTG (Invitrogen). The incubation temperature was then reduced to 16 °C and bacteria were grown overnight.
Soluble recombinant protein was purified using the following methodology. Bacteria were harvested by centrifugation at 3000 g, resuspended in binding buffer (60 mM NaH2PO4, 500 mM NaCl and 20 mM imidazole, pH 8.0) and lysed via sonication (3×30 s) in the presence of a protease inhibitor cocktail [Protease Inhibitor Cocktail Set III; 100 mM 4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride (AEBSF), 80 µM aprotinin, 5 mM bestatin, 1.5 mM E-64, 2 mM leupeptin, 1 mM pepstatin A] (Calbiochem). The lysate was subsequently centrifuged at 20 000 g at 4 °C for 30 min and the supernatant filtered through a 0.45 µm pore-size filter. Purification was performed on an ÄKTAprime Plus fast protein liquid chromatography system (GE Healthcare) fitted with a 1 ml HisTrap HP Column (GE Healthcare). All steps were performed at a flow rate of 1 ml min−1 and a pressure limit of 0.3 MPa. The run parameters included a 12 ml equilibration step with binding buffer, application of 30 ml of the filtered supernatant (protein sample), a 40 ml wash with binding buffer, and a gradient elution of 0–100 % elution buffer (60 mM NaH2PO4, 500 mM NaCl and 500 mM imidazole, pH 8.0) over a 50 ml volume with fractions collected at 0.5 ml increments. Fractions containing recombinant OmpL32 (rOmpL32) (identified via A280 readings of fractions in combination with SDS-PAGE) were then combined and concentrated using a 10K Amicon Ultra-4 centrifugal filter unit (Millipore).
Desalting was achieved in HEPES buffered saline (20 mM HEPES, 150 mM NaCl, pH 7.4), over 90 ml, via an ÄKTAprime Plus fast protein liquid chromatography apparatus fitted with a HiLoad 16/60 Superdex 75 prep grade gel filtration column (GE Healthcare). Purified rOmpL32 was quantified using a BCA Protein Assay (Pierce).
Antibodies.
Polyclonal rabbit antiserum was prepared against recombinantly expressed LIC11848 (rLIC11848) by ProSci, Inc. Polyclonal rabbit antisera against OmpL36 (Eshghi et al., 2009), FlaA1 (Cullen et al., 2005) and LipL32 (Haake et al., 2000) have been described previously.
Immunoblot analysis.
Approximately 1×108 Leptospira were harvested at 10 000 g and washed twice with wash buffer (PBS and 5 mM MgCl2). The pellet was resuspended in SDS-PAGE loading buffer, boiled for 10 min and separated on a 15 % SDS-polyacrylamide gel. Proteins were electrophoretically transferred to an Immobilon PVDF membrane (Millipore). Immunoblots were blocked with 2.5 % milk powder in Tris-buffered saline, pH 7.4, with 0.05 % Tween 20 (TBST) for 1 h at room temperature. Membranes were washed 2×5 min with TBST, followed by incubation for 90 min at room temperature with a 1 : 1500 dilution of rLIC11848-specific rabbit antiserum or preimmune serum in 2.5 % milk powder/TBST. The membrane was washed with TBST 4×5 min, incubated with a 1 : 5000 dilution of goat-anti-rabbit IgG (whole molecule) F(ab′)2 fragment–peroxidase (Sigma) secondary antibody in 2.5 % milk powder/TBST for 60 min at room temperature, followed by rinsing twice and washing 1×15 min and 3×5 min with TBST. Enhanced chemiluminescence reagent (GE Healthcare) was applied as described by the manufacturer’s protocol with the modification of incubating the membrane with the reagent for 1 min. The membrane was exposed to X-ray film for 1 min and developed with a Kodak X-OMAT 2000A processor (Carestream Health).
Immunofluorescence assay (IFA).
IFAs were conducted as previously described (Cullen et al., 2005; Pinne & Haake, 2011, 2009). Briefly, L. interrogans was harvested at 2000 g to maintain outer-membrane integrity, resuspended in PBS/5 mM MgCl2 to a density of 5×108 cells ml−1, applied in 1 ml aliquots to Nunc four-well Lab-Tek II Chamber Slides (Thermo Fisher Scientific) and incubated at 29.5 °C for 80 min for the purpose of adhering cells to the glass slides. All analyses were done in duplicate. Chamber slides were carefully aspirated, and bacteria were fixed using 1 ml per well of 2 % (w/v) paraformaldehyde in PBS for 40 min at 29.5 °C. For the purpose of demonstrating an intact outer membrane, replicate chamber slides were permeabilized with 100 % ice-cold methanol for 20 min at −20 °C. Chamber slides were then aspirated and blocked with 1 ml EMJH medium for 90 min at 29.5 °C. Rabbit antisera specific for OmpL36, rLIC11848 and FlaA1 were diluted 1 : 200, 1 : 100 and 1 : 600, respectively, and preimmune serum was diluted 1 : 100 in EMJH. Sera at the specified dilutions were added in 1 ml volumes to chamber slides and incubated for 60 min at 29.5 °C. Chamber slides were washed three times with PBS. Alexa Fluor 568 goat anti-rabbit IgG (H+L) (Invitrogen) and DAPI stain (Invitrogen) were diluted to 1 : 1500 and 0.25 µg ml−1, respectively, in EMJH and applied to chamber slides in 1 ml volumes followed by a 45 min incubation at 29.5 °C in the dark. Chambers were then washed twice with PBS and once with water. Chambers were removed and glass slides were air-dried in the dark. ProLong Gold antifade reagent (Invitrogen) diluted 1 : 3 in PBS (20 µl) was added to each slide, coverslips were mounted, and slides were incubated at room temperature overnight in the dark.
Fluorescence was achieved with an Eclipse 80i microscope fitted with an X-Cite 120 illuminator and a DS-U1 camera (all from Nikon Canada). Images were processed using ACT-2U imaging software (Excel Technologies).
Immunofluorescence of tissue sections.
Immunofluorescence of tissue sections was measured as previously described (Matsunaga et al., 2006). All animal studies were approved by the local institutional review boards and conducted in accordance with standard accepted principles. Briefly, golden Syrian hamsters were inoculated with L. interrogans, serovar Pomona strain RM211. Moribund and healthy uninfected hamsters were euthanized, and liver and kidney tissues were removed, fixed in 10 % buffered zinc formalin, and paraffin-embedded. Serial 4 µm sections of hamster tissue were cut. Paraffin was removed from sections with xylene and ethanol, using standard procedures. Antigen retrieval was performed using 10 mM citrate buffer with boiling for 20 min, cooling for 20 min and rinsing with PBS. Slides were permeabilized with 0.5 % Tween-20 in PBS for 10 min. Non-specific staining of tissue sections was blocked using 10 % normal goat serum in PBS at room temperature for 60 min, prior to incubation overnight at 4 °C with primary antibody. Anti-rLIC11848, anti-LipL46 (Matsunaga et al., 2006) and anti-LipL32 (Haake et al., 2000) antisera were used at 1 : 50, 1 : 100 and 1 : 200 dilutions, respectively. Normal goat sera block was used as a negative control (no primary antibody) on all sections from both infected and uninfected hamsters. Sections were washed with PBS to remove unbound antibody, and then incubated for 60 min at room temperature in the dark with a 1 : 5000 dilution of Alexa Fluor 488 F(ab′)2 goat anti-rabbit secondary antibody and 0.4 µg DAPI ml−1 (Invitrogen). Slides were mounted with ProLong Gold antifade reagent (Invitrogen). All images were captured on a Spot RT colour CCD camera mounted on a Nikon Eclipse E800. All immunohistochemistry images were captured under the same exposure conditions.
Results
L. interrogans cells differentially express isoforms of a putative outer-membrane protein
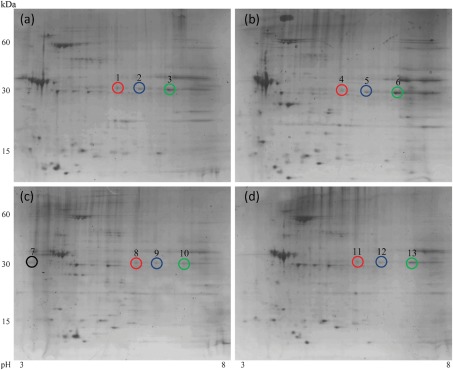
To detect changes in protein expression resulting from exposure to varying environmental conditions, L. interrogans cells were grown at 37 °C in EMJH medium, in EMJH medium depleted of iron, in EMJH medium supplemented with 10 % FBS or in EMJH medium supplemented with 10 % FBS and depleted of iron. Total protein was separated by 2DGE, gels were stained with colloidal Coomassie stain, and protein spots that were observed to vary in intensity between the compared conditions (see below) were selected and subjected to trypsin digestion and subsequent MALDI-TOF MS. Comparative analysis of triplicate gels of Leptospira grown under these varied culture conditions, using conventional growth at 37 °C in normal EMJH medium as the comparator, revealed consistently altered expression of proteins corresponding to ORFs LIC10361 (electron transfer flavoprotein EtfB), LIC13050 (putative glycosyl hydrolase), LIC13166 (putative coagulase), LIC11336 (GroES), LIC12210 (IbpA-1) and LIC11890 (periplasmic flagellin FlaB1), as previously reported (Eshghi et al., 2009). An additional protein identified through this study, corresponding to the ORF LIC11848, was deemed to be of particular interest due to the identification of multiple LIC11848 isoforms that displayed altered intensity and differed in their respective isoelectric points over the pI range 3.5–7.5 (Fig. 1). Visual comparison of triplicate gels prepared from each of the comparative growth conditions confirmed that the individual LIC11848 protein spots remained constant between replicates of a particular growth condition (Supplementary Fig. S1), indicating that altered protein intensities did not result from gel-to-gel variation.
Fig. 1.
Comparative proteome analysis of L. interrogans exposed to differing growth conditions. Total protein from L. interrogans was separated using 2DGE following growth of L. interrogans under the following comparative conditions: 37 °C in EMJH (a), EMJH supplemented with 10 % FBS and depleted of iron (b), EMJH supplemented with 10 % FBS (c) and EMJH depleted of iron (d). Representative protein spots selected for subsequent proteomic analyses via MALDI-TOF MS and LC-ESI-MS/MS are indicated. Identified identical isoforms arising within the different growth conditions are distinguished by different coloured circles; refer to main text for further descriptions.
Due to the complexity associated with analysis of the comparative 2DGE patterns, a brief description of how the different LIC11848 isoforms were identified is provided. A visual identification of similar LIC11848 isoforms was made by locating protein spots that migrated to the same location in each of the comparative growth conditions. This allowed for the designation of representative spots that constituted each of the identified isoforms and corresponded to spot 1 [deemed to be the same as spots 4, 8 and 11, indicated by red circles in Fig. 1, panels (a), (b), (c) and (d), respectively], spot 3 [deemed to be the same as spots 6, 10 and 13, indicated by green circles in Fig. 1, panels (a), (b), (c) and (d), respectively] and spot 2 [deemed to be the same as spots 5, 9 and 12, indicated by blue circles in Fig. 1, panels (a), (b), (c) and (d), respectively]. Representative spots 1, 3 and 7, as well as spots 6 and 13 to ensure accurate prediction of identical isoforms, were subjected to in-gel tryptic digestion, the resulting peptides were analysed via MALDI-TOF MS, and PMFs were used to perform mascot database searches to allow protein identification.
The mass to charge (m/z) ratios, peptide coverage and expect values obtained for each of the representative spots (1, 3, 6, 7 and 13) used for identification via PMF are summarized in Supplementary Table S1. A high degree of peptide mass coverage was obtained for the representative spots, allowing for a definitive identification of each of the five protein spots as LIC11848. Spot 2, which is representative of spots 5, 9 and 12, was definitively identified as a LIC11848 isoform through subsequent MS/MS analyses (see below).
LIC11848 is differentially methylated on glutamic acid residues
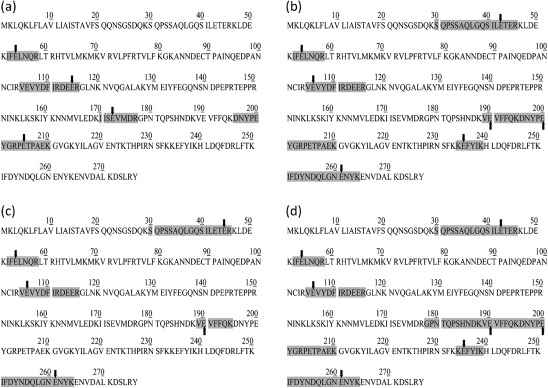
To identify a plausible cause for the observed pI shift in LIC11848, PMF data were further analysed using the ExPASy FindMod tool (Wilkins et al., 1999) to predict potential PTMs. This analysis predicted methylation of a total of 11 glutamic acid residues within LIC11848; specifically, methylation was predicted for four peptides for spot 1, five peptides for spot 3, three peptides for spot 6, six peptides for spot 7 and eight peptides for spot 13. The number of predicted methylated peptides was proportional to the number of m/z ratios used to conduct searches, with a higher degree of peptide coverage correlating with the identification of an increased number of methyl-ester adducts. To test these in silico predictions, tryptic digests of LIC11848 protein from representative spots 1, 2, 6 and 7 (Fig. 1a–c) from replicate second-dimension gels were subjected to online high-performance LC-ESI-MS/MS. These analyses confirmed in silico predictions, and revealed that a total of 11 glutamic acid residues were methylated within LIC11848, with experimental confirmation of methylation of four peptides in spot 1 (Fig. 2a and Supplementary Fig. S2), seven peptides in spot 2 (Fig. 2b and Supplementary Fig. S3), five peptides in spot 7 (Fig. 2c and Supplementary Fig. S4) and seven peptides from spot 6 (Fig. 2d and Supplementary Fig. S5). The glutamic acid residue at amino acid residue position 54 showed methylation in all four spots (Fig. 2 and Supplementary Figs S2–S5). Similar to the PMF analyses, the number of methyl esters identified was proportional to the number of m/z values present in the MS/MS searches, with a higher peptide coverage correlating with identification of more extensive methylation. Of particular note was the observation that methyl group location varied among glutamic acid residues within an identical peptide identified between comparative growth conditions. The peptide spanning amino acids 105–116 (VEVYDFIRDEER) was represented in all isoforms, with three of four isoforms displaying methylation on the most N-terminal glutamic acid residue (residue 2, counting from the N terminus of the peptide; Fig. 2b–d) and one of four isoforms displaying methylation on the most C-terminal glutamic acid (residue 11, counting from the N terminus of the peptide; Fig. 2a).
Fig. 2.
Differential glutamic acid methylation of the LIC11848 protein. Glutamic acid methyl ester peptides detected from representative protein spots 1 (a), 2 (b), 7 (c) and 6 (d). Peptides containing methyl ester glutamic acids are highlighted and the methylated glutamic acids are indicated by arrows. Spectral evidence can be accessed in Supplementary Figs S2–S5.
Correlation between the observed LIC11848 protein methylation pattern and predicted B cell epitope locations
Comparison of the identified LIC11848 methylated peptides with B cell epitopes predicted using the BCPreds online prediction tool revealed all 11 methylated glutamic acid residues to be contained within regions of the protein predicted as possible B cell epitopes (Table 1). Further, four of the predicted B cell epitopes correlated with methylated peptides contained within all four of the protein spots subjected to methylation profile analysis (predicted epitopes 43–54, 89–110, 103–122 and 173–192). With the exception of two predicted B cell epitopes, the scores for these analyses were highly significant (≥0.926).
Table 1. Predicted B cell epitopes present within the LIC11848 protein (OmpL32) and correlation with the observed methylation profile.
| Predicted B cell epitope* | Probability | Predicted epitope length (aa) | Amino acid range within LIC11848 | Methylated amino acids (residue numbers) | Protein spots |
| ETERKLDEKIFE | 0.28 | 12 | 43–54 | 43, 45, 54 | 1, 2, 6, 7 |
| CTPAINQEDPANNCIRVEVYDF | 0.992 | 22 | 89–110 | 106 | 1, 2, 6, 7 |
| IRVEVYDFIRDEERGLNKNV | 0.959 | 20 | 103–122 | 106, 115 | 1, 2, 6, 7 |
| EVMDRGPNTQPSHNDKVEVF | 0.961 | 20 | 173–192 | 173, 190 | 1, 2, 6, 7 |
| FQKDNYPEYGRPETPAEKGVGK | 0.996 | 20 | 195–214 | 200, 205 | 1, 2, 6 |
| SFKKEFYIKHLDQF | 0.741 | 14 | 231–244 | 235 | 2, 6 |
| TKIFDYNDQLGNENYKENVDAL | 0.926 | 22 | 249–270 | 261 | 2, 6, 7 |
Bold type indicates methylated glutamic acid residues.
LIC11848 is a surface-exposed protein
To predict the cellular location of LIC11848, the amino acid sequence was analysed using the signal peptide prediction tools LipoP 1.0 (Rahman et al., 2008) and SignalP 3.0 (Emanuelsson et al., 2007). Both tools predicted the presence of an N-terminal SpI signal peptide, with LipoP predicting a potential cleavage site between serine 20 and glutamine 21, and SignalP predicting a potential cleavage site between glycine 24 and serine 25, a finding which is consistent with a potential outer-membrane location for this protein. Secondary structure analysis conducted using GOR predicted the secondary structure composition of the protein to be 15 % β-sheet. To further assess the predicted location of LIC11848, the amino acid sequence was searched against the OMPdb database for Gram-negative bacterial β-barrel-containing outer-membrane proteins (Tsirigos et al., 2011). This search identified significant homology between LIC11848 and the C-terminal residues 633–752 from the Bacteroides fragilis outer-membrane receptor (OMR-TonB-dependent receptor) family (27.13 % sequence identity and 44.97 % sequence similarity over 129 aa). Further evidence suggesting a membrane association for this protein was obtained via identification of nine transmembrane domains as predicted by PRED-TMBB analysis, although the generated score for this analysis of 3.020 was slightly higher than the outer-membrane protein threshold cut-off value of 2.965 for this prediction program.
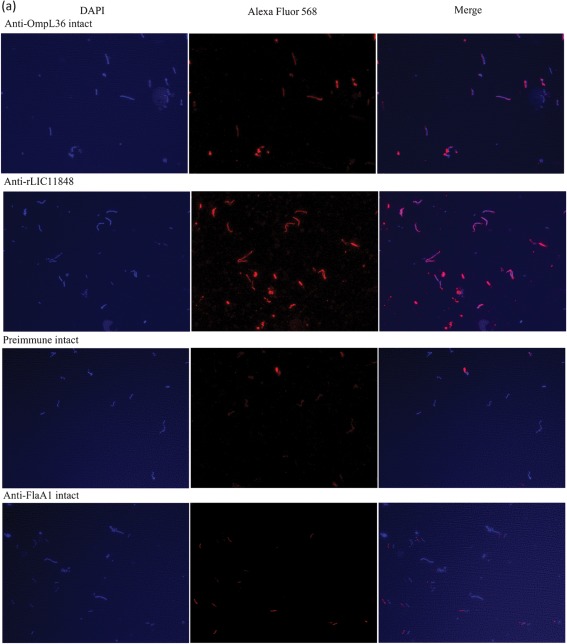
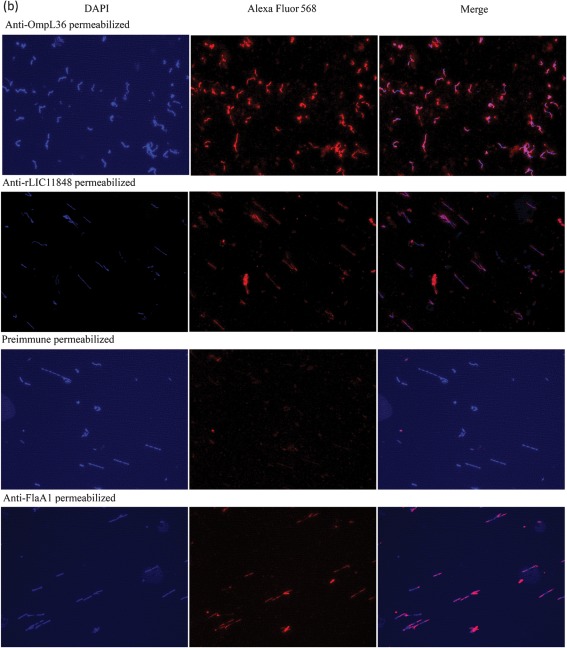
To experimentally evaluate whether LIC11848 protein is present on the surface of L. interrogans, IFA (Cullen et al., 2005; Pinne & Haake, 2009, 2011) was conducted using intact and methanol-permeabilized L. interrogans with rLIC11848-specific serum, serum specific for the known surface-exposed protein OmpL36 (positive control) (Pinne & Haake, 2009) and serum specific for the periplasmic endoflagellar protein FlaA1 (negative control). Immunofluorescence microscopy revealed reactivity to intact L. interrogans with serum specific for rLIC11848 and OmpL36, but not with preimmune serum or serum specific for FlaA1 (Fig. 3a). Immunofluorescence was also detected in permeabilized Leptospira using serum specific for LIC11848, OmpL36 and FlaA1, but not preimmune serum (Fig. 3b). The specificity of the rLIC11848-specific antiserum was tested via immunoblot analysis. This analysis revealed reactivity to a protein band migrating at a molecular mass corresponding to the 32 kDa predicted molecular mass for LIC11848 (Supplementary Fig. S6). Immunoblot analysis using preimmune serum showed an absence of reactivity.
Fig. 3.
Immunofluorescence microscopy identifies the LIC11848 protein as a surface-exposed, outer-membrane protein. Intact (a) and methanol-permeabilized (b; following page) L. interrogans was probed with rabbit preimmune serum or rabbit serum specific for OmpL36 (outer-membrane protein positive control), rLIC11848 or FlaA1 (periplasmic negative control protein). The observed fluorescence corresponds to either Alexa Fluor 568-labelled leptospires or DAPI-stained leptospiral DNA. Merged images reveal the overlap of DAPI-stained and Alexa Fluor 568-labelled Leptospira.
Leptospira express LIC11848 during colonization of hamster kidneys and liver
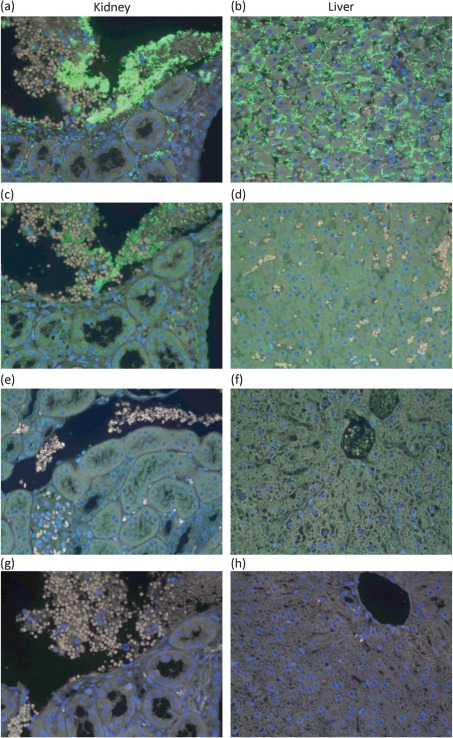
To assess expression of LIC11848 during the infection process, immunofluorescence microscopy was conducted on kidney and liver sections of healthy uninfected and Leptospira-infected hamsters. Use of antiserum specific for LipL32 (positive control) or rLIC11848 in these experiments resulted in leptospiral fluorescence in both kidney and liver sections of hamsters infected with Leptospira (Fig. 4a–d). Although leptospires were observed throughout the tissue of infected hamsters, the level of LIC11848 protein expression was reduced in comparison with the highly conserved and abundant outer-membrane lipoprotein LipL32. This observation was in agreement with absolute protein numbers per Leptospira reported using selective reaction monitoring (Malmström et al., 2009), and more similar in protein expression levels to LipL46 (data not shown) in serially sectioned tissue. In kidney sections, the spirochaetes were mainly observed disseminated in the blood vessels and in the interstitium between tubules. In liver sections, the leptospires were observed within the intercellular spaces between hepatocytes. Use of antiserum specific for rLIC11848 on healthy (not infected with Leptospira) hamster kidney and liver sections did not result in fluorescence (Fig. 4e, f). Sections of Leptospira-infected hamster kidney and liver tissue incubated with secondary antibody alone did not display any fluorescence, confirming that the detected fluorescence was specific for the leptospiral proteins and did not result from non-specific staining from the fluorescent secondary antibody (Fig. 4g, h).
Fig. 4.
Leptospira cells express LIC11848 protein during colonization of hamster kidneys and liver, as evidenced by immunofluorescence microscopy. Leptospira-infected tissue samples from golden Syrian hamsters were probed with LipL32 (positive control) or rLIC11848 antisera and viewed by indirect fluorescence microscopy (Matsunaga et al., 2006). Fluorescence (green areas) was detected using both LipL32 and rLIC11848 antisera with infected kidney [(a) and (c), respectively] and liver [(b) and (d), respectively] tissue sections but not in healthy uninfected kidney (e) or liver (f) tissue sections using rLIC11848 antiserum. Use of secondary antibody alone did not result in fluorescence in kidney (g) or liver (h) tissue sections.
Discussion
The results presented herein enhance our understanding of L. interrogans biology and serve as a platform for the study of leptospiral virulence. In this study, proteome analyses conducted on L. interrogans grown under the varying environmental conditions of media depleted in iron, media supplemented with 10 % serum or media depleted of iron and containing 10 % serum (in vivo-like growth conditions) identified the novel protein LIC11848. Immunofluorescence experiments revealed reactivity of serum specific for the 32 kDa LIC11848 protein product with intact L. interrogans, thus providing evidence for the exposure of this protein on the surface of viable L. interrogans. This finding, combined with the bioinformatic predictions that this protein possesses a cleavable SpI signal sequence and may have a propensity to form a β-barrel structure, prompted our designation of this protein as OmpL32. The present study also demonstrated by immunohistochemistry the expression of OmpL32 during leptospiral infection of the kidneys and liver of hamsters, correlating with a prior report of OmpL32 expression in virulent Leptospira freshly cultured from the liver and kidney of infected hamsters (Vieira et al., 2009). The level of OmpL32 expression compared with LipL32 in vivo was in agreement with an earlier proteomic study that utilized selective reaction monitoring and demonstrated an 11-fold higher expression level of LipL32 (32 190 copies per cell) compared with OmpL32 (2740 copies per cell) (Malmström et al., 2009) in Leptospira grown under laboratory conditions. Finally, and most importantly, proteomic analysis of multiple isoforms of OmpL32 revealed the interesting observation of differential glutamic acid methylation within this surface-exposed protein.
To date, glutamic acid carboxymethylation in bacteria has been discussed within the context of chemotaxis (Ahlgren & Ordal, 1983; Kehry et al., 1984; Kleene et al., 1977; Nishiyama et al., 1999). Specific carboxymethylation on glutamic acid residues serves to modulate protein function from an active to an inactive conformation or vice versa. Activation via methylation results in conformational changes of the methylated protein, leading to activation of downstream proteins that eventually activate flagella for directional motility (Clarke, 1993). The discovery that OmpL32 displays differential methylation suggests that a related methylation-induced regulation may occur with this leptospiral protein. However, two observations provide evidence suggesting that OmpL32 does not play a role in bacterial chemotaxis. First, the OmpL32 amino acid sequence does not reveal homology to proteins involved in either chemotaxis or motility, which in general are highly homologous to one another. And second, glutamic acid methylation of chemotaxis proteins has been demonstrated to occur on only a few targeted amino acid residues (Rice & Dahlquist, 1991), which is in distinct contrast to the widespread methylation pattern observed for OmpL32. These observations suggest that the glutamic acid-specific methylation pattern detected in OmpL32 has an alternative functional role.
Multiple studies have established a requirement for methylation in bacterial pathogenesis. A requirement for methylation has been demonstrated in Y. pseudotuberculosis, where an insertion mutant deficient in the VagH protein is avirulent in mice (Garbom et al., 2004). In Y. pseudotuberculosis, the VagH protein is homologous to the E. coli HemK protein (Garbom et al., 2007), an N5-methyltransferase (Heurgué-Hamard et al., 2002; Nakahigashi et al., 2002). The Y. pseudotuberculosis vagH insertion mutant demonstrated repression of secretion of the virulence determinant YopD via the type III secretion system (Garbom et al., 2007). Additionally, this mutant displayed characteristics similar to a type III secretion mutant during pathogenesis, suggesting a role for protein methylation in type III secretion function (Garbom et al., 2007). Within the context of outer-membrane protein methylation and bacterial pathogenesis, a link has been demonstrated in the bacterial pathogen R. prowazekii. The avirulent Madrid E strain has a null mutation in the methyltransferase-encoding rp027 gene (Chao et al., 2007), and consequently shows hypomethylation of the immunodominant outer-membrane protein B (OmpB). In comparison, the virulent R. prowazekii Breinl strain, which expresses the Rp027 protein product, exhibits hypermethylation of OmpB (Chao et al., 2004), suggesting a role in virulence for outer-membrane protein methylation in R. prowazekii (Chao et al., 2007).
Methylation of bacterial outer-membrane proteins could contribute to virulence in at least two ways. First, methylation could potentially be used by bacteria to regulate protein function, in a manner analogous to that seen for chemotaxis proteins (Wadhams & Armitage, 2004) and the Y. pseudotuberculosis type III secretion system (Garbom et al., 2007). In this scenario, methylation could act to regulate the function of leptospiral virulence factors present on the spirochaetal surface by providing a post-translationally controlled switch between an active/inactive state, and in this way ensure optimal timing of virulence protein function within the infection process. Also, the presence or absence of protein methylation could be critical for real-time phase variation as a method of altering the antigenicity of bacterial outer-membrane proteins, thereby modulating the host immune response recognition of these proteins. Support for the existence of effects on antigenicity and their influence on cellular and humoral immune responses to bacterial pathogens comes from studies conducted with the pathogens M. tuberculosis and R. typhi. In M. tuberculosis, correlation of the level of protein methylation with host immune response modulation has been demonstrated (Temmerman et al., 2004), with native methylation of the surface-exposed M. tuberculosis heparin-binding haemagglutinin protein being required for an effective T cell-mediated immune response. The effect of protein methylation on the host humoral immune response has been studied in R. typhi using methylated and non-methylated protein fragments of the outer-membrane protein OmpB (Chao et al., 2008). In that study experiments conducted using patient sera in an ELISA-based format revealed higher titres to the methylated OmpB fragment when compared with the unmethylated fragment. Together these studies provide evidence that methylation of surface-exposed proteins in bacteria directly affects pathogen recognition by the host immune response.
In order for OmpL32 to function in a methylation-dependent, immune evasion capacity within Leptospira, the protein must be present on the surface of the bacterium. Immunofluorescence microscopy experiments confirmed surface exposure of this protein in L. interrogans serovar Copenhageni strain Fiocruz L1-130. The presentation of this protein on the leptospiral surface would facilitate direct recognition by the host immune response. The capacity of the bacterium to differentially methylate OmpL32 could contribute to the ability of this bacterium to persist in the presence of the generated host immune response, and thus establish a chronic infection. In this scenario OmpL32 methylation could serve to modulate the level of the host immune response in a manner similar to that documented for M. tuberculosis (Temmerman et al., 2004) and R. typhi (Chao et al., 2008). In support of this we have observed different isoforms within an individual growth condition, suggesting that the extent of methylation differs between OmpL32 isoforms. Further, the direct identification of multiple different locations of methylation on amino acid residues within OmpL32 provides an additional indication that this process may represent a novel mechanism that would alter the surface of the bacterium. A definitive answer as to whether Leptospira cells alter the number of methylated residues within OmpL32 upon exposure to a host environment must await further investigation. However, in this study we have provided a foundation for future studies by showing that an outer-membrane protein is methylated and that the methylation pattern of this protein varies, resulting in multiple isoforms. An alternative question arising from these observations is whether the position of methylation alone rather than the absolute number of methylations leads to altered recognition by the host.
Consistent with a purported functional role for differential methylation of OmpL32 in evasion of a host-generated humoral immune response, the entire repertoire of differentially methylated glutamic residues observed within OmpL32 in our study is located within predicted OmpL32-specific B cell epitopes. Also of note is the observation that the gene encoding OmpL32 (lic11848) resides in a putative three-component operon with a gene encoding a hypothetical protein (lic11849) and a gene encoding a putative methyltransferase (lic11850). Since co-expressed proteins are routinely functionally linked, this suggests that OmpL32 serves as a methylation substrate for the downstream putative methyltransferase. Combined, these observations suggest that all the required components for a rapid and effective immune evasion strategy focused around OmpL32 may be in place. In this model, exposure to altered environmental conditions would promote methylation of OmpL32 by the simultaneously expressed methyltransferase on residues contained within B cell epitopes, thus leading to an altered immune recognition of this protein on the leptospiral surface.
In summary, the current study has identified a novel L. interrogans protein designated OmpL32, which we have shown to be surface-exposed, expressed during the course of infection and differentially methylated on glutamic acid residues. Further investigation into the methylation status of leptospiral surface-exposed proteins and the potential role that this PTM plays in the leptospiral infection process is warranted, and may lead to the identification of a novel immune evasion strategy within Leptospira.
Acknowledgements
The authors would like to thank Dr Martin Boulanger, University of Victoria, Victoria, BC, Canada, for his gift of the pET28a vector and assistance with construct design, Anna von Rossum and Marcus Barron for assistance with recombinant protein production, and Dr Christoph Borchers, Derek Smith, Jen Proc, Leanne Ohlund, Darryl Hardie and the members of the University of Victoria-Genome BC Proteomics Centre for their support and guidance with the proteomic analyses. This work was supported by a University of Victoria Fellowship, Graduate Scholarship and the Pacific Century Graduate Scholarship (A. E.), a Discovery Grant from the Natural Sciences and Engineering Research Council of Canada (327186; C. E. C.), the British Columbia Proteomics Network (C. E. C.), the Canada Foundation for Innovation (C. E. C.), the British Columbia Knowledge Development Fund (C. E. C.), VA Medical Research Funds (D. A. H.), and Public Health Service grant AI-034431 (D. A. H.) from the National Institute of Allergy and Infectious Diseases. C. E. C. is a Canada Research Chair in Molecular Pathogenesis and a Michael Smith Foundation for Health Research Scholar.
Abbreviations:
- 2DGE
2D gel electrophoresis
- IFA
immunofluorescence assay
- LC-ESI-MS/MS
liquid chromatography-electrospray ionization-tandem MS
- PMF
peptide mass fingerprint
- PTM
post-translational modification
Footnotes
Six supplementary figures and a supplementary table are available with the online version of this paper.
References
- Ahlgren J. A., Ordal G. W. (1983). Methyl esterification of glutamic acid residues of methyl-accepting chemotaxis proteins in Bacillus subtilis. Biochem J 213, 759–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artiushin S., Timoney J. F., Nally J., Verma A. (2004). Host-inducible immunogenic sphingomyelinase-like protein, Lk73.5, of Leptospira interrogans. Infect Immun 72, 742–749. 10.1128/IAI.72.2.742-749.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagos P. G., Liakopoulos T. D., Spyropoulos I. C., Hamodrakas S. J. (2004). PRED-TMBB: a web server for predicting the topology of β-barrel outer membrane proteins. Nucleic Acids Res 32 (Web Server issue), W400–W404. 10.1093/nar/gkh417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck M., Malmström J. A., Lange V., Schmidt A., Deutsch E. W., Aebersold R. (2009). Visual proteomics of the human pathogen Leptospira interrogans. Nat Methods 6, 817–823. 10.1038/nmeth.1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharti A. R., Nally J. E., Ricaldi J. N., Matthias M. A., Diaz M. M., Lovett M. A., Levett P. N., Gilman R. H., Willig M. R. & other authors (2003). Leptospirosis: a zoonotic disease of global importance. Lancet Infect Dis 3, 757–771. 10.1016/S1473-3099(03)00830-2 [DOI] [PubMed] [Google Scholar]
- Bourhy P., Louvel H., Saint Girons I., Picardeau M. (2005). Random insertional mutagenesis of Leptospira interrogans, the agent of leptospirosis, using a mariner transposon. J Bacteriol 187, 3255–3258. 10.1128/JB.187.9.3255-3258.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X. J., Dai J., Xu H., Nie S., Chang X., Hu B. Y., Sheng Q. H., Wang L. S., Ning Z. B. & other authors (2010). High-coverage proteome analysis reveals the first insight of protein modification systems in the pathogenic spirochete Leptospira interrogans. Cell Res 20, 197–210. 10.1038/cr.2009.127 [DOI] [PubMed] [Google Scholar]
- Chao C. C., Wu S. L., Ching W. M. (2004). Using LC-MS with de novo software to fully characterize the multiple methylations of lysine residues in a recombinant fragment of an outer membrane protein from a virulent strain of Rickettsia prowazekii. Biochim Biophys Acta 1702, 145–152. [DOI] [PubMed] [Google Scholar]
- Chao C. C., Chelius D., Zhang T., Mutumanje E., Ching W. M. (2007). Insight into the virulence of Rickettsia prowazekii by proteomic analysis and comparison with an avirulent strain. Biochim Biophys Acta 1774, 373–381. [DOI] [PubMed] [Google Scholar]
- Chao C. C., Zhang Z., Wang H., Alkhalil A., Ching W. M. (2008). Serological reactivity and biochemical characterization of methylated and unmethylated forms of a recombinant protein fragment derived from outer membrane protein B of Rickettsia typhi. Clin Vaccine Immunol 15, 684–690. 10.1128/CVI.00281-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke S. (1993). Protein methylation. Curr Opin Cell Biol 5, 977–983. 10.1016/0955-0674(93)90080-A [DOI] [PubMed] [Google Scholar]
- Cullen P. A., Cordwell S. J., Bulach D. M., Haake D. A., Adler B. (2002). Global analysis of outer membrane proteins from Leptospira interrogans serovar Lai. Infect Immun 70, 2311–2318. 10.1128/IAI.70.5.2311-2318.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen P. A., Xu X., Matsunaga J., Sanchez Y., Ko A. I., Haake D. A., Adler B. (2005). Surfaceome of Leptospira spp. Infect Immun 73, 4853–4863. 10.1128/IAI.73.8.4853-4863.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deitsch K. W., Lukehart S. A., Stringer J. R. (2009). Common strategies for antigenic variation by bacterial, fungal and protozoan pathogens. Nat Rev Microbiol 7, 493–503. 10.1038/nrmicro2145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Manzalawy Y., Dobbs D., Honavar V. (2008). Predicting linear B-cell epitopes using string kernels. J Mol Recognit 21, 243–255. 10.1002/jmr.893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellinghausen H. C., Jr, McCullough W. G. (1965). Nutrition of Leptospira pomona and growth of 13 other serotypes: fractionation of oleic albumin complex and a medium of bovine albumin and polysorbate 80. Am J Vet Res 26, 45–51. [PubMed] [Google Scholar]
- Emanuelsson O., Brunak S., von Heijne G., Nielsen H. (2007). Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc 2, 953–971. 10.1038/nprot.2007.131 [DOI] [PubMed] [Google Scholar]
- Eshghi A., Cullen P. A., Cowen L., Zuerner R. L., Cameron C. E. (2009). Global proteome analysis of Leptospira interrogans. J Proteome Res 8, 4564–4578. 10.1021/pr9004597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbom S., Forsberg A., Wolf-Watz H., Kihlberg B. M. (2004). Identification of novel virulence-associated genes via genome analysis of hypothetical genes. Infect Immun 72, 1333–1340. 10.1128/IAI.72.3.1333-1340.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbom S., Olofsson M., Björnfot A.-C., Srivastava M. K., Robinson V. L., Oyston P. C. F., Titball R. W., Wolf-Watz H. (2007). Phenotypic characterization of a virulence-associated protein, VagH, of Yersinia pseudotuberculosis reveals a tight link between VagH and the type III secretion system. Microbiology 153, 1464–1473. 10.1099/mic.0.2006/000323-0 [DOI] [PubMed] [Google Scholar]
- Giacomodonato M. N., Sarnacki S. H., Llana M. N., Cerquetti M. C. (2009). Dam and its role in pathogenicity of Salmonella enterica. J Infect Dev Ctries 3, 484–490. 10.3855/jidc.465 [DOI] [PubMed] [Google Scholar]
- Guerreiro H., Croda J., Flannery B., Mazel M., Matsunaga J., Galvão Reis M., Levett P. N., Ko A. I., Haake D. A. (2001). Leptospiral proteins recognized during the humoral immune response to leptospirosis in humans. Infect Immun 69, 4958–4968. 10.1128/IAI.69.8.4958-4968.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haake D. A., Matsunaga J. (2002). Characterization of the leptospiral outer membrane and description of three novel leptospiral membrane proteins. Infect Immun 70, 4936–4945. 10.1128/IAI.70.9.4936-4945.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haake D. A., Chao G., Zuerner R. L., Barnett J. K., Barnett D., Mazel M., Matsunaga J., Levett P. N., Bolin C. A. (2000). The leptospiral major outer membrane protein LipL32 is a lipoprotein expressed during mammalian infection. Infect Immun 68, 2276–2285. 10.1128/IAI.68.4.2276-2285.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensel M., Shea J. E., Gleeson C., Jones M. D., Dalton E., Holden D. W. (1995). Simultaneous identification of bacterial virulence genes by negative selection. Science 269, 400–403. 10.1126/science.7618105 [DOI] [PubMed] [Google Scholar]
- Heurgué-Hamard V., Champ S., Engström A., Ehrenberg M., Buckingham R. H. (2002). The hemK gene in Escherichia coli encodes the N5-glutamine methyltransferase that modifies peptide release factors. EMBO J 21, 769–778. 10.1093/emboj/21.4.769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R. C., Harris V. G. (1967). Differentiation of pathogenic and saprophytic letospires. I. Growth at low temperatures. J Bacteriol 94, 27–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehry M. R., Doak T. G., Dahlquist F. W. (1984). Stimulus-induced changes in methylesterase activity during chemotaxis in Escherichia coli. J Biol Chem 259, 11828–11835. [PubMed] [Google Scholar]
- Kleene S. J., Toews M. L., Adler J. (1977). Isolation of glutamic acid methyl ester from an Escherichia coli membrane protein involved in chemotaxis. J Biol Chem 252, 3214–3218. [PubMed] [Google Scholar]
- Kloczkowski A., Ting K. L., Jernigan R. L., Garnier J. (2002). Combining the GOR V algorithm with evolutionary information for protein secondary structure prediction from amino acid sequence. Proteins 49, 154–166. 10.1002/prot.10181 [DOI] [PubMed] [Google Scholar]
- Ko A. I., Galvão Reis M., Ribeiro Dourado C. M., Johnson W. D., Jr, Riley L. W., Salvador Leptospirosis Study Group (1999). Urban epidemic of severe leptospirosis in Brazil. Lancet 354, 820–825. [DOI] [PubMed] [Google Scholar]
- Ko A. I., Goarant C., Picardeau M. (2009). Leptospira: the dawn of the molecular genetics era for an emerging zoonotic pathogen. Nat Rev Microbiol 7, 736–747. 10.1038/nrmicro2208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinska E., Bhatnagar S., Sweet L., Chatterjee D., Schorey J. S. (2005). Mycobacterium avium 104 deleted of the methyltransferase D gene by allelic replacement lacks serotype-specific glycopeptidolipids and shows attenuated virulence in mice. Mol Microbiol 56, 1262–1273. 10.1111/j.1365-2958.2005.04608.x [DOI] [PubMed] [Google Scholar]
- Levett P. N. (2001). Leptospirosis. Clin Microbiol Rev 14, 296–326. 10.1128/CMR.14.2.296-326.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao S., Sun A., Ojcius D. M., Wu S., Zhao J., Yan J. (2009). Inactivation of the fliY gene encoding a flagellar motor switch protein attenuates mobility and virulence of Leptospira interrogans strain Lai. BMC Microbiol 9, 253–262. 10.1186/1471-2180-9-253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo M., Cordwell S. J., Bulach D. M., Adler B. (2009). Comparative transcriptional and translational analysis of leptospiral outer membrane protein expression in response to temperature. PLoS Negl Trop Dis 3, e560. 10.1371/journal.pntd.0000560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y., Liu Y., Sun D., Ojcius D. M., Zhao J., Lin X., Wu D., Zhang R., Chen M. & other authors (2011). InvA protein is a Nudix hydrolase required for infection by pathogenic Leptospira in cell lines and animals. J Biol Chem 286, 36852–36863. 10.1074/jbc.M111.219931 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Ma B., Zhang K., Hendrie C., Liang C., Li M., Doherty-Kirby A., Lajoie G. (2003). peaks: powerful software for peptide de novo sequencing by tandem mass spectrometry. Rapid Commun Mass Spectrom 17, 2337–2342. 10.1002/rcm.1196 [DOI] [PubMed] [Google Scholar]
- Malmström J., Beck M., Schmidt A., Lange V., Deutsch E. W., Aebersold R. (2009). Proteome-wide cellular protein concentrations of the human pathogen Leptospira interrogans. Nature 460, 762–765. 10.1038/nature08184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga J., Werneid K., Zuerner R. L., Frank A., Haake D. A. (2006). LipL46 is a novel surface-exposed lipoprotein expressed during leptospiral dissemination in the mammalian host. Microbiology 152, 3777–3786. 10.1099/mic.0.29162-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga J., Lo M., Bulach D. M., Zuerner R. L., Adler B., Haake D. A. (2007). Response of Leptospira interrogans to physiologic osmolarity: relevance in signaling the environment-to-host transition. Infect Immun 75, 2864–2874. 10.1128/IAI.01619-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride A. J., Athanazio D. A., Reis M. G., Ko A. I. (2005). Leptospirosis. Curr Opin Infect Dis 18, 376–386. 10.1097/01.qco.0000178824.05715.2c [DOI] [PubMed] [Google Scholar]
- Merien F., Truccolo J., Rougier Y., Baranton G., Perolat P. (1998). In vivo apoptosis of hepatocytes in guinea pigs infected with Leptospira interrogans serovar icterohaemorrhagiae. FEMS Microbiol Lett 169, 95–102. 10.1111/j.1574-6968.1998.tb13304.x [DOI] [PubMed] [Google Scholar]
- Monahan A. M., Callanan J. J., Nally J. E. (2008). Proteomic analysis of Leptospira interrogans shed in urine of chronically infected hosts. Infect Immun 76, 4952–4958. 10.1128/IAI.00511-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray G. L., Srikram A., Henry R., Puapairoj A., Sermswan R. W., Adler B. (2009). Leptospira interrogans requires heme oxygenase for disease pathogenesis. Microbes Infect 11, 311–314. 10.1016/j.micinf.2008.11.014 [DOI] [PubMed] [Google Scholar]
- Murray G. L., Srikram A., Henry R., Hartskeerl R. A., Sermswan R. W., Adler B. (2010). Mutations affecting Leptospira interrogans lipopolysaccharide attenuate virulence. Mol Microbiol 78, 701–709. 10.1111/j.1365-2958.2010.07360.x [DOI] [PubMed] [Google Scholar]
- Nakahigashi K., Kubo N., Narita S.-i., Shimaoka T., Goto S., Oshima T., Mori H., Maeda M., Wada C., Inokuchi H. (2002). HemK, a class of protein methyl transferase with similarity to DNA methyl transferases, methylates polypeptide chain release factors, and hemK knockout induces defects in translational termination. Proc Natl Acad Sci U S A 99, 1473–1478. 10.1073/pnas.032488499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nally J. E., Artiushin S., Timoney J. F. (2001a). Molecular characterization of thermoinduced immunogenic proteins Q1p42 and Hsp15 of Leptospira interrogans. Infect Immun 69, 7616–7624. 10.1128/IAI.69.12.7616-7624.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nally J. E., Timoney J. F., Stevenson B. (2001b). Temperature-regulated protein synthesis by Leptospira interrogans. Infect Immun 69, 400–404. 10.1128/IAI.69.1.400-404.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nally J. E., Whitelegge J. P., Bassilian S., Blanco D. R., Lovett M. A. (2007). Characterization of the outer membrane proteome of Leptospira interrogans expressed during acute lethal infection. Infect Immun 75, 766–773. 10.1128/IAI.00741-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama S. I., Umemura T., Nara T., Homma M., Kawagishi I. (1999). Conversion of a bacterial warm sensor to a cold sensor by methylation of a single residue in the presence of an attractant. Mol Microbiol 32, 357–365. 10.1046/j.1365-2958.1999.01355.x [DOI] [PubMed] [Google Scholar]
- Parra M., Pickett T., Delogu G., Dheenadhayalan V., Debrie A. S., Locht C., Brennan M. J. (2004). The mycobacterial heparin-binding hemagglutinin is a protective antigen in the mouse aerosol challenge model of tuberculosis. Infect Immun 72, 6799–6805. 10.1128/IAI.72.12.6799-6805.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinne M., Haake D. A. (2009). A comprehensive approach to identification of surface-exposed, outer membrane-spanning proteins of Leptospira interrogans. PLoS ONE 4, e6071. 10.1371/journal.pone.0006071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinne M., Haake D. (2011). Immuno-fluorescence assay of leptospiral surface-exposed proteins. J Vis Exp (53), 2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman O., Cummings S., Harrington D., Sutcliffe I. (2008). Methods for the bioinformatic identification of bacterial lipoproteins encoded in the genomes of Gram-positive bacteria. World J Microb Biot 24, 2377–2382. 10.1007/s11274-008-9795-2 [DOI] [Google Scholar]
- Rice M. S., Dahlquist F. W. (1991). Sites of deamidation and methylation in Tsr, a bacterial chemotaxis sensory transducer. J Biol Chem 266, 9746–9753. [PubMed] [Google Scholar]
- Ristow P., Bourhy P., da Cruz McBride F. W., Figueira C. P., Huerre M., Ave P., Girons I. S., Ko A. I., Picardeau M. (2007). The OmpA-like protein Loa22 is essential for leptospiral virulence. PLoS Pathog 3, e97. 10.1371/journal.ppat.0030097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakolvaree Y., Maneewatch S., Jiemsup S., Klaysing B., Tongtawe P., Srimanote P., Saengjaruk P., Banyen S., Tapchaisri P. & other authors (2007). Proteome and immunome of pathogenic Leptospira spp. revealed by 2DE and 2DE-immunoblotting with immune serum. Asian Pac J Allergy Immunol 25, 53–73. [PubMed] [Google Scholar]
- Sassetti C. M., Boyd D. H., Rubin E. J. (2001). Comprehensive identification of conditionally essential genes in mycobacteria. Proc Natl Acad Sci U S A 98, 12712–12717. 10.1073/pnas.231275498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seguro A. C., Lomar A. V., Rocha A. S. (1990). Acute renal failure of leptospirosis: nonoliguric and hypokalemic forms. Nephron 55, 146–151. 10.1159/000185943 [DOI] [PubMed] [Google Scholar]
- Sen T. Z., Jernigan R. L., Garnier J., Kloczkowski A. (2005). GOR V server for protein secondary structure prediction. Bioinformatics 21, 2787–2788. 10.1093/bioinformatics/bti408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setubal J. C., Reis M. G., Matsunaga J., Haake D. A. (2006). Lipoprotein computational prediction in spirochaetal genomes. Microbiology 152, 113–121. 10.1099/mic.0.28317-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temmerman S., Pethe K., Parra M., Alonso S., Rouanet C., Pickett T., Drowart A., Debrie A. S., Delogu G. & other authors (2004). Methylation-dependent T cell immunity to Mycobacterium tuberculosis heparin-binding hemagglutinin. Nat Med 10, 935–941. 10.1038/nm1090 [DOI] [PubMed] [Google Scholar]
- Thiermann A. B., McClellan R. D., Hill H. T. (1984). Improved techniques for the isolation of leptospires from swine abortion cases. American Assn of Veterinary Laboratory Diagnosticians 27, 233–244. [Google Scholar]
- Tsirigos K. D., Bagos P. G., Hamodrakas S. J. (2011). OMPdb: a database of β-barrel outer membrane proteins from Gram-negative bacteria. Nucleic Acids Res 39 (Database issue), D324–D331. 10.1093/nar/gkq863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velineni S., Asuthkar S., Sritharan M. (2006). Iron limitation and expression of immunoreactive outer membrane proteins in Leptospira interrogans serovar icterohaemorrhagiae strain lai. Indian J Med Microbiol 24, 339–342. 10.4103/0255-0857.29414 [DOI] [PubMed] [Google Scholar]
- Verma A., Rathinam S. R., Priya C. G., Muthukkaruppan V. R., Stevenson B., Timoney J. F. (2008). LruA and LruB antibodies in sera of humans with leptospiral uveitis. Clin Vaccine Immunol 15, 1019–1023. 10.1128/CVI.00203-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira M. L., Pimenta D. C., de Morais Z. M., Vasconcellos S. A., Nascimento A. L. (2009). Proteome analysis of Leptospira interrogans virulent strain. Open Microbiol J 3, 69–74. 10.2174/1874285800903010069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadhams G. H., Armitage J. P. (2004). Making sense of it all: bacterial chemotaxis. Nat Rev Mol Cell Biol 5, 1024–1037. 10.1038/nrm1524 [DOI] [PubMed] [Google Scholar]
- WHO (1999). Leptospirosis worldwide, 1999. Wkly Epidemiol Rec 74, 237–242. [PubMed] [Google Scholar]
- Wilkins M. R., Gasteiger E., Gooley A. A., Herbert B. R., Molloy M. P., Binz P. A., Ou K., Sanchez J. C., Bairoch A. & other authors (1999). High-throughput mass spectrometric discovery of protein post-translational modifications. J Mol Biol 289, 645–657. 10.1006/jmbi.1999.2794 [DOI] [PubMed] [Google Scholar]