Abstract
We previously observed that the HERV type K (HERV-K) envelope (env) protein was expressed in the majority of human breast tumors from a U.S. cohort of women from Texas. We also made the preliminary observation that the expression of HERV-K env transcripts was associated with markers of disease progression. In this follow-up study, env protein expression was evaluated immunohistochemically in an additional 195 paraffin-embedded breast tumors from a second U.S. patient cohort (Baltimore, Maryland) and in 110 tumors from Chinese patients. Moreover, we compared env transcript expression between fresh-frozen normal and cancerous breast tissues. We observed that while env mRNA and protein expression was undetectable in normal breast tissue and in a subset of uninvolved normal-appearing tissue adjacent to the tumor epithelium, it was overexpressed in most tumors. Furthermore, env expression was associated with breast cancer progression. In Baltimore cohort women, HERV-K tumor positivity was significantly associated with disease stage and lymph node metastasis. In Chinese women, HERV-K env positivity was significantly associated with tumor size, TNM stage, and lymph node metastases, which is consistent with the observations in the U.S. cohort. We also found that Chinese breast cancer patients with a high expression of HERV-K had a decreased overall survival compared with patients who had either a moderate or low HERV-K expression in their tumors (P = 0.049, χ2 log rank test). In conclusion, the HERV-K env gene is expressed in the majority of breast cancers from U.S. or Chinese women but not in normal breast tissue. High expression of HERV-K env protein in breast cancer patients is associated with markers of disease progression and poor disease outcome, indicating that HERV-K env protein is a novel candidate prognostic marker for breast cancer.
Keywords: human endogenous retrovirus K (HERV-K), breast cancer, envelope (env) protein, prognosis, survival
Introduction
Breast cancer is the most common cancer among women in the United States and has become the second leading cause of death from cancer for women worldwide.1 Its incidence has been rising in recent years in the developing world, especially in Chinese women.2 Previous studies have revealed that aberrant expression of tumor-specific antigens can generate novel biomarkers for prognosis and targets for therapy. Thus, searching for new tumor-associated antigens and analyzing their mechanism of action in the development and progression of breast cancer can potentially improve diagnosis and treatment of breast cancer. In this regard, human endogenous retrovirus type K (HERV-K) is perhaps an ideal tumor marker and therapeutic target, because HERVs are reactivated in tumor biology and are suspected to influence human cancer biology.3
Retroviruses are regarded as an etiological agent in mammary carcinogenesis, based on molecular epidemiology findings and animal experiments.4 Mouse mammary tumor virus (MMTV), discovered by Bittner5 more than half a century ago, serves as an insertion mutagen that causes mammary carcinoma by activation of cellular proto-oncogenes such as int, wnt, and fgf in certain strains of mice.6 HERVs generally reside in the human chromosome and are the remnants of germline infections of human ancestors by retroviruses millions of years ago. The human genome harbors many distinct families of HERVs. Most HERVs are defective because of multiple termination codons and deletions.7 In contrast to defective sequences in many HERV families, the HERV-K family shows a conservation of seemingly intact retroviral genes.8 Thus, it is the most likely of all the families to encode for complete and possibly infections retroviruses.9
HERV-K was originally identified by its homology to MMTV10 and also contains members that are transcriptionally active in several human cancer tissues11-14 as well as in tumor cell lines, notably the human cancer cell line T47D.15,16 Previously, we demonstrated the expression of HERV-K envelope (env) messenger RNA (mRNA) and protein in breast cancer tissues obtained from U.S. women.11,12,17 In this study, we evaluated the expression of HERV-K env at the level of protein and messenger RNA (mRNA) in breast cancer tissues obtained from both U.S. and Chinese women and analyzed the relationship between HERV-K env expression and disease severity in these women using a quantitative PCR technology and a previously validated anti-HERV-K monoclonal antibody. Our findings suggest that env is a novel prognostic marker in human breast cancer.
Results
Detection of HERV-K env mRNA Transcripts in Breast Tissues by RT-PCR
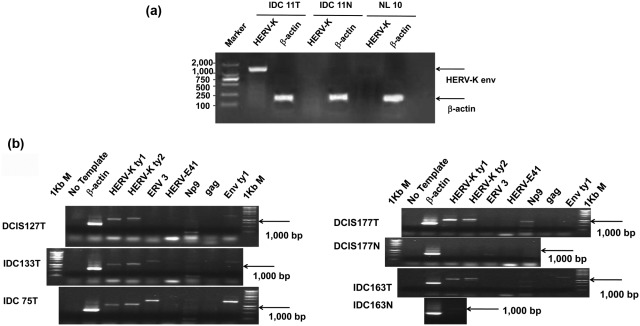
One hundred and twenty tissues from Chinese women, including 40 breast cancer tissues, 40 matched adjacent uninvolved breast tissues, and 40 breast tissues from women with benign breast diseases but without cancer, were used for evaluating the expression of HERV-K env mRNA by RT-PCR (Table 1). A representative RT-PCR result is shown in Figure 1A. HERV-K env RNA (1,494 bp) was detected in 70% of breast cancer samples and in 20% of adjacent, nontumor breast tissues derived from the breast cancer patients, indicating that env expression is significantly upregulated in the cancerous breast when compared with surrounding, normal appearing breast tissue (P < 0.0001, χ2 test). There was no detectable expression of HERV-K env mRNA in normal breast tissues derived from women who did not have a breast cancer diagnosis (Table 1), indicating that env expression is absent in the normal breast but induced during breast carcinogenesis. HERV-K env protein expression in the cancerous breast was also significantly higher than in the adjacent uninvolved breast tissues (Table 1).
Table 1.
HERV-K env mRNA and Protein Expression in Breast Tissues from Chinese women
| Sample description | Negative | Positive | % Positive | Total | P value (χ2 test) | ||
|---|---|---|---|---|---|---|---|
| HERV-K mRNA expression assessed by RT-PCR | |||||||
| Breast cancer tissue | 12 | 28 | 70.0 | 40 | |||
| Uninvolved breast tissue | 32 | 8 | 20.0 | 40 | |||
| Normal breast tissue |
40 |
0 |
0.0 |
40 |
<0.0001a |
||
| Sample description |
– |
+ |
++ |
+++ |
% Positive |
Total |
P value (χ2 test) |
| HERV-K protein expression assessed by IHC | |||||||
| Benign tissues | 18 | 12 | 0 | 0 | 40.0 | 30 | |
| Cancer tissues | 15 | 11 | 25 | 59 | 86.4 | 110 | <0.0001b |
Comparison of the positive expression rate of the 3 groups using contingency table analysis χ2 (and Fisher’s exact) test by GraphPad Prism 5.
Comparison of the positive expression rate of the 2 groups using contingency table analysis χ2 (and Fisher’s exact) test by GraphPad Prism 5.
Figure 1.
Expression of HERV-K env mRNA in breast tissues. (A) The expression of HERV-K was detected in a breast cancer tumor biopsy obtained from a Chinese breast cancer patient diagnosed with invasive ductal carcinoma IDC (IDC 11T) but not in a matched uninvolved breast biopsy (IDC 11N) or in a benign breast tissue obtained from a Chinese women diagnosed with hyperplasia (NL10). β-actin expression in the corresponding breast tissue samples was also measured. Marker: DNA Marker DL2000. (B) Expression of HERV-K (type 1 and type 2), ERV3, HERV-E41, Np9, and gag and full-length env mRNAs was evaluated in various breast tissues by RT-PCR using corresponding primer pairs. NC in each set is a no-template control. β-actin was used as a control. The expression of HERV-K type 1 (Ktp1:1,104 bp), type 2 (Ktp2: 1,194 bp), Rec (437bp), Np9 (256 bp), and full length env (1,813bp) was detected in 3 breast cancer tissues (ductal carcinoma in situ [DCIS], DCIS127T, invasive ductal carcinomas [IDC], IDC133T, and IDC75T). Importantly, the expression of each transcript was compared in tumor tissues versus uninvolved breast tissues obtained from the same patient. The expression of HERV-K type 1, type 2, and full length type 1 env mRNA was demonstrated in DCIS 177T and IDC 163T but not in matched uninvolved breast tissues (DCIS 177N or IDC 163N). The expression of a splice variant of HERV-K env mRNA, was detected in 3 breast cancer biopsies but not in matched uninvolved adjacent breast tissues. The expression of ERV3 env mRNA (1,700 bp) was detected in 2 IDC cancer tissues (IDC133T and IDC75T), however expression of HERV-E env mRNA was not detected in any of these tissues.
The expression of mRNA of HERV-K and the HERV families HERV-E and ERV-3 was examined in U.S. breast cancer tissues (n = 32), uninvolved breast tissues (n = 30), and noncancer breast tissues (n = 8) by RT-PCR, as described previously.15,16 One representative result is shown in Figure 1B. Type 1 and type 2 surface (SU) of HERV-K env mRNA were characterized based on the presence (type 2) or absence (type 1) of a 293 bp insert between the pol and env region, as described previously.13 The expression of HERV-K SU env mRNA type 1 and type 2 was detected in tumors (Fig. 2B). Importantly, the expression of HERV-K was demonstrated in cancer tissues but not in matched uninvolved normal breast tissues. A no-template control demonstrated lack of contamination, and β-actin was used to show equal RNA loading. The expression of HERV-K full-length env mRNAs or spliced env mRNAs (Np9) was detected in most breast cancer biopsies, whereas the expression of ERV3 env mRNA was detected in some breast cancer tissues. There was no expression of HERV-K type 1 and type 2, HERV-E, or ERV3 env mRNAs in adjacent matched uninvolved breast tissues. Three bands were detected in some tumor tissues using Np9 primers. One band corresponds to Np9 (205 bp), one corresponds to Rec (400 bp), and one is an unidentified band of approximately 600 bp (DCIS 177T and IDC 163T; Figure 2B), which will be further characterized in future studies.
Figure 2.
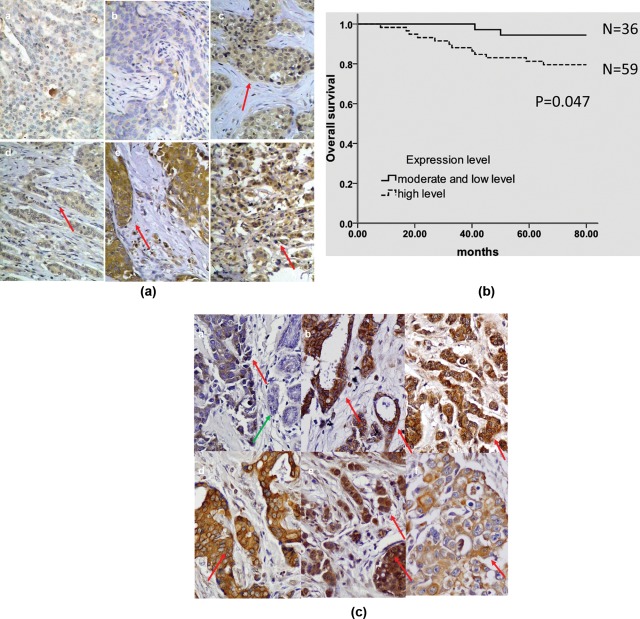
Detection of HERV-K env protein expression in various breast tissues. (A) The expression of HERV-K in Chinese breast cancer biopsies. No HERV-K expression was detected in benign breast tissue (a) or in one sample of breast cancer (b). Positive expression (red arrow) of HERV-K env protein was detected in tumor epithelial cells. The expression of HERV-K env protein was detected in DCIS (c and e) and in IDC (d and f). (B) Kaplan-Meier survival curve. Chinese breast cancer patients with a high expression of HERV-K had a significantly lower survival rate compared with patients with moderate and low levels of expression of HERV-K (P = 0.047), using the log-rank test. (C) The expression of HERV-K in U.S. breast cancer biopsies. Positive expression (red arrow) of HERV-K env protein in various breast cancer tissues; a. IDC4852: stage I, grade 3, no node metastasis; b. mIDC1043: stage N/A, grade 1, 1 node metastasis; c. mIDC2623: stage IIIA, grade 2, 2 node metastasis; d. IDC+DCIS1336: stage I, grade 2, no node metastasis; e. IDC1285: stage IIB, grade 1, no node metastasis; f. IDC1068: stage IIA, grade 3, no node metastasis. No expression of HERV-K env protein was detected in adjacent uninvolved epithelial cells (green arrow; a).
Expression Profiles of HERV-K env Protein in Breast Biopsies Assessed by Immunohistochemistry
Significantly higher HERV-K env protein expression was observed in cancer tissues (n = 110) in comparison to benign breast tumor tissues in Chinese women (n = 30, P < 0.0001, χ2 test), as shown in Table 1. Medium to strong expression of HERV-K env protein was found in 84 of 110 (76.4%) breast cancer samples. Photomicrographs of stained samples (Fig. 2A) revealed medium (c and d) or strong expression (e and f) of HERV-K in most breast cancer samples, whereas mostly negative or sometimes weak expression was found in adjacent benign breast samples (a) and in one breast cancer (b).
Relationship between Expression Level of HERV-K env Protein and Clinicopathological Characteristics of Breast Cancer
Among the Chinese breast cancer patients who had highly expressed HERV-K env protein in their tumors (+++), 82.9% had a tumor size greater than 3 cm and only 17.1% had a tumor size 3 cm or smaller (P = 0.002, χ2 test, Table 2). HERV-K env also correlated positively with clinical stage (P = 0.010) and the number of lymph node metastases (P = 0.023). Thus, HERV-K env protein expression was significantly associated with 3 markers of disease progression but not with age, ER or PR protein expression, or histological grade (Table 2). In addition, the rate of recurrence and metastasis was increased in the patient group with a highly elevated env expression in their tumors (72.7% with +++) compared with the patient group with lower expression of env (27.3% with +/++), although this difference was not statistically significant. The overall survival rate was 14.3% in the group with a strong HERV-K expression (+++), whereas it was 85.7% in the group with medium or weak expression, and this difference between the 2 groups was significant (P = 0.049). Furthermore, a Kaplan-Meier analysis showed that the patients whose tumors highly expressed HERV-K env protein had a significantly shorter overall survival in comparison with patients whose tumor expressed HERV-K env protein in moderate or low levels (P = 0.047; Fig. 2B). In summary, these data indicate that the expression of HERV-K in breast tumors of Chinese patients is a candidate prognostic factor.
Table 2.
Association between HERV-K env Expression Levels and Clinicopathological Characteristics in Chinese Breast Cancer Patients
| Clinical parameter | Total | +/++ (%) | +++ (%) | P value (χ2 test) |
|---|---|---|---|---|
| Age | 95 | 0.866 | ||
| ≤60 | 73 | 28 (38.4) | 45 (61.6) | |
| >60 | 22 | 8 (36.4) | 14 (63.6) | |
| Tumor size | 95 | 0.002 | ||
| ≤3cm | 60 | 30 (50.0) | 30 (50.0) | |
| >3 cm | 35 | 6 (17.1) | 29 (82.9) | |
| Histologic grade | 0.086 | |||
| I or II | 72 | 31 (43.1) | 41 (56.9) | |
| III | 23 | 5 (21.7) | 18 (78.3) | |
| Clinical stage | 0.010 | |||
| I | 8 | 6 (75.0) | 2 (25.0) | |
| II | 57 | 24 (42.1) | 33 (57.9) | |
| III | 30 | 6 (20.0) | 24 (80.0) | |
| Number of lymph node metastases | 0.023 | |||
| 0 | 39 | 19 (48.7) | 20 (51.3) | |
| 1-3 | 21 | 10 (47.6) | 11 (52.4) | |
| ≥4 | 35 | 7 (20.0) | 28 (80.0) | |
| ER | 0.257 | |||
| <15% | 44 | 14 (31.8) | 30 (68.2) | |
| ≥15% | 51 | 22 (43.1) | 29 (56.7) | |
| PR | 0.796 | |||
| <15% | 57 | 21 (36.8) | 36 (63.2) | |
| ≥15% | 38 | 15 (39.5) | 23 (60.5) | |
| Recurrence and metastasis | 0.241 | |||
| No | 73 | 30 (41.1) | 43 (58.9) | |
| Yes | 22 | 6 (27.3) | 16 (72.7) | |
| Death | 0.049 | |||
| No | 81 | 34 (42.0) | 47 (58.0) | |
| Yes | 14 | 2 (14.3) | 12 (85.7) |
Note: Comparison of the 2 groups using contingency table analysis χ2 (and Fisher’s exact) test by GraphPad Prism 5. ER = estrogen receptor; PR = progesterone receptor.
We previously observed that the HERV-K env protein was expressed in a majority of human breast tumors from U.S. women.11,12,17 In a larger follow-up study of 195 breast tumors from patients in the greater Baltimore area (see description of these patients in Supplementary Fig. S1), expression of HERV-K env protein was detected in tumor epithelial cells but not in uninvolved normal epithelial cells, as shown in Figure. 2C. Expression of HERV-K was significantly associated with a positive node status (P = 0.006). Additionally we examined the association of HERV-K expression with 10-year breast cancer specific survival in these patients (Table 3). Although HERV-K was not an independent predictor of breast cancer specific survival in this patient cohort, like TNM stage, nodal status, and p53 mutation accumulation, we did find that HERV-K status in the stratified analysis modified the ability of stage, nodal status, and p53 mutation to predict patient survival. After stratification by HERV-K status, the association of TNM stage, p53, mutations and nodal status with poor survival was strongest in patients who were HERV-K positive. The percentage of cases with greater than 50% of tumor cells staining positive for HERV-K increased with stage (Table 4).
Table 3.
Effect of HERV-K Status on the Association of TNM Stage, Tumor Grade, Nodal Status, and p53 Mutations with Patient Survival in U.S. (Baltimore) Breast Cancer Patients
| Univariate analysis |
Multivariable analysisa |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| HR | 95% | P | n | HR | 95% | P | n | ||
| TNM stage | All patients | 4.44 | 2.63-7.47 | <0.001 | 197 | 4.29 | 2.47-7.47 | <0.001 | 167 |
| HERV-K neg | 2.97 | 1.17-7.49 | 0.021 | 65 | 3.11 | 1.08-8.97 | 0.036 | 51 | |
| HERV-K pos | 5.58 | 2.94-10.6 | <0.001 | 132 | 5.35 | 2.73-10.5 | <0.001 | 116 | |
| Grade | All patients | 1.61 | 0.98-2.65 | 0.060 | 188 | 1.16 | 0.59-2.25 | 0.665 | 167 |
| HERV-K neg | 1.17 | 0.50-2.72 | 0.711 | 59 | 2.82 | 0.62-17.8 | 0.179 | 51 | |
| HERV-K pos | 1.9 | 1.02-3.54 | 0.042 | 129 | 1.08 | 0.50-2.46 | 0.794 | 116 | |
| P53 mutations | All patients | 2.01 | 1.18-3.40 | 0.010 | 213 | 1.6 | 0.88-2.90 | 0.121 | 167 |
| HERV-K neg | 1.56 | 0.61-4.00 | 0.354 | 70 | 0.74 | 0.24-2.24 | 0.596 | 51 | |
| HERV-K pos | 2.3 | 1.21-4.36 | 0.011 | 143 | 2.04 | 0.94-4.39 | 0.070 | 116 | |
| Node status | All patients | 3.68 | 2.14-6.31 | <0.001 | 198 | 2.98 | 1.58-5.62 | 0.001 | 163 |
| HERV-K neg | 3.1 | 1.27-7.56 | 0.013 | 64 | 1.78 | 0.44-7.16 | 0.420 | 50 | |
| HERV-K pos | 4.72 | 2.28-9.78 | <0.001 | 134 | 4.1 | 1.78-9.42 | 0.001 | 113 | |
Note: Cox regression analysis was used to calculate the hazard ratio, 95% confidence interval, (95%), and the P-value for the individual association of TNM stage, grade, p53 mutations, and node status with breast cancer specific survival (all patients). Stratification based on HERV-K status was used to determine the effect of HERV-K positive or negative status on the association of TNM stage, grade, p53 mutations, and node status with breast cancer specific survival (HERV-K neg and HERV K-pos). A P value of <0.05 indicated a statistically significant association with breast cancer specific survival. HR = hazard ratio; 95% = 95% confidence interval.
Adjusted for age at diagnosis, ER status, race, TNM stage, tumor grade, chemotherapy, p53 mutations.
Table 4.
Association of HERV-K Positivity with Disease Stage
| Stage | % HERV-K positive d(n +/N total) | % Tumor with <50% tumor cell positivitya (n) | % Tumor with >50% tumor cell positivityb (n) |
|---|---|---|---|
| DCIS | 53 (9/17) | 33 (3) | 67 (6) |
| I | 52 (17/33) | 35 (6) | 65 (11) |
| II | 62 (64/103) | 15 (10) | 85 (54) |
| III/IV | 62 (26/42) | 11 (3) | 89 (23) |
| cTotal | 59 (116/195) | 19 (22) | 81 (94) |
DCIS = ductal carcinoma in situ.
Less than 50% of tumor cells stained positive for HERV-K.
Greater than 50% of tumor cells stained positive for HERV-K.
Total = DCIS + I + II+ II/IV.
N +/N total = number of positive samples/ number of total cases.
These data, together with the data from the Chinese patient cohort and previous data from our group analyzing breast tumors from cancer patients at the MD Anderson Cancer Center, provide strong evidence that HERV-K env protein expression is highly upregulated in human breast cancer cells but not in adjacent noncancerous tissue or normal breast tissue from women without a cancer diagnosis.
Discussion
Viruses are thought to have an etiological role in 25% of human cancers. The viral cause of human breast cancer has never been fully established, although evidence from cancer epidemiology and animal studies suggested that viruses may play an important role in some breast cancers, including inflammatory breast cancer.18 In this regard, endogenous retroviruses and specifically HERV-K reactivation during early stage mammary carcinogenesis may have a previously unrecognized contribution to breast cancer development and disease aggressiveness, because our genome harbors many endogenous retroviral sequences and some of them may continue to perform various retroviral functions, including tumor induction. HERVs have been long suspected to have a functional role in human cancer biology.3 For example, investigations demonstrated the existence of RNAs that encode for ORFs of retroviral genes in human breast cancer cell lines,15,19,20 and some have shown the selective expression of pol and env viral proteins in human breast cancer.14,17 Moreover, a few studies have established a candidate relationship between the prognosis of breast cancer and viral enzymes or polymorphisms of HERV-K.14,21 In contrast, HERVs are silenced under normal conditions.
In an effort to uncover novel tumor antigens, we demonstrated previously that transcripts of HERV-K env mRNA, with coding potential for the env region of the gene, are expressed frequently in breast cancer.11,12 We further reported that HERV-K env protein was expressed in 85% of breast cancers in U.S. women but not in normal breast tissues from this population.17 Importantly, HERV-K was able to trigger an antigen-specific immune response in only breast cancer patients.17 In the current study, we detected the expression of HERV-K env in various breast tissues and analyzed the association of HERV-K env protein expression level with clinicopathological characteristics and prognosis. We detected expression of both HERV-K SU env mRNA type 1 and type 2 in breast tumors. We also observed both type 1 and type 2 expression of HERV-K env mRNA in breast tumors and breast cancer cells in an earlier study,11 further confirming that both variants are present in breast cancer. A key unanswered question is which factors are responsible for directing type 1 or type 2 expression of HERV-K env mRNA in breast cancer. HERV-K type 1 encodes Np9, a protein expressed in tumors and transformed cell lines, whereas type 2 encodes Rec, a nuclear export factor and functional homologue of the HIV and HTLV encoded accessory proteins Rev and Rex. Since Rec and Np9 are both putative oncogenes, the factors responsible for expression of these variant HERV-K env mRNAs could be of critical importance.
We found that envelope expression was significantly associated with disease stage and a positive node status, suggesting that envelope expression may enhance metastatic spread in Chinese and U.S. women diagnosed with breast cancer. These findings support the results previously observed in cohorts obtained from women diagnosed with breast cancer in Birmingham, Alabama, and Houston, Texas.11,12,17 In addition, the expression of HERV-K env and Np9 was detected in only tumor tissues but not in matched uninvolved normal breast tissues. Since the expression of viral RNAs was demonstrated in various areas of the United States as well as in China, activation of HERVs appears to be a common event in breast cancer irrespective of geographic location.
The current study provided us with an opportunity to directly compare HERV-K expression in breast cancer patients from the United States and China. This comparison is important because it demonstrates that HERV-K expression affects breast cancer patients of different ethnic background and also provides important cross-validation of our findings that HERV-K is highly expressed in most human breast tumors. Several older publications have documented that the expression of HERV-K RNA and protein was higher in breast cancer cell lines treated with estradiol and progesterone than in cells without treatment.11,14 In our study, we did not find an association of HERV-K env protein expression with the tumor ER/PR status among Chinese or U.S. patients. We previously reported that 8 completely sequenced complementary DNAs from human breast tumors showed greater than 97% nucleotide homology to HERV-K102,11,12 indicating that this locus drives HERV-K env expression in human tumors. Our current study further confirms this finding by showing that the mRNAs encoding env in the breast tumors are almost all HERV-K type-1 based, consistent with this locus.
HERV-K is reactivated in breast tumors by mechanisms that may include the stress response pathway and, as part of this pathway, may affect disease progression. We indeed found in both the Chinese and U.S. breast cancer patients an association between the expression level of HERV-K env protein and disease stage and number of axillary node metastases. For the patients from China, we also obtained preliminary evidence that tumor HERV-K env protein expression is associated with patient survival. These data are consistent with previously published results showing that HERV-K-T47D reverse transcriptase expression was associated with the prognosis of breast cancer patients and thus could be used as a novel independent prognostic marker for disease outcome.14
In summary, the expression of HERV-K env was upregulated in nearly all breast cancer tissues of Chinese and U.S. women and was found to be associated with disease prognosis. Thus, evaluation of HERV-K expression could prove useful in clinical early stage disease detection and in the diagnosis of breast cancer.
Materials and Methods
Study Subjects
Cohort 1
Paraffin-embedded tissues (n = 110 breast cancer cases and n = 30 benign breast tissues) were obtained from the Department of Breast Cancer in the Cancer Institute and Hospital of Tianjin Medical University with patient consent. These tissues were obtained from female patients who underwent breast surgery in 2004 and 2005. The average age was 53 and the average diameter of the tumor was 3.2 cm. For HERV-K env expression evaluation using RT-PCR, additional, freshly resected breast tumors (n = 40) and matched uninvolved adjacent tissues (n = 40) were obtained from surgeries between July and December of 2009 in the Department of Breast Cancer in the Cancer Institute and Hospital of Tianjin Medical University. Informed consent was obtained from these patients prior to surgery. None of the patients had undergone preoperative chemotherapy or radiotherapy. Normal breast tissues (n = 40) were obtained from female patients who did not have cancer but had benign breast diseases, necessitating removal of the diseased area and some surrounding normal tissue, which was analyzed by a pathologist. The breast tissues used in this study were diagnosed as normal by a pathologist to confirm that the tissues were without any malignant cells. Tissues were stored in liquid nitrogen until RNA isolation.
Cohort 2
Fresh-frozen primary tumors (n = 32) and uninvolved adjacent breast tissues (n = 30) were obtained from breast cancer patients at the MD Anderson Cancer Center according to an approved Institutional Review Board protocol (LAB04-0083). Patients signed a consent form. Clinical and pathological information was obtained from medical records and pathology reports. Patient 75 is a 45-year-old African American female diagnosed with invasive ductal carcinoma (IDC) who is ER negative, PR negative, Her-2/neu negative; has no lymph node involvement; and is nuclear grade 3. Patient 127 is a 46-year-old Caucasian female diagnosed with DCIS who is ER positive, PR positive, Her-2/neu negative; has no lymph node involvement; and is nuclear grade 2. Patient 133 is a 60-year-old Caucasian female diagnosed with IDC who is ER negative, PR negative, Her-2/neu negative; has no lymph node involvement; and is nuclear grade 3. Patient 163 is a 63-year-old Caucasian female diagnosed with IDC who is ER positive, PR positive, Her-2/neu negative; has no lymph node involvement; and is nuclear grade 3. Patient 177 is a 60-year-old Caucasian female diagnosed with DCIS who is ER positive, PR positive, Her-2/neu negative; has no lymph node involvement; and is nuclear grade 3.
Cohort 3
Paraffin-embedded breast tumor specimens were obtained for immunohistochemical analysis from 223 breast cancer patients who resided in the greater Baltimore area between 1993 and 2003, as described.22
RT-PCR
The TRIzol single stage method was used to isolate total RNA from breast tissue samples of Chinese women. RNA extraction was carried out under conditions that were RNase-free. The HERV-K env PCR primers were designed by Sangon Biotech Co. Ltd. (Shanghai) and produced by Takara Biotechnology Co. Ltd. (Dalian), according to the HERV-K102 env sequence (GenBank accession no. AF164610.1). The human β-actin primer was purchased from Takara Biotechnology Co. Ltd. (Dalian). Primer sequences were as follows: HERV-K env sense, 5-ACAAA AAGAAGGGGGAGACAG-3; HERV-K env anti-sense (6151-6172bp), 5′-TGAACAGAAGAGTGCAATGCA-3′ (7,644-7,624 according to HERV-K102, AF164610): β-actin sense, 5-TGGCACCCAGCACAATGAA3-; β-actin anti-sense, 5-CTAAGTCATAGTCCGCCTAGAAGCA-3. First strand cDNA was synthesized using 2 µg total RNA and M-MuLV reverse transcriptase plus an oligo-dT primer. The reverse transcribed samples were amplified in a volume of 20 µL containing 0.5 µL of first-strand cDNA synthesis mixture, 2 µL of 10× buffer, 0.25 µL of rTaq DNA polymerase, and 1.5 µL of dNTP (2.5 mmol). Each sample was analyzed in parallel with a human β-actin primer, and samples were normalized to this housekeeping gene. PCR reactions were initially denatured at 94°C for 5 minutes, followed by 35 cycles of denaturation (94°C for 30 seconds), annealing (53°C for 45 seconds), and extension (72°C for 30 seconds), with a final extension of 72°C for 10 minutes. Amplified products were analyzed on a 1% agarose gel.
For the samples obtained at the MD Anderson Cancer Center (cohort 2), RNA isolation and RT-PCR were used as described previously.11,12 Primers used for RT-PCR included HERV-K SU env type 1 and type 2 primers, HERV-E env primers, ERV-3 primers, Np9 primers, and β-actin primers, which were described previously.13 Primer sequences for HERV-K full-length env gene or gag gene were as follows: HERV-K full-length env sense (6,471-6,493 bp), 5-AGAAAAGGGCCTCCACGGAGA-3; HERV-K full-length env anti-sense (8,277-8,258 bp), 5′-TCTCCTATGTC TACTTCTTT-3′ (according to HERV-K102, AF164610); HERV-K gag sense (1,083-1,102 bp), 5-TCGGAAGAAGCTAGGGTGAT-3; HERV-K env anti-sense (3,138-3,119 bp), 5′-TGGAAGCAGAGAGACTGCTT-3′ (according to HERV-K115, AY037929.1). No template and β-actin were used as negative and positive controls, respectively.
Immunohistochemistry
Anti-HERV-K monoclonal antibody (mAb) 6H5 was produced and purified as described previously.17 6H5 was used for immunohistochemistry (IHC) of breast biopsies from Chinese women as described previously.17 The secondary antibody and other reagents were purchased from Beijing Zhongshan Goldenbridge Biotechnology Co. Ltd. Tissue sections (4 µm) were cut from paraffin blocks and transferred to silanized glass slides. They were then dewaxed by incubation in xylene and rehydrated using a graded series of ethanol solutions. The endogenous peroxidase was blocked with 3% H2O2 in methanol. Antigen retrieval was performed with citrate buffer by high temperature and pressure for 15 minutes. The primary antibody was diluted 1:100. The color reaction was developed in 3,3′ diaminobenzidine for 10 minutes and the slides were then hematoxylin counterstained. For the assessment of expression in the tumors from the Chinese patients, the staining intensity was scored as 0 (negative), 1+ (weak), 2+ (medium) or 3+ (strong). Ten high power fields (400×) were randomly chosen, and the percentage of positive staining cells in every high power field was calculated. The percentage of tumor cells staining positive for HERV-K env was classified as 0 (no staining or <25% positive staining of tumor cells), 1+ (25%-39%), 2+ (40%-59%), or 3+ (≥60%), according to the average percentage of positive staining cells calculated in 10 randomly-selected high power fields.
For the assessment of HERV-K expression in tumors from U.S. patients (cohort 3), 6H5 IHC was performed on formalin-fixed, paraffin-embedded tissue sections using standard protocols and VECTASTAIN ABC reagents as described previously.17 HERV-K env expression was categorized by intensity (0 = absent; 1 = weak; 2 = moderate; 3 = strong) and distribution (percentage of tumors staining positive for env). A sum score of intensity × distribution was generated for each tumor. The immunohistochemistry produced good staining with low background for 195 of 223 tumors. Only these tumors were included in the analysis.
Follow-Up Status
The 110 Chinese breast cancer patients with available paraffin-embedded tumor specimens were followed up for overall survival from the time of surgery until September 30, 2010. The median follow-up duration was 73 (range 8-80) months.
Statistical Analysis
Statistical analyses were performed using the statistical software package SPSS (version 17.0). The χ2 test was used to analyze for differences between groups. A difference was regarded as significant if the calculated probability P < 0.05. Analyses of overall survival were performed by Kaplan-Meier analysis.
Footnotes
J. Zhao and K. Rycaj contributed equally to this work. Dr. Lin Gu was the leader of the China team.
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported in part by grant BCTR0402892 from Susan G. Komen for the Cure, Avon Foundation grant 07-2007-070 01, and the National Breast Cancer Foundation. We thank the Cancer Institute and Hospital of Tianjin Medical University for providing us with funds for this research (Innovative Research Team in University of China; IRT0743).
Supplementary material for this article is available on the Genes & Cancer website at http://ganc.sagepub.com/supplemental.
References
- 1. Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69-90 [DOI] [PubMed] [Google Scholar]
- 2. Porter P. “Westernizing” women’s risks? Breast cancer in lower-income countries. N Engl J Med. 2008;358:213-6 [DOI] [PubMed] [Google Scholar]
- 3. Zur Hausen H. The search for infectious causes of human cancers: where and why. Virology. 2009;392:1-10 [DOI] [PubMed] [Google Scholar]
- 4. Lawson JS, Gunzburg WH, Whitaker NJ. Viruses and human breast cancer. Future Microbiol. 2006;1:33-51 [DOI] [PubMed] [Google Scholar]
- 5. Bittner JJ. Some possible effects of nursing on the mammary gland tumor incidence in mice. Science. 1936;84:162. [DOI] [PubMed] [Google Scholar]
- 6. Matsuzawa A, Nakano H, Yoshimoto T, Sayama K. Biology of mouse mammary tumor virus (MMTV). Cancer Lett. 1995;90:3-11 [DOI] [PubMed] [Google Scholar]
- 7. Larsson E, Kato N, Cohen M. Human endogenous proviruses. Curr Top Microbiol Immunol. 1989;148:115-32 [DOI] [PubMed] [Google Scholar]
- 8. Mayer J, Sauter M, Racz A, Scherer D, Mueller-Lantzsch N, Meese E. An almost-intact human endogenous retrovirus K on human chromosome 7. Nat Genet. 1999;21:257-8 [DOI] [PubMed] [Google Scholar]
- 9. Hanke K, Kramer P, Seeher S, Beimforde N, Kurth R, Bannert N. Reconstitution of the ancestral glycoprotein of human endogenous retrovirus k and modulation of its functional activity by truncation of the cytoplasmic domain. J Virol. 2009;83:12790-800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ono M, Yasunaga T, Miyata T, Ushikubo H. Nucleotide sequence of human endogenous retrovirus genome related to the mouse mammary tumor virus genome. J Virol. 1986;60:589-98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang-Johanning F. Quantitation of HERV-K env gene expression and splicing in human breast cancer. Oncogene. 2003;22:1528-35 [DOI] [PubMed] [Google Scholar]
- 12. Wang-Johanning F, Frost AR, Johanning GL, et al. Expression of human endogenous retrovirus k envelope transcripts in human breast cancer. Clin Cancer Res. 2001;7:1553-60 [PubMed] [Google Scholar]
- 13. Wang-Johanning F, Liu J, Rycaj K, et al. Expression of multiple human endogenous retrovirus surface envelope proteins in ovarian cancer. Int J Cancer. 2007;120:81-90 [DOI] [PubMed] [Google Scholar]
- 14. Golan M, Hizi A, Resau JH, et al. Human endogenous retrovirus (HERV-K) reverse transcriptase as a breast cancer prognostic marker. Neoplasia. 2008;10:521-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Seifarth W, Baust C, Murr A, et al. Proviral structure, chromosomal location, and expression of HERV-K-T47D, a novel human endogenous retrovirus derived from T47D particles. J Virol. 1998;72:8384-91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Etkind PR, Lumb K, Du J, Racevskis J. Type 1 HERV-K genome is spliced into subgenomic transcripts in the human breast tumor cell line T47D. Virology. 1997;234:304-8 [DOI] [PubMed] [Google Scholar]
- 17. Wang-Johanning F, Radvanyi L, Rycaj K, et al. Human endogenous retrovirus K triggers an antigen-specific immune response in breast cancer patients. Cancer Res. 2008;68:5869-77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Levine PH, Steinhorn SC, Ries LG, Aron JL. Inflammatory breast cancer: the experience of the surveillance, epidemiology, and end results (SEER) program. J Natl Cancer Inst. 1985;74:291-7 [PubMed] [Google Scholar]
- 19. Ono M, Kawakami M, Ushikubo H. Stimulation of expression of the human endogenous retrovirus genome by female steroid hormones in human breast cancer cell line T47D. J Virol. 1987;61:2059-62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Patience C, Simpson GR, Colletta AA, Welch HM, Weiss RA, Boyd MT. Human endogenous retrovirus expression and reverse transcriptase activity in the T47D mammary carcinoma cell line. J Virol. 1996;70:2654-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Burmeister T, Ebert AD, Pritze W, Loddenkemper C, Schwartz S, Thiel E. Insertional polymorphisms of endogenous HERV-K113 and HERV-K115 retroviruses in breast cancer patients and age-matched controls. AIDS Res Hum Retroviruses. 2004;20:1223-9 [DOI] [PubMed] [Google Scholar]
- 22. Prueitt RL, Boersma BJ, Howe TM, et al. Inflammation and IGF-I activate the Akt pathway in breast cancer. Int J Cancer. 2007;120:796-805 [DOI] [PubMed] [Google Scholar]